SUMMARY
Serum starvation stimulates cilia growth in cultured cells, yet, serum factors associated with ciliogenesis are unknown. Previously, we showed that starvation induces rapid Rab11-dependent vesicular trafficking of Rabin8, a Rab8 GEF, to the mother centriole leading to Rab8 activation and cilium growth. Here, we demonstrate that through the LPA receptor 1 (LPAR1), serum lysophosphatidic acid (LPA) inhibits Rab11a-Rabin8 interaction and ciliogenesis. LPA/LPAR1 regulates ciliogenesis initiation via downstream PI3K/Akt activation, independent of effects on cell cycle. Akt stabilizes Rab11a binding to its effector, WDR44, and a WDR44-pAkt-phosphomimetic mutant blocks ciliogenesis. WDR44 depletion promotes Rabin8 preciliary trafficking and ciliogenesis-initiating events at the mother centriole. Our work suggests disruption of Akt signaling causes a switch from Rab11-WDR44 to the ciliogenic Rab11-FIP3-Rabin8 complex. Finally, we demonstrate Akt regulates downstream ciliogenesis processes associated with Rab8-dependent cilia growth. Together, this study uncovers a mechanism whereby serum mitogen signaling regulates Rabin8 preciliary trafficking and ciliogenesis initiation.
Keywords: Ciliogenesis, preciliary trafficking, Rabin8, Rab11 effector switch, lysophosphatidic acid (LPA), Akt, WDR44, FIP3, phosphorylation, mother centriole (MC)
eTOC
Cilia assembly is inhibited by activation of the PI3K/Akt pathway by the serum factor lysophosphatidic acid, independent of cell cycle alterations. Walia et al. show that activated Akt phosphorylates WDR44, stabilizing its effector-based binding to Rab11 and preventing Rabin8 preciliary trafficking and ciliogenesis initiation.
INTRODUCTION
The primary cilium functions in signal transduction pathways, including phototransduction, chemosensation, mechanosensation, and specific morphogen signaling (reviewed in Reiter and Leroux, 2017). Defects in cilia function and formation have been linked to a number of human genetic diseases and certain cancers (Waters and Beales, 2011). Ciliogenesis is cell cycle-dependent, with cilia growth observed in G1 or following cell cycle exit in G0 phase and resorption occurring before mitosis (Izawa et al., 2015). Serum deprivation triggers ciliogenesis in cultured fibroblasts and other cell lines, such as hTERT-immortalized retinal pigment epithelial (RPE-1) cells, presumably by interrupting cell cycle progression (Pugacheva et al., 2007; Tucker et al., 1979). However, the mechanism regulating the initiation of ciliogenesis upon serum deprivation is unknown.
The cilium assembles from the mother centriole (MC), which, unlike the daughter centriole, acquires appendage structures and becomes the basal body (Izawa et al., 2015; Sorokin, 1968). Depending on the cell type, ciliogenesis follows two distinct pathways, the intracellular or extracellular pathway (Molla-Herman et al., 2010; Sorokin, 1962; Sorokin, 1968). During intracellular ciliogenesis, membrane vesicles are transported to the distal appendages where they assemble into a larger ciliary vesicle (CV) promoting the uncapping of the MC via removal of a complex of proteins that includes CP110 and CEP97 (Sanchez and Dynlacht, 2016). This uncapping step leads to accumulation of intraflagellar transport proteins (IFT) and transition zone (TZ) proteins, followed by IFT-dependent growth of the microtubule-based axoneme (Lu et al., 2015). The developing intracellular cilium membrane subsequently fuses with the plasma membrane to extend the cilium from the cell surface. In extracellular ciliogenesis, the MC migrates to and docks with the plasma membrane, resulting in its distal uncapping. This pathway has been proposed to function in polarized epithelial cells, whereas intracellular ciliogenesis occurs in photoreceptors, fibroblasts, and other mesenchymal-like cells, including RPE-1 cells (Molla-Herman et al., 2010; Sedmak and Wolfrum, 2011; Sorokin, 1962; Sorokin, 1968).
Vesicle transport regulators including certain Rab small GTPases, SNAREs, and the exocyst complex are required for ciliogenesis (Feng et al., 2012; Knodler et al., 2010; Lobo et al., 2017; Lu et al., 2015; Nachury et al., 2007; Sato et al., 2013; Westlake et al., 2011; Yoshimura et al., 2007). The function of these proteins is best understood in the intracellular ciliation pathway. Our group and others have shown that a Rab11-Rab8 membrane trafficking cascade functions in the docking of distal appendage vesicles (DAV) to the MC, the reorganization of these membranes to form a CV, and ciliary growth (Knodler et al., 2010; Westlake et al., 2011). Rab11a and −11b, two regulators of the endocytic recycling compartment (ERC), bind to Rabin8, a guanine-nucleotide exchange factor (GEF) for Rab8, and transport this protein to the centrosome where it activates Rab8 to assemble the ciliary membrane from the CV (Lu et al., 2015; Westlake et al., 2011). Rabin8 is also a GEF for Rab10, a protein required for ciliogenesis in Rab8 knockout murine embryonic fibroblasts (Homma and Fukuda, 2016; Sato et al., 2013). Rabin8 also interacts with other ciliogenesis-associated membrane trafficking regulators including Sec15, an exocyst subunit, and the BBSome (Chiba et al., 2013; Feng et al., 2012; Nachury et al., 2007).
Rab11-dependent preciliary vesicular trafficking of Rabin8 to the MC is an early step in ciliogenesis (Westlake et al., 2011). Using live-cell imaging, rapid Rab11-dependent recruitment of GFP-Rabin8 to vesicles was observed within a few minutes of serum starvation and resulted in robust centrosomal accumulation of Rabin8 vesicles after one hour of serum withdrawal. Following ciliogenesis, Rabin8 preciliary trafficking and centrosomal accumulation ceases, presumably to control ciliary length by limiting Rab8 activation (Chiba et al., 2013; Westlake et al., 2011). This regulation of Rabin8 centrosomal trafficking is associated with the phosphorylation of Ser272 by NRD2 kinase (Chiba et al., 2013). However, the mechanism involved in the initiation of Rabin8 preciliary trafficking is unknown. Given the acute response of preciliary trafficking during nutrient starvation, we hypothesized that serum factor-dependent signaling is important.
In the present study, we investigate the mechanism underlying growth factor regulation of ciliogenesis and its effect on Rab11-dependent preciliary trafficking of Rabin8 to the centrosome. We discover that lysophosphatidic acid (LPA) blocks serum starvation-induced ciliogenesis in several human cell lines. LPA regulates ciliogenesis via the LPAR1 receptor and downstream PI3K-Akt kinase pathway, blocking Rab11a-Rabin8 binding and preciliary transport. Interestingly, inhibition of these signaling pathways promotes Rabin8 preciliary trafficking and ciliogenesis initiation independent of changes to the cell cycle and cell growth. We demonstrate that Akt phosphorylates Rabin8 near the GEF domain at Serine147/149, and that mutation of these residues to Alanine does not affect binding to Rab11a, but affects Rab8a activation. Examining the role of other Rab11 effectors in Akt inhibition of ciliogenesis led to the discovery that WDR44 knockdown promotes Rabin8 preciliary trafficking and modifications to the MC that precede cilia growth. We show that WDR44 is phosphorylated by Akt at Serine342/344 in the Rab11 binding domain, and a phosphomimetic mutant binds more strongly to Rab11 than the phosphoinactive mutant and blocks ciliogenesis in human cells and zebrafish embryos. Lastly, we provide evidence that another Rab11 effector, FIP3, is involved in the switch from an inhibitory Rab11-WDR44 to a ciliogenic Rab11-FIP3-Rabin8 complex. Together, our results indicate that LPA/LPAR1 activation of Akt prevents Rabin8 preciliary trafficking and ciliogenesis initiation at the MC by promoting Rab11 effector-based interaction with WDR44.
RESULTS
Lysophosphatidic acid blocks Rabin8 preciliary trafficking and ciliogenesis-initiating events at the mother centriole
We hypothesized that serum starvation triggers ciliogenesis in cultured cells by affecting signaling pathways associated with mitogenic factors, and, to test this theory, we examined the effects of common growth factors on ciliation in RPE-1 cells. Strikingly, none of the protein-based growth factors blocked ciliogenesis in serum-starved cells (Fig 1A). Importantly, the cell cycle profiles of growth factor-treated cells were similar to serum starved cells with most cells in G0/G1, whereas the serum-fed cells showed a normal cell cycle distribution (Figure S1A). Interestingly, cells treated with boiled serum failed to ciliate and showed a cell cycle profile consistent with serum-fed cells (Fig 1B), suggesting that a non-protein mitogen could regulate ciliogenesis. Based on these results, we investigated lysophosphatidic acid (LPA), a lipid mitogen reported to be present at high concentrations of up to ~30μM in bovine serum (Tokumura et al., 1994). Remarkably, we show that LPA treatment prevents ciliation as effectively as serum (Fig 1A, S1B, S1C). Consistent with the effects observed on ciliogenesis, LPA-treated cells displayed similar cell cycle profiles as serum-fed cells (Fig 1B) and had higher cell numbers than serum starved cells (Fig S1D), indicating that LPA-treated cells were proliferating. The LPA precursors, phosphatidic acid (PA) and lysophosphatidyl choline (LPC), had no effect on ciliation (Fig 1C). Similarly, LPA treatment blocked ciliogenesis induced by serum starvation in normal human pancreatic nestin expressing (hTERT-HPNE) cells (Fig S1E), neonatal human dermal fibroblasts (NeoHDF) (Fig 1D), breast epithelial MCF12A cell lines (Fig S1F), and murine embryonic fibroblasts (MEF) (Fig S1G). In contrast, LPA treatment had no effect on ciliation in murine inner medullary collecting duct (IMCD3) cells, which can ciliate in the presence of serum (Fig S1H). Together, these results demonstrate that LPA inhibits serum-dependent ciliogenesis in cultured cells.
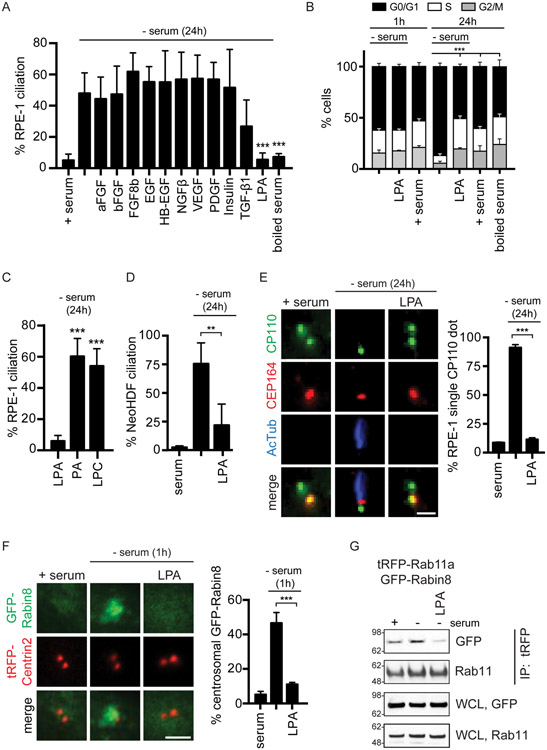
Figure 1: LPA inhibits serum starvation-induced ciliogenesis and Rab11-dependent Rabin8 preciliary trafficking.
(A) Quantification of ciliation in RPE-1 cells grown in 2% serum, 2% boiled serum, or treated with indicated growth factors (25ng/ml) for 24h in the absence of serum followed by staining with acetylated a-tubulin (Actub) and pericentrin antibodies. 5μM of LPA was used with 0.1% BSA vehicle . Mean ± s.e.m from three independent experiments is shown for serum, starvation, TGF-β1, boiled serum, and LPA; Mean ± s.d. are shown for rest of the growth factors two independent experiments. n>65 cells were counted for EGF, HB-EGF, and insulin. All other treatments n >150 cells counted. ***P < 0.001.
(B) Cell cycle analysis was performed on cells treated for 1h and 24h as described in (A). Representative plot (mean ± s.d.) of G0/G1, S, and G2/M cells determined by analyzing nuclear DAPI staining from images captured using a Celigo Image Cytometer from two (1h) and n=3 (24h) independent experiments. P <0.001 (***) for G0/G1 are shown.
(C, D) Ciliation quantification in RPE-1 (C) and NeoHDF (D) cells stained as described in (A). 5μM of LPA or its precursors were used. Mean ± s.e.m from three independent experiments. n>150 cells counted ***P < 0.001, **P < 0.01.
(E) (Left) Representative images of RPE-1 cells grown in serum or treated with or without LPA for 24h as in (A) in the absence of serum and stained with CP110 and CEP164 antibodies. (Right) Corresponding quantification of RPE-1 cells with single CP110 dot. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001. Scale bar = 2μm.
(F) (Left) Representative images of RPE-1 GFP-Rabin8 + tRFP-Centrin2 cells grown in serum or treated with or without LPA as in (A) for 1h in the absence of serum and imaged by live epifluorescence microscopy. (Right) Corresponding quantification of GFP-Rabin8 centrosomal accumulation. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001. Scale bar = 2μm.
(G) Immunoprecipitation of tRFP-Rab11a from RPE-1 cells stably-expressing GFP-Rabin8 and tRFP-Rab11a following incubation in serum, 1h starvation, or 1h starved and LPA-treated (5μM). tRFP antibody was used for IP. Blots were probed with GFP and Rab11 antibodies. Representative image from three independent experiments is shown.
See also Figure S1.
We investigated the effects of LPA on membrane-dependent modifications at the distal end of the MC by examining the removal of CP110. LPA treatment prevented CP110 removal from the MC (Fig 1E). Next, we examined Rab11-dependent preciliary centrosomal trafficking of Rabin8. In contrast to the ciliation and CP110 localization studies performed at 24h following treatment, GFP-Rabin8 trafficking was monitored in live cells after 1h treatments. Interestingly, unlike serum-starved cells, which showed rapid GFP-Rabin8 vesicular trafficking and centrosomal accumulation (Fig 1F), this protein remained cytoplasmic in LPA-treated serum-starved cells, similar to what is observed with serum-fed cells. Together, our results indicate that LPA inhibits Rabin8 preciliary trafficking and ciliogenesis-initiating processes at the MC. Furthermore, in contrast to the findings with ciliogenesis, the effect of LPA treatment on Rabin8 preciliary trafficking was not associated with changes to the cell cycle (Fig 1B).
Because Rabin8 preciliary trafficking requires association with Rab11 vesicles (Knodler et al., 2010; Westlake et al., 2011), we next examined the effects of LPA on Rab11a-Rabin8 binding using cells expressing GFP-Rabin8 and tRFP-Rab11a. Starvation-induced interaction between tRFP-Rab11a and GFP-Rabin8 was demonstrated via live cell microscopy and immunoprecipitation studies (Fig 1G, S1I). Consistent with disruption of Rabin8 preciliary trafficking via LPA treatment, we show that GFP-Rabin8 binding to tRFP-Rab11a is also reduced (Fig 1G). The effect of LPA on Rabin8 preciliary trafficking was not associated with changes in GFP-Rab11a localization (Fig S1J). Together, our findings support a model whereby LPA prevents Rabin8 association with Rab11a, which is needed for preciliary vesicle transport to the MC for ciliogenesis.
LPA blocks preciliary trafficking and ciliogenesis via the LPAR1 receptor
Since LPA acts as a negative regulator of ciliogenesis, we hypothesized that the G-protein-coupled LPA receptors (LPAR), LPAR1-5 (Choi et al., 2010), may be required for inhibiting ciliation. Because LPAR1 was found to be the predominant LPAR in RPE-1 cells, with mRNA levels > 100 times higher than the other isoforms (Fig 2A), we further investigated this protein’s function in ciliogenesis by RNAi in serum-fed cells. LPAR1 knockdown promoted ciliation in ~40-60% of RPE-1 cells, while only ~8% of siControl-treated cells displayed cilia (Fig 2B, 2C). Off-target effects were ruled out by expressing an siRNA-non-targetable (NT) GFP-tagged LPAR1 protein (Fig 2C), which did not affect ciliation induced by serum starvation (Fig S2A). Notably, siLPAR1 treatment reduced cell numbers without affecting the cell cycle profile based on comparisons to siControl treatments (Fig 2D, S2B), suggesting that LPAR1 may be important for cell survival in RPE-1 cells. Together, these results and the observation that the percentage of siLPAR1-treated cells with cilia was similar to the number of cells in G0/G1 suggest that ciliogenesis is occurring independent of cell cycle effects. To further investigate LPA-LPAR1 ciliogenesis regulation, we tested the effects of the LPAR1-3 antagonist Ki16425 (Choi et al., 2010). Treatment with Ki16425 for 24h promoted ciliation in the presence of serum and in LPA-treated starved RPE-1 and NeoHDF cells (Fig 2E, 2F, S2C-E). Importantly, serum-fed Ki16425-treated cells have cell cycle profiles and cell numbers similar to the control cells with serum alone (Fig 2G, S2F), suggesting that cilia may be developing in cycling cells, likely G1, in the absence of LPA signaling. In contrast, starved cells treated with LPA and Ki16425, which also develop cilia, displayed a higher percentage of G0/G1 cells (Fig 2G), as well as reduced cell numbers (Fig 2H). Together, these results demonstrate that other factors in serum are able to promote cell proliferation and cell cycling in the absence of LPA signaling.
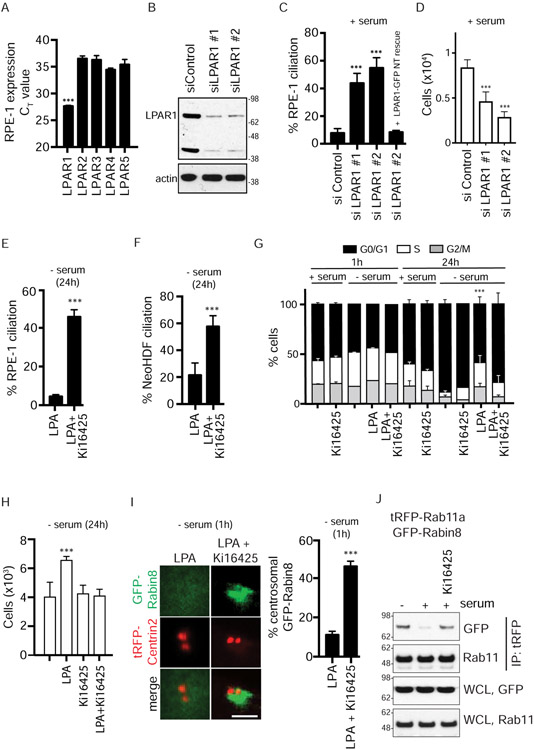
Figure 2: LPAR1 signaling mediates inhibition of ciliogenesis and Rab11a-Rabin8 binding.
(A) Measurement of LPAR1-5 expression using TaqMan analysis from RPE-1 cell RNA. LPAR1 mRNA is highly expressed in RPE-1 cells. The probes for LPAR1-5 were normalized using the LPAR-GFP plasmids and TaqMan probes against GFP. Mean ± s.e.m from three independents are shown. ***P < 0.001.
(B) Immunoblot of lysate from RPE-1 cells after LPAR1 siRNA knockdown for 72h probed with LPAR1 and actin antibodies.
(C) RPE-1 cells fed with 10% serum were treated with LPAR1 siRNA as described in (B) and stained for cilia as described in Fig 1A. GFP-tagged, non-targetable (NT)-LPAR1 was used to test the specificity of LPAR1 siRNA. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001.
(D) Representative plot (mean ± s.d.) of cell counts were determined following treatments described in (C) from DAPI and propidium iodide (PI) stained images of cells from two independent experiments. P < 0.01 (***P < 0.001) are shown.
(E) Quantification of ciliation in RPE-1 cells as described in Fig 1A after treatment with LPA (5μM) and DMSO or LPA and LPAR1 inhibitor, Ki16425 (10μM), for 24h in the absence of serum. Mean ± s.e.m from three independent experiments. ***P < 0.001.
(F) Quantification of ciliation in NeoHDF cells treated as described in (E). Mean ± s.e.m from three independent experiments. ***P < 0.001.
(G) Representative plot (mean ± s.d.) of cell cycle analysis performed as described in Fig 1B for the treatments in (E) for 1h and 24h with serum (2%) and without serum from two (1h) and three (24h) independent experiments. P <0.001 (***) for G0/G1 are shown.
(H) Representative plot (mean ± s.d.) of cell counts determined as described in (D) from three independent experiments. P < 0.01 (***P < 0.001) are shown.
(I) (Left) Representative images showing effects of LPA (5μM) alone or LPA and Ki16425 on GFP-Rabin8 preciliary vesicle trafficking. (Right) Corresponding quantification of the GFP-Rabin8 centrosomal accumulation in RPE-1 cells as described in Fig 1F. ***P < 0.001. Scale bar = 2μm.
(J) Immunoprecipitation of tRFP-Rab11a as described in Fig 1G in the presence and absence of Ki16425. Representative images from three independent experiments is shown.
See also Figure S2.
We next tested whether Rabin8 preciliary transport was affected by the LPA antagonist. Strikingly, Ki16425 treatment promoted Rabin8-Rab11a interaction and Rabin8 vesicular accumulation at the centrosome in LPA-treated starved cells (Fig 2I, 2J). These effects were not associated with changes in the cell cycle status nor cell proliferation (Fig 2G, S2G). Together, our findings demonstrate that LPA regulates Rab11-dependent Rabin8 preciliary membrane trafficking and ciliogenesis via the LPAR1 receptor and, further, suggests that the LPA regulation of Rabin8 preciliary trafficking is not associated with changes in cell cycling and health.
PI3K-Akt signaling regulates Rab11a-Rabin8 interaction and preciliary trafficking needed for cilium assembly
Because LPA-LPAR1 activates PI3K-Akt, Ras-MAPK, PLC-IP3, and Rho-ROCK signaling (Fig 3A) (Blazer-Yost et al., 2011; Choi et al., 2010; Lin et al., 2010), we investigated whether these downstream pathways affected ciliogenesis. Using pharmacological inhibitors against these pathways, we demonstrated that blocking PI3K and Akt function promotes ciliation in serum-fed RPE-1 and NeoHDF cells (Fig 3B, 3C, S3A), whereas inhibitors of the other pathways downstream of LPAR1 showed no effect. Ciliation induced by the PI3K inhibitor LY294002 may be associated with effects on the cell cycle and cell proliferation as there was an increase in the percentage of G0/G1 cells and a reduction in cell number compared to control-treated cells (Fig 3D, 3E). In contrast, cell counts and cell cycle profiles were unaffected by treatment with the Akt inhibitors AKTiIV and MK-2206 (Fig 3D, 3E, S3B, S3C). As PI3K signaling leads to activation of Akt, a serine-threonine protein kinase (Hers et al., 2011), we further investigated Akt’s ciliogenesis regulation. Strikingly, LPA promoted Akt activation similar to serum conditions based on immunoblotting with pAKTS473 antibodies (Fig 3F). Other growth factors that failed to block ciliogenesis in serum starved cells could only partially activate or had no detectable effect on Akt (Fig 3F), including EGF, which strongly activates Akt when added to pre-starved cells (Fig S3D). Consistent with Akt inhibitor studies, depletion of Akt1 and Akt2, but not Akt3, promoted ciliogenesis in serum-fed RPE-1 cells (Fig 3G), whereas Akt1 overexpression prevented ciliation in serum-starved cells (Fig 3H). Similar to LPAR1 RNAi treatments, sipanAkt reduced cell numbers without affecting cell cycle profiles (Fig 3I, S3E), and the percentage of cells that ciliated was consistent with cell numbers identified as either in G0 or G1. Together, these findings suggest that LPA-LPAR1 prevents ciliogenesis via downstream activation of Akt, independent of cell cycle effects.
Figure 3: PI3K/Akt signaling downstream of LPA/LPAR1 regulates ciliogenesis and Rabin8 preciliary trafficking.
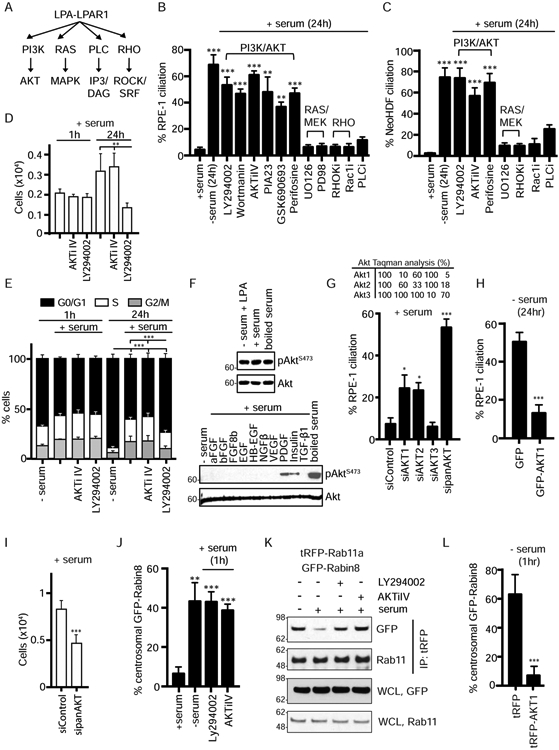
(A) Schematic representation of LPA/LPAR1 downstream signaling pathways.
(B) Quantification of ciliation in RPE-1 cells grown in presence of 2% serum, absence of serum, or upon incubation with various inhibitors for 24h in presence of serum and treated as described in Fig 1A. LY294002 (150μM), Wortmannin (15μM), AktiIV (50nM), PIA23 (10μM), Perifosine (10μM), GSK690693 (10μM), UO126 (100μM), PD98059 (50μM), RHOKi (2μM), Rac1i (2μM), PLCi (0.4μM). n>150 cells counted per treatment. Mean ± s.e.m from n=3 independent experiments is shown. ***P < 0.001, **P < 0.01.
(C) Analysis of ciliation in NeoHDF cells as described in (B). ***P < 0.001.
(D) Representative plot (mean ± s.d.) of cell counts determined as described in Fig 2D following 1h and 24h treatments as in (B) from two (1h) and three (24hr) independent experiments. P < 0.01 (**) are shown.
(E) Representative plot (mean ± s.d.) of cell cycle analysis performed as described in Fig 1B for the treatments in (B) from two (1h) and three (24h) independent experiments. P < 0.001 (***) for G0/G1 are shown.
(F) RPE-1 cells were treated for 24h as described in Fig 1A and western analysis was performed on cell lysate, immunoblotted with pAKTS473 and Akt antibodies.
(G) Quantification of ciliation in RPE-1 cells treated with Akt1, Akt2, Akt3, or panAKT siRNAs for 72h and stained as described in Fig 1A. Mean ± s.e.m from three independent experiments is shown. (Top) Relative expression of Akt isoforms resulting from knockdown with specific siRNAs as determined by TaqMan analysis. ***P < 0.001, *P < 0.05.
(H) Quantification of ciliation in RPE-1 cells overexpressing GFP alone or GFP-AKT1 after 24h starvation and stained as described in Fig 1A. Mean ± s.e.m from three independent experiment is shown. n>100 cells counted per treatment. ***P < 0.001.
(I) Representative plot (mean ± s.d.) of cell counts determined as described in Fig 2D following treatments described in (G) from two independent experiments. P < 0.01 (***P < 0.001) are shown.
(J) Quantification of GFP-Rabin8 trafficking in RPE-1 cells as described in Fig 1F with treatments as described in (B). >100 cells counted per treatment. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001, **P < 0.01.
(K) Immunoprecipitation of tRFP-Rab11a as described in Fig 1G in the presence and absence of PI3K or Akt inhibitors in (B). Representative image from three independent experiments is shown.
(L) Quantification of the GFP-Rabin8 centrosomal accumulation in RPE-1 GFP-Rabin8 cells after transient expression of tRFP alone or tRFP-AKT1 for 48 h and serum starvation for 1h. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001. Scale bar = 2μm.
See also Figure S3.
We next investigated the effects of PI3K-Akt signaling pathway on preciliary trafficking regulation. Unlike 24h treatments, no changes in cell numbers and cell cycle profiles were observed after incubating cells with LY294002 for 1h (Fig 3D, 3E), results similar to AKTiIV-treated cells. Importantly, these PI3K and Akt inhibitors stimulated centrosomal accumulation of GFP-Rabin8 and Rab11a-Rabin8 binding (Fig 3J, 3K). Likewise, Akt1 overexpression prevented preciliary trafficking of Rabin8 (Fig 3L). Together, these results link LPA-LPAR1 signaling to PI3K and its downstream target Akt in the regulation of Rab11-dependent Rabin8 preciliary trafficking.
Rabin8 Akt phosphorylation site does not affect Rab11a or Rab8 interaction, but may alter Rab8 GEF activity
Next, we hypothesized that Rab11a, −11b and/or Rabin8, whose association is regulated by Akt, may be substrates of this enzyme. Using in vitro Akt phosphorylation studies with 32P-incorporation, we demonstrated that GFP-Rabin8, but not GFP-Rab11a or −11b, is an Akt substrate (Fig 4A). Consistent with this result, overexpressed GFP-Rabin8 immunoprecipitated from 293T cells was more strongly detected by pAkt substrate antibodies when incubated with recombinant Akt (Fig 4B). Moreover, we show GFP-Rabin8 stably expressed in RPE-1 cells is detected by an Akt substrate antibody in serum-fed cells, and this signal is strongly reduced after treatments with LY294002, AKTiIV, and serum starvation (Fig 4C). Using 32P-labelled GFP-Rabin8 peptides, we mapped the Akt phosphorylation sites on Rabin8 to the Ser147 and Ser149 residues located proximal to the NH2-terminal end of the minimal GEF domain (Fig 4D). To investigate whether Rabin8’s Akt phosphorylation sites affect binding to Rab11a, we generated phosphomimetic (serine to aspartic acid) and phosphoinactive (serine to alanine) double mutants of Ser147 and Ser149. Co-immunoprecipitation binding studies performed with the GFP-Rabin8 wild-type and the serine mutants and constitutively-active Rab11a-Q70L showed no differences in binding to Rabin8 (Fig S4A). Likewise, Rab11a interaction was not affected by truncating the NH2-terminal region of Rabin8 to amino acid 150 (Fig S4B, S4C), which removes the mapped Akt phosphorylation site. Because Rabin8 contains a predicated Akt site at Ser272 in the Rab11-binding domain, that was not detected by 32P-peptide sequencing (Fig 4D), we tested binding to Rabin8 triple-serine aspartic acid or alanine mutants at residues 147, 149, and 272, and likewise did not observe effects on Rab11-Rabin8 interaction (Fig 4E). Together, these studies demonstrate that Akt phosphorylation of Rabin8 does not directly affect its interaction with Rab11a.
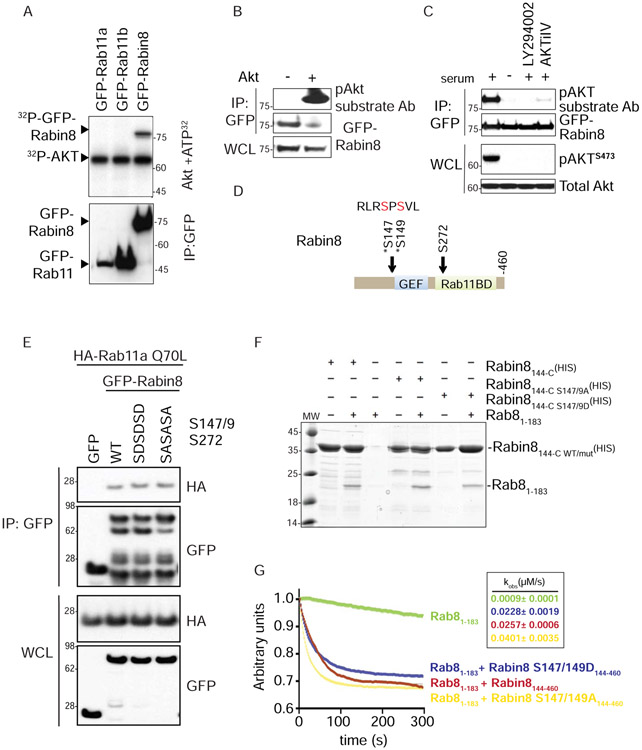
Figure 4: Rab11a and Rab8 binding to Rabin8 is unaffected by Rabin8 Akt phosphorylation mutation, but these residues influence GEF activity.
(A) In vitro kinase assay of immunoprecipitated GFP-fusion proteins from 293T cells incubated with Akt and 32P-ATP.
(B) Western blot analysis of immunoprecipitated GFP-Rabin8 transiently transfected in 293T cells treated with and without Akt. Immunoprecipitation (IP) and cell lysate (WCL) were probed with GFP and pAkt substrate antibodies as indicated.
(C) Western blot analysis of immunoprecipitated GFP-Rabin8 from RPE-1 cells grown in 2% serum, serum-starved for 3h, and treated with LY294002 (150 μM) or AKTiIV (50 nM) in the presence of 2% serum for 3h. IP blots were probed with GFP and pAkt substrate antibodies. WCL blots were probed with Akt and pAktS473 antibodies. Representative blot from two experiments is shown.
(D) Domain structure of Rabin8 showing that Akt phosphorylation sites are proximal to the GEF domain and in the Rab11-binding domain. Mapped Akt-phosphorylation sites from (A) are Ser147 and Ser149.
(E) Immunoprecipitation analysis of HA-Rab11aQ70L with wild-type, phosphomimetic (S147D/S149D/S272D), and phosphoinactive (S147A/S149A/S272A) mutants of GFP-Rabin8 expressed in 293T cells. Immunoblots were probed with GFP and HA antibodies. Representative blot from three experiments is shown.
(F) Coomassie-stained, in vitro binding assay between Rab8 and the Rabin8 wild-type, S147/149A, and S147/149D mutants. Rabin8144-C represents N-terminally-truncated Rabin8 purified using the His-tag. Rabin8144-C S147/149A and S147/149D represent truncated Rabin8 with phosphoinactive and phosphomimetic mutations, respectively. Rabin8144-C WT/mut represents both Rabin8 mutants. Rab81-183 represents purified C-terminally-truncated Rab8.
(G) In vitro Rab8 GEF activity assay using wild-type, phosphomimetic, and phosphoinactive Rabin8. The exchange of fluorescent Mant-GDP by unlabeled GTP was followed over 300s, and the exchange rates calculated using unrestrained single-exponential decay kinetics (Kobs mean and ± SD are indicated in the box). Using a 3X SD cutoff, the S147/149A mutant has statistically significant higher activity than the wild-type protein.
See also Figure S4.
Given that the Rabin8 Akt phosphorylation site is near the GEF domain and within a region that affects the oligomeric state and activity of the GEF (Vetter et al., 2018), we investigated whether these residues could affect Rab8 binding and/or activation. Recombinant protein pull-down and immunoprecipitation studies showed that binding of the GDP-form of Rab8a (T22N) to Rabin8 is not affected by phosphomutation of Ser147/Ser149 (Fig 4F, S4D, S4E). Next, we tested if these residues influenced Rabin8 GEF activity on Rab8. Both Rabin8 wild-type and phosphomimetic Ser147/149Asp mutant proteins promoted similar levels of Rab8 GDP to GTP exchange rates. However, the phosphoinactive Ser147/149Ala mutant showed a statistically significant 1.6 fold increase in Rab8 GTPase activity over the wild-type protein and a 1.8-fold increase compared to the Ser147/149Asp mutant (Fig 4G). Together, these results show that Rabin8 Ser147/149 residues are dispensable for Rab8 binding, but affect Rabin8 GEF activity.
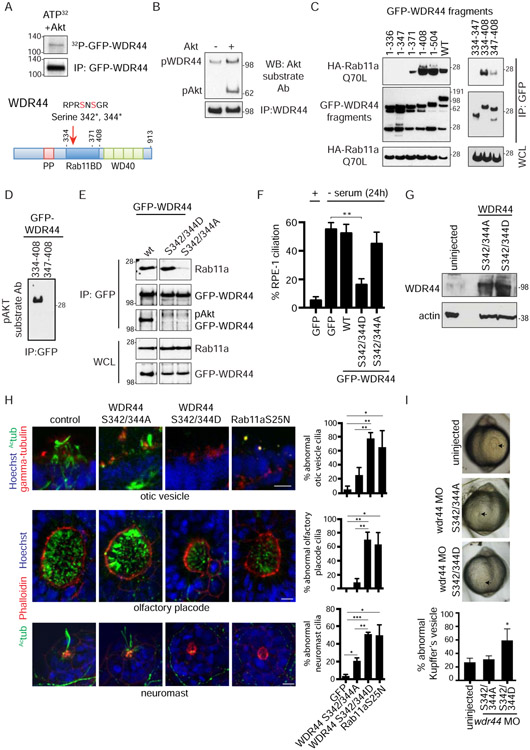
Depletion of the Rab11 effector WDR44 promotes Rabin8 preciliary trafficking and early ciliogenesis processes at the mother centriole
We next hypothesized that another Rab11-binding protein could regulate serum-dependent Rab11-Rabin8 association and preciliary trafficking and, as such, would be expected to display reciprocal binding behavior to Rabin8 in serum-fed and starved cells. To identify candidate Rab11-interactors, which have this serum-dependent binding behavior, we performed mass spectrometry analysis on immunoprecipitated GFP-Rab11a from cells grown with and without serum for 1h (Fig S5A). Consistent with our prediction, peptides detected for three Rab11 effectors (FIP1, FIP2, and WDR44) were reduced in serum-starved cells (Fig 5A), while peptides for the Rab effector FIP5 and Rab-GDP binding protein, RabGDI, were similar in both conditions. The absence of Rabin8 peptides is consistent with our previous findings (Westlake et al., 2011). Based on these results, we tested whether FIP1, FIP2, and WDR44 could regulate Rabin8 preciliary trafficking. SiRNA knockdowns of these proteins were performed and GFP-Rabin8 trafficking was monitored in live cells grown in the presence of serum. We also included well-characterized Rab11 effectors, FIP3 and FIP4, in these studies which were not observed in our LC/MS analysis. Remarkably, all three siRNAs against WDR44 stimulated Rabin8 centrosomal accumulation by more than 3-fold, with two of the oligos (#2 and #3) enhancing Rabin8 trafficking by ~6-fold (Fig 5B). In contrast, only a single FIP1 siRNA (#3) promoted Rabin8 transport to the centrosome, while FIP2, FIP3, and FIP4 siRNAs had no effect. Based on these results, WDR44 was further investigated for its possible function in the Rab11-regulation of preciliary trafficking. Endogenous WDR44 immunoprecipitation studies confirmed that binding to GFP-Rab11a is reduced by 3h serum starvation (Fig 5C). Importantly, PI3K and Akt inhibitors prevented WDR44 binding to GFP-Rab11a in serum-fed cells (Fig 5C). Consistent with these findings, GFP-WDR44 co-localization with endogenous Rab11-positive membrane vesicles was reduced by 1h serum starvation treatment (Fig 5D), and GFP-WDR44 showed stronger peripheral cell targeting, possibly to the plasma membrane. We confirmed that WDR44 binds in a GTP-dependent manner to Rab11a and −11b (Fig S5B, S5C) (Zeng et al., 1999), and does not bind to other Rabs tested (Fig S5B). Together, these studies demonstrate that WDR44 specifically binds to Rab11a/b and that this interaction requires serum-dependent PI3K and Akt signaling.
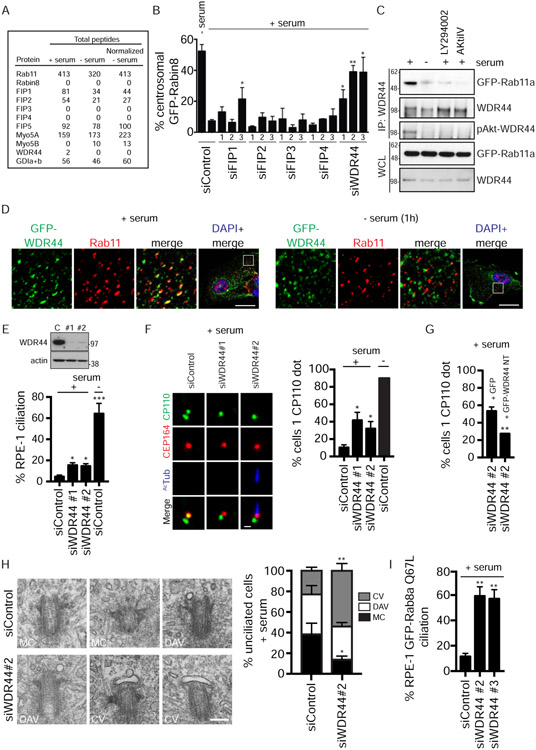
Figure 5: WDR44 displays serum- and Akt-dependent binding to Rab11a, and its knockdown promotes ciliogenesis to the CV stage.
(A) Total peptide count from mass spectrometry analysis of Rab11a-binding proteins immunoprecipitated with GFP antibodies from stable GFP-Rab11a RPE-1 cells grown in the presence or absence of 10% serum for 1h. Normalized peptide count was obtained by multiplying peptide count in - serum lane by a factor of 1.29 obtained after normalizing Rab11a peptides.
(B) Quantification of RPE-1 GFP-Rabin8 centrosomal trafficking in presence of 10% serum following a 72h siRNA treatment and imaged as described in Fig 1F. Trafficking after 1h in absence of serum for siControl is also shown. Mean ± s.e.m from three independent experiments is shown. **P < 0.01, *P < 0.05.
(C) Immunoprecipitation of endogenous WDR44 from RPE-1 GFP-Rab11a cells. Cells were grown in the presence of 2% serum, in serum-starved conditions, or treated with LY294002 (150μM) or AKTiIV (50nM) in 2% serum before immunoprecipitation with WDR44 antibody. Immunoblotting was performed with GFP, WDR44, and pAkt-substrate antibodies. Representative blot from three experiments is shown.
(D) Spinning-disk confocal imaging of transiently-expressed GFP-WDR44 in RPE-1 cells grown with or without serum following PFA fixation and staining with anti-Rab11 and secondary Alexa-568 antibodies. White box in full-cell, “DAPI + merged” images shows magnified area of the cell. Scale bar = 10μm.
(E) (Top) Western blot corresponding to cells treated with control [C] or WDR44 siRNAs [#1 and #2] for 72h in serum and probed with WDR44 and actin antibodies. (Bottom) Quantification of ciliation in cells treated with control or WDR44 siRNAs for 72h in 10% serum and stained as described in Fig 1A. Cells were also treated with control siRNAi for 72h and serum-starved for 24h prior to fixation. Mean ± s.e.m from three independent experiments is shown. n>100 cells counted per treatment. ***P < 0.001, *P < 0.05.
(F) (Left) Representative images showing CP110 loss upon WDR44 knockdown in serum-fed RPE-1 cells as described in (E) and stained and quantified (Right) as described in Fig 1D. Mean ± s.e.m from three independent experiments is shown. *P < 0.05. Scale bar = 2μm.
(G) GFP or non-targetable (NT) GFP-WDR44 plasmids were transfected into RPE-1 cells after 24h of siRNA transfection. 48h later, cells were stained, and CP110 loss was quantified as described in (F). Mean ± s.e.m from three independent experiments is shown. **P < 0.01.
(H) Electron micrographs (Left) of serum-fed RPE-1 cells treated with siControl and siWDR44 for 72h. (Right) Quantification of ciliary vesicle (CV), distal appendage vesicles (DAV), and mother centriole (MC) in EM images. Mean ± s.e.m from three independent experiments is shown for siControl n=62 and siWDR44 n=59 treated cells where the mother centriole distal ends could be observed. For MC, * P <0.05; for CV, ** P < 0.001. Scale bar = 200nm.
(I) Quantification of ciliation in GFP Rab8aQ67L RPE-1 cells treated with WDR44 siRNAs in serum for 72h. Mean ± s.e.m from three independent experiments is shown. **P < 0.01.
See also Figure S5.
Because depletion of WDR44 promotes Rabin8 preciliary trafficking, we investigated the effects of its knockdown on ciliogenesis. Compared to siControl, a 3–fold enhancement in ciliation was observed in serum-fed siWDR44-treated cells (Fig 5E). However, only ~15-17% of cells developed cilia, which is a modest effect compared to the >60% ciliation levels occurring in serum-starved siControl-treated cells. This promotion of ciliogenesis may explain why GFP-Rabin8 preciliary trafficking levels were lower in WDR44 siRNA-treated serum-fed cells than serum-starved conditions (Fig 5B), since Rabin8 trafficking ceases after ciliogenesis (Chiba et al., 2013; Westlake et al., 2011). To test if other early stages of ciliogenesis were affected in WDR44-depleted cells, we examined CP110 loss from the MC. Indeed, siWDR44 had a strong effect on this stage of ciliogenesis with ~40% of cells losing CP110 compared to ~10% of siControl cells (Fig 5F). This phenotype was rescued by expressing an siRNA-NT form of GFP-WDR44, validating the specificity of the knockdown (Fig 5G). To determine whether siWDR44-mediated loss of CP110 in serum-fed cells is associated with vesicular membrane attachment to the distal ends of the MC, we performed transmission electron microscopy (TEM) analysis. Compared to siControl, WDR44 depletion resulted in higher levels of membrane vesicles on the distal appendages (Fig 5H). Finally, we tested whether these effects were associated with changes in cell proliferation. There were no differences observed in cell cycle and cell numbers between siControl and siWDR44-treated, serum-fed cells (Fig S5D, S5E), effects similar to treatments with Ki16425 and Akt inhibitors that promoted ciliogenesis under serum conditions. Together, these results indicate that WDR44 depletion promotes Rabin8 preciliary trafficking and ciliogenesis initiation at the MC in cycling cells.
Interestingly, TEM studies showed that the majority (55±4%) of siWDR44-treated unciliated cells displayed a CV structure (Fig 5H). Previously, we showed that Rab8 functions at the CV stage to initiate the growth of the developing cilium (Lu et al., 2015). Based on these observations and our finding that Rabin8 GEF activity is affected by mutation of the Akt phosphorylation site at Ser147/149 near the GEF domain (Fig 4G), we reasoned that serum-fed WDR44-ablated cells may be unable to fully activate Rab8 to the GTP-bound state, thereby preventing ciliogenesis progression at the CV stage. To test this theory, an RPE-1 cell line expressing the GTP-locked Rab8a (Q67L) mutant was generated, and siWDR44 treatments were performed in serum-fed cells. Remarkably, ~60% of these cells developed cilia (Fig 5I), comparable to ciliation levels observed in siControl-treated serum-starved cells (Fig 1A, 3B, 5E). Thus, these findings support a role for Akt in regulating Rab8 activation downstream of preciliary trafficking at the CV stage.
Akt phosphorylation of WDR44 stabilizes its interaction with Rab11 and is important for blocking ciliogenesis
Because GFP-Rab11a interaction with WDR44 is reduced following Akt inhibition, we investigated whether WDR44 is a substrate of Akt. We show that immunoprecipitated native WDR44 and overexpressed GFP-WDR44 can be phosphorylated by Akt using immunodetection with a pAkt substrate antibody and in vitro kinase assays with 32P-autoradiography (Fig 6A, 6B). The WDR44 Akt phosphorylation site was mapped to residues Ser342 and Ser344, within the Rab11 binding domain, after sequencing GFP-WDR44 tryptic 32P-peptides (Fig 6A). Importantly, we immunodetected endogenous pAkt WDR44 from serum-fed cells, but not from serum-starved cells or serum-fed cells treated with PI3K and Akt inhibitors (Fig 5C). To test if Ser342/344 are important for Rab11-WDR44 interactions, binding studies were performed using WDR44 fragments and mutant proteins. Using WDR44 fragments, we defined the Rab11-binding domain between residues 334-408 and showed that removal of residues 334-347 containing the Akt phosphorylation site strongly reduced its binding to Rab11aQ70L (Fig 6C, 6D). Consistent with these results, mutation of Ser342/344 to Alanine and Aspartic acid prevented detection with the p-Akt substrate antibody (Fig 6E). Finally, we demonstrated that overexpressed wild-type, which is phosphorylated at Ser342/344, and the phosphomimetic S342/344D protein bind more strongly to endogenous Rab11 and GFP-Rab11aQ70L than the phosphoinactive S342/344A mutant protein (Fig 6E, S6A). Together, these results demonstrate that the WDR44 Akt phosphorylation site is important for binding to Rab11.
Figure 6: The WDR44 AKT phosphomimetic mutation promotes binding to Rab11 and inhibits ciliation in human cells and zebrafish embryos.
(A) (Top) In vitro kinase assay of immunoprecipitated GFP-WDR44 from RPE-1 cells showing 32P incorporation in WDR44 by Akt. (Bottom) Domain map of WDR44. Mapped Akt phosphorylation sites at Serine-342 and Serine-344 are in the Rab11-binding domain.
(B) Immunoprecipitated endogenous WDR44 from RPE-1 cells incubated with and without recombinant Akt and immunoblotted with Akt substrate antibody.
(C) GFP antibody immunoprecipitation of GFP-WDR44 wild-type and truncated fragments co-expressed with constitutively-active mutant HA-Rab11aQ70L in 293T cells for 48h. Blots were probed with HA and GFP antibodies. Representative blot from three independent experiments is shown.
(D) Immunoblot of immunoprecipitated GFP-WDR44-truncated proteins containing the pAKT site (334-408) or deleted for the pAKT site (347-408) blotted with an Akt substrate antibody. The input immunoblot is shown in (C). Representative image of three independent experiments is shown.
(E) Immunoprecipitation of GFP-WDR44 wild-type, phosphomimetic (Asp) and phosphoinactive (Ala) mutants expressed in 293T cells (corresponding to the mapped Akt phosphorylation sites) and immunoblotted with GFP and a Rab11a/b antibodies. Blots shown are from the same experiment and exposure and are representative from two independent experiments.
(F) Quantification of ciliation in RPE-1 cells transiently expressing GFP alone, GFP-WDR44 wild-type, and phosphomutants after 24h starvation as described in Fig 1A. Mean ± s.e.m from three independent experiments are shown. Counted n>120 cells. **P< 0.01.
(G) Western blot analysis of lysate from zebrafish embryos injected with mRNA for GFP, WDR44 S342/344D, or WDR44 S342/344A mutant proteins and probed with WDR44 and actin antibodies.
(H) (Right) Quantification of abnormal otic vesicles, olfactory placodes, and neuromasts from 2-day-old wild-type zebrafish embryos injected with mRNA for GFP, WDR44 mutants, or Rab11a S25N (dominant-negative mutant). (Left) Representative images of embryos stained with anti-acetylated tubulin and anti-γ tubulin (otic vesicles) or Phalloidin (olfactory placodes and neuromasts) antibodies, and Hoechst. Embryos injected with Rab11a S25N mRNA were used as positive controls. Total number of organelles (n) counted in all experiments: GFP OP=13, OV=15, neuromast=80; WDR44 S432/344D OP n=16, OV=14, neuromast=38; WDR44 S432/344A OP=21, OV=24, neuromast=48; Rab11aS25N OP=13, OV=13, neuromast=58. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001, **P < 0.01, *P < 0.05. Scale bar = 10μm.
(I) Brightfield images of Kupffer's vesicles (KV, black arrows) from live embryos injected with wdr44 morpholino and rescued with WDR44 mutants (upper panel). Quantification of abnormal KVs (lower panel). Mean ± s.e.m is shown for n>50 KVs counted in four independent experiments. *P < 0.05.
See also Figure S6.
We next examined if the expression of the WDR44 phosphomutants affects ciliogenesis. Strikingly, the S342/344D mutant reduced ciliogenesis in 24h serum-starved RPE-1 cells, whereas both wild-type and phosphoinactive mutants had no effect (Fig 6F). Similar results were observed in 2 dfp zebrafish embryos with the phosphomimetic protein, which reduced cilia formation in the olfactory placode, otic vesicle, and neuromasts (Fig 6G, 6H). Abnormal formation of the Kupffer’s vesicle (KV), a ciliated organ forming at the 6-8 somite stage, was also observed in embryos injected with WDR44 S342/344D, but only following depletion of the endogenous wdr44 using morpholino treatment (Fig 6I, S6B). In contrast, this treatment with the phosphoinactive mutant did not affect KV formation as compared to control injections. Interestingly, treatment with the morpholino and WDR44 S342/344D caused phenotypes similar to those observed in embryos expressing the Rab11a dominant-negative protein (Rab11a-S25N) (Fig S6B) (Westlake et al., 2011), which blocks the function of this Rab. Given these findings, we theorized that WDR44 phosphorylation at Ser342/344 stabilizes interaction with Rab11 and prevents Rab11-Rabin8 complex formation and ciliogenesis.
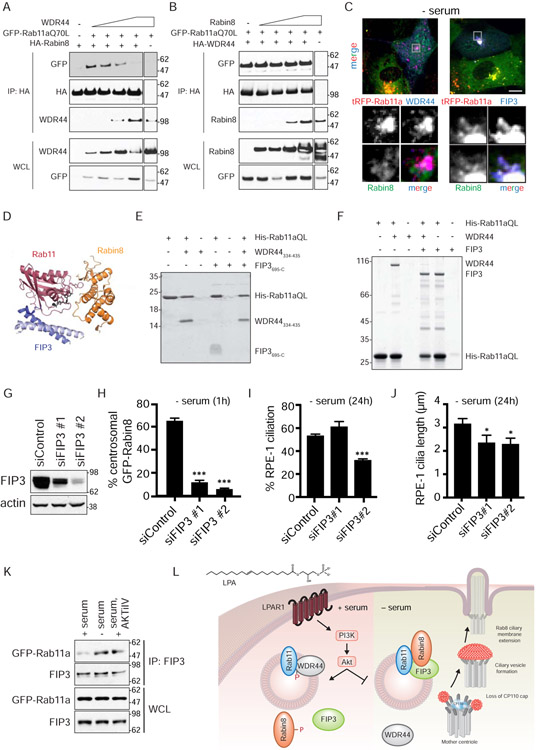
FIP3 functions in a Rab11 effector switch required for Rabin8 preciliary trafficking and ciliogenesis
Based on our observations, we hypothesized that Rab11 binding switches from a Rab11-WDR44 interaction to a Rab11-Rabin8 ciliogenesis complex upon serum starvation and Akt inactivation. To further investigate this Rab11-effector switch theory, we performed binding studies with co-expressed Rab11a, WDR44, and Rabin8 proteins. Consistent with our prediction, increasing amounts of the WDR44 protein displaced GFP-Rab11aQ70L from HA-Rabin8 (Fig 7A). In contrast, Rabin8, which has a KD of 40μM for Rab11 (Vetter et al., 2015), did not affect GFP-Rab11aQ70L binding to HA-WDR44 (Fig 7B), suggesting that WDR44 has higher affinity for Rab11 than Rabin8. Consistent with these results, GFP-Rabin8 did not co-localize with HA-WDR44 and tRFP-Rab11a (Fig 7C). To investigate WDR44-Rab11 binding behavior, we performed isothermal titration calorimetry (ITC) analysis and show that the WDR44334-435 fragment containing the Rab11 binding domain has >70-fold higher affinity (KD=0.54 ± 0.04 μM) for Rab11a than Rabin8 (Fig S7A), a KD similar to other Rab11 effectors (Eathiraj et al., 2006). These findings, together with the observation that the WDR44 phosphoinactive mutants display some binding to Rab11 (Fig 6E, S6A), suggested that changes in WDR44 phosphorylation alone may not explain the switch to a Rab11-Rabin8 complex required for ciliogenesis.
Figure 7: FIP3 functions in Rab11-dependent Rabin8 preciliary trafficking and ciliogenesis and competes with WDR44 for binding to Rab11a.
(A) Immunoblot showing co-immunoprecipitation of GFP-Rab11a Q70L with HA-Rabin8 in the presence of increasing amounts of WDR44 transiently-expressed in 293T cells grown in serum for 48h. Immunoblots were probed with GFP, HA, and WDR44 antibodies. Representative blot from two independent experiments is shown.
(B) Immunoblot showing co-immunoprecipitation of GFP-Rab11a Q70L with HA-WDR44 in the presence of increasing amount of Rabin8 as described in (A). Immunoblots were probed as described in (A). Representative blot from two independent experiments is shown.
(C) Immunostaining of RPE-1 cells stably-expressing GFP-Rabin8, tRFP-Rab11a, and HA-WDR44 or HA-FIP3 fixed with PFA and stained with HA antibodies. Grey box in top panels corresponds to the region of the cell in the bottom panels. Cells were serum-starved for 1h to promote Rabin8 trafficking and imaged by epifluorescence microscopy. Scale bar = 5μm.
(D) Structure of ternary complex containing Rab11, Rabin8, and FIP3 (Vetter et al., 2015).
(E) Coomassie-stained gel showing a Ni-NTA pull-down of WDR44334-435 (Rab11-binding domain of WDR44) and FIP3695-C (Rab11-binding domain of FIP3) by His-tagged Rab11aQ70L6-186 (HIS) (constitutively-active GTPase domain of Rab11a). Ni-NTA His-Rab11a Q70L6-186 (HIS) beads were incubated with and without excess FIP3695-C (308.7μM) and/or WDR44334-435 (354.4μM).
(F) Coomassie-stained gel showing a Ni-NTA pull-down as described in (E) using full-length WDR44 and FIP3. Ni-NTA His-Rab11a Q70L6-186 (HIS) beads were incubated with and without excess FIP3 (46μM) and/or WDR44334-435 (45μM).
(G) Western blot analysis of lysates from 293 cells treated with FIP3 siRNAs. Lysates were collected 72h after siRNA transfection. FIP3 and actin antibodies were used for immunoblotting.
(H) Quantification of GFP-Rabin8 centrosomal accumulation in RPE-1 cells upon treatment with FIP3 siRNA after 72h as described in Fig 1F. Mean ± s.e.m from three independent experiment are shown. >60 cells counted per treatment. ***P < 0.001.
(I) Quantification of ciliation in cells treated with FIP3 siRNAs for 72h and serum-starved for the final 24h. Cells were stained and imaged as described in Fig 1A. Mean ± s.e.m from three independent experiments is shown. ***P < 0.001.
(J) Quantification of cilia length in siControl and siFIP3-treated RPE-1 cells imaged as described in (H). *P < 0.05.
(K) Immunoprecipitation of endogenous FIP3 from GFP-Rab11a RPE-1 cells grown in 2% serum, 2% serum with AKTiIV (50nM), or serum-starved for 3h. FIP3 and GFP antibodies were used for immunoblotting. Representative immunoblots from three independent experiments for + serum and − serum, and two independent experiments for AKTiIV treatment are shown.
(L) Model for signaling regulation of the ciliogenesis Rab11-effector switch.
See also Figure S7.
Rabin8 is known to form a complex with Rab11 and its effector FIP3. FIP3 occupies the canonical effector site in the switch II region of Rab11, and Rabin8 interacts with a nearby site (Fig 7D) (Vetter et al., 2015). Consistent with this finding, FIP3 colocalized with Rab11 and Rabin8 in RPE-1 cells (Fig 7C). Based on these observations, we investigated FIP3 effects on Rab11-WDR44 interaction. We could not test Rab11 binding competition in mammalian cells with FIP3 as we had done with WDR44 and Rabin8 (Fig 7A, 7B) due to much stronger FIP3 expression, as compared to WDR44. Nevertheless, we demonstrated that FIP3 prevents WDR44 interaction with Rab11aQ70L (Fig S7B). Using bacterially-expressed fragments of WDR44 and FIP3 containing the Rab11 interaction domains, we demonstrated that WDR44 blocks FIP3 interaction with the GTP-locked form of Rab11a (Fig 7E). Similar Rab11 binding competition was observed using recombinant full-length proteins, albeit the results were opposite with FIP3 preventing WDR44 interaction with Rab11aQ70L (Fig 7F). Similar concentrations of WDR44 and FIP3 full-length and fragment proteins were used in these studies indicating that these proteins have different binding behaviors with Rab11. Nevertheless, these studies demonstrate that WDR44 and FIP3 have mutually exclusive and spatially overlapping binding sites on Rab11.
Based on these results and evidence that the Rab11-FIP3-Rabin8 complex functions in ciliary-associated trafficking in photoreceptor cells (Wang and Deretic, 2015), we investigated the involvement of FIP3 in preciliary trafficking and ciliogenesis in RPE-1 cells. Following knockdown with two independent FIP3 siRNAs, we show that GFP-Rabin8 trafficking induced by serum starvation is reduced in cells (Fig 7G, 7H). SiFIP3#2 resulted in a stronger trafficking defect and a greater depletion of the protein as compared to siFIP3#1. These RNAi oligos also affected ciliation (Fig 7I, 7J). siFIP3#2 reduced cilia numbers (Fig 7I), while the less efficient siFIP3#1 caused cilia shortening compared to control siRNA-treated cells (Fig 7J). Likewise, the cilia that did form in siFIP3#2-treated cells were also shorter (Fig 7J). Together, these results indicate that FIP3 is important for regulating preciliary trafficking and ciliogenesis. Based on these observations, we investigated FIP3’s biochemical association with a ciliogenic Rab11-effector switch. Endogenous FIP3 displayed stronger binding to GFP-Rab11a following serum starvation and in serum-fed cells treated with the Akt inhibitor, as compared to serum-fed cells alone (Fig 7K). These results together with our other findings suggest a model whereby ciliogenesis initiation is controlled by an Akt-dependent effector switch from Rab11-WDR44 to a complex with Rab11-FIP3-Rabin8 (Fig 7L).
Discussion
Serum starvation is commonly used to promote ciliogenesis in cultured cell lines. However, the molecular mechanism by which serum factor(s) inhibit ciliogenesis was unknown. Rab11-dependent Rabin8 preciliary trafficking to the MC is stimulated minutes after serum starvation, which, to our knowledge, is the earliest reported marker of ciliogenesis initiation (Westlake et al., 2011). Here, we describe a serum factor, LPA, and its receptor, LPAR1, as key regulators of an early stage of ciliogenesis. LPA/LPAR1 activates Akt which inhibits ciliogenesis by stabilizing Rab11 effector-based binding to WDR44, thus preventing the formation of the ciliogenic Rab11-FIP3-Rabin8 complex, and also may regulate Rabin8 GEF activity towards Rab8, important for cilia growth.
Our findings showed that treatment of serum-starved cells with protein-based growth factors, but not boiled serum, failed to prevent ciliation and led us to examine the involvement of the lipid mitogen LPA in ciliogenesis regulation. Treatment with LPA blocked ciliogenesis induced by serum starvation in several human cell lines with the maximal effects observed at 10μM. This concentration is consistent with the LPA levels found in fetal bovine serum. Not surprisingly, LPA had no effect on ciliation in mouse IMCD3 epithelial cells, which ciliate in the presence of serum and have been proposed to use the extracellular ciliogenesis pathway. In contrast, fibroblasts and mesenchymal cells have been observed to utilize the intracellular pathway (Molla-Herman et al., 2010). Hence, differences in ciliation induction cues could be cell-type-specific. The effect of LPA on ciliogenesis is consistent with a recent report (Kim et al., 2015), where 24h-LPA treatment activates YAP/TAZ transcriptional activator function via modulation of the cytoskeleton, thus suppressing axonemal growth. Here, we show acute LPA-mediated blockade of early ciliogenesis processes, including preciliary trafficking of Rab11a-Rabin8 vesicles and CP110 uncapping of the MC. Importantly, we demonstrate that Rab11a-Rabin8 interaction and Rabin8 preciliary trafficking is not associated with changes in the cell cycle. Moreover, given the acute effects of the serum withdrawal, it is unlikely that preciliary trafficking induction is mediated by changes in transcription.
LPAR1 is the predominant LPA receptor in RPE-1 cells, and treatment with a LPAR1/LPAR3 antagonist, Ki16425, promoted preciliary trafficking and ciliogenesis. However, the implications for ciliogenesis regulation by LPA/LPAR1 in vivo are not clear. In zebrafish, overactivation of the LPA pathway either by expression of the LPA-producing enzyme, enpp2/atx, or the Lpar1 receptor results in KV cilia defects (Frisca et al., 2016). This effect can be rescued by treating embryos with Ki16425. However, another group showed that depletion of Lpar3 and enpp2, but not Lpar1, caused defects in KV formation (Lai et al., 2012). LPAR1-null mice are viable, but these animals have lower weights and increased mortality attributed to a compromised suckling response (Contos et al., 2000), which corresponds to phenotypes associated with olfaction defects in ciliopathy mutant mice (Norris and Grimes, 2012). Interestingly, injection of LPA into E13.5 mice brains causes hydrocephalus in 100% of pups, an effect reversed by Ki16425 (Yung et al., 2011). In certain ciliopathies, there is susceptibility to hydrocephalus, a condition attributed to defects in motile ependymal cilia function (Reiter and Leroux, 2017). Based on these observations, a regulatory role for LPA and LPAR receptors in mammalian ciliogenesis in vivo merits further investigation, although redundancy of the receptors could complicate the study. Moreover, the phenotypes of these animals may not correlate with those of proteins needed for ciliogenesis, as LPAR1 depletion may be expected to promote aberrant induction of ciliation.
By screening downstream regulators of LPA-LPAR1 signaling, we demonstrated that PI3K and Akt inhibitors promote Rab11a-Rabin8 interaction and preciliary trafficking. An Akt substrate and Rab11 effector, WDR44, was identified and shown to prevent ciliogenesis-initiating events via its Akt phosphorylation site in the Rab11-binding domain. WDR44/Rabphillin11/Rab11BP functions with Rab11 to regulate the recycling of endocytosed transferrin receptor back to the plasma membrane (Mammoto et al., 1999; Zeng et al., 1999). Notably, Rabin8 preciliary vesicles co-localize with internalized transferrin, supporting an endosomal origin of these membranes (Westlake et al., 2011). Our findings point to an Akt-dependent repurposing of Rab11-positive endosomes that directs a switch in vesicular function from recycling molecules to the plasma membrane in association with WDR44 to preciliary trafficking to the MC distal end. In support of this model, knockdown of WDR44 was sufficient to promote CP110 loss from the MC and CV assembly in serum-fed cells. Absence of strong ciliary growth in WDR44-ablated cells suggested that Akt regulates additional downstream ciliogenesis processes. Because Rabin8, a substrate for Akt, is phosphorylated near the GEF domain at Ser147/149, and Rab8 functions at the CV stage in ciliary axonemal growth (Lu et al., 2015), we hypothesize that Akt regulates Rab8 activation. This theory is supported by the findings that expression of constitutively-active Rab8a strongly promoted ciliogenesis in WDR44-ablated cells, and mutating the Akt phosphorylation sites to alanine reduced Rab8 activation. Paradoxically, Akt kinase activity is required for establishing proper ciliary length (Suizu et al., 2016) and depends on ciliary activation of the platelet-derived growth factor receptor alpha-pathway (Clement et al., 2013). These studies suggest that Akt has both positive and negative ciliary regulatory functions and that the spatial and temporal regulation of Akt activity is important. Consistent with this idea, activated Akt has been shown to be associated with Rab11 recycling endosomes (Garcia-Regalado et al., 2008), where it could directly participate in WDR44 phosphorylation. Moreover, Akt ciliary functions are likely mediated by specific phosphatase modulators that remain to be identified. Together, our work indicates that LPA and other factors that modulate Akt signaling regulate the function of multiple ciliogenic factors. Given the links between these pathways and human disease, cancer in particular (Martini et al., 2014; Riaz et al., 2016), further investigation of these signaling mechanisms and their associations with ciliogenesis dysfunction in ciliopathies and cancer is warranted.
Rabin8 centrosomal trafficking is strongly diminished following ciliary growth (Chiba et al., 2013; Westlake et al., 2011). This may play an important role in controlling cilia length, an idea supported by the observation that overexpression of Rab8 promotes longer cilia. Consequently, Akt and WDR44 may also participate in reducing Rabin8 centrosomal trafficking following ciliogenesis. Another kinase, NRD2, has been reported to function in retention of Rabin8 at the MC. NDR2 phosphorylates Rabin8 at Ser272 and promotes binding to the exocyst subunit Sec15, present at the MC (Chiba et al., 2013). Together, this work suggests that different kinases may regulate discrete Rabin8 trafficking processes associated with the cilium. Tight regulation of preciliary trafficking may also be important in the delivery of other proteins that function at distinct steps in the ciliogenesis pathway. For example, EHD1/3 are thought to be trafficked on membranes to the MC where they function in ciliary CV assembly, a process requiring CP110 removal from the MC (Lu et al., 2015). Thus, it may be interesting to examine how these ciliogenesis regulators are affected by Akt signaling.
The observation that Rabin8 was unable to compete with WDR44 for binding to Rab11a suggested that other factors are involved in repurposing Rab11a vesicles for ciliary assembly. The Rab11 effector FIP3 forms a ternary complex with Rab11 and Rabin8 (Vetter et al., 2015), and a ciliary connection with this complex has been previously established in the transport of rhodopsin to the outer segment of photoreceptor cells (Wang and Deretic, 2015), which are modified primary cilia. FIP3 was previously shown to be dispensable for ciliogenesis in IMCD3 cells (Wang and Deretic, 2015), suggesting that its ciliogenic function may be restricted to the intracellular ciliogenesis pathway. Here, we show that FIP3 ablation blocked Rabin8 preciliary trafficking and biochemical studies showed that WDR44 and FIP3 display opposite binding to Rab11 in serum-fed and Akt-inhibited cells. As shown using WDR44 Ser342/344 mutants, reduction in affinity for Rab11 is associated with the phosphorylation status of WDR44. The observation that the phosphomimetic mutant of WDR44 blocks ciliogenesis in human cells and zebrafish embryos supports a role for Akt in regulating initiation of ciliogenesis by stabilizing a Rab11-WDR44 interaction, thus preventing Rab11 association with FIP3 and Rabin8. FIP3 increases Rabin8 affinity for Rab11 by 4-fold due to direct contacts observed between Rabin8 and FIP3 in the structure of the ternary complex (Vetter et al., 2015), whereas Rabin8 binding to Rab11 is not enhanced by WDR44 (KD= 37.2 ± 9.7 μM) (Fig S7C). These observations suggest that WDR44 likely has an indirect effect on Rabin8 interaction with Rab11 due to competition with FIP3 for the canonical effector binding site, which subsequently lowers Rabin8 affinity for Rab11. Thus, our work extends the ciliary function of FIP3 and suggests that an Akt-dependent effector switch functions at a nexus of the Rab11-dependent endosome recycling pathway and preciliary trafficking to the MC.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Futher information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher J Westlake (chris.westlake@nih.gov)
EXPERIMENTAL MODEL DETAILS
Zebrafish
Wildtype male and female AB zebrafish were used in this study. Embryos were maintained at 28°C in incubator and analyzed at the Long-Pec stage, 2 days post fertilization (dpf). Animal procedures, care and husbandry were carried out in accordance with animal use and guidelines at NIH and approved by Animal Care and Use Committee of NIH/NCI- Frederick (Study Proposal 17-416).
CELL LINES
hTERT-RPE cells (RPE-1)
Parental hTERT-RPE cells were purchased from ATCC and originate from a male. Parental and stable expressing RPE-1 cell lines were cultured as per the suppliers instructions at 37°C with 5% CO2 in DMEM-F12 media supplemented with 10% FBS.
Neonatal Human Dermal Fibroblast (NeoHDF)
Cells were purchased from ATCC and were cultured as per the suppliers instructions at 37°C with 5% CO2 in Fibroblast Basal Medium and 10% FBS.
MCF-12A
MCF-12A cells of female origin were purchased from ATCC. Cells were cultured according to the manufactures instructions at 37°C in 5% CO2 in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 medium, 20 ng/ml hEGF and 5% horse serum.
hTERT-HPNE
hTERT-HPNE cells are derived from a male and were purchased from ATCC. Cells were cultured as per the manufactures instructions at 37°C with 5%CO2 in 75% DMEM without glucose, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, and 25% Medium M3 Base with 5% FBS, 10 ng/ml hEGF, 5.5 mM D-glucose.
mIMCD3
Mouse IMCD3 cells were purchased from ATCC and cultured at 37 °C in 5% CO2 in DMEM-F12 media supplemented with 10% FBS.
PRIMARY CELL CULTURE
MEFs
Murine embryonic fibroblast (MEF) were a kind gift from Lino Tessarollo of unknown sex. Cells were cultured in 37°C in 5%CO2 in DMEM and 10% FBS.
METHODS DETAILS
Plasmids and Reagents
cDNAs for LPAR1 (Insinna et al., 2019), Akt1 (Catalog no. 9005, Addgene), WDR44 (Catalog no. OHS5894-202499323, Thermo Fisher), and FIP3 (gift from Rytis Prekeris) were cloned into the Gateway compatible vectors. WDR44-truncated constructs (1-336, 1-347, 1-371, 1-408, 1-504, 334-347, 334-408, 347-408) were generated by PCR using Phusion polymerase (Catalog no. M0530L, NEB). PCR-purified product was cloned into pENTR plasmids, sequenced, and cloned into pDest vectors. Rab constitutively-active, LPAR1 rescue, Rabin8 phosphomimetic (Rabin8 S147D/S149D/S272D, Rabin8 S147D/S149D), Rabin8 phosphoinactive (Rabin8 S147A/S149A/S272A, Rabin8 S147A/S149A), WDR44 phosphomimetic (WDR44 S342D/S344D), WDR44 phosphoinactive (WDR44 S342A/S344A), and WDR44 rescue plasmids were generated using QuickChange mutagenesis kit (Catalog no. 200524, Agilent Technologies). LPAR1-GFP was generated by Gateway cloning into the GLAP7 vector (Torres et al., 2009). Rab mutants were cloned into the pDEST53 plasmid unless otherwise indicated. WDR44-GFP fragments were generated by Gateway cloning into GLAP7 vector. GFP- and tRFP-Akt and GFP-Rab11a constructs were generated by Gateway cloning into GLAP1 vector (Torres et al., 2009). Untagged WDR44, Rabin8, and FIP3 plasmids were cloned into pCS2+ vector. GFP-Rabin8, GFP-Rab11a, GFP-Rab11b, GFP-Rab11aQ70L, GFP-Rab8aT22N, HA-Rab11aQ70L, and Rab11aS25N have been described previously (Lu et al., 2015; Westlake et al., 2011). HA-WDR44 and HA-FIP3 were generated by Gateway cloning into the pDest-HA vector. pPGK HA-FIP3 and pPGK HA-WDR44 were generated using Multisite Gateway cloning from vectors that include pDest686, hPGK in pENTR (attL4/attL5), no. C453-E17 and HA in pENTR (attR5/attR1), no. C514-E10 (Protein Expression Laboratory, Frederick National Lab for Cancer Research) (Wall et al., 2014). All siRNA target sequences used in this study are listed in Key Resource Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-acetylated-tubulin | Sigma | T7451, RRID:AB_609894 |
| Rabbit polyclonal anti-pericentrin | Novus | NB100-61071, RRID:AB_925559 |
| Goat polyclonal anti-CEP164 | SantaCruz | N-40 SC240226, RRID:AB_10841981 |
| Rabbit polyclonal anti-CP110 | Proteintech | 12780-1-AP, RRID:AB_10638480 |
| Rabbit polyclonal anti-LPAR1/EDG2 | ThermoFisher | PA5-28768, RRID:AB_2546244 |
| Rabbit polyclonal anti-tRFP | Evrogen | AB233, RRID:AB_2571743 |
| Rabbit polyclonal anti-GFP (for immunoprecipiation) | In house | N/A |
| Mouse monoclonal anti-GFP-HRP | Miltenyi Biotec | 130-091-833, RRID:AB_247003 |
| Mouse monoclonal anti-Rab11 | BD Biosciences | 610657, RRID:AB_397984 |
| Rabbit monoclonal anti-Phospho Akt substrate (RXRXXS*/T*) 23C8D2 | Cell Signaling | 10001S, RRID:AB_10950819 |
| Rabbit monoclonal anti-Phospho-AKT S473 | Cell Signaling | 4060S, RRID:AB_2315049 |
| Rabbit monoclonal anti-Akt pan C67E7 | Cell Signaling | 4691S, RRID:AB_915783 |
| Rat monoclonal anti-HA-HRP | Roche/Sigma | 12013819001, RRID:AB_390917 |
| Rabbit polyclonal anti-WDR44 | Bethyl | A301-440A, RRID:AB_961125 |
| Mouse monoclonal anti-β-Actin-HRP | Sigma-Aldrich | A3854, RRID:AB_262011 |
| Rabbit polyclonal anti-RAB3IP/Rabin8 | Proteintech | 12321-1-AP, RRID:AB_2177510 |
| Mouse monoclonal anti-HA (for immunofluorescence) | Roche/Sigma | 11867423001, RRID:AB_10094468 |
| Rabbit polyclonal anti-RAB11FIP3 (FIP3) | Proteintech | 25843-1-AP RRID:AB N/A |
| Mouse monoclonal anti-gamma tubulin | Sigma | T6557, RRID:AB_477584 |
| Mouse monoclonal anti-Acetylated tubulin T6793 | Sigma | T6793, RRID:AB_477585 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| aFGF | R&D Systems | 232-FA |
| bFGF | R&D Systems | 233-FB |
| EGF | Sigma | E9644 |
| FGF-8b | R&D Systems | 423-F8 |
| HB-EGF | R&D Systems | 259-HE |
| NGFβ | R&D Systems | 256-GF |
| VEGF | Sigma | V7259 |
| PDGF | R&D Systems | 120-HD |
| Insulin | Sigma | 19278 |
| TGF-β1 | Cell Signaling | 8915 |
| Lysophosphatidic Acid | Sigma | 1372050 |
| Phosphatidic Acid | Sigma | P9511 |
| LY294002 | Cell Signaling | 9901S |
| Wortmanin | Sigma | W1628 |
| AKTiIV | Calbiochem/MilliporeSigma | 124015 |
| PIA23 | Zhang et al., 2011 | N/A |
| Perifosine | Selleckchem | S1037 |
| GSK690693 | Sigma | SML0428 |
| UO126 | Cell Signaling | 9903 |
| PD98059 | Cell Signaling | 9900 |
| RHOKi | Calbiochem/MilliporeSigma | 555550 |
| Rac1i | Calbiochem/MilliporeSigma | 553511 |
| PLCi/ U73122 | Tocris | 1268 |
| MK-2206 | Cayman Chemical Company | 1032350-13-2 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human: hTERT- RPE-1 cells (RPE-1) | ATCC | CRL-4000 |
| Human: Neonatal Human Dermal Fibroblast (NeoHDF) | ATCC | PCS201-010 |
| Human: hTERT-HPNE | ATCC | CRL-4023 |
| Human:MCF12A | ATCC | CRL-10782 |
| Mouse: Murine embryonic fibroblast cells (MEF) | gift from Lino Tessarollo | N/A |
| Mouse: mIMCD3 | ATCC | CRL-2123 |
| Experimental Models: Organisms/Strains | ||
| zebrafish AB line | ZIRC | ZL-1 |
| Oligonucleotides | ||
| siRNA targeting sequence: siLPAR1 #1 CCACAGUGCUUCUACAACG |
Dharmacon | J-003656-07 |
| siRNA targeting sequence: siLPAR1 #2 GCAUGCAGCUCCACACACG |
Dharmacon | J-003656-08 |
| siRNA targeting sequence: siAKT1 GGCUCCCCUCAACAACUUC |
Dharmacon Santi and Lee, 2010 |
Custom/ OTP |
| siRNA targeting sequence: siAKT2 GGAUGAAGUCGCUCACACA |
Dharmacon Santi and Lee, 2010 |
Custom/ OTP |
| siRNA targeting sequence: siAKT3 GGACCGCACACGUUUCUAU |
Dharmacon Santi and Lee, 2010 |
Custom/ OTP |
| siRNA targeting sequence: sipanAKT sequence not available from company |
Cell Signaling | 6211 |
| siRNA targeting sequence: siFIP1 #1 AAACAGAAGGAAACGAUAA |
Dharmacon | J-015968-08 |
| siRNA targeting sequence: siFIP1 #2 GGAAAGAUGUAAAUCAGCA |
Dharmacon | J-015968-09 |
| siRNA targeting sequence: siFIP1 #3 AGUGAGAACUUGAACAAUG |
Dharmacon | J-015968-10 |
| siRNA targeting sequence: siFIP2 #1 AGGAACAAUAUGACCGCAA |
Dharmacon | J-020474-09 |
| siRNA targeting sequence: siFIP2 #2 GCAUAUAUUUAGCGGGUAU |
Dharmacon | J-020474-10 |
| siRNA targeting sequence: siFIP2 #3 GGUGAAUUGUGUUUCGGAA |
Dharmacon | J-020474-11 |
| siRNA targeting sequence: siFIP3 #1 CGGACGAGUUCGAUGACUU |
Dharmacon | J-021079-17 |
| siRNA targeting sequence: siFIP3 #2 AGGCCAACAUUGAGCGUCU |
Dharmacon | J-021079-20 |
| siRNA targeting sequence: siFIP3 #3 GAUUAAAGCUGUUACUGCA |
Dharmacon | J-021079-18 |
| siRNA targeting sequence: siFIP4 #1 UGUUAGUGAUGUCGGGCAA |
Dharmacon | J-018023-09 |
| siRNA targeting sequence: siFIP4 #2 CCGAAGACCUGUUCCGGGA |
Dharmacon | J-018023-10 |
| siRNA targeting sequence: siFIP4 #3 UCAUCGAGGACUUGCGGAA |
Dharmacon | J-018023-11 |
| siRNA targeting sequence: siWDR44 #1 GUAUAAGGGUUACGUCAAU |
Dharmacon | J-018913-09 |
| siRNA targeting sequence: siWDR44 #2 CUUCAGAAAGUACGGUUAA |
Dharmacon | J-018913-10 |
| siRNA targeting sequence: siWDR44 #3 ACUGUUAGCCAUAGAUCAA |
Dharmacon | J-018913-12 |
| morpholino oligonucleoide sequence: wdr44 GCTGCATACAGAGGCCGCCGCCACT |
Genetools | Custom-synthesized |
| Recombinant DNA | ||
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Other | ||
Growth factors used in the study are Fibroblast Growth Factor-Acidic (aFGF) (Catalog no. 232-FA, R&D Systems); Fibroblast Growth Factor-Basic (bFGF) (Catalog no. 233-FB, R&D Systems); Epidermal Growth Factor (EGF) (Catalog no. E9644, Sigma); FGF-8b (Catalog no. 423-F8, R&D Systems); Heparin-Binding EGF-like growth factor (HB-EGF) (Catalog no. 259-HE, R&D Systems); Nerve Growth Factor beta (NGFβ) (Catalog no. 256-GF, R&D Systems); Vascular Endothelial Growth Factor human (VEGF) (Catalog no.V7259, Sigma); Platelet-derived Growth Factor (PDGF) (Catalog no. 120-HD, R&D Systems); Insulin (Catalog no. 19278, Sigma); Human Transforming Growth Factor β1 (TGF-β1) (Catalog no. 8915, Cell Signaling); Lysophosphatidic Acid (LPA) (Catalog no. 62215, Cayman Chemical Company); Lysophosphatidylcholine (LPC) (Catalog no. 1372050, Sigma); and Phosphatidic Acid (PA) (Catalog no. P9511, Sigma). LPA was added, in concentrations as noted, to media containing an equal volume of 0.1% BSA.
Chemical inhibitors LY294002 (Catalog no. 9901S, Cell Signaling), Wortmanin (Catalog no. W1628, Sigma), AKT Inhibitor IV (AKTiIV) (Catalog no. 124015, Calbiochem/MilliporeSigma), MK-2206 (Catalog no 1032350-13-2, Cayman Chemical Company), PIA23 was a kind gift from Peter Johnson (Zhang et al., 2011), Perifosine (Catalog no. S1037, Selleckchem), GSK690693 (Sigma, SML0428), UO126 (Catalog no. 9903, Cell Signaling), PD98059 (Catalog no. 9900, Cell Signaling) (Referred as PD98), RHOKi (Catalog no. 555550, Calbiochem/MilliporeSigma), Rac1i (Catalog no. 553511, Calbiochem/MilliporeSigma), PLCi/U73122 (Catalog no. 1268, Tocris). Protease inhibitors (Catalog no. 4693132001, Roche/Millipore Sigma) and phosphatase inhibitors (Catalog no. 4906845001, Roche//Millipore Sigma) were used in immunoprecipitation and whole cell lysate analysis as per manufacturer’s recommendations.
Antibodies
Commercial antibodies were used for acetylated a-tubulin (Actub) (Catalog no. T7451, Sigma, RRID:AB_609894), pericentrin (Catalog no. NB100-61071, Novus, RRID:AB_925559), CEP164 (Catalog no. N-40 SC240226, SantaCruz, RRID:AB_10841981), anti-gamma tubulin (Catalog no. T6557, Sigma), CP110 (Catalog no., 12780-1-AP, Proteintech, RRID:AB_10638480), LPAR1/EDG2 (Catalog no. PA5-28768, ThermoFisher, RRID:AB_2546244), tRFP (Catalog no. AB233, Evrogen, RRID:AB_2571743), GFP for IP (custom), GFP-HRP (Catalog no. 130-091-833, Miltenyi Biotec, RRID:AB_247003), Rab11 (Catalog no. 610657, BD Biosciences, RRID:AB_397984), Phospho Akt substrate (RXRXXS*/T*) 23C8D2 (Catalog no. 10001S, Cell Signaling, RRID:AB_10950819), Phospho-AKT S473 (Catalog no. 4060S, Cell Signaling, RRID:AB_2315049), Akt pan C67E7 (Catalog no. 4691S, Cell Signaling, RRID:AB_915783), HA-HRP (Catalog no. 12013819001, Roche/Sigma, RRID:AB_390917), WDR44 for IP and Western (Catalog no. A301-440A, Bethyl, RRID:AB_961125), β-Actin-HRP (Catalog no. A3854, Sigma-Aldrich, RRID:AB_262011), RAB3IP/Rabin8 (Catalog no. 12321-1-AP, Proteintech, RRID:AB_2177510), HA for IF (Catalog no. 11867423001, Roche/Sigma, RRID:AB_10094468), RAB11FIP3 (Catalog no. 25843-1-AP, Proteintech).
Cell lines and cell line generation
GFP-Rabin8 and GFP-Rabin8 + Centrin2-tRFP RPE-1 cells were previously described (Westlake et al., 2011). Stable GFP-Rabin8/ tRFP-Rab11a RPE-1 cells were generated by infecting GFP-Rabin8 cells with tRFP-Rab11a lentivirus under the control of doxycycline-inducible promoter (pHUSH vector). GFP-Rab11a RPE-1 cells were generated using the GLAP1 vector transfected into hTERT RPE-1 FlpIn cells as previously described (Lu et al., 2015). GFP-Rab8aQ67L RPE-1 cell line was generated using pHUSH-LAP-Rab8Q67L lentivirus. Lentivirus were produced as described previously (Lu et al., 2015). HA-FIP3 and HA-WDR44 lentiviruses were used for infection of Rab11-tRFP/Rabin8-GFP RPE-1 cells generating triple-expressing, stable cell lines. Infected cells were maintained in puromycin (2ug/ml) for a week and induced with 1ug/ml of doxycycline (Catalog no. D3072, Sigma) before being used for analysis.
Immunofluorescence and live cell microscopy
Live and fixed cell imaging was performed using a Zeiss Axio Scan Z1 inverted epifluorescence microscope equipped with a CoolSNAP HQ2 camera. Image acquisition and analysis used Slidebook software (Intelligent Imaging Innovations). For immunofluorescence studies, cells were fixed with 4% PFA and stained with antibodies as described (Westlake et al., 2011). For ciliation determinations, cells were stained with acetylated a-tubulin (Actub) and pericentrin antibodies and cilia numbers determined in > 150 cells (unless otherwise indicated) from images taken using a 40x 1.4 numerical aperture (NA) oil objective. CP110 levels at the MC was quantified in >100 cells using a 63x 1.3 NA objective as described (Lu et al., 2015). GFP-Rabin8 centrosomal accumulation in live RPE-1 cells was monitored as previously described (Westlake et al., 2011). Briefly, cells expressing GFP-Rabin8 alone and with tRFP-Centrin2, tRFP-Rab11a, or tRFP-Akt were imaged live (37°C, 5% CO2) every 1s for 5-10s using an X-Cite 120 lamp at 50% power and a 63x 1.3 NA oil objective. Quantification of GFP-Rabin8 centrosomal accumulation was performed in >100 cells unless otherwise indicated.
Immunoprecipitation and immunoblotting
RPE-1 or 293T cells were transfected with indicated constructs. After 48h of transfection, cells were washed with PBS, lysed in low-salt triton buffer (30mM Tris HCl pH 8.0, 75mM NaCl, 10% Glycerol, 1% Triton X-100) supplemented with 5mM MgCl2, and protease and phosphatase cocktail inhibitors (Complete Tablets, Catalog no. 04693159001 and PhosSTOP, 04906837001, Roche/Millipore Sigma) followed by immunoprecipitation with Protein A beads conjugated with GFP antibody, magnetic HA beads (Catalog no. 88836, Thermo Fisher), or tRFP antibody-conjugated Protein A/G magnetic beads (Catalog no. 88803, ThermoFisher). Samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane, and probed with appropriate antibodies.
For identifying Rab11-Rabin8 binding, stable GFP-Rabin8 + tRFP-Rab11a RPE-1 cells were grown to ~70% confluency, incubated in media without serum for 3h, and lysed in low-salt triton buffer supplemented with 5mM MgCl2. Immunoprecipitation was performed by incubation with tRFP antibody (5ug) for 3h followed by addition of Protein A/G magnetic beads for 1h. Beads were washed with low-salt triton buffer and PBS. Samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane, and probed with indicated antibodies.
For Rab11-Rabin8 mutant binding experiments, we generated phosphomimetic and phosphoinactive mutants of Rabin8. 293T cells were transfected with HA-Rab11aQ70L and GFP-Rabin8 mutants or with GFP control. After 48h of transfection, cells were lysed in low-salt triton buffer supplemented with 5mM MgCl2, followed by immunoprecipitation for 3h with Protein A beads conjugated with GFP antibody. Beads were washed with low-salt triton buffer and twice with cold PBS. Samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane, and probed with indicated antibodies.
For identifying Rab11-WDR44 and Rab11-FIP3 interactions, stable GLAP:Rab11a RPE-1 cells were grown to ~70% confluency, incubated in media without serum or in 2% serum-containing media with or without AKTiIV (25nM) for 3h, and lysed in low-salt triton buffer supplemented with 5mM MgCl2. WDR44 (5 μg) or FIP3 antibody (8 μg) was added to the lysates for immunoprecipitation and incubated while inverting overnight (4°C) followed by magnetic bead incubation for an additional 3h (4°C).
For identifying Rab11-binding site in WDR44, we generated WDR44-truncated constructs by sequentially deleting the conserved domains in WDR44. 293T cells were transfected with GFP-WDR44 full-length (1ug) and truncated constructs along with HA-Rab11QL mutant (1ug) in a 10-cm2 dish. After 48h of transfection, cells were lysed in low-salt triton buffer supplemented with 5mM MgCl2, followed by immunoprecipitation for 3h with Protein A beads conjugated with GFP antibody. Beads were washed with low-salt triton buffer and twice with PBS. Samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane, and probed with indicated antibodies.
For immunoblots showing the phosphorylation status of Akt and WDR44, RPE-1 cells or 293T cells were transiently transfected with wild-type and mutant GFP-fusion constructs of WDR44. Cells were lysed under stringent conditions in RIPA buffer (1% NP-40 buffer containing 0.5% sodium deoxycholate and 0.1% SDS). For WDR44 phospho-analysis, WDR44 or GFP antibody beads were used to immunopurify proteins followed by SDS-PAGE electrophoresis. Phosphorylation status of immunoprecipitated protein was determined using Akt substrate antibody. For Akt phospho-analysis, SDS-PAGE electrophoresis was performed on cell lysate followed by immunoblotting with AKT S473 antibodies.
Cell cycle analysis and cell counting
RPE-1 cells were seeded in 96-well plate (Greiner) and allowed to grow in complete medium for 24h to reach 50-60% confluency. They were washed in PBS twice and treated with relevant drug or inhibitor concentrations with either serum free-, 2% serum-, or 10% serum-condition for 1h or 24h as indicated in the text/figure legend. The effects of siRNAs on cell cycle were evaluated at 72h. Cells were fixed in 4% PFA, washed with PBS twice, and stained with 4,6-diamidino-2-phenylindole (DAPI) in PBS (1μM, Catalog no.1757, AAT Bioquest) for 30min. After PBS washes, cell images were captured using the Celigo Image Cytometer (Nexcelom Bioscience) system, and the integrated nuclear staining intensities were used to analyze cell numbers and cell cycle profiles (G0/G1, S and G2/M) following the manufacturer’s instructions. In parallel, cells were stained with propidium iodide (PI, 0.5 μg/ml) to mark the dead cells and absolute cell number was measured by quantifying the difference between DAPI and PI counts. For each condition, 3-5 wells were analyzed.
In vitro kinase assay and phosphopeptide mapping
For 32P kinase assay and phosphopeptide sequencing, 293T cells were transiently transfected with GFP-fusion constructs of Rab11a, Rab11b, Rabin8, and WDR44. Cells were starved for 24h, washed with cold PBS, and lysed under stringent conditions in ice-cold RIPA buffer (1% NP-40 buffer containing 0.5% sodium deoxycholate and 0.1% SDS) and protease and phosphatase inhibitors (0.15 U/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium vanadate, 20 μM leupeptin). GFP antibody-conjugated beads were used to immunopurify GFP-tagged proteins, washed two-times with lysis buffer and one-time with kinase buffer without ATP and active AKT, and were incubated in kinase buffer (30 mM Tris pH 7.4, 10 mM MnCl2, 5 mM MgCl2, 1 mM DTT, 16.5 μM ATP) and kinase-active Akt (100 ng) (Catalog no. 14-276, EMD-Millipore) with 20μM ATP32 per reaction for 30 mins at 25°C. Samples were resolved using SDS-PAGE and transferred to nitrocellulose membrane before exposing. 32P-labeled proteins were isolated following SDS-PAGE electrophoresis, transferred to nitrocellulose membrane, and digested with trypsin. Tryptic peptides were separated by reversed-phase HPLC and the radioactivity collected in each fraction was plotted to identify the fractions containing the phosphorylated peptides. The phosphorylated peptides were then subjected to Edman degradation and phosphoamino acid analysis to determine the sites of phosphorylation. The level of total protein present in the immunoprecipitate was quantified by immunoblot analysis.
For p-Akt substrate immunodetection, RPE-1 cells, RPE-1 GFP-Rabin8, and RPE-1 GFP-Rab11a cells were lysed, immunoprecipitated with WDR44 or GFP antibodies, and treated with Akt as described above. Samples were resolved using SDS-PAGE electrophoresis, transferred to nitrocellulose membrane, and immunoblotted with pAkt substrate antibodies.
Mass spectrometry
RPE-1 cells stably-expressing GFP-Rab11a were used for identifying binding partners of Rab11 in presence of serum and upon starvation for 1h. RPE-1 cells were grown in a set of ten 15-cm2 dishes to a confluency of 70% and were washed with PBS before starvation for 1h. Cells were lysed on ice in low-salt triton buffer (30mM Tris HCl pH 8.0, 75mM NaCl, 10% Glycerol, 1% Triton X-100) supplemented with 5mM MgCl2, followed by immunoprecipitation for 3h with Protein A beads conjugated with GFP antibody. Beads were washed with low-salt triton buffer and twice with PBS. Samples were resolved using SDS-PAGE, stained with Coomassie dye, and bands were excised for in-gel trypsin digestion to extract the peptides. For LC-MS, each sample was loaded on an Agilent 1100 nanocapillary HPLC system (Agilent Technologies, Santa Clara, CA) and MS/MS analysis was performed on LTQ XP mass spectrometer (ThermoFisher Scientific, Waltham, MA). The MS/MS data were searched using BioWorks 3.3.1 SP1 interfaced SEQUEST from ThermoFisher Scientific.
Zebrafish analysis
RNA injections of zebrafish embryos and analysis of 2-day-old ciliated organs were performed as described previously (Lu et al., 2015). Briefly, full-length sequences of GFP, human WDR44 mutants or Rab11aSN were cloned into pCS2+ vectors and messenger RNAs were transcribed using the mMESSAGE mMACHINE kit (Ambion) according to manufacturer's instructions. Embryos were injected with 100 pg/nl of capped mRNAs at the one-cell stage. For morpholino knockdown and rescue experiments, 250 μM of wdr44 morpholino (5’-GCTGCATACAGAGGCCGCCGCCACT-3’) custom-synthesized by Genetools (see Key Resource Table) was injected in combination with mutant RNAs. Ciliated organ assessment at 48hpf was performed in embryos fixed with 4% PFA and stained with Phalloidin conjugated with Alexa 488 (Catalog no A12379, Molecular Probes Life Technologies) or anti-gamma tubulin (Catalog no. T6557, Sigma), anti-Acetylated tubulin (Catalog no. T6793, Sigma), antibodies and Hoechst (Catalog no. H3570, Molecular Probes Life Technologies). Embryos were imaged by spinning disk confocal (SDC) microscopy using a 40x 1.4 NA or 63x 1.3 NA oil objective on a Marianas SDC (Intelligent Imaging Innovations). For Kupffer’s vesicles analysis, embryos at the 6-8 somites stage were mounted on a custom made slotted petri dish with KVs facing upward and imaged live using a 4X water immersion objective (0.13 N.A) and a Nikon Eclipse Ni-E upright microscope equipped with a DS-Ri2 camera. Western blotting analysis was performed as previously described (Lu et al., 2015). Briefly, zebrafish embryos were homogenized in lysis buffer (20mM Tris at pH 8, 137mM NaCl, 10% glycerol, 1% Triton X-100) with protease inhibitor cocktail (Roche) and centrifuged at 13,000 r.p.m for 10 min. Supernatants were collected and sample buffer was added prior to boiling.
Purification of recombinant proteins
Human full-length Rabin8 (residues 1-460), Rabin8 144-C, Rabin8 144-C S147/S149A, Rabin8 144-C S147/S149D, Rabin8C (residues 270-460), full-length Rab11a (residues 1-216, Q70L constitutively-active mutant), Rab11aQ70L 6-186, full-length FIP3 (residues 1-756), FIP3 695-C (695-756, Rab11-binding domain), C-terminal truncated Rab8 1-183 and WDR44 334-435 (Rab11-binding domain) constructs containing tobacco etch virus (TEV)-cleavable N-terminal 6xHistidine tags were recombinantly-expressed in Escherichia coli using BL21 (DE3) cells. Full-length FIP3 (residues 1-756) has been expressed in Rosetta (DE3) pLyS cells.
Human full-length WDR44 (residues 1-913) was cloned into a pFL vector with a N-terminal tobacco etch virus (TEV) cleavable 6xHistidine tag and expressed using the baculovirus and insect cell expression system. Plasmid was transformed into DH10Bac Escherichia coli cells (Invitrogen). Obtained viral genome was used to transfect Sf21 (Spodoptera frugiperda) cells to produce recombinant baculovirus. High Five insect cells (Invitrogen) were infected with the recombinant virus and harvested after 72h by centrifugation (2000 rpm, 20 min).
Cell pellets were resuspended in lysis buffer (50 mM phosphate pH 7.5, 150 mM NaCl, 10% Glycerol, 5 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM PMSF, 25 ug/mL DNase I) and subjected to sonication. Lysates were clarified by centrifugation (25000 rpm, 40min) and filtration through 0.45 micron filter and were loaded onto a HisTrap HP column (GE Healthcare) in lysis buffer supplemented with 10 mM imidazole. Column was then washed extensively with lysis buffer, followed by 5 CV of lysis buffer supplemented with 1 M NaCl. For purification of Rabin8 constructs, 10 CV of heat shock protein removal buffer (50 mM Tris pH 7.5, 50 mM KCL, 10 mM MnCl2 and 2 mM ATP) was applied to the column followed by a final wash of lysis buffer. Bound proteins were eluted with a gradient from 10-500 mM imidazole. Fractions, which contained the protein, were pooled and dialyzed overnight at 4°C in buffer A (20 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2 and 5 mM DTT) with or without the addition of TEV protease. Protein solution was again passed over a HisTrap HP column to remove cleaved tag and TEV protease. Subsequently, eluate was loaded onto a HiTrapQ-Sepharose anion-exchange chromatography column (GE Healthcare) and equilibrated in buffer A, followed by a linear gradient from 50 mM to 1000 mM NaCl. Fractions containing the protein of interest were pooled, concentrated, and loaded onto a suitable column for size-exclusion chromatography (superdex200 or superdex75, GE Healthcare) in a SEC buffer containing 10 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT or TCEP.
Transmission electron microscopy
Electron microscopy imaging and quantification of MC structures was performed as described (Lu et al., 2015).
Nucleotide exchange assays
GEF activity assays were performed as described previously (Vetter et al., 2018). Prior to the measurements, samples of Rabin8 144-C, phosphoinactive Rabin8 144-C S147A/S149A, and phosphomimetic Rabin8 144-C S147D/S149D mutants were applied to a SEC column to separate dimeric and tetrameric forms. Only fractions of the dimeric form of Rabin8 were pooled and used for GEF activity measurements. 0.5 μM of mant-GDP (mant-GDP, Jena Bioscience)-loaded Rab8 1-183 was incubated with 2 μM of Rabin8 144-C, Rabin8 144-C S147A/S149A or Rabin8 144-C S147D/S149D for 30 min on ice in a buffer containing 30 mM Tris pH 7.5, 5 mM MgCl2, 3 mM DTT and 10 mM potassium phosphate, pH 7.4. The reaction was carried out in a 50 μL quartz Hellmann cuvette using a fluorescence spectrometer (PerkinElmer). 1 mM of GTP was added to start the nucleotide-exchange reaction and dissociation of mant-GDP from Rab8 was monitored every 2s for a total of 300s at 20°C. The data was analyzed using GraphPad Prism 6.0 software and fitted into a one-phase exponential-decay equation without constraints using non-linear regression to obtain rate constants (kobs).
Isothermal titration calorimetry (ITC) experiments
All ITC experiments were carried out using a VP-ITC MicroCal iTC 200 calorimeter (Malvern). Prior to analysis, protein solutions were dialyzed against a buffer containing 10mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP. For the WDR44-Rab11 experiment, 20μM of WDR44334-435 sample was loaded into the reaction cell and 300μM of Rab11Q70L 6-186 into the syringe. Aliquots of 2 uL were injected every 180s for a total of 20 injections. For the WDR44-Rab11-Rabin8 experiment, the syringe was loaded with 1500 μM Rabin8C (270-C) and the cell with 150 μM of Rab11Q70L 6-186-WDR44 334-435 complex. 19 injections of a volume of 2 μL at 120s intervals were injected into the reaction cell. The experimental temperature was 20°C, and all experiments were carried out under constant stirring at 600 rpm. Data was analyzed using Origin 7.0 software provided by MicroCal. A minimum of three independent experiments was performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical data analyses were performed using GraphPad Prism 6 and means ± s.e.m or s.d. are specified in the figure legends. Unpaired two-tailed Student's t-test was applied for comparisons with siControl or as indicated in figure legends. For cell cycle analyses, means from the G0/G1 phase were compared using this test. Where indicated n equals the number of cells counted or zebrafish embryo organelles. Cell numbers counted for cilia determination are indicated in methods or figure legends. For randomization of zebrafish experiments, each injected group was assigned a random number, which was not revealed to the investigator until final outcome assessment.
Supplementary Material
Highlights:
Serum LPA signaling through PI3K/Akt regulates the Rab11-Rab8 ciliogenesis cascade
Akt phosphorylates Rabin8 near its GEF domain affecting Rab8-dependent cilia assembly
Akt stabilizes Rab11-effector binding to WDR44 by phosphorylating WDR44 at Ser342/344
Effector switch from Rab11-WDR44 to Rab11-FIP3 initiates Rabin8 preciliary trafficking
ACKNOWLEDGEMENTS
We thank Dr. Peter Johnson for Akt inhibitors. We are grateful to Joseph Meyer for assistance in preparing the pathway model. We also thank Dr. Ming Zhou, Dr. Thorkel Andressen, and Dr. Sudipto Das for their assistance with LC-MS analysis.
Footnotes
DECLARATION OF INTERESTS
No competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Blazer-Yost BL, Blacklock BJ, Flaig S, Bacallao RL, and Gattone VH (2011). Lysophosphatidic acid is a modulator of cyst growth in autosomal dominant polycystic kidney disease. Cell Physiol Biochem 28, 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Amagai Y, Homma Y, Fukuda M, and Mizuno K (2013). NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. EMBO J 32, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, et al. (2010). LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol 50, 157–186. [DOI] [PubMed] [Google Scholar]
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MP, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, et al. (2013). TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep 3, 1806–1814. [DOI] [PubMed] [Google Scholar]
- Contos JJ, Fukushima N, Weiner JA, Kaushal D, and Chun J (2000). Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci U S A 97, 13384–13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eathiraj S, Mishra A, Prekeris R, and Lambright DG (2006). Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. Journal of molecular biology 364, 121–135. [DOI] [PubMed] [Google Scholar]
- Feng S, Knodler A, Ren J, Zhang J, Zhang X, Hong Y, Huang S, Peranen J, and Guo W (2012). A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J Biol Chem 287, 15602–15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisca F, Colquhoun D, Goldshmit Y, Anko ML, Pebay A, and Kaslin J (2016). Role of ectonucleotide pyrophosphatase/phosphodiesterase 2 in the midline axis formation of zebrafish. Sci Rep 6, 37678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Regalado A, Guzman-Hernandez ML, Ramirez-Rangel I, Robles-Molina E, Balla T, Vazquez-Prado J, and Reyes-Cruz G (2008). G protein-coupled receptor-promoted trafficking of Gbeta1gamma2 leads to AKT activation at endosomes via a mechanism mediated by Gbeta1gamma2-Rab11a interaction. Mol Biol Cell 19, 4188–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I, Vincent EE, and Tavare JM (2011). Akt signalling in health and disease. Cell Signal 23, 1515–1527. [DOI] [PubMed] [Google Scholar]
- Homma Y, and Fukuda M (2016). Rabin8 regulates neurite outgrowth in both GEF activity-dependent and -independent manners. Mol Biol Cell 27, 2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Lu Q, Teixeira I, Harned A, Semler EM, Stauffer J, Magidson V, Tiwari A, Kenworthy AK, Narayan K,Westlake CJ (2019). Investigation of F-BAR domain PACSIN proteins uncovers membrane tubulation function in cilia assembly and transport. Nat Commun 10, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I, Goto H, Kasahara K, and Inagaki M (2015). Current topics of functional links between primary cilia and cell cycle. Cilia 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jo H, Hong H, Kim MH, Kim JM, Lee JK, Heo WD, and Kim J (2015). Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nature communications 6, 6781. [DOI] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, and Guo W (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A 107, 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Yao WL, Tsao KC, Houben AJ, Albers HM, Ovaa H, Moolenaar WH, and Lee SJ (2012). Autotaxin/Lpar3 signaling regulates Kupffer's vesicle formation and left-right asymmetry in zebrafish. Development 139, 4439–4448. [DOI] [PubMed] [Google Scholar]
- Lin ME, Herr DR, and Chun J (2010). Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat 91, 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo GP, Fulmer D, Guo L, Zuo X, Dang Y, Kim SH, Su Y, George K, Obert E, Fogelgren B, et al. (2017). The exocyst is required for photoreceptor ciliogenesis and retinal development. J Biol Chem 292, 14814–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al. (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol 17, 531. [DOI] [PubMed] [Google Scholar]
- Mammoto A, Ohtsuka T, Hotta I, Sasaki T, and Takai Y (1999). Rab11BP/Rabphilin-11, a downstream target of rab11 small G protein implicated in vesicle recycling. J Biol Chem 274, 25517–25524. [DOI] [PubMed] [Google Scholar]
- Martini M, De Santis MC, Braccini L, Gulluni F, and Hirsch E (2014). PI3K/AKT signaling pathway and cancer: an updated review. Ann Med 46, 372–383. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A, et al. (2010). The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci 123, 1785–1795. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- Norris DP, and Grimes DT (2012). Mouse models of ciliopathies: the state of the art. Disease models & mechanisms 5, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, and Golemis EA (2007). HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, and Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz A, Huang Y, and Johansson S (2016). G-Protein-Coupled Lysophosphatidic Acid Receptors and Their Regulation of AKT Signaling. Int J Mol Sci 17, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez I, and Dynlacht BD (2016). Cilium assembly and disassembly. Nat Cell Biol 18, 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi SA, and Lee H The Akt isoforms are present at distinct subcellular locations. (2010). Am J Physiol Cell Physiol 298, C580–C591. [DOI] [PubMed] [Google Scholar]
- Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, Jung Y, Hagiwara H, Yoshihara Y, Yuzaki M, Harada R, et al. (2013). Rab8a and Rab8b are essential for multiple apical transport pathways but insufficient for ciliogenesis. J Cell Sci. 127, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak T, and Wolfrum U (2011). Intraflagellar transport proteins in ciliogenesis of photoreceptor cells. Biol Cell 103, 449–466. [DOI] [PubMed] [Google Scholar]
- Sorokin S (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol 15, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP (1968). Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3, 207–230. [DOI] [PubMed] [Google Scholar]
- Suizu F, Hirata N, Kimura K, Edamura T, Tanaka T, Ishigaki S, Donia T, Noguchi H, Iwanaga T, and Noguchi M (2016). Phosphorylation-dependent Akt-Inversin interaction at the basal body of primary cilia. EMBO J 35, 1346–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumura A, Iimori M, Nishioka Y, Kitahara M, Sakashita M, and Tanaka S (1994). Lysophosphatidic acids induce proliferation of cultured vascular smooth muscle cells from rat aorta. Am J Physiol 267, C204–210. [DOI] [PubMed] [Google Scholar]
- Torres JZ, Miller JJ, and Jackson PK (2009). High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9, 2888–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, and Fujiwara K (1979). Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 17, 527–535. [DOI] [PubMed] [Google Scholar]
- Vetter M, Boegholm N, Christensen A, Bhogaraju S, Andersen MB, Lorentzen A, and Lorentzen E (2018). Crystal structure of tetrameric human Rabin8 GEF domain. Proteins. 86, 405–413. [DOI] [PubMed] [Google Scholar]
- Vetter M, Stehle R, Basquin C, and Lorentzen E (2015). Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11. Nat Struct Mol Biol 22, 695–702. [DOI] [PubMed] [Google Scholar]
- Wall VE, Garvey LA, Mehalko JL, Procter LV, and Esposito D (2014). Combinatorial assembly of clone libraries using site-specific recombination. Methods in molecular biology (Clifton, N.J.) 1116, 193–208. [DOI] [PubMed] [Google Scholar]
- Wang J, and Deretic D (2015). The Arf and Rab11 effector FIP3 acts synergistically with ASAP1 to direct Rabin8 in ciliary receptor targeting. J Cell Sci 128, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AM, and Beales PL (2011). Ciliopathies: an expanding disease spectrum. Pediatr Nephrol 26, 1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al. (2011). Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A 108, 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, and Barr FA (2007). Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 178, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung YC, Mutoh T, Lin ME, Noguchi K, Rivera RR, Choi JW, Kingsbury MA, and Chun J (2011). Lysophosphatidic acid signaling may initiate fetal hydrocephalus. Sci Transl Med 3, 99ra87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Ren M, Gravotta D, De Lemos-Chiarandini C, Lui M, Erdjument-Bromage H, Tempst P, Xu G, Shen TH, Morimoto T, et al. (1999). Identification of a putative effector protein for rab11 that participates in transferrin recycling. Proc Natl Acad Sci U S A 96, 2840–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Elkahloun AG, Liao H, Delaney S, Saber B, Morrow B, Prendergast GC, Hollander MC, Gills JJ, and Dennis PA (2011). Expression signatures of the lipid-based Akt inhibitors phosphatidylinositol ether lipid analogues in NSCLC cells. Mol Cancer Ther 10, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.