Abstract
Molecular biotechnology has made it possible to explore the potential of plants for different purposes. The 3’ regulatory regions have a great diversity of cis-regulatory elements directly involved in polyadenylation, stability, transport and mRNA translation, essential to achieve the desired levels of gene expression. A complex interaction between the cleavage and polyadenylation molecular complex and cis-elements determine the polyadenylation site, which may result in the choice of non-canonical sites, resulting in alternative polyadenylation events, involved in the regulation of more than 80% of the genes expressed in plants. In addition, after transcription, a wide array of RNA-binding proteins interacts with cis-acting elements located mainly in the 3’ untranslated region, determining the fate of mRNAs in eukaryotic cells. Although a small number of 3’ regulatory regions have been identified and validated so far, many studies have shown that plant 3’ regulatory regions have a higher potential to regulate gene expression in plants compared to widely used 3’ regulatory regions, such as NOS and OCS from Agrobacterium tumefaciens and 35S from cauliflower mosaic virus. In this review, we discuss the role of 3’ regulatory regions in gene expression, and the superior potential that plant 3’ regulatory regions have compared to NOS, OCS and 35S 3’ regulatory regions.
Keywords: molecular biotechnology, plant 3’ regulatory regions, plant cis-regulatory elements, increase gene expression in plants, cleavage and polyadenylation factors in plants
Introduction
In eukaryotes, nuclear processing of pre-messenger 3’ RNA (pre-mRNA 3’) influences several subsequent stages of gene expression, that include, but are not limited to mRNA splicing, stability, transport and translation (Zhao et al., 1999; Millevoi and Vagner, 2010). A highly efficient surveillance system degrades any pre-mRNA that has not been properly processed (Jensen et al., 2001). In mammalian and yeast, a molecular complex composed of more than 20 proteins, interacts with cis-elements present in the pre-mRNA 3’ to cleave and polyadenylate the newly transcribed mRNA (Mandel et al., 2008; Chan et al., 2011). These cis-elements are polyadenylation signals, which define where the molecular complex should cleave and polyadenylate the pre-mRNA (Loke et al., 2005).
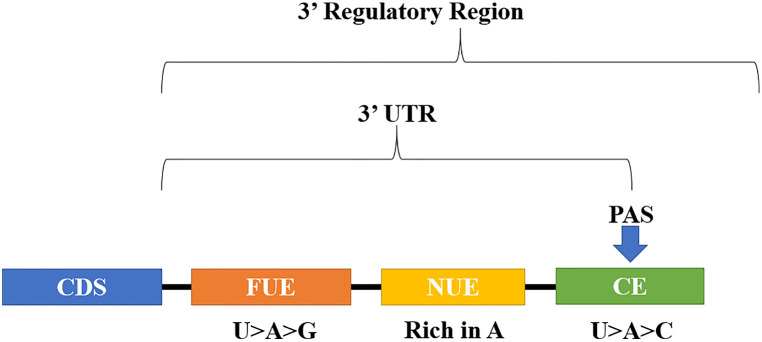
In plants, a similar molecular complex has been identified (Hunt, 2008; Hunt et al., 2012), and although less conserved, plants also have polyadenylation signals: far upstream element (FUE), near upstream element (NUE) and the cleavage element (CE) (Xing and Li, 2011). The polyadenylation site (PAS), point from which the pre-mRNA is cleaved and polyadenylated, is defined by surrounding cis-elements (Tian and Manley, 2016). Furthermore, the strength of a given PAS is also defined, in part, by the cis-elements (Neve et al., 2017). Polyadenylation is essential for the stability of the transcript, preventing the mRNA from being the target of posttranscriptional gene silencing (PTGS) via RNA-dependent RNA polymerase 6 (RDR6) in plants (Luo and Chen, 2007). Besides, polyadenylation mediates transcriptional processes such as initiation, elongation, and termination (Mapendano et al., 2010), as well as post-transcriptional processes, such as transport of mRNA into the cytoplasm and start of translation (Millevoi and Vagner, 2010; Chan et al., 2011).
Eukaryotes have a mechanism termed alternative polyadenylation (APA), which allows the selective use of PAS in genes containing multiple PAS. The APA allows fine regulation of gene expression, being recognized as one of the main regulatory mechanisms of expression (Hunt, 2012; Tian and Manley, 2016). It is estimated that approximately half of the eukaryotic genes have multiple PAS (Tian et al., 2005; Shen et al., 2008a; Hunt, 2012). Through APA, a single gene containing multiple PAS can generate a considerable number of transcript isoforms, thereby producing a highly diversified transcriptome (Tian and Manley, 2013). The availability of multiple PAS in the same 3’ regulatory region enables the inclusion or exclusion of 3’ untranslated region (3’UTR) sequences, resulting in transcripts that may differ in particularities involving post-transcriptional processes such as stability, transport and translation (Mayr, 2016), and even protein localization (Berkovits and Mayr, 2015). Moreover, several studies have shown that APA activity can vary according to the cell state or cycle, as well as with the cell type, being involved in different biological processes in plants (Xing and Li, 2011; Deridder et al., 2012; Tian and Manley, 2016; Ji et al., 2018).
The non-coding regions downstream of coding sequences (CDS) is usually termed terminators. However, we believe 3’ regulatory region is a more appropriate term, as it will be referred to here. This is because transcription termination is only one of the roles of the 3’ regulatory regions, which, in many cases, can have profound effects on gene expression, as it will be discussed in this review (Menossi et al., 2003; Yang et al., 2009; Nagaya et al., 2010; Hirai et al., 2011; Hiwasa-Tanase et al., 2011; Matsui et al., 2014; Diamos and Mason, 2018; Pérez-González and Caro, 2018; Rosenthal et al., 2018; Yamamoto et al., 2018).
3’ Regulatory Regions
Eukaryotes have non-coding DNA sequences located downstream of the CDS, termed 3’ regulatory regions ( Figure 1 ), which are involved in important processes of gene transcription termination, such as cleavage and polyadenylation (Ingelbrecht et al., 1989; Huang and Carmichael, 1996; Luo and Chen, 2007; Rosenthal et al., 2018). Although they do not encode polypeptides, 3’ regulatory regions have cis-elements that guide the CPMC during cleavage and polyadenylation (Yang et al., 2009; Xing et al., 2010; Hiwasa-Tanase et al., 2011; Hunt et al., 2012; Matsui et al., 2014; Rosenthal et al., 2018). Indeed, 3’ regulatory regions have a significant weight on gene expression levels, as shown by the use of different 3’ regulatory regions in expression cassettes (Ingelbrecht et al., 1989; Mitsuhara et al., 1996; Richter et al., 2000; Nagaya et al., 2010; Hirai et al., 2011; Diamos et al., 2016; Wei et al., 2017; Diamos and Mason, 2018; Rosenthal et al., 2018). Some 3’ regulatory regions have multiple PAS, being largely responsible for the diversity of the eukaryotic transcriptome (Xing et al., 2010; Xing and Li, 2011; Tian and Manley, 2013). The presence of multiple PAS in the same 3’ regulatory region allows fine regulation of gene expression through the APA mechanism, with consequent effects on mRNA metabolism and metabolic pathways (Wei et al., 2017; Hong et al., 2018; Turner et al., 2018; Wang et al., 2019). However, the mechanism that controls the use of a particular PAS over another is a complex process that is far from being wholly understood (Neve et al., 2017; Turner et al., 2018).
Figure 1.
Structure of the tripartite polyadenylation signal in plants: FUE, far upstream element; NUE, near upstream element; CE, cleavage element; PAS, polyadenylation site; CDS, coding sequence; 3’UTR, 3’ untranslated region.
Cis-Elements in Plants
Plants, as well as mammals, yeast and algae have regulatory sequences, termed cis-elements that guide pre-mRNA polyadenylation (Loke et al., 2005; Hunt, 2008; Mandel et al., 2008; Shen et al., 2008b). In plants, a tripartite model has been suggested, composed by the cis-elements, far upstream element (FUE), near upstream element (NUE), and cleavage element CE, which together constitute the signal of polyadenylation ( Figure 1 ). Rich in U>A>G, the FUE element has 6-18 nucleotides (nt) in size, distributed within a region of approximately 125 nt, which in general starts at approximately 30 nt upstream of PAS. The NUE element is A-rich, with a size ranging from 6–10 nt, and is located approximately between 10 to 40 nt upstream of the PAS. The CE element is composed of a region rich in U>A>C, having as part of its sequence the cleavage site itself, where after the RNA cleavage the polyadenine tail is added, in a process called polyadenylation. The cleavage site is formed by 2 nt, defined by some authors as YA, where Y = U or C (Li and Hunt, 1997; Loke et al., 2005; Hunt, 2008; Xing and Li, 2011).
Plants have NUE sequences similar to the dominant A(A/U)UAAA polyadenylation signal found in mammals. Although found in more than 50% of mammalian genes, AAUAAA, the most common variant of hexamer is found in only 10% of transcripts from Arabidopsis thaliana and rice (Oryza sativa) (Zarudnaya et al., 2003; Chan et al., 2011; Xing and Li, 2011). Also, a recent analysis of bioinformatics found that AAUAAA is also very poorly conserved in mosses (Selaginella moellendorffii and Physcomitrella patens), not exceeding 8%, with the first 2 nt being highly degenerate (Zhao et al., 2019). Also, the second most frequent variant AUUAAA hexamer in mammals, present in about 16% of transcripts, was found in only 2.17% and 2.08% of A. thaliana and S. moellendorffii, respectively (Zhao et al., 2019). Unlike mammals, point mutations in the AAUAAA variant have little influence on the efficiency of polyadenylation in plants, with some mutations even increasing, which justifies the low frequency of this exact sequence in plants (Rothnie et al., 1994). However, in vitro assays showed that the deletion of NUE or FUE results in the choice of unusual PAS (Zhao et al., 2011), consistent with findings from other earlier studies (Mogen et al., 1990; Rothnie et al., 1994).
The sequence complementarity profile that cis-elements present can generate secondary structures in the 3’ end region of the pre-mRNAs that appear to positively influence the functionality of the cis-elements itself (Loke et al., 2005). The formation of these secondary structures are important for the interaction between proteins from polyadenylation complex and mRNA (Zarudnaya et al., 2003). The efficiency of recognition and choice of a given PAS is partially determined by the signal strength of the cis-elements (Loke et al., 2005; Guo et al., 2016; Neve et al., 2017). More importantly, these cis-elements can be found in different regions of the genes, such as 5’UTR, exons, introns, and mainly in 3’ regulatory regions (Simpson et al., 2003; Hunt, 2012; Hoque et al., 2013; Rosenthal et al., 2018).
Cleavage and Polyadenylation Molecular Complex
During the final steps of the transcription process in eukaryotes, a multiprotein complex composed of more than 20 proteins, here named cleavage and polyadenylation molecular complex (CPMC), indispensable for the biogenesis of mRNA, recognizes and interacts with the cis-elements to cleave and polyadenylate the pre-mRNA (Chan et al., 2011). Most of these proteins have already been identified in animals and yeasts (Mandel et al., 2008), and most of their homologs have also been identified in plants (Hunt, 2008; Zhao et al., 2009; Zhao et al., 2011). This molecular complex is formed by subcomplexes, also called factors, and can be divided basically into four large subcomplexes: Cleavage and Polyadenylation Specificity Factor (CPSF), Cleavage Stimulatory Factor (CstF), Cleavage Factor I e II (CF I e II) (Millevoi and Vagner, 2010; Chan et al., 2011). Each factor is formed by protein subunits that interact with each other in the form of even more complex heterodimers, heterotrimers or oligomers. The interaction between the protein subunits and the cis-elements is crucial to maintain the cohesion of the complex (Chan et al., 2011). The assembly of the molecular complex takes place after binding of these factors to the cis-elements, which generally takes around 10 seconds, with this response time being influenced by the strength of the PAS (Chao et al., 1999).
The best and richest description of the molecular polyadenylation complex among eukaryotes is by far that of mammals. In mammals, the CPSF factor mediates the cleavage and polyadenylation process. After recognizing the canonical hexamer A(A/U)UAAA, or its more frequent variant, AAUAAA, CPSF recruits the other factors to cleave and polyadenylate the 3’ pre-mRNA region. There is evidence indicating that CPSF160 is the subunit responsible for recognizing A(A/U)UAAA (Murthy and Manley, 1995) and recently it has been shown that CPSF30 and WDR33 interact directly with the AAUAAA hexamer (Chan et al., 2014). A cryogenic electron microscopy study reported that in humans, CPSF160 does not interact with the AAUAAA, but functions as a scaffold to preorganize two other subunits, CPSF30 and WDR33, which synergistically bind to the hexamer with high affinity (Sun et al., 2018).
Indeed, AtCPSF30 has also been shown to be an RNA-binding protein (RBP) with an affinity for U-rich sequences such as FUE (Hunt, 2008). Assays with orthologs of AtCPSF30, encoding CPSF160 and WDR33 in A. thaliana (AtCPSF160 and AtFY, respectively), found an interaction between these subunits, demonstrating that a similar complex may form in plants during the processing of the 3’ end pre-mRNA (Zhao et al., 2009). AtCPSF30 is involved in the choice of canonical NUE from a large number of genes in A. thaliana, and mutations of this subunit result in the choice of unusual PAS (Thomas et al., 2012). AtFY was found to be involved in the 3’ end processing of mRNA in A. thaliana (Simpson et al., 2003) and recently, the role of AtFY in the recognition of canonical NUE has been demonstrated using fy mutants in A. thaliana (Yu et al., 2019). Indeed, the choice of canonical NUE appears to rely on the interaction between AtCPSF30 and AtFY, with double mutants being able to generate up to 50% more APA events, which had some interference on processes such as the control of flowering time (Yu et al., 2019). These data are in agreement with previous studies (Jiang et al., 2012; Chakrabarti and Hunt, 2015). Besides, AtCPSF160 and AtCPSF30 have been shown to possess nuclear localization (Delaney et al., 2006; Xu et al., 2006), the nuclear location of AtCPSF30 appears to depend on its interaction with AtCPSF160 (Rao et al., 2009).
In A. thaliana, At1g30460 is the gene that encodes AtCPSF30, which interestingly is the target of alternative splicing events, resulting in the production of another protein containing an additional motif related to pre-mRNA splicing in mammals. These two proteins may form different complexes, connecting mRNA splicing and polyadenylation in plants (Delaney et al., 2006). AtCPSF30 has been shown to possess endonuclease activity, which appears to be inhibited by AtFip1, a mammalian hFip1 orthologous protein (Addepalli and Hunt, 2007). Furthermore, AtCPSF30 mutants of A. thaliana are more tolerant to oxidative stress, confirming a likely role in the regulation of gene expression (Zhang et al., 2008).
As previously demonstrated, CPSF73 has endoribonuclease activity and may be the subunit responsible for pre-mRNA cleavage in humans (Mandel et al., 2006). Two mammalian CPSF73-like proteins, AtCPSF73(I) e AtCPSF73(II), are found in A. thaliana (Xu et al., 2004). Although very similar, AtCPSF73(I), and AtCPSF73(II) have distinct roles in plants (Xu et al., 2004; Xu et al., 2006). An in vitro cleavage assay showed that the AtCPSF73(I) subunit has endonuclease activity (Zhao et al., 2011).
In mammals, in vitro experiments suggest that after cleavage of the pre-mRNA, CPSF160 and hFip1 act directly in the recruitment of poly-A polymerase (PAP), protein responsible for polyadenylation (Barabino et al., 1997; Kaufmann et al., 2004). The expression of hFip1 changes according to the degree of cell differentiation, and it has been shown to be a potent regulator of APA. hFip1 promotes stem cell maintenance by activating APA profiles specific to embryonic stem cells (ESC), and also restores APA profiles similar to those found in ESC during the reprogramming of somatic cells (Lackford et al., 2014). Intriguingly, it has recently been proposed that CPSF can mediate cytoplasmic polyadenylation of mRNAs (Dai et al., 2019).
In A. thaliana, there is an interaction between the counterparts of hFip1 and PAP, shown by a yeast two-hybrid assay between AtFip1 with AtPAP. Also, AtFip1 stimulates AtPAP activity, and as well as hFip1, AtFip1 is also an RBP with a preference for G-rich sequences (Forbes et al., 2006). PABPN1 (poly-A polymerase-binding nuclear protein) is another critical piece of PAP activity. This protein stimulates the reaction of PAP catalysis during the synthesis of the polyadenine tail and also dictates its size by regulating the interaction between CPSF and PAP (Kühn et al., 2009). In A. thaliana, at least three isoforms of PABPN, AtPABN1, 2, and 3 are found, and interaction between AtPABN and AtPAP isoforms is reported (Hunt et al., 2008). Also, the interaction between AtPABN1, AtPAP4, and AtCPSF30 has been reported (Forbes et al., 2006).
In mammals, the CstF factor contributes decisively to the processing of the 3’ regulatory region of the pre-mRNA. However, the stable binding of the CstF subcomplex to the 3’ regulatory region of the pre-mRNA is dependent on the interaction with the CPSF subcomplex. An interaction between CstF64 and CPSF160 proved to be necessary to define the cleavage site (Chan et al., 2011). CstF64 is directly involved in the recognition of PAS and in the global regulation of APA, being able to binding to G/U-rich sequences downstream of the PAS, and its specificity dependent of the interaction with CPSF (Yao et al., 2012; Masoumzadeh et al., 2020). Also, CstF77 increases the affinity of CstF64 for RNA targets, recruiting CstF50, that is involved in the recognition of G/U-rich sequences (Yang et al., 2018). Once in the cytoplasm, the transport of CstF64 to the nucleus is dependent on its interaction with CstF77, essential for the both cleavage and polyadenylation (Grozdanov et al., 2018b).
It was demonstrated in vitro that CstF64 is essential for an adequate differentiation of embryonic stem cells into endodermal lines, and its absence may result in dysfunctional cardiomyocytes (Youngblood and Macdonald, 2014). Furthermore, co-expression of CstF50 or CstF77 with CstF64 promoted an increase in cleavage and polyadenylation rates of a reporter gene in vitro (Grozdanov et al., 2018b). CstF64/tau is encoded by Cstf2t, a paralog of the Cstf2 gene that encodes CstF64. Cstf2t is expressed in germ cells, also has a role on polyadenylation, being indispensable for spermatogenesis in rats (Harris et al., 2016; Grozdanov et al., 2018a). CstF64/tau binds to sequences rich in U>G also downstream of the PAS (Mandel et al., 2008). In addition, CstF64/tau promotes the use of non-canonical distal PAS, an important regulator of the APA (Hwang H. et al., 2016).
A. thaliana has orthologs of subunits CstF64 and CstF77, encoding AtCstF64 and AtCstF77, that interact in vitro and have the ability to bind RNA, as shown for mammalian counterparts (Yao et al., 2002; Bell and Hunt, 2010). In Glycine max there is gene duplication for CstF50 and CstF64, probably due to recent genomic duplication. In contrast, S. moellendorffii presents two orthologs for CstF64. The presence of orthologs for CstF50 in some plants and the absence in others, such as Chlamydomonas reinhardii and Populus trichocarpa, suggests that there may be functional redundancy (Hunt et al., 2012). Interestingly, A. thaliana also has a protein, enhanced silencing phenotype 1 (ESP1), that has a degree of similarity with mammalian CstF64. Unlike AtCstF64, ESP1 does not have the canonical RNA (RRM) recognition domain present in CstF64. However, similarly to AtCstF64, ESP1 presents the domain that allows an interaction with CstF77 and other factors involved in cleavage and polyadenylation (Herr et al., 2006).
The subcomplex CFIm is indispensable for the cleavage step of pre-mRNA in mammals (Ryan, 2007; Chan et al., 2011). CFIm also seems to be involved in the choice of PAS, according to APA events presented by HeLa cells knocked down for CFIm25 (Kubo et al., 2006), what seems to be happening also for CFIm68 (Kim et al., 2010). In mammals, CFIm presents itself as a heterotetrametric complex composed of two CFIm25, one CFIm59 and one CFIm68 subunits. However, it was demonstrated in vitro that the complex presents activity only with a CFIm25 dimer and two CFIm68 subunits, suggestive of functional redundancy between CFIm59 and CFIm68 (Ruëgsegger et al., 1998), although mutants for CFIm68 have effects on APA while CFIm59 does not (Kim et al., 2010). CFIm assists in the interaction of PAP with hFip1 and CPSF160 during the cleavage process. CFIm also assists in the definition of PAS and potentiates the recognition of noncanonical cis-elements (Chan et al., 2011), being involved in the recruitment of hFip1 and PAP (Venkataraman et al., 2005). CFIm binds specifically to the cis-elements UGUA, with CFIm25 being the subunit responsible for the recognition and interaction with UGUA (Yang et al., 2010).
As demonstrated by mutation studies, the RRM of CFIm68 increases the affinity of CFIm25 for UGUA (Yang et al., 2011a; Yang et al., 2011b). It was demonstrated that CFIm59 and CFIm68 also potentiate the use of PAS that have the polyadenylation signal UGUA, being the position of UGUA able to affect this activity in vitro. This CFIm activity requires an interaction with hFip1, mediated by the serine-arginine (RS) repeat domain present in CFIm59 and CFIm68. In addition, the binding of CFIm to UGUA promotes the recruitment of CPSF and CstF (Zhu et al., 2018). In A. thaliana, an interaction between AtFip1 with AtPAP, AtCFI25, AtCPSF30 and AtPABN1 was reported, suggesting that AtFip1 and AtPAP may also be recruited by AtCFI during the processing of the 3’ end pre-mRNA region (Forbes et al., 2006). A. thaliana has at least two genes encoding CFIm25 orthologs, At4g25550 and At4g29820, and at least four CFIm68 orthologs have been found in plants (Hunt et al., 2012).
In contrast, little is known about the subcomplex CFIIm, which has two subunits, Clp1 and Pcf11, required for the cleavage process (Chan et al., 2011). Recently it was demonstrated that Pcf11 has a role in the global regulation of APA events, since it promotes the use of proximal PAS, and its depletion increases the use of distal PAS, which seems to be true also for Clp1 (Li et al., 2015; Ogorodnikov et al., 2018). In A. thaliana, at least two orthologs were identified for Pcf11: At4g04885 and At2g36480. In particular, At4g04885 presents two of the three functional domains found in Pcf11 (Hunt et al., 2012). The depletion of AtPCFS4, a homologue of the yeast Pcf11p, resulted in a delay in flowering time in A. thaliana. It was shown that AtPCFS4 is an APA regulator, promoting the use of more proximal PAS, within the intron 3 from the FCA gene (Xing et al., 2008b).
Orthologs for the gene encoding the Clp1 subunit also appear to be present in plants, being in A. thaliana encoded by two genes, At3g04680 and At5g39930 (Hunt et al., 2012). An interaction between AtPCFS4 and AtCLPS3 (At3g04680) has already been demonstrated (Xing et al., 2008b). The suspicion that AtCLPS3 is also an APA regulator was raised by the fact that the overexpression of AtCLPS3 promotes the use of a regular PAS in FCA, which results in the functional FCA isoform, causing early flowering in A. thaliana (Xing et al., 2008a). Also, direct interactions between AtCLPS3 and AtFY, AtCPSF30, AtCPSF100 e AtCPSF160, as well as between AtCLPS5 (At5g39930), AtFip3 and AtFip5 were observed (Hunt et al., 2008; Xing et al., 2008a; Xing et al., 2008b).
Although plants exhibit homologous proteins and patterns of interaction between subunits similar to their counterparts in mammals and yeasts, their functions may be different. Another point to consider is that unlike the CPMC of mammals and yeasts, where the subunits are encoded by a single gene, some subunits of A. thaliana are encoded by gene families, and, the same gene may encode at least two isoforms of the same subunit. Also, the possibility of gene duplication, mainly highly conserved subunits, may incur functional redundancy or even functional specialization (Hunt, 2008; Zhao et al., 2009; Hunt et al., 2012).
Polyadenylation
Polyadenylation is the process of synthesis of a polymer of adenine, having been observed for the first time in the 60’s (Edmonds and Abrams, 1960). The polyadenylation process is oriented by cis-elements, and involves all CPMC proteins and PAP (Bardwell et al., 1990; Misra and Green, 2016). After PAS recognition and pre-mRNA cleavage by CPMC, the PAP protein initiates the synthesis of the adenine polymer at the 3’ end of the pre-mRNA, from the cleavage site, resulting in a tail of polyadenines (tail poly-A) ranging from 70 to 250 nt between eukaryotes (Kühn et al., 2009). Most mature eukaryotic mRNAs are polyadenylated. It is estimated that less than 5% is not, most of which are histone mRNAs (Tian et al., 2005; Djebali et al., 2012).
A. thaliana has at least three canonical nuclear PAPs, AtPAPS1, AtPAPS2 and AtPAPS4, and one cytoplasmic, AtPAPS3 (Addepalli et al., 2004; Hunt et al., 2008; Meeks et al., 2009). Several studies have investigated whether there is functional redundancy between these AtPAPS isoforms (Hunt et al., 2008; Vi et al., 2013; Trost et al., 2014; Kappel et al., 2015; Czesnick and Lenhard, 2016). Encoded by the At3g06560 gene, AtPAPS3 is a truncated protein, and is involved in the development of pollen (Hunt et al., 2008). AtPAPS1 (At1g17980) is responsible for the polyadenylation of a restricted group of pre-mRNAs, involved in the development of the male gametophyte, leaves and flowers, as well as in response to pathogens, ribosome biogenesis and redox homeostasis. In addition, depletion of AtPAPS1 results in shortening of the poly-A tail of a specific group of transcripts from the SMALL AUXIN UP RNA (SAUR) family, with an evident reduction in the abundance of these transcripts (Trost et al., 2014; Kappel et al., 2015). In cases of AtPAPS2 (At2g25850) and AtPAPS4 (At4g32850), single or double mutants show normal development, suggesting that there may be functional redundancy between these isoforms and AtPAPS1 (Vi et al., 2013). Interestingly, a more recent study has shown that while AtPAPS2 and AtPAPS4 promote flowering, and AtPAPS1 causes delays in the transition to flowering (Czesnick and Lenhard, 2016).
The polyadenylation process seems to be necessary for the nuclear export of mRNAs (Huang and Carmichael, 1996). Polyadenylation also seems to be extremely important for translation, since the high affinity of poly-A-binding protein (PABP) for A-rich sequences, promotes its association with the poly-A tail, allowing the association of PABP with EIF4G, which then associates with EIF4E, in a sequence of interactions that are crucial for the recruitment of the 40S ribosomal subunit (Jacobson and Favreau, 1983; Tarun and Sachs, 1996; Wells et al., 1998; Cho et al., 2019). More importantly, long poly-A tail mRNAs have higher translation rates than short poly-A tail mRNAs (Beilharz and Preiss, 2007). In addition to mediating transcription termination, the polyadenylation process also likely mediates processes such as initiation, promoting the recycling of proteins involved in transcription (Mapendano et al., 2010), and elongation, as demonstrated by the stable interaction of CPSF with the transcription factor TFIID and with the Pol II elongation complex (Dantonel et al., 1997).
Poly-A tail is also essential for mRNA stability since non-polyadenylation mRNAs are targets of PTGS via RNA-dependent RNA polymerase 6 (RDR6) in plants. It has been reported that non-polyadenylated mRNAs are used as templates by RDR6 to produce long double-stranded RNAs (dsRNAs) in plants. These molecules are subsequently used as a substrate by dicer-like enzymes (DCL2 and DCL4), resulting in the production of siRNA by argonaute enzymes and consequently in gene silencing mediated by the RNA-induced silencing complex (RISC) (Dalmay et al., 2000; Baeg et al., 2017). In vitro assays have demonstrated that the poly-A tail inhibits the initiation step, not the RDR6 elongation step and that the poly-A tail size is important for this inhibition (Baeg et al., 2017). Indeed, polyadenylated mRNAs have different proteins attached to their 3’ end and poly-A tail. For example, AtPABN acts as an obstacle to RDR6 binding, while other proteins are involved in nuclear export, thereby evading RDR6 action (Luo and Chen, 2007). In fact, non-polyadenylated mRNAs accumulate in the nucleus (Huang and Carmichael, 1996).
Once in the cytoplasm, mRNA is targeted by different exoribonucleases that shorten the poly-A tail, a process called deadenylation, taken as a starting point that leads to two different pathways of mRNA degradation: 5’ deadenylation-dependent decapping and 3’ decay (Meyer et al., 2004). The main pathway of mRNA decay in yeast is the deadenylation-dependent decapping pathway, which involves a molecular complex composed of different proteins, including two deadenylases, CCR4p/POP2p (Tucker et al., 2001). Interestingly, CCR4-POP2 complex can be recruited by Pumilio homologs (PUMs), resulting in increased deadenylation (Weidmann et al., 2014). After deadenylation, mRNA can be targeted by decapping enzymes, such as Dcp1 and Dcp2, responsible for cleavage of the 5’cap, which makes degradation of the 5’ to 3’ mRNA possible by the hydrolytic activity of the Xrn1p exoribonuclease. Once deadenylated, mRNA may also be degraded in the 3’ to 5’ direction by the exoribonucleolytic activity of a multiprotein complex called the exosome, which does not require cleavage of the 5’cap (Coller and Parker, 2004; Meyer et al., 2004). In addition, mRNAs that have lost the poly-A tail are direct targets of repression by PUMs (Etten et al., 2012). As expected, most of the protein involved in the two different pathways of degradation following mRNA deadenylation are also found in plants. Many of these proteins are encoded by gene families, suggesting specialization or functional redundancy (Chiba and Green, 2009).
The size of the poly-A tail may influence the lifetime of the mRNA due to the continuous shortening that the poly-A tail undergoes in the cytoplasm by the action of deadenylases (Eckmann et al., 2011; Weill et al., 2012). However, a recent study presented data that are in contrast to other studies regarding the correlation between poly-A tail size and stability. Analyzing the data set on somatic cell poly-A tail length, it was found that transcripts with short poly-A tail showed higher levels of stability and translation, while transcripts with long poly-A tail showed lower levels of stability and translation (Lima et al., 2017). Changes in the size of the poly-A tail of certain genes during the cell cycle, such as the shortening of the poly-A tail, preventing translation during the M phase (Park et al., 2016). Nevertheless, no correlation was found between the size of the poly-A tail with translation, not even with accumulation of mRNA at steady-state, but, in contrast, transcripts with longer poly-A tails presented greater stability (Chang et al., 2014). As we will see later, other mechanisms than just the length of the poly-A tail are involved with mRNA metabolism.
Alternative Polyadenylation
Alternative polyadenylation (APA) is a mechanism whereby it is possible to generate transcript isoforms with different 3’UTR or CDS from the same gene, directly reflecting in the diversity of the transcriptome and proteome, and therefore, in the fate of these biomolecules in eukaryotes (Tian and Manley, 2013; Tian and Manley, 2016). APA occurs due to the presence of multiple PAS in the same gene and has been recognized by many authors as one of the main mechanisms of gene regulation (Lutz and Moreira, 2010; Hunt, 2012; Tian and Manley, 2016). Although we have only just begun to unravel the mechanism, we know that APA has temporal (cell and developmental cycle) and tissue specificity, in addition to being involved in different biological processes, such as embryogenesis, gametogenesis, morphogenesis, control of flowering time in plants and control of oncogenes expression in animals (Xing and Li, 2011; Deridder et al., 2012; Tian and Manley, 2016; Ji et al., 2018). Moreover, APA events are also involved with growth and development (Hong et al., 2018), circadian rhythm (Yang et al., 2020), cell signaling (Chakrabarti and Hunt, 2015; Li et al., 2017; Conesa et al., 2020), immunity (Lyons et al., 2013; Ye et al., 2019) and stress response in plants (Zheng et al., 2018; Conesa et al., 2020).
In fact, APA is a potent regulatory agent for gene expression, can affect more than 80% of the genes expresses in a plant (Hunt, 2012). About half of mammalian genes have multiple PAS (Tian et al., 2005), which has also been shown to be true for plants (Shen et al., 2008a; Wu et al., 2011; Hunt, 2012; Wu et al., 2014). In mouse, most PAS are found in the 3’UTR region, but a considerable portion can also be found within intronic regions (Hoque et al., 2013). A similar scenario occurs in plants, and, interestingly, PAS are found even in 5’UTR and coding regions (Hunt, 2012). New bioinformatics approaches based on RNA-seq and other sources, have enabled significant advances in the identification of PAS, which will facilitate the study of the regulation of APA-mediated expression in different plant species (Guo et al., 2016; Wu et al., 2016; Chen et al., 2017; Ye et al., 2018). There is currently a database with PAS sets of at least five different plant species, including A. thaliana, O. sativa, Medicago truncatula, Trifolium pratense and Phyllostachys edulis (Zhu et al., 2020).
The choice of a PAS is determined by the signal strength given by the cis-elements, but not only (Cheng et al., 2006; Shi, 2012; Tian and Manley, 2016; Rosenthal et al., 2018). Mammalian genes that present strong PAS have less frequent or even absent APA events. Furthermore, there is evidence of a relationship with gene function, since genes involved in cell metabolism, morphology, and proliferation are more often targets of the APA mechanism (Wang et al., 2018). However, the distance between competing PAS (Li et al., 2015), the availability and affinity of CPMC for cis-elements, as well as protein interference from other pathways can contribute to choosing one given PAS over another (Shen et al., 2008a; Hornyik et al., 2010; Neve et al., 2017).
In mice, high CstF64 expression results in the choice of weaker PAS from IgM mRNA, resulting in the expression of IgM secreted isoform, suggesting involvement in mouse B cell maturation (Takagaki et al., 1996). Still, in mouse cells, hPcf11 and hFip1 promote the use of proximal PAS. On the other hand, CFIm25 and CFIm68, as well as PABPN1 and poly-A-binding protein C1 (PABPC1) promote the use of distal PAS (Li et al., 2015). In humans, a negative feedback mechanism comes into play in the presence of high levels of CstF77, which results in the selection of PAS in an intronic region near the promoter, leading to the generation of truncated transcripts and consequently downregulation of CstF77 (Luo et al., 2013).
In A. thaliana, the interaction between AtFY and FCA, a RBP involved in flowering promotion, results in the selection of a PAS within intron 3, leading to the production of a truncated and dysfunctional transcript (Simpson et al., 2003). The selection of PAS in intron 3 is promoted by FCA itself in a manner dependent on high cellular levels, acting as a negative feedback, since the removal of intron 3 results in high levels of FCA and early flowering (Quesada et al., 2003). In addition, AtPCFS4 seems to mediate the choice of a PAS in intron 3 in A. thaliana (Xing et al., 2008b). More interesting, the differential use of PAS from the same mRNA encoding AtCPSF30 can generate two distinct proteins. For example, the use of a PAS within an intron results in the production of AtCPSF30, and the use of a more distal PAS results in the production of a mammalian splicing factor-like protein (Delaney et al., 2006). Similarly, the use of two different weak PAS within an intron results in short monofunctional lysine ketoglutarate reductase transcripts in Gossypium hirsutum, and is likely to occur also in A. thaliana, Zea mays and Lycopersicon esculentum (Tang et al., 2002).
The modulation of 3’UTR by APA allows changes in the translational regulation, localization, and stability of mRNAs (Meyers et al., 2004; Tushev et al., 2018). Shortening 3’UTR can make the translation of specific mRNAs more efficient by promoting polysome formation (Chang et al., 2015). Differences in the size of 3’UTR can change the location of mRNAs in the cell (Tushev et al., 2018). Interestingly, the size of 3’UTRs can also directly influence the location of proteins. Short 3’UTRs from CD47 have been shown to promote the localization in the endoplasmic reticulum and long 3’UTRs promote a membrane localization in a HuR-SET-RAC1-dependent manner. This change in location incurred in different functions for the two CD47 isoforms (Berkovits and Mayr, 2015). In mouse, the production of 3’UTR short through the APA allows greater stability of transcripts, since the shortening results in a smaller number of microRNA target sites (Sandeberg et al., 2008). In A. thaliana, the shortening of 3’UTRs of mRNA encoding Rubisco Activase (RCA) in response to heat stress promoted greater transcript stability (Deridder et al., 2012), results that were also found for cotton RCA (Deridder and Salvucci, 2007). On the other hand, two studies found that long 3’UTRs were less targeted by microRNAs (Kim et al., 2014; Agarwal et al., 2015). Similarly, Tushev et al. (2018) found that longer 3’UTRs promote higher stability of mRNAs than short 3’UTRs. Whereas, no difference in stability was found between short and long 3’UTR isoforms of most mouse fibroblast mRNAs (Spies et al., 2013). Intriguingly, long 3’UTRs seem to prevent degradation of mRNAs that have uncommon codons (Mishima and Tomari, 2016).
Although the results are controversial, in addition to the presence or absence of microRNA sites, we must consider that APA generates different 3’UTRs with different cis-acting elements that can vary in their composition and diversity according to the size of the 3’UTR, sometimes with stabilizing elements, sometimes with elements destabilizing or both. Moreover, long 3’UTRs can form secondary structures (stem-loop) with stabilizing effects, which perhaps short 3’UTRs do not form. Furthermore, APA events and the diversity of regulatory factors may vary according to cell type or state, tissue type or even with environmental stimulus, as well as the availability and performance of trans-regulatory factors (Deridder et al., 2012; Ulitsky et al., 2012; Lackford et al., 2014; Díaz-muñoz et al., 2015; Kim et al., 2016).
Cis-Acting Elements in 3’UTRs
During the final stages of the eukaryotic transcription, the CPMC promotes cleavage and polyadenylation of the pre-mRNA in its 3’ end. These two processes are guided by cis-elements, and occurs mainly in 3’ regulatory regions. After cleavage, the part of the 3’ regulatory region that remained in the mRNA is now called 3’UTR ( Figure 1 ), comprising from the cleavage site to the stop codon of the CDS. Compared to less complex organisms, 3’UTRs of higher organisms have expanded, reaching an average size that can be almost ten times larger (Jan et al., 2011). 3’UTRs are involved in important post-transcriptional processes, including, but not limited to stability, transport and mRNA translation (Mayr, 2016; Mayr, 2017), in addition to mediating protein localization, and therefore, its function (Berkovits and Mayr, 2015). The effects of 3’UTRs on these processes are mediated by RBPs that bind to a wide variety of specific sequences or cis-acting elements present in these regions (Cho et al., 2019). Interestingly, some RBPs may be their expression regulated by the 3’UTRs themselves, perhaps involving the presence of cis-acting elements (Tian et al., 2019). It is estimated that the human genome can encode at least 1,500 RBPs (Baltz et al., 2012; Gerstberger et al., 2014). In A. thaliana, studies point to somewhere between 1,145 and 1,408 RBPs (Marondedze et al., 2016; Marondedze et al., 2019). Once bound to the mRNA, these RBPs recruit effector proteins that will then determine the fate of the transcript (Mayr, 2016).
The AU-rich element (ARE), has between 50 and 150 nt in size and can present multiple copies of the AUUUA motif, normally found in the 3’UTR (Gutiérrez et al., 1999). In mammals, the ARE element is involved in both the 3’ to 5’ and 5’ to 3’ mRNA decay. Some ARE-binding RBPs can promote recruitment complexes involved in translational decay or silencing, others can inhibit degradation by preventing RBPs involved in decay from interacting with ARE (Garneau et al., 2007; Allen et al., 2013). Site-directed mutagenesis of ARE present in 3’UTRs from her1 was enough to prolong the half-life of reporter mRNAs (Tietz et al., 2020). On the other hand, deletion of a sequence in the 3’UTR of BCL2 with a high content of AREs promoted instability, resulting in low BCL2 levels in murine B cells (Díaz-muñoz et al., 2015). Interestingly, ARE has the potential to induce exosomal degradation of mRNAs without the need for mediation of other proteins, since RNase domains present in exosomal complex proteins have high affinity for AU-rich sequences (Anderson et al., 2006).
Unlike yeast, mammalian transcripts that present the ARE element in their 3’UTR are rapidly degraded, which is also true for plants. The reason for this is due to a poly-A ribonuclease (PARN), a protein with deadenylase activity present only in multicellular eukaryotes (Gutiérrez et al., 1999; Chiba and Green, 2009). PARN promotes rapid deadenylation of mRNA containing multiple copies of the AUUUA pentamer (Lai et al., 2003). It has been reported in HeLa cells that an interaction between CUG-binding protein 1 (CUGBP1) and PARN, promoted the deadenylation of mRNAs that presented ARE in their sequences (Moraes et al., 2006). Consistent with these findings, mutants for AtPARN showed hyperadenylation of embryonic development-specific mRNA, resulting in slow development, that was completely disrupted at the cotyledon stage (Reverdatto et al., 2004). In addition to its role on the stability and translation of transcripts, ARE also has effects on transport and subcellular location (García-mauriño et al., 2017).
Found in 3’UTRs of short half-life transcripts, the sequence UGUUUGUUUGU (GU-rich element, GRE) is involved in the decay of mRNAs also mediated by CUGBP1 (Vlasova et al., 2008; Lee et al., 2010; Rattenbacher et al., 2010; Louis and Bohjanen, 2011). More importantly, Lee et al. (2010) demonstrated that GREs and AREs effects depend on the type of cell, suggesting a cell-specific context, and perhaps a cell-specific stage, since the effects of AREs were more significant in stem cells. This is probably due to different levels of CUGBP1 expression in each cell type, as well as APA events on target transcripts, suggesting that the same may occur for other cis-acting elements. Indeed, according to Kalsotra et al. (2008), the Elav-like family to which CUGBP1 belongs, is down-regulated during the development of the heart. More interestingly, the shortening of the 3’UTRs of CUGBP1 target mRNAs promoted by APA during the activation of human T cells resulted in a lower presence of GREs and increased levels of expression (Beisang et al., 2014).
Also found in 3’UTRs, a CU-rich sequence similar to ARE, seems to promote the instability of transcripts encoding the protein MARCKS (myristoylated alanine-rich C kinase substrate), possibly mediated by CUGBP1. However, the overexpression of two proteins of the Elav family, HuD and HuR, which bind with high affinity to CU-rich sequence promoted high levels of MARCKS expression (Wein et al., 2003). Similar results were found involving the ARE and HuD element in long 3’UTRs of the transcript from brain-derived neurotrophic factor (BDNF) in mouse neurons (Allen et al., 2013), and also for ARE and HuR in HEK293 cells, but an interaction between ARE and ZFP36 promoted degradation of transcripts (Mukherjee et al., 2014). This suggests that like ARE, the CU-rich cis-acting element is also the target of stabilizing and destabilizing RBPs. Despite this, 3’UTRs can have both stabilizing and destabilizing sequences. For example, the KRAS 3’UTR has sequences that bind stabilizing factors, such as HuR, but also has inhibitory sequences that are targets of microRNAs (Kim et al., 2016).
The downstream element (DST), highly conserved in a special gene family encoding small auxin up RNA (SAUR) from plants, consists of repetitions of the ATAGAT and GTA motifs located in the 3’UTR of mRNA (McClure et al., 1989; Newman et al., 1993). Studies involving mutation in these regions have shown that any change in one of these different motifs is enough to increase the stability of the mRNA (Sullivan and Green, 1996; Johnson et al., 2000). Mutations of the Pumilio response element (PRE) and ARE present in 3’UTRs of her1 also dramatically increased the expression levels of the reporter gene. Mutations in ARE alone showed a slight increase in expression, compared to the double mutant PRE and ARE, suggesting that the two elements mediate the decay of mRNAs in parallel (Tietz et al., 2020). The G3A element, GA-rich, located in the 3’UTRs of the chicken elastin mRNA, and confirmed also for other animal species, showed stabilizing effects on the transcripts (Hew et al., 2000). Consistent with this finding, the deletion of a GA-rich sequence upstream of a NUE from the extensin 3’ regulatory region resulted in a reduction of up to 60% in the expression of the target gene in tobacco (Rosenthal et al., 2018).
As revealed by Geisberg et al. (2014), mRNAs with U-rich 3’UTRs also have high stability. Furthermore, it was found that the interaction of the poly-A tail with these U-rich sequences results in the formation of secondary structures (stem-loop and others double-stranded structures) with positive effects on the stability of transcripts. Recently, it was demonstrated that the AT-rich interactive domain-containing protein 5a (Arid5a) stabilizes the OX40 (TNFR) transcripts in Th17 cells, through its interaction with a stem-loop formed by the GU-rich element (ADE-like) present in 3 ‘UTRs of OX40 mRNA (Hanieh et al., 2018). A recent extensive analysis of Zebrafish 3’UTRs using UTR-seq has shown that U-rich (poly-U) and UUAG sequences are involved with stability and GC-rich sequences with instability (Rabani et al., 2017). The C-rich element (CRE) has also been shown to mediate transcript stability. The presence of CRE in 3’UTRs of the mu-opioid receptor (MOR) proved to be determinant for the stabilization of MOR transcripts. Depletion of Poly (rC) binding protein 1 (PCBP1), a CRE ligand, had negative effects on MOR mRNAs half-life. It has been suggested that when interacting with CRE, PCBP1 recruits other RPBs, such as AUF1 and PABP, with this complex being responsible for stabilizing MOR transcripts (Hwang C. K. et al., 2016).
Some RBPs are involved in translational repression. PUMs, are ligands of the PRE element, and are present in different organisms. After inserting PRE into the 3’UTR of the reporter gene for expression in HEK293 cells, PUMs have been shown to promote translational repression by a highly conserved deadenylation pathway, involving the recruitment of the CCR4-NOT (CNOT) complex. Interestingly, there was also a repression by PUMs independent of deadenylation (Etten et al., 2012). In addition, PUMs anchored to 3’UTRs can recruit argonaut and repress the translational activity of eEF1A (Friend et al., 2012). PUMs may also promote translational repression of reporter mRNAs in a PABP-dependent manner, probably preventing the interaction between PABP and EIF4G (Weidmann et al., 2014).
TIAR, an ARE-binding, has been shown to promote translational repression of eIF4A, eIF4E, eEF1B and c-Myc, being found in the 3’UTRs of all these factors (Mazan-mamczarz et al., 2006). A translational repression mediated by the cytoplasmic polyadenylation element (CPE) was also observed. Interestingly, the effects on translational repression are dependent on the proximity of CPE to PAS in the 3’UTRs (Dai et al., 2019). The presence of a CU-rich sequence in the 3’UTR of 15-lipoxygenase (LOX) gene has been identified as a mediator of translational silencing. The interaction of hnRNPK and hnRNPE1 with these sequences prevents the initiation of translation of LOX mRNAs by blocking the assembly of the 80S ribosome during the erythropoiesis process (Ostareck et al., 1997). Other forms of translational repression involving preventing the recruitment of subunits or blocking the assembly of ribosomal complexes, mediated by cis-acting elements and RBPs, have been reported in the literature (Duncan et al., 2006; Deng et al., 2008; Hussey et al., 2011). In fact, cis-acting elements are also found in 5’UTRs, which can act alone and even overlap effects conferred by cis-acting elements from 3’UTRs, and vice versa (Theil et al., 2018).
In addition, cis-acting elements and RBPs are also involved in transport, determining the location of mRNAs (Kislauskis et al., 1994). Equally, cis-acting elements are involved in determining tissue location during developmental stages (Bullock and Ish-horowicz, 2001). This allows for fine regulation of gene expression by promoting an asymmetric distribution of mRNAs, which is essential for cell polarization, division and motility, especially during embryonic development (Martin and Ephrussi, 2009; Zappulo et al., 2017). The construction of chimeric reporters demonstrated that 3’UTR sensorin promotes the localization of mRNAs to distal sensory neurites, although cis-acting elements have not been identified in the 3’UTR (Meer et al., 2012). The presence of localization elements (LEs) in 3’UTRs of approximately one-third of known dendritic mRNAs have been shown to be potent regulators of the location of PSD-95 and CaMKIIa mRNAs (Subramanian et al., 2011).
Plants also have cis-acting elements involved in determining the location of mRNAs. Rice prolamine transcripts are preferentially located in the protein body of the endoplasmic reticulum (ER), mediated by a putative LE present in its 3’UTR. The construction of a chimeric construct showed that the 3’UTR of the prolamine is sufficient to direct the reporter mRNA to protein body ER (Hamada et al., 2003). Rice glutelin also has a putative LE in its 3’UTR that determine its location in the cisternal endoplasmic reticulum (ER), since the 3’UTR of glutelin was sufficient to promote ER cisternal localization of the reporter mRNA (Washida et al., 2009). More recently, it has been demonstrated that the determination of the location of prolamine and glutelin is dependent on two RBPs, RBP-P and RBP-L (Tian et al., 2018; Tian et al., 2019). Indeed, cis-acting elements can also predispose mRNAs to degradation by promoting a cytoplasmic or even tissue localization that is not suitable for the stability or translation of a particular transcript (Ding et al., 1993; Semotok et al., 2005; Tadros et al., 2007).
Additionally, it was observed that cis-acting elements and RBPs are involved in determining protein localization. Berkovits and Mayr (2015) proposed that during the translation process 3’UTRs can function as a scaffold, promoting the interaction of HuR with SET and the nascent amino acids. This interaction between SET and nascent amino acids determines the location of the newly synthesized protein in a RAC1-dependent manner. HuR depletion led to a reduction in surface CD47, CD44, ITGA1, and TNFRSF13C expression. Isoforms with long 3’UTRs from these four proteins showed U-rich sequences, which are probable targets for HuR.
Plant 3’ Regulatory Regions for Expression of Target Genes
A careful selection of modulators of gene expression, such as 3’ regulatory regions, has proven to be an indispensable strategy when the goal is to maximize expression. Although 3’ regulatory regions are extremely important for gene expression, they are still poorly studied compared to other regulatory sequences (Ingelbrecht et al., 1989; Luo and Chen, 2007; Yang et al., 2009; Hirai et al., 2011; Hiwasa-Tanase et al., 2011; Rosenthal et al., 2018). Due to their great potential, 3’ regulatory regions such as NOS and OCS of A. tumefaciens and 35S of cauliflower mosaic virus (CaMV) are widely used in plant expression vectors. Similar to NOS and OCS, the 3’ regulatory region of 35S has cis-elements involved in cleavage and polyadenylation (Mogen et al., 1990; Macdonald et al., 1991; Sanfaçon et al., 1991). Other viral 3’ regulatory regions, such as the figwort mosaic virus (FMV) and the rice tungro bacilliform virus (RTBV), also have efficient polyadenylation signals, although they are less used in plant molecular biotechnology (Hay et al., 1991; Sanfaçon, 1994).
Indeed, 35S has greater potential in regulating expression than NOS and OCS, both in monocot (rice) or dicot (tobacco) plants (Mitsuhara et al., 1996; Nagaya et al., 2010). However, although the number of 3’ regulatory regions identified and validated in plants is still reduced, several studies have shown that plant 3’ regulatory regions have a higher potential to increase expression compared to NOS, OCS or 35S 3’ regulatory regions ( Table 1 ) (Richter et al., 2000; Weeks et al., 2008; Yang et al., 2009; Nagaya et al., 2010; Hirai et al., 2011; Hiwasa-Tanase et al., 2011; Schaart et al., 2011; Kurokawa et al., 2013; Limkul et al., 2015; Diamos et al., 2016; Diamos and Mason, 2018; Yamamoto et al., 2018; Pérez-González and Caro, 2018; Rosenthal et al., 2018).
Table 1.
Expression levels of reporter genes regulated by 3’ regulatory regions from plant genes compared to 3’ regulatory regions of NOS, OCS and 35S widely used in plant molecular biotechnology.
| Plant 3’ regulatory regions | Expression levels of the reporter gene | Organism | Transformation Method | Expression System | References |
|---|---|---|---|---|---|
| Flaveria bidentisMe1 | 64 to 440-fold higher than OCS | F. bidentis | A. tumefaciens | Stable | Marshall et al., 1997 |
| Potato pinII | ~ 10-fold higher than NOS | Potato | A. tumefaciens | Stable | Richter et al., 2000 |
| F. bidentisMe1 | 2 to 26-fold higher than OCS | Tobacco | A. tumefaciens | Stable | Ali and Taylor, 2001 |
| F. bidentisMe1 | ~ 4-fold higher than NOS | Tobacco | A. tumefaciens | Stable | Ali and Taylor, 2001 |
| F. bidentisMe1 | ~ 3-fold higher than OCS | Tobacco | A. tumefaciens | Stable | Schünmann et al., 2003 |
| Rice GluB-1 | 4-fold higher than NOS | Rice | A. tumefaciens | Stable | Yang et al., 2009 |
|
A. thaliana
HSP |
~ 2,5-fold higher than OCS | Arabidopsis and Rice | A. tumefaciens | Transient | Nagaya et al., 2010 |
|
A. thaliana
HSP |
~ 2-fold higher than NOS and 35S | Arabidopsis and Rice | A. tumefaciens | Transient | Nagaya et al., 2010 |
|
A. thaliana
ADH |
~ 1,4-fold higher than NOS | Arabidopsis | A. tumefaciens | Transient | Nagaya et al., 2010 |
|
A. thaliana
UBQ5 |
~ 1,4-fold higher than NOS | Arabidopsis | A. tumefaciens | Transient | Nagaya et al., 2010 |
|
A. thaliana
HSP |
~ 8-fold higher than NOS | Tomato | A. tumefaciens | Stable | Hirai et al., 2011 |
| Apple rbcS | 3 to 11-fold higher than NOS | Apple and tobacco | A. tumefaciens | Stable | Schaart et al., 2011 |
| Richadella dulcificaMIR | 1.5-fold higher than NOS | Tomato | A. tumefaciens | Stable | Hiwasa-Tanase et al., 2011 |
| Rice GluA-2 | 2,45-fold higher than NOS |
Rice | A. tumefaciens | Transient and Stable | Li et al., 2012 |
| Rice GluB-5 | 3,12-fold higher than NOS |
Rice | A. tumefaciens | Transient and Stable | Li et al., 2012 |
| Rice GluC | 2,14-fold higher than NOS |
Rice | A. tumefaciens | Transient and Stable | Li et al., 2012 |
|
A. thaliana
HSP |
~7-fold higher than NOS | Tomato | A. tumefaciens | Stable | Kurokawa et al., 2013 |
|
A. thaliana
HSP |
~2-fold higher than NOS | Nicotiana benthamiana | A. tumefaciens | Stable | Limkul et al., 2015 |
| Potato pinII | 8.5-fold higher than NOS | N. benthamiana | A. tumefaciens | Stable | Diamos and Mason, 2018 |
| Pea rbcS | 5.4-fold higher than NOS | N. benthamiana | A. tumefaciens | Stable | Diamos and Mason, 2018 |
|
A. thaliana
HSP |
2.5-fold higher than NOS | N. benthamiana | A. tumefaciens | Stable | Diamos and Mason, 2018 |
| N. benthamianaNbHSP | 6.3-fold higher than NOS | N. benthamiana | A. tumefaciens | Stable | Diamos and Mason, 2018 |
| N. benthamianaNbACT3 | 8.9-fold higher than NOS | N. benthamiana | A. tumefaciens | Stable | Diamos and Mason, 2018 |
| Tobacco Ext without native intron | 13.9-fold higher than NOS and 2.8-fold higher than 35S | N. benthamiana | A. tumefaciens | Transient | Rosenthal et al., 2018 |
3’ regulatory regions: NOS (nopaline synthase), 35S (cauliflower mosaic virus), OCS (octopine synthase), HSP (heat shock protein), MIR (miraculin), rbcS (ribulose-1,5-biphosphate carboxylase), Glu (glutelin), Me1 (malic enzyme 1), Ext (extensin), pinII (proteinase inhibitor II), NbHSP (N. benthamiana heat shock protein), NbACT3 (N. benthamiana actin 3), ADH (alcohol dehydrogenase), UBQ5 (ubiquitin 5).
In alfalfa seedlings, constructions of the ribulose-1,5-bisphosphate carboxylase (rbcS) 3’ regulatory region was able to regulate higher levels of expression of the GUS reporter gene than the NOS using the FMV 35S promoter of the FMV (Weeks et al., 2008). Tobacco plants transformed with different combinations of promoters and 3’ regulatory regions, also demonstrated that the rbcS 3’ regulatory region results in higher levels of expression compared to NOS (Schaart et al., 2011). Similar results were also found for the pea rbcS 3’ regulatory region in Nicotiana benthamiana (Diamos and Mason, 2018). The regulation ensured by suitable 3’ regulatory regions has also been shown to be efficient for application in new genomic editing technologies, such as the Clustered Regularly Interspaced Short Palindromic Repeat Associated Cas9 Nuclease (CRISPR/Cas9) system. The use of the Pisum sativum rbcS E9 3’ regulatory region showed higher Cas9 levels in A. thaliana egg cells than NOS (Wang et al., 2015). In particular, it has already been demonstrated that the rbcS E9 3’ regulatory region has multiple putative PAS downstream of the canonical PAS (Hunt, 1988). Sequences identified mainly upstream of these PAS in the rbcS E9 3’ regulatory region, FUE and NUE elements, were determinant for the functionality and choice of these PAS. Interestingly, a single FUE appears to be involved in choosing three out of four different PAS (Hunt and Macdonald, 1989). Similar results were found for the 3’ regulatory region of the maize gene encoding (Wu et al., 1993) and of a wheat gene encoding a histone 3 (Ohtsubo and Iwabuchi, 1994).
In rice seeds, the accumulation of a modified house dust mite allergen (mDer f 2) was 4 times higher in constructions with the 3’ regulatory region from glutelin B-1 (GluB-1), compared to the NOS (Yang et al., 2009). Similarly, GluB-5, GluA-2 and GluC 3’ regulatory regions, also resulted in high levels of expression compared to NOS in rice (Li et al., 2012). As revealed by Yang et al. (2009), mRNAs extracted from seeds and leaves regulated by NOS showed a higher diversification in the choice of the PAS than the GluB-1 3’ regulatory region. This intense APA observed in NOS may be because the availability and/or diversity of subunits of the CPMC may vary according to the cell type, degree of cell differentiation, stage of development or in response to environmental changes (Hunt et al., 2008; Rao et al., 2009; Thomas et al., 2012; Lackford et al., 2014; Yu et al., 2019). Interestingly, Hiwasa-Tanase et al. (2011), studying recombinant GUS expression in tomatoes under the regulation of the R. dulcifica MIR 3’ regulatory region (miraculin), found that the PAS used were close to those that occur in native MIR mRNA. In this study, the levels of GUS expression using the MIR 3’ regulatory region were also higher compared to NOS.
In rice cells, the GUS or Renilla Luciferase (Rluc) expression driven respectively by the constitutive CaMV 35S promoter and the elongation factor 1α promoter, were 2-fold higher, when the NOS was replaced by the heat shock protein (HSP) 3’ regulatory region. This demonstrates that expression regulation by the HSP 3’ regulatory region is not affected by the promoter or reporter gene, although studies have shown that there seems to be an ideal combination between 3’ regulatory region and promoters. In addition, the HSP 3’ regulatory region showed higher levels of expression of Rluc, both in monocot and dicot, compared to the NOS, OCS or 35S 3’ regulatory regions (Nagaya et al., 2010). Similar results were also found by Kurokawa et al. (2013) and Limkul et al. (2015).
In tomato fruits, the expression of recombinant MIR using the 35S promoter was 6 to 8-fold higher when the NOS was replaced by the HSP 3’ regulatory region from A. thaliana. (Hirai et al., 2011). Interestingly, the concentration of recombinant MIR varied widely from tissue to tissue, both for NOS and for the HSP 3’ regulatory regions (Hirai et al., 2011). These differences probably also reflect the availability and tissue diversity of CPMC and other RBPs factors involved in post-transcriptional processes. Perhaps it is also due to APA events, allowing fine regulation of expression (Takagaki et al., 1996; Tang et al., 2002; Quesada et al., 2003; Simpson et al., 2003; Delaney et al., 2006; Dong et al., 2007; Ji et al., 2009; Tian and Manley, 2013). Matsui et al. (2014) demonstrated that the use of a longer version of the HSP 3’ regulatory region results in higher levels of expression compared to a smaller version. According to the authors, the high levels of expression achieved in the longer version occurred due to the presence of a matrix attachment region (MAR), AT-rich DNA sequences that assist in the chromatin structural organization, being involved in transcriptional control (Abranches et al., 2005; Tetko et al., 2006).
As revealed by Pérez-González and Caro (2018), increases in expression of firefly luciferase (LUC) regulated by the HSP 3’ regulatory region compared to the 35S, in part, occurred due to less promoter methylation, through a phenomenon called RNA-directed DNA methylation (RdDM). It has been proposed that siRNA produced by PTGS events triggers RdDM, probably involving non-polyadenylated transcripts, resulting from readthrough or improperly terminated mRNA. In N. benthamiana, Diamos and Mason (2018) found that the expression directed by some plant 3’ regulatory regions were 2.5- to 8.9-fold higher than the NOS ( Table 1 ). Similarly, the use of other A. thaliana 3’ regulatory regions, alcohol dehydrogenase (ADH), histone H4 (H4), and ubiquitin 5 (UBI5), also resulted in higher levels of GUS activity compared to NOS (Nagaya et al., 2010). Potato plants transformed to express recombinant hepatitis B surface antigen (HBsAg) showed higher levels of HBsAg mRNA when soybean VSP gene or potato pinII genes 3’ regulatory regions were used, compared to the NOS (Richter et al., 2000).
Also, the Me1 gene 3’ regulatory region from F. bidentis was able to increase GUS expression several times compared to the 35S or OCS 3’ regulatory regions in leaves of F. bidentis plants (Ali and Taylor, 2001). The use of the tobacco extensin 3’ regulatory region (Ext) to express different recombinant reporter proteins in N. benthamiana leaves resulted in high levels of expression compared to the NOS and 35S, even higher than the VSP 3’ regulatory region. The Ext 3’ regulatory region has been shown to prevent readthrough, resulting in high concentrations of recombinant mRNAs and proteins (Diamos et al., 2016). It is worth to note that the presence of a native intron in the Ext tobacco 3’ regulatory region appears to have deleterious effects on expression (Rosenthal et al., 2018).
Rosenthal et al. (2018), demonstrated that the removal of this intron results in levels of expression up to 3-times higher compared to Ext in its native form, and much higher than NOS (13.5 x), VSP (11.9 x) and 35S, although to a lesser degree (2.8 x). Constructions with Ext without intron showed low or undetectable readthrough, and among the 5 NUEs, about 75% of the polyadenylation events observed occurred from the fourth element. These results are in agreement with the expression levels of a shorter version of Ext without intron that lost this main NUE. In fact, a version of Ext without intron, lacking its first 465 nt deleted, showed no expression, probably due to the loss of a putative FUE, demonstrating the importance of this cis-element for gene expression. On the other hand, the presence of a native intron in the maize Hrgp 3’ regulatory region had positive effects on the regulation of expression in maize compared to the 3’ regulatory region in its native conformation (Menossi et al., 2003).
Furthermore, the use of double 3’ regulatory regions has resulted in considerable increases in expression levels compared to the use of a single 3’ regulatory region (Nagaya et al., 2010). According to Luo and Chen (2007), the cloning of two 3’ regulatory regions, 35S and NOS downstream from GUS, was enough to reduce the levels of readthrough transcripts and siRNA, as well as increased GUS expression. Similar results were found by Beyene et al. (2011). In the same way, the use of two 3’ regulatory regions, Ext without a native intron and NbACT3, resulted in 2.8-4 times higher expression levels compared to the use of a single 3’ regulatory region (Diamos and Mason, 2018). Likewise, the combination of two downstream 3’ regulatory regions of GFP, Ext, and HSP in N. benthamiana leaves, resulted in expression levels of 1.7-2.2-fold higher compared to the use of a single 3’ regulatory region. On the other hand, the use of three 3’ regulatory regions appears to result in low levels of expression (Yamamoto et al., 2018).
Finally, some examples of the application of the 3’ regulatory region in plant biotechnology that have revolutionized agriculture are listed. The first transgenic Bt (Bacillus thuringiensis) plant produced, tomato, in 1987, used the NOS 3’ regulatory region (Fischhoff et al., 1987). The 35S has been used in the generation of Bt maize (Koziel et al., 1993). The rice event GR2E (Golden Rice) plants express three transgenes to increase beta-carotene levels, and all the genes use the NOS regulatory region (Paine et al., 2005). The Bollgard Cotton plants have a gene encoding a Bt protein, using a plant 3´regulatory region from the α’ subunit of β-conglycinin gene (Perlak et al., 1990; Biosafety Clearing House, 2006). Likewise, the NOS 3’ regulatory region was used in the production of the first glyphosate-resistant plants, such as soybeans and wheat (Guilan et al., 2002; Parrot and Clemente, 2004). More recently, the rbcS E9 3’ regulatory region has also been used in the generation of glyphosate resistant soybeans (Malven et al., 2015). In addition, transgenic plants have been developed to obtain biomolecules for therapeutic purposes (Chen and Davis, 2016), including, but not limited to antigens, antibodies, epitopes of antigens, coagulation factors and antimicrobial, employing different 3’ regulatory regions, such as NOS, VSP, pinII, and psbA from lettuce (Richter et al., 2000; Giritch et al., 2006; Rajabi-Memari et al., 2006; Schulz et al., 2015; Su et al., 2015a; Su et al., 2015b).
Conclusions and Perspectives
Molecular biotechnology has enabled new ways to exploit the potential of plants. Firstly, the genetic improvement of several species has allowed significant increases in food production (Duvick, 1996; Gepts and Hancock, 2006). With rapid population growth, it is essential to create new strategies to increase food production, their nutritional content, and to reduce environmental impacts. The development of plants more resistant to abiotic and biotic stresses have allowed considerable increases in production, as well as a reduction in the use of pesticides and fertilizers, and even water. Secondly, the possibility of transforming plants into bioreactors has allowed the production of proteins for industrial applications, research, diagnosis, or therapeutic purposes, among others (Sharma and Sharma, 2009; Desai et al., 2010). Besides the production cost being much lower, there are several other advantages and even limitations to producing proteins in non-plant biological systems, such as microorganisms or animal cells (Streatfield, 2007; Desai et al., 2010; Shinmyo and Kato, 2010; Egelkrout et al., 2012; Chen and Davis, 2016).
The use of plant 3’ regulatory regions in the construction of vectors has shown to be able to not only optimize, but also to make possible a fine regulation of gene expression, presenting potential superior to 3’ regulatory regions, NOS, OCS and 35S, commonly used in the production of genetically modified organisms (Mitsuhara et al., 1996; Yang et al., 2009; Nagaya et al., 2010; Diamos and Mason, 2018; Rosenthal et al., 2018), therefore, very useful in plant molecular biotechnology. However, the molecular mechanism behind optimization in expression remains poorly understood. We know that 3’ regulatory regions can have multiple cis-elements directly involved in the cleavage and polyadenylation steps, and, as observed in mammals, polyadenylation in plants certainly requires a very well-orchestrated interaction between cis-elements and CPMC.
Although the canonical NUE AAUAAA is present in more than half of mammalian genes, it is found in only 10% of plant genes, certainly due to the high NUE degeneration that plants tolerate (Rothnie et al., 1994; Loke et al., 2005), consistent with the findings of Hiwasa-Tanase et al. (2011). Analyzing the effects of four different lengths of the MIR 3’ regulatory region (46, 287, 508 and 1085 nt), these authors found almost the same expression levels for the transcripts containing 508 and 1085 nt. Interestingly, only the longest version had canonical NUEs, confirming that plant CPMC can recognize sequences similar to AAUAAA with a high degree of variation. Plant 3’ regulatory regions commonly have several NUE and PAS in their sequences (Nagaya et al., 2010; Hiwasa-Tanase et al., 2011; Rosenthal et al., 2018). The interaction of CPMC with these different NUE and PAS can lead to APA events, and, consequently produce mRNAs with different 3’UTR from the same 3’ regulatory region, resulting in different levels of expression, since 3’UTR can present cis-acting elements involved in the stability, transport and translation of mRNAs. In addition, weak PAS 3’ regulatory regions can promote APA events within exons, resulting in non-stop codon mRNAs, which are direct targets for silencing via nonstop decay (NSD) (Frischmeyer et al., 2002; Szádeczky-Kardoss et al., 2018).
In fact, transformed plants with constructions without a 3’ regulatory region have low transcriptional and translational transgene levels, or even no expression (Ingelbrecht et al., 1989; Luo and Chen, 2007). The absence of the 3’ regulatory region results in the choice of random PAS contained within the plants own genomic DNA, resulting in longer 3’UTR, which generally have cis-acting elements involved in mRNA decay (Ingelbrecht et al., 1989; Shi and Manley, 2015). Furthermore, the absence of 3’ regulatory region may lead to readthrough mRNA or abortive elongation, resulting in incorrectly terminated and non-polyadenylated transcripts, direct targets of RDR6-mediated PTGS (Luo and Chen, 2007; Baeg et al., 2017). In addition, mutated PAS can generate readthrough events, with RNA Polymerase II (RNAPII) sequestering factors involved in initiation and stretching, with negative effects on expression, consistent with the findings by Wei et al. (2017). Pérez-González and Caro (2018), also demonstrated that the absence of a 3’ regulatory region results in high methylation of the promoter through RdDM events, with negative effects on the expression of the target gene. It is likely that the use of 3’ regulatory regions with weak polyadenylation signals may also promote RdDM. It would be interesting to have a better understanding of the mechanisms that regulate these processes, starting with a more detailed exploration of the effects of pathways involved in gene silencing such as PTGS-RDR6 and PTGS-RdDM.
The effects of two 3’ regulatory regions for the expression of the target gene has also generated curious results, worthy of more attention. The use of double 3’ regulatory regions for expression of target genes not only increases expression, but also reduces readthrough events and prevents depletion via PTGS-RDR6, prolonging the half-life of mRNAs (Luo and Chen, 2007; Beyene et al., 2011). Intriguingly, reversing the position of the 3’ regulatory regions, 35S-NOS to NOS-35S, there was a 40% loss in expression levels (Diamos and Mason, 2018). Possibly, this reduction occurred due to the positioning of strong PAS further downstream, when the inversion was made, resulting in longer 3’UTRs that maybe has cis-acting elements involved in the degradation of transcripts (Gutiérrez et al., 1999; Hui et al., 2003; Wein et al., 2003; Garneau et al., 2007; Vlasova et al., 2008; Lee et al., 2010; Allen et al., 2013; Mukherjee et al., 2014; Rabani et al., 2017; Tietz et al., 2020).
Additionally, it would be interesting to explore the effects of the presence of other regulatory sequences on expression, as in the case of MARs (matrix attachment regions) elements. The presence of MARs can interfere with the chromosomal DNA conformation, making genes more accessible for transcription, thus increasing expression rates. As revealed by Mlynárová et al. (2003), MARs can prevent the silencing of transgenes. Corroborating this finding, Matsui et al. (2014) reported that increases in expression levels shown by the long version of the HSP 3’ regulatory region were possibly due to the presence of a MAR, absent in the smaller version of the HSP. Similar results were found by Diamos and Mason (2018), using tobacco MARs RB7 and TM6, downstream of different 3’ regulatory regions, resulting in a 60-fold increase in expression compared to using only the 3’ regulatory region.
Another interesting point for further studies is related to the fact that in some cases the presence of introns in 3’ regulatory regions may have either positive or negative effects on expression (Chung et al., 2006; Bicknell et al., 2012). Probably, intronic regions can, as well as 3’UTRs, have cis-acting elements involved in mRNA metabolism, which may explain this controversy. The repetitive presence of dinucleotide CA in intronic sequences of nitric oxide endothelial synthase (eNOS) was linked to transcript stability, involving heterogeneous nuclear ribonucleoprotein L (hnRNP L). Interestingly, hnRNP L depletion results in reduced eNOS expression, as well as leads to APA in eNOS, suggesting that this CA-rich sequence it is also the target of pro-decay RBPs (Hui et al., 2003). Moreover, the presence of introns in 3’UTRs can promote the degradation of mRNA via nonsense-mediated decay (NMD) (Kertész et al., 2006; Schweingruber et al., 2013). Also, another point that may explain this controversy would be tissue specificity that some introns present (Sieburth and Meyerowitz, 1997; Menossi et al., 2003; Kooiker et al., 2005; Showalter et al., 2010).
Last, but not least, it would interesting be to investigate the effects of the ideal combination between 3’ regulatory regions and promoters. The expression levels of target genes can vary according to the combination of 3’ regulatory regions and promoters. The combination of strong 3’ regulatory regions with strong promoters reduces expression, whereas a combination of weak 3’ regulatory regions with strong promoters, or the opposite, increased expression (Li et al., 2012; Wei et al., 2017). Also, it would be interesting to identify whether and which 3’ regulatory regions present tissue specificity (Debode et al., 2013), widely observed in promoters, as noted by Yang et al. (2009); Hirai et al. (2011); Kurokawa et al. (2013). Indeed, APA events are involved in the regulation of tissue-specific development (Chakrabarti et al., 2018), which may present a higher frequency than splicing events (Wang et al., 2008). This regulation is likely to involve tissue diversity of CPMC factors, as well as inhibitors. However, to test this hypothesis, it will be essential to identify all subunits of the plant CPMC, as well as to know its interaction network, how this network interacts with cis-elements and with possible inhibitors or stimulators. It would also be important to know whether, in fact, the levels and diversity of CPMC subunits change from tissue to tissue, with the stage of the cell cycle or development and also with the cellular state. These and other important questions remain to be answered in future studies.
Author Contributions
Drafting the work and manuscript writing: WB. Critical review and manuscript writing: MM.
Funding
This work was funded by São Paulo Research Foundation (FAPESP), research grant 2013/15576-5 and 2014/50884-5 (MM). WB received a fellowship from Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES, Brazil; 88882.329484/2019-01) and MM was recipient of a Research Fellowship from National Council for Scientific and Technological Development (CNPq, Brazil; Grant #333270/2018-7).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank the FAPESP, CNPq and CAPES.
References
- Abranches R., Shultz R. W., Thompson W. F., Allen G. C. (2005). Matrix attachment regions and regulated transcription increase and stabilize transgene expression. Plant Biotechnol. J. 3, 535–543. 10.1111/j.1467-7652.2005.00144.x [DOI] [PubMed] [Google Scholar]
- Addepalli B., Hunt A. G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35, 4453–4463. 10.1093/nar/gkm457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B., Meeks L. R., Forbes K. P., Hunt A. G. (2004). Novel alternative splicing of mRNAs encoding poly (A) polymerases in Arabidopsis. Biochim. Biophys. Acta - Gene Regul. Mech. 1679, 117–128. 10.1016/j.bbaexp.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J., Bartel D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, 1–38. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Taylor W. C. (2001). The 3′ non-coding region of a c4 photosynthesis gene increases transgene expression when combined with heterologous promoters. Plant Mol. Biol. 46, 325–333. 10.1023/A:1010669204137 [DOI] [PubMed] [Google Scholar]
- Allen M., Bird C., Feng W., Liu G., Li W., Perrone-bizzozero N., II (2013). HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3 9 UTR mRNA. PloS One 8, 9. 10.1371/journal.pone.0055718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. R., Mukherjee D., Muthukumaraswamy K., Moraes K. C. M. (2006). Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. Rna 12, 1810–1816. 10.1261/rna.144606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeg K., Iwakawa H. O., Tomari Y. (2017). The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat. Plants 3, 1–4. 10.1038/nplants.2017.36 [DOI] [PubMed] [Google Scholar]
- Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., et al. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barabino S. M. L., Hiibner W., Jenny A., Minvielle-Sebastia L., Keller W. (1997). The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 387, 1703–1716. 10.1101/gad.11.13.1703 [DOI] [PubMed] [Google Scholar]
- Bardwell V. J., Zarkower D., Edmonds M., Wickens M. (1990). The enzyme that adds poly(A) to mRNAs is a classical poly(A) polymerase. Mol. Cell. Biol. 10, 846–849. 10.1128/mcb.10.2.846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T. H., Preiss T. (2007). Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA J. 13, 982–997. 10.1261/rna.569407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisang D., Reilly C., Bohjanen P. R. (2014). Alternative polyadenylation regulates CELF1/CUGBP1 target transcripts following T cell activation. Gene 550, 93–100. 10.1016/j.gene.2014.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. A., Hunt A. G. (2010). The Arabidopsis ortholog of the 77 kDa subunit of the cleavage stimulatory factor (AtCstF-77) involved in mRNA polyadenylation is an RNA-binding protein. FEBS Lett. 584, 1449–1454. 10.1016/j.febslet.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Berkovits B. D., Mayr C. (2015). Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. 10.1038/nature14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene G., Buenrostro-Nava M. T., Damaj M. B., Gao S. J. (2011). Unprecedented enhancement of transient gene expression from minimal cassettes using a double terminator. Plant Cell Rep. 30, 13–25. 10.1007/s00299-010-0936-3 [DOI] [PubMed] [Google Scholar]
- Bicknell A. A., Cenik C., Chua H. N., Roth F. P., Moore M. J. (2012). Introns in UTRs: Why we should stop ignoring them. Bioessays 34, 1025–1034. 10.1002/bies.201200073 [DOI] [PubMed] [Google Scholar]
- Biosafety Clearing House (2006). Available at: http://bch.cbd.int/database/record.shtml?documentid=14775 (Accessed July 13, 2020)
- Bullock S. L., Ish-horowicz D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611–616. 10.1038/414611a [DOI] [PubMed] [Google Scholar]
- Chakrabarti M., Hunt A. G. (2015). CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 5, 1151–1168. 10.3390/biom5021151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M., Dinkins R. D., Hunt A. G. (2018). Genome-wide atlas of alternative polyadenylation in the forage legume red clover. Sci. Rep. 8, 1–14. 10.1038/s41598-018-29699-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Choi E. A., Shi Y. (2011). Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA 2, 321–335. 10.1002/wrna.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. L., Huppertz I., Yao C., Weng L., Moresco J. J., Iii J. R. Y., et al. (2014). CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3’ processing. Genes Dev. 28, 2370–2380. 10.1101/gad.250993.114.these [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Lim J., Ha M., Kim V. N. (2014). TAIL-seq: Genome-wide determination of poly(A) tail length and 3’ end modifications. Mol. Cell 53, 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Chang J.-W., Zhang W., Yeh H.-S., Jong E. P. D., Jun S., Kim K.-H., et al. (2015). mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6, 1–9. 10.1038/ncomms8218 [DOI] [PubMed] [Google Scholar]
- Chao L. C., Jamil A., Kim S. J., Huang L., G. M. H. (1999). Assembly of the cleavage and Polyadenylation apparatus requires about 10 seconds In Vivo and is faster for strong than for weak poly (A) sites. Mol. Cell. Biol. 19, 5588–5600. 10.1128/MCB.19.8.5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Davis K. R. (2016). The potential of plants as a system for the development and production of human biologics. F1000Research 5, 912. 10.12688/f1000research.8010.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jia Q., Song Y., Fu H., Wei G., Ni T. (2017). Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinf. 15, 287–300. 10.1016/j.gpb.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Miura R. M., Tian B. (2006). Prediction of mRNA polyadenylation sites by support vector machine. Genome Anal. 22, 2320–2325. 10.1093/bioinformatics/btl394 [DOI] [PubMed] [Google Scholar]
- Chiba Y., Green P. J. (2009). mRNA degradation machinery in plants. J. Plant Biol. 52, 114–124. 10.1007/s12374-009-9021-2 [DOI] [Google Scholar]
- Cho H., Cho H. S., Hwang I. (2019). Emerging roles of RNA-binding proteins in plant development. Curr. Opin. Plant Biol. 51, 51–57. 10.1016/j.pbi.2019.03.016 [DOI] [PubMed] [Google Scholar]
- Chung B. Y. W., Simons C., Firth A. E., Brown C. M., Hellens R. P. (2006). Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7, 1–13. 10.1186/1471-2164-7-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. (2004). Eukaryotic mRNA Decapping. Annu. Rev. Biochem. 73, 861–890. 10.1146/annurev.biochem.73.011303.074032 [DOI] [PubMed] [Google Scholar]
- Conesa C. M., Saez A., Navarro-neila S., Lorenzo L., Hunt A. G., Sep E. B., et al. (2020). Alternative Polyadenylation and Salicylic acid nitrogen availability. Plants 9, 16. 10.3390/plants9020251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesnick H., Lenhard M. (2016). Antagonistic control of flowering time by functionally specialized poly(A) polymerases in Arabidopsis thaliana. Plant J. 88, 570–583. 10.1111/tpj.13280 [DOI] [PubMed] [Google Scholar]
- Dai X., Jiang J., Sha Q., Jiang Y., Ou X., Fan H. (2019). A combinatorial code for mRNA 3 -UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 47, 328–340. 10.1093/nar/gky971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D. C. (2000). An RNA-dependent RNA polymerase gene in arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. 10.1016/S0092-8674(00)80864-8 [DOI] [PubMed] [Google Scholar]
- Dantonel J. C., Murthy K. G. K., Manley J. L., Tora L. (1997). Transcription factor TFIID recruits factor CPSF for formation of 3’ end of mRNA. Nature 389, 399–402. 10.1038/38763 [DOI] [PubMed] [Google Scholar]
- Debode F., Janssen E., Berben G. (2013). Development of 10 new screening PCR assays for GMO detection targeting promoters (pFMV, pNOS, pSSuAra, pTA29, pUbi, pRice actin) and terminators (t35S, tE9, tOCS, tg7). Eur. Food Res. Technol. 236, 659–669. 10.1007/s00217-013-1921-1 [DOI] [Google Scholar]
- Delaney K. J., Xu R., Zhang J., Li Q. Q., Yun K.-Y., Falcone D. L., et al. (2006). Calmodulin Interacts with and Regulates the RNA-Binding Activity of an Arabidopsis Polyadenylation factor subunit. Plant Physiol. 140, 1507–1521. 10.1104/pp.105.070672.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Singer R. H., Gu W. (2008). Translation of ASH1 mRNA is repressed by Puf6p – Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 22, 1037–1050. 10.1101/gad.1611308.tion [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deridder B. P., Salvucci M. E. (2007). Modulation of Rubisco activase gene expression during heat stress in cotton (Gossypium hirsutum L.) involves post-transcriptional mechanisms. Plant Sci. 172, 246–254. 10.1016/j.plantsci.2006.08.014 [DOI] [Google Scholar]
- Deridder B. P., Shybut M. E., Dyle M. C., Kremling K. A. G., Shapiro M. B. (2012). Changes at the 3’-untranslated region stabilize Rubisco activase transcript levels during heat stress in Arabidopsis. Planta 236, 463–476. 10.1007/s00425-012-1623-0 [DOI] [PubMed] [Google Scholar]
- Desai P. N., Shrivastava N., Padh H. (2010). Production of heterologous proteins in plants : Strategies for optimal expression. Biotechnol. Adv. 28, 427–435. 10.1016/j.biotechadv.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Diamos A. G., Mason H. S. (2018). Chimeric 3’ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J. 16, 1971–1982. 10.1111/pbi.12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamos A. G., Rosenthal S. H., Mason H. S. (2016). 5′ and 3′ Untranslated Regions Strongly Enhance Performance of Geminiviral Replicons in Nicotiana benthamiana Leaves. Front. Plant Sci. 7:200. 10.3389/fpls.2016.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-muñoz M. D., Bell S. E., Turner M. (2015). Deletion of AU-Rich Elements within the Bcl2 3’UTR Reduces Protein Expression and B Cell Survival In Vivo. PloS One 10, 1–19. 10.1371/journal.pone.0116899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Parkhurst S. M., Halsell S. R., Lipshitz H. D. (1993). Dynamic Hsp83 RNA Localization during Drosophila Oogenesis and Embryogenesis. Mol. Cell. Biol. 13, 3773–3781. 10.1128/MCB.13.6.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Deng Y., Chen J., Wang S., Peng S., Dai C., et al. (2007). An exploration of 3’-end processing signals and their tissue distribution in Oryza sativa. Gene 389, 107–113. 10.1016/j.gene.2006.10.015 [DOI] [PubMed] [Google Scholar]
- Duncan K., Grskovic M., Strein C., Beckmann K., Niggeweg R., Abaza I., et al. (2006). Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3’ UTR : translational repression for dosage compensation. Genes Dev. 20, 368–379. 10.1101/gad.371406.form [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D. N. (1996). Plant breeding, an evolutionary concept. Crop Sci. 36, 539. 10.2135/cropsci1996.0011183x003600030001x [DOI] [Google Scholar]
- Eckmann C. R., Rammelt C., Wahle E. (2011). Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2, 348–361. 10.1002/wrna.56 [DOI] [PubMed] [Google Scholar]
- Edmonds M., Abrams R. (1960). Polynucleotide Adenylate biosynthesis: formation of a sequence units from Adenosine triphosphate by an enzyme from Thymus nuclei. J. Biol. Chem. 235, 1142–1149. [PubMed] [Google Scholar]
- Egelkrout E., Rajan V., Howard J. A. (2012). Overproduction of recombinant proteins in plants. Plant Sci. 184, 83–101. 10.1016/j.plantsci.2011.12.005 [DOI] [PubMed] [Google Scholar]
- Etten J. V., Schagat T. L., Hrit J., Weidmann C. A., Brumbaugh J., Coon J. J., et al. (2012). Human Pumilio proteins recruit multiple Deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 287, 36370–36383. 10.1074/jbc.M112.373522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhoff D. A., Bowdish K. S., Perlak F. J., Marrone P. G., McCormick S. M., Niedermeyer J. G., et al. (1987). Insect tolerant transgenic tomato plants. Nat. Biotechnol. 5, 807–813. 10.1038/nbt0887-807 [DOI] [Google Scholar]
- Forbes K. P., Addepalli B., Hunt A. G. (2006). An Arabidopsis Fip1 Homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281, 176–186. 10.1074/jbc.M510964200 [DOI] [PubMed] [Google Scholar]
- Friend K., Campbell Z. T., Cooke A., Kroll-conner P., Wickens M. P., Kimble J. (2012). A conserved PUF – Ago – eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19, 176–183. 10.1038/nsmb.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P. A., Van Hoof A., O’Donnell K., Guerrerio A. L. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Sci. (80-.). 295, 2258–2261. 10.1126/science.1067338 [DOI] [PubMed] [Google Scholar]
- García-mauriño S. M., Rivero-rodríguez F., Velázquez-cruz A., Hernández-vellisca M., Díaz-quintana A., Rosa M. A. D., et al. (2017). RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate. Front. Mol. Biosci. 4:71. 10.3389/fmolb.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Geisberg J. V., Moqtaderi Z., Fan X., Ozsolak F., Struhl K. (2013). Global Analysis of mRNA Isoform Half-Lives Reveals Stabilizing and Destabilizing Elements in Yeast. Cell 156, 812–824. 10.1016/j.cell.2013.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P., Hancock J. (2006). The future of plant breeding. Crop Sci. 46, 1630–1634. 10.2135/cropsci2005-12-0497op [DOI] [Google Scholar]
- Gerstberger S., Hafner M., Tuschl T. (2014). A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritch A., Marillonnet S., Engler C., Eldik G. V., Botterman J., Klimyuk V., et al. (2006). Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. PNAS 103, 14701–14706. 10.1073/pnas.0606631103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov P. N., Li J., Yu P., Yan W., MacDonald C. C. (2018. a). Cstf2t Regulates expression of histones and histone-like proteins in male germ cells. Andrology 6, 1–11. 10.1111/andr.12488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov P. N., Masoumzadeh E., Latham M. P., Macdonald C. (2018. b). The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity. Nucleic Acids Res. 46, 12022–12039. 10.1093/nar/gky862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilan C., Catherine M. H., Hua-Ping Z. (2002). Glyphosate tolerant wheat plant 33391 and compositions and methods for detection thereof 35. [Google Scholar]
- Guo C., Spinelli M., Liu M., Li Q. Q., Liang C. (2016). A Genome-wide Study of “non-3UTR” Polyadenylation Sites in Arabidopsis thaliana. Sci. Rep. 6, 1–10. 10.1038/srep28060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R. A., Macintosh G. C., Green P. J. (1999). Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 4, 429–438. 10.1016/S1360-1385(99)01484-3 [DOI] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Sakulsingharoj C., Choi S., Wu Y. (2003). Dual regulated RNA Transport pathways to the cortical region in developing rice endosperm. Plant Cell 15, 2265–2272. 10.1105/tpc.013821.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanieh H., Hanieh H., Masuda K., Metwally H., Chalise J. P., Mohamed M., et al. (2018). Molecular immunology and signaling Arid5a stabilizes OX40 mRNA in murine CD4 + T cells by recognizing a stem-loop structure in its 3 UTR. Eur. J. Immunol. 48, 593–604. 10.1002/eji.201747109 [DOI] [PubMed] [Google Scholar]
- Harris J. C., Martinez J. M., Grozdanov P. N., Bergeson S. E., Grammas P., Macdonald C. C. (2016). The Cstf2t Polyadenylation gene plays a sex-specific role in learning behaviors in mice. PloS One 11, 1–19. 10.1371/journal.pone.0165976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J. M., Jones M. C., Blakebrough M. L., Dasgupta I., Davies J. W., Hull R. (1991). An analysis of the sequence of an infectious clone of rice tungro bacilliform virus, a plant pararetrovirus. Nucleic Acids Res. 19, 2615–2621. 10.1093/nar/19.10.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Molnar A., Jones A., Baulcombe D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis 103, 14994–15001. 10.1073/pnas.0606536103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew Y., Lau C., Grzelczak Z., Keeley F. W. (2000). Identification of a GA-rich sequence as a protein-binding site in the 3’-untranslated region of chicken elastin mRNA with a potential role in the developmental regulation of elastin mRNA stability. J. Biol. Chem. 275, 24857–24864. 10.1074/jbc.M002776200 [DOI] [PubMed] [Google Scholar]
- Hirai T., Kurokawa N., Duhita N., Hiwasa-Tanase K., Kato K., Kato K., et al. (2011). The HSP terminator of Arabidopsis thaliana induces a high level of miraculin accumulation in transgenic tomatoes. J. Agric. Food Chem. 59, 9942–9949. 10.1021/jf202501e [DOI] [PubMed] [Google Scholar]
- Hiwasa-Tanase K., Nyarubona M., Hirai T., Kato K., Ichikawa T., Ezura H. (2011). High-level accumulation of recombinant miraculin protein in transgenic tomatoes expressing a synthetic miraculin gene with optimized codon usage terminated by the native miraculin terminator. Plant Cell Rep. 30, 113–124. 10.1007/s00299-010-0949-y [DOI] [PubMed] [Google Scholar]
- Hong L., Ye C., Lin J., Fu H., Wu X., Li Q. Q. (2018). Alternative polyadenylation is involved in auxin-based plant growth and development. Plant J. 93, 246–258. 10.1111/tpj.13771 [DOI] [PubMed] [Google Scholar]
- Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., et al. (2013). Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10, 133–139. 10.1038/nmeth.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornyik C., Duc C., Rataj K., Terzi L. (2010). Alternative polyadenylation of antisense RNAs and flowering time control. RNA UK 38, 1077–1081. 10.1042/BST0381077 [DOI] [PubMed] [Google Scholar]
- Huang Y., Carmichael G. G. (1996). Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16, 1534–1542. 10.1128/mcb.16.4.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Reither G., Bindereif A. (2003). Novel functional role of CA repeats and hnRNP L in RNA stability. RNA 9, 931–936. 10.1261/rna.5660803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. G., Macdonald M. H. (1989). Deletion analysis of the polyadenylation signal of a pea ribulose-1,5-bisphosphate carboxylase small-subunit gene. Plant Mol. Biol. 13, 125–138. 10.1007/BF00016132 [DOI] [PubMed] [Google Scholar]
- Hunt A. G., Xu R., Addepalli B., Rao S., Forbes K. P., Meeks L. R., et al. (2008). Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 56, 1–15. 10.1186/1471-2164-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. G., Xing D., Li Q. Q. (2012). Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants. BMC Genomics 13, 1–12. 10.1186/1471-2164-13-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. G. (1988). Identification and characterization of Cryptic Polyadenylation sites in the 3’ region of a Pea Ribulose-1,5-Bisphosphate carboxylase small subunit gene. DNA 7, 329–336. 10.1089/dna.1.1988.7.329 [DOI] [PubMed] [Google Scholar]
- Hunt A. G. (2008). Messenger mRNA 3’-end formation in plants. Nucl. pre-mRNA Process. Plants Curr. Top. Microbiol. Immunol. 326, 151–176. 10.1007/978-3-540-76776-3 [DOI] [PubMed] [Google Scholar]
- Hunt A. G. (2012). RNA regulatory elements and polyadenylation in plants. Front. Plant Sci. 2:109. 10.3389/fpls.2011.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey G. S., Chaudhury A., Dawson A. E., Lindner D. J., Knudsen C. R., Wilce M. C. J., et al. (2011). Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41, 419–431. 10.1016/j.molcel.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C. K., Wagley Y., Law P., Wei L., Loh H. H. (2016). Phosphorylation of poly (rC) binding protein 1 (PCBP1) contributes to stabilization of mu opioid receptor (MOR) mRNA via interaction with AU-rich element RNA-binding protein 1 (AUF1) and poly A binding protein (PABP). Gene 1, 18. 10.1016/j.gene.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H., Park C. Y., Goodarzi H., Moore M. J., Saito Y., Darnell R. B., et al. (2016). PAPERCLIP identifies MicroRNA targets and a role of CstF64/64tau in promoting non-canonical poly (A) site usage. CellReports 15, 1–13. 10.1016/j.celrep.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelbrecht I. L. W., Herman L. M. F., Dekeyser R. A., Montagu M. C. V., Depicker A. G. (1989). Different 3’ end regions strongly influence the level of gene expression in plant cells. Plant Cell 1, 671. 10.2307/3868957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A., Favreau M. (1983). Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 11, 6353–6368. 10.1093/nar/11.18.6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P. (2011). Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 469, 97–101. 10.1038/nature09616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T. H., Patricio K., Mccarthy T., Rosbash M. (2001). A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7, 887–898. 10.1016/S1097-2765(01)00232-5 [DOI] [PubMed] [Google Scholar]
- Ji Z., Ju Y. L., Pan Z., Jiang B., Tian B. (2009). Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development (Proceedings of the National Academy of Sciences of the United States of Americ ) 106, (7028-7033) DOI 10.1073/pnas.090. Proc. Natl. Acad. Sci. U. S. A. 106, 17, 7028–7033. 10.1073/pnas.0904454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Chen M., Ye W., Zhu S., Ye C., Su Y., et al. (2018). TSAPA: Identification of tissue-specific alternative polyadenylation sites in plants. Bioinformatics 34, 2123–2125. 10.1093/bioinformatics/bty044 [DOI] [PubMed] [Google Scholar]
- Jiang S., Kumar S., Eu Y. J., Jami S. K., Stasolla C., Hill R. D. (2012). The Arabidopsis mutant, fy-1, has an ABA-insensitive germination phenotype. J. Exp. Bot. 63, 2693–2703. 10.1093/jxb/err452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. A., Pérez-Amador M. A., Lidder P., Green P. J. (2000). Mutants of Arabidopsis defective in a sequence- specific mRNA degradation pathway. PNAS 97, 13991–13996. 10.1073/pnas.240354097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A., Xiao X., Ward A. J., Castle J. C., Johnson J. M., Burge C. B., et al. (2008). A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U. S. A. 105, 20333–20338. 10.1073/pnas.0809045105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel C., Trost G., Czesnick H., Ramming A., Kolbe B., Vi S. L., et al. (2015). Genome-wide analysis of PAPS1-dependent polyadenylation identifies novel roles for functionally specialized Poly(A) Polymerases in Arabidopsis thaliana. PloS Genet. 11, 1–30. 10.1371/journal.pgen.1005474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I., Martin G., Friedlein A., Langen H., Keller W. (2004). Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626. 10.1038/sj.emboj.7600070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertész S., Kerényi Z., Mérai Z., Bartos I. (2006). Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34, 6147–6157. 10.1093/nar/gkl737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yamamoto J., Chen Y., Aida M., Wada T., Handa H., et al. (2010). Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes to Cells 15, 1003–1013. 10.1111/j.1365-2443.2010.01436.x [DOI] [PubMed] [Google Scholar]
- Kim D., Kim J., Baek D. (2014). Global and local competition between exogenously introduced microRNAs and endogenously expressed microRNAs. Mol. Cells 37, 412–417. 10.14348/molcells.2014.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Kogan N., Slack F. J. (2016). Cis-acting elements in its 3 ′ UTR mediate post-transcriptional regulation of KRAS. Oncotarget 7, 14. 10.18632/oncotarget.7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis E. H., Zhu X., Singer R. H. (1994). Sequences responsible for intracellular localization of -Actin messenger RNA also affect cell phenotype. J. Cell Biol. 127, 441–451. 10.1083/jcb.127.2.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiker M., Airoldi C. A., Losa A., Manzotti P. S., Finzi L., Kater M. M., et al. (2005). Basic Pentacysteine1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene Seedstick. Plant Cell 17, 722–729. 10.1105/tpc.104.030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel M. G., Beland G., Evola S. (1993). Field performance of elite trangenic mazie plants expressing an insecticidal protein derived from Bt. Nat. Biotechnol. 11, 194–200. 10.1038/nbt0293-194 [DOI] [Google Scholar]
- Kubo T., Wada T., Yamaguchi Y., Shimizu A., Handa H. (2006). Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3’-UTRs. Nucleic Acids Res. 34, 6264–6271. 10.1093/nar/gkl794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U., Gündel M., Knoth A., Kerwitz Y., Rüdel S., Wahle E. (2009). Poly(A) tail length is controlled by the nuclear Poly(A)-binding protein regulating the interaction between Poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814. 10.1074/jbc.M109.018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa N., Hirai T., Takayama M., Hiwasa-Tanase K., Ezura H. (2013). An E8 promoter-HSP terminator cassette promotes the high-level accumulation of recombinant protein predominantly in transgenic tomato fruits: A case study of miraculin. Plant Cell Rep. 32, 529–536. 10.1007/s00299-013-1384-7 [DOI] [PubMed] [Google Scholar]
- Lackford B., Yao C., Charles G. M., Weng L., Zheng X., Choi E., et al. (2014). Fip 1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33, 878–889. 10.1002/embj.201386537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W. S., Kennington E. A., Blackshear P. J. (2003). Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23, 3798–3812. 10.1128/mcb.23.11.3798-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Lee J. Y., Wilusz J., Tian B., Wilusz C. J. (2010). Systematic analysis of Cis-elements in unstable mRNAS demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PloS One 5, 14. 10.1371/journal.pone.0011201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Hunt A. G. (1997). The Polyadenylation of RNA in Plants. Plant Physiol. 115, 321–325. 10.1104/pp.115.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. J., Dai L. L., Chai Z. J., Yin Z. J., Qu L. Q. (2012). Evaluation of seed storage protein gene 3’-untranslated regions in enhancing gene expression in transgenic rice seed. Transgenic Res. 21, 545–553. 10.1007/s11248-011-9552-4 [DOI] [PubMed] [Google Scholar]
- Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., et al. (2015). Systematic profiling of Poly(A)+ transcripts modulated by Core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PloS Genet. 11, 1–29. 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang R., Gao Y., Wang C., Zhao L., Xu N., et al. (2017). The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. New Phytol. 216, 1205–1222. 10.1111/nph.14743 [DOI] [PubMed] [Google Scholar]
- Lima S. A., Chipman L. B., Nicholson A. L., Chen Y. H., Yee B. A., Yeo G. W., et al. (2017). Short Poly(A) tails are a conserved feature of highly expressed genes. Physiol. Behav. 176, 1057–1063. 10.1016/j.physbeh.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limkul J., Misaki R., Kato K., Fujiyama K. (2015). The combination of plant translational enhancers and terminator increase the expression of human glucocerebrosidase in Nicotiana benthamiana plants. Plant Sci. 240, 41–49. 10.1016/j.plantsci.2015.08.018 [DOI] [PubMed] [Google Scholar]
- Loke J. C., Stahlberg E. A., Strenski D. G., Haas B. J., Wood P. C., Li Q. Q. (2005). Compilation of mRNA Polyadenylation Signals in Arabidopsis Revealed a New Signal Element and Potential Secondary Structures. Plant Physiol. 138, 1457–1468. 10.1104/pp.105.060541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis I. V. S., Bohjanen P. R. (2011). Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Genet. Dev. 21, 444–451. 10.1016/j.gde.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Chen Z. (2007). Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell Online 19, 943–958. 10.1105/tpc.106.045724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Ji Z., Pan Z., You B., Hoque M., Li W., et al. (2013). The conserved intronic cleavage and Polyadenylation Site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PloS Genet. 9, 1–14. 10.1371/journal.pgen.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C. S., Moreira A. (2010). Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2, 22–31. 10.1002/wrna.47 [DOI] [PubMed] [Google Scholar]
- Lyons R., Iwase A., Gänsewig T., Sherstnev A., Duc C., Barton G. J., et al. (2013). The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci. Rep. 3, 10. 10.1038/srep02866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald M. H., Mogen B. D., Hunt A. G. (1991). Characterization of the polyadenylation signal from the T-DNA-encoded octopine synthase gene. Nucleic Acids Res. 19, 5575–5581. 10.1093/nar/19.20.5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malven M., Rinehart J., Taylor N., Dickinson E. (2015). Soybean event MON89788 and methods for detection thereof 24. [Google Scholar]
- Mandel C. R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J. L., et al. (2006). Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956. 10.1038/nature05363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C. R., Bai Y., Tong L. (2008). Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65, 1099–1122. 10.1007/s00018-007-7474-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano C. K., Lykke-Andersen S., Kjems J., Bertrand E., Jensen T. H. (2010). Crosstalk between mRNA 3′-end processing and transcription initiation. Mol. Cell 40, 410–422. 10.1016/j.molcel.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Marondedze C., Thomas L., Serrano N. L., Lilley K. S., Gehring C. (2016). The RNA-binding protein repertoire of Arabidopsis thaliana. Nat. Publ. Gr. 6, 1–13. 10.1038/srep29766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marondedze C., Thomas L., Gehring C., Lilley K. S. (2019). Changes in the Arabidopsis RNA-binding proteome reveal novel stress response mechanisms. BMC Plant Biol. 19, 1–11. 10.1186/s12870-019-1750-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. S., Stubbs J. D., Chitty J. A., Surin B., Johnson J. M., Taylor W. C. (1997). Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5’ and 3’ sequences. Plant Cell 9, 1515–1525. 10.1105/tpc.9.9.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. C., Ephrussi A. (2009). mRNA localization : gene expression in the spatial dimension. Cell 136, 719–730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumzadeh E., Latham M., Macdonald C., Grozdanov P. (2020). Study of self-association of human CstF-64 RNA recognition motif. Biophys J 118, 520a. 10.1016/j.bpj.2019.11.2862 [DOI] [Google Scholar]
- Matsui T., Sawada K., Takita E., Kato K. (2014). The longer version of Arabidopsis thaliana heat shock protein 18.2 gene terminator contributes to higher expression of stably integrated transgenes in cultured tobacco cells. Plant Biotechnol. 31, 191–194. 10.5511/plantbiotechnology.14.0117b [DOI] [Google Scholar]
- Mayr C. (2016). Evolution and biological roles of alternative 3’UTRs. Trends Cell Biol. 26, 1–11. 10.1016/j.tcb.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. (2017). Regulation by 3 -Untranslated Regions. Annu. Rev. Genet. 51, 171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Mazan-mamczarz K., Lal A., Martindale J. L., Kawai T., Gorospe M. (2006). Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26, 2716–2727. 10.1128/MCB.26.7.2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. A., Hagen G., Brown C. S., Gee M. A., Guilfoyle T. J. (1989). Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1, 229–239. 10.2307/3869039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks L. R., Addepalli B., Hunt A. G. (2009). Characterization of genes encoding Poly(A) Polymerases in plants: evidence for duplication and functional specialization. PloS One 4, 10. 10.1371/journal.pone.0008082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer E. J., Ohtan D., Kim S., Barr I., Guo F., Martin K. C. (2012). Identification of a cis-acting element that localizes mRNA to synapses. PNAS 109, 4639–4644. 10.1073/pnas.1116269109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menossi M., Rabaneda F., Puigdomènech P., Martínez-Izquierdo J. A. (2003). Analysis of regulatory elements of the promoter and the 3’ untranslated region of the maize Hrgp gene coding for a cell wall protein. Plant Cell Rep. 21, 916–923. 10.1007/s00299-003-0602-0 [DOI] [PubMed] [Google Scholar]
- Meyer S., Temme C., Wahle E. (2004). Messenger RNA Turnover in Eukaryotes : pathways and enzymes. Crit. Rev. Biocherm. Mol. Biol. 39, 197–216. 10.1080/10409230490513991 [DOI] [PubMed] [Google Scholar]
- Meyers B. C., Vu T. H., Tej S. S., Ghazal H., Matvienko M., Agrawal V., et al. (2004). Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 22, 1006–1011. 10.1038/nbt992 [DOI] [PubMed] [Google Scholar]
- Millevoi S., Vagner S. (2010). Molecular mechanisms of eukaryotic pre-mRNA 3’ end processing regulation. Nucleic Acids Res. 38, 2757–2774. 10.1093/nar/gkp1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Tomari Y. (2016). Codon usage and 3’UTR length determine maternal mRNA stability in Zebrafish Article Codon Usage and 3’UTR length determine maternal mRNA stability in Zebrafish. Mol. Cell 61, 874–885. 10.1016/j.molcel.2016.02.027 [DOI] [PubMed] [Google Scholar]
- Misra A., Green M. R. (2016). From polyadenylation to splicing: dual role for mRNA 3’ end formation factors. RNA Biol. 13, 259–264. 10.1080/15476286.2015.1112490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I., Ugaki M., Hirochika H., Ohshima M., Murakami T., Goto Y., et al. (1996). Efficient promoter cassettes or enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37, 49–59. 10.1093/oxfordjournals.pcp.a028913 [DOI] [PubMed] [Google Scholar]
- Mlynárová L., Hricová A., Loonen A., Nap J. P. (2003). The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell 15, 2203–2217. 10.1105/tpc.012070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B. D., Macdonald M. H., Graybosch R., Hunt A. G. (1990). Upstream sequences other than AAUAAA are required for efficient messenger RNA 3 ‘ -end formation in plants. Plant Cell 2, 1261–1272. 10.1105/tpc.2.12.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes K. C. M., Wilusz C. J., Wilusz J. (2006). CUG-BP binds to RNA substrates and recruits PARN deadenylase. Rna 12, 1084–1091. 10.1261/rna.59606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N., Jacobs N. C., Hafner M., Kennington E. A., Nusbaum J. D., Tuschl T., et al. (2014). Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 15, 1–16. 10.1186/gb-2014-15-1-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K. G. K., Manley J. L. (1995). The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3’-end formation. Genes Dev. 9, 2672–2683. 10.1101/gad.9.21.2672 [DOI] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. (2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332. 10.1093/pcp/pcp188 [DOI] [PubMed] [Google Scholar]
- Neve J., Patel R., Wang Z., Louey A., Furger A. M. (2017). Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 14, 865–890. 10.1080/15476286.2017.1306171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T. C., Ohme-Takagi M., Taylor C. B., Green P. J. (1993). DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5, 701. 10.2307/3869812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorodnikov A., Levin M., Tattikota S., Tokalov S., Hoque M., Scherzinger D., et al. (2018). Transcriptome 3’-end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat. Commun. 9, 1–16. 10.1038/s41467-018-07580-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo N., Iwabuchi M. (1994). The conserved 3’-flanking sequence, AATGGAAATG, of the wheat histone H3 gene is necessary for the accurate 3’-end formation of mRNA. Nucleic Acids Res. 22, 1052–1058. 10.1093/nar/22.6.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D. H., Ostareck-lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. (1997). mRNA silencing in Erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-Lipoxygenase translation from the 3’ End. Cell 89, 597–606. 10.1016/S0030-6657(08)70226-9 [DOI] [PubMed] [Google Scholar]
- Paine J. A., Shipton C. A., Chaggar S., Howells R. M., Kennedy M. J., Vernon G., et al. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487. 10.1038/nbt1082 [DOI] [PubMed] [Google Scholar]
- Park J., Yi H., Kim Y., Chang H. (2016). Regulation of Poly (A) tail and translation during the somatic cell cycle. Mol. Cell 62, 462–471. 10.1016/j.molcel.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Parrot W. A., Clemente T. E. (2004). “Transgenic Soybean,” in Soybeans: Improvement, Production, and Uses of the Agronomy Monographs book series, 265–302. 10.2134/agronmonogr16.3ed.c7 [DOI] [Google Scholar]
- Pérez-González A., Caro E. (2018). Effect of transcription terminator usage on the establishment of transgene transcriptional gene silencing. BMC Res. Notes 11, 1–8. 10.1186/s13104-018-3649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlak F. J., Deaton R. W., Armstrong T. A., Fuchs R. L., Sims S. R., Greenplate J. T., et al. (1990). Insect resistant cotton plants. Bio/Technology 8, 939–943. 10.1038/nbt1090-939 [DOI] [PubMed] [Google Scholar]
- Quesada V., Macknight R., Dean C., Simpson G. G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22, 3142–3152. 10.1093/emboj/cdg305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M., Pieper L., Chew G., Schier A. F. (2017). A massively parallel reporter assay of 3’UTR sequences identifies In Vivo rules for mRNA A massively parallel reporter assay of 3’UTR sequences identifies In Vivo rules for mRNA degradation. Mol. Cell 68, 1083–1094. 10.1016/j.molcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi-Memari H., Jalali-Javaran M., Rasaee M. J., Rahbarizadeh F. (2006). Expression and Characterization of a Recombinant Single-Domain Monoclonal Antibody Against MUC1 Mucin in Tobacco Plants. Hybridoma 25, 209–215. 10.1089/hyb.2006.25.209 [DOI] [PubMed] [Google Scholar]
- Rao S., Dinkins R. D., Hunt A. G. (2009). Distinctive interactions of the Arabidopsis homolog of the 30 kD subunit of the cleavage and polyadenylation specificity factor (AtCPSF30) with other polyadenylation factor subunits. BMC Cell Biol. 10, 1–12. 10.1186/1471-2121-10-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenbacher B., Beisang D., Wiesner D. L., Jeschke J. C. (2010). Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol. Cell. Biol. 30, 3970–3980. 10.1128/mcb.00624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto S. V., Dutko J. A., Chekanova J. A., Hamilton D. A., Belostotsky D. A. (2004). mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA J. 10, 1200–1214. 10.1261/rna.7540204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter L. J., Thanavala Y., Arntzen C. J., Mason H. S. (2000). Production of hepatitis B surface antigen in transgenic plants for oral immunization. Nat. Biotechnol. 18, 1167–1171. 10.1038/81153 [DOI] [PubMed] [Google Scholar]
- Rosenthal S. H., Diamos A. G., Mason H. S. (2018). An intronless form of the tobacco extensin gene terminator strongly enhances transient gene expression in plant leaves. Plant Mol. Biol. 96, 429–443. 10.1007/s11103-018-0708-y [DOI] [PubMed] [Google Scholar]
- Rothnie H. M., Reid J., Hohn T. (1994). The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3’-end formation in plants. EMBO J. 13, 2200–2210. 10.1002/j.1460-2075.1994.tb06497.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruëgsegger U., Blank D., Keller W. (1998). Human pre-mRNA cleavage Factor I m is related to Spliceosomal SR proteins and can be reconstituted In Vitro from recombinant subunits. Mol. Cell 1, 243–253. 10.1016/S1097-2765(00)80025-8 [DOI] [PubMed] [Google Scholar]
- Ryan K. (2007). Pre-mRNA 3′ cleavage is reversibly inhibited in vitro by cleavage factor dephosphorylation. RNA Biol. 4, 26–33. 10.4161/rna.4.1.4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008). Proliferating cells express mRNAs with shortened 3 UTRs and fewer microRNA target sites. Electr. Eng. 320, 1643–1647. 10.1126/science.1155390.Proliferating [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H., Brodmann P., Hohn T. (1991). A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 5, 141–149. 10.1101/gad.5.1.141 [DOI] [PubMed] [Google Scholar]
- Sanfaçon H. (1994). Analysis of Figwort Mosaic Virus (Plant Pararetrovirus) Polyadenylation Signal. Virology 198, 39–49. 10.1006/viro.1994.1006 [DOI] [PubMed] [Google Scholar]
- Schaart J. G., Tinnenbroek-Capel I. E. M., Krens F. A. (2011). Isolation and characterization of strong gene regulatory sequences from apple, Malus × domestica. Tree Genet. Genomes 7, 135–142. 10.1007/s11295-010-0320-z [DOI] [Google Scholar]
- Schulz S., Stephan A., Hahn S., Bortesi L. (2015). Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc. Natl. Acad. Sci. 112, E5454–E5460. 10.1073/pnas.1513311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C., Rufener S. C., Zünd D., Yamashita A., Mühlemann O. (2013). Nonsense-mediated mRNA decay — Mechanisms of substrate mRNA recognition and degradation in mammalian cells. BBA - Gene Regul. Mech. 1829, 612–623. 10.1016/j.bbagrm.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Schünmann P. H. D., Llewellyn D., Surin B., Boevink P. (2003). A suite of novel promoters and terminators for plant biotechnology. Funct. Plant Biol. 30, 443–452. 10.1071/fp02167 [DOI] [PubMed] [Google Scholar]
- Semotok J. L., Cooperstock R. L., Pinder B. D., Vari H. K., Lipshitz H. D., Smibert C. A. (2005). Smaug recruits the CCR4/POP2/NOT Deadenylase complex to trigger maternal transcript localization in the early drosophila embryo. Curr. Biol. 15, 284–294. 10.1016/j [DOI] [PubMed] [Google Scholar]
- Sharma A. K., Sharma M. K. (2009). Plants as bioreactors : recent developments and emerging opportunities. Biotechnol. Adv. 27, 811–832. 10.1016/j.biotechadv.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Ji G., Haas B. J., Wu X., Zheng J., Reese G. J., et al. (2008. a). Genome level analysis of rice mRNA 3 ‘ -end processing signals and alternative polyadenylation. Nucleic Acids Res. 36, 3150–3161. 10.1093/nar/gkn158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Liu Y., Liu L., Liang C., Li Q. Q. (2008. b). Unique features of nuclear mRNA Poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics 179, 167–176. 10.1534/genetics.108.088971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Manley J. L. (2015). The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev. 29, 889–897. 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2012). Alternative polyadenylation: new insights from global analyses. Rna 18, 2105–2117. 10.1261/rna.035899.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmyo A., Kato K. (2010). Molecular farming: production of drugs and vaccines in higher plants. J. Antibiot. (Tokyo) 63, 431–433. 10.1038/ja.2010.63 [DOI] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513. 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L. E., Meyerowitz E. M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365. 10.1105/tpc.9.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., Dijkwel P. P., Quesada V., Henderson I., Dean C. (2003). FY Is an RNA 3’-end processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787. 10.1016/S0092-8674(03)00425-2 [DOI] [PubMed] [Google Scholar]
- Spies N., Burge C. B., Bartel D. P. (2013). 3′ UTR-Isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 23, 2078–2090. 10.1101/gr.156919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield S. J. (2007). Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol. J. 5, 2–15. 10.1111/j.1467-7652.2006.00216.x [DOI] [PubMed] [Google Scholar]
- Su J., Sherman A., Doerfler P. A., Byrne B. J., Herzog R. W., Daniell H. (2015. a). Oral delivery of Acid Alpha Glucosidase epitopes expressed in plant chloroplasts suppresses antibody formation in treatment of Pompe mice. Plant Biotechnol. J. 13, 1023–1032. 10.1111/pbi.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Zhu L., Sherman A., Wang X., Lin S., Kamesh A., et al. (2015. b). Biomaterials Low cost industrial production of coagulation factor IX bioencapsulated in lettuce cells for oral tolerance induction in hemophilia B. Biomaterials 70, 84–93. 10.1016/j.biomaterials.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Rage F., Tabet R., Flatter E., Mandel J., Moine H. (2011). G–quadruplex RNA structure as a signal for neurite mRNA targeting. Eur. Mol. Biol. Organ. 12, 697–704. 10.1038/embor.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. L., Green P. J. (1996). Mutational analysis of the DST element in tobacco cells and transgenic plants: Identification of residues critical for mRNA instability. RNA 2, 308–315. [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang Y., Hamilton K., Manley J. L., Shi Y., Walz T., et al. (2018). Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl. Acad. Sci. U. S. A. 115, E1419–E1428. 10.1073/pnas.1718723115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szádeczky-Kardoss I., Csorba T., Auber A., Schamberger A., Nyikó T., Taller J., et al. (2018). The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 46, 4632–4648. 10.1093/nar/gky279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Goldman A. L., Babak T., Menzies F., Vardy L., Orr-weaver T., et al. (2007). SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU Kinase. Dev. Cell 12, 143–155. 10.1016/j.devcel.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt R. L., Peterson M. L., Manley J. L. (1996). The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87, 941–952. 10.1016/S0092-8674(00)82000-0 [DOI] [PubMed] [Google Scholar]
- Tang G., Zhu X., Gakiere B., Levanony H., Kahana A., Galili G. (2002). The Bifunctional LKR/SDH locus of plants also encodes a highly active monofunctional Lysine-Ketoglutarate Reductase using a Polyadenylation signal located within an Intron. Plant Physiol. 130, 147–154. 10.1104/pp.005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S. Z., Sachs A. B. (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15, 7168–7177. 10.1002/j.1460-2075.1996.tb01108.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetko I. V., Haberer G., Rudd S., Meyers B., Mewes H. W., Mayer K. F. X. (2006). Spatiotemporal expression control correlates with intragenic scaffold matrix attachment regions (S/MARs) in Arabidopsis thaliana. PloS Comput. Biol. 2, 136–145. 10.1371/journal.pcbi.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil K., Herzog M., Rajewsky N. (2018). Post-transcriptional regulation by 3’ UTRs can be masked by regulatory elements in 5’ UTRs. Cell Rep. 22, 3217–3226. 10.1016/j.celrep.2018.02.094 [DOI] [PubMed] [Google Scholar]
- Thomas P. E., Wu X., Liu M., Gaffney B., Ji G., Li Q. Q., et al. (2012). Genome-Wide Control of Polyadenylation Site Choice by CPSF30 in Arabidopsis. Plant Cell 24, 4376–4388. 10.1105/tpc.112.096107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Manley J. L. (2013). Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem. Sci. 38, 312–320. 10.1016/j.tibs.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Manley J. L. (2016). Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Hu J., Zhang H., Lutz C. S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212. 10.1093/nar/gki158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Chou H.-L., Zhang L., Hwang S.-K., Starkenburg S. R., Doroshenk K. A., et al. (2018). RNA-Binding Protein RBP-P is required for Glutelin and Prolamine mRNA localization in rice Endosperm cells. Plant Cell 30, 2529–2552. 10.1105/tpc.18.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Chou H.-L., Zhang L., Okita T. W. (2019). Targeted Endoplasmic reticulum localization of storage protein mRNAs requires the RNA-binding protein RBP-L. Plant Physiol. 179, 1111–1131. 10.1104/pp.18.01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz K. T., Gallagher T. L., Mannings M. C., Morrow Z. T., Derr N. L., Amacher S. L. (2020). Pumilio response and AU-rich elements drive rapid decay of Pnrc2-regulated cyclic gene transcripts. Dev. Biol. 462, 129–140. 10.1016/j.ydbio.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost G., Vi S. L., Czesnick H., Lange P., Holton N., Giavalisco P., et al. (2014). Arabidopsis poly(A) polymerase PAPS1 limits founder-cell recruitment to organ primordia and suppresses the salicylic acid-independent immune response downstream of EDS1/PAD4. Plant J. 77, 688–699. 10.1111/tpj.12421 [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., Parker R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. 10.1016/S0092-8674(01)00225-2 [DOI] [PubMed] [Google Scholar]
- Turner R. E., Pattison A. D., Beilharz T. H. (2018). Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin. Cell Dev. Biol. 75, 61–69. 10.1016/j.semcdb.2017.08.056 [DOI] [PubMed] [Google Scholar]
- Tushev G., Glock C., Heumüller M., Biever A., Jovanovic M., Schuman E. M. (2018). Alternative 3′ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Cell Press 98, 495–511. 10.1016/j.neuron.2018.03.030 [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C. H., Subtelny A. O., Koppstein D., Bell G. W., et al. (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res. 22, 2054–2066. 10.1101/gr.139733.112.Freely [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K., Brown K. M., Gilmartin G. M. (2005). Analysis of a noncanonical poly (A) site reveals a tripartite mechanism for vertebrate poly (A) site recognition. Genes Dev. 19, 1315–1327. 10.1101/gad.1298605.least [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi S. L., Trost G., Lange P., Czesnick H., Rao N., Lieber D., et al. (2013). Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. U. S. A. 110, 13994–13999. 10.1073/pnas.1303967110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova I. A., Tahoe N. M., Fan D., Larsson O. (2008). Conserved GU-rich elements mediate mRNA decay by binding to CUG-Binding Protein 1. Mol. Cell 29, 263–270. 10.1016/j.molcel.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xing H., Dong L., Zhang H., Han C.-Y., Wang X.-C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 1–12. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zheng D., Yehia G., Tian B. (2018). A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res. 28, 1427–1441. 10.1101/gr.237826.118.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wei L., Sheng Y., Zhang G. (2019). Yeast synthetic terminators: fine regulation of strength through linker sequences. ChemBioChem 20, 2383–2389. 10.1002/cbic.201900163 [DOI] [PubMed] [Google Scholar]
- Washida H., Kaneko S., Crofts N., Sugino A., Wang C., Okita T. W. (2009). Identification of cis- localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol. 50, 1710–1714. 10.1093/pcp/pcp103 [DOI] [PubMed] [Google Scholar]
- Weeks J. T., Ye J., Rommens C. M. (2008). Development of an in planta method for transformation of alfalfa (Medicago sativa). Transgenic Res. 17, 587–597. 10.1007/s11248-007-9132-9 [DOI] [PubMed] [Google Scholar]
- Wei L., Wang Z., Zhang G., Ye B. (2017). Characterization of terminators in Saccharomyces cerevisiae and an exploration of factors affecting their strength. ChemBioChem 18, 2422–2427. 10.1002/cbic.201700516 [DOI] [PubMed] [Google Scholar]
- Weidmann C. A., Raynard N. A., Blewett N. H., Etten J. V. A. N., Goldstrohm A. C. (2014). The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA 20, 1298–1319. 10.1261/rna.046029.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L., Belloc E., Bava F. A., Méndez R. (2012). Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 19, 577–585. 10.1038/nsmb.2311 [DOI] [PubMed] [Google Scholar]
- Wein G., Rössler M., Klug R., Herget T. (2003). The 3’ -UTR of the mRNA coding for the major protein kinase C substrate MARCKS contains a novel CU-rich element interacting with the mRNA stabilizing factors HuD and HuR. Eur. J. Biochem. 270, 350–365. 10.1046/j.1432-1033.2003.03396.x [DOI] [PubMed] [Google Scholar]
- Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140. 10.1016/S1097-2765(00)80122-7 [DOI] [PubMed] [Google Scholar]
- Wu L., Ueda T., Messing J. (1993). 3’-end processing of the maize 27 KDa zein mRNA. Plant J. 4, 535–544. 10.1046/j.1365-313X.1993.04030535.x [DOI] [PubMed] [Google Scholar]
- Wu X., Liu M., Downie B., Liang C., Ji G., Li Q. Q., et al. (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. PNAS 108, 12533–12538. 10.1073/pnas.1019732108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Gaffney B., Hunt A. G., Li Q. Q. (2014). Genome-wide determination of poly(A) sites in Medicago truncatula: evolutionary conservation of alternative poly(A) site choice. BMC Genomics 15, 11. 10.1186/1471-2164-15-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Y., Li Q. Q. (2016). PlantAPA: a portal for visualization and analysis of alternative polyadenylation in plants. Front. Plant Sci. 7, 889. 10.3389/fpls.2016.00889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Li Q. Q. (2011). Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2, 445–458. 10.1002/wrna.59 [DOI] [PubMed] [Google Scholar]
- Xing D., Zhao H., Li Q. Q. (2008. a). Arabidopsis CLP1-SIMILAR PROTEIN3, an Ortholog of Human Polyadenylation Factor CLP1, Functions in Gametophyte, Embryo, and Postembryonic Development. Plant Physiol. 148, 2059–2069. 10.1104/pp.108.129817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Zhao H., Xu R., Li Q. Q. (2008. b). Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 54, 899–910. 10.1111/j.1365-313X.2008.03455.x [DOI] [PubMed] [Google Scholar]
- Xing A., Moon B. P., Mills K. M., Falco S. C., Li Z. (2010). Revealing frequent alternative polyadenylation and widespread low-level transcription read-through of novel plant transcription terminators. Plant Biotechnol. J. 8, 772–782. 10.1111/j.1467-7652.2010.00504.x [DOI] [PubMed] [Google Scholar]
- Xu R., Ye X., Li Q. Q. (2004). AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene 324, 35–45. 10.1016/j.gene.2003.09.025 [DOI] [PubMed] [Google Scholar]
- Xu R., Zhao H., Dinkins R. D., Cheng X., Carberry G., Li Q. Q. (2006). The 73 kD Subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol. Biol. 61, 799–815. 10.1007/s11103-006-0051-6 [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Hoshikawa K., Ezura K., Okazawa R., Fujita S., Takaoka M., et al. (2018). Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 8, 1–10. 10.1038/s41598-018-23024-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wakasa Y., Kawakatsu T., Takaiwa F. (2009). The 3′-untranslated region of rice glutelin GluB-1 affects accumulation of heterologous protein in transgenic rice. Biotechnol. Lett. 31, 1625–1631. 10.1007/s10529-009-0056-8 [DOI] [PubMed] [Google Scholar]
- Yang Q., Gilmartin G. M., Doublié S. (2010). Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3’ processing. Proc. Natl. Acad. Sci. 107, 10062–10067. 10.1073/pnas.1000848107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Coseno M., Gilmartin G. M., Doublié S. (2011. a). Crystal structure of a human cleavage factor CFI m 25/CFI m 68/RNA complex provides an insight into Poly (A) Site recognition and RNA looping. Cell Press 19, 368–377. 10.1016/j.str.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Gilmartin G. M., Doublié S. (2011. b). The structure of human cleavage factor Im hints at functions beyond UGUA-specific RNA binding. RNA Biol. 8, 748–753. 10.4161/rna.8.5.16040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hsu P. L., Yang F., Song J. E. (2018). Reconstitution of the CstF complex unveils a regulatory rolefor CstF-50 in recognition of 3-end processing signals. Nucleic Acids Res. 46, 493–503. 10.1093/nar/gkx1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li Y., Sancar A., Oztas O. (2020). The circadian clock shapes the Arabidopsis transcriptome by regulating alternative splicing and alternative polyadenylation. J. Biol. Chem. 295, 7608–7619. 10.1074/jbc.RA120.013513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Song L., Katz Y., Galili G. (2002). Cloning and characterization of Arabidopsis homologues of the animal CstF complex that regulates 3’ mRNA cleavage and polyadenylation. J. Exp. Bot. 53, 2277–2278. 10.1093/jxb/erf073 [DOI] [PubMed] [Google Scholar]
- Yao C., Biesinger J., Wan J., Weng L., Xing Y., Xie X., et al. (2012). Transcriptome-wide analyses of CstF64 – RNA interactions in global regulation of mRNA alternative polyadenylation. PNAS 109, 18773–18778. 10.1073/pnas.1211101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Long Y., Ji G., Li Q. Q., Wu X. (2018). APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics 34, 1841–1849. 10.1093/bioinformatics/bty029 [DOI] [PubMed] [Google Scholar]
- Ye C., Zhou Q., Wu X., Ji G., Li Q. Q. (2019). Genome-wide alternative polyadenylation dynamics in response to biotic and abiotic stresses in rice. Ecotoxicol. Environ. Saf. 183, 12. 10.1016/j.ecoenv.2019.109485 [DOI] [PubMed] [Google Scholar]
- Youngblood B. A., Macdonald C. C. (2014). CstF-64 is necessary for endoderm differentiation resulting in cardiomyocyte defects. Stem Cell Res. 13, 413–421. 10.1016/j.scr.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Lin J., Li Q. Q. (2019). Transcriptome analyses of Fy mutants reveal its role in mRNA alternative polyadenylation. Plant Cell 31, 2332–2352. 10.1105/tpc.18.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulo A., Bruck D., Mattioli C. C., Franke V., Imami K., Mcshane E., et al. (2017). RNA localization is a key determinant of neurite-enriched proteome. Nat. Commun. 8, 1–12. 10.1038/s41467-017-00690-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarudnaya M., II, Kolomiets I. M., Potyahaylo A. L., Hovorun D. M. (2003). Downstream elements of mammalian pre-mRNA polyadenylation signals: Primary, secondary and higher-order structures. Nucleic Acids Res. 31, 1375–1386. 10.1093/nar/gkg241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Addepalli B., Yun K., Hunt A. G., Xu R., Rao S., et al. (2008). A Polyadenylation Factor Subunit Implicated in Regulating Oxidative Signaling in Arabidopsis thaliana. PloS One 3, 12. 10.1371/journal.pone.0002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman L., Moore C. (1999). Formation of mRNA 3’-ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445. 10.1007/s13146-011-0050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Xing D., Li Q. Q. (2009). Unique features of plant cleavage and polyadenylation specificity factor revealed by proteomic studies. Plant Physiol. 151, 1546–1556. 10.1104/pp.109.142729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zheng J., Li Q. Q. (2011). A Novel Plant in Vitro Assay System for Pre-mRNA Cleavage during 3’-end Formation. Plant Physiol. 157, 1546–1554. 10.1104/pp.111.179465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Wu X., Ji G., Liang C., Li Q. Q. (2019). Genome-wide comparative analyses of Polyadenylation signals in eukaryotes suggest a possible origin of the AAUAAA Signal. Int. J. Mol. Sci. 20, 15. 10.3390/ijms20040958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Wang R., Ding Q., Wang T. (2018). Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat. Commun. 9, 1–14. 10.1038/s41467-018-04730-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang X., Forouzmand E., Jeong J. (2018). Molecular mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation. Mol. Cell 69, 62–74. 10.1016/j.molcel.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Ye W., Ye L., Fu H., Ye C., Xiao X., et al. (2020). PlantAPAdb: a comprehensive database for alternative polyadenylation sites in plants. Plant Physiol. 182, 228–242. 10.1104/pp.19.00943 [DOI] [PMC free article] [PubMed] [Google Scholar]