Abstract
We established a unique platform for proteomic analysis of cultured three-dimensional (3D) neurons and brain tissue from Alzheimer’s disease (AD) patients. We collected peripheral blood mononuclear cells (PBMC), converted PBMC to induced pluripotent stem cell (iPSC) lines, and differentiated the iPSC into human 3D neuro-spheroids. The postmortem brain tissue from the superior frontal cortex, inferior frontal cortex and cerebellum area of the AD patients was compared to the same regions from the control subjects. Proteomic analysis of 3D neuro-spheroids derived from AD subjects revealed the alteration of a number of proteins involved in axon growth, mitochondrial function, and antioxidant defense. Similar analysis of post-mortem AD brain tissue revealed significant alteration in proteins involved in oxidative stress, neuro-inflammation, along with proteins related to axonal injury. These results clearly indicate that the dysfunction of 3D neurons from AD patients in our in vitro environment is comparable to the post-mortem AD brain tissue in vivo. In conclusion, our study revealed a number of candidate proteins that have important implications in AD pathogenesis and supports the notion that the iPSC-derived 3D neuronal system functions as a model to examine novel aspects of AD pathology.
Significance
In this study, we present a unique platform for proteomic analysis of induced pluripotent stem cell-derived three dimensional (3D) neurons and compare the results to those from three regions of post-mortem brain tissue from Alzheimer’s disease patients and normal control subjects. Our results show that the dysfunction of 3D neurons from AD patients in our in vitro environment is comparable to the post-mortem AD brain tissue in vivo. Our results revealed several candidate proteins that have important implications in AD pathogenesis.
Keywords: Alzheimer, Bioinformatic, Proteomics, iPSC, 3D, Neuro-spheroid
1. Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia and is characterized by multiple cognitive deficits including early memory loss, impaired language skills, and a compromised ability to focus and reason [1,2]. The classic AD pathological features include the extracellular deposition of misfolded amyloid-β (Aβ) peptide, the accumulation of hyperphosphorylated tau containing neurofibrillary tangles, and massive neuronal cell and synapse loss [3–5]. Extensive work has attempted to correlate the aforementioned AD cognitive impairments with these biological processes [6–13]; however, this relationship remains inconclusive.
Efforts have been made to model AD brains in cultured dishes. One major limitation of the conventional 2D cell cultures is that secreted Aβ peptides diffuse quickly from the cells to the media and are removed once the media is changed [14]. With the intention of improving cell culture models of disease and solving this limitation, a three-dimensional (3D) neuronal system was created which aims to mimic the aged AD brain environment in an effort to better reproduce the full AD pathology [15]. Due to the heterogeneity and multifactorial nature of AD, a 3D culture may represent a better model to study molecular pathways contributing to AD in vitro. It was hypothesized that 3D hydrogels would provide suitable conditions for accelerating Aβ deposition by limiting the circulation into the cell culture medium [15]. Choi et al. have reported the presence of Aβ aggregates and phosphorylated Tau accumulation in 3D neuronal cultures, indicating that both of these processes are accelerated in 3D conditions [15]. One source of cells for developing 3D neurons is induced pluripotent stem cells (iPSC) [16]. Continuous advancements in molecular technologies have allowed us to successfully reprogram cells from peripheral blood mononuclear cells (PBMCs) derived from AD patients to generate iPSCs and subsequently, 3D human neurons [16]. Previous work has proven the efficacy of this new technology by showing that fibroblast-derived iPSCs from AD patients can be used as an in vitro model to mimic AD pathology [17–26], and pathological proteins, like tau, have been characterized in iPSC-derived 3D neuronal culture [27,28].
The use of proteomic profiling on AD pathologic processes could serve as an indicator of disease presence and progression. The development of mass spectrometry (MS)-based proteomics has been driven by the growth of new technologies for peptides/protein fractionation, advancements in MS instruments, and new labeling reagents. MS-based proteomics possess the advantage of having no requirement of prior knowledge of the proteins being identified, allowing for unbiased, hypothesis-free biomarker discovery in complex biological samples such as plasma and tissue extracts. This approach also meets the requirement for discovery-level proteomics, which is to measure multiple targets simultaneously in a multiplexing manner. Therefore, MS has been increasingly applied to the study of neurodegenerative diseases including AD. Previous MS-based proteomic studies in plasma from AD patients have revealed alterations in proteins linked to inflammation, vascular dysfunction, disturbed metal homeostasis and lipid metabolism [29–31]. Further, profiling of post-mortem brain tissues from AD patients suggested abnormal phosphorylation and O-GlcNAcylation of many proteins [32–34].
The present study is the first reporting global quantitative MS-based proteome analysis of human 3D neurons derived from AD subjects. So far, there are only a few MS-based proteome studies using 2D cultured mouse neurons [35–37]. Here we describe a proteomic assessment of isobaric tags for relative quantitation using 3D cultures and postmortem brain tissues from AD patients with the goal of exploring disease mechanisms and discovering proteins associated with AD pathology. Specifically, proteomic profiles of 3D neurons were obtained, and the ratio changes in protein expression associated with each AD relative to the average of the healthy subjects were calculated. Similarly, the changes in protein expression associated with the three regions of post-mortem brain tissues from AD subjects were analyzed in the same way. Differentially expressed proteins were analyzed using the bioinformatics tools Database for Annotation, Visualization, and Integrated Discovery (DAVID) and STRING (a biological database and web resource of known and predicted protein–protein interactions) to determine functional relationships altered in AD. Changes in protein expression between AD and control subject-derived 3D neurons and brain tissues were compared, allowing us to evaluate the similarities in proteomic profiles from in vitro 3D neurons and in vivo brain tissue.
2. Materials and methods
2.1. Materials and reagents
Reagents used for biochemical methods and cell culture preparation were purchased from Sigma–Aldrich (St Louis, MO, USA) unless otherwise indicated. Reagents for BCA protein assay and sample preparation kits for liquid chromatograph (LC)-mass spectrometer (MS) analysis were purchased from Thermo Scientific (Rockford, IL, USA), including Pierce top twelve abundant protein depletion spin columns, Pierce protein concentrators (3 kDa) and Tandem Mass Tag (TMT) reagent 10-plex kits.
2.2. Subjects
The human study was approved by the Bedford VA Hospital Institutional Review Board, and the signed informed consents were obtained before the initiation of the study. Ten subjects including 5 healthy controls and 5 diagnosed with AD were enrolled from the Bedford VA Hospital Dementia Care Special Unit. The average ages of the AD patients were 69.4 ± 11.8 years (two female and three male subjects) and the controls were 71.2 ± 4.9 years (five male subjects). Controls scored over 27 on the Montreal Cognitive Assessment, which is a validated screening tool for subjects with mild cognitive impairment or AD [38]. Blood from these 10 subjects was obtained for iPSC-differentiated 3D neuronal culture.
The postmortem brain tissue from 5 AD patients and 5 healthy controls were obtained from the Bedford Brain Bank. Among all subjects, two of the five AD cases were the same subjects described above who donated blood for the generation of 3D neuronal cultures. The areas collected were the superior frontal cortex, inferior frontal cortex and cerebellum. The post-mortem interval between the donor’s death and brain autopsy/specimen collection was ≤24 h. The average age of patients from whom we had postmortem brain tissue was 75.6 ± 8.8 for AD patients and 83.0 ± 6.0 for the controls (three males and two females for both groups). Alzheimer’s disease “ABC” scoring system was used for neuropathological assessment of the post-mortem human brains, showing a high degree of Alzheimer’s neuropathological change according to the NIA Alzheimer Association’s guidelines and is sufficient to account for the patient’s dementia [39].
2.3. Generation of human 3D neuro-spheroids from blood-derived iPSCs
Blood was collected in Vacutainer cell tubes (CPT, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and centrifuged at 1500×g for 20 min at room temperature shortly after collection. After centrifugation, the plasma was separated and kept at −80 °C and the peripheral blood mononuclear cell (PBMC) layer was transferred to a new 15 mL Falcon tube with 10 mL of sterile PBS. The PBMCs were centrifuged at 300×g for 10 min at room temperature and the supernatant was discarded. The cell pellet was resuspended with 6 mL of PMBC medium with 10% DMSO (Invitrogen), aliquoted in 2 mL tubes, and frozen at a controlled rate of−1 °C/min in −80 °C then transferred to a liquid nitrogen cryogenic tank for storage.
Induced pluripotent stem cells (iPSC) were derived from PBMC following the integration-free CytoTune-iPS Sendai Reprogramming Kit (Invitrogen) [40,41] and characterized, as previously reported [16]. Generation of 3D neuro-spheroids from iPSCs was accomplished using a modified protocol [42]. Briefly, the iPSCs in E8 medium with a ROCK inhibitor (Thiazovivin, 1 uM, MiltenyiBiotec, San Diego, CA) were transferred into 100mm ultra-low-attachment plastic plates (Corning, Tewksbury, MA). On the day following formation of the spheroid, the medium was replaced with neural induction medium (Invitrogen) for 6 days. After the sixth day, the media on the floating spheroids was exchanged with Neural Medium (NM) containing Neurobasal (Invitrogen), B-27 serum substitute without vitamin A (Invitrogen), GlutaMax (Invitrogen), penicillin and streptomycin (Invitrogen). The NM was supplemented with 20 ng/mL FGF2 and 20 ng/mL EGF (R&D Systems, Minneapolis, MN). The cells were grown in this medium for 21 days with daily replacement during the first 10 days, and replacement every other day for the subsequent 11 days. To promote differentiation of the neural progenitors into neurons, FGF2 and EGF were replaced with 20 ng/mL BDNF and 20 ng/mL NT3 (Peprotech, Rocky Hill, NJ) starting at day 27. From day 48 onwards, NM without growth factors was used and replaced every 4 days.
2.4. Sample preparation for MS
3D neurons were collected after a total of 62 days of differentiation from iPSC’s spheroid formation. Each sample was prepared by first washing the cells 2–3 times with 1× PBS to remove cell culture media and then, lysing the cells by adding approximately five cell-pellet-volume of Lysis Buffer (20mM TrisHCl, 150mM NaCl and 1% NP-40). The lysate was centrifuged at 16,000 ×g for 10 min at 4 °C, and the supernatant was then transferred into a new tube and stored at 4 °C until processing for Mass Spectrometry. Preparation of tryptic peptides for TMT 10-plex labeling was carried out according to the manufacturer’s instructions. After measuring the protein concentrations in each 3D cell lysate sample by the BCA method, 100 μg protein of each sample was transferred into a new vial and adjusted to a final volume of 100 μL with tetraethylammonium bicarbonate (TEAB) and reduced with TCEP at 55 °C for 1 h, and then alkylated with iodoacetamide for 30 min in the dark. Proteins were precipitated by pre-chilled (−20 °C) acetone and proceeded overnight. Acetone-precipitated protein was obtained by centrifuging at 8000×g for 10 min at 4 °C. Protein pellets were resuspended with 100 μL of 50mM TEAB and digested with trypsin overnight at 37 °C.
Areas of the superior frontal cortex, inferior frontal cortex and cerebellum were dissected from AD and control subjects’ frozen postmortem brain tissue. Brain tissue was broken into pieces in mortar by pestle in liquid nitrogen. 600 μL of sample buffer (2% SDS, 0.5M TEAB, protease inhibitor cocktail) was added to each tissue piece (200 mg) and then homogenized by Tissue Lyser LT (Qiagen, Valencia, CA). Tissue homogenates were centrifuged at 17,000×g for 20 min at 4 °C. The supernatant was transferred into a new vial for protein concentration measurement by BCA assay. Each sample containing 100 μg proteins was reduced and alkylated as described above. Methanol–chloroform precipitation was performed prior to protease digestion. To summarize, four parts methanol was added to each sample and vortexed, then one part chloroform was added to the sample and vortexed, and then three parts water was added to the sample and vortexed. The sample was centrifuged at 14,000×g for 4 min at room temperature and the aqueous phase was removed. The organic phase with protein precipitate at the surface was subsequently washed twice with four parts methanol and centrifuged with supernatant being removed subsequently. After air-drying, precipitated protein pellets were re-suspend with 100 μL of 50mM TEAB and digested with trypsin overnight at 37 °C.
2.5. TMT-labeling and sample clean up
TMT enables relative quantitation of proteins present in multiple samples by labeling peptides with isobaric stable isotope tags that fragment upon collision-induced dissociation into reporter ions used for quantitation. In this study, 10-plex TMT tags used for 3D neuron culture and brain tissue samples from five AD patients and five controls were analyzed simultaneously, avoiding run-to-run variation.
Tryptic digested peptides from 3D cell culture and brain samples were labeled with TMT 10-plex reagents. Labelling of tryptic peptides was carried out according to the manufacturer’s instructions (Thermo Fisher). Briefly, the TMT reagents (0.8 mg) were dissolved in 41 μL of anhydrous acetonitrile. Aliquots of samples were incubated with TMT reagents for 1 h at room temperature. The reactions were quenched by 8 μL of 5% hydroxylamine solution and reacted for 15 min. In TMT 10-plex labeling, samples from AD patient 1 to 5 and control 1 to 5 were labeled with Reagent 126, 127N, 127C, 128N, 128C, 129N, 129C, 130N, 130C and 131, respectively. The combined TMT labeled samples were dried under Speed Vac, and then reconstituted by trifluoroacetic acid solution followed by desalting using Oasis HLB 96-well μElution plate (Waters) prior to LC-MS/MS analysis. For enrichment of phosphopetides, samples were enriched with the Fe-NTA phosphopeptide enrichment kit and followed by graphite spin columns according to the manufacturer’s instructions (Thermo Scientific Pierce) before LC-MS/MS analysis.
2.6. LC-MS/MS analysis
LC MS/MS was performed on a Q Exactive Orbitrap Mass Spectrometer (Thermo Fisher Scientific) coupled with a Dionex ultimate 3000 HPLC system equipped with a nano-ES ion source. The TMT labeled peptides were separated on a C18 reverse-phase capillary column (PepMap,75 μm x150 mm, Thermo Fisher) with linear gradients of 2%–35% acetonitrile in 0.1% formic acid, at a constant flow rate of 300 nL/min for 220 min. The instrument was operated in the positive-ion mode with the ESI spray voltage set at 1.8 kV. A full scan MS spectra (300–1800 m/z) was acquired in the Orbitrap at a mass resolution of 70,000 with an automatic gain control target (AGC) of 3e6. Fifteen peptide ions showing the most intense signal from each scan were selected for higher energy collision-induced dissociation (HCD)-MS/MS analysis (normalized collision energy 32) in the Orbitrap at a mass resolution of 35,000 and AGC value of 1e5. Maximal filling times were 100 ms in full scans and 120 ms in HCD for the MS/MS scans. Ions with unassigned charge states and single charged species were rejected. The dynamic exclusion was set to 50 s and a relative mass window of 10 ppm. The data were acquired using ThermoXcalibur 3.0.63. The phosphopeptides were analyzed under the same conditions as above, except that separation was done with linear gradients of 1%–30% acetonitrile in 0.1% formic acid, and full scan MS ranging 300–2000 m/z with NCE at 25.
2.7. Protein identification and quantification
Raw data were processed using Proteome Discoverer (Version 2.1, Thermo Fisher Scientific). Data were searched against the Homo sapiens Universal Protein Resource sequence database (UniProt, August 2013). The searches were performed with the following guidelines: trypsin digestion with two missed cleavage allowed; fixed modification, carbamidomethyl of cysteine; variable modification, oxidation of methionine, TMT 10plex (peptide labeled) for N-terminus and Lys; MS tolerance, 5 ppm; MS/MS tolerance, 0.02 Da; false discovery rate (FDR) at peptide and protein levels, < 0.01; and required peptide length, ≥6 amino acids. Phosphorylation of serine, threonine and tyrosine was defined as variable modification. Phosphorylation sites were localized by phosphoRS. Protein grouping was enabled, therefore, if one protein was equal to or completely contained within the set of peptides of another protein, these two proteins were put into the same protein group. At least one unique peptide per protein group was required for identifying proteins. The relative protein abundance ratios (fold changes) between each AD and the average of controls were calculated from at least three analytical runs in each experiment. The mean and standard deviation (SD) for relative ratios of each protein were computed and a student t-test was used to compare these repeated measures with the threshold of 1.2 (H1: μ > 1.2) or 0.8 (H1: μ < 0.8). P-values from these tests were recorded. For 3D data, we use the threshold of 1.2 and 0.9 in similar tests for the repeated measures. The protein ratios with a p-value < 0.05 from both independent experiments are considered to be significant. Proteins with the relative ratios below the low range (0.8 for brain regions or 0.9 for 3D) were considered to be down-regulated, whereas those above the high range (1.2) were considered to be up-regulated.
2.8. ELISA assay of S100B and vimentin
Frozen brain tissue was homogenized with 5M GnHCl2 (1 g tissue/5 mL of GnHCl2) using a TissueLyser LT (Qiagen) at 50× speed for 4 min. The homogenates were mixed in 5M GnHCl2 overnight at room temperature. 3D lysate samples were prepared as described above. The plates were coated with the corresponding primary antibodies. The homogenates were centrifuged at 16,000×g for 15 min at 4 °C before being loaded onto the ELISA plates specific for human proteins Vimentin (Abcam, Cambridge, MA) and S100B (LifeSpan Biosciences, Seattle, WA). The protocol for these ELISA kits was carried out according to the manufacturers’ instructions.
2.9. Data analysis
The list of differentially regulated proteins was submitted to STRING Version 10.0 to identify protein networks which are based on currently known associations among proteins, indicated by the scientific literature. However, in the absence of literature-based evidence for specific connections, some proteins may miss inclusion in a STRING cluster. Therefore, the DAVID tool, an open-source software Database for Annotation, Visualization, and Integrated Discovery, was also used to explore enrichment of biological processes, molecular functions, cellular components and KEGG pathways. Significance of enrichment was based on a p-value of < 0.05 using FDR correction. An enrichment analysis was performed by submitting the UniProt accession numbers containing the list of differentially regulated proteins to DAVID v6.7 [43,44]. DAVID is able to extract biological features/meanings associated with large gene lists. The gene names of the proteins were entered to analyze Gene Ontology (GO) biological process (GOBP), GO molecular function (GOMF), and GO cellular components (GOCCs) and KEGG pathways. Statistically significant differences (p-value < 0.05) were identified using an EASE score test, which was provided by the DAVID gene bioinformatics online resource. The cellular and molecular process networks of the dysregulated proteins in three brain regions of AD patients were analyzed using Metacore. Metacore uses these proteins and their identifiers to navigate the curated literature database and extract the biologically relevant information among the candidate proteins. Associated biofunctions were generated, along with a score representing the log probability of a particular process network.
3. Results
3.1. Alteration of protein expression in 3D neuro-spheroids derived from AD patients
We quantified levels of proteins from AD and normal control subject-derived 3D neuro-spheroids by LC-MS/MS. We established 10 lines of iPSC using the PBMC from 5 AD patients (AD1–5) and 5 control subjects, and then differentiated these 10 iPSC lines into 3D neuro-spheroids [16]. A comparative proteomic analysis of the 3D lines was performed using TMT labeling and LC-MS/MS. A total of 1855 proteins from 3D neuro-spheroids with at least one unique peptide were detected. The TMT labeling efficiency was up to 99.8%. Only 0.3% of peptides missed two cleavages, and < 6% missed one cleavage. The average number of unique peptide (having a confidence level of > 95%) per protein was 4.4 and > 26% of the proteins had ≥5 unique peptides. Many neuronal markers of 3D neuro-spheroids derived from both AD and control subjects were labeled, such as nestin (NSE), neuronal migration protein doublecortin (DCX), calrentinin, calbindin, neurofilament protein (NFP), and microtubule-associated protein 2 (MAP 2).
We compared ratios of proteins from each AD patient-derived 3D neuro-spheroids to the average levels of proteins from the control group. We calculated the fold of ratio changes (ratio ≥ 1.2 or ≤0.9) of proteins detected from 3D neuronal line derived from each individual AD to the average of the control group. One advantage of this approach over the conventional approach of comparing the average of the AD group to that from the control group is that the later method could be influenced by one subject having a protein with extremely high or low ratios impacting the average values, especially in a study with a small sample size. Among 3D neuronal lines from each AD subject, we have identified 76 and 178 up/down-regulated proteins from two AD patient-derived 3D neurons when compared to the average of the controls, but only 10, 15, and 8 such proteins from the other three AD patient-derived 3D neurons, respectively. We grouped the common proteins that were significantly differentially regulated among the five AD patients. We found a total of 21 commonly differentially regulated proteins in at least two AD subjects (Table 1), including 8 up-regulated and 13 down-regulated proteins.
Table 1.
Commonly altered proteinsa in 3D neuro-spheroids derived from AD patients relative to those from control subjects.
| Accession | Description | Ratio | Coverage | Unique peptides | MW (kDa) |
|---|---|---|---|---|---|
| P21796 | Voltage-dependent anion-selective channel protein 1 (VDAC1) | 1.74 | 13 | 2 | 30.8 |
| P07196 | Neurofilament light polypeptide (NEFL) | 1.65 | 17 | 7 | 61.5 |
| P59768 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 (GNG2) | 1.64 | 32 | 2 | 7.8 |
| P17677 | Neuromodulin (GAP43) | 1.58 | 72 | 14 | 24.8 |
| O43602 | Neuronal migration protein doublecortin (DCX) | 1.49 | 29 | 8 | 49.3 |
| P80723 | Brain acid soluble protein 1 (BASP1) | 1.46 | 82 | 15 | 22.7 |
| P05141 | ADP/ATP translocase 2 (SLC25A5) | 1.46 | 11 | 2 | 32.8 |
| P06576 | ATP synthase subunit beta, mitochondrial (ATP5B) | 1.38 | 32 | 12 | 56.5 |
| P09211 | Glutathione S-transferase P (GSTP1) | 0.84 | 43 | 6 | 23.3 |
| P18621 | 60S ribosomal protein L17 (RPL17) | 0.84 | 23 | 3 | 21.4 |
| P15880 | 40S ribosomal protein S2 (RPS2) | 0.82 | 20 | 5 | 31.3 |
| Q15121 | Astrocytic phosphoprotein PEA-15 (PEA15) | 0.82 | 45 | 5 | 15.0 |
| O00299 | Chloride intracellular channel protein 1 (CLIC1) | 0.81 | 41 | 8 | 26.9 |
| P07602 | Prosaposin (PSAP) | 0.81 | 26 | 12 | 58.1 |
| P06396 | Gelsolin (GSN) | 0.80 | 10 | 7 | 85.6 |
| P21333 | Filamin-A (FLNA) | 0.80 | 12 | 21 | 280.6 |
| Q96DG6 | Carboxymethylenebutenolidase homolog (CMBL) | 0.80 | 20 | 4 | 28.0 |
| P07355 | Annexin A2 (ANXA2) | 0.79 | 15 | 4 | 38.6 |
| P48681 | Nestin (NES) | 0.79 | 40 | 49 | 177.3 |
| P53985 | Monocarboxylate transporter 1 (SLC16A1) | 0.77 | 7 | 2 | 53.9 |
| P08670 | Vimentin (VIM) | 0.76 | 66 | 31 | 53.6 |
The proteins in the table are differentially regulated 3D proteins in at least 2 out of 5 AD patients when comparing each AD relative to the average of 5 controls.
3.2. Alteration of protein functions in 3D neuro-spheroids derived from AD patients
To analyze up/down-regulated proteins, we used the online bioinformatics resource DAVID, which revealed several functional categories related to phosphorylation, acetylation, and Ubl conjugation. GO enrichment analysis was applied to the 8 up-regulated and 13 down-regulated proteins separately to describe the functions of these dysregulated proteins, which were classified into three major categories: biological process (BP), cellular component (CC) and molecular function (MF) (Table 2). Several representative processes were identified. The biological category indicated that the up-regulated proteins were commonly involved in axon development and cellular response to glucagon stimulus, whereas down-regulated proteins were involved in platelet aggregation, glutathione metabolic processes, RNA and translation related processes. In the cellular component category, the up-regulated proteins found were mainly associated with myelin sheath and mitochondrial nucleoid, whereas down-regulated proteins were found to be associated with cell–cell junction and focal adhesion, and cytosolic large ribosomal subunit. The molecular functions of the up-regulated proteins were commonly associated with transmembrane transporter activity and protein binding, whereas the down-regulated proteins were mostly associated with cadherin binding involved in cell–cell adhesion, glutathione transferase activity and poly(A) RNA binding. The GO terms (Table 2) from individual AD cases showed detailed functions of the dysregulated protein. The KEGG pathway analysis (Table 3) for 3D neuro-spheroids from each AD case showed that proteins in the ribosome pathway were commonly down-regulated in three out of five AD patients. A number of proteins related to pathways of neuronal functions or neurodegenerative diseases, such as Gap junction, GABAergic synapse/dopaminergic synapse, and oxidative phosphorylation, were up-regulated in two AD cases. Bile secretion pathway was also found to be up-regulated in one AD case but not in the other AD cases. ATP5B, SLC25A5, NEFL and VDAC1 are four proteins up-regulated in AD samples that are commonly associated with myelin sheath (Table 1). SLC25A5 and VDAC1 are commonly associated with mitochondria function. GNG2 is the protein commonly involved in cellular response to glucagon stimulus, and ATP5B protein functions as a transmembrane transporter.
Table 2.
Up- and down-regulated elements from AD patient-derived 3D neuron-spheroids.
| Regulation | Category | Terms | p-Value |
|---|---|---|---|
| UP | BP | Cellular response to glucagon stimulus | ≤ 0.028 |
| MF | Transmembrane transporter activity | ≤ 0.017 | |
| Protein binding | ≤ 0.020 | ||
| CC | Mitochondrion | ≤ 0.026 | |
| Myelin sheath | ≤ 0.001 | ||
| DOWN | BP | Glutathione metabolic process | ≤ 0.043 |
| Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | ≤ 0.003 | ||
| rRNA processing | ≤ 0.011 | ||
| SRP-dependent cotranslational protein targeting to membrane | ≤ 0.002 | ||
| translation | ≤ 0.016 | ||
| Translational initiation | ≤ 0.005 | ||
| MF | Cadherin binding involved in cell-cell adhesion | ≤ 0.001 | |
| mRNA binding | ≤ 0.044 | ||
| poly(A) RNA binding | ≤ 0.009 | ||
| Structural constituent of ribosome | ≤ 0.012 | ||
| CC | Cell–cell junction | ≤ 0.001 | |
| Cytosolic large ribosomal subunit | ≤ 0.047 | ||
| Cytosolic small ribosomal subunit | ≤ 0.012 | ||
| Small ribosomal subunit | ≤ 0.027 |
BP-Biological process, MF-Molecular function, CC-Cellular component.
Note: The table doesn’t include the general cellular components, such as cytoplasm, cytosol, extraocular exosome, and membrane.
Table 3.
The main KEGG pathways in up- and down-regulated 3D proteins of each AD analyzed by DAVID.
| Regulation | AD case | KEGG pathways | p-Value |
|---|---|---|---|
| UP | AD1 | Bile secretion | 3.00E-02 |
| UP | AD2 | Gap junction | 2.80E-08 |
| Pathogenic Escherichia coli infection | 3.80E-07 | ||
| Phagosome | 2.20E-04 | ||
| GABAergic synapse | 1.40E-03 | ||
| Dopaminergic synapse | 6.10E-03 | ||
| Glycolysis/gluconeogenesis | 6.70E-03 | ||
| DOWN | Ribosome | 1.30E-02 | |
| UP | AD3 | Parkinson’s disease | 1.80E-09 |
| Huntington’s disease | 8.20E-07 | ||
| Oxidative phosphorylation | 9.50E-03 | ||
| cGMP-PKG signaling pathway | 1.50E-02 | ||
| Alzheimer’s disease | 1.50E-02 | ||
| DOWN | AD4 | Ribosome | 9.60E-07 |
| Protein processing in endoplasmic reticulum | 2.40E-04 | ||
| RNA transport | 4.00E-03 | ||
| DOWN | AD5 | Ribosome | 7.50E-06 |
3.3. Enhanced inflammation in postmortem brain tissues from AD patients
To determine alterations in relative protein abundance in postmortem brain tissue from AD and control subjects, we conducted proteomic analysis in the three brain regions, superior frontal cortex (SF), inferior frontal cortex (IF) and cerebellum (CRLM), from five AD patients and five controls. SF and IF regions have more AD pathological lesions compared to the less affected CRLM region. A total of 1902, 1837 and 1896 proteins were identified with FDR < 1% and at least one unique peptide was detected for identified proteins from SF, IF and CRLM regions. The average labeling efficiency was 99.9%.
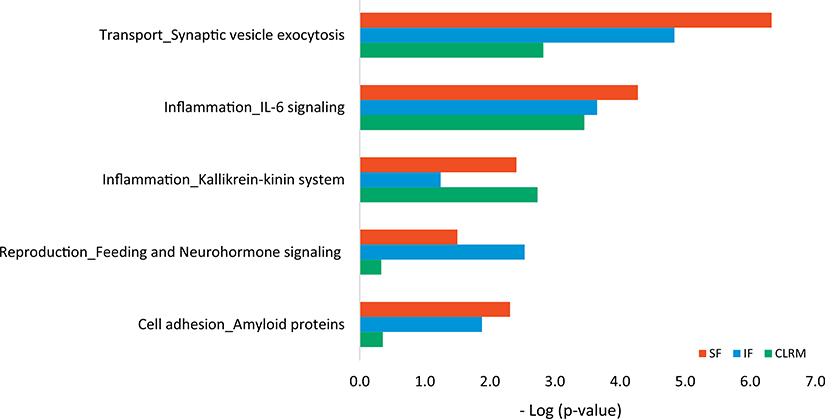
The differentially regulated proteins were identified by comparing proteins from each AD subject relative to those from the average of the control subjects. We grouped the commonly differentially regulated proteins (at least two out of five AD patients relative to the same brain region of the control group) in each brain region. There were 28 commonly dysregulated proteins in IF region, 48 in SF region and 27 in CRLM region. Among them, 17 proteins were found to be common in at least two brain regions (Table 4), and the notable proteins involved in inflammation include Protein S100-B and Glial fibrillary acid protein. We put this list of dysregulated proteins from each of these three brain regions into Metacore enrichment analysis. Fig. 1 shows the comparative distribution of cellular and molecular process networks of dysregulated proteins from SF, IF, and CRLM in AD relative to control subjects. The top common networks enriched in all three brain regions are in transport and inflammation, which include synaptic vesicle exocytosis, IL-6 signaling, and Kallikrein-kinin system. Cell adhesion of amyloid proteins are enriched in SF and IF regions.
Table 4.
Change of proteins in three brain regions from AD patients.a
| Accession | Description | Ratio IF | Ratio SF | Ratio CRLM |
|---|---|---|---|---|
| P04271 | Protein S100-B | 2.03 | 1.94 | 0.71 |
| P05067 | Amyloid beta A4 protein | 1.89 | 1.81 | n/a |
| P14136 | Glial fibrillary acidic protein | 1.82 | 1.59 | n/a |
| P00738 | Haptoglobin | 1.82 | 1.94 | 0.63 |
| P60201 | Myelin proteolipid protein | 1.80 | 1.78 | n/a |
| P13611 | Versican core protein | 1.74 | 1.56 | n/a |
| P01857 | Ig gamma-1 chain C region | 1.61 | 0.64 | 0.59 |
| P01876 | Ig alpha-1 chain C region | 1.57 | 1.60 | n/a |
| P01024 | Complement C3 | 1.43 | 1.42 | n/a |
| P15104 | Glutamine synthetase | 0.68 | n/a | 0.75 |
| P08670 | Vimentin | 0.67 | 0.69 | n/a |
| P11137 | Microtubule-associated protein 2 | 0.64 | 0.58 | n/a |
| Q6PUV4 | Complexin-2 | 0.54 | 0.49 | 0.59 |
| O14810 | Complexin-1 | 0.48 | 0.51 | n/a |
| P01009 | Alpha-1-antitrypsin | n/a | n/a | 0.66 |
| P02671 | Fibrinogen alpha chain | n/a | 1.45 | 0.64 |
| P01871 | Ig mu chain C region | n/a | 0.59 | 0.64 |
n/a: The protein was not differentially regulated in this brain region.
IF – Inferior frontal, SF – Superior frontal, CRLM – Cerebellum.
The proteins in the table are differentially regulated proteins from three brain areas in at least 2 out of 5 AD patients when comparing each AD relative to the average of 5 controls.
Fig. 1.
The cellular and molecular process networks of the dysregulated proteins in three brain regions of AD patients. (SF: Superior frontal cortex; IF: Inferior frontal cortex; CRLM: Cerebellum.)
3.4. Quantification of proteins in multi-region postmortem brain tissue illustrates innate immune responses and complement activation in AD vulnerable brain regions
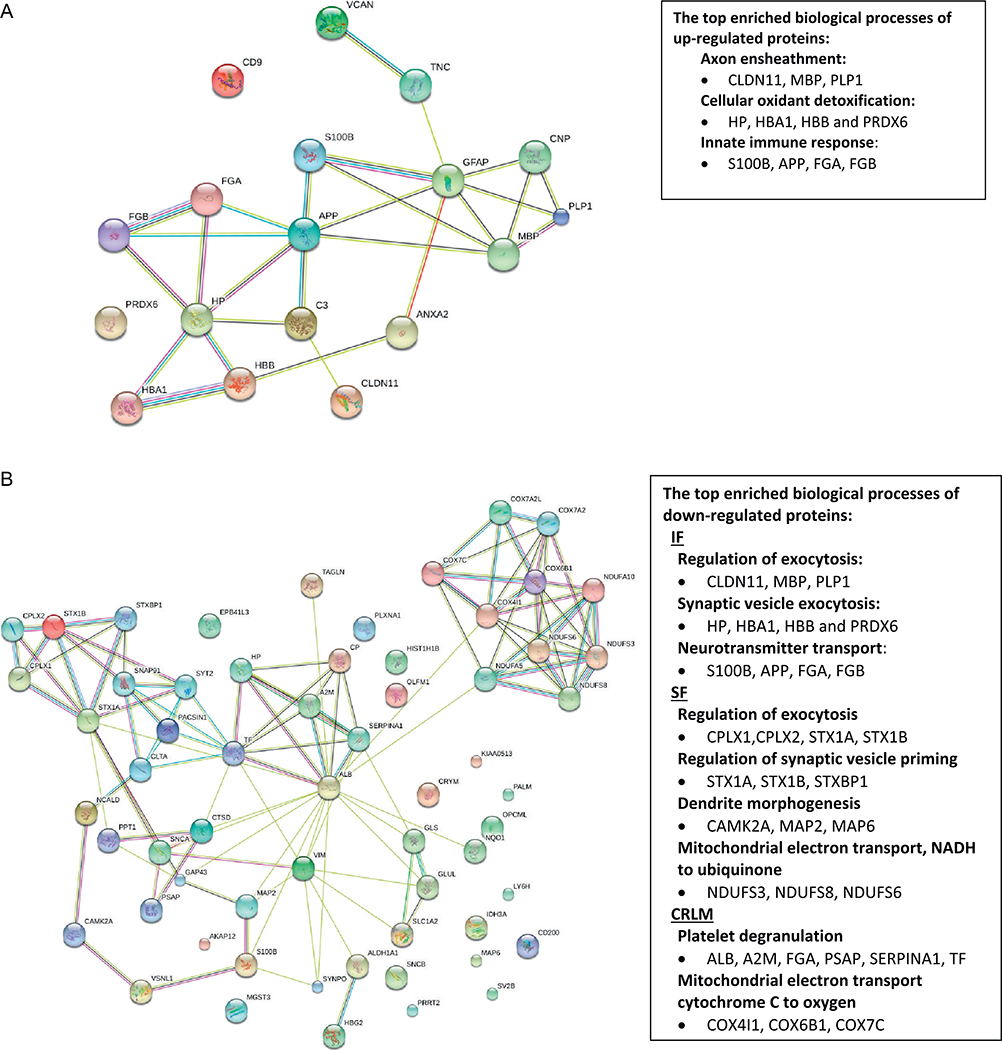
The up- and down-regulated proteins in all three brain regions were analyzed separately. DAVID analysis results (Fig. 2A) show that the top enriched biological process among up-regulated proteins in IF and SF regions include axon ensheathment (14%, p = 2.80E-5), cellular oxidant detoxification (20%, p = 7.50E-5) and innate immune response (29%, p = 1.20E-4). The protein networks from STRING illustrated the clusters of proteins mainly involved in these processes, such as in axon ensheathment (CLDN11, MBP, PLP1), cellular oxidant detoxification (HP, HBA1, HBB, and PRDX6) and innate immune response (S100B, APP, FGA, FGB). DAVID analysis of down-regulated proteins shows that the top enriched biological process is regulation of exocytosis in both IF and SF regions (p = 1.9E-5, p = 1.7E-2, respectively) and platelet degranulation in the cerebellum region (Fig. 2B). The clusters of proteins mainly involved in the biological processes of platelet degranulation, respiratory electron transport chain, neurotransmitter secretion, glutamate secretion (CPLX1, STX1A and STXBP1) were illustrated by STRING (Fig. 2B).
Fig. 2.
Protein network analysis of up- (A) and down-regulated (B) proteins in three brain regions (SF, IF and CRLM) from AD patients relative to control subjects using the web-based tool STRING 10.0. The enriched biological process and dysregulated proteins were analyzed by DAVID. (SF: Superior frontal cortex; IF: Inferior frontal cortex; CRLM: Cerebellum.)
The KEGG pathway analysis of up- and down-regulated proteins show that complement and coagulation cascades pathway (p = 5.8-E-4) is enriched in up-regulated proteins from the SF region (including FGA, FGB,) as well as in down-regulated proteins from CRLM region (including A2M, FGA, and SERPINA1) (Fig. 2). The synaptic vesicle cycle pathway is enriched in the down-regulated proteins in both IF and SF regions, with proteins CPLX1 and CPLX2 in the IF region, and CPLX1, CPLX2, STX1A, STX1B, and STXBP1 in the SF region. The oxidative phosphorylation pathway was enriched in the down-regulated proteins from both the SF and CRLM regions (p < 0.05), with proteins NDUFS3, NDUFS8, NDUFS6 from the SF region and NDUFA10, COX4I1, COX6B1, COX7A2, and COX7C from the CRLM region.
Classical complement activation and platelet degranulation are enriched in both up- and down-regulated proteins. Among differentially regulated proteins in the three brain regions, APP, C3, S100B, IGHA1 and IGHG1 are involved in innate immune response and/or complement activation. They are up-regulated in SF and IF, but either down-regulated or show no changes in CRLM, indicating the activation of immune responses and inflammatory processes in these AD vulnerable brain regions. C3 (Complement 3) has a central role in the complement cascade, and its proteolytic fragments aid in stimulation of pro-inflammatory responses.
3.5. Alteration of axon proteins in both 3D neuro-spheroids and the brain tissue from AD patients
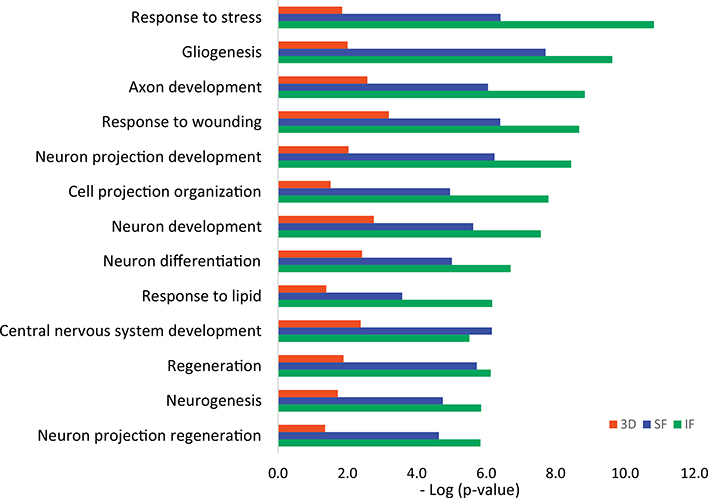
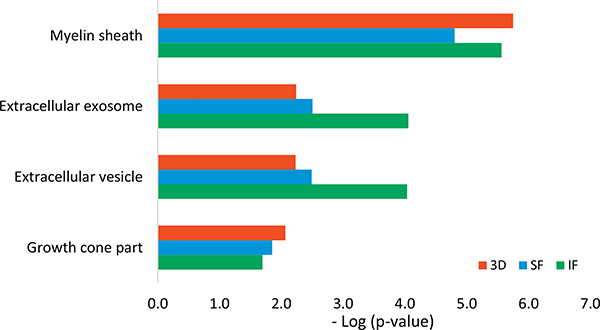
Since SF and IF regions are vulnerable during AD pathogenesis while CRLM regions have less AD pathology, we conducted comparisons of differentially expressed proteins between 3D neurons and the SF and IF brain regions from AD patients. The GO process networks showed that proteins involved in response to stress, gliogenesis, axon development, response to wounding and neuron projection developments were significantly up-regulated in both 3D as well as in brain regions of IF and SF from AD subjects (Fig. 3), demonstrating a consistency of biological processes between 3D and the brain area of SF and IF. From the network objects ranking in GO process among up-regulated 3D proteins, the top four proteins are neuromodulin (GAP43), doublecortin (DCX), Neurofilament light polypeptide (NEFL), and mitochondrial ATP synthase subunit beta (ATP5B). Among them, GAP43, DCX and NEFL are all involved in the process of axon development. GAP43 is also involved in cell and glial cell differentiation, while NEFL is involved in response to toxic substances. The comparison of GO localizations between up-regulated proteins from 3D and brain regions of SF and IF showed the most common cellular component enriched was myelin sheath (Fig. 4). This is consistent with the result that the top enriched biological process among up-regulated proteins in the IF and SF regions is axon ensheathment. Axon ensheathment is a process in which the axon of a neuron is insulated, and myelin is the fatty sheath that coats the axons of the nerves, allowing for efficient conduction of nerve impulses. Other cellular components that were commonly enriched in 3D neuro-spheroids and the two cortex regions of brain were extracellular exosome, extracellular vesicle and growth cone. The Venn diagram illustrates the number of common proteins shared among 5 AD subjects when values from individual AD subject were compared to the average protein levels from all 5 control samples (Supplementary Fig. 1).
Fig. 3.
Comparison of GO Process Networks of up-regulated proteins from 3D neuro-spheroid and AD vulnerable brain regions (SF and IF) from AD patients using Metacore. Sorting is carried out for the statistically significant networks only. (SF: Superior frontal cortex, IF: Inferior frontal cortex.)
Fig. 4.
Comparison of GO Localization of up-regulated proteins from AD-derived 3D neuro-spheroid and brain regions (IF and SF) from AD patients using Metacore. (SF: Superior frontal cortex; IF: Inferior frontal.)
Phosphopeptides enrichment analysis in 3D neuro-spheroids and brain regions of IF and SF showed several differentially regulated phosphorylated proteins when comparing the average of AD group to the average of the control group (Table 5). Phosphorylated GAP43 was found up-regulated whereas phosphorylated NES was down-regulated in 3D neuro-spheroids. Phosphorylated proteins related to regulation of synaptic vesicle priming (STX1B and STX1A) and structure molecule activity (NES and MAP1B) were found down-regulated.
Table 5.
Differentially regulated phophorylated proteins when the average of AD group is compared to the average of the control group.
| Sample type | Description | Ratio | Phospho positions |
|---|---|---|---|
| 3D | Nestin (NES) | 0.86 | Phospho [S352(96.8); S680(100); S768(100)] |
| 3D | Neuromodulin (GAP43) | 1.22 | Phospho [T181(99.4)] |
| IF | Microtubule-associated protein 1B (MAP1B) | 0.84 | Phospho [S1501(99.3); S1779(100)] |
| IF | Syntaxin-1B (STX1B) | 0.81 | Phospho [S14(100)] |
| SF | Syntaxin-1A (STX1A) | 0.77 | Phospho [S14(100)] |
3.6. Postmortem brain tissues from AD patients carry more S100B and less vimentin, key elements in immune responses
The top three network objects/proteins ranking in the molecular functions are vimentin, annexin II, and filamin A in 3D neuro-spheroids, and vimentin, NAD(P)H dehydrogenase-[quinone]-1 (NQO1), and microtubule-associated protein 2 (MAP2) in IF and SF brain regions. Vimentin ranks on the top of the common molecular function of down-regulated proteins in both 3D neuro-spheroids and SF and IF brain regions. Vimentin and nestin are involved in the intermediate filament binding which provides mechanical support to the cell.
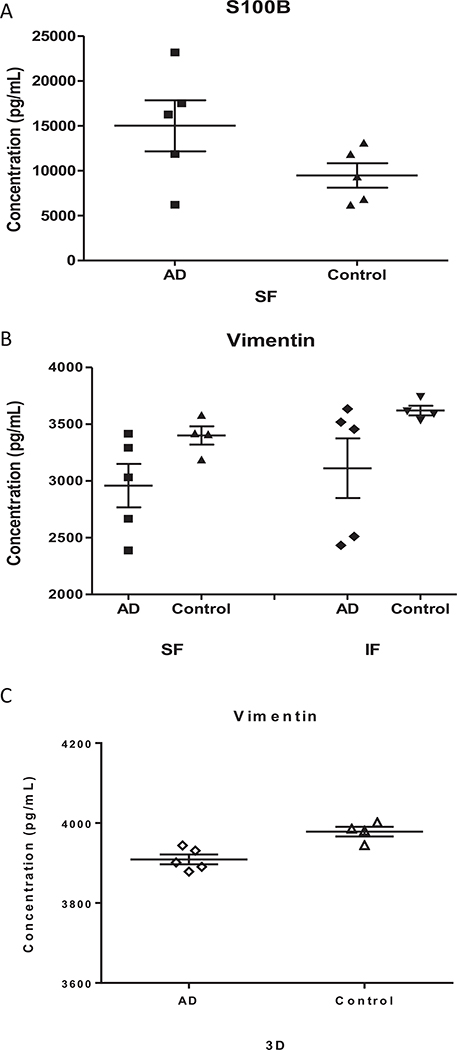
In order to confirm the MS-based quantitative measurements, ELISA quantification of vimentin and S100B in 3D neuro-spheroids and IF/SF brain regions of AD and control subjects were carried out. S100B was not detectable in 3D neuro-spheroids. Vimentin in 3D neuro-spheroids from AD subjects was slightly reduced compared to that from control subjects. A significant (p < 0.05) increase of S100B in the brain SF region and a decrease of vimentin in SF and IF regions in the brain of AD subjects relative to the controls were revealed by ELISA (Fig. 5). These values correlated well with those obtained by quantitative MS; the ratio between AD and normal control subjects were around 1.9 and 0.7 for S100B and vimentin, respectively.
Fig. 5.
Levels of S100B and Vimentin in the brain tissue and 3D neuro-spheroids from AD and control subjects. ELISA was performed to quantify levels of S100B (A) and Vimentin in brain tissue (B) and 3D neuro-spheroids (C). The difference between two groups of brain samples (A and B) is statistically significant, p < 0.05. The standard error of means (short lines) and means (long middle line) are illustrated for each group. (SF: Superior frontal cortex; IF: Inferior frontal cortex.)
We also compared dysregulated proteins from 3D neuron culture to the two brain regions, IF and SF, in two AD cases whose 3D neurons and postmortem brain tissues were derived from the same subject (Table 6). Several ribosomal proteins and proteins localized in focal adhesions (FLNA, RPL4, RPS2 and VIM) were commonly down-regulated in both 3D neuro-spheroids and SF region in AD2. Annexin A2, VIM and ADH5 were found down-regulated in 3D as well as in both IF and SF regions in AD2. However, no common up and down-regulated proteins were found between 3D neuro-spheroids and the two brain regions in AD1, except for protein HIST1H2BK.
Table 6.
The common dysregulated proteins with ratio changes in the same directions between 3D and Brain regions.
| AD | Accession | Protein descriptions | Sample | Ratio |
|---|---|---|---|---|
| AD1 | O60814 | Histone H2B type 1-K (HIST1H2BK) | 3D | 1.21 |
| SF | 1.24 | |||
| AD2 | P15880 | 40S ribosomal protein S2 (RPS2) | 3D | 0.81 |
| SF | 0.64 | |||
| P36578 | 60S ribosomal protein L4 (RPL4) | 3D | 0.84 | |
| SF | 0.63 | |||
| P18621 | 60S ribosomal protein L17 (RPL17) | 3D | 0.86 | |
| SF | 0.63 | |||
| P11766 | Alcohol dehydrogenase class-3 (GN = ADH5) | 3D | 0.83 | |
| SF | 0.72 | |||
| IF | 0.74 | |||
| P07355 | Annexin A2 (ANXA) | 3D | 0.82 | |
| SF | 0.62 | |||
| IF | 0.65 | |||
| Q15121 | Astrocytic phosphoprotein PEA-15 (PEA15) | 3D | 0.82 | |
| SF | 0.69 | |||
| P21333 | Filamin-A (FLNA) | 3D | 0.83 | |
| SF | 0.76 | |||
| P08670 | Vimentin (VIM) | 3D | 0.85 | |
| SF | 0.67 | |||
| IF | 0.65 |
Note: There is no common dysregulated proteins between 3D and IF with the same direction of changes in AD1.
4. Discussion
Proteomics examines changes in protein levels and protein alterations that result from or foster specific diseases. Given the complexity of AD, a panel of proteins rather than single protein candidate may have greater implications in disease onset and may better collectively describe and characterize the disease pathology. In this study, our proteomic analysis of iPSC-derived 3D neuro-spheroids and post-mortem brain tissue revealed cellular components, subcellular structure and specific pathways that have important implications in AD pathogenesis. We established a novel platform to explore pathological proteins in AD. Specifically, we acquired a proteomic profile of a 3D neuro-spheroids that we have previously reported [16]. For the first time, an iPSC-derived 3D human neural cell culture was used for proteomic profiling. Importantly, 3D neurons and post-mortem brain tissues from two AD cases are derived from the same subject, and these sample pairs (3D neuro-spheroids and brain tissue) carry identical genetic information, although we have not defined to which brain region our 3D neuro-spheroids are mostly related. We compared proteomic profiles of both 3D neurons and brain tissues from AD patients. By comparing individual AD cases to the control subjects, even with a small sample size, we identified a list of proteins, some of which have been reported in previously published studies [45–48].
4.1. 3D neuro-spheroids share proteomic profiles related to changes in axon proteins and immune response pathways with those from postmortem AD brain tissue
Our comparative proteomic analysis of 3D neuro-spheroids and postmortem brain tissues revealed specific groups of proteins that represent several common processes that may be dysregulated in the brains of AD patients. Proteins involved in axon development were found to be up-regulated in both 3D neuro-spheroids and SF and IF AD brain regions, where axonal injury has been detected. Several brain proteins involved in axon development and/or localized in myelin sheath, such as myelin basic protein (MBP), S100B, and glial fibrillary acidic protein (GFAP), were found up-regulated in SF and IF regions of AD patients. Previous studies suggest that MBP and degraded myelin basic protein complex (dMBP) could participate in the formation of amyloid plaques in AD brain [49]. Increased levels of MBP proteins in the AD cortex compared to controls have been reported [50,51]. It is possible that myelin injury is associated with re-myelination which increases total MBP in AD brain. Protein S100B is a calcium binding protein that can stimulate neurite extension. GFAP is an intermediate filament protein which can help to maintain astrocyte mechanical strength and the shape of cells. Both S100B and GFAP are well documented brain injury biomarkers [52]. In neurons, S100B triggers trophic or toxic effects, depending on its concentration. A nanomolar concentration of S100B is neuroprotective, induces neurite outgrowth, and triggers glial cell proliferation in a RAGE dependent manner, whereas a micromolar concentration of S100B is neurotoxic [53]. S100B was found highly expressed by reactive astrocytes in close vicinity of β-amyloid deposits [54,55], and over expression of S100B may accelerate AD-like pathology [53,56]. High doses of S100B were found to stimulate astrocytes and microglia to produce pro-inflammatory cytokines including IL-1β, and modulate the expression and processing of the amyloid precursor protein (APP) to generate Aβ [57]. Evidence also suggests a role of S100B in the formation of neurofibrillary tangles by promoting the hyperphosphorylation of tau protein [58]. GFAP is commonly found to be increasingly expressed by reactive astrocytes in many brain areas suffering injury [52] and in AD brain [59]. Up-regulated S100B and GFAP found in SF and IF regions in AD brain in the current study confirmed neuronal dysfunction and neuro-inflammation involvement in AD brain.
4.2. AD-related neurofilament light chain (NEFL) and Neuromodulin (GAP43) are increased in 3D neuro-spheroids from AD patients
A number of up-regulated proteins from AD-derived 3D neuro-spheroids are localized in myelin sheath, such as NEFL, GAP43, VDAC1 and ATP5. These proteins have been linked to AD pathogenesis in previous studies [45–48,60,61]. Neurofilament protein is a component of the mature neuronal cytoskeleton and functionally maintains the neuronal caliber. Neurofilaments are a type IV intermediate filament of heteropolymers composed of light, medium, and heavy chains. A previous report suggests that NEFL might be a good biomarker for neuronal function [45]. Elevated CSF NEFL levels were found in subjects at the early clinical stage of AD, whereas levels of CSF NEFL were lowest in cognitively normal control group [45]. Neurons carrying dephosphorylated NF triplet proteins appear to be vulnerable to neurofibrillary tangle formation in AD [62]. Neurofilament polypeptides were also found to be localized to reactive axonal structures in physically damaged neurons in experimental trauma models [60]. GAP43 plays a role in axonal and dendritic filopodia induction. It is expressed at high levels in neuronal growth cones during development and axonal regeneration. This synaptic protein is considered a crucial component of an effective regenerative response in the nervous system. CSF proteomic profiling in two independent cohorts revealed elevated levels of GAP43 in AD patients [46]. Therefore, the up-regulation of NEFL and GAP43 from our AD-derived 3D neuro-spheroids is similar to those found in AD brains.
4.3. Mitochondrial dysfunction and oxidative stress are conserved in 3D neuro-spheroid and postmortem brain tissue from AD patients
Proteins related to mitochondria are well represented in both 3D neuro-spheroids and postmortem brain tissue from AD patients. These proteins appear to be essential for neuron homeostasis [63]. VDAC1 and ATP5B are components of the mitochondrial nucleoid and participate in mitochondrial transport. VDAC1 is central to mitochondria-mediated apoptosis. Altered VDAC1 activity has been linked to demyelination [64], and VDAC1 was found overexpressed in postmortem brains of AD patients [47]. It has been suggested that Aβ-mediated toxicity involves mitochondrial membrane VDAC1, leading to mitochondrial dysfunction and apoptosis induction [65]. Mitochondrial ATP synthase is a multiprotein complex that synthesizes ATP from ADP, utilizing an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation. The α-subunit of the ATP-synthase has been reported as one of the key targets of oxidative insult in AD brains [66]. The ATP synthase subunit β was found up-regulated in the hippocampus of APP-transgenic mice [48]. Therefore, the up-regulation of these mitochondrial proteins in our 3D neurons and brain tissue from AD patients reflects a mitochondrial dysfunction found in AD patients.
Mitochondrial dysfunction and oxidative stress constitute the most prominent features found in AD. Growing evidence has demonstrated that oxidative stress is an important factor contributing to the initiation and progression of AD. An increase in free radical production has been observed in several studies by treating cells with Aβ or by overexpressing APP [67,68]. In the present analysis of postmortem brain tissue, APP was found consistently up-regulated in the IF and SF region of the AD patients. Furthermore, several proteins (HP, HBA1, HBB and PRDX6) involved in cellular oxidant detoxification processes were found to be up-regulated in our analysis. However, the increase in inflammatory products during the process of conferring resistance to oxidative stress could have an opposing effect on AD pathology [69]. Cytochrome C oxidases were found to be down-regulated in the AD patients, e.g., four in the CRLM (COX, COX7C, COX4I1and COX6B1) and COX7A2L in IF region. Oxidative stress induced by β-amyloid can impair the activity of the mitochondrial respiratory chain and reduce COX activities, leading to neuronal dysfunction [70,71]. Oxidative stress can also lead to the alteration of glutathione metabolism and glutathione related enzymes [72]. In the present study, two proteins involved in the glutathione metabolic process, glutathione S-transferase P (GSTP1) and chloride intracellular channel protein 1 (CLIC1), were found down-regulated in 3D neurons derived from AD patients. Glutathione is the most abundant endogenous antioxidant and plays a significant role in combating oxidative stress. Previous studies have shown that glutathione S-transferase (GST) activity is significantly decreased in all areas of brain as well as in ventricular CSF of AD patients in comparison with healthy controls [73]. GSTP1 was proposed to be involved in critical regulation of a cyclin dependent kinase-5 (CDK5) activity, elimination of oxidative stress, and prevention of neurodegeneration in human AD brains [74]. Consistent with these reports, our results indicate that proteins related to oxidative stress are altered in both 3D neuro-spheroids and postmortem brain tissue, supporting a notion that our 3D neuro-spheroid culture can be an in vitro model to screen oxidative stress suppressors with a goal of slowing down AD progression.
4.4. Neuroinflammation in 3D neuro-spheroids without microglia exhibit different profiles from those of postmortem AD brain tissue
We have compared inflammatory responses in 3D neuro-spheroids and postmortem brain tissue from AD patients. Although inflammatory responses and oxidative stress are independent phenomena, they do overlap in some regards. Reactive oxygen species (ROS) can activate redox-sensitive transcription factors in glial cells, which can produce pro-inflammatory cytokines, leading to potentially neurotoxic reactive oxygen species and excitotoxins. Both microglia and astrocytes produce multiple pro-inflammatory factors, including cytokines (such as IL-6) [75]. Furthermore, enhanced production of pro-inflammatory cytokines such as IL-1β, IL-6, IL-18, and the up-regulation of their related receptors have been reported in the AD brains [76,77]. Consistent with these previous reports, our results indicate that proteins involved in inflammation, such as the IL-6 signaling pathway, were abnormal in post-mortem AD brain tissue. However, we did not observe similar inflammation proteins in the 3D neuro-spheroids. Since reactive astrocytes are induced by activated microglia [78], we believe that the difference is attributable to the absence of microglia cells in our 3D neuro-spheroids culture. In accord with this, a previous study on a glial-depleted culture system demonstrated that the presence of microglial cells is required to mediate the effects of neuroinflammation [79].
4.5. Vimentin is decreased in both AD-derived 3D neuro-spheroids and postmortem AD brain tissue
Vimentin was found decreased in both 3D neuron and brain regions (SF and IF) from AD patients in this study. Vimentin is known to provide protection against cellular stress, and expression of vimentin is decreased in AD [80]. In AD brains, co-localization of vimentin and plaques is found in the hippocampus, cerebral cortex and cerebellum [81]. Expression of vimentin may represent a cellular response to injury while maintaining dendrites and synaptic connections. Vimentin is involved in the maintenance of structural integrity of cells. It has been suggested that vimentin could participate in mitochondrial function; knock-down of vimentin expression results in mitochondrial fragmentation, swelling, and disorganization [82]. Vimentin is important for maintaining neuronal homeostasis, and a reduction in vimentin expression weakens neuronal response to stress and damages that are commonly present in the brains of AD patients.
4.6. Gaps in comparing proteomic profiles between 3D neuro-spheroid and postmortem AD brain tissue
A key drawback of the current study is the small sample size, which is in part limited by the number of samples that could be tagged by the 10-plex TMT system. To minimize the negative effect of this limitation, we calculated the fold changes of AD cases relative to the average values of the control subjects. This approach is more appropriate for small sample size studies versus comparing proteins using pooled samples. One advantage of this approach over the conventional approach of comparing the average of the AD group to that from the control group is that the later method could be biased by one subject having a protein with an extremely high or low ratio impacting the average values, especially in a study with a small sample size. A second limitation with this study is the difference in age between the AD and control groups at death; the average age of the AD subjects at time of postmortem brain collection was 76 while it was 83 for the non-AD controls. Age-related pathological alterations likely occurs in older subjects, the fact that our AD patients were younger rather than older than the controls should not introduce false positive outcomes despite the 7 year of difference in age. The age difference between the AD and control subjects donating blood for 3D neurons was 2 years. Third, we realize that it is difficult to reconstitute full neurodegenerative disease conditions in 3D neuro-spheroids in vitro over a relative short period of time as compared to the slow progressive pathogenic changes in brains of AD patients over several decades. It is also difficult to depict the pathological variations among brain regions most affected in AD patients [14]. Finally, we do not have a methodology set up to differentiate and co-culture microglia along with 3D neuro-spheroids. Given the importance of microglia cells in triggering an inflammation-induced neurodegeneration, it is necessary to incorporate microglia cells into the 3D neuron culture system in future studies.
4.7. Summary
We found alterations in axon proteins and immune response pathways in both 3D neuro-spheroids and brain tissues from AD patients compared to those from controls. These proteins have been implicated in specific pathological process of AD. The alteration of these proteins essentially affects homeostatic neural functions, such as axon growth and antioxidant defense, in both 3D neuro-spheroids and brains of AD patients. Therefore, proteomic profiles of our AD-derived 3D neuro-spheroid model illustrate certain levels of similarity when compared to those of human brain tissue. This correlation between in vivo and in vitro models demonstrates that candidate proteins identified from iPSC-derived 3D neuro-spheroids could be useful markers to reflect the status and progression of AD pathogenesis.
Supplementary Material
5. Acknowledgements
We thank Dr. Peter Morin, Dr. John Wells, Dr. Guy Surpris and Benjamin Morris-Eppolito for critical discussions. This study was supported by the award I21BX002215 and IO1 BX003527 from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development (WX) and the Cure Alzheimer’s Fund (WX). The views expressed in this article are those of the authors and do not represent the views of the US Department of Veterans Affairs or the US Government.
Abbreviation
- AD
Alzheimer’s disease
- CRLM
cerebellum
- DAVID
the Database for Annotation Visualization and Integrated Discovery
- ELISA
the enzyme-linked immunosorbent assay
- FDR
false discovery rate
- GO
Gene Ontology
- IF
inferior frontal cortex
- iPSC
induced pluripotent stem cells
- LC-MS/MS
liquid chromatography mass spectrometry
- PBMC
peripheral blood mononuclear cells
- SF
superior frontal cortex
- TMT
tandem mass tag
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jprot.2018.04.032.
References
- [1].Burns A, Iliffe S, Alzheimer’s disease, BMJ 338 (2009) b158. [DOI] [PubMed] [Google Scholar]
- [2].Tanzi RE, Bertram L, Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective, Cell 120 (2005) 545–555. [DOI] [PubMed] [Google Scholar]
- [3].Querfurth HW, LaFerla FM, Alzheimer’s disease, N. Engl. J. Med 362 (2010) 329–344. [DOI] [PubMed] [Google Scholar]
- [4].Selkoe DJ, Alzheimer’s disease is a synaptic failure, Science 298 (2002) 789–791. [DOI] [PubMed] [Google Scholar]
- [5].van der Zee J, Sleegers K, Van Broeckhoven C, Invited article: the Alzheimer disease-frontotemporal lobar degeneration spectrum, Neurology 71 (2008) 1191–1197. [DOI] [PubMed] [Google Scholar]
- [6].Berg L, McKeel DW Jr., Miller JP, Storandt M, Rubin EH, Morris JC, Baty J, Coats M, Norton J, Goate AM, Price JL, Gearing M, Mirra SS, Saunders AM, Clinicopathologic studies in cognitively healthy aging and Alzheimer’s disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype, Arch. Neurol. 55 (1998) 326–335. [DOI] [PubMed] [Google Scholar]
- [7].Gold G, Bouras C, Kovari E, Canuto A, Glaria BG, Malky A, Hof PR, Michel JP, Giannakopoulos P, Clinical validity of Braak neuropathological staging in the oldest-old, Acta Neuropathol. 99 (2000) 579–582 (discussion 583–4). [DOI] [PubMed] [Google Scholar]
- [8].Kraybill ML, Larson EB, Tsuang DW, Teri L, McCormick WC, Bowen JD, Kukull WA, Leverenz JB, Cherrier MM, Cognitive differences in dementia patients with autopsy-verified AD, Lewy body pathology, or both, Neurology 64 (2005) 2069–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Blessed G, Tomlinson BE, Roth M, The association between quantitative measures of dementia and senile change in the cerebral grey matter of elderly subjects, Br. J. Psychiatry 114 (1968) 797–811. [DOI] [PubMed] [Google Scholar]
- [10].Mungas D, Reed BR, Ellis WG, Jagust WJ, The effects of age on rate of progression of Alzheimer disease and dementia with associated cerebrovascular disease, Arch. Neurol 58 (2001) 1243–1247. [DOI] [PubMed] [Google Scholar]
- [11].Robinson JL, Geser F, Corrada MM, Berlau DJ, Arnold SE, Lee VM, Kawas CH, Trojanowski JQ, Neocortical and hippocampal amyloid-beta and tau measures associate with dementia in the oldest-old, Brain 134 (2011) 3708–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, Castellani RJ, Crain BJ, Davies P, Del Tredici K, Duyckaerts C, Frosch MP, Haroutunian V, Hof PR, Hulette CM, Hyman BT, Iwatsubo T, Jellinger KA, Jicha GA, Kovari E, Kukull WA, Leverenz JB, Love S, Mackenzie IR, Mann DM, Masliah E, McKee AC, Montine TJ, Morris JC, Schneider JA, Sonnen JA, Thal DR, Trojanowski JQ, Troncoso JC, Wisniewski T, Woltjer RL, Beach TG, Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature, J. Neuropathol. Exp. Neurol 71 (2012) 362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Blom ES, Giedraitis V, Zetterberg H, Fukumoto H, Blennow K, Hyman BT, Irizarry MC, Wahlund LO, Lannfelt L, Ingelsson M, Rapid progression from mild cognitive impairment to Alzheimer’s disease in subjects with elevated levels of tau in cerebrospinal fluid and the APOE epsilon4/epsilon4 genotype, Dement. Geriatr. Cogn. Disord 27 (2009) 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].D’Avanzo C, Aronson J, Kim YH, Choi SH, Tanzi RE, Kim DY, Alzheimer’s in 3D culture: challenges and perspectives, BioEssays 37 (2015) 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY, A three-dimensional human neural cell culture model of Alzheimer’s disease, Nature 515 (2014) 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee HK, Sanchez C, Chen M, Morin P, Wells J, Hanlon E, Xia W, Three dimensional human neuro-spheroid model of Alzheimer’s disease based on differentiated induced pluripotent stem cells, PLoS One (2016), 10.1371/journal.pone.0163072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS, Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells, Nature 482 (2012) 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N, Modeling familial Alzheimer’s disease with induced pluripotent stem cells, Hum. Mol. Genet 20 (2011) 4530–4539. [DOI] [PubMed] [Google Scholar]
- [19].Duan L, Bhattacharyya BJ, Belmadani A, Pan L, Miller RJ, Kessler JA, Stem cell derived basal forebrain cholinergic neurons from Alzheimer’s disease patients are more susceptible to cell death, Mol. Neurodegener 9 (2014) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB, Saurat N, McGlade A, Kirwan P, Blennow K, Hardy J, Zetterberg H, Livesey FJ, APP metabolism regulates tau proteostasis in human cerebral cortex neurons, Cell Rep. 11 (2015) 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, Young-Pearse TL, The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons, Hum. Mol. Genet 23 (2014) 3523–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M, Santa-Maria I, Zimmer M, Aubry S, Steele JW, Kahler DJ, Dranovsky A, Arancio O, Crary JF, Gandy S, Noggle SA, Characterization and molecular profiling of PSEN1 familial Alzheimer’s disease iPSC-derived neural progenitors, PLoS One 9 (2014) e84547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, Hibino S, Choshi T, Nakahata T, Hioki H, Kaneko T, Naitoh M, Yoshikawa K, Yamawaki S, Suzuki S, Hata R, Ueno SI, Seki T, Kobayashi K, Toda T, Murakami K, Irie K, Klein WL, Mori H, Asada T, Takahashi R, Iwata N, Yamanaka S, Inoue H, Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness, Cell Stem Cell 5909 (2013) 009. [DOI] [PubMed] [Google Scholar]
- [24].Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, Li Z, Yuan SH, Zhang K, Goldstein LS, The presenilin-1 DeltaE9 mutation results in reduced gamma-secretase activity, but not total loss of PS1 function, in isogenic human stem cells, Cell Rep. 5 (2013) 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Koch P, Tamboli IY, Mertens J, Wunderlich P, Ladewig J, Stuber K, Esselmann H, Wiltfang J, Brustle O, Walter J, Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of gamma-secretase activity in endogenous amyloid-beta generation, Am. J. Pathol 180 (2012) 2404–2416. [DOI] [PubMed] [Google Scholar]
- [26].Choi SH, Tanzi RE, iPSCs to the rescue in Alzheimer’s research, Cell Stem Cell 10 (2012) 235–236. [DOI] [PubMed] [Google Scholar]
- [27].Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH, Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes, PLoS One 11 (2016) e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Seo J, Kritskiy O, Watson LA, Barker SJ, Dey D, Raja WK, Lin YT, Ko T, Cho S, Penney J, Silva MC, Sheridan SD, Lucente D, Gusella JF, Dickerson BC, Haggarty SJ, Tsai LH, Inhibition of p25/Cdk5 attenuates tauopathy in mouse and iPSC models of frontotemporal dementia, J. Neurosci 37 (2017) 9917–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shen L, Liao L, Chen C, Guo Y, Song D, Wang Y, Chen Y, Zhang K, Ying M, Li S, Liu Q, Ni J, Proteomics analysis of blood serums from Alzheimer’s disease patients using iTRAQ labeling technology, J. Alzheimers Dis 56 (2017) 361–378. [DOI] [PubMed] [Google Scholar]
- [30].Song F, Poljak A, Kochan NA, Raftery M, Brodaty H, Smythe GA, Sachdev PS, Plasma protein profiling of mild cognitive impairment and Alzheimer’s disease using iTRAQ quantitative proteomics, Proteome Sci. 12 (2014) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Muenchhoff J, Poljak A, Song F, Raftery M, Brodaty H, Duncan M, McEvoy M, Attia J, Schofield PW, Sachdev PS, Plasma protein profiling of mild cognitive impairment and Alzheimer’s disease across two independent cohorts, J. Alzheimers Dis 43 (2015) 1355–1373. [DOI] [PubMed] [Google Scholar]
- [32].Bai B, Tan H, Peng J, Quantitative phosphoproteomic analysis of brain tissues, Methods Mol. Biol 1598 (2017) 199–211. [DOI] [PubMed] [Google Scholar]
- [33].Wang S, Yang F, Petyuk VA, Shukla AK, Monroe ME, Gritsenko MA, Rodland KD, Smith RD, Qian WJ, Gong CX, Liu T, Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer’s disease, J. Pathol 243 (2017) 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lv J, Ma S, Zhang X, Zheng L, Ma Y, Zhao X, Lai W, Shen H, Wang Q, Ji J, Quantitative proteomics reveals that PEA15 regulates astroglial Abeta phagocytosis in an Alzheimer’s disease mouse model, J. Proteome 110 (2014) 45–58. [DOI] [PubMed] [Google Scholar]
- [35].Lee JS, Jeremic A, Shin L, Cho WJ, Chen X, Jena BP, Neuronal porosome proteome: molecular dynamics and architecture, J. Proteome 75 (2012) 3952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lovell MA, Xiong S, Markesbery WR, Lynn BC, Quantitative proteomic analysis of mitochondria from primary neuron cultures treated with amyloid beta peptide, Neurochem. Res 30 (2005) 113–122. [DOI] [PubMed] [Google Scholar]
- [37].Jensen P, Myhre CL, Lassen PS, Metaxas A, Khan AM, Lambertsen KL, Babcock AA, Finsen B, Larsen MR, Kempf SJ, TNFalpha affects CREB-mediated neuroprotective signaling pathways of synaptic plasticity in neurons as revealed by proteomics and phospho-proteomics, Oncotarget 8 (2017) 60223–60242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ng A, Chew I, Narasimhalu K, Kandiah N, Effectiveness of Montreal cognitive assessment for the diagnosis of mild cognitive impairment and mild Alzheimer’s disease in Singapore, Singap. Med. J 54 (2013) 616–619. [DOI] [PubMed] [Google Scholar]
- [39].Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT, National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach, Acta Neuropathol. 123 (2012) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L, Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures, Cell Res. 21 (2011) 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee HK, Morin P, Wells J, Hanlon EB, Xia W, Induced pluripotent stem cells (iPSCs) derived from frontotemporal dementia patient’s peripheral blood mononuclear cells, Stem Cell Res. 15 (2015) 325–327. [DOI] [PubMed] [Google Scholar]
- [42].Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Pasca SP, Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture, Nat. Methods 12 (2015) 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [44].Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA, The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists, Genome Biol. 8 (2007) R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zetterberg H, Skillback T, Mattsson N, Trojanowski JQ, Portelius E, Shaw LM, Weiner MW, Blennow K, Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression, JAMA Neurol. 73 (2016) 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Remnestal J, Just D, Mitsios N, Fredolini C, Mulder J, Schwenk JM, Uhlen M, Kultima K, Ingelsson M, Kilander L, Lannfelt L, Svenningsson P, Nellgard B, Zetterberg H, Blennow K, Nilsson P, Haggmark-Manberg A, CSF profiling of the human brain enriched proteome reveals associations of neuromodulin and neurogranin to Alzheimer’s disease, Proteom. Clin. Appl 10 (2016) 1242–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cuadrado-Tejedor M, Vilarino M, Cabodevilla F, Del Rio J, Frechilla D, Perez-Mediavilla A, Enhanced expression of the voltage-dependent anion channel 1 (VDAC1) in Alzheimer’s disease transgenic mice: an insight into the pathogenic effects of amyloid-beta, J. Alzheimers Dis 23 (2011) 195–206. [DOI] [PubMed] [Google Scholar]
- [48].Takano M, Yamashita T, Nagano K, Otani M, Maekura K, Kamada H, Tsunoda S, Tsutsumi Y, Tomiyama T, Mori H, Matsuura K, Matsuyama S, Proteomic analysis of the hippocampus in Alzheimer’s disease model mice by using two-dimensional fluorescence difference in gel electrophoresis, Neurosci. Lett 534 (2013) 85–89. [DOI] [PubMed] [Google Scholar]
- [49].Zhan X, Jickling GC, Ander BP, Liu D, Stamova B, Cox C, Jin LW, DeCarli C, Sharp FR, Myelin injury and degraded myelin vesicles in Alzheimer’s disease, Curr. Alzheimer Res 11 (2014) 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhan X, Jickling GC, Ander BP, Stamova B, Liu D, Kao PF, Zelin MA, Jin LW, DeCarli C, Sharp FR, Myelin basic protein associates with AbetaPP, Abeta1–42, and amyloid plaques in cortex of Alzheimer’s disease brain, J. Alzheimers Dis 44 (2015) 1213–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Selkoe DJ, Brown BA, Salazar FJ, Marotta CA, Myelin basic protein in Alzheimer disease neuronal fractions and mammalian neurofilament preparations, Ann. Neurol 10 (1981) 429–436. [DOI] [PubMed] [Google Scholar]
- [52].Liao CW, Fan CK, Kao TC, Ji DD, Su KE, Lin YH, Cho WL, Brain injury-associated biomarkers of TGF-beta1, S100B, GFAP, NF-L, tTG, AbetaPP, and tau were concomitantly enhanced and the UPS was impaired during acute brain injury caused by Toxocara canis in mice, BMC Infect. Dis 8 (2008) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Leclerc E, Sturchler E, Vetter SW, The S100B/RAGE Axis in Alzheimer’s disease, Cardiovasc. Psychiatry Neurol. 2010 (2010) 539–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mori T, Koyama N, Arendash GW, Horikoshi-Sakuraba Y, Tan J, Town T, Overexpression of human S100B exacerbates cerebral amyloidosis and gliosis in the Tg2576 mouse model of Alzheimer’s disease, Glia 58 (2010) 300–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mrak RE, Sheng JG, Griffin WS, Correlation of astrocytic S100 beta expression with dystrophic neurites in amyloid plaques of Alzheimer’s disease, J. Neuropathol. Exp. Neurol 55 (1996) 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Petzold A, Jenkins R, Watt HC, Green AJ, Thompson EJ, Keir G, Fox NC, Rossor MN, Cerebrospinal fluid S100B correlates with brain atrophy in Alzheimer’s disease, Neurosci. Lett 336 (2003) 167–170. [DOI] [PubMed] [Google Scholar]
- [57].Anderson PJ, Watts HR, Jen S, Gentleman SM, Moncaster JA, Walsh DT, Jen LS, Differential effects of interleukin-1beta and S100B on amyloid precursor protein in rat retinal neurons, Clin. Ophthalmol 3 (2009) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Businaro R, Leone S, Fabrizi C, Sorci G, Donato R, Lauro GM, Fumagalli L, S100B protects LAN-5 neuroblastoma cells against Abeta amyloid-induced neurotoxicity via RAGE engagement at low doses but increases Abeta amyloid neurotoxicity at high doses, J. Neurosci. Res 83 (2006) 897–906. [DOI] [PubMed] [Google Scholar]
- [59].Hoozemans JJ, Rozemuller AJ, van Haastert ES, Eikelenboom P, van Gool WA, Neuroinflammation in Alzheimer’s disease wanes with age, J. Neuroinflammation 8 (2011) 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dickson TC, Chuckowree JA, Chuah MI, West AK, Vickers JC, alpha-Internexin immunoreactivity reflects variable neuronal vulnerability in Alzheimer’s disease and supports the role of the beta-amyloid plaques in inducing neuronal injury, Neurobiol. Dis 18 (2005) 286–295. [DOI] [PubMed] [Google Scholar]
- [61].Beck SJ, Guo L, Phensy A, Tian J, Wang L, Tandon N, Gauba E, Lu L, Pascual JM, Kroener S, Du H, Deregulation of mitochondrial F1FO-ATP synthase via OSCP in Alzheimer’s disease, Nat. Commun 7 (2016) 11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vickers JC, Morrison JH, Friedrich VL Jr., Elder GA, Perl DP, Katz RN, Lazzarini RA, Age-associated and cell-type-specific neurofibrillary pathology in transgenic mice expressing the human midsized neurofilament subunit, J. Neurosci 14 (1994) 5603–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Funfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, Brinkmann BG, Kassmann CM, Tzvetanova ID, Mobius W, Diaz F, Meijer D, Suter U, Hamprecht B, Sereda MW, Moraes CT, Frahm J, Goebbels S, Nave KA, Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity, Nature 485 (2012) 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gonzalez S, Berthelot J, Jiner J, Perrin-Tricaud C, Fernando R, Chrast R, Lenaers G, Tricaud N, Blocking mitochondrial calcium release in Schwann cells prevents demyelinating neuropathies, J. Clin. Invest 127 (2017) 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Smilansky A, Dangoor L, Nakdimon I, Ben-Hail D, Mizrachi D, Shoshan-Barmatz V, The voltage-dependent anion channel 1 mediates amyloid beta toxicity and represents a potential target for Alzheimer disease therapy, J. Biol. Chem 290 (2015) 30670–30683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Terni B, Boada J, Portero-Otin M, Pamplona R, Ferrer I, Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer’s disease pathology, Brain Pathol. 20 (2010) 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Keil U, Bonert A, Marques CA, Scherping I, Weyermann J, Strosznajder JB, Muller-Spahn F, Haass C, Czech C, Pradier L, Muller WE, Eckert A, Amyloid beta-induced changes in nitric oxide production and mitochondrial activity lead to apoptosis, J. Biol. Chem 279 (2004) 50310–50320. [DOI] [PubMed] [Google Scholar]
- [68].Schilling T, Eder C, Amyloid-beta-induced reactive oxygen species production and priming are differentially regulated by ion channels in microglia, J. Cell. Physiol 226 (2011) 3295–3302. [DOI] [PubMed] [Google Scholar]
- [69].Yun HM, Jin P, Han JY, Lee MS, Han SB, Oh KW, Hong SH, Jung EY, Hong JT, Acceleration of the development of Alzheimer’s disease in amyloid beta-infused peroxiredoxin 6 overexpression transgenic mice, Mol. Neurobiol 48 (2013) 941–951. [DOI] [PubMed] [Google Scholar]
- [70].Askanas V, McFerrin J, Baque S, Alvarez RB, Sarkozi E, Engel WK, Transfer of beta-amyloid precursor protein gene using adenovirus vector causes mitochondrial abnormalities in cultured normal human muscle, Proc. Natl. Acad. Sci. U. S. A 93 (1996) 1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yao J, Brinton RD, Targeting mitochondrial bioenergetics for Alzheimer’s prevention and treatment, Curr. Pharm. Des 17 (2011) 3474–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mazzetti AP, Fiorile MC, Primavera A, Lo Bello M, Glutathione transferases and neurodegenerative diseases, Neurochem. Int 82 (2015) 10–18. [DOI] [PubMed] [Google Scholar]
- [73].Lovell MA, Xie C, Markesbery WR, Decreased glutathione transferase activity in brain and ventricular fluid in Alzheimer’s disease, Neurology 51 (6) (1998) 1562. [DOI] [PubMed] [Google Scholar]
- [74].Sun KH, Chang KH, Clawson S, Ghosh S, Mirzaei H, Regnier F, Shah K, Glutathione-S-transferase P1 is a critical regulator of Cdk5 kinase activity, J. Neurochem 118 (2011) 902–914. [DOI] [PubMed] [Google Scholar]
- [75].Heneka MT, O’Banion MK, Inflammatory processes in Alzheimer’s disease, J. Neuroimmunol 184 (2007) 69–91. [DOI] [PubMed] [Google Scholar]
- [76].Sastre M, Klockgether T, Heneka MT, Contribution of inflammatory processes to Alzheimer’s disease: molecular mechanisms, Int. J. Dev. Neurosci 24 (2006) 167–176. [DOI] [PubMed] [Google Scholar]
- [77].Ojala J, Alafuzoff I, Herukka SK, van Groen T, Tanila H, Pirttila T, Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients, Neurobiol. Aging 30 (2009) 198–209. [DOI] [PubMed] [Google Scholar]
- [78].Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, Stevens B, Barres BA, Neurotoxic reactive astrocytes are induced by activated microglia, Nature 541 (2017) 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hui CW, Zhang Y, Herrup K, Non-neuronal cells are required to mediate the effects of neuroinflammation: results from a neuron-enriched culture system, PLoS One 11 (2016) e0147134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Shafi O, Inverse relationship between Alzheimer’s disease and cancer, and other factors contributing to Alzheimer’s disease: a systematic review, BMC Neurol. 16 (2016) 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Levin EC, Acharya NK, Sedeyn JC, Venkataraman V, D’Andrea MR, Wang HY, Nagele RG, Neuronal expression of vimentin in the Alzheimer’s disease brain may be part of a generalized dendritic damage-response mechanism, Brain Res. 1298 (2009) 194–207. [DOI] [PubMed] [Google Scholar]
- [82].Tang HL, Lung HL, Wu KC, Le AH, Tang HM, Fung MC, Vimentin supports mitochondrial morphology and organization, Biochem. J 410 (2008) 141–146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.