Abstract
IFIH1 gain-of-function has been reported as a cause of a type I interferonopathy encompassing a spectrum of autoinflammatory phenotypes including Aicardi–Goutières syndrome and Singleton Merten syndrome. Ascertaining patients through a European and North American collaboration, we set out to describe the molecular, clinical and interferon status of a cohort of individuals with pathogenic heterozygous mutations in IFIH1. We identified 74 individuals from 51 families segregating a total of 27 likely pathogenic mutations in IFIH1. Ten adult individuals, 13.5% of all mutation carriers, were clinically asymptomatic (with seven of these aged over 50 years). All mutations were associated with enhanced type I interferon signaling, including six variants (22%) which were predicted as benign according to multiple in silico pathogenicity programs. The identified mutations cluster close to the ATP binding region of the protein. These data confirm variable expression and nonpenetrance as important characteristics of the IFIH1 genotype, a consistent association with enhanced type I interferon signaling, and a common mutational mechanism involving increased RNA binding affinity or decreased efficiency of ATP hydrolysis and filament disassembly rate.
Keywords: Aicardi–Goutières syndrome, IFIH1, MDA5, Singleton Merten syndrome, Type I interferonopathy
1 ∣. INTRODUCTION
In 2014, heterozygous gain-of-function mutations in IFIH1 were reported to cause a spectrum of neuroimmune phenotypes including classical Aicardi–Goutières syndrome (AGS; Oda et al., 2014; Rice et al., 2014). IFIH1 encodes interferon-induced helicase C domain-containing protein 1 (IFHI1; also known as melanoma differentiation associated gene 5 protein: MDA5) which senses viral double-stranded (ds) RNA in the cytosol, leading to the induction of a type I interferon-mediated antiviral response. Consequent to Mendelian determined gain-of-function, it is suggested that IFIH1 inappropriately senses self-derived nucleic acid as viral, leading to an autoinflammatory state classified as a type I interferonopathy (Ahmad et al., 2018; Crow & Manel, 2015). In 2015, a p.Arg822Gln substitution in IFIH1 was shown to cause Singleton Merten syndrome (SMS), an autosomal dominant trait variably characterized by a deforming arthropathy, abnormal tooth development and cardiac valve calcification, again in association with enhanced type I interferon signaling (Rutsch et al., 2015). Although it was initially considered that SMS was a distinct, mutation-specific disorder, subsequent reports indicate that SMS and the neuroinflammatory phenotypes seen in the context of IFIH1 gain-of-function constitute part of the same disease spectrum (Buers, Rice, Crow, & Rutsch, 2017; Bursztejn et al., 2015).
Type I interferonopathy associated IFIH1 mutations are either absent from control databases, or only present at very low frequency. However, we have noted previously that in silico algorithms are not always reliable in differentiating IFIH1 disease-causing variants from benign polymorphisms (Ruaud et al., 2018). Such difficulty in assigning molecular pathogenicity is compounded by marked variability in disease expression, sometimes even within the same family, and the observation of complete non-penetrance in certain pedigrees (Rice et al., 2014). Given this background, we considered it important to provide an update of our experience of sequencing individuals for pathogenic IFIH1 mutations associated with a type I interferonopathy state. In total, we describe molecular and clinical data relating to 74 individuals from 51 families, identifying 27 likely pathogenic mutations that cluster close to the ATP binding region of the protein. Our data confirm variable expression and nonpenetrance as important characteristics of these mutant genotypes, and the consistent association with enhanced type I interferon signaling as assessed by interferon-stimulated gene (ISG) expression, referred to as the interferon score.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Subjects
Patients were ascertained through direct contact and/or collaborating physicians across clinical research laboratories in the UK and France (Crow), the USA (Vanderver), and Italy (Orcesi). The study was approved by the Leeds (East) Research Ethics Committee (10/H1307/132), the Comite de Protection des Personnes (ID-RCB/EUDRACT: 2014-A01017-40), IRB study protocol (Myelin Disorders Bioregistry Project: IRB# 14-011236) and the local ethics committee of the IRCCS Mondino Foundation, Pavia, Italy (3549/2009 of 30/9/2009 and 11/12/2009; n.20170035275 of 23/10/2017). Amino acid substitutions were considered as pathogenic mutations when they were seen in the context of a neuroimmune/autoinflammatory state (including AGS, a spastic-dystonic syndrome, nonsyndromic spastic paraparesis or SMS), and when two or more of the following applied: observation of the same variant in an unrelated family; de novo occurrence; documented increase in ISG expression; in vitro data consistent with IFIH1 gain-of-function.
2.2 ∣. Mutational analysis
Mutations were identified on a variety of next-generation sequencing platforms. Where Sanger sequencing was undertaken, primers were designed to amplify the coding exons of IFH1, with mutation annotation based on the reference cDNA sequence NM_022168.2. Variants were assessed using the in silico programs SIFT (http://sift.jcvi.org), Polyphen2 (http://genetics.bwh.harvard.edu/pph2/), and CADD (https://cadd.gs.washington.edu), summarized in VarCards (http://varcards.biols.ac.cn/). Population allele frequencies were obtained from the gnomAD database (http://gnomad.broadinstitute.org).
2.3 ∣. Protein modeling
Molecular graphics figures were generated with PyMOL (Schrödinger) using the PDB coordinates (4GL2).
2.4 ∣. Interferon score
Interferon scores were calculated on the basis of the expression of ISGs according to previously published protocols. In brief, this involved either a quantitative reverse transcription-polymerase chain reaction (qPCR) analysis using TaqMan probes (Crow laboratory: Rice et al., 2013), or testing on a Nanostring platform (Vanderver laboratory: Adang et al., 2018+). In the former, the relative abundance of IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1) transcripts was normalized to the expression levels of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1). The median fold change of the six genes, compared to the median of 29 previously collected healthy controls, was then used to create an interferon score for each individual, with an abnormal interferon score being defined as greater than +2 standard deviations above the mean of the control group that is 2.466. Alternatively, the copy number of mRNA transcripts of the six ISGs listed above, and four housekeeping genes (ALAS1, HPRT1, TBP, and TUBB), was quantified using a Nanostring nCounter™ Digital Analyzer. The raw copy number of mRNA transcripts of each ISG was standardized using the geometric mean of the four housekeeping genes for each individual, and the six-gene interferon signature for each individual calculated using the median of the Z scores, with the result considered positive if ≥1.96 (>98th centile; one tail analysis).
2.5 ∣. Interferon reporter assay
The pFLAG-CMV4 plasmid encoding IFIH1 has been described elsewhere (Rice et al., 2014). Indicated mutations were introduced using Phusion HiFi DNA polymerase. HEK 293T cells (ATCC) were maintained in 48-well plates in DMEM (Cellgro) supplemented with 10% fetal bovine serum and 1% L-glutamine. At 80% confluence, cells were cotransfected with pFLAGCMV4 plasmids encoding wild-type or mutant IFIH1 (5 ng, unless indicated otherwise), interferon β (IFNb) promoter-driven firefly luciferase reporter plasmid (100 ng), and a constitutively expressed Renilla luciferase reporter plasmid (pRL-TK, 10 ng), by using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. The medium was changed 6 hr after transfection, and cells were subsequently incubated for 18 hr with or without stimulation with poly(I-C) (500 ng; InvivoGen) using Lipofectamine 2000. Cells were lysed with Passive Lysis Buffer (Promega), and IFNb promoter activity was measured using a Dual-Luciferase Reporter Assay (Promega) and a Synergy 2 plate reader (BioTek). Firefly luciferase activity was normalized to Renilla luciferase activity Each experiment was performed in triplicate and data are presented as mean ± standard mean of error. Statistical significance was determined by two-tailed, unpaired Student’s t-test with *, **, and *** indicating p values <.05, <.01, and <.001, respectively. Expression levels of individual constructs were tested by western blot analysis.
3 ∣. RESULTS
3.1 ∣. Molecular data
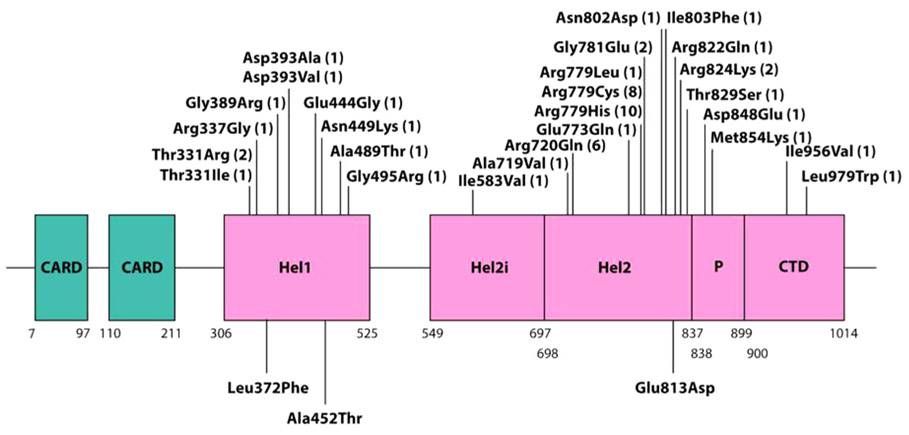
We collected data on 74 individuals from 51 families, identifying 27 distinct mutations in total (Figure 1; Table 1). Fourteen mutations were recorded in a single proband, seven in more than one individual belonging to a single-family, and six in more than one family. Of these six recurrent mutations, the p.Arg720Gln, p.Arg779Cys, and p.Arg779His substitutions were observed most frequently (6, 8, and 10 times, respectively). Twenty-two mutations were recorded to have occurred de novo in at least one individual, whilst four mutations were only ascertained in familial cases demonstrating autosomal dominant transmission (two mutations, p.Ala489Thr and p.Gly495Arg, were transmitted from a father in whom the mutation arose de novo). Three mutations, p.Thr331Arg, p.Arg779Cys, and p.Arg779His, was documented to have occurred both de novo, in association with severe, AGS-like, neurological disease, and in families with transmission across two or more generations.
FIGURE 1.
Schematic showing the positions of protein domains and their amino acid boundaries within the 1,025-residue IFIH1 protein. The 27 mutations ascertained in the present study are annotated, with the numbers in brackets indicating the number of families in which each mutation was observed. Three previously published mutations (p.Leu372Phe; p.Ala452Thr; p.Glu813Asp), not ascertained in our series, are also denoted (below the cartoon). CARD, caspase activation recruitment domain; Hel, helicase domain, where Hel1 and Hel2 are the two conserved core helicase domains and Hel2i is an insertion domain that is conserved in the RIG-I-like helicase family; P, pincer or bridge region connecting Hel2 to the C-terminal domain (CTD) involved in binding double stranded RNA
TABLE 1.
Details of individual IFIH1 mutations identified in the families included in the present data set
| cDNA change | Protein change | Families (de novo inheritance; or, number of symptomatic and non- penetrant individuals where familial) |
Associated phenotypes (‘/’ within family)(‘;’ between families) |
Upregulation of interferon signalling |
Assessment by interferon reporter assay |
gnomAD | SIFT | Polyphen2 | CADD score |
Var- cards |
|---|---|---|---|---|---|---|---|---|---|---|
| c.992C>G | p.Thr331Arg | AGS674 (de novo); AGS1972 (2;0) | AGS-SMS; SMS | Yes | Yes (de Carvalho et al., 2017) | Novel | Deleterious 0 | Probably damaging 1.000 | 29.7 | 22:23 |
| c.992C>T | p.Thr331Ile | AGS1938 (3;0) | SMS | Yes | Yes (de Carvalho et al., 2017) | Novel | Deleterious 0 | Probably damaging 1.000 | 31 | 22:23 |
| c.1009A>G | p.Arg337Gly | AGS237 (de novo) | NR | Yes | Yes (Rice et al., 2014) | Novel | Tolerated 0.12 | Probably damaging 1.000 | 26.8 | 17:23 |
| c.1165G>A | p.Gly389Arg | AGS848 (2;1) | AGS/SP/CNP | Yes | Yes (this paper) | Novel | Tolerated 0.88 | Benign 0.108 | 5.325 | 01:23 |
| c.1178A>T | p.Asp393Val | AGS626 (de novo) | NR | Yes | Yes (Rice et al., 2014) | Novel | Deleterious 0.01 | Probably damaging 0.998 | 28.6 | 16:23 |
| c.1178A>C | p.Asp393Ala | AGS2586 (de novo) | AGS | Yes | No | Novel | Deleterious 0.03 | Possibly damaging 0.913 | 24.8 | 12:23 |
| c.1331A>G | p.Glu444Gly | AGS2669 (de novo) | AGS | Yes | Yes (this paper) | Novel | Deleterious 0 | Probably damaging 1 | 31 | 23.23 |
| c.1347C>G | p.Asn449Lys | AGS1001 (de novo) | SP | Yes | Yes (this paper) | Novel | Tolerated 0.64 | Benign 0.163 | 13.91 | 03:23 |
| c.1465G>A | p.Ala489Thr | AGS755 (3;0)a | CLL/AGS-SMS/SMS | Yes | Yes (Bursztejn et al., 2015) | Novel | Deleterious 0 | Probably damaging 1.000 | 32 | 21:23 |
| c.1483G>A | p.Gly495Arg | AGS524 (2;0)a | SP-LLD/SP | Yes | Yes (Rice et al., 2014) | Novel | Deleterious 0.01 | Probably damaging 0.982 | 23.3 | 14:23 |
| c.1747A>G | p.Ile583Val | AGS2369 (de novo) | AGS | Yes | Yes (this paper) | Novel | Tolerated 0.48 | Benign 0.00 | 0.573 | 5.23 |
| c.2156C>T | p.Ala719Val | Hm_1 (de novo) | AGS | Yes | No | Novel | Tolerated 0.07 | Possibly damaging 0.949 | 27.1 | 09:23 |
| c.2159G>A | p.Arg720Gln | AGS102 (de novo); AGS647 (de novo); AGS1504 (de novo); AGS2422 (NPDT); AGS2548 (de novo); LD_0982.0 (de novo) | AGS; SP | Yes | Yes (Rice et al., 2014) | Novel | Deleterious 0 | Probably damaging 0.992 | 34 | 17:23 |
| c.2317G>C | p.Glu773Gln | AGS2399 (de novo) | NR | NA | Yes (this paper) | Novel | Tolerated 0.27 | Possibly damaging 0.743 | 24.8 | 13:23 |
| c.2335C>T | p.Arg779Cys | AGS376 (NPDT); AGS723 (NPDT); AGS1004 (de novo); AGS1156 (de novo); AGS2154 (1;1); AGS2180 (de novo); AGS2507 (de novo); LD_1030.0 (de novo) |
AGS-LLD; SP-ICC; NR; unilateral white matter disease/CNP; AGS |
Yes | Yes (Rice et al., 2014) | Novel | Deleterious 0.01 | Probably damaging 1.000 | 34 | 21:23 |
| c.2336G>A | p.Arg779His | AGS163 (de novo); AGS259 (3;2); AGS1351 (de novo); AGS1509 (de novo); AGS2177 (1;2); Berg_1 (de novo); Orc_0098 (de novo); LD_1199.0 (de novo); LD_1381 (3;1); LD_1585.0 (de novo) | AGS; CNP; NR; SP | Yes | Yes (Rice et al., 2014) | 1/244230 | Tolerated 0.05 | Probably damaging 0.994 | 28.9 | 19:23 |
| c.2336G>T | p.Arg779Leu | LD_1067.0 (de novo) | AGS | Yes | No | Novel | Tolerated 0.06 | Probably damaging 1.000 | 35 | 21:23 |
| c.2342G>A | p.Gly781Glu | LD_0940.0 (de novo); LD_0943.0 (de novo) | NR; SP | Yes | No | Novel | Deleterious 0 | Probably damaging 1.000 | 32 | 19:23 |
| c.2404A>G | p.Asn802Asp | AGS2662 (de novo) | NR | Yes | No | Novel | Tolerated 0.22 | Probably damaging 1.000 | 28.1 | 18:23 |
| c.2407A>T | p.Ile803Phe | LD_1488.0 (de novo) | AGS | Yes | Yes (this paper) | Novel | Tolerated 0.24 | Benign 0.043 | 11.8 | 04:23 |
| c.2465G>A | p.Arg822Gln | AGS1514 (de novo) | SD-ICC | Yes | Yes (Rutsch et al., 2015) | 6/244096 | Deleterious 0 | Probably damaging 1.000 | 35 | 23:23 |
| c.2471G>A | p.Arg824Lys | AGS735 (de novo); AGS2222 (de novo) | NR; Isolated liver disease | Yes | No | Novel | Deleterious 0 | Probably damaging 1.000 | 34 | 22:23 |
| c.2486C>G | p.Thr829Ser | AGS1290 (2 siblings and NPDT) | AGS | Yes | No | Novel | Tolerated 0.73 | Possibly damaging 0.512 | 16.61 | 12:23 |
| c.2544T>G | p.Asp848Glu | AGS531 (3;2) | SP-ICC/CNP | Yes | Yes (Ruaud et al., 2018) | Novel | Tolerated 0.4 | Benign 0.004 | 10.08 | 02:23 |
| c.2561T>A | p.Met854Lys | AGS2081 (de novo) | AGS/SMS | Yes | No | Novel | Deleterious 0 | Probably damaging 1.000 | 31 | 18:23 |
| c.2866A>G | p.Ile956Val | AGS1430 (2;1) | SP-ICC/CNP | Yes | Yes (this paper) | Novel | Tolerated 0.77 | Benign 0.004 | 3.576 | 06:23 |
| c.2936T>G | p.Leu979Trp | LD_1346.0 (de novo) | AGS | Yes | Yes (this paper) | Novel | Deleterious 0.01 | Probably damaging 1.000 | 26.6 | 16:23 |
Note: IFIH1 mutation annotation based on the reference complementary DNA sequence NM_022168.2.
Abbreviations: AGS, Aicardi–Goutières syndrome; CLL, Chilblain-like lesions; CNP, clinical nonpenetrance; ICC, Intracranial calcification; LLD, Lupus-like disease; NPDT, no parental DNA testing; NR, neuro-regression; SD, spastic dystonia; SP, spastic paraparesis; SMS, Singleton Merten syndrome.
This mutation was shown to have been paternally inherited by the proband and to have occurred de novo in the proband’s father.
For six putative mutations (p.Gly389Arg; p.Asn449Lys; p.Ile583Val; p.Ile803Phe; p.Asp848Glu; p.Ile956Val), in silico predictions using both SIFT and Poyphen2 suggested that the substitutions were benign, with relatively poor evolutionary conservation (Figure S1). However, all of these variants were novel (i.e., not recorded in gnomAD), and assays of interferon signaling (ISG expression and in vitro testing) indicate that they represent pathogenic mutations conferring gain-of-function (Table S1; Figure S2). Of note, four of these variants were seen in the context of a spastic paraparesis phenotype with no or minimal cognitive impairment. Clinical nonpenetrance was observed in three of these families (the other three variants arising in the proband de novo).
3.2 ∣. Clinical phenotype
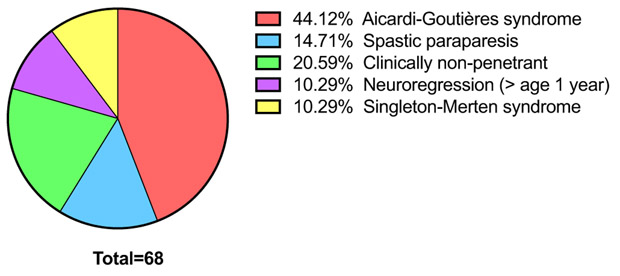
Consistent with previous data, we observed a spectrum of phenotypes in our cohort, encompassing classical AGS, less easily defined rapid neuroregression, a spastic-dystonic syndrome, spastic paraparesis, SMS, and clinical nonpenetrance (Figure 2; Table 2; Table S2). A single individual, AGS2222, experienced neonatal hepatitis and then developed chronic fibrotic liver disease in the absence of any other clinical features (note that this same variant was seen in another proband, AGS735, presenting with neuroregression at age 1 year). Unequivocal episodes of rapid neuroregression were noted in at least 20 patients, in seven of whom an acute loss of skills occurred after the age of 1 year on a background of completely normal development. Recognition/onset of symptoms was frequently later in patients with a spastic paraparesis phenotype, with one patient experiencing the development of lower limb spasticity beginning at 13 years of age (AGS531_P4). Six symptomatic patients were recorded to have died. Five of these individuals demonstrated a severe AGS phenotype with features obvious at, or soon after, birth that is indicating prenatal onset. One further deceased patient presented with neuroregression at age 15 months, and died suddenly of a cardiorespiratory arrest at 16 years of age, with pulmonary hypertension documented on postmortem examination. Ten individuals were reported as asymptomatic mutation carriers, across five mutations (p.Gly389Arg, p.Arg779Cys, p.Arg779His p.Asp848Glu, and p.Ile956Val), with seven aged over 50 years.
FIGURE 2.
Overview of phenotypes observed in the IFIH1-mutation-positive cohort. Classification of 68 of 74 individuals according to phenotype. For clarity, six individuals displaying characteristics difficult to classify were omitted from this analysis
TABLE 2.
Molecular and clinical data by family
| Family | Individual | Sex | cDNA | Protein | Inheritance (number of mutation-positive individuals) |
Previously reported (reference) |
Clinical phenotype | Status at last contact (age in years) |
|---|---|---|---|---|---|---|---|---|
| AGS102 | P1 | M | c.2159G>A | p.Arg720Gln | De novo | Rice et al. (2014) | AGS | Deceased (2) |
| AGS163 | P1 | M | c.2336G>A | p.Arg779His | De novo | Rice et al. (2014) | AGS | Alive (13) |
| AGS237 (LD_0762) | P1 | M | c.1009A>G | p.Arg337Gly | De novo | Rice et al. 2014; Adang et al., 2018 | Neuroregression and SD starting at age 15 months | Deceased (16) |
| AGS259 | P1 | M | c.2336G>A | p.Arg779His | Familial (3) | Rice et al. (2014) | AGS | Alive (13) |
| P2 (father of P1) | M | Clinically nonpenetrant | Alive (54) | |||||
| P3 (mother of P2) | F | Clinically nonpenetrant | Deceased (84) | |||||
| AGS376 | P1 | M | c.2335C>T | p.Arg779Cys | No parental testing | Rice et al. (2014) | AGS with LLD | Deceased (3) |
| AGS524 | P1 | F | c.1483G>A | p.Gly495Arg | Familial (2)(shown to have occurred de novo in P2) | Rice et al. (2014); Hacohen et al. 2015; Crow et al. 2015; McLellan et al. 2018 | SP with LLD and AQP4 + TM | Alive (10) |
| P2 (father of P1) | M | Pure SP | Alive (39) | |||||
| AGS531 | P1 | F | c.2544T>G | p.Asp848Glu | Familial (5) | Ruaud et al. (2018) | SP with ICC | Alive (13) |
| P2 (brother of P1) | M | Clinically nonpenetrant | Alive (13) | |||||
| P3 (father of P1 and P2) | M | SP with ICC | Alive (40) | |||||
| P4 (brother of P3) | M | SP with ICC | Alive (38) | |||||
| P5 (father of P3 and P4) | M | Clinically non-penetrant | Alive (66) | |||||
| AGS626 | P1 | M | c.1178A>T | p.Asp393Val | De novo | Rice et al. (2014) | Neuroregression and SD starting at 13 months | Alive (13) |
| AGS647 | P1 | M | c.2159G>A | p.Arg720Gln | De novo | Rice et al. (2014) | AGS | Alive (2) |
| AGS674 | P1 | M | c.992C>G | p.Thr331Arg | De novo | Unreported | SP-SMS overlap | Alive (14) |
| AGS723 | P1 | F | c.2335C>T | p.Arg779Cys | Mother negative; no paternal DNA | Unreported | SP with ICC | Alive (19) |
| AGS735 | P1 | M | c.2471G>A | p.Arg824Lys | De novo | Galli et al. 2018 | Neuroregression and SD starting at 12 months | Alive (19) |
| AGS755 | P1 | M | c.1465G>A | p.Ala489Thr | Familial (3) | Bursztejn et al. (2015) | CLL | Alive (4) |
| P2 (brother of P1) | M | AGS-SMS overlap | Alive (3) | |||||
| P3 (father of P1 and P2) | M | SMS-like | Alive (41) | |||||
| AGS848 | P1 | M | c.1165G>A | p.Gly389Arg | Familial (3) | Unreported | AGS | Alive (8) |
| P2 (father of P1) | M | SP | Alive (42) | |||||
| P3 (maternal grandmother of P2) | F | Clinically nonpenetrant | Alive (84) | |||||
| AGS1001 | P1 | M | c.1347C>G | p.Asn449Lys | De novo | Unreported | SP | Alive (19) |
| AGS1004 | P1 | F | c.2335C>T | p.Arg779Cys | De novo | Unreported | AGS (neuroregression with onset at age 8 months) | Alive (8) |
| AGS1156 | P1 | M | c.2335C>T | p.Arg779Cys | De novo | Kothur et al. 2018 | AGS (neuroregression with onset at age 8 months) | Alive (5) |
| AGS1290 | P1 | M | c.2486C>G | p.Thr829Ser | 2 affected (no parental DNA) | Unreported | AGS | Alive (6) |
| P2 (brother of P1) | M | AGS | Alive (4) | |||||
| AGS1351 | P1 | F | c.2336G>A | p.Arg779His | De novo | Unreported | AGS | Deceased (2) |
| AGS1430 | P1 | M | c.2866A>G | p.Ile956Val | Familial (3) | Unreported | SP with ICC with onset at age 6 years | Alive (14) |
| P2 (father of P1) | M | SP with onset at age 2 years | Alive (50) | |||||
| P3 (father of P2) | M | Clinically non-penetrant | Alive (72) | |||||
| AGS1504 (LD_1175) | P1 | F | c.2159G>A | p.Arg720Gln | De novo | Unreported | AGS | Alive (10) |
| AGS1509 | P1 | M | c.2336G>A | p.Arg779His | De novo | Unreported | AGS | Alive (8) |
| AGS1514 | P1 | M | c.2465G>A | p.Arg822Gln | De novo | Buers et al. (2017) | SD with ICC | Alive (6) |
| AGS1938 | P1 | F | c.992C>T | p.Thr331Ile | Familial (3) | de Carvalho et al. (2017) | SMS | Alive (18) |
| P2 (mother of P1) | F | SMS | Alive (45) | |||||
| P3 (sister of P2) | F | SMS | Alive (27) | |||||
| AGS1972 | P1 | F | c.992C>G | p.Thr331Arg | Familial (2) | de Carvalho et al. (2017) | SMS | Alive (9) |
| P2 (father of P1) | M | SMS | Alive (47) | |||||
| AGS2081 | P1 | M | c.2561T>A | p.Met854Lys | De novo | Unreported | SP-SMS overlap | Alive (12) |
| AGS2154 (LD_1240) | P1 | M | c.2335C>T | p.Arg779Cys | Familial (2) | Unreported | Unilateral white matter disease with normal development | Alive (13) |
| P2 (mother of P1) | F | Clinically nonpenetrant | Alive (40) | |||||
| AGS2177 | P1 | M | c.2336G>A | p.Arg779His | Familial (3) | Neuroregression and SD starting at age 12 months | Alive (29) | |
| P2 (mother of P1) | F | Clinically nonpenetrant | Alive (62) | |||||
| P3 (sister of P1) | F | Clinically nonpenetrant | Alive (33) | |||||
| AGS2180 | P1 | F | c.2335C>T | p.Arg779Cys | De novo | Unreported | AGS | Alive (4) |
| AGS2222 | P1 | M | c.2471G>A | p.Arg824Lys | De novo | Unreported | Isolated liver disease | Alive (9) |
| AGS2369 | P1 | M | c.1747A>G | p.Ile583Val | De novo | Unreported | AGS | Alive (10) |
| AGS2399 | P1 | M | c.2317G>C | p.Glu773Gln | De novo | Unreported | Neuroregression and SD starting at age 16 months | Alive (8) |
| AGS2422 | P1 | F | c.2159G>A | p.Arg720Gln | No parental testing | Unreported | SP | Alive (38) |
| AGS2507 | P1 | F | c.2335C>T | p.Arg779Cys | De novo | Unreported | AGS | Alive (1) |
| AGS2548 | P1 | M | c.2159G>A | p.Arg720Gln | De novo | Unreported | AGS | Alive (3) |
| AGS2586 | P1 | M | c.1178A>C | p.Asp393Ala | De novo | Unreported | AGS-like with frank regression at age 21 months | Alive (3) |
| AGS2662 (LD_1640) | P1 | F | c.2404A>G | p.Asn802Asp | De novo | Unreported | Neuroregression and SD starting at age 11 months | Alive (1) |
| AGS2669 | P1 | M | c.1331A>G | p.Glu444Gly | De novo | Unreported | AGS | Deceased (0.5) |
| Hm_1 | P1 | F | c.2156C>T | p.Ala719Val | De novo | Unreported | AGS | Alive (2) |
| Berg_1 | P1 | F | c.2336G>A | p.Arg779His | De novo | Unreported | Neuroregression and SD starting at age 9 months | Alive (7) |
| Orc_0098 | P1 | M | c.2336G>A | p.Arg779His | De novo | Unreported | AGS | Alive (4) |
| LD_0940.0 | P1 | M | c.2342G>A | p.Gly781Glu | De novo | Unreported | Neuroregression and SD starting at age 15 months | Alive (5) |
| LD_0943.0 | P1 | F | c.2342G>A | p.Gly781Glu | De novo | Unreported | SP | Alive (14) |
| LD_0982.0 | P1 | M | c.2159G>A | p.Arg720Gln | De novo | Adang et al. (2018); Case 2 | AGS | Alive (9) |
| LD_1030.0 | P1 | F | c.2335C>T | p.Arg779Cys | De novo | Unreported | AGS | Alive (5) |
| LD_1067.0 | P1 | M | c.2336G>T | p.Arg779Leu | De novo | Unreported | AGS | Alive (8) |
| LD_1199.0 | P1 | F | c.2336G>A | p.Arg779His | De novo | Unreported | AGS | Alive (4) |
| LD_1346.0 | P1 | M | c.2936T>G | p.Leu979Trp | De novo | Adang et al. (2018); Case 3 | AGS | Deceased (0.4) |
| LD_1381 (Hart) | P1 | F | c.2336G>A | p.Arg779His | Familial (4) | Unreported | SP | Alive (4) |
| P2 (brother of P1) | M | SP | Alive (3) | |||||
| P3 (father of P1 and P2) | M | SP | Alive (32) | |||||
| P4 (father of P3) | M | Clinically nonpenetrant | Alive (68) | |||||
| LD_1488.0 | P1 | F | c.2407A>T | p.Ile803Phe | De novo | Unreported | AGS | Alive (2) |
| LD_1585.0 | P1 | F | c.2336G>A | p.Arg779His | De novo | Unreported | AGS | Alive (5) |
Note: IFIH1 mutation annotation based on the reference complementary DNA sequence NM_022168.2.
Abbreviations: AGS, Aicardi–Goutières syndrome; CLL, Chilblain-like lesions; F, Female; ICC, intracranial calcification; LLD, Lupus-like disease; M, Male; SD, spastic dystonia; SP, spastic paraparesis; SMS: Singleton Merten syndrome; TM, transverse myelitis
3.3 ∣. Interferon status
Where tested, all mutations (i.e., 26 of 27) were associated with increased expression of ISGs in peripheral blood (Table 1). Samples were unavailable for the single patient carrying the p.Glu773Gln substitution. This variant is not recorded in gnomAD, occurring de novo in the context of a phenotype compatible with IFIH1 upregulation, and conferring a gain-of-function in our in vitro assay (Figure S2). Considering all (51) mutation-positive individuals tested for ISG expression in the Crow laboratory (given that a direct comparison of results across laboratories is not possible), 109 of 117 values were positive (Table S3; Figure S3). Only one clinically symptomatic patient (AGS2154_1) demonstrated a negative interferon signature (on two of three occasions tested). The phenotype, in this case, was unusual; a child with white matter disease confined to the right cerebral hemisphere on MRI and no abnormal neurological signs on examination, having presented at age 8 years with headaches. We leave open the possibility that these two normal results, and three normal results from his mother, might be due to technical artifact, given that the samples had been stored for many months before testing. Sixteen samples from seven clinically nonpenetrant subjects exhibited an upregulation of interferon signaling, with two asymptomatic mutation carriers demonstrating normal interferon signatures (each tested on three occasions).
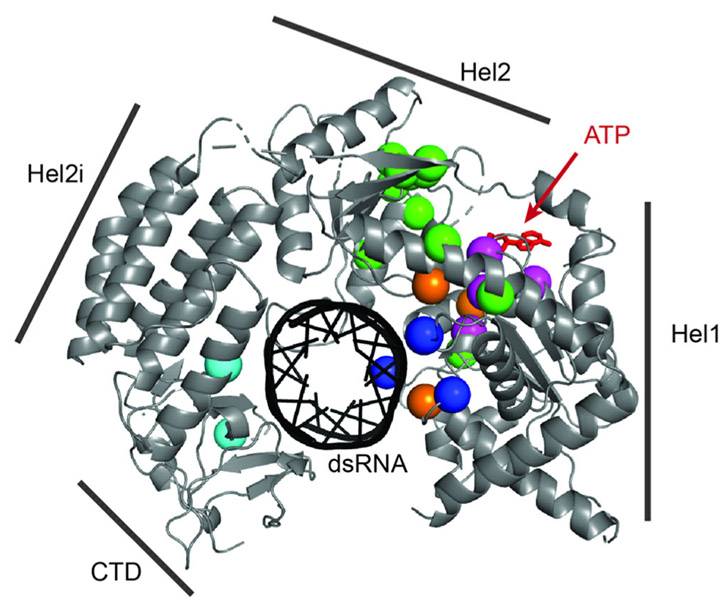
3.4 ∣. Modeling of IFIH1 gain-of-function mutations
Modeling of the 27 mutations described here showed that most residues cluster near the ATP binding site within the helicase domain (Figure 3). Three mutations, p.Ileu583Val, p.Ileu956Val, and p.Leu979Trp were the only residues not situated in the cluster (colored cyan; only p.Ileu583Val and p.Leu979Val are shown since residue p.Ileu956 is disordered in the crystal structure). Within this main cluster, residues can be further categorized into three groups: those at the ATP binding pocket (magenta spheres), those in the double stranded RNA (dsRNA) binding surface (colored blue) and those not directly involved in either ATP or RNA binding (colored green). Three published mutations (p.Leu372phe; p.Ala452Thr; p.Glu813Asp; Table S4) not ascertained in our cohort are also located within the main cluster (colored orange), further supporting the importance of this region in the regulation of IFIH1 signaling activity.
FIGURE 3.
Mutation mapping. Structure of human IFIH1 (4GL2) in complex with double stranded RNA (dsRNA; blue stick model in the center). Only the RNA binding domain (helicase domain and C-terminal domain, CTD) are included in the crystal structure. Note that the helicase domain consists of Hel1, Hel2i, and Hel2. Mutations are indicated by spheres using the following color code: residues in the ATP binding pocket (magenta), residues in the dsRNA binding surface (blue), residues within the main cluster but not directly involved in RNA binding or ATP binding (green), residues outside the main cluster (cyan), and residues previously reported by others but not in our cohort (orange). We considered all 27 mutations reported here plus three previously published mutations (p.Leu372Phe; p.Ala452Thr; p.Glu813Asp) not ascertained in our series. Residues p.Arg822, p.Arg824, and p.Ile956 are not shown because they are disordered in the crystal structure, but are expected to be located in the ATP binding (p.Arg822 and p.Arg824) and RNA binding (p.Ile956) pockets
4 ∣. DISCUSSION
Here we present data on 74 individuals, 41 previously unreported, from 51 families, with a putative gain-of-function mutation in IFIH1. Consistent with previous descriptions, we observed a spectrum of phenotypes, encompassing AGS, spastic-dystonia, spastic paraparesis, SMS and clinical nonpenetrance. Phenotypic variability was common, both in the context of familial inheritance and mutations seen recurrently across families so that no obvious genotype–phenotype correlations could be ascertained.
Acute regression was noted in almost one-third of symptomatic mutation carriers, occurring after the age of 1 year in seven patients demonstrating completely normal development to that time. Beyond acute regression, a slower onset of disease, and subsequent progression, was seen in patients demonstrating a spastic paraparesis phenotype. Together with the observation of clinical nonpenetrance (10:13.5% of 74 mutation-positive individuals in our series), with seven individuals identified to be apparently disease-free beyond the age of 50 years, these data suggest the importance of additive genetic factors and/or environmental triggers in determining phenotypic status. Although we did not formally record neuroimaging features in our cohort, white matter disease and intracranial calcification were observed frequently. Such imaging characteristics can be seen in the absence of overt neurological signs (see Bursztejn et al., 2015 and de Carvalho et al., 2017). Conversely, significant neurological disease, most typically spastic paraparesis, can occur in the context of normal brain and spinal imaging (e.g., the father in family AGS524).
Clinically manifest extraneurological illness was uncommon in our series, but there appears to be a real association between IFIH1 gain-of-function and lupus-like illness, autoimmune hepatitis, and hypothyroidism. Furthermore, psoriatic-like skin disease is a well-recognized feature of the SMS phenotype. As recently described (Adang et al., 2018), two patients included here were diagnosed with pulmonary hypertension, a feature which was not searched for in most patients and may be under-recognized.
We observed a strong association of mutation status with an enhanced expression of ISGs, with 109 of 117 samples from 51 patients being positive in the experience of one laboratory. A similar conclusion can be drawn from in vitro testing. As such, upregulated interferon signaling represents a reliable biomarker of IFIH1 gain-of-function, and can serve as an indicator of variant pathogenicity where doubt exists as to the significance of a molecular lesion. This is important given that we show here that in silico algorithms do not always accurately predict pathogenicity (involving 22% of the mutations that we recorded). Where tested, clinical nonpenetrance was also associated with a persistent upregulation of interferon signaling, with only two of nine such individuals nonpenetrant on ISG testing in blood. Whether these individuals demonstrate fluctuations in ISG expression is not known at this time.
Despite documented clinical nonpenetrance in some cases, all putative IFIH1 gain-of-function substitutions are rare, with only two of the 30 discrete mutations described here and in previous reports recorded in gnomAD. Furthermore, all ascertained type I interferonopathy associated mutations are missense variants, likely conferring increased sensitivity to a self-derived nucleic acid. Although premature termination mutations in the helicase domain are seen in control populations as common polymorphisms, none has been associated with a type I interferonopathy phenotype, further supporting the role of nucleic acid binding by the helicase domain in disease pathogenesis. Substitutions of the arginine residues at positions 720 and 779 were seen in six and 19 probands, respectively, in our series. Given the focus of our laboratories on pediatric neurological disease, our data are likely to subject to ascertainment bias. Indeed, although only observed once by us, the p.Arg822Gln mutation has been reported in an additional five pedigrees demonstrating a classical SMS phenotype (Pettersson et al., 2017; Rutsch et al., 2015).
IFIH1 is a member of the retinoic acid-inducible gene I (RIG-I) receptor family (del Toro Duany, Wu, & Hur, 2015). Recognition of cytoplasmic viral dsRNA by IFIH1 induces filament assembly along the dsRNA axis, with the helicase domains and C terminal domain responsible for RNA recognition. Filament formation then induces oligomerization of the tandem CARD domains (2CARD) of IFIH1, leading to the interaction with mitochondrial MAVS and subsequent induction of interferon and other proinflammatory cytokines. IFIH1 filament stability is intrinsically regulated by ATP hydrolysis, which is stimulated upon dsRNA binding. Mutations that impair ATP hydrolysis generally increase filament stability and, often, but not always, confer gain-of-function signaling activity. The clustering of mutations that we ascertained, and of a further three unique published mutations, near the ATP binding region likely highlights common mechanisms, perhaps increasing RNA binding affinity or decreasing the efficiency of ATP hydrolysis and the rate of filament disassembly.
Summarizing, IFIH1 gain-of-function is associated with a spectrum of phenotypes, occurring due to de novo mutations or transmitted as an autosomal dominant trait. Testing for an interferon signature in blood represents a useful biomarker in this context, which can aid in the interpretation of identified sequence variants.
Supplementary Material
ACKNOWLEDGMENTS
Yanick J. Crow acknowledges The University of Maryland Brain and Tissue Bank of the NIH NeuroBioBank. Yanick J. Crow acknowledges the European Research Council (786142-E-T1IFNs), a state subsidy managed by the National Research Agency (France) under the “Investments for the Future” program bearing the reference ANR-10-IAHU-01 and the MSDAvenir fund (DEVO-DECODE Project). Tracy A. Briggs acknowledges the National Institute for Health Research (NIHR; NIHR Transitional Research Fellowship, TRF-2016-09-002; with the views expressed were those of the author and not necessarily those of the NHS, the NIHR or the Department of Health). Adeline L. Vanderver is supported by the Kamens endowed chair for Translational Neurotherapeutics and the Myelin Disorders Bioregistry Project. Adeline L. Vanderver and Laura A. Adang acknowledge the CURE Pennsylvania Frontiers in Leukodystrophy grant and U01HD082806. Laura A. Adang also acknowledges the National Center for Advancing Translational Sciences of the National Institutes of Health under award number KL2TR001879. Lien Van Eyck received funding from Research Foundation Flanders (FWO).
Funding information
Public Health Research Programme, Grant/Award Number: TRF-2016-09-002; National Center for Advancing Translational Sciences of the National Institutes of Health, Grant/Award Number: KL2TR001879; CURE Pennsylvania Frontiers in Leukodystrophy, Grant/Award Number: U01HD082806; H2020 European Research Council, Grant/Award Number: 786142-E-T1IFNs; Agence Nationale de la Recherche, Grant/Award Number: ANR-10-IAHU-01; MSDAvenir fund, Grant/Award Number: DEVO-DECODE Project; NIHR Transitional Research Fellowship, Grant/Award Number: TRF-2016-09-002
Footnotes
CONFLICT OF INTERESTS
Y. J. Crow has undertaken consultancy work with Biogen on behalf of the University of Edinburgh.
DATA AVAILABILITY STATEMENT
Data available on request due to privacy/ethical restrictions. Identified variants submitted to ClinVar (Submission ID: SUB6667166; Organization ID: 507341).
Gillian I. Rice and Sehoon Park equally contributed to this study.
REFERENCES
- Adang LA, Frank DB, Gilani A, Takanohashi A, Ulrick N, Collins A, & Vanderver AL (2018). Aicardi goutieres syndrome is associated with pulmonary hypertension. Molecular Genetics and Metabolism, 125(4), 351–358. 10.1016/j.ymgme.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, & Hur S (2018). Breaching self-tolerance to alu duplex RNA underlies MDA5-mediated inflammation. Cell, 172(4), 797–810. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buers I, Rice GI, Crow YJ, & Rutsch F (2017). MDA5-associated neuroinflammation and the Singleton-Merten syndrome: Two faces of the same type I interferonopathy spectrum. Journal of Interferon and Cytokine Research, 37(5), 214–219. 10.1089/jir.2017.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursztejn AC, Briggs TA, del Toro Duany Y, Anderson BH, O'Sullivan J, Williams SG, & Crow YJ (2015). Unusual cutaneous features associated with a heterozygous gain-of-function mutation in IFIH1: Overlap between Aicardi-Goutieres and Singleton-Merten syndromes. British Journal of Dermatology, 173(6), 1505–1513. 10.1111/bjd.14073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho LM, Ngoumou G, Park JW, Ehmke N, Deigendesch N, Kitabayashi N, & Crow YJ (2017). Musculoskeletal disease in MDA5-related type I interferonopathy: A Mendelian mimic of Jaccoud's arthropathy. Arthritis Rheumatol, 69, 2081–2091. 10.1002/art.40179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, & Manel N (2015). Aicardi-Goutieres syndrome and the type I interferonopathies. Nature Reviews Immunology, 15(7), 429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- del Toro Duany Y, Wu B, & Hur S (2015). MDA5-filament, dynamics and disease. Curr Opin Virol, 12, 20–25. 10.1016/j.coviro.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli J, Gavazzi F, De Simone M, Giliani S, Garau J, Valente M, … Fazzi E (2018). AGS study group. Sine causa tetraparesis: A pilot study on its possible relationship with interferon signature analysis and Aicardi Goutiéres syndrome related genes analysis. Medicine Baltimore, 97(52), e13893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacohen Y, Zuberi S, Vincent A, Crow YJ, & Cordeiro N (2015). Neuromyelitis optica in a child with Aicardi-Goutieres syndrome. Neurology, 85, 381–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothur K, Bandodkar S, Chu S, Wienholt L, Johnson A, Barclay P, … Dale (2018). An open-label trial of JAK 1/2 blockade in progressive IFIH1-associated neuroinflammation. Neurology, 90, 289–291. [DOI] [PubMed] [Google Scholar]
- McLellan KE, Martin N, Davidson JE, Cordeiro N, Oates BD, Neven B, … Crow YJ (2018). JAK 1/2 Blockade in MDA5 Gain-of-Function. Journal Clinic Immunology, 38, 844–846. [DOI] [PubMed] [Google Scholar]
- Oda H, Nakagawa K, Abe J, Awaya T, Funabiki M, Hijikata A, & Heike T (2014). Aicardi-Goutieres syndrome is caused by IFIH1 mutations. American Journal of Human Genetics, 95(1), 121–125. 10.1016/j.ajhg.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M, Bergendal B, Norderyd J, Nilsson D, Anderlid BM, Nordgren A, & Lindstrand A (2017). Further evidence for specific IFIH1 mutation as a cause of Singleton-Merten syndrome with phenotypic heterogeneity. American Journal of Medical Genetics. Part A, 173(5), 1396–1399. 10.1002/ajmg.a.38214 [DOI] [PubMed] [Google Scholar]
- Popp B, Ekici AB, Thiel CT, Hoyer J, Wiesener A, Kraus C, … Zweier C (2017). Exome Pool-Seq in neurodevelopmental disorders. European Journal Human Genetics, 25(12), 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, & Crow YJ (2013). Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: A case-control study. Lancet Neurology, 12(12), 1159–1169. 10.1016/S1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, & Crow YJ (2014). Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature Genetics, 46(5), 503–509. 10.1038/ng.2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud L, Rice GI, Cabrol C, Piard J, Rodero M, van Eyk L, & Van Maldergem L (2018). Autosomal-dominant early-onset spastic paraparesis with brain calcification due to IFIH1 gain-of-function. Human Mutation, 39(8), 1076–1080. 10.1002/humu.23554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, MacDougall M, Lu C, Buers I, Mamaeva O, Nitschke Y, & Hennekam RC (2015). A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. American Journal of Human Genetics, 96(2), 275–282. 10.1016/j.ajhg.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.