Abstract
Objectives
There is a clear need for better therapeutics and diagnostics for pancreatic cancer. We aimed to discover plasma membrane-associated proteins overexpressed in pancreatic cancer using quantitative proteomics and apply RNA interference (RNAi) to uncover proteins associated with cancer cell survival.
Methods
Cell surface glycoproteins from 5 pancreatic cancer cell lines were isolated, and differential analyses were performed using mass spectrometry and the "normoid" cell line Hs766T as the comparator. For validation, immunohistochemistry was performed on tissues from 10 independent patients and 2 normal donors. Correlation of protein and mRNA expression level was determined, and functional activity characterized using RNAi.
Results
Integrin β6, CD46, tissue factor, and a novel protein, chromosome 14 open reading frame 1, were identified as overexpressed on pancreatic cancer cell lines. Immunohistochemistry demonstrated the 4 targets were overexpressed in 20% to 70% of primary pancreatic tumor specimens. Small interfering RNA knockdown resulted in a reduction of cellular proliferation by inhibiting DNA synthesis, blocking S-phase progression or induction of apoptosis.
Conclusions
By combining a mass spectrometry identification platform and an RNAi validation platform, we have identified a panel of cell surface glycoproteins that not only are overexpressed, but also play a functional role in pancreatic tumor cell survival.
Keywords: proteomics, RNAi, integrin β6, tissue factor, C14ORF1, CD46
Pancreatic adenocarcinoma is one of the most aggressive human malignancies with a clear need for additional therapeutic strategies. Currently, the only way to cure this disease is through complete surgical removal of the tumor. A lack of early detection combined with the propensity of pancreatic cancer to rapidly disseminate to the lymphatic system has meant that more than 80% of patients at the time of diagnosis already have locally advanced disease or distant metastasis.1 For the majority of pancreatic cancer patients, the tumor is therefore no longer at a stage that is resectable. Palliative therapies such as radiation and chemotherapeutic interventions remain a first-line treatment. However, a low response rate even to the criterion standard, gemcitabine, has meant that only 26% of patients will survive 1 year, and 5% will survive 5 years beyond diagnosis.2 Indeed, there has been no improvement in the survival rate for pancreatic cancer patients in the last 25 years.2
Of 120 known protein targets of marketed drugs, two thirds are represented by plasma membrane proteins.3,4 Their functional significance highlights their importance in drug discovery and development; they not only play a vital role in the regulation of cell signaling, but are also important for cell-cell interactions as well as metabolite and ion transport. The ability to target cell surface proteins using antibody-based approaches, irrespective of the existence of a druggable domain, further contributes to their amenability to therapeutic targeting. However, because of their hydrophobic nature and low abundance, it has been difficult to identify new candidates for drug therapy. This difficulty has recently been circumvented by combining cell membrane labeling strategies with liquid chromatography (LC)—mass spectrometry (MS)/MS methodology.3,5 For example, it has been possible to study surface membrane proteins of the human epidermis6 and chronic and acute leukemia cell lines.7 By utilizing quantitative MS as a method to identify proteins that are differentially expressed on malignant tumor cells compared with corresponding normal tissue cells, it has also been possible to discover glycoproteins up-regulated in colon and ovarian cancer.8,9
In addition to displaying elevated expression, demonstration of a functional dependence is a major consideration when selecting a target for drug development. The functional dissection of a broad range of gene families is now feasible with the development of RNA interference (RNAi).10 It is now possible to down-regulate the expression of any gene in mammalian cells by using small interfering (si) RNAs that are complementary in sequence to the target of choice.11 This method has been applied to many aspects of cancer research, including the identification of roles for proteins in tumor progression, apoptosis, and cell invasion.12–14
In this study, we combined an MS-based strategy to identify cell surface glycoproteins up-regulated in pancreatic cancer with an RNAi-based validation platform to identify a functional role for these proteins. For target identification, we used pancreatic cell lines that differ in their differentiation status, namely, PANC-1 (poorly differentiated), BXPC-3 (moderately differentiated), HPAC (moderately towell differentiated), and Capan-2 (well differentiated), and comparisons were made to the normoid cell line Hs766T.15–17 Tissue factor (TF), integrin β6 (ITGB6), CD46, and chromosome 14 open reading frame 1 (C14ORF1), a novel protein homologous to yeast protein Yer044c that belongs to the ergosterol biosynthetic protein 28 family,18,19 were among the proteins identified from our MS-based discovery. Expression analyses in primary tumor tissues from 10 patients using immunohistochemistry (IHC) confirmed upregulation in pancreatic cancer. Using a panel of siRNAs, we discovered a functional role for each selected protein in the growth and survival of pancreatic cancer cells.
METHODS
Cell Lines and Culture
The pancreatic cell lines Hs766T, PANC-1, HPAC, BXPC-3, ASPC-1, and MPANC96 were obtained from American Type Culture Collection (Manassas, Va). Each cell line was cultured as monolayers at 37°C with 5% CO2. Hs766T, PANC-1, and HPAC were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, Calif) supplemented with 10% fetal bovine serum (Invitrogen), whereas BXPC-3, MPANC96, and ASPC-1 were maintained in RPMI-1640 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen).
Cell Surface Glycoprotein Enrichment and LC/MS Analysis of Cys-Peptides
Protein enrichment and mass spectrometric analyses were carried out as described previously.8,9,20 Viable cells were incubated with 1 mM sodium periodate for 10 minutes to oxidize glycoproteins.21 After incubation, the cells were rechecked for viability by using trypan blue. If the viability remained greater than 80%, the cells were washed with phosphate-buffered saline and lysed, and the protein concentration was then determined by DC assay (Bio-Rad, Hercules, Calif ). Oxidized glycoproteins were conjugated to hydrazide resin (Bio-Rad) at 4°C overnight.22,23 After removal of nonglycoproteins via washing (sequentially with 2 M NaCl, 2% sodium dodecyl sulphate solution, 200 mM propanol amine [in 0.1 M NaOAc, pH 5.5], 40% ethanol, and 80% ethanol), bound proteins were reduced with dithiothreitol and alkylated with isotope-coded affinity tag (ICAT) reagent (Applied Biosystems, Framingham, Mass). Alkylated proteins were digested with trypsin followed by purification of cysteine-containing peptides according to the vendor’s recommendations. Cysteine-containing peptides were separated over a C18 monomeric column (150 mm, 150-μm internal diameter, Grace Vydac 238EV5, 5 μm) on a capillary HPLC system (Agilent, Santa Clara, Calif ) as described.20 Eluted peptides were analyzed online using a QSTAR Pulsar or XL system (MDS/Sciex, Toronto, Ontario, Canada). Peptide ion peaks from the LC/MS map were detected with RESPECT software (Positive Probability Ltd, Isleham, ham, Cambridgeshire, UK), and MASCOT (Matrix Science, London, UK) was used to search MS/MS spectra for peptide/ protein identification.
Mass Spectrometric Data Alignment and Expression Analysis
Peptide ion peaks of LC/MS maps from normal and tumor samples were aligned based on mass-to-charge ratio (m/z), corrected retention time, and charge state (z).20 The intensities of ions in each map were compared with the mean intensities of those ions across all maps in the alignment. A normalization factor was generated (using ions with mean intensities in the 10th-90th percentiles) for each map, using unconstrained nonlinear optimization, minimizing the sum of the differences between intensities of each ion and the mean intensity for that ion across all maps. After intensity normalization, the list of aligned peptide ions was loaded into Spotfire (TIBCO Spotfire, Somerville, Mass) to produce scatter plots, calculate differential ratios, and select ions to send to MS/ MS for identification. Peptide ions with a differential expression of greater than 3-fold between tumor and normal samples were manually verified before LC-MS/MS—based peptide sequencing. Database searching (MSDB) of the resulting MS/MS spectra for peptide/protein identification was accomplished using MASCOT (Matrix Science). Significance within Mascot was set at 0.05, indicating a false-positive rate of 5%.
Immunofluorescence
Immunofluorescence was performed by adding 2.5 μg/mL (final) wheat germ agglutinin conjugated to Alexa-Fluor 594 to the growth media of live HPAC cells grown on chamber slides. After 10 minutes, the media was removed, and the cells were fixed with 1% ultraparaformaldehyde. The cells were then washed and stained with rabbit polyclonal anti-C14ORF1 antibody generated by LifeSpan Biosciences (Seattle, Wash) (LP8850, 10 μg/mL) for 1 hour. After further washing, the cells were stained with anti-rabbit Alexa-Fluor 488 (Pierce, Thermo Fisher Scientific, Rockford, Ill). The cells were then permeabilized with 0.1% Triton X-100 and stained with 1 μg/mL DAPI for 5 minutes before analysis on a Nikon Eclipse i-Series 80i microscope and by using Image-Pro Plus software version 5.1 (Media Cybernetics, Bethesda, Md).
Immunohistochemistry
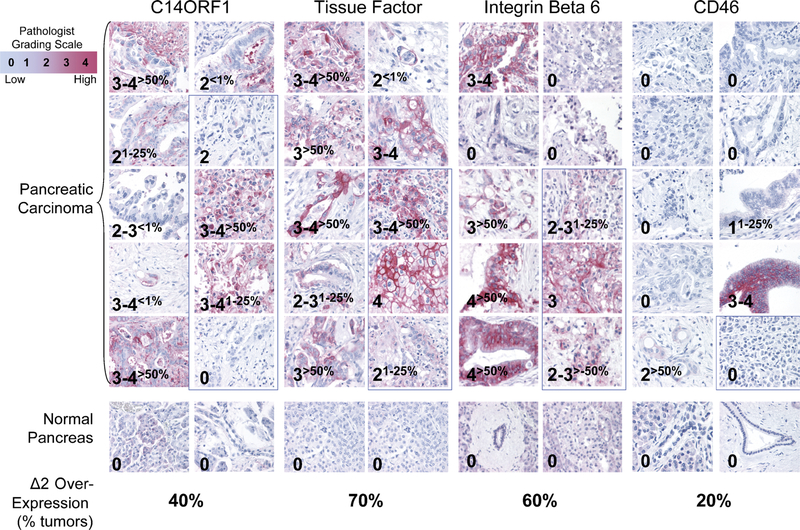
Immunohistochemistry was performed by LifeSpan Biosciences on formalin-fixed paraffin-embedded tissues using tissue microarrays. The cancer array consisted of homogeneous cancer cells. The tissues were deparaffinized, and antigen retrieval was done using EZ-retriever (Biogenex, San Ramon, Calif). Samples were preblocked with non—serum protein block (DAKO A/S, Carpinteria, Calif) for 15 minutes. Primary antibodies, either mouse monoclonal anti-CD142 antibody (HTF-1; BD Biosciences, San Jose, Calif), mouse monoclonal anti-ITGB6 antibody (442.5C4, EMD Biosciences, San Diego, Calif), rabbit polyclonal anti-C14ORF1 antibody generated by LifeSpan Biosciences (LP8849), and mouse monoclonal anti-CD46 antibody (13/42; Accurate Chemical, Westbury, NY) were incubated overnight at room temperature. Envision Plus system HRP (DAKO) was used for detection with 3,3'-diaminobenzidine as substrate for horseradish peroxidase—conjugated secondary antibodies. Slides were then manually scored by one pathologist and were based on (i) staining intensity and (ii) staining frequency modifiers. The negative staining intensity was assigned the score of "0," blush staining a score of "1," faint staining a score of "2," moderate staining a score of "3," and strong staining a score of "4." In addition, when 0.1% to 30% cells showed positive staining, the staining was quantified as rare or occasional, 30% to 75% of cells showing positive staining were quantified as "many or frequent," and greater than 75% of cells staining positive were designated "most." Hematoxylin was used as a counterstain. Representative images were acquired using 40 × objective (400 × magnification).
RNA Isolation and Real-Time Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from cell lines using the RNEasy kit (Qiagen, Valencia, Calif ) with RNase-free DNase (Qiagen) treatment per manufacturer’s instructions. Expression of mRNA was quantitated in a 1-step RT-PCR reaction using TaqMan Gene Expression Assays (Applied Biosystems, Life Technologies, Carlsbad, Calif ). Each sample was assayed in triplicate and included a control well without reverse transcriptase. Quantitative reverse transcriptase—polymerase chain reaction (RT-PCR) was performed using the ABI Prism 7900HT Sequence Detection System (ABI). The following assays were tested: C14orf1 (Hs00200371_m1), CD142 (Hs00175225_m1), ITGB6 (Hs00168458_m1), and CD46 (Hs00611257_m1). Gene expression was quantitated relative to 18S rRNA expression, and copy number was estimated assuming 5 × 106 copies of 18S rRNA per cell.
Quantitative Flow Cytometry
Cell surface expression levels were quantified using the Quantum Simply Cellular System (Bangs Laboratories, Fishers, Ind) according to the manufacturer’s instructions. Standard curve beads and cell lines were stained with TF (BD Biosciences) or CD46 antibodies (Ancell, Bayport, Minn), and analysis was performed on an LSR I (BD Biosciences) flow cytometer. Relative copy number was established by determining the antibody-binding capacity and was calculated using geometric means and linear regression.
RNA Interference
Pooled synthetic siRNA (SMARTpool siRNA) specific for ribonucleotide reductase M2 polypeptide (RRM2) (D-010379), X-linked inhibitor of apoptosis protein (XIAP) (D-004098), and scrambled negative control (D-001216–13) were purchased from Dharmacon (Lafayette, Colo). Knockdown experiments were performed with TF siRNA (Dharmacon) with siRNA1 directed against the sequence GGCAAGGACTTAATTTATA, siRNA2 against GA ACAACACTTTCCTAAGC, siRNA3 against AGTCTACACTGT TCAAATA; C14ORF1 siRNA (Qiagen) with siRNA 1 directed against sequence CTCCATCCAAGTCAAGAAA, siRNA2 directed against GGGACAGGCAGTTCTTCTA; ITGB6 siRNA (Qiagen) with siRNA 1 directed against GAAGGTGGATTTGA TGCAA, siRNA 2 directed against GGAATTACTTGTCAGC CCA, siRNA 3 directed against CCCTTGCAGTAGTATTCCA; CD46 siRNA (Dharmacon) with siRNA 1 directed against GGA GAGAGCACGATTTATT, siRNA 2 directed against AGAGAA ACATGTCCATATA, siRNA 3 directed against GGACTTACGA GTTTGGTTA, siRNA 4 directed against CTATGGAGCTC ATTGGTAA.
For siRNA transfection, MPANC96 cells were seeded into 96-well tissue culture plates at a density of 10,000 cells/well. Transfections with MPANC96 were performed using Lipofectamine 2000 and Plus reagent (Invitrogen) according to the manufacturer’s protocol.
mRNA knockdown of TF, C14ORF1, ITGB6, and CD46 mRNA levels was monitored by quantitative RT-PCR 1 day after transfection using TaqMan Gene Expression Assay for C14orf1 (Hs00200371_m1), CD142 (Hs00175225_m1), ITGB6 (Hs00168458_m1), and CD46 (Hs00611257_m1, Applied Biosystems). Percent mRNA expression for knockdown validation was calculated using the ΔΔCt method.24 RPLP0 (Hs99999902_m1) served as the endogenous control to normalize for template load, and cells transfected with negative control siRNA were the reference sample.
Cell growth was assessed 3 days after transfection using alamar blue reagent (Invitrogen) and SPA-thymidine incorporation kit from GE Healthcare (Piscataway, NJ). Cell cycle analysis was performed using the fluorescein isothiocyanate BrdU Flow Kit from BD Biosciences and analyzed on an LSR I (BD Biosciences) flow cytometer. Caspase 3 analysis by flow cytometry was performed by using a PE-conjugated antibody from BD Biosciences.
RESULTS
C14ORF1, TF, ITGB6, and CD46 Discovered by Proteomics to Be Elevated in Pancreatic Cancer Cell Lines
In a previous study, we described use of an MS-based proteomics platform to identify proteins overexpressed on the surface of primary colon adenocarcinoma specimens compared with adjacent normal tissue.9 A major problem with the application of this technique to pancreatic cancer is the presence of secreted proteases in pancreatic tissues and the difficulty in obtaining sufficient neoplastic pancreatic ductal cells. To circumvent this, we performed analysis on cell line models that represent different stages of differentiation. Hs766Twas the chosen normal comparator cell line as it exhibits a gene expression profile highly similar to normal pancreatic ductal epithelial cells.15–17
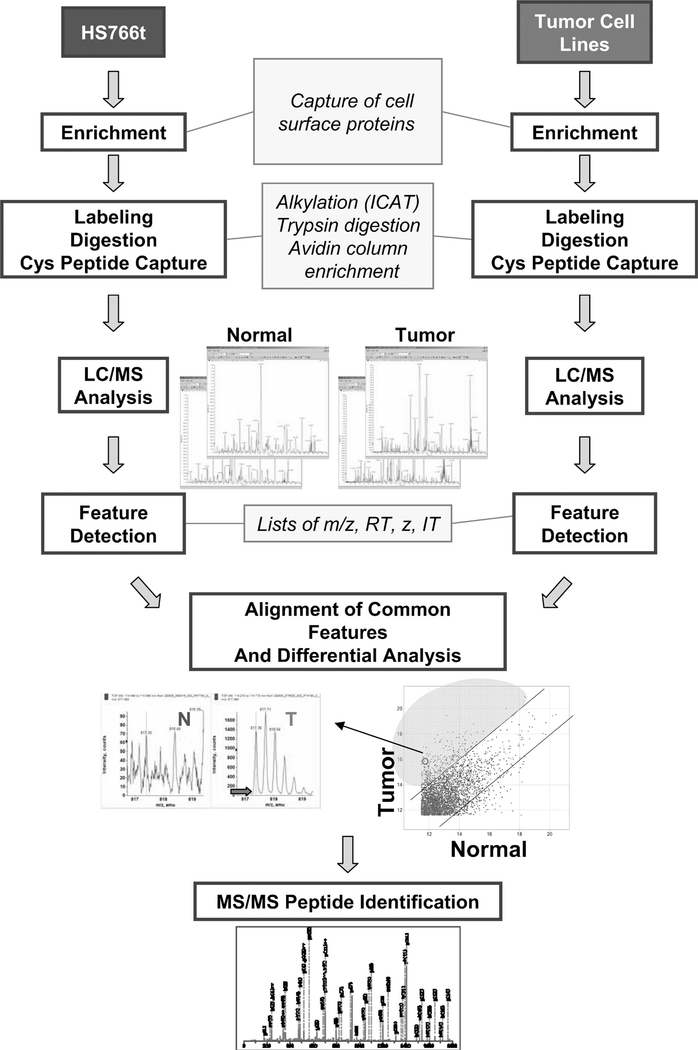
Figure 1 outlines the proteomics identification process used. By maintaining cells viable before and after the sodium periodation step, it was possible to oxidize only cell surface glycoproteins. Capture of the oxidized proteins was achieved by using a hydrazide resin, and the samples were alkylated with an ICAT light reagent for selectively enriching cysteine-containing peptides and reducing sample complexity.25 Subsequent to labeling, samples were digested with trypsin, and decoupled LC/MS analysis was performed for relative quantitation of cysteine-containing peptides.20 For each cell line, the reproducibility of intensities of common features between process replicates is greater than that of a tumor versus normal comparison, indicating that the differences observed between tumor and normal samples are significant. For example, 99.4% of the common features’ intensities are within 3-fold for BXPC3 replicates compared with 80.0% for that of BXPC3 versus Hs766T (Fig. 1). Peptides demonstrating an intensity of expression 3-fold or greater relative to that observed in Hs766T were selected for tandem MS analysis and identification using Mascot. Using the label-free (or decoupled) method described by Kim et al20 and through the construction of a quantitative peptide reference map in silico, it was possible for peptide/protein abundance(s) across multiple samples to be analyzed. This is in contrast to the ICAT light and heavy labeling method, which may provide only pairwise comparisons.20 Thus, by using the decoupled method, it became possible to confirm the extent of overexpression in multiple specimens and reveal patterns of expression across multiple samples.26
FIGURE 1.
Schematic of proteomic identification process. Hs766t normoid and tumor cell lines were oxidized, and the activated glycoproteins were captured, labeled with ICAT, and digested with trypsin. The cysteine-containing peptide fraction was collected, and relative quantitation was performed using LC/MS. Overexpressed proteins were identified using MS/MS analysis.
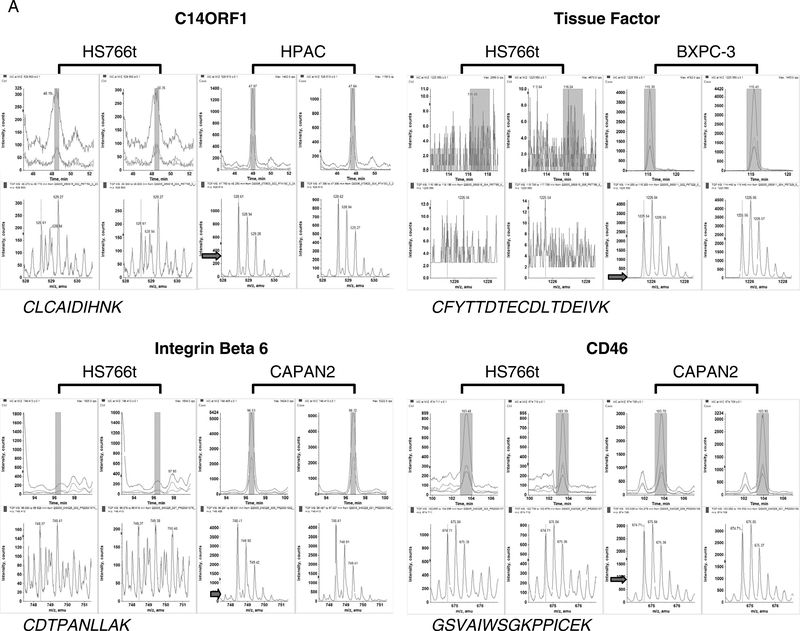
Figure 2 and Table 1 show 4 example proteins, namely, C14ORF1, TF, ITGB6, and CD46, which were identified from our proteomic analysis and were subsequently validated by IHC and RNAi. C14ORF1, TF, ITGB6, and CD46 represent an interesting set of targets as they include proteins known to play a role in pancreatic tumorigenesis (TF), those known to play a role in tumorigenesis but as yet have no evidence for a role in pancreatic cancer (ITGB6 and CD46) and those with no known functional activity (ie, C14ORF1). Two additional proteins, namely, elongation of very long chain fatty acids protein 5 (ELOVL5 or HELO1) and transmembrane protease serine 4 (TMPRSS4), were identified from our proteomic analysis (data not shown). Although expression of ELOVL5 and HELO1 was confirmed by IHC, no functionality in pancreatic cancer was observed following RNAi analysis.
FIGURE 2.
Mass spectrometry analysis reveals overexpression of C14ORF1, TF, ITGB6, and CD46 in pancreatic cancer cell lines. Examples of MS relative quantification data for peptides with highest-intensity-level differences for tumor cell lines compared with Hs766t. The arrow indicates the intensity level of the peptide ion in Hs766t.
TABLE 1.
Proteins Identified as Overexpressed in Pancreatic Tumor Cell Lines Compared With Hs766T “Normoid” Cell Line
| Average Ratio (SD) |
|||||
|---|---|---|---|---|---|
| Protein | Peptide | PANC-1 | BXPC-3 | HPAC | Capan-2 |
| C14ORF1 | CLCAIDIHNK | 0.9 (0.2) | 2.7 (0.2) | 4.9 (0.7) | 1.6 (0.04) |
| TF | CFYTTDTECDLTDEIVK* | 1.9 (0.6) | 341 (150) | 14.1 (4.8) | 6.2 (0.5) |
| ITGB6 | LAGIVIPNDGLCHLDSK | 1.3 (0.3) | 2.2 (0.7) | 5.1 (2.4) | 14.3 (6.8) |
| CDTPANLLAK | |||||
| ISANIDTPEGGFDAIMQAAVCK | |||||
| CD46 | GSVAIWSGKPPICEK | 3.8 (1.0) | 1.1 (0.1) | 2.0 (0.2) | 3.3 (0.09) |
Peptide sequenced with varying charge states.
Figure 2 displays the MS analysis for a peptide ion from C14ORF1, TF, ITGB6, and CD46. The top panels are the extracted ion chromatograms for the peptides in 2 process replicate LC-MS experiments for HS766t (left) and tumor cell lines (right). The highlighted portion of the extracted ion chromatograms indicates the mass spectra that are summed and shown in the lower panels. In each case, the peptide ion intensity observed in the tumor cell lines is greater than that of the normal cell line, HS766t. Peptide ratios (tumor vs normal) are calculated using the areas of the first 3 isotopic peaks (if unobstructed) of the ion in the mass spectrum. Table 1 shows the average ratio of the peptides identified for each target in each pancreatic cell line. Of the 4 targets, TF exhibited the highest level of overexpression with an average ratio of greater than 100 for BXPC-3 and approximately 5 to 15 for HPAC and Capan-2. Three peptides were overexpressed for ITGB6 in HPAC (5-fold) and Capan-2 (14-fold) cell lines, whereas the average ratio for CD46, represented by 1 peptide, was 3 to 4 in PANC-1 and Capan-2. For C14ORF, we identified the only cysteine-containing tryptic peptide and determined that this peptide was overexpressed 3- to 5-fold in BXPC-3 and HPAC. These proteins therefore represent proteins with relatively strong proteomics data (ie, ITGB6, a 788-amino-acid protein with multiple corresponding peptides identified in multiple cell lines) and those with limited data (ie, C14ORF1, a protein of 140 amino acids in length, identified by only 1 peptide). Taking these selected proteins further down the validation path provides a guide to the depth of proteomics identification data necessary for the selection of candidates for biomarker and therapeutic development.
FACS and Immunofluorescence Studies Confirms Cell Surface Expression
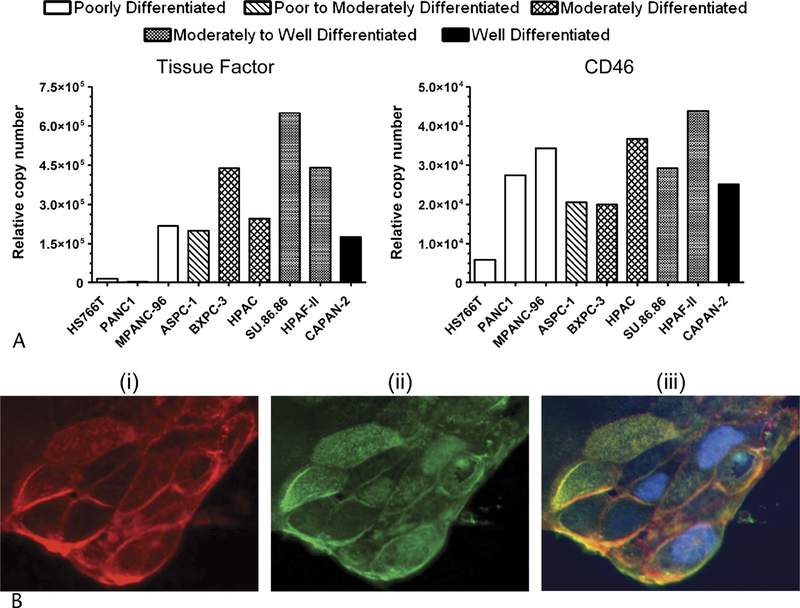
As an initial validation step, we determined if proteins detected overexpressed by MS were also observed overexpressed using an orthogonal protein-based methodology. We wanted to not only confirm cell surface expression in pancreatic cancer, but also evaluate expression levels in additional cell lines. As shown in Figure 3A, flow cytometry analysis confirmed cell surface expression of both TF and CD46 in pancreatic cancer, consistent with what has previously been demonstrated in other cell types.27,28 Flow cytometry results also confirmed overexpression of each protein on the more differentiated cell lines compared with Hs766t. For TF, 3-fold or greater increase in expression was observed in 7 of 8 cell lines. Only PANC-1 did not show overexpression by FACS, which is consistent with the MS data. For CD46, all cell lines tested by FACS appeared to be overexpressed compared with Hs766t. This is in contrast to the MS data where CD46 was not detected overexpressed in BXPC-3. This discrepancy may, in part, be attributed by the fact that the CD46 peptide identified by MS mapped to the SCR2 domain (virus and complement binding region), whereas the FACS antibody was specific to the SCR1 domain (virus-binding site).29
FIGURE 3.
FACS and immunofluorescence studies confirm cell surface expression. A, Quantitative flow cytometry analysis of TF and CD46 using a bead-based standard. Multiparameter gating was used to determine epitope copy number. B, HPAC pancreatic carcinoma cells, original magnification 100×. (i) Staining of wheat germ agglutinin on live cells (in red), (ii) staining of C14ORF1 (in green), (iii) merged image and staining of nucleus (in blue).
While C14ORF1 protein contains a region predicted to be a transmembrane domain, considering the subcellular localization of C14ORF1 in mammalian cells has not previously been determined we wanted to confirm our MS observation that it is located on the cell surface. To this end, antiYwheat germ agglutinin antibody was added to the culture media of live HPAC pancreatic cancer cells, permitting the staining of glycosylated proteins located on the cell surface (Fig. 3Bi). Figure 3Bii shows staining with an anti-C14ORF1 antibody. As shown in Figure 3Biii, C14ORF-1 colocalized with wheat germ agglutinin confirming that C14ORF1 is located on the cell surface of pancreatic cancer cells.
C14ORF1, TF, ITGB6, and CD46 mRNA Expression in Pancreatic Cancer Cell Lines
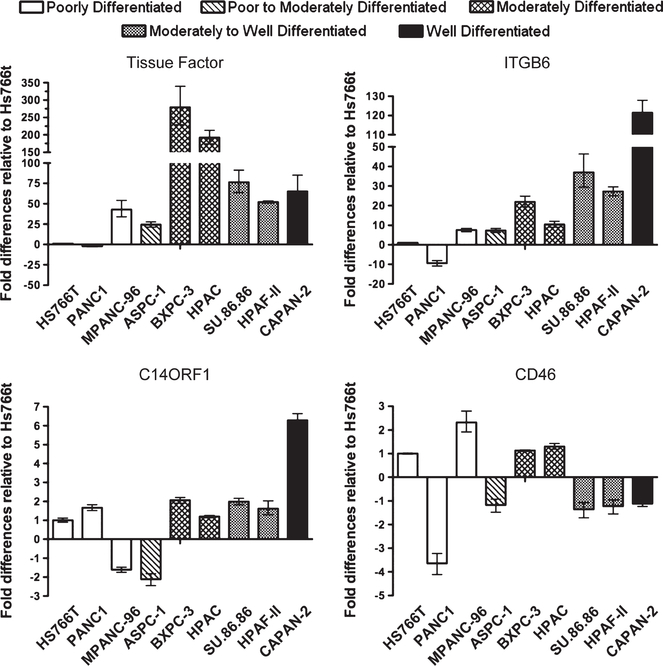
To understand the mechanism of up-regulated expression, we examined the level of mRNA expression for the targets across a panel of pancreatic cancer cell lines, including those evaluated by MS. As shown in Figure 4, TF and ITGB6 showed an increase in mRNA expression levels in all cell lines tested, as compared with Hs766t, except in the poorly differentiated cell line PANC-1. These data directly correlate our MS findings and, for TF, correlated with our FACS findings. Interestingly, TF showed highest expression in the moderately differentiated cell lines BXPC-3 and SU.86.86, whereas ITGB6 showed highest expression in the well-differentiated cell line Capan-2.
FIGURE 4.
Expression validation by quantitative PCR analysis. mRNA expression levels of C14ORF1, TF, ITGB6, and CD46 in pancreatic cancer cell lines.
In contrast, no clear correlation was observed between the cell line mRNA data for both CD46 and C14ORF1 with the MS data or FACS data. For CD46, no significant increase was observed at the mRNA level for all cell lines tested. Similarly for C14ORF1, no significant increase was observed for the majority of cell lines tested, including HPAC, the cell line we observed to have a 5-fold increase in expression by MS.
IHC Confirms Target Overexpression in Pancreatic Cancer Tissues
To determine the expression pattern of TF, ITGB6, C14ORF, and CD46 on pancreatic cancer tissue specimens, IHC was performed on 10 pancreatic adenocarcinoma specimens including 1 to 4 metastatic samples (Fig. 5). The staining intensities were compared with ductal epithelium tissues from normal donors, and the scores represent grading by a pathologist. For all 4 targets, no staining was observed in the normal ductal epithelium. For TF, strong staining was observed in 7 of 10 cancer tissues, with 2 samples representing metastatic tissues. All other remaining tissues showed staining but in fewer number of cells. For ITGB6, strong staining was observed in 6 of 10 cancer tissues, with 2 samples representing metastatic tissues. One other metastatic tissue sample showed staining in less than 25% of the section. Strong staining for C14ORF1 was observed in 4 of 10 pancreatic cancer tissues, 2 of which represent metastatic cancer tissues. Except for 1 sample, all remaining tissues showed staining but in a fewer number of cells. For CD46, staining in 2 of 10 tissues was observed.
FIGURE 5.
Validation of differential expression by IHC. C14ORF1, TF, ITGB6, and CD46 staining in pancreatic carcinomas (top), original magnification ×40. No staining was observed for normal epithelium (middle). Images highlighted with blue box represent specimens from metastatic tissues. Percentages of tumors that overexpress each target compared with normal ductal epithelium by 2 pathology grades or more are indicated (bottom). Percentage overexpression was calculated from samples with greater than 50% of section showing staining, and staining must be at least 2 pathology grades or higher than staining observed for normal ductal epithelium. Samples with staining in less than 100% of tissue sections are indicated by percentages shown in subscript.
RNAi Knockdown of C14ORF1, TF, ITGB6, and CD46 mRNA Inhibits Growth of Pancreatic Cancer Cells
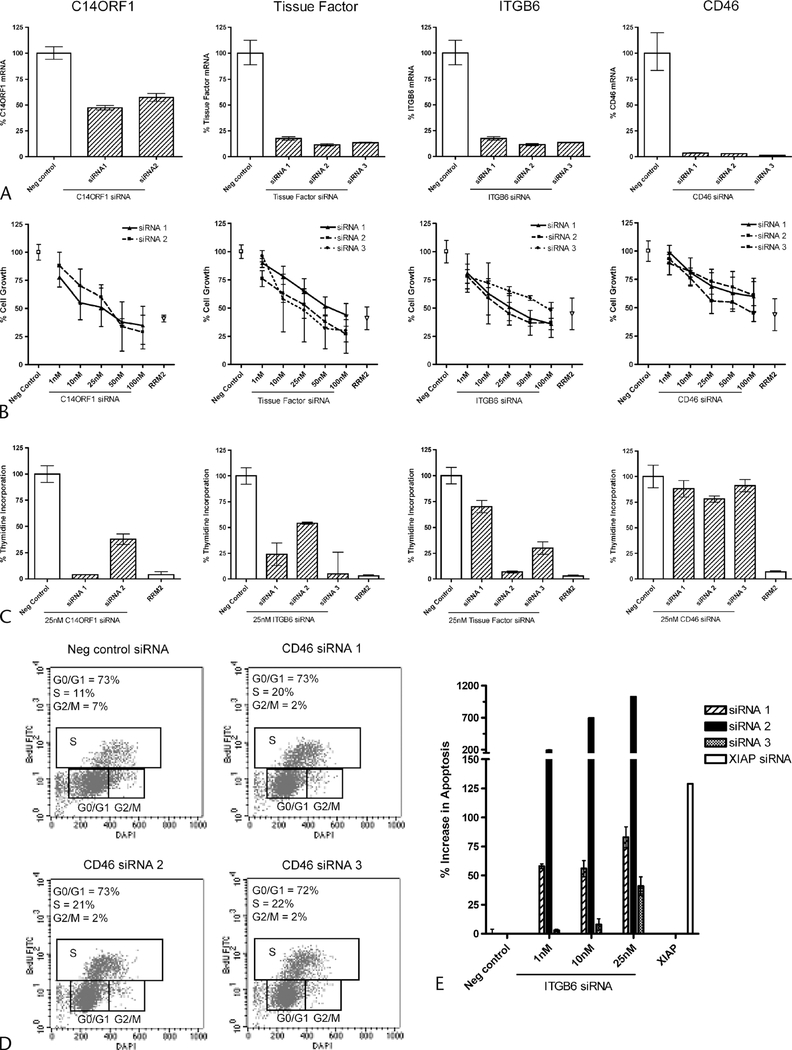
To determine if C14ORF1, TF, ITGB6, and CD46 play a functional role in cell survival, MPANC96 cell line was treated with multiple independent siRNA duplexes targeting C14ORF1, TF, ITGB6, or CD46. MPANC96 was chosen for our RNAi studies based on our ability to transfect siRNA efficiently (>990% efficient, data not shown) and on the low toxicity profile given following transfection with a panel of negative controls, including scrambled negative control siRNA, nontargeting firefly luciferase siRNA, and transfection reagent-alone control (<10% toxicity, data not shown). Twenty-four hours following C14ORF1, TF, ITGB6 siRNA transfection, ~40% to 50% knockdown at the mRNA level was observed for C14ORF1, and more than 70% knockdown was observed for TF, ITGB6, and CD46, respectively (Fig. 6A). As shown in Figure 6B, 3 days following siRNA transfection, a dose-dependent inhibition of cell growth, as measured by cellular metabolic activity, was observed for C14ORF1, TF, ITGB6, and CD46 with the phenotypic effects observed using independent siRNAs targeting different portions of each candidate mRNA. Similar inhibitory effects were observed for our positive control RRM2, which is known to inhibit proliferation of cancer cells in vitro and in vivo.30 As anticipated, no inhibitory effects were observed for the negative control siRNA.
FIGURE 6.
RNAi knockdown in MPANC96 pancreatic cell lines. A, Quantitative RT-PCR analysis of C14ORF1, TF, ITGB6, and CD46 mRNA levels 24 hours after transfections. Results show % difference in mRNA levels compared with scrambled negative control siRNA. B, Alamar blue cell proliferation following titration of C14ORF1, TF, ITGB6, and CD46 siRNA. Data are plotted as a percentage of the scrambled negative control. The positive control, RRM2 siRNA, is also shown. C, Relative thymidine incorporation following transfection of 25 nM of C14ORF1, TF, ITGB6, and CD46 siRNA. Data are plotted as a percentage of the scrambled negative control. D, Cell cycle analysis by flow cytometry following transfection of 25 nM CD46 or scrambled negative control siRNA. Percentages of cells in each phase of the cell cycle are indicated in the top left corner of each dot plot. E, Apoptotic cell death, as measured by caspase 3/7 activity, following transfection of ITGB6 siRNA. Results shown are relative to scrambled negative control. Positive control, XIAP, is also indicated.
Additional studies addressed whether inhibition of cell growth was due to a decreased number of cells undergoing DNA synthesis or was the result of an S-phase or G2/M cell cycle arrest. As shown in Figure 6C, siRNA targeting of C14ORF-1, TF, and ITGB6 inhibits thymidine incorporation. The observed inhibition of DNA synthesis was evident for all independent duplexes as compared with the scrambled negative control. In contrast, no significant inhibition of DNA synthesis was observed for knockdown of CD46. Instead, as shown in Figure 6D, siRNA targeting of CD46 induced an S-phase cell cycle arrest, thereby inhibiting the number of cells that passes on to G2/M phase of the cell cycle. Inhibition of ITGB6 by RNAi dramatically induced apoptosis in MPANC96 cells. At 25-nM siRNA concentration, all 3 siRNA duplexes induced a 40% to greater than 1000% increase in caspase 3/7 activity as compared with negative siRNA control. The level of caspase 3/7 activity increase was similar to that observed with the knockdown of XIAP. At the lower siRNA concentrations of 1 nM, the 2 most potent duplexes still induced a 50% to 200% increase in caspase 3/7 activity. Flow cytometry analysis using a caspase 3 antibody revealed that 40% to 80% of the total population of cells was apoptotic following ITGB6 knockdown (Supplementary Figure 1, Supplemental Digital Content 1, http://links.lww.com/MPA/A89). No significant apoptotic induction was observed upon transfection with C14ORF1, CD46, or TF siRNA (data not shown).
DISCUSSION
Proteomics is an emerging tool that offers opportunity to discover novel diagnostic and therapeutic markers for cancer. Considering variation inthe rates of mRNA translation and protein stability exhibited by different genes, it is anticipated that proteomics will identify differentially expressed targets independent of those revealed by mRNA expression studies. Indeed, from the limited comparisons in this study and from previously published global analysis of pancreatic cancer secretome and lung adenocarcinoma, there appears to be little correlation between protein and mRNA levels.31,32
The proteomics strategy we used utilizes LC-MS for identification and quantitation of glycoproteins.9,20,26 The method we describe is high throughput and can be performed on an industrial scale.34,35 Previous proteomic studies in pancreatic tumors include those that utilized laser capture microdissection combined with 2-dimensional electrophoresis or 2-dimensional electrophoresis combined with MS analysis.33–35 By incorporating our identification strategy with an IHC expression and RNAi functional validation platform, we confirmed overexpression in tissues and ascertained a functional role for selected targets in pancreatic cancer.
Four examples of proteins identified from our proteomics analysis are described (Table 1; Fig. 2). We include the identification of TF, a protein previously reported to be up-regulated in pancreatic tumor tissues by IHC studies.36,37 We also identified 2 targets, which have previous association with other cancer types but not pancreatic cancer: CD46, which has been reported to be associated with melanoma and colon, breast, lung, and gastric cancers,38–40 and ITGB6, which has been reported to be up-regulated in epithelial ovarian cancer and oral and cervical squamous cancers.41–43 Furthermore, we report and confirm the cell surface location of C14ORF1 in pancreatic cancer, a protein with no known function or previous disease association (Fig. 3). The confirmation of cell surface overexpression in the pancreatic cancer cell lines used in the proteomic analyses was shown for CD46 and TF (for which there were available reagents) demonstrating the reliability of the MS-based approach to identify meaningful associations (Fig. 3). Flow cytometry also demonstrated that CD46 and TF cell surface overexpression was not restricted to the pancreatic cancer cell lines examined by MS. Interestingly, analysis of target mRNA expression did not necessarily correlate with cell surface overexpression. Whereas TF and ITGB6 did show up-regulated expression at the mRNA level in pancreatic cancer consistent with cell surface overexpression, this was not observed for CD46 or C14ORF1. This emphasizes the importance of applying a protein-based approach and not just mRNA profiling to identify the spectrum of overexpressed cell-associated proteins.
Significantly, IHC analysis of a set of pancreatic cancer tissue and normal pancreatic ductal epithelium revealed that overexpression of all 4 targets was not restricted to just pancreatic cancer cell lines. Although no expression was detectable in normal pancreatic epithelium, each of the 4 targets demonstrated expression in the subset of the primary pancreatic cancer tissue specimens examined (Fig. 5). The observation that TF was found overexpressed in 70% of specimens in pancreatic cancer is consistent with a larger study linking TF expression with poor patient outcome.37 With the shortcomings of existing markers (CA-19–9 and CEA) to reliably distinguish differences between benign and malignant pancreatic conditions,44,45 examination of TF, ITGB6, C14ORF1, and CD46 expression across additional panels of pancreatic cancer specimens will be of interest, particularly to determine if overexpression is associated with early-stage pancreatic cancer where there is a dire need for improved diagnostics.46,47 Based on our MS analysis, there is evidence to suggest the 4 targets are differentially expressed across pancreatic cancer tumor development: Whereas ITGB6 was highly expressed in the well-differentiated cell line Capan-2, TF and C14ORF1 were highly expressed in the moderately differentiated cell lines BXPC-3 and/or HPAC, and CD46 was the only target overexpressed in the least differentiated cell line PANC-1. Considering the amenability of a serum- or plasma-based diagnostic, the prior observation that TF and CD46 can be shed supports evaluation of their circulating levels in patient serum samples. Indeed, prior data have already demonstrated overexpression of TF in cancer serum.37
In addition to the diagnostic opportunities provided from the MS analysis, through use of RNAi, we demonstrate that inhibition of TF, ITGB6, C14ORF1, or CD46 leads to a reduction in pancreatic cancer cell proliferation. This is a first demonstration of such a functional role for each protein in pancreatic cancer and, for C14ORF1, the first report of its functional activity in mammalian cells. For TF, our studies complemented previous studies in colon and pancreatic cancer. Whereas RNAi knockdown of TF in HCT116 colon cells was shown to inhibit tumor growth in nude mice, knockdown in BXPC-3 pancreatic cancer suppressed invasiveness in vitro.37 Consistent with our findings that siRNA targeting of ITGB6 leads to inhibition of cell proliferation and apoptosis was the prior observation that overexpression of ITGB6 leads to increased proliferation of SW480 colon cells in nude mice.48 Our observation that knockdown of CD46 leads to S-phase arrest and growth inhibition of MPANC96 cells indicates CD46 not only acts as an inhibitor of complement activation49,50 but may also play a direct role in tumor cell growth providing additional rationale for its therapeutic targeting.
In summary, our proteomic analysis has enabled us to identify cell surface glycoproteins with no previous associations with pancreatic cancer. Our validation strategy has allowed us to not only confirm overexpression, but by incorporating an RNAi validation strategy, we have also been able to assign novel functional roles to the proteins identified. The work reported here will not only act as a template toward a strategy that would allow us to validate more proteins, but we also hope that the proteins reported here may serve as novel diagnostic markers and/or therapeutic targets for pancreatic cancer.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Deborah Mesmer, Jim Norton, Erin Brand, Anh Nguyen, Brian Feild, Roxanne Armstrong, Katherine Paweletz, and Ian McCaffrey for their technical assistance. They also thank Jim Duff and Eric Whitener for their assistance in data management; a special thanks also goes to Charlie Birse for his critical reading of the manuscript.
Footnotes
Supplemental digital contents are available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.pancreasjournal.com).
REFERENCES
- 1.von Wichert G, Seufferlein T, Adler G. Palliative treatment of pancreatic cancer. J Dig Dis. 2008;9(1):1–7. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. [DOI] [PubMed] [Google Scholar]
- 3.Bledi Y, Inberg A, Linial M. PROCEED: a proteomic method for analysing plasma membrane proteins in living mammalian cells. Brief Funct Genomics Proteomics. 2003;2(3):254–265. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. [DOI] [PubMed] [Google Scholar]
- 5.Jang JH, Hanash S. Profiling of the cell surface proteome. Proteomics. 2003;3(10):1947–1954. [DOI] [PubMed] [Google Scholar]
- 6.Blonder J, Terunuma A, Conrads TP, et al. A proteomic characterization of the plasma membrane of human epidermis by high-throughput mass spectrometry. J Investig Dermatol. 2004;123(4):691–699. [DOI] [PubMed] [Google Scholar]
- 7.Lee SJ, Kim KH, Park JS, et al. Comparative analysis of cell surface proteins in chronic and acute leukemia cell lines. Biochem Biophys Res Commun. 2007;357(3):620–626. [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal S, He T, Fitzhugh W, et al. Immune modulator CD70 as a potential cisplatin resistance predictive marker in ovarian cancer. Gynecol Oncol. 2009;115(3):430–437. [DOI] [PubMed] [Google Scholar]
- 9.Van Orden KL, Birse CE, He T, et al. Proteomic Analysis of Colorectal Tumor Cells Indeitifies CD133/Prominin-1, a Cancer Stem Cell Marker, as a Monocolonal Antibody Therapeutic Candidate. Los Angeles, CA: AACR; 2007. [Google Scholar]
- 10.Bartz S, Jackson AL. How will RNAi facilitate drug development? Sci STKE. 2005;2005(295):pe39. [DOI] [PubMed] [Google Scholar]
- 11.Kurreck J. Expediting target identification and validation through RNAi. Expert Opin Biol Ther. 2004;4(3):427–429. [DOI] [PubMed] [Google Scholar]
- 12.Schlabach MR, Luo J, Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319(5863): 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan FL, Yin JQ. Application of RNAi to cancer research and therapy. Front Biosci. 2005;10:1946–1960. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Elledge SJ. Cancer: deconstructing oncogenesis. Nature. 2008;453(7198):995–996. [DOI] [PubMed] [Google Scholar]
- 15.Iwamura T, Hollingsworth MA. Pancreatic tumors In: Masters JR, Palsson B, eds. Cancer Cell Lines Part I. New York, NY: Springer; 1999:107–122. [Google Scholar]
- 16.Sipos B, Moser S, Kalthoff H, et al. A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch. 2003;442(5):444–452. [DOI] [PubMed] [Google Scholar]
- 17.Ryu B, Jones J, Blades NJ, et al. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62(3):819–826. [PubMed] [Google Scholar]
- 18.Veitia RA, Ottolenghi C, Bissery MC, et al. A novel human gene, encoding a potential membrane protein conserved from yeast to man, is strongly expressed in testis and cancer cell lines. Cytogenet Cell Genet. 1999;85(3–4):217–220. [DOI] [PubMed] [Google Scholar]
- 19.Veitia RA, Hurst LD. Accelerated molecular evolution of insect orthologues of ERG28/C14orf1: a link with ecdysteroid metabolism? J Genet. 2001;80(1):17–21. [DOI] [PubMed] [Google Scholar]
- 20.Kim YJ, Zhan P, Feild B, et al. Reproducibility assessment of relative quantitation strategies for LC-MS based proteomics. Anal Chem. 2007;79(15):5651–5658. [DOI] [PubMed] [Google Scholar]
- 21.Bobbitt JM. Periodate oxidation of carbohydrates. Adv Carbohydr Chem. 1956;48(11):1–41. [DOI] [PubMed] [Google Scholar]
- 22.Bayer EA, Ben-Hur H, Wilchek M. Biocytin hydrazide—a selective label for sialic acids, galactose, and other sugars in glycoconjugates using avidin-biotin technology. Anal Biochem. 1988;170(2):271–281. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Li XJ, Martin DB, et al. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. [DOI] [PubMed] [Google Scholar]
- 24.Tuzmen S, Kiefer J, Mousses S. Validation of short interfering RNA knockdowns by quantitative real-time PCR. Methods Mol Biol (Clifton, NJ). 2007;353:177–203. [DOI] [PubMed] [Google Scholar]
- 25.Gygi SP, Rist B, Griffin TJ, et al. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res. 2002;1(1):47–54. [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, Feild B, Fitzhugh W, et al. Reference map for liquid chromatography—mass spectrometry—based quantitative proteomics. Anal Biochem. 2009;393(2):155–162. [DOI] [PubMed] [Google Scholar]
- 27.Forster Y, Meye A, Albrecht S, et al. Tissue factor and tumor: clinical and laboratory aspects. Clin Chim Acta. 2006;364(1–2):12–21. [DOI] [PubMed] [Google Scholar]
- 28.Kim DD, Song WC. Membrane complement regulatory proteins. Clin Immunol (Orlando, Fla). 2006;118(2–3):127–136. [DOI] [PubMed] [Google Scholar]
- 29.Casasnovas JM, Larvie M, Stehle T. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 1999;18(11):2911–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avolio TM, Lee Y, Feng N, et al. RNA interference targeting the R2 subunit of ribonucleotide reductase inhibits growth of tumor cells in vitro and in vivo. Anticancer Drugs. 2007;18(4):377–388. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1(4):304–313. [DOI] [PubMed] [Google Scholar]
- 32.Gronborg M, Kristiansen TZ, Iwahori A, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157–171. [DOI] [PubMed] [Google Scholar]
- 33.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129(5):1454–1463. [DOI] [PubMed] [Google Scholar]
- 34.Chen R, Yi EC, Donohoe S, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129(4):1187–1197. [DOI] [PubMed] [Google Scholar]
- 35.Lu Z, Hu L, Evers S, et al. Differential expression profiling of human pancreatic adenocarcinoma and healthy pancreatic tissue. Proteomics. 2004;4(12):3975–3988. [DOI] [PubMed] [Google Scholar]
- 36.Khorana AA, Ahrendt SA, Ryan CK, et al. Tissue factor expression, angiogenesis, and thrombosis in pancreatic cancer. Clin Cancer Res. 2007;13(10):2870–2875. [DOI] [PubMed] [Google Scholar]
- 37.Nitori N, Ino Y, Nakanishi Y, et al. Prognostic significance of tissue factor in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2005;11(7):2531–2539. [DOI] [PubMed] [Google Scholar]
- 38.Niehans GA, Cherwitz DL, Staley NA, et al. Human carcinomas variably express the complement inhibitory proteins CD46 (membrane cofactor protein), CD55 (decay-accelerating factor), and CD59 (protectin). Am J Pathol. 1996;149(1):129–142. [PMC free article] [PubMed] [Google Scholar]
- 39.Weichenthal M, Siemann U, Neuber K, et al. Expression of complement regulator proteins in primary and metastatic malignant melanoma. J Cutan Pathol. 1999;26(5):217–221. [DOI] [PubMed] [Google Scholar]
- 40.Inoue T, Yamakawa M, Takahashi T. Expression of complement regulating factors in gastric cancer cells. Mol Pathol. 2002;55(3):193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazelbag S, Kenter GG, Gorter A, et al. Overexpression of the alpha v beta 6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J Pathol. 2007;212(3):316–324. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed N, Riley C, Rice GE, et al. Alpha(v)beta(6) integrin-A marker for the malignant potential of epithelial ovarian cancer. J Histochem Cytochem. 2002;50(10):1371–1380. [DOI] [PubMed] [Google Scholar]
- 43.Breuss JM, Gallo J, DeLisser HM, et al. Expression of the beta 6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J Cell Sci. 1995;108(pt 6):2241–2251. [DOI] [PubMed] [Google Scholar]
- 44.Jewkes AJ, Macdonald F, Downing R, et al. Labelled antibody imaging in pancreatic cancer, cholangiocarcinoma, chronic pancreatitis and sclerosing cholangitis. Eur J Surg Oncol. 1991;17(4):354–357. [PubMed] [Google Scholar]
- 45.Duraker N, Hot S, Polat Y, et al. CEA, CA 19–9, and CA 125 in the differential diagnosis of benign and malignant pancreatic diseases with or without jaundice. J Surg Oncol. 2007;95(2):142–147. [DOI] [PubMed] [Google Scholar]
- 46.Goggins M. Identifying molecular markers for the early detection of pancreatic neoplasia. Semin Oncol. 2007;34(4):303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrell JJ, van Rijnsoever M, Elsaleh H. Early detection markers in pancreas cancer. Cancer Biomarkers. 2005;1(2–3):157–175. [DOI] [PubMed] [Google Scholar]
- 48.Agrez M, Chen A, Cone RI, et al. The alpha v beta 6 integrin promotes proliferation of colon carcinoma cells through a unique region of the beta 6 cytoplasmic domain. J Cell Biol. 1994;127(2):547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russell SM, Johnstone RW, Wilton A, et al. Molecular characterization of the polymorphic expression of CD46: a cell surface molecule protecting cells from complement attack. Transplant Proc. 1992;24(1):211–213. [PubMed] [Google Scholar]
- 50.Blok VT, Daha MR, Tijsma OM, et al. A possible role of CD46 for the protection in vivo of human renal tumor cells from complement-mediated damage. Lab Invest. 2000;80(3):335–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.