Abstract
Few cell signals match the impact of the TGF-β family in metazoan biology. TGF-β cytokines regulate cell fate decisions during development, tissue homeostasis and regeneration, and are major players in tumorigenesis, fibrotic disorders, immune malfunctions, and various congenital diseases. The effects of the TGF-β family are mediated by a combinatorial set of ligands and receptors, and a common set of receptor-activated SMAD transcription factors. Yet, the effects can dramatically differ depending on the cell type and the conditions. Recent progress has illuminated a model of TGF-β action in which SMADs bind genome-wide in partnership with lineage-determining transcription factors and additionally integrate inputs from other pathways and the chromatin to trigger specific cellular responses. The new insights clarify the operating logic of the TGF-β pathway in physiology and disease.
Even the simplest multicellular organisms require a high level of intercellular coordination. Cells communicate in part via small, secreted proteins variously known as cytokines, growth factors, or polypeptide hormones depending on the nature of their effects and the distance between their source and physiologic targets. There is no cytokine family in metazoans with more widespread and diverse effects than the transforming growth factor (TGF)-β family. Essentially all cells in the developing embryo and the adult can perceive TGF-β family signals, and respond with effects on cell proliferation, differentiation, communication, adhesion, movement, metabolism, and death that are as varied as the target cell types1. Collectively these cytokines have long captured the interest of the biomedical field because of their numerous roles in development and adult physiology, as well as the many developmental, immune and fibrotic diseases that result from their malfunctions2-8, not the least of which is cancer9-12.
The molecular basis for the contextual nature of TGF-β action, an open question for decades13, has recently gained unprecedented clarity. At the transcriptional level, recent progress illuminates a unifying theme –the collaboration between SMADs, which are signal-driven transcription factors [G] (SDTFs) directly activated by TGF-β receptors, and lineage-determining transcription factors that specify cell identity and determine the binding of SMADs to loci genome wide. By additionally sensing the chromatin status at these loci and the cooperating with other SDTFs, SMADs integrate these multiple determinants to yield strikingly contextual responses to TGF-β family inputs.
Here we cover this progress by focusing on three contexts that best reveal the operating logic of the TGF-β pathway. These three contexts are early embryogenesis14-16, immune regulation17,18, and tumor progression9-12. The biological effects of the TGF-β family in these settings have been extensively discussed (op. cit.). Our purpose here is to discuss the current model for how the TGF-β cytokines trigger specific responses in these biological contexts.
Basics of TGF-β signaling
The TGF-β pathway is a paradigm of membrane-to-nucleus signaling by receptor-mediated activation of transcription factors19. As ligands the cytokines bind to two pairs of transmembrane serine/threonine protein kinases, triggering receptor activation and phosphorylation of SMAD transcription factors20,21 (Fig. 1). In the basal state, the SMAD proteins shuttle between the cytoplasm and the nucleus22,23. Upon being phosphorylated by the receptors, SMADs reenter the nucleus ready to bind throughout the genome and activate or repress hundreds of genes. The core components of the TGF-β–SMAD pathway are highly conserved throughout metazoan evolution24. X-ray crystal structures have been determined for most of these components and complexes, providing key insights into the structural basis for their function and specificity19,25-27.
Figure 1. The TGF-β–SMAD pathway in the basal and activated states.
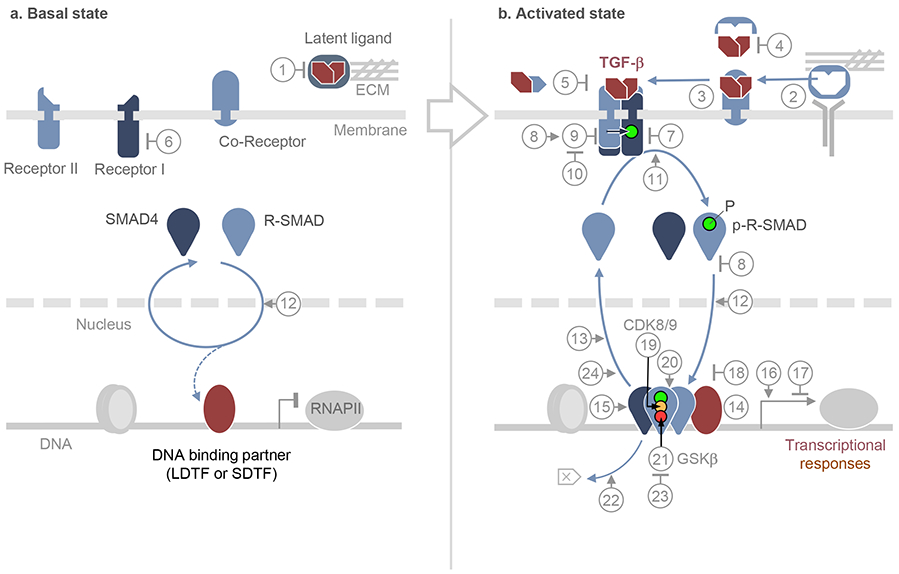
a. In the basal state, TGF-β family ligands are either absent or present as latent factors, sequestered by binding components of the extracellular matrix (ECM) or the membrane of other cells [1]. Allosteric inhibitors may enforce the inactive state of the receptors, all of which are transmembrane serine/threonine protein kinases [6]. SMAD proteins that serve as receptor substrates (R-SMADs), and their shared partner SMAD4, continuously transit between cytoplasm and nucleus by direct interaction with nuclear pore components [12]. SMAD partners including lineage-determining transcription factors (LDTF), other signal-driven transcription factors (SDTF), and the histone binding protein TRIM33, may already be pre-bound to the genome. Numbers in circles refer to these and other pathway regulators, as detailed in Box 1.
b. In the activated state, ligand from cells in the microenvironment or released from latent complexes allosterically by integrins or enzymatically by proteases [2], binds to pairs of type I and type II receptors. Ligands can access the receptors directly or, in the case of TGF-β, Nodal and BMP9/10, with assistance of accessory co-receptors [3]. Ligand traps [4] and antagonistic ligands [5] block access to the receptors. In the ligand-induced receptor complex, the type II receptor subunits primarily phosphorylate and activate the type I receptors, which phosphorylate R-SMADs, with the assistance of adaptor proteins [11]. Receptor-phosphorylated R-SMADs form heterotrimers with SMAD4. Pseudo-receptors [7], inhibitory SMADs [8], and SKI proteins [18] inhibit the formation of receptor and SMAD complexes and provide negative feedback in the pathway. Inhibitory SMADs recruit ubiquitin ligases that target the receptor [9], which are opposed by deubiquitylases [10]. In the nucleus, activated SMAD complexes bind to hundreds of genomic loci as dictated by context-defining LDTFs and SDTFs [14]. SMADs contact DNA using a highly conserved β-hairpin structure located in the N-terminal domain (or MH1) domain. The SMAD C-terminal domain (or MH2 domain) binds not only LDTFs and SDTFs but also chromatin binding proteins [15], co-activators and co-repressors [16,17]. The nuclear kinases CDK8/9 phosphorylate the SMAD interdomain linker region [19] for recruitment of additional cofactors [20]. Subsequent phosphorylation by GSK3 targets SMADs for ubiquitin- and proteasome-dependent degradation [21,22]. Alternatively, SMADs are dephosphorylated [13, 23] and dissociated from DNA [24] for recycling.
The signaling steps summarized here are reviewed in detail elsewhere13,19,25,26,28,30,93.
Although TGF-β can also activate phosphatidylinositol 3-kinase (PI3K), mitogen-activated protein kinases (MAPK) and other pathways that have their own potent agonists13,28-30, the effects mediated by SMADs fully recapitulate the context-dependent nature of TGF-β action. The apparent simplicity of this two-step signal transduction process belies the diversity of cellular responses that it elicits in different cellular contexts. For example, TGF-β-activated SMADs promote differentiation of embryonic stem (ES) cells16,31,32, proliferation of chondrocyte precursors33, growth inhibition in epithelial progenitors34,35, apoptosis in premalignant cells36-38, and metastatic invasion in carcinoma cells39,40.
Ligands
Structural and functional considerations separate the 32 members of the family into TGF-β and bone morphogenetic protein (BMP) subfamilies, with a few outliers (Table 1). The TGF-β subfamily in mammals comprises TGF-β1, TGF-β2 and TGF-β3; Activins A and B; Nodal, Myostatin, and several members known as GDFs (growth and differentiation factors). Broadly speaking, Nodal14,16 and GDF3 41-43 are active during early embryo development, whereas TGF-β, Activin, GDF8 (also known as Myostatin) and other GDFs are critical for organogenesis, tissue homeostasis4,15 and immune regulation17,18. The BMP subfamily6,44,45 includes over a dozen factors, which function just as broadly as the TGF-β subfamily, frequently in contraposition to it. The most extensively studied members are BMPs 2, 4, 6, 7, 9, and 15, GDFs 5 and 9, and Anti-Muellerian Hormone [G] 2. The bioactive factors are disulfide-linked dimers cleaved from the C-terminal portion of a biosynthetic precursor by furin and other pro-protein convertases. The canonical ligands are homodimeric, but heterodimers such as TGF-β1.2 46, Activin AB 47, BMP2.7 48,49 Nodal-Vg1 (Nodal-GDF3)42 and others exist that further diversify the repertoire.
Table 1.
Mammalian TGF-β family members and their receptors
| Family member | Type I Receptor | Type II Receptor | Co-receptor | SMAD |
|---|---|---|---|---|
| TGFβ-1 | ALK5 * | TGFBR2 | Betaglycan | SMAD2/3 |
| TGFβ-2 | ALK5 | TGFBR2 | Betaglycan | SMAD2/3 |
| TGFβ-3 | ALK5 | TGFBR2 | Betaglycan | SMAD2/3 |
| Activin-A | ALK4, ALK7 | ACVR2, ACVR2B | SMAD2/3 | |
| Activin-B | ALK4, ALK7 | ACVR2, ACVR2B | SMAD2/3 | |
| Nodal | ALK4, ALK7 | ACVR2, ACVR2B | Cripto, Cryptic | SMAD2/3 |
| GDF1 | ALK4, ALK7 | ACVR2, ACVR2B | Cripto, Cryptic | SMAD2/3 |
| GDF3 | ALK4, ALK7 | ACVR2, ACVR2B | Cripto, Cryptic | SMAD2/3 |
| Myostatin (GDF8) | ALK4, ALK5 | ACVR2 | SMAD2/3 | |
| GDF9 | ALK4 | BMPR2 | SMAD2/3 | |
| GDF11 | ALK4, ALK5 | ACVR2, ACVR2B | SMAD2/3 | |
| Inhibin ** | — | ACTR2 | Betaglycan | |
| Lefty-1 ** | — | — | Cripto, Cryptic | |
| Lefty-2 ** | — | — | Cripto, Cryptic | |
| BMP2 | ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | RGM | SMAD1/5 |
| BMP4 | ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| BMP5 | ALK2, ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| BMP6 | ALK2, ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | RGM | SMAD1/5 |
| BPM7 | ALK2, ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| BPM8 | ALK2, ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| BMP9 | ALK1 | ACVR2, BMPR2 | Endoglin | SMAD1/5 |
| BMP10 | ALK1 | ACVR2, BMPR2 | Endoglin | SMAD1/5 |
| BMP15 | ALK6 | BMPR2 | SMAD1/5 | |
| GDF5 (BMP14) | ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| GDF6 | ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| GDF7 | ALK3, ALK6 | ACVR2, ACVR2B, BMPR2 | SMAD1/5 | |
| AMH | ALK2, ALK3 | AMHR2 | SMAD1/5 | |
| BMP3 ** | — | ACVR2B | ||
| Activin C | Unknown | Unknown | ||
| Activin E | Unknown | Unknown | ||
| BMP3b (GDF10) | Unknown | Unknown | ||
| GDF15 | *** |
Alternative names are indicated in parentheses, except
ALK5 is also known as TβRI or TGFBR1; ALK2 as ActR1A or ACVR1A; ALK4 as ActR1B or ACVR1B; ALK7 as ACVR1C; ALK3 and ALK6 as BMPR1A and BMPR1B, respectively; and, ACTVR2 and ACVR2B as ActRII and ActRIIB, respectively.
Antagonistic ligands: Inhibin antagonizes Activins, Lefty antagonizes Nodal, and BMP3 antagonizes other BMPs.
GDF15 is a distant member of the TGF-β family and signals through glial-derived neurotrophic factor receptor α-like (GFRAL)
Receptors
Mammalian genomes encode seven type I and five type II receptors19 (Table 1). Among the type II receptors, ACVR2 (or ACTR2, Activin Receptor Type II) and ACVR2B are widely shared by ligands from both subfamilies (Activins, Nodal, BMPs and GDFs); BMPR2 (BMP Receptor Type II) is shared by ligands of the BMP subfamily, whereas TGFBR2 (or TβR-II, TGF-β Receptor Type II) and AMHR2 (AMH Receptor Type II) only bind TGF-β and AMH, respectively. Among the type I receptors for the TGF-β subfamily, TGFBR1 (or TβR-I; also known as ALK5, Activin Receptor-Like Kinase 5) is restricted to TGF-β and GDF8/11, whereas ACVR1B (ActR1B/ALK4) and ACVR1C/ALK7 are shared by the other members of the TGF-β subfamily. In the BMP subfamily, BMP9 and BMP10 share ALK1, and the other BMP subfamily members share ACVR1A/ALK2, BMPR1A/ALK3 and BMPR1B/ALK6 (Table 1). In the ligand-induced receptor complex, the type II receptors phosphorylate the type I receptors, which then recognize and phosphorylate SMAD proteins. In addition to the type I receptors, TGFBR2 and BMPR2 directly regulate the cell polarity protein PAR6 50 and the cytoskeletal regulatory kinase LIMK1 (LIM domain kinase 1) 51,52, respectively.
Combinatorial ligand perception
The contextual nature of TGF-β signaling is not only determined by the intrinsic transcriptional responsiveness of the target cells, which the following sections cover, but also by the repertoires of ligands and membrane receptor that interact in the extracellular space, which we briefly highlight here. The large number of ligands in the TGF-β and BMP subfamily interact with a small set of receptors that signal through an even smaller set of messenger SMADs. The diversity of ligands provides opportunities for differential regulation of ligand access to receptors by ligand-binding proteins and membrane co-receptors (Box 1). Filipodial extensions53 and endocytosis54 are involved in BMP delivery and diffusion, and participate in the formation of DMP morphogen gradients54.
Box 1: Regulation of TGF-β–SMAD signaling.
Ligand regulators.
TGF-β, GDFs 7~11 and BMP9 remain tightly bound to their biosytnthetic prodomain portion as a latency associated protein (LAP), forming a complex in which the ligand cannot bind to receptors [1] (Numbers in brackets are as in Figure 1). The LAP:TGF-β complex binds to latent TGF-β binding proteins LTBP1~4 in the extracellular matrix, or the membrane protein GARP in T cells. Allosteric and enzymatic mechanisms release latent TGF-β [2]. Integrin αVβ6 binds LAP to allosterically release TGF-β from the latent complex. Thrombospondin-1 (TSP-1) also activates latent TGF-β by an allosteric mechanism, whereas matrix metalloprotease 9 activates by cleaving LAP.
Membrane co-receptors assist TGF-β family members in binding to signaling receptors [3]. Betaglycan (also known as type III receptor) is a co-receptor for TGF-β, Endoglin for BMP9/10, Cripto and Cryptic for Nodal, and RGMs (Repulsion Guidance Molecules) for BMP2, BMP6 and probably other BMPs. Noggin, Follistatin, Chordin, Gremlin, Cerberus and Coco, bind bioactive Activins and BMPs to bar their access to receptors [4]. Inhibin, Lefty and BMP3 interfere with signaling by Activin, Nodal, and BMP receptors, respectively (see Table 1) [5].
Receptor regulators.
FKBP12 locks the basal state TGF-β type I receptor in an inactive conformation [6]. BAMBI is a membrane-bound inhibitor of BMP and Activin receptors [7]. SMAD6 and SMAD7 bind to activated TGF-β and BMP receptors for inhibition [8]. SMAD7 recruits the E3 ligase SMURF2 for TGF-β receptor ubiquitylation and turnover [9]. SMAD7 expression is upregulated by TGF-β and BMP, providing negative feedback. SMAD6 also inhibits SMAD4 binding to SMAD1. The deubiquitylating enzymes USP4, USP11 and USP15 remove ubiquitylation from type I receptors [10] and R-SMADs, and USP9X from SMAD4.
Regulators of SMAD activation.
Endosomal proteins SARA and Endofin serve as adaptors for SMAD phosphorylation by TGF-β and BMP receptors, respectively [11]. R-SMADs and SMAD4 interact with nuclear pore components NUP153 and NUP214, Importin β1 and nuclear export factor CRM1 for nucleo-cytoplasmic shuttling [12]. Phosphatases PPM1A and PP2A can dephosphorylate C-terminal sites of R-SMADs [13], though their relevance is not yet settled.
Transcriptional complex regulators.
LDTFs bound to DNA [14] and TRIM33 bound to H3K9me3:K18ac histone marks [15] regulate SMAD binding to DNA. Histone acetylases p300 and CBP [16] and deacetylatase complexes formed by TGIF and HDAC1/2 [17] act as SMAD co-activators and co-repressors, respectively. SKI and the related protein SKIL disrupt R-SMADs binding to SMAD4 [18].
Nuclear kinases CDK8/9 phosphorylate Ser-Pro sites in the linker region [19] to recruit transcriptional cofactors YAP and PIN1 [20]. But these pSer-Pro marks prime the linker for GSK3β-mediated phosphorylation [21], which recruits SMURF1/2 and NEDD4L to SMAD1/5 and SMAD2/3, respectively, for SMAD poly-ubiquitylation and degradation [22]. SCP1~3 antagonize this fate by dephosphorylating the linker region [23]. These sites are also phosphorylated by ERK in response to RAS and by CDK2/4 during the cell cycle. PARP1 mediated ADP-ribosylation releases SMADs from DNA [24], to end SMAD signaling. Other regulatory inputs include TGF-β receptor neddylation [G] and SMAD sumoylation.
For more information on these regulators refer to specialized reviews 13,19,25,26,184-192
Cells in vivo may be exposed to multiple members of the TGF-β and BMP subfamilies at once, resulting in additive, synergistic as well as antagonistic combinations of ligand inputs55-58. As shown in cell culture models exposed to combinations of different BMP ligands, cells can use their repertoire of BMP receptors to perceive mixed ligand milieus and turn them into specific signaling input patterns59. Moreover, heterodimeric ligands may assemble complexes including more than one kind of type I receptor. For example, BMP2.7 heterodimers assemble complexes that include both ALK2 and ALK3 during dorsoventral patterning in zebrafish48.
SMAD transcription factors
SMAD proteins consist of globular N-terminal and C-terminal domains, known as the MH1 and MH2 domains respectively, separated by a flexible linker region19,24. The MH1 domain binds DNA, and the MH2 domain interacts with other SMADs, different lineage-determining transcription factors (see below), chromatin readers such as TRIM3360, co-activators such as p300 and CBP61-63 and co-repressors such as TGIF64. The linker region of R-SMADs contains sites for regulatory phosphorylation by mitogen-activated protein kinases (MAPKs)65-67, cyclin-dependent kinases (CDKs)68,69, and glycogen synthase kinase 3β (GSK3β)67.
Five SMAD proteins serve as receptor substrates (R-SMADs) in mammals. The type I receptors for the TGF-β subfamily (ALK4, ALK5 and ALK7) primarily phosphorylate SMAD2 and SMAD3, whereas the type I receptors for the BMP subfamily (ALK1, ALK2, ALK3 and ALK6) primarily phosphorylate SMADs 1, 5 and 8. SMAD8 (also known as SMAD9) has been reported to possess weak transcriptional activity and may act as a dominant negative regulator of SMAD1/5 in BMP signaling70. Receptor-mediated phosphorylation targets two serine residues at the C-terminus of R-SMADs, generating an acidic tail (pSer-X-pSer-COO−) that mediates MH2-MH2 oligomerization with R-SMADs and SMAD419.
SMAD4 is not a receptor substrate and is not required for movement of activated R-SMADs into the nucleus. However, SMAD4 is essential for the assembly of R-SMAD-SMAD4 complexes that trigger most TGF-β family gene responses19 (Fig. 1). What specific function SMAD4 brings to these transcriptional complexes remains unknown. Certain TGF-β responses, such as the upregulation of the transcription factor SOX4 in pancreatic epithelial progenitors, require just SMAD2/3, not SMAD438 (Fig. 1), in line with the requirement for SMAD2/371 but not SMAD436 in pancreas development in mice. TGF-β induction of fibronectin, a major extracellular matrix component, also requires SMAD2/3 but not SMAD438. Two additional family members, SMAD6 and SMAD7, act as inhibitory SMADs that mediate negative feedback in the pathway (Box 1).
The SMAD transcriptional cycle.
Receptor-activated SMAD1/5 and SMAD2/3 that enter the nucleus undergo phosphorylation at Ser-Pro motifs in the linker region 68,72. These phosphorylations are mediated by CDK8 and CDK9, which also phosphorylate the C-terminal domain (CTD) of RNA polymerase II (RNAPII) for transcriptional initiation and elongation. In R-SMADs, CDK8/9-mediated phosphorylation creates sites for binding of YAP and PIN1, which enhance the transcriptional activity of the SMAD complex. But this phosphorylation also primes the linker region for GSK3β-mediated phosphorylation, which creates binding sites for the E3 ubiquitin ligases SMURF1/2 in SMAD1/5 and NEDD4L in SMAD2/3, for SMAD degradation68,72,73. This degradation fate can be averted by SCP1/2 (small CTD phosphatases), which dephosphorylate the linker region and spare SMADs for new rounds of signaling74. Poly(ADP-ribose) polymerase-1 (PARP-1) participates in dissociating SMAD complexes from DNA by ADP-ribosylating SMAD3 and SMAD475. The involvement of related protein kinases and phosphatases in the regulation of SMADs and RNAPII suggests close coordination between these transcription factors and RNA polymerase.
Core pathway regulators
Much has been learned about the regulation of the core pathway components (Box 1). Regulators include factors that trap the ligands in a latent state, and factors that release the active ligands from these latent complexes. Co-receptors for TGF-β, Nodal and BMP, and family members that competitively antagonize Nodal and Activin modulate ligand access to the receptors. Regulators of SMAD stability and nucleocytoplasmic shuttling, and transcriptional co-activators and co-repressors interact with SMADs to enable their function in the nucleus. Negative feedback regulators, mediators of receptor and SMAD turnover, and mediators of regulatory inputs from other pathways, have also been identified. Box 1 provides a brief summary of 24 distinct classes of TGF-β-SMAD pathway regulators.
Context-dependent interpretation of TGF-β and BMP signals
The identification and structural characterization of the core components of the TGF-β pathway was important for understanding signal transduction process but insufficient to explain how TGF-β and BMP activate different genes in the same cell, or how the same SMADs activate different genes in different contexts. For example, the growth inhibitory effect of TGF-β in epithelial76,77 and hematopoietic progenitors78, and the role of TGF-β in promoting wound healing responses79, both a focus of this field since the early days, are mediated by transcriptional activation of such disparate components as CDK inhibitors and extracellular matrix proteins, respectively. Eventually, genome-wide transcriptomic analysis showed that different cell types respond to TGF-β-activated SMADs with largely non-overlapping gene expression profiles, and the same holds true for BMP-activated SMADs. Thus, the contextual nature of TGF-β and BMP action has long been apparent but has defied a unifying explanation.
SMADs bind a common DNA motif
TGF-β and BMP trigger direct responses by activating different R-SMADs19. However, SMAD2/3 and SMAD1/5 do not regulate different genes by having different DNA binding specificities. Oligonucleotide binding assays80 showed that SMAD3 and SMAD4 bind the palindromic duplex 5′-GTCTAGAC-3′. The X-ray crystal structures of MH1 domains of SMADs 1, 3, and 4 bound to this sequence demonstrated that all three SMADs similarly recognize the GTCT motif or its complementary extended sequence CAGAC 81,82. SMAD2 is an exception: its most abundant isoform contains an insert that is thought to inhibit binding to DNA19,80. SMAD1 and SMAD4 were also found to bind GC-rich motifs in BMP target gene promoters83-85, an observation that led to the proposal that GC-rich motifs function as BMP response elements (BRE) whereas the CAGAC motifs serves as SMAD binding elements (SBE) for the TGF-β subfamily. However, this dichotomy was puzzling, given the sequence identity of the MH1 domain β-hairpin, which is the structural element that binds DNA26. Moreover, analysis of SMAD4 binding to promoter sequences of the mesendoderm specification gene goosecoid (Gsc) 84, and of genome-wide SMAD3 and SMAD4 binding in ES cells86,87 showed that these SMADs also bind GC-rich regions.
Addressing these discrepancies, recent work showed that SMADs 1, 3, 4 and 5 recognize the common consensus sequence GGC(GC)∣(CG) 24. These 5-bp GC-rich sites are referred to as the 5GC SBE motif. X-ray crystal structures of the different SMAD MH1 domains bound to 5GC motifs demonstrate a high degree of conformational flexibility of the β-hairpin. The SMAD-binding cis-regulatory elements of TGF-β and BMP target genes are significantly enriched in clusters of 5GC SBEs, which are more prevalent than the CAGAC motif. In sum, TGF-β-activated SMAD3 and BMP-activated SMAD1/5 recognize common DNA motifs yet regulate different sets of target genes (Fig. 2a).
Figure 2. LDTFs and SDTFs as contextual determinants of TGF-β–SMAD action.
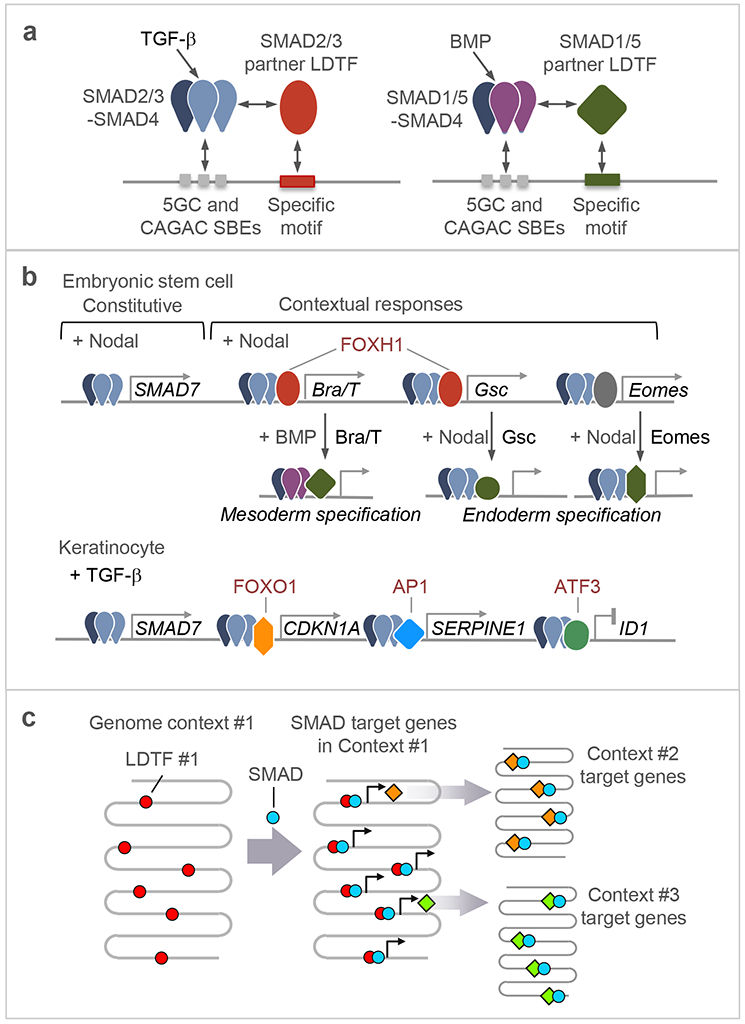
a. All R-SMADs recognize a common set of DNA elements including GGCGC and related 5bp motifs (5GC motifs), as well as the CAGAC motif. TGF-β-activated SAMAD2/3 and BMP-activated SMAD1/5 recognize different partner LDTFs, thereby achieving specificity in target gene recognition.
b. Examples of interactions between SMAD complexes and DNA binding cofactors. In ES cells poised to undergo mesendodermal differentiation, the LDTF FOXH1 is pre-bound to loci throughout the genome and recruits Nodal-activated SMAD2/3:SMAD4 complexes to these loci. The target genes encode LDTFs like BRA/T and GSC. These and the related LDTF EOMES, interact in turn with Nodal-activated or BMP-activated SMADs to mediate further specification of the mesoderm and endoderm lineages (refer also to Table 2). In keratinocyte precursors, TGF-β-activated SMAD2/3:SMAD4 complexes bind the genome in partnership with different SDTFs to regulate the expression of different subsets of target genes. Although SMADs target largely different gene sets in different cell types as a function of context-specific LDTFs and SDTFs, certain SMAD-mediated gene responses appear to be constitutive to the pathway and common to many cell types, as in the case of the negative feedback regulator SMAD7.
c. Schematic representation of successive waves of SMAD-LDTF driven gene expression programs. An LDTF that defines a particular phenotypic context (Context #1) within a given cell lineage recruits signal-driven SMADs to activate the expression of other LDTFs, which in turn interact with SMADs to mediate transitions into new differentiated states (Context #2, #3).
Cell-specific LDTFs dictate SMAD binding genome-wide
If the DNA binding activity of SMAD1/5 and SMAD2/3 proteins is not a major determinant of TGF-β versus BMP subfamily specificity in target gene recognition, then other factors must determine this specificity. Time and again the genetic dissection of developmental processes has led to the identification of transcription factors that determine lineage commitment and cell differentiation within this lineage. These master regulators are referred to as lineage-determining transcription factors, LDTFs 88. Early work on two LDTFs, FOXH1 and ZFP423, provided pioneering examples of the crucial role of these factors as contextual determinants of TGF-β and BMP responses. FOXH1 (previously known as Fast1), a member of the forkhead transcription factor family, directs nodal-activated SMAD2/3 to target promoters during mesendoderm formation87,89,90 (Fig. 2b). The zinc-finger transcription factor ZFP423 (previously known as OAZ) directs BMP-activated SMAD1 to target promoters during Xenopus ventral mesoderm specification91. FOXH1 and ZFP423 discriminately bind SMAD2/3 and SMAD1/5, respectively. The resulting complexes target gene regulatory regions that contain FOXH1 or ZFP423 binding elements alongside SBE, thereby defining the specificity.
Another stride towards answering the question of how cells read TGF-β family signals came from analysis of genome-bound SMADs in various cell types. The binding pattern of signal-activated SMAD2/3 was different in ES cells, myoblast precursors, and pro-B cells, and it closely aligned with the binding of highly expressed LDTFs in each lineage31. This paradigm was extended to BMP-regulated SMAD192. In sum, LDTFs determine the cell type- and pathway-specific activity of SMADs, a model further validated by more examples below.
Although LDTFs determine the genome binding patterns of SMADs, this does not mean that the SMADs are only passive participants in cellular transcriptional networks, merely following LDTFs around the genome to adjust gene expression as dictated by these master regulators. In ES cells and progenitor cells, LDTF-guided SMADs frequently regulate the transcription of other LDTFs, which in turn trigger differentiation along lineage fates (Fig. 2b,c). Moreover, SMADs do not always form exclusive partnerships with a dominant transcription factor, and LDTFs are not obligatory partners of SMADs. SMADs can also form partnerships with other SDTFs (Table 2, and see below). Thus, SMAD signaling is both context dependent, guided by LDTFs, and context determining, through SMAD regulation of LDTF expression.
Table 2.
Transcription factors (TF) that interact with R-SMADs to regulate developmental processes.
| TF | Signal | SMAD | Cell Context | Target genes | Process (species) |
|---|---|---|---|---|---|
| Oct4 | BMP | SMAD1 | ES cells | Sox2, Nanog, Oct4 | Pluripotency (m) |
| Oct4 | Nodal | SMAD2 | Epiblast ES cells | Sox2, Nanog, Oct4 | Pluripotency (h,m) |
| FOXH1 | Nodal | SMAD2/3 | ES cells | Bra/T, Gsc, Mixl1 | Mesendoderm formation (h,m) |
| EOMES | Nodal | SMAD2/3 | Mesendoderm progenitors | Sox17, Fzd8, Cer, Foxa2 | Endoderm specification (h,m) |
| BRA/T | BMP | SMAD1/5 | Mesendoderm progenitors | Cdx2, Cbr, Tbx6 | Mesoderm specification (h) |
| ZFP423 | BMP | SMAD1/5 | Mesoderm progenitors | Xvent2 | Ventral mesoderm specification (x) |
| GATA1/2 | BMP | SMAD1/5 | Erythroid progenitors | Hemgn | Erythroid differentiation (h) |
| C/EBPα | BMP | SMAD2/3 | Myeloid progenitors | Cxcr4 | Myeloid differentiation (h) |
| MYOD1 | TGF-β | SMAD2/3 | Myogenic progenitors | Adora1 | Myoblast differentiation (m) |
| PU.1 | TGF-β | SMAD2/3 | Pro-B cells | Vpreb2 | B-cell differentiation (m) |
| RUNX3 | TGF-β | SMAD2/3 | B cells | Ig locus | Immunoglobulin class switching (m) |
| STAT5* | TGF-β | SMAD2/3 | Naïve CD4+ T cells | Foxp3 | Treg cell differentiation (m) |
| RORγt | TGF-β | SMAD2/3 | Naïve CD4+ T cells | IL-17, IL-23R | TH17 cell differentiation (m) |
| ATF1 | TGF-β | SMAD2/3 | CD8+ T cells | Prf1, GzmB | CTL inhibition (m) |
| NFkB* | TGF-β | SMAD2/3 | Osteoclast precursors | Nfatc1 | Osteoclast differentiation (m) |
| DLX3 | BMP | SMAD1/5 | Hair follicle progenitors | Gata3 | Hair follicle development (m) |
| ? (RAS)* | TGF-β | SMAD2/3 | Epithelial progenitors | Snail | EMT (h,m) |
| SNAIL | TGF-β | SMAD2/3 | Epithelial progenitors | (Cdh1), (Klf5) | EMT (h,m) |
| ATF3 | TGF-β | SMAD2/3 | Epithelial progenitors | (Id1) | Differentiation (h) |
| FOX1~3* | TGF-β | SMAD2/3 | Epithelial progenitors | Cdkn1a, Cdkn2b | Cell cycle inhibition (h,m) |
| E2F4/5* | TGF-β | SMAD2/3 | Epithelial progenitors | (Myc) | Cell cycle inhibition (h) |
| AP1* | TGF-β | SMAD2/3 | Epithelial progenitors | SerpinE1 | Extracellular matrix modulation (h) |
TFs include LDTFs and
SDTFs. Target genes in parenthesis are repressed. EMT, epithelial-mesenchymal transition. Examples are from human (h) and mouse (m). .
SMAD cooperation with other signal-driven transcription factors
SMADs integrate inputs from other pathways. These inputs can be broadly separated into two categories. One includes signal-dependent post-translational modification that regulate the strength and duration of SMAD signaling activity, as has been reviewed elsewhere13,93,94 (refer to Box 1). The other category involves SDTFs that respond to other signals and cooperate with SMADs alongside LDTFs at target gene loci. In partnerships with other SDTFs SMADs control cell cycle progression, extracellular matrix production, and other effects (Table 2). Classical examples include partnerships of SMAD2/3 with AP1 in the regulation of extracellular matrix proteolysis95, with FOXO1~4 in the induction of CDK inhibitors34, with RUNX3 in B-cell immunoglobulin class switching96, and with E2F4 and ATF3 in the repression of MYC and ID1, which are transcriptional regulators of cell growth and differentiation35,97.
The convergence of signal-activated SMADs and other SDTFs on LDTF genes as targets ties to the recent emergence of the concept of “super-enhancers”, clusters of transcription factor-binding elements that capture entire networks of SDTFs and are massively enriched with machinery for transcription activation98. Compared to regular enhancers, super-enhancers are small in number, and are strongly enriched at LDTF loci99. Examination of the DNA elements that comprise super-enhancers showed enrichment for binding sites for both, LDTFs and SDTFs of multiple pathways100. This suggests that the control of cell identity in general is a cooperative affair involving LDTFs and multiple SDTFs, with SMADs prominent among them.
TGFβ action in developmental contexts
The regulation of cell identity by the TGF-β pathway during development provides key examples of SMADs working in partnership with LDTFs and other SDTFs to generate specific responses that orchestrate phenotypic transitions of different stem and progenitor cells.
Stem cell differentiation control by SMADs and LDTFs
The dependence of activated SMAD on LDTFs was identified in studies of early vertebrate development. Treatment of early-stage Xenopus frog embryos comprising pluripotent ES cells with Activin (a mimic of endogenous Nodal) induced the formation of mesoderm-endoderm progenitor cells (mesendoderm). SMAD2/3 in complex with SMAD4 required FOXH1 to activate transcription of the homeobox gene MIXL1 (also known as Mix.2 in mammals; Mixer in zebrafish) in this context89. SMADs and FOXH1 jointly bind to regulatory elements at key developmental loci90. Mix.2 was later found to be part of a larger network of SMAD-induced LDTFs that regulate mesendoderm formation101. In mouse and human ES cells Nodal signaling stimulates differentiation through extensive upregulation of mesendoderm LDTFs including Mixl1, Eomesodermin (Eomes), Brachyury (T), Goosecoid (Gsc) and Foxa2102 (Fig. 2c, and Table 2). FOXH1 occupies cis-regulatory elements of most of these genes from the start of zygotic gene expression, prior to SMAD activation by Nodal signals103, and Nodal-activated SMAD2/3 are recruited to FOXH1-occupied regions genome-wide104 (Fig. 1). FOXH1 therefore is an archetype of LDTF that dictates SMAD binding genome-wide, including loci that encode LDTFs for differentiation into the next developmental stage. This work also supports the concept that LDTFs controlling mesendoderm development form gene regulatory networks akin to those that control pluripotency105.
Mesendoderm LDTFs are silenced in ES cells by the action of histone binding proteins that maintain a repressive chromatin state. To gain access to such sites, SMAD2/3 bind to TRIM33, a member of the “chromatin reader” [G] class of proteins60. The SMAD2/3-TRIM33 complex is alternative to the SMAD2/3-SMAD4 complex, and the two complexes cooperate in binding to these obstructive chromatin territories (Fig. 3a). TRIM33 contains PHD domain [G] and Bromo domain [G]. These domains bind to histone marks H3K9me3 and H3K18ac, respectively, to displace a repressor, heterochromatin protein 1 (HP1) 60. How the SMAD2/3-SMAD4 and SMAD2/3-TRIM33 complexes cooperate60, and how SMADs interact with chromatin in different contexts106 are open questions. Additionally, TRIM33 can act as a SMAD4 ubiquitin ligase to limit SMAD transcriptional action107.
Figure 3. SMAD cooperation with different DNA binding partners.
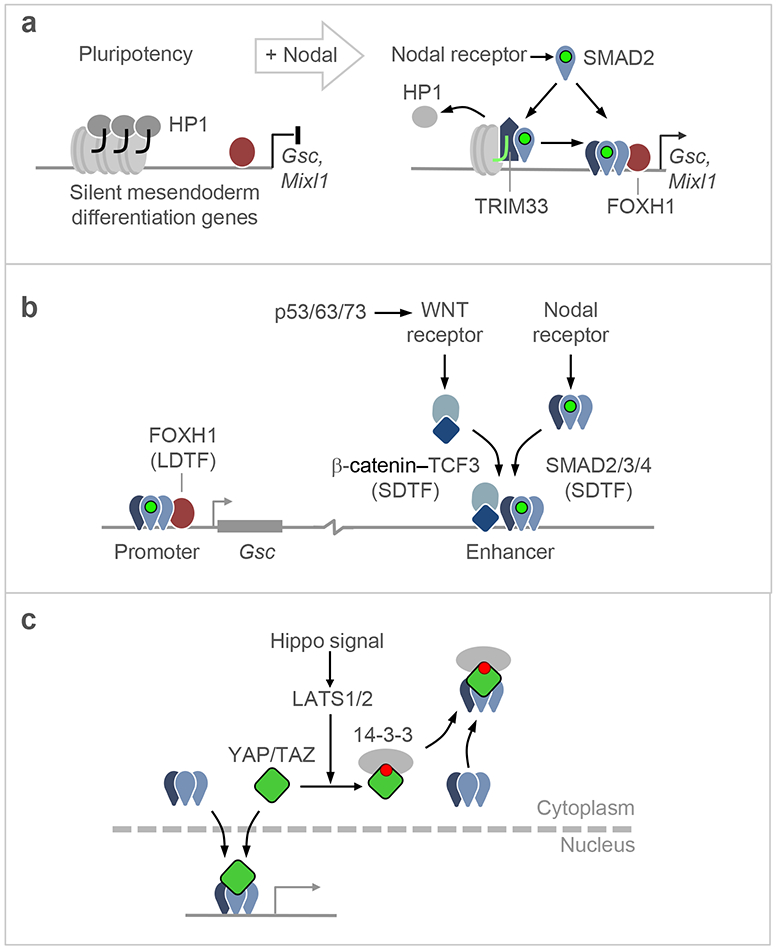
a. Certain gene responses, including the induction of LDTF-encoding genes by Nodal-driven SMAD2/3:SMAD4 in ES cells, require the participation of TRIM33. Through its PHD and Bromo domains, TRIM33 binds to histone H3 N-terminal tails that have repressive chromatin marks (H3K4me0, H3K9me3 and H3K18ac), displacing heterochromatin protein 1 (HP1) from these sites. Nodal-activated SMAD2/3 binds to TRIM33, an interaction that allows the recruitment of SMAD2/3:SMAD4 complexes by FOXH1 to induce gene expression and drive mesendodermal differentiation. TRIM33 can also act as a SMAD4 ubiquitin ligase to limit SMAD transcriptional action (not shown).
b. Interactions of Nodal-activated SMAD2/3:SMAD4 with LDTFs and SDTFs on a target gene. In addition to SMAD2/3:SMAD4 recruitment by FOXH1 to the Gsc promoter, the activation of this gene requires WNT signals. A WNT-driven β-catenin:TCF3 complex and a Nodal-driven SMAD2/3:SMAD4 cooperate in binding to a distal enhancer required for activation of Gsc transcription. The WNT input is delivered by WNT3, whose expression is induced by p53 and its family members p63 and p73. The p53 family, WNT and Nodal are all essential for gastrulation, providing evidence for their involvement in SMAD-driven mesendodermal specification.
c. Representation of YAP-mediated eviction of Nodal-activated SMADs from the nucleus. When YAP is phosphorylated by LATS1/2kinases in response to Hippo pathway activation. The phosphorylation sites act as binding site for 14-3-3 proteins, which drag SMADs into the nucleus and prevents TGF-β signaling.
SMAD-LDTF interplay in committed progenitors
Instances of an LDTF dictating SMAD binding genome-wide, and in turn SMADs driving expression of other LDTFs, are also found in lineage-restricted progenitors (Table 2). Genome-wide analysis of SMAD2/3 binding in these cells helped establish the general principle that LDTFs determine the genome-wide binding landscape of SMADs. The LDTFs MYOD1 in myogenic progenitors and PU.1 in pre-B cells direct TGF-β-activated SMAD3 to co-occupy the genome and trigger the differentiation of these cells31. Similarly, BMP-activated SMAD1 co-occupies the genome with the myeloid LDTF C/EBPα and the erythroid LDTFs GATA1 and GATA2, to drive differentiation of hematopoietic progenitors along these two lineages, respectively92. In hair follicle progenitor cells, BMP-activated SMAD1/5 induce expression of GATA3 for differentiation along the hair-generating lineage108,109.
After partnering with one set of LDTFs in one developmental stage and inducing the expression of LDTFs for transition into the next, SMADs can partner with this second wave of LDTFs to further regulate lineage differentiation (Fig. 2b,c, and Table 2). SMAD-mediated expression of a transcription factor that then serves as a SMAD co-regulator of other genes is called “self-enabling” regulation97. For example, Nodal-activated SMAD2/3 and SMAD4 cooperate with FOXH1 to induce BRA and GSC for mesendodermal differentiation of ES cells. GSC then interacts with activated SMAD2/3 to further regulate mesoderm differentiation, and BRA interacts with BMP-activated SMAD1 to regulate endoderm differentiation110-112. Another FOXH1 target, MIXL1, functions in a similar manner in zebrafish113.
SMAD crosstalk with other stem cell differentiation pathways
In addition to interacting with LDTFs to set genome-wide transcriptional response programs, SMADs engage in extensive crosstalk with SDTFs regulated by different signals. A notable example is provided by the connection between the TGF-β and WNT signaling that occurs in development and regeneration13,15,114. Nodal and WNT signals critically cooperate in the induction of mesendoderm115. Effector transcription factor complexes for each pathway –Nodal-driven SMAD2/3:SMAD4:FOXH1, and WNT-driven β-catenin:TCF3– bind to neighboring sites in mesendoderm LDTFs enhancers116 (Fig. 3b). Combinatorial WNT and BMP signaling, with co-occupancy of hundreds of cis-regulatory DNA elements by SMAD1 and β-catenin-TCF, guides gastrointestinal organogenesis in frogs117. Many other examples of cooperation between the TGF-β and WNT families are known13,15,114.
In mesendoderm development, the SMAD-TCF cooperation additionally integrates inputs from p53 (Fig. 3b). In the adult p53 functions as a tumor suppressor transcription factor that induces cell cycle arrest, senescence, and apoptosis in response to DNA damage. However, p53 is highly expressed during early embryogenesis in frogs and mice, and is required for mesendoderm differentiation of human and mouse ES cells118 and mesendoderm formation in frogs119,120. In ES cells, p53 does not co-bind the genome with SMAD2/3 but directly induces WNT3 expression, which activates TCF for cooperation with Nodal-activated SMADs86. p53 activity is essential for gastrulation in the mouse embryo86. Mammals have two additional family members, p63 and p73, which are functionally redundant with p53 in this context. This explains why p53-null mice develop normally whereas p53-depleted frog embryos do not119,120.
Crosstalk between SMADs and other SDTFs can also be antagonistic. Binding of transcription factor YAP to SMAD2/3 at promoters of mesendoderm specification genes in ES cells establishes both cooperative121,122 and competitive interactions123 (Fig. 3c). In this context, the Crumbs cell polarity complex relays cell density information via the Hippo pathway, inducing the kinase LATS to phosphorylate YAP and the related factor TAZ. Protein 14-3-3 binds to phosphorylated YAP/TAZ and retains these proteins in the cytoplasm. To the extent that YAP/TAZ are bound to SMADs, this process retains SMADs in the cytoplasm and inhibits their transcriptional function123.
Another antagonistic input is provided by the PI3K pathway, which in contrast to p53 inhibits ES cell differentiation by suppressing WNT pathway activation124. Crosstalk between SMADs and other SDTFs occurs in adult stem cells as well. For example, TGF-β-activated SMAD2/3 collaborates with RANK signaling (receptor activator of nuclear factor kB, NFkB) to induce expression of the LDTF NFATC1 (nuclear factor of activated T cells, cytoplasmic 1) for osteoclast differentiation125.
Determinants of TGFβ-induced EMT
Cells in developing embryos undergo multiple rounds of a phenotypic change known as the epithelial-to-mesenchymal transition (EMT) and its reverse, the mesenchymal-to-epithelial transition (MET)126. During EMT, cells lose apicobasal polarity and down-regulate key epithelial cell adhesion molecules such as E-cadherin, resulting in loss of cell contacts with neighboring cells. EMT is driven by a set of transcriptional repressors including SNAIL and ZEB, which are generally united in their ability to inhibit the expression of the cell adhesion molecule E-cadherin126. TGF-β has long been known to be an inducer of EMT in normal and oncogenically transformed cells127,128. TGF-β is among the most potent EMT inducers in epithelial cell cultures from mammary, lung, pancreatic and other tissues126,129,130. During the initiation of cardiac valve formation131, TGF-β2 and -β3 activate expression of SNAIL and the related SLUG to drive this process132. Transient EMT-like processes, including forms called “partial EMT”, are frequently observed in association with TGF-β at the interface between tumors and the surrounding stroma133,134. The biology of EMT and MET and the regulation of these transitions by TGF-β and BMP have been extensively reviewed13,126,129,135. Of relevance here, the induction of EMT by TGF-β requires inputs from other pathways, most prominently the RAS–MAPK pathway136,137. This phenomenon is particularly manifest in the presence of oncogenic RAS signaling, which enables TGF-β–SMAD to strongly induce SNAIL expression38,138,139. The mechanistic basis for the synergy between the TGF-β–SMAD and RAS–MAPK pathways remains unclear.
TGFβ action in immune regulation
Proper function of the immune system requires a balance between immunity and tolerance, which depends on the dynamic regulation of numerous cell fate transitions to produce various lymphocyte subtypes in adequate numbers. TGF-β plays many critical roles in setting this balance, perhaps more so than any other cytokine140. The regulation of phenotypic plasticity in this setting again is mediated in large part by SMAD cooperation with LDTFs.
Multiple functions in regulating lymphocyte pools and their effector functions
TGF-β plays a pleiotropic role in lymphocyte regulation, and this is particularly apparent in T cells18. This role goes beyond controlling cell differentiation into specific lineages (see below for details), and also includes effects on cell survival, proliferation, and effector functions. TGF-β supports the development of multiple T cell lineages in the thymus by promoting survival of progenitor cells141. It also promotes the maintenance of autoreactive CD4+ and CD8+ T cells, which are characterized by high T cell receptor (TCR) signaling and undergo negative selection in the thymus to build immune tolerance. TGF-β modulates this process by inhibiting TCR activity in autoreactive T cells that escape this negative selection18.
In contrast to its role in supporting the maintenance of naïve T cell pools, TGFβ markedly restrains the activity of effector T cells derived from these precursors. TGF-β limits CD4+ T cell proliferation by suppressing autocrine IL-2 production142. In CD8+ cytotoxic T cells (TC cells), TGF-β-activated SMADs cooperate with ATF1 to suppress the expression of interferon-γ and the cytolytic factors Perforin, Granzyme, and Fas ligand143, thus inhibit the anti-tumor activity of these cells. TGF-β carried by T regulatory (Treg) cells as a cell-surface latent complex can exert this effect in direct contacts between Treg cells and CD8+ T cells144. TGF-β also modulates B-cell immunoglobulin class switching96 (Fig. 4), and the proliferation and other effector functions of B cells, natural killer (NK) cells, monocytes, macrophages and granulocytes18.
Figure 4. Transcriptional determinants of TGF-β regulation of immune cell fate.
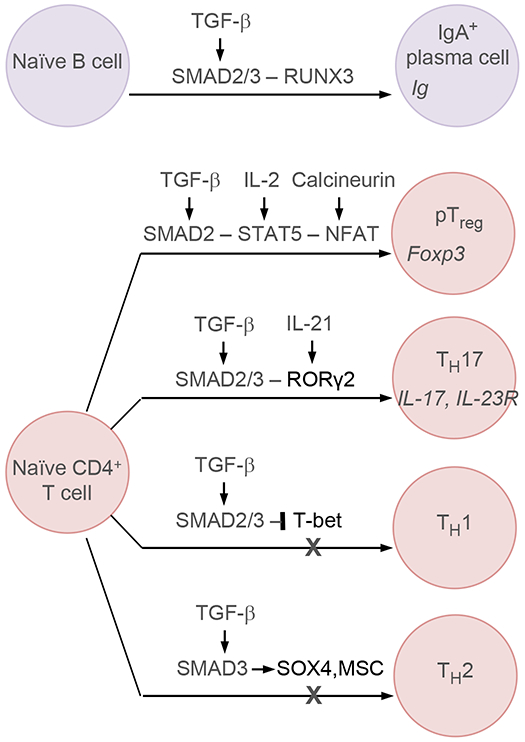
TGF-β-activated SMADs cooperate with RUNX3 to promote class switch recombination during B-cell maturations. TGF-β-activated SMAD2/3 also regulate the specification of different cell subtypes in naïve CD4+ T cells. In this context, TGF-β-activated SMADs in Treg cells cooperate with signal-activated STAT5 and NFAT to induce Foxp3 expression for Treg differentiation. Similarly, TGF-β-activated SMAD2/3 cooperate with RORγ2 to induce a TH17 phenotype. In contrast, TGF-β-activated SMAD2/3 inhibit the specification of TH1 cell fate by repressing T-bet transcription factor, and the specification of TH2 cell fate by inducing the expression of SOX4 and MSC.
SMAD-LDTF cooperation in the regulation of T cell subtype specification
TGF-β promotes the differentiation of naïve CD4+ T cells into Treg cells, a key mediator of immune tolerance145. Treg cells develop from naïve CD4+ T cells both in the thymus and the periphery. The LDTF FOXP3 is a major determinant of the Treg state146, and its induction by TGF-β in CD4+ T cells promotes Treg differentiation147. In Treg cells that develop in the periphery (pTreg) this regulation involves SMAD3 binding to a Foxp3 enhancer in cooperation with IL-2-activated STAT5 and calcineurin-activated NFAT148, providing evidence for the importance of cooperation with other SDTFs in TGF-β-family-mediated regulation of cell fate (Fig. 4). A more recent examination of TGF-β control of Treg differentiation implicated an expanded repertoire of SMAD-regulated LDTFs, including BATF3, FOSL2 and Musculin (MSC), in this process149. In Treg cells that develop in the thymus, TGF-β may be dispensable for FOXP3 induction but contributes to Treg formation by preventing negative selection of these cells (see also above)150.
TGF-β can promote adoption of an alternative fate by naïve CD4+ cells, the T helper (TH) 17 cell fate151 (Fig. 4). TH17 cells are a subtype of pro-inflammatory T helper cells that express IL-17 and function as mediators of mucosal immunity against pathogens. TH17 cells are specified by RORγ2 in mice (retinoic acid receptor-related receptor gamma 2; RORC in human)152. RORγ2 expression is induced by IL-21153, and TGF-β-activated SMAD2 cooperates with RORγt to drive TH17 differentiation154.
TGF-β additionally favors Treg differentiation by restricting the specification of TH1 and TH2 cells (Fig. 4), which produce cytokines that support the growth and functions of TC cells, B cells, and macrophages. TH1 lineage specification occurs through T-bet transcription factor, and TGF-β represses T-bet expression155. In the TH2 lineage, TGF-β-activated SMAD3 induces expression of MSC and SOX4, which prevent the expression of TH2 specific cytokines149,156. It remains to be seen whether SMADs directly contribute to these repression events by binding to loci of these LDTFs. In any case, these instances add to the examples of regulation of crucial cell identity transcription factors by TGF-β signaling, including cases in which TGF-β cooperates with, and others in which it inhibits LDTF functions.
Determinants of TGFβ action in cancer
If TGF-β drives cell lineage specification and differentiation, and cancer is a disease of failed cell differentiation, it should come as no surprise that TGF-β is important in tumor progression.
TGF-β is famously dual-natured, functioning as both a tumor suppressor in premalignant cells and a tumor promoter in overtly malignant ones9. The suppressive pressure that TGF-β exerts on premalignant cell populations selects for cancer cell clones with an inactivated or subverted SMAD pathway. By avoiding the tumor suppressive effects of TGF-β these clones can use other effects of the cytokine for invasiveness and immune evasion, thus turning TGF-β from inhibitory impediment into stimulus for metastasis.
Much is known about the incidence of TGF-β pathway mutations in human cancer, the tumorigenic effects of TGF-β pathway knockouts in mice, the effects of TGF-β on cancer cells lines, and other related aspects. However, this work has remained largely disconnected from the question of whether and how the roles of TGF-β as a tumor suppressor or a metastasis promoter are related to its normal role as a regulator of developmental fate. Recent insights into the interplay between SMADs, LDTFs and cell differentiation during tumorigenesis provided some answers to these questions. Though largely confined to gastrointestinal cancers, these insights have implications for the broader role of TGF-β in cancer.
TGF-β tumor suppressive effects
The tumor suppressive capacity of TGF-β became apparent with the identification of inactivating mutations in TGFBR2 in colorectal cancer157 and SMAD4 in pancreatic ductal adenocarcinoma (PDA)158,159. These gastrointestinal tissues with the highest incidence of TGF-β pathway mutations are derived from endoderm160, suggesting that the shared sensitivity to TGF-β has a developmental underpinning. A loss of TGF-β signaling components is seldom sufficient for tumor initiation but allows cells harboring oncogenic mutations to progress to invasive carcinoma36,37,161. Thus, TGF-β plays its tumor suppressive role mostly by blocking the transition of premalignant cells to a more overtly malignant phenotype.
In principle, TGF-β could suppress tumor growth by inhibiting cancer cell proliferation. In normal epithelial and hematopoietic cells, TGF-β is an archetypical growth inhibitor that reversibly halts cell cycle progression through increased expression of the CDK inhibitors p15INK4, p21CIP1, p27KIP1 and/or p57KIP2 162-165. However, oncogenically transformed cells frequently harbor strong CDK activating signals, which diminishes the effectiveness of these CDK inhibitors.
If the cytostatic activity of TGF-β is not an effective tumor suppressive mechanism, the ability of TGF-β to induce apoptosis in premalignant cells certainly is. Oncogenic mutations increase the sensitivity of premalignant cells to TGF-β-induced apoptosis, as documented in several genetically engineered mouse models of cancer. TGF-β-SMAD signaling induces apoptosis In pancreatic premalignant progenitors that contain KRAS oncogenic mutations36. Mouse skin and mucosal epithelia with conditional deletion of TGFBR2 are mildly hyperplasic, whereas epithelia harboring HRAS mutations show hyperproliferation that is offset by TGF-β-dependent apoptosis; if TGFBR2 is deleted in this HRAS-mutant context, apoptosis is limited and tumors emerge37 (Fig. 5a). Mutant RAS [G] is not the only oncogenic driver that turns TGF-β into a tumor suppressive signal. In mouse prostate epithelium TGF-β–SMAD signaling inhibits tumor progression when the tumor suppressor PTEN [G] suffers inactivation166,167.
Figure 5. Determinants of TGF-β tumor suppression and its subversion in cancer.
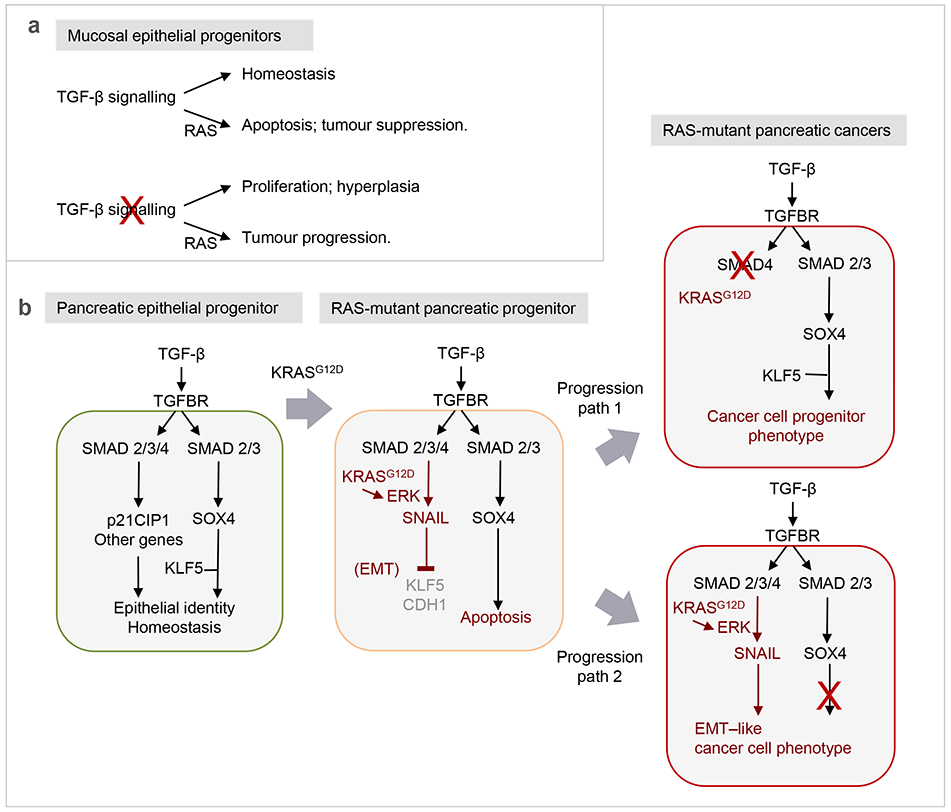
a. In mouse skin and mucosal epithelia, loss of TGF-β receptor signaling leads to hyperplasia, whereas gain of oncogenic HRAS mutations leads to TGF-β-associated apoptosis. Loss of TGF-β signaling in a HRAS-mutant background leads to the emergence of tumors. Thus, TGF-β enforces homeostasis in the wild type background, and tumor suppression in a RAS-mutant background.
b. The switch of TGF-β from a signal for lineage maintenance and homeostasis to a signal for apoptosis that cancer cells must disable, as observed in pancreatic epithelial progenitors. In normal progenitors TGF-β signaling through SMAD2/3 with SMAD4 activates the expression of CDK inhibitors, SOX4, and other genes. CDK inhibitors tone down cell proliferation, whereas SOX4 associates with KLF5 to co-occupy the genome and enforce epithelial progenitor identity. Stimulation of SOX4 expression requires SMAD2/3 but not SMAD4. In premalignant pancreatic progenitors harboring oncogenic KRAS mutations, a hyperactive ERK MAPK pathway enables SMAD2/3:SMAD4 to strongly induce the expression of the EMT master regulator SNAIL. SNAIL represses KLF5 expression. In the absence of KLF5, SOX4 induces pro-apoptotic genes BIM and BMF for elimination of the cell. Nearly one half of human pancreatic ductal adenocarcinomas (PDA) progress through this bottleneck by selecting for clones that harbor SMAD4 inactivating mutations. Loss of SMAD4 does still allows TGF-β to support SOX4 expression and the SOX4:KLF5 mediated pancreatic progenitor state. The other half of PDA tumors progress with a functionally intact TGF-β signaling system but accumulate unknown alterations that prevent the pathway from triggering apoptosis.
TGF-β tumor suppression through altered regulation of LDTFs
How does TGF-β become a potent inducer of apoptosis? TGF-β receptors are not directly coupled of apoptosis activating pathways. The induction of apoptosis by TGF-β in susceptible cell cultures is SMAD dependent and occurs days after TGF-β addition, arguing that it is a secondary response. Recent work on premalignant pancreatic progenitors harboring mutant KRAS revealed that TGFβ induces apoptosis as a result of an altered regulation of the LDTFs KLF5 and SOX4 38 (Fig. 5b). KLF5 (Krueppel-like factor 5) is essential for proliferation, survival and maintenance of progenitor cells in the intestinal mucosa168 prior to their differentiation into KLF5-negative progeny. KLF5 enforces the epithelial state in intestinal and pancreatic progenitors by up-regulating specific cytokeratins169, E-cadherin38, and the pro-epithelial microRNA miR-200 170. In pancreatic progenitors, KLF5 co-binds the genome with SOX4 to enforce epithelial progenitor identity, and TGF-β supports this function by increasing SOX4 expression38. The role of KLF5 as an enforcer of the epithelial progenitor state also applies to carcinoma cells. KLF5 is highly expressed in gastrointestinal cancers and implicated in pancreatic 38,169 and gastric tumorigenesis171.
Strong MAPK signaling in RAS-mutant pancreatic, mammary and keratinocyte progenitors turns TGF-β into an inducer of EMT 38,128,137. in premalignant pancreatic cells, KRAS–MAPK signaling enables TGF-β-activated SMADs to induce the transcriptional repressor SNAIL, which represses KLF5 expression38. While TGF-β–SMAD signaling down-regulates KLF5 to impose a mesenchymal phenotype, it still enforces the expression of the pro-epithelial SOX4 (Fig. 5b). On being left without KLF5 as its genome-binding partner, SOX4 activates a different set of genes including the pro-apoptotic BIM and BMF, causing cell death. Thus, TGF-β–SMAD signaling in KRAS-mutant pancreatic progenitors delivers conflicting pro- and anti-EMT commands, which triggers apoptosis –a process termed lethal EMT38.
Given that KLF5 is an essential component of epithelial progenitor cells in the gastrointestinal tract, an EMT involving its repression would oppose the genesis of tumors that depend on KLF5. In line with this, using a bioinformatics scoring system for quantification of EMT-like characteristics in human tumors, colorectal, gastric and pancreatic cancers are consistently among the lowest EMT scores of all tumor types172. A role of TGFβ-induced EMT in eliminating neoplastic cells is also suggested by observations in other cell types. TGF-β-induced EMT precedes apoptosis in murine mammary epithelial cells173.
Pro-metastatic subversion of SMAD transcriptional programs
Much evidence shows that EMT can support tumor invasion and metastasis126,134,135,174. The selective pressure imposed by TGF-β results in the accumulation of clones that can undergo a TGF-β-induced EMT without its apoptotic effects137,175 (Figs. 5b, 6). Mutational inactivation of TGF-β pathway components allows KRAS-mutant pancreatic cells to avoid the lethal rearrangement of core transcriptional networks that occur with EMT. SMAD4 is mutationally inactivated in half of PDA cases in the clinic, preventing TGF-β from inducing SNAIL expression (a SMAD4 dependent effect) but still allowing TGF-β to induce SOX4 expression (a SMAD4-independent effect) and supporting tumor progression via SOX4 with KLF538.
Figure 6.
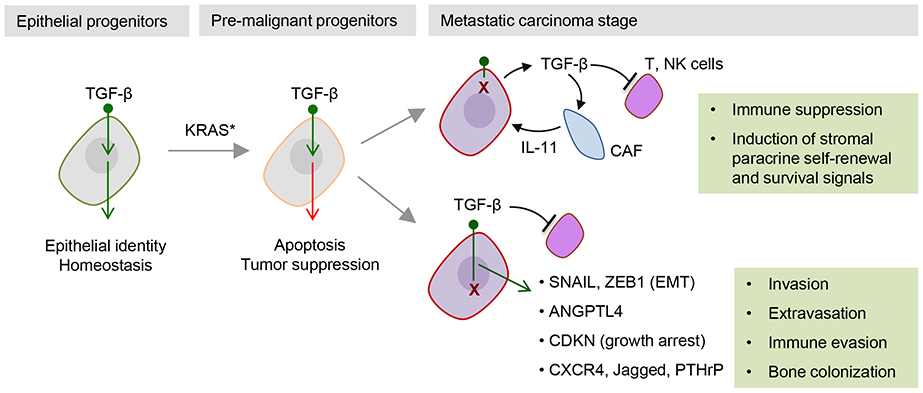
Cancer cells can traverse the tumor suppressive TGF-β bottleneck by accumulating mutations (“X”) that disable the signal transduction pathway. In many gastrointestinal cancers, TGF-β receptor and SMAD mutations deeply disable the pathway, and this now allows stromal TGF-β to exert pro-tumorigenic effects including immune suppressive action on T and NK cells and activation of stromal paracrine loops such as the production of tumorigenic cytokines (e.g. IL-11) by cancer-associated fibroblasts (CAF). In many other types of cancer, the TGF-β receptors and SMADs remain intact but downstream alterations prevent the pathway from triggering tumor suppressive effects. Thus corrupted, the TGF-β–SMAD pathway can drive a non-lethal EMT as well the production of factors that promote metastatic extravasation, immune evasion, and colonization of target tissues.
Even in a tumor type with this high propensity to accumulate TGF-β pathway mutations, one half of all PDA cases retain a functional TGF-β pathway, indicating that these tumors can successfully circumvent the lethality associated with a TGF-β-induced EMT (Fig. 5b). It is unclear what allows epithelial-mesenchymal plasticity to emerge in populations that are, in bulk, dependent on epithelial transcriptional programs. One candidate is PI3K/AKT signaling, which is activated by growth factors of the tumor stroma and can inhibits apoptosis by induction of the anti-apoptotic protein BCL-XL38. The existence of a TGF-β-imposed bottleneck from which resistant cell clones can eventually emerge has also been documented in a mouse mammary epithelial cells, in which TGF-β induced EMT-like changes followed by widespread cell death and, after prolonged exposure (>2 weeks), the emergence of mesenchymal cells resistant to TGF-β induced apoptosis173.
Malignant cells that evade TGF-β tumor suppression and still keep a functional TGF-β pathway can use this pathway to advantage, not only to profit from EMT but also from other SMAD-mediated effects (Fig. 6). Examples of these effects include the production of the autocrine stem cell mitogens LIF and PDGF in glioblastoma176, expression of angiopoietin-like 4 as a mediator of infiltration of circulating breast cancer cells into the lungs177, expression of osteoclast stimulating genes (PTHrP, CXCR4, IL-11, CTGF and JAGGED1) for bone colonization by breast cancer cells39,40,178, and other pro-metastatic effects179. Moreover, TGF-β contributes to the entry of disseminated cancer stem cells into anon-proliferative state that evades surveillance and elimination of metastatic seeds by NK cells180.
Even those tumors that have a fully disabled TGF-β pathway can still benefit from TGF-β through its effects on the tumor microenvironment (Fig. 6). Colorectal carcinomas (CRC) frequently sustain mutational inactivation of the TGF-β pathway157 yet show high TGF-β levels and TGF-β signaling activity in mesenchymal and immune components of the tumor stroma, in association with a high risk of relapse181. In mouse models of CRC, tumor-derived TGF-β stimulates the production of IL-11 in cancer-associated fibroblasts [G] (CAFs), which in turn supports the survival of CRC metastasis-initiating cells in the liver. Treatment of these mice with TGFBR1 inhibitory drugs suppressed metastasis182. Considering the strong capacity of TGF-β to suppress immune surveillance, TGF-β can likely support tumor progression by protecting malignant cells from immune-mediated clearance. The possibility of combining immunotherapy with TGF-β blockade is under active consideration144,183.
Conclusions and perspectives
The present analysis has focused on the partnership of ligand-activated SMADs with LDTFs and other SDTF for three reasons. First, these interactions are at the core of some of the most important functions of the TGF-β family in development, immunity and cancer. Second, the elucidation of these interactions illuminates the molecular basis for the context-dependent nature of TGF-β action and provides a blueprint for the dissection of SMADs transcriptional mechanisms in other developmental and disease contexts. And third, knowledge on how cells read TGF-β signals will inform efforts to pharmacologically target these factors in diseases that involve malfunctions of the TGF-β pathway, such as fibrosis, inflammation, autoimmunity, and cancer.
Based on this blueprint, future work may solve long-standing questions such as: how the pleiotropic effects of TGF-β family members during embryo development or of TGF-β itself in immune homeostasis are kept in balance; how the persistent TGF-β signaling in fibrosis originates and how could be selectively blocked; how oncogenic signals turn TGF-β into an EMT inducer; how cancer cells avert the tumor suppressive lethality associated with this and other effects; and, what role TGF-β plays in immune evasion of cancer cells and in limiting the effectiveness of immunotherapy.
SMADs transcriptional mechanisms are at the core of many TGF-β family responses, and so the biochemical processes that regulate to SMAD activation are also of crucial importance. The components of the pathway are identified and well characterized, but their combinatorial interplay is rather incompletely understood. There is a growing appreciation for the question of how a microenvironments that contains various TGF-β family members, cognate receptors, regulators, and coexisting signals is translated by the cell into SMAD activation pulses of defined intensity, duration, and shape. These variables are likely critical in cells that must choose between several SMAD-dependent fates during rapidly proceeding developmental processes. The interplay between ligands, receptors, SMAD partners, and the chromatin, analysed at a systems level, merits additional research.
This knowledge is also necessary for work at a more applied level. Advances on the structural basis for ligand activation and receptor binding of TGF-β family members are currently empowering the development of compounds that target ligand-receptor interactions in order to treat diseases that result from excessive or defective activity of a particular TGF-β family member. Achieving selectivity in targeting modulators of such pleiotropic factors is key.
Supplementary Material
Supplementary Figure 1. Mutational inactivation of TGF-β–SMAD tumor suppression
cBioPortal query 196 of available TCGA datasets for non-synonymous mutations and deep deletions in TGFBR1, TGFBR2, SMAD2, SMAD3 and SMAD4. a. Frequency of pathway mutations in different types of cancer. All studies included at least 30 samples. b. Frequency of non-synonymous mutations and deep deletions in TGFBR1, TGFBR2, SMAD2, SMAD3 and SMAD4 in individual human pancreatic tumors. SMAD4 is at chromosome 18q21.2 and SMAD2 is located closely at 18q21.1. The two are frequently co-deleted. However, SMAD4 is more commonly deleted than SMAD2, and SMAD2 deletions do not occur without SMAD4 deletions. This and the presence of truncating mutations in SMAD4, suggests that a unique function of SMAD4 is selected against in tumors.
Box 2. TGF-β–MAD pathway deregulation in cancer.
The central components of the TGF-β pathway display a high-degree of lineage specificity in the distribution of loss-of-function mutations (Suppl Fig. 1a). Overall, SMAD4 is the most frequently altered gene in the pathway193 across all tumor types and in the particular case of pancreatic cancer (Suppl. Fig. 1b). Missense mutations frequently affect residues in the SMAD2/3:SMAD4 trimer interface26,159. There are multiple possible explanations for why SMAD4 loss, rather than TGF-β receptor loss, is the preferred route to escape the tumor suppressive effects of the pathway. One possibility is that, because SMAD4 also functions downstream of Activin and BMP, its loss may simultaneously protect from tumor suppressive effects of multiple TGF-β superfamily cytokines. Alternatively, SMAD4 loss may preserve partial TGF-β receptor signaling, allowing SMAD4-independent responses such as SOX4 expression that increase cancer cell fitness38. In line with this possibility, high levels of TGF-β and its receptors in PDA cells are clinically associated with decreased survival194,195. The tumor suppressive action of TGF-β may be more widespread than the frequency of these mutations suggests, as it may be lost without mutational inactivation of the pathway. Indeed, even in human PDA, the tumor type with the highest proportion of TGF-β pathway mutations, half of cases in the clinic escape tumor suppression with the core pathway intact.
Key points.
The TGFβ family of cytokines plays myriad roles throughout metazoan biology, and is characterized by highly cell type specific responses to pathway activation. One of the most prevalent roles of TGFβ signaling is the control cell identity in a variety of different contexts, which underlies some of the family’s most important biological effects.
TGFβ signals through cell surface receptors that directly activate SMAD transcription factors. These factors then enter the nucleus and cooperate with additional transcription factors to control gene expression.
Context-specific effects of TGFβ family members occur through the influence of highly expressed lineage-determining transcription factors (LDTFs) on SMAD genome-wide binding. In turn, the TGFβ family orchestrates cell identity changes by controlling the expression of key LDTFs.
SMAD control of key identity transcription factors during cellular differentiation is highly cooperative, requiring critical crosstalk with additional signaling pathways and LDTFs.
These themes play out in a wide variety of contexts. Well-studied examples from early embryonic development and in the control of the adult immune system are highlighted here.
The powerful ability of SMAD signaling to affect cell identity change mediates the pathway’s tumor suppressive effects. This again hinges on regulation of key LDTFs, and its disruption turns TGFβ from a tumor suppressor to an instigator of metastasis.
Glossary terms
- Signal-driven transcription factors.
Transcription factors that are often ubiquitously expressed, but are only switched on in response to activation of a given signaling pathway.
- Anti-Muellerian Hormone.
A developmentally restricted TGFβ superfamily member expressed at high levels in male Sertoli cells to guide testicular development, and to a lesser extent in female granulosa cells where it regulates follicle development.
- Chromatin reader.
A protein containing one or more domains that interact specifically with modified histones, providing readout of and often enforcing the transcriptional status (active vs. silent) of nearby genes.
- PHD domain.
A 50-80 amino acid, Cys4-His-Cys3-containing domain found in some chromatin readers, usually displaying a binding preference for lysine-methylated histone tails.
- Bromo domain.
An approximately 100 amino acid domain frequently found in chromatin readers, displaying a preference for acetylated lysine residues.
- Ras.
A family of small GTPases involved in signal transduction activated in response to a number of extracellular signals, often functioning as oncogenes upon constitutive activation by point mutations.
- PTEN.
A tumor suppressor that restrains activity of the PI3K/AKT pathway through dephosphorylation of phosphatidylinositol (3,4,5)-trisphosphate.
- Cancer-associated fibroblasts (CAFs).
Fibroblasts present in the tumor stroma that play an important role in promoting tumor growth, invasion, and metastasis.
- Neddylation.
A process analogous to ubiquitylation in which the ubiquitin-like protein NEDD8 is conjugated to target proteins.
Biography
Charles J. David is an Assistant Professor at the Tsinghua University School of Medicine in Beijing, China. Dr. David completed his Ph.D. training at Columbia University in the laboratory of James L. Manley and his postdoctoral training at Memorial Sloan Kettering Cancer Center in the laboratory of Joan Massagué.
Joan Massagué received his Ph.D. from the University of Barcelona in 1978. A member of the Sloan-Kettering Institute since 1989, he is currently its Director. His laboratory studies stem cell regulation and cancer metastasis. Contributions include the delineation of the TGF-β pathway and the principles of metastatic latency and outbreak.
Citations
- 1.Massagué J How cells read TGF-beta signals. Nat Rev Mol Cell Biol 1, 169–178 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Josso N, Belville C, di Clemente N & Picard JY AMH and AMH receptor defects in persistent Mullerian duct syndrome. Hum Reprod Update 11, 351–356 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lindsay ME & Dietz HC The genetic basis of aortic aneurysm. Cold Spring Harb Perspect Med 4, a015909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matzuk MM & Burns KH Genetics of mammalian reproduction: modeling the end of the germline. Annu Rev Physiol 74, 503–528 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Meng XM, Nikolic-Paterson DJ & Lan HY TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 12, 325–338 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Salazar VS, Gamer LW & Rosen V BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol 12, 203–221 (2016). [DOI] [PubMed] [Google Scholar]
- 7.van der Kraan PM The changing role of TGFbeta in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol 13, 155–163 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Cai J, Pardali E, Sanchez-Duffhues G & ten Dijke P BMP signaling in vascular diseases. FEBS Lett 586, 1993–2002 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Massagué J TGFbeta in cancer. Cell 134, 215–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickup M, Novitskiy S & Moses HL The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer 13, 788–799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauriello DVF & Batlle E Targeting the Microenvironment in Advanced Colorectal Cancer. Trends Cancer 2, 495–504 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Wakefield LM & Hill CS Beyond TGFbeta: roles of other TGFbeta superfamily members in cancer. Nat Rev Cancer 13, 328–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massagué J TGFbeta signalling in context. Nat Rev Mol Cell Biol 13, 616–630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullen AC & Wrana JL TGF-beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb Perspect Biol 9 (2017).This paper highlighted the tendency of Smads to bind the genome in close association with lineage-specific master transcription factor.
- 15.Oshimori N & Fuchs E The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell 11, 751–764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauklin S & Vallier L Activin/Nodal signalling in stem cells. Development 142, 607–619 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Chen W & Ten Dijke P Immunoregulation by members of the TGFbeta superfamily. Nat Rev Immunol 16, 723–740 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sanjabi S, Oh SA & Li MO Regulation of the Immune Response by TGF-beta: From Conception to Autoimmunity and Infection. Cold Spring Harb Perspect Biol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y & Massagué J Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Wrana JL et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Wrana JL, Attisano L, Wieser R, Ventura F & Massagué J Mechanism of activation of the TGF-beta receptor. Nature 370, 341–347 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Inman GJ, Nicolas FJ & Hill CS Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell 10, 283–294 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Kang Y, Col S & Massague J Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol Cell 10, 271–282 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Martin-Malpartida P et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nature Communications (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinck AP, Mueller TD & Springer TA Structural Biology and Evolution of the TGF-beta Family. Cold Spring Harb Perspect Biol 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macias MJ, Martin-Malpartida P & Massagué J Structural determinants of Smad function in TGF-beta signaling. Trends Biochem Sci 40, 296–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolan K & Thompson TB The DAN family: modulators of TGF-beta signaling and beyond. Protein Sci 23, 999–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata A & Chen YG TGF-beta Signaling from Receptors to Smads. Cold Spring Harb Perspect Biol 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamouille S & Derynck R Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-beta-induced epithelial-mesenchymal transition. Cells Tissues Organs 193, 8–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heldin CH & Moustakas A Signaling Receptors for TGF-beta Family Members. Cold Spring Harb Perspect Biol 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen AC et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576 (2011).This paper highlighted the tendency of Smads to bind the genome in close association with lineage-specific master transcription factor.
- 32.Sun LT et al. Nanog co-regulated by Nodal/Smad2 and Oct4 is required for pluripotency in developing mouse epiblast. Dev Biol 392, 182–192 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wang W et al. Smad2 and Smad3 Regulate Chondrocyte Proliferation and Differentiation in the Growth Plate. PLoS Genet 12, e1006352 (2016).This work highlights the role of p53 in promoting ME differentiation by coordinating crosstalk between the Wnt and Nodal/activin pathways.
- 34.Seoane J, Le HV, Shen L, Anderson SA & Massagué J Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Chen CR, Kang Y, Siegel PM & Massague J E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110, 19–32 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Bardeesy N et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev 20, 3130–3146 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guasch G et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12, 313–327 (2007).The authors carefully dissected the in vivo effects of Tgfbr2 in normal and neoplastic skin, demonstrating the key role of TGF-β-induced apoptosis in tumor suppression.
- 38.David CJ et al. TGF-beta tumor suppression through a lethal EMT. Cell 164, 1015–1030 (2016).Demonstration that in PDA cells, a TGF-β-induced EMT can result in disruption of an essential lineage-specific transcriptional network, leading to apoptosis.
- 39.Kang Y et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A 102, 13909–13914(2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethi N, Dai X, Winter CG & Kang Y Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19, 192–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bisgrove BW, Su YC & Yost HJ Maternal Gdf3 is an obligatory cofactor in Nodal signaling for embryonic axis formation in zebrafish. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montague TG & Schier AF Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelliccia JL, Jindal GA & Burdine RD Gdf3 is required for robust Nodal signaling during germ layer formation and left-right patterning. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brazil DP, Church RH, Surae S, Godson C & Martin F BMP signalling: agony and antagony in the family. Trends Cell Biol 25, 249–264 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Wang RN et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 1, 87–105 (2014).This work highlights the role of p53 in promoting ME differentiation by coordinating crosstalk between the Wnt and Nodal/activin pathways.
- 46.Cheifetz S et al. The transforming growth factor-beta system, a complex pattern of cross-reactive ligands and receptors. Cell 48, 409–415 (1987). [DOI] [PubMed] [Google Scholar]
- 47.Ling N et al. Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature 321, 779–782 (1986). [DOI] [PubMed] [Google Scholar]
- 48.Little SC & Mullins MC Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11, 637–643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimmi O, Umulis D, Othmer H & O'Connor MB Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120, 873–886 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozdamar B et al. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307, 1603–1609 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Kashima R et al. Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of fragile X syndrome. Sci Signal 9, ra58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee-Hoeflich ST et al. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J 23, 4792–4801 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiung F, Ramirez-Weber FA, Iwaki DD & Kornberg TB Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature 437, 560–563 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Kicheva A et al. Kinetics of morphogen gradient formation. Science 315, 521–525 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Keller B et al. Interaction of TGFbeta and BMP signaling pathways during chondrogenesis. PLoS One 6, e16421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klammert U et al. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol 13, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piscione TD et al. BMP-2 and OP-1 exert direct and opposite effects on renal branching morphogenesis. Am J Physiol 273, F961–975 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Ying Y, Qi X & Zhao GQ Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc Natl Acad Sci U S A 98, 7858–7862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Antebi YE et al. Combinatorial Signal Perception in the BMP Pathway. Cell 170, 1184–1196 e1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xi Q et al. A poised chromatin platform for TGF-beta access to master regulators. Cell 147, 1511–1524 (2011).The authors demonstrate that early developmental master regulator genes are accessed by Smads with the assistance of a Trim33, a chromatin reader that recognizes silencing-associated histone marks.
- 61.Feng XH, Zhang Y, Wu RY & Derynck R The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev 12, 2153–2163 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janknecht R, Wells NJ & Hunter T TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev 12, 2114–2119 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pouponnot C, Jayaraman L & Massague J Physical and functional interaction of SMADs and p300/CBP.J Biol Chem 273, 22865–22868 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Wotton D, Lo RS, Lee S & Massagué J A Smad transcriptional corepressor. Cell 97, 29–39 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Kretzschmar M, Doody J & Massague J Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature 389, 618–622 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Pera EM, Ikeda A, Eivers E & De Robertis EM Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev 17, 3023–3028 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH & Massague J Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25, 441–454 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Alarcon C et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139, 757–769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuura I et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430, 226–231 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Tsukamoto S et al. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci Rep 4, 7596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiater E & Vale W Roles of activin family in pancreatic development and homeostasis. Mol Cell Endocrinol 359, 23–29 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Gao S et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 36, 457–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aragon E et al. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev 25, 1275–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sapkota G et al. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J Biol Chem 281, 40412–40419 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Lonn P et al. PARP-1 attenuates Smad-mediated transcription. Mol Cell 40, 521–532 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Roberts AB et al. Type beta transforming growth factor: a bifunctional regulator of cellular growth. Proc Natl Acad Sci U S A 82, 119–123 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tucker RF, Shipley GD, Moses HL & Holley RW Growth inhibitor from BSC-1 cells closely related to platelet type beta transforming growth factor. Science 226, 705–707 (1984). [DOI] [PubMed] [Google Scholar]
- 78.Ohta M, Greenberger JS, Anklesaria P, Bassols A & Massagué J Two forms of transforming growth factor-beta distinguished by multipotential haematopoietic progenitor cells. Nature 329, 539–541 (1987). [DOI] [PubMed] [Google Scholar]
- 79.Sporn MB et al. Polypeptide transforming growth factors isolated from bovine sources and used for wound healing in vivo. Science 219, 1329–1331 (1983). [DOI] [PubMed] [Google Scholar]
- 80.Zawel L et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1, 611–617 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Shi Y et al. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 94, 585–594 (1998). [DOI] [PubMed] [Google Scholar]
- 82.BabuRajendran N et al. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res 38, 3477–3488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kusanagi K et al. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol Biol Cell 11, 555–565 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Labbe E, Silvestri C, Hoodless PA, Wrana JL & Attisano L Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2, 109–120 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Morikawa M et al. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res 39, 8712–8727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Q et al. The p53 family coordinates Wnt and nodal Inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20, 70–86 (2017).This work highlights the role of p53 in promoting ME differentiation by coordinating crosstalk between the Wnt and Nodal/activin pathways.
- 87.Yoon SJ, Foley JW & Baker JC HEB associates with PRC2 and SMAD2/3 to regulate developmental fates. Nat Commun 6, 6546 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Zhang DX & Glass CK Towards an understanding of cell-specific functions of signal-dependent transcription factors.J Mol Endocrinol 51, T37–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen X, Rubock MJ & Whitman M A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383, 691–696 (1996). [DOI] [PubMed] [Google Scholar]
- 90.Chen X et al. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389, 85–89 (1997). [DOI] [PubMed] [Google Scholar]
- 91.Hata A et al. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100, 229–240 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Trompouki E et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Attisano L & Wrana JL Signal integration in TGF-beta, WNT, and Hippo pathways. F1000Prime Rep 5, 17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo X & Wang XF Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res 19, 71–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qing J, Zhang Y & Derynck R Structural and functional characterization of the transforming growth factor-beta -induced Smad3/c-Jun transcriptional cooperativity.J Biol Chem 275, 38802–38812 (2000). [DOI] [PubMed] [Google Scholar]
- 96.Park SR, Seo GY, Choi AJ, Stavnezer J & Kim PH Analysis of transforming growth factor-beta1-induced Ig germ-line gamma2b transcription and its implication for IgA isotype switching. Eur J Immunol 35, 946–956 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Kang Y, Chen CR & Massagué J A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell 11, 915–926 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Hnisz D et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).This work showed that lineage-defining super-enhancers are constructed through the cooperative actions of LDTFs and SDTFs.
- 99.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hnisz D et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58, 362–370 (2015).This work showed that lineage-defining super-enhancers are constructed through the cooperative actions of LDTFs and SDTFs.
- 101.Loose M & Patient R A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol 271, 467–478 (2004). [DOI] [PubMed] [Google Scholar]
- 102.Gaarenstroom T & Hill CS TGF-beta signaling to chromatin: how Smads regulate transcription during self-renewal and differentiation. Semin Cell Dev Biol 32, 107–118 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Charney RM et al. Foxh1 Occupies cis-Regulatory Modules Prior to Dynamic Transcription Factor Interactions Controlling the Mesendoderm Gene Program. Dev Cell 40, 595–607 e594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim SW et al. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357, 492–504 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Nelson AC et al. In Vivo Regulation of the Zebrafish Endoderm Progenitor Niche by T-Box Transcription Factors. Cell Rep 19, 2782–2795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coda DM et al. Distinct modes of SMAD2 chromatin binding and remodeling shape the transcriptional response to NODAL/Activin signaling. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dupont S et al. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121, 87–99 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Genander M et al. BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15, 619–633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hwang M, Mehrani T, Millar SE & Morasso MI Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development 135, 3149–3159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Faial T et al. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development 142, 2121–2135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Izzi L et al. Foxh1 recruits Gsc to negatively regulate Mixl1 expression during early mouse development. EMBO J 26, 3132–3143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teo AK et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25, 238–250 (2011).Demonstration that the early ME master regulator Eomes dictates Smad genome-wide binding during ES cell differentiation, an early demonstration of the concept further elucidated by Mullen et al.
- 113.Germain S, Howell M, Esslemont GM & Hill CS Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14, 435–451 (2000). [PMC free article] [PubMed] [Google Scholar]
- 114.Sancho E, Batlle E & Clevers H Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol 20, 695–723 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Watabe T et al. Molecular mechanisms of Spemann's organizer formation: conserved growth factor synergy between Xenopus and mouse. Genes Dev 9, 3038–3050 (1995). [DOI] [PubMed] [Google Scholar]
- 116.Reid CD, Zhang Y, Sheets MD & Kessler DS Transcriptional integration of Wnt and Nodal pathways in establishment of the Spemann organizer. Dev Biol 368, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stevens ML et al. Genomic integration of Wnt/beta-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs. Development 144, 1283–1295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shigeta M et al. Maintenance of pluripotency in mouse ES cells without Trp53. Sci Rep 3, 2944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cordenonsi M et al. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113, 301–314 (2003). [DOI] [PubMed] [Google Scholar]
- 120.Wallingford JB, Seufert DW, Virta VC & Vize PD p53 activity is essential for normal development in Xenopus. Curr Biol 7, 747–757 (1997). [DOI] [PubMed] [Google Scholar]
- 121.Beyer TA et al. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5, 1611–1624 (2013).These authors demonstrate important crosstalk between Hippo and TGF-β–Smad signaling in determining the transition from self-renewal into ME differentiation of human embryonic stem cells.
- 122.Estaras C, Benner C & Jones KA SMADs and YAP compete to control elongation of beta-catenin:LEF-1-recruited RNAPII during hESC differentiation. Mol Cell 58, 780–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varelas X et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell 19, 831–844 (2010). [DOI] [PubMed] [Google Scholar]
- 124.Singh AM et al. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10, 312–326 (2012).This work highlights key cross-pathway interactions that determine the outcome of TGFβ/Smad signaling during early development..
- 125.Omata Y et al. Genomewide comprehensive analysis reveals critical cooperation between Smad and c-Fos in RANKL-induced osteoclastogenesis.J Bone Miner Res 30, 869–877 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Nieto MA, Huang RY, Jackson RA & Thiery JP Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 127.Miettinen PJ, Ebner R, Lopez AR & Deiynck R TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol 127, 2021–2036 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oft M et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10, 2462–2477 (1996). [DOI] [PubMed] [Google Scholar]
- 129.Moustakas A & Heldin CH Mechanisms of TGFbeta-induced epithelial-mesenchymal transition.J Clin Med 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang J & Weinberg RA Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 131.Boyer AS et al. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev Biol 208, 530–545 (1999). [DOI] [PubMed] [Google Scholar]
- 132.Romano LA & Runyan RB Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol 223, 91–102 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Oshimori N, Oristian D & Fuchs E TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Puram SV et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624 e1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ye X & Weinberg RA Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gotzmann J et al. Hepatocytes convert to a fibroblastoid phenotype through the cooperation of TGF-beta1 and Ha-Ras: steps towards invasiveness.JCell Sci 115, 1189–1202 (2002). [DOI] [PubMed] [Google Scholar]
- 137.Oft M, Akhurst RJ & Balmain A Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol 4, 487–494 (2002). [DOI] [PubMed] [Google Scholar]
- 138.Horiguchi K et al. Role of Ras signaling in the induction of snail by transforming growth factor-beta. J Biol Chem 284, 245–253 (2009). [DOI] [PubMed] [Google Scholar]
- 139.Peinado H, Quintanilla M & Cano A Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem 278, 21113–21123 (2003). [DOI] [PubMed] [Google Scholar]
- 140.Sanjabi S, Oh SA & Li MO Regulation of the Immune Response by TGF-beta: From Conception to Autoimmunity and Infection. Cold Spring Harb Perspect Biol 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stritesky GL, Jameson SC & Hogquist KA Selection of self-reactive T cells in the thymus. Annu Rev Immunol 30, 95–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brabletz T et al. Transforming growth factor beta and cyclosporin A inhibit the inducible activity of the interleukin-2 gene in T cells through a noncanonical octamer-binding site. Mol Cell Biol 13, 1155–1162 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thomas DA & Massagué J TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005). [DOI] [PubMed] [Google Scholar]
- 144.Budhu S et al. Blockade of surface-bound TGF-beta on regulatory T cells abrogates suppression of effector T cell function in the tumor microenvironment. Sci Signal 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Josefowicz SZ, Lu LF & Rudensky AY Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hori S, Nomura T & Sakaguchi S Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 147.Chen W et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tone Y et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol 9, 194–202 (2008). [DOI] [PubMed] [Google Scholar]
- 149.Wu C et al. The transcription factor musculin promotes the unidirectional development of peripheral Treg cells by suppressing the TH2 transcriptional program. Nat Immunol 18, 344–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ouyang W, Beckett O, Ma Q & Li MO Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 32, 642–653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou L et al. TGF-beta-induced Foxp3 inhibits T(H) 17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ivanov II et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 153.Takimoto T et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development.J Immunol 185, 842–855 (2010). [DOI] [PubMed] [Google Scholar]
- 154.Martinez GJ et al. Smad2 positively regulates the generation of Th17 cells. Biol Chem 285, 29039–29043 (2010).This work showed that Smad2 phyiscally interacts with the Th17 master regulator Rorγt and promotes its lineage-defining function .
- 155.Gorelik L, Constant S & Flavell RA Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J Exp Med 195, 1499–1505 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kuwahara M et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat Immunol 13, 778–786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Markowitz S et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268, 1336–1338 (1995). [DOI] [PubMed] [Google Scholar]
- 158.Hahn SA et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271, 350–353 (1996). [DOI] [PubMed] [Google Scholar]
- 159.Shi Y, Hata A, Lo RS, Massagué J & Pavletich NP A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 388, 87–93 (1997). [DOI] [PubMed] [Google Scholar]
- 160.Zorn AM & Wells JM Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ijichi H et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev 20, 3147–3160 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hannon GJ & Beach D p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371, 257–261 (1994). [DOI] [PubMed] [Google Scholar]
- 163.Polyak K et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66 (1994). [DOI] [PubMed] [Google Scholar]
- 164.Reynisdóttir I, Polyak K, Iavarone A & Massagué J Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev 9, 1831–1845(1995). [DOI] [PubMed] [Google Scholar]
- 165.Scandura JM, Boccuni P, Massagué J & Nimer SD Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 upregulation. Proc Natl Acad Sci U S A 101, 15231–15236 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ding Z et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bjerke GA, Yang CS, Frierson HF, Paschal BM & Wotton D Activation of Akt signaling in prostate induces a TGFbeta-mediated restraint on cancer progression and metastasis. Oncogene 33, 3660–3667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nandan MO, Ghaleb AM, Bialkowska AB & Yang VW Kruppel-like factor 5 is essential for proliferation and survival of mouse intestinal epithelial stem cells. Stem Cell Res 14, 10–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Diaferia GR et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 35, 595–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang B et al. KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial-mesenchymal transition in epithelial cells. Mol Cell Biol 33, 4919–4935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chia NY et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut 64, 707–719 (2015). [DOI] [PubMed] [Google Scholar]
- 172.Tan TZ et al. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 6, 1279–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gal A et al. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 27, 1218–1230 (2008).Demonstration that the acute response of mammary epithelial cells induced to undergo an EMT by TGF-β is widespread cell death, while a proliferative EMT-competent subpopulation can be selected after chronic exposure to the cytokine.
- 174.Diepenbruck M & Christofori G Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol 43, 7–13 (2016). [DOI] [PubMed] [Google Scholar]
- 175.Tiwari N et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013). [DOI] [PubMed] [Google Scholar]
- 176.Penuelas S et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15, 315–327 (2009). [DOI] [PubMed] [Google Scholar]
- 177.Padua D et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 133, 66–77 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kakonen SM et al. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J Biol Chem 277, 24571–24578 (2002). [DOI] [PubMed] [Google Scholar]
- 179.Massague J & Obenauf AC Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Malladi S et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 165, 45–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Calon A et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47, 320–329 (2015). [DOI] [PubMed] [Google Scholar]
- 182.Calon A et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Terabe M et al. Blockade of only TGF-beta 1 and 2 is sufficient to enhance the efficacy of vaccine and PD-1 checkpoint blockade immunotherapy. Oncoimmunology 6, e1308616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bruce DL & Sapkota GP Phosphatases in SMAD regulation. FEBS Lett 586, 1897–1905 (2012). [DOI] [PubMed] [Google Scholar]
- 185.Dong X et al. Force interacts with macromolecular structure in activation of TGF-beta. Nature 542, 55–59 (2017).Remarkable paper providing evidence for a force-dependent mechanism of activation for latent extracellular TGF-β.
- 186.Dupont S, Inui M & Newfeld SJ Regulation of TGF-beta signal transduction by mono- and deubiquitylation of Smads. FEBS Lett 586, 1913–1920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Herhaus L & Sapkota GP The emerging roles of deubiquitylating enzymes (DUBs) in the TGFbeta and BMP pathways. Cell Signal 26, 2186–2192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Luo K Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev 14, 65–70 (2004). [DOI] [PubMed] [Google Scholar]
- 189.Miyazawa K & Miyazono K Regulation of TGF-beta Family Signaling by Inhibitory Smads. Cold Spring Harb Perspect Biol 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Parrow NL & Fleming RE Bone morphogenetic proteins as regulators of iron metabolism. Anna Rev Nutr 34, 77–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Saito T et al. Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1. Cell Rep 19, 1917–1928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Siebold C, Yamashita T, Monnier PP, Mueller BK & Pasterkamp RJ RGMs: Structural Insights, Molecular Regulation, and Downstream Signaling. Trends Cell Biol 27, 365–378(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ryan DP, Hong TS & Bardeesy N Pancreatic adenocarcinoma. N Engl J Med 371, 1039–1049 (2014). [DOI] [PubMed] [Google Scholar]
- 194.Friess H et al. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology 105, 1846–1856(1993). [DOI] [PubMed] [Google Scholar]
- 195.Wagner M, Kleeff J, Friess H, Buchler MW & Korc M Enhanced expression of the type II transforming growth factor-beta receptor is associated with decreased survival in human pancreatic cancer. Pancreas 19, 370–376 (1999). [DOI] [PubMed] [Google Scholar]
- 196.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Mutational inactivation of TGF-β–SMAD tumor suppression
cBioPortal query 196 of available TCGA datasets for non-synonymous mutations and deep deletions in TGFBR1, TGFBR2, SMAD2, SMAD3 and SMAD4. a. Frequency of pathway mutations in different types of cancer. All studies included at least 30 samples. b. Frequency of non-synonymous mutations and deep deletions in TGFBR1, TGFBR2, SMAD2, SMAD3 and SMAD4 in individual human pancreatic tumors. SMAD4 is at chromosome 18q21.2 and SMAD2 is located closely at 18q21.1. The two are frequently co-deleted. However, SMAD4 is more commonly deleted than SMAD2, and SMAD2 deletions do not occur without SMAD4 deletions. This and the presence of truncating mutations in SMAD4, suggests that a unique function of SMAD4 is selected against in tumors.


