Abstract
Background
Human immunodeficiency virus (HIV) infection causes impairment of the gastrointestinal barrier, with substantial depletion of CD4+ T cells in the gut. Antiretroviral therapy (ART) restores CD4+ counts and may have beneficial effects on gut microbiota in adults. Little is known about effect of long-term ART on gut microbiome in HIV-infected children. We investigated composition of gut microbiota in HIV-infected and -uninfected children and assessed associations between gut microbiota and patient characteristics.
Methods
In a cross-sectional study, rectal swabs were collected from 177 HIV-infected and 103 HIV-uninfected controls. Gut microbial composition was explored using 16S ribosomal ribonucleic acid sequencing.
Results
Human immunodeficiency virus-infected children had significantly lower alpha-diversity and higher beta-diversity compared to HIV-uninfected. No association was observed between microbiome diversity and CD4+ T-cell count, HIV viral load, or HIV-associated chronic lung disease. We found enriched levels of Corynebacterium (P < .01), Finegoldia (P < .01), and Anaerococcus (P < .01) in HIV-infected participants and enrichment of Enterobacteriaceae (P = .02) in participants with low CD4+ counts (<400 cells/mm3). Prolonged ART-treatment (≥10 years) was significantly associated with a richer gut microbiota by alpha diversity.
Conclusions
Human immunodeficiency virus-infected children have altered gut microbiota. Prolonged ART may restore the richness of the microbiota closer to that of HIV-uninfected children.
Keywords: Africa, antiretroviral therapy, children, gut microbiota, HIV infection
HIV-infected African children and adolescents have altered gut microbiota compared to HIV uninfected. ART was significantly associated with a higher alpha diversity, and prolonged ART may restore richness of the microbiota closer to that of HIV-uninfected children.
The gastrointestinal (GI) tract plays an important role in the pathogenesis of human immunodeficiency virus (HIV) infection, with the majority of CD4+ T cells residing in the GI tract and associated lymphatic tissue [1]. Human immunodeficiency virus-induced depletion of CD4+ T cells causes structural impairment of the GI epithelial barrier, systemic microbial translocation, and ultimately alteration of the gut microbial community composition [2].
Recent evidence indicates that HIV-associated gut dysbiosis is characterized by decreased abundance of commensal (protective) bacteria and enrichment of potentially pathogenic taxa [3]. For example, the genera Pseudomonas, Enterobacteriaceae, Acinetobacter, and Campylobacter are thought to have infectious and inflammatory properties and are enriched in adults with HIV [3, 4].
Studies show that altered gut microbiota is associated with elevated circulating inflammatory markers such as C-reactive protein and interleukin-6 [5–8] as well as markers of microbial translocation such as lipopolysaccharide and lipopolysaccharide binding protein [9, 10]. Furthermore, studies suggest that antiretroviral therapy (ART) may only partially restore the gut microbiota towards levels observed in HIV-uninfected populations, and patients continue to suffer from dysbiosis even when HIV infection is controlled [1, 11, 12].
Moreover, gut dysbiosis, and associated microbial translocation may drive systemic chronic inflammation, which increases the risk of chronic noninfectious HIV complications, such as cardiovascular disease and lung complications [13–16].
Few studies have investigated the gut microbiome in sub-Saharan African children and its relation to the development of HIV-associated chronic complications. Most studies to date have been performed in adult populations and potentially confounded by sexual preference and are therefore not directly comparable to our study. The overall aim of our study was to investigate the gut microbiota in HIV-infected and HIV-uninfected children in Harare, Zimbabwe and to evaluate the association between gut microbial composition and clinical and laboratory parameters (chronic lung disease, CD4+ T-cell count, viral load [VL]).
MATERIALS AND METHODS
Study Population
This study investigated bacterial profiles of rectal swabs collected from participants enrolled to the Bronchopulmonary Function in Response to Azithromycin Treatment for Chronic Lung Disease in HIV-infected Children (BREATHE) trial [17] (clinicaltrials.gov identifier NCT02426112). Chronic lung disease was defined as forced expiratory volume in 1 second (FEV1) z-score less than −1.0 with no reversibility (<12% improvement in FEV1 after 200 μg of salbutamol inhaled using a spacer). The detailed study protocol has been described previously [17]. For the present substudy, only participants enrolled in Harare, Zimbabwe were included. HIV-infected children aged 6–16 years without chronic lung disease, defined as no prior history of heart/lung diseases, tuberculosis (TB), no chronic cough, reported chest pain or shortness of breath during exercise, and HIV-uninfected participants were recruited at the same outpatient clinic. These were recruited as comparison groups and not randomized into the trial. The route of HIV transmission was likely perinatal for most of the HIV-infected participants. Human immunodeficiency virus-infected participants had to be stable on ART for at least 6 months, to meet eligibility criteria. All study participants completed a detailed questionnaire regarding demographic, socioeconomic characteristics and clinical history.
The study was approved by the following: London School of Hygiene and Tropical Medicine Ethics Committee; Harare Central Hospital Ethics Committee; Medical Research Council of Zimbabwe; The Regional Committee for Medical and Health Research Ethics REC North 2015/1650; and University of Cape Town Human Research Ethics Committee. All participants and/or legal guardians gave written informed consent to participate in the study.
Sample Collection
Rectal swabs were collected from all participants at enrollment into the trial by study nurses. Swabs were immediately preserved in 1.5 mL of transport medium PrimeStore MTM (Longhorn Diagnostics, Bethesda, MD), directly stored on ice for maximum 1 hour, and then frozen at −80°C before shipment on dry ice to the laboratory at the University of Cape Town.
Deoxyribonucleic Acid Extraction
The Zymo Research Quick-DNA Fecal/Soil Microbe Microprep kit (Zymo Research, Irvine, CA) was used for deoxyribonucleic acid (DNA) extractions. Deoxyribonucleic acid was extracted according to the manufacturer's description, with modifications. In brief, a 400-μL aliquot of each sample was mixed with 400 μL BashingBead Buffer in a ZR BashingBead Lysis Tube. Mechanical lysis (bead beating) was performed using the TissueLyser LT (QIAGEN, Hilden, Germany) set to 50 Hz for 5 minutes. Supernatant (500 μL) was transferred to a Zymo-Spin III-F Filter (Zymo Research, Irvine, CA) and centrifuged at 8000 ×g for 1 minute. Chemical lysis was done by adding Genomic Lysis Buffer. All other procedures were done according to manufacturer's protocol.
16S Library Preparation and Gene Sequencing
To assess DNA quality and total bacterial load, a real-time quantitative polymerase chain reaction (PCR) was performed as previously described [18]. Subsequently, 2 PCR sets targeting the V4 hypervariable region of the 16S ribosomal ribonucleic acid (rRNA) gene were performed according to previously described protocols [19, 20] (Supplementary Data).
Samples were sequenced on an Illumina Miseq instrument using the Miseq Reagent v3 kit (600 cycles) (Illumina, San Diego, CA). The final library was diluted to a 6-pM concentration, and a 25% PhiX library spike-in was added as internal control [21]. The preprocessing of sequence reads was done using the H3ABioNet 16S rDNA diversity analysis package (https://github.com/h3abionet/h3abionet16S) [20], with the exception that taxonomy of representative reads was assigned using SILVA version 132. Raw sequence files have been submitted to the European Nucleotide Archive, accession number PRJEB32077.
Data Analysis
Statistical analyses were performed in STATA 14 (StataCorp LLC, College Station, TX) and R Statistical software (http://www.r-project.org/). Characteristics between study groups were compared using Fisher's exact test (for categorical parameters) and Kruskal-Wallis or Wilcoxon rank-sum test (for continuous parameters).
Richness of bacterial taxa within a single sample was represented by the number of operational taxonomic units (OTUs) and Chao1 index [22]. Chao1 index uses mark-release-recapture-like ratio to estimate richness by adding a correction factor to the observed number of species. Richness and evenness (relative abundances of the different species) were characterized by Shannon's index [23]. Alpha diversity measures were calculated at sampling depth 4000 reads to include 95% of samples.
Interindividual differences, beta diversity, were determined using Bray-Curtis dissimilarity index [24] with sampling depth set at 2000 reads to include 99% of samples. Beta diversity comparisons were explored using Principal Coordinate plots generated by the stats package in R (version 3.4.4). Comparisons were made using Wilcoxon rank-sum test where not specified otherwise. We also used Kruskal-Wallis test in cases with more than 2 groups. The same groups were compared using permutational multivariate analysis of variance in QIIME2 (version 2018.4) [25], with number of permutations set to 999. P values were adjusted for multiple testing using the Benjamini-Hochberg method [26].
Relative Abundance
To assess relative abundance, a linear discriminant analysis was performed using linear discriminant analysis effect size [27] with default settings (alpha values for the statistical test 0.05). To reduce the number of markers, the effect size threshold was set to 1.0 for the plots. Relative abundance comparison plots were generated using the MicrobiomeAnalyst web-based software tool with standard feature filtering [28]. Heatmaps for comparing relative abundance of specific taxa between groups were generated using only the taxa found to be significantly different by linear discriminant analysis effect size comparison (Supplementary Figures 1–3). The average fraction of each taxa was calculated from all samples within each group. The data were transformed to fractional abundance (Phyloseq) before performing the linear discriminant analysis effect size analysis. All P values reported are corrected for multiple testing using false discovery rate (FDR).
Alpha diversity indices between study groups were compared using Wilcoxon rank-sum test. P values were corrected for multiple testing using FDR. Spearman's rank correlation with Bonferroni correction was used to assess the association between alpha diversity indices and continuous parameters. We fitted a linear regression model to estimate the association between HIV status and alpha diversity indices. Body mass index (BMI), age, and sex were adjusted for a priori. An interaction term between HIV status and antibiotics the 3 previous months (co-trimoxazole prophylaxis for HIV-infected participants) was included into the regression model to determine whether antibiotics modify the effect of HIV status on alpha diversity estimates. The association between other participant characteristics and alpha diversity indices was further evaluated in regression analysis stratified by HIV status and adjusted for BMI, age, and sex. A 2-tailed significance level of 0.05 was used.
RESULTS
Study Population
In total, 149 HIV-infected participants with chronic lung disease, 28 HIV-infected participants without chronic lung disease, and 103 HIV-uninfected participants were enrolled. All HIV-infected participants were on ART, for a median of 6.6 years for those with chronic lung disease and 8.0 for those without. Eighty-nine percent of HIV-infected participants were taking co-trimoxazole prophylaxis as per World Health Organization guidelines [29]. No HIV-uninfected participants were taking co-trimoxazole. The study group characteristics are presented in Table 1.
Table 1.
Characteristics of Study Participants
| Parameter | HIV− (N = 103) | HIV+ Chronic Lung Disease+ (N = 149) | HIV+ Chronic Lung Disease− (N = 28) |
|---|---|---|---|
| Age, median (IQR) | 9.9 (7.4–12.7) | 15.5 (12.8–17.7) | 16.7 (11.7–18.1) |
| Male, N (%) | 53 (52) | 84 (56) | 8 (29) |
| BMI-for-age z score, median (IQR)a | −0.24 (−0.69 to 0.35) | −1.19 (−1.80 to −0.62) | −0.11 (−0.73 to 0.61) |
| Stunted (height-for-age z-score ≤2), N (%)a | 5 (5) | 66 (44) | 7 (25) |
| Underweight (weight-for-age z-score ≤2), N (%)a | 5 (5) | 78 (52) | 2 (7) |
| Took antibiotics the 3 previous months for HIV-uninfected group or co-trimoxazole prophylaxis for HIV-infected group, N (%) | 2 (2) | 133 (89) | 25 (89) |
| Episodes of diarrhea during the last 3 months, N (%)b | 3 (3) | 11(13) | 1 (4) |
| Residential Area, N (%)c | |||
| High density | 107 (95) | 83 (98) | 24 (86) |
| Medium density | 4 (4) | 1 (1) | 3 (11) |
| Low density | 2 (2) | 1 (1) | 1 (4) |
| HIV-Related Parameters | |||
| ART Regimen, N (%) | |||
| NNRTI-based regimen | - | 93 (62) | 24 (86) |
| PI-based regimen | - | 56 (38) | 4 (14) |
| CD4 count ≤400 cells/mm, N (%)d | - | 40 (27) | 9 (32) |
| VL suppression (VL <1000 copies/mL), N (%) | - | 87 (58) | 17 (61) |
| Age at ART initiation, median (IQR)d | - | 8.2 (5.2–11.4) | 8.6 (5.0–9.9) |
| Years spent on ART, median (IQR)d | - | 6.6 (4.4–8.4) | 8.0 (5.0–9.1) |
| ART Duration Categories, N (%)d | |||
| <5 years | - | 46 (31) | 7 (25) |
| 5–10 years | - | 83 (56) | 17 (61) |
| ≥10 years | - | 19 (13) | 4 (14) |
| Previously treated for TB, N (%) | - | 54 (36) | 2 (7) |
Abbreviations: ART, antiretroviral therapy; BMI, body mass index; HIV, human immunodeficiency virus; IQR, interquartile range; NNRTI, nonnucleoside reverse-transcriptase inhibitor; PI, protease inhibitor; TB, tuberculosis; VL, viral load.
aParameters were calculated using British 1990 Growth Reference Curves.
bData on episodes of diarrhea during the last 3 months were missing for 64 participants.
cData on residential area were missing for 64 participants.
dData was missing for 1 participant.
Human immunodeficiency virus-infected participants were older compared to the HIV-uninfected participants (15.6 years, interquartile range [IQR] = 12.8–17.7 vs 9.9 years, IQR = 7.4–12.7; P < .001) and were more likely to be stunted and underweight compared to HIV-uninfected participants (stunted 41% vs 5%, P < .001; underweight 45% vs 5%, P < .001). The proportion of participants who experienced diarrhoeal episodes during the last 3 months before enrollment was also higher in the HIV-infected group than in the HIV-uninfected group (11% vs 3%, P = .03).
Alpha Diversity
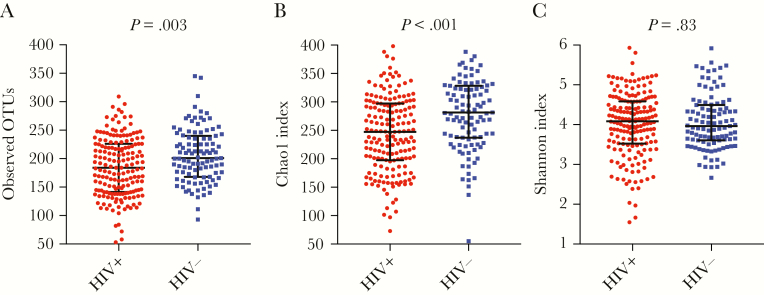
Species richness (OTUs, Chao1) was significantly higher in HIV-uninfected participants compared to HIV-infected participants. There was no difference in Shannon index between these 2 study groups (Figure 1, Supplementary Table 1). After adjustment for BMI, age, and sex using linear regression analysis, the negative association between richness indices and HIV status remained significant (P = .02 for OTUs, P = .001 for Chao1 index). The use of antibiotics during the 3 previous months did not change the significant effect of positive HIV status for the Chao1 index (Supplementary Table 2). Human immunodeficiency virus-infected participants with suppressed VL had borderline higher OTUs (median 192.5 [IQR, 145.5–228.5] vs 176 [IQR, 138–220], P = .18) and higher Chao1 index (median 259.3 [IQR, 201.2–302.1] vs 233.2 [IQR, 175–276], P = .05) compared to nonsuppressed participants in regression analysis adjusted for BMI, age, and sex (Supplementary Tables 3 and 4).
Figure 1.
Alpha diversity indices in human immunodeficiency virus (HIV)-infected and there was found a significantly lower richness in HIV infected participants compared to HIV uninfected by (a) Observed OTUs and (b) Chao1 index. Mid line showing median and error bars showing the interquartile range. OTUs, operational taxonomic units.
We stratified HIV-infected participants based on their time spent on ART (ART <5 years [n = 53]; ART 5–10 years [n = 100]; ART ≥10 years [n = 23]). When comparing HIV-infected participants based on these subgroups, we found that participants who had been on ART ≥10 years had an alpha diversity similar to the HIV-uninfected study group (Table 2).
Table 2.
Alpha Diversity in HIV-Infected Participants Stratified by Years on ART and in HIV-Uninfected Participants
| Alpha diversity indices | HIV+, <5 Years on ART (N = 53) [Median (IQR)] |
HIV+, 5–10 Years on ART (N = 100) [Median (IQR)] |
HIV+, ≥10 Years on ART (N = 23) [Median (IQR)] |
HIV− Group (N = 103) [Median (IQR)] |
HIV+, <5 Years on ART vs HIV− (P Values*) |
HIV+, 5–10 Years on ART vs HIV− (P Values*) |
HIV+, ≥10 Years on ART vs HIV− (P Values*) |
|---|---|---|---|---|---|---|---|
| Observed OTUs | 176 (138–214) | 186.5 (143–223.5) | 204 (162–242) | 201 (168–240) | .001 | .10 | .28 |
| Chao1 | 229.4 (175.0–277.9) | 249.6 (200.2–299.6) | 268.9 (224.4–306) | 281.3 (237.2–328.4) | <.001 | .02 | .08 |
| Shannon index | 4.03 (3.48–4.39) | 4.12 (3.52–4.58) | 4.23 (3.82–4.84) | 4.0 (3.6–4.5) | .20 | .75 | .86 |
Abbreviations: ART, antiretroviral therapy; BMI, body mass index; HIV, human immunodeficiency virus; IQR, interquartile range; OTUs, operational taxonomic units.
*, Presented P values were obtained from regression model adjusted for BMI, age, and sex where HIV status with years of ART was introduced as an independent variable and alpha diversity estimates as a dependent (outcome) variable.
There was no difference in alpha diversity indices between HIV-infected participants with and without chronic lung disease (Supplementary Table 5). The same was observed after adjusting for BMI, age, and sex using regression analysis. The associations between participant characteristics and alpha diversity indices in HIV-infected participants is presented in Supplementary Table 4.
Prolonged ART treatment was the only parameter significantly associated with richer gut microbiota after adjustment for BMI, age and sex, suggesting a positive effect of prolonged ART. No parameters were found to be significantly associated with alpha diversity estimates in the HIV-uninfected group (Supplementary Table 6).
Beta Diversity
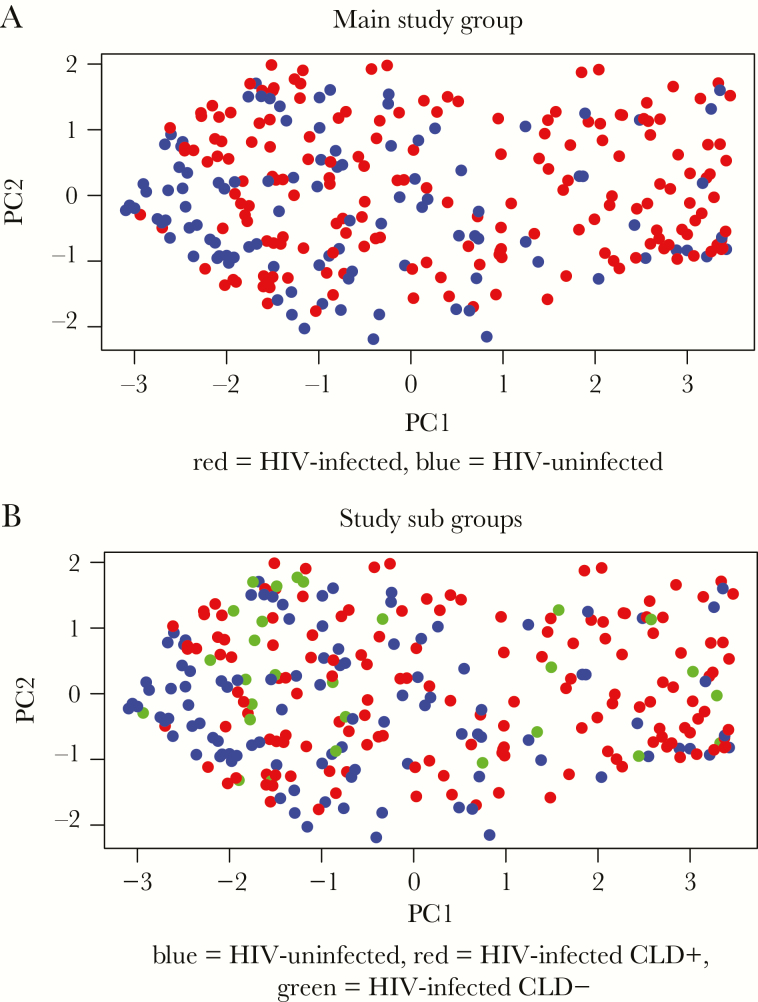
We found significantly higher beta diversity amongst HIV-infected participants compared to HIV-uninfected participants (P < .01) (Figure 2a). Antiretroviral therapy duration had no impact on beta diversity when stratified by years spent on ART. There was no association between beta diversity and VL suppression, type of ART regimen, time on ART, or prior TB in HIV-infected participants (Supplementary Table 7).
Figure 2.
Beta-diversity comparison between study groups. Principal cooridnate (PC) analysis plot showing beta-diversity by Bray-Curtis dissimilarity comparing (a) human immunodeficiency virus (HIV)-infected (red) and HIV-uninfected (blue) participants (P < .01) and (b) HIV-infected with chronic lung disease (red), HIV-infected without chronic lung disease (green), and HIV-uninfected (blue) participants. P value obtained using Wilcoxon rank-sum test.
Human immunodeficiency virus-infected participants with chronic lung disease had higher beta diversity compared to both HIV-uninfected (P < .01) and HIV-infected participants without chronic lung disease (P = .03). There was no significant difference between HIV-infected participants without chronic lung disease and HIV-uninfected participants (P = .74) (Figure 2b). Unweighted UniFrac analysis showed similar results.
Relative Abundance
We identified 26 different phyla in the rectal swabs from all participants. Only 5 phyla contributed more than 1% of the total sequences of the entire dataset. Firmicutes (43.9%), Bacteroidetes (33.9%), Epsilonbacteraeota (9%) (previously within the phylum Proteobacteria), Actinobacteria (5.3%) and Proteobacteria (7.7%) accounted for 99.8% of the bacteria present.
Human Immunodeficiency Virus (HIV)-Infected Versus HIV-Uninfected Participants
At phylum level, HIV-infected participants had significantly lower abundance of Epsilonbacteraeota (7%) (P < .01) and Bacteroidetes (32%) (P < .01) compared to HIV-uninfected participants (with 13% and 38%, respectively) (Supplementary Figure 4).
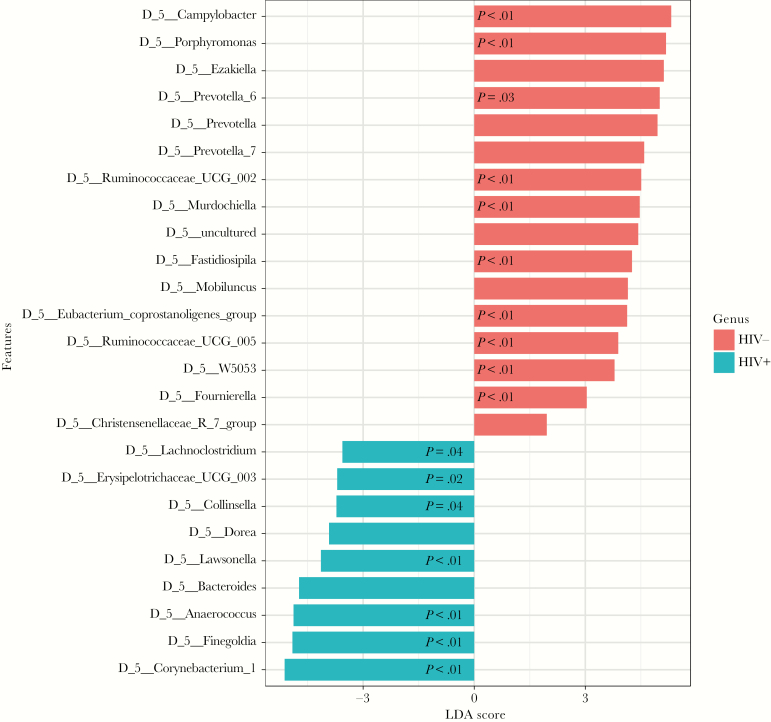
At genus level, HIV-infected participants had enriched Corynebacterium (P < .01), Lawsonella (P < .01), and Collinsella (P = .04), belonging to the Actinobacteria phylum; whereas in the Firmicutes phylum, Finegoldia (P < .01), Anaerococcus (P < .01), Erysipelotrichaceae (P = .02), and Lachnoclostridium (P = .04) were enriched when compared to HIV-uninfected participants.
Human immunodeficiency virus-uninfected participants, compared to HIV-infected participants, were enriched in Campylobacter (P < .01), phylum Epsilonbacteraeota; Porphyromonas (P < .01) and Prevotella (P = .03), phylum Bacteroidetes; and Eubacterium coprostanoligenes_group (P < .01), Ruminococcaceae (P < .01), Fastidiosipila (P < .01), Fournierella (P < .01), W5053 (P < .01), Coprococcus (P = .02), and Murdochiella (P < .01), phylum Firmicutes (Figure 3).
Figure 3.
Linear discriminant analysis effect size plot. This plot shows enriched taxa that are significantly different between human immunodeficiency virus (HIV)-infected (blue) and HIV-uninfected (red) participants. Only taxa meeting a significant level of 0.05 and effect size threshold of 1.0 are included. P values shown are only those significant after adjustment for false discovery rate.
Human immunodeficiency virus-infected participants with chronic lung disease had a higher abundance of the genus Faecalibacterium (P = .05), phylum Firmicutes, compared to participants without chronic lung disease. Participants without chronic lung disease had higher abundance of genus W5053 (P < .01), phylum Firmicutes and Prevotella (P = .05), phylum Bacteroidetes, compared to participants with chronic lung disease.
Characteristics of Human Immunodeficiency Virus-Infected Participants and Gut Microbiota
When we stratified HIV-infected participants based on CD4 count (CD4 ≤400 cells/mm3 vs >400 cells/mm3), we found no statistically significant differences at genus level. However, we found higher proportions at family level of Enterobacteriaceae (P = .02) and Burkholderiaceae (P = .04) in those with CD4 counts ≤400 cells/mm3, whereas Succinivibrionaceae (P = .04) was higher in those with CD4 counts >400 cells/mm3. No differences in relative abundance were found at any taxonomic level between virally suppressed and nonsuppressed participants (<1000 vs ≥1000 copies/mL).
We compared HIV-infected participants based on ART duration subgroups to HIV-uninfected participants using linear discriminant analysis effect size. Genera such as Bacteroides, Prevotella, Porphyromonas, Blautia, and Roseburia were similarly abundant in HIV-uninfected and HIV-infected participants who have been on ART ≥10 years (Supplementary Figures 1–3 and 5–8). This finding suggests that prolonged ART helps shift microbial composition towards that of HIV uninfected, but this result needs further investigation.We found no differences in relative abundance when comparing HIV-infected participants on ART <5 years to those on ART for 5–10 years or for ≥10 years.
DISCUSSION
Our study showed that gut microbiota in HIV-infected, ART-treated children was less diverse compared to HIV-uninfected children. Children who had been taking ART for 10 years or more had a more diverse microbiota resembling that of HIV-uninfected children. Our results suggest that prolonged ART may minimize differences in gut microbiota between HIV-infected and -uninfected children.
Impact of Human Immunodeficiency Virus on Gut Microbiota
Several studies in adults demonstrated that untreated HIV infection is associated with intestinal dysbiosis, reduced alpha diversity, and increased beta diversity [9, 30, 31]. These changes may persist despite ART [5, 6, 10, 32, 33]. Our results of overall lower alpha diversity and higher beta diversity in HIV-infected, ART-treated children support these findings.
Published data are less consistent with regards to relative abundance of specific taxa in HIV-infected individuals. Types of specimens used, study populations, geographical area, sequencing method, and false discovery may explain these conflicting results. For example, rectal swab analysis from HIV-infected, ART-treated adults in Nigeria found higher abundance of Finegoldia and Anaerococcus in HIV-infected individuals [34], which is consistent with our findings. However, in the same study, Campylobacter was significantly enriched in HIV-infected participants, whereas we found enriched Campylobacter in the HIV-uninfected group.
Several studies showed enriched levels of Proteobacteria in HIV-infected, ART-naive individuals [6, 7, 9], but only 1 study showed similar findings in ART-treated individuals [35]. We found enrichment of Proteobacteria in HIV-infected individuals, but this was not statistically significant.
Impact of Antiretroviral Therapy on Gut Microbiota
At least 2 studies have found a negative impact of ART on gut microbiota diversity [9, 34]. In a longitudinal study, Nowak et al [9] found a significant decrease in the number of observed species and the Shannon index after ART introduction. However, Nowak et al [9] investigated the effect of ART initiation, with a relatively short follow-up of 10 months. In our population, we had no ART-naive participants, and minimum duration of ART was 1 year. We observed lower alpha diversity in those on ART <10 years compared to HIV-uninfected participants.
Previous studies that investigated the gut microbiome in individuals on long-term ART reported similar alpha diversity profiles in HIV-infected, ART-treated and HIV-uninfected individuals [30, 35]. For example, Dinh et al [35] found no significant difference in alpha diversity measures between HIV-infected participants on ART for a median of 13.3 years and HIV-uninfected controls. This is similar to our findings for participants who received ART for 10 or more years. The impact of ART duration on gut microbiota was also noted by Lozupone et al [36] who found that individuals with longer ART duration showed closer resemblance to HIV-uninfected individuals than to subjects with untreated HIV infection. These studies support our findings of the possibility that long-term ART may restore HIV-associated dysbiotic gut microbiota.
We did not observe an association between immunological or virological markers (VL and CD4 count) and gut microbiome diversity measures. In contrast, other studies showed significantly lower microbiome diversity in those with more severe HIV status [9, 37, 38]. Findings of previous studies may have been affected by sample size and ART duration. A longitudinal study with repeated measurements of VL, CD4, and microbiome profiles is needed to uncover the relationship between these parameters.
We found enriched levels of Enterobacteriaceae in HIV-infected participants with low CD4+ T-cell counts (≤400 cells/mm3). Enterobacteriaceae may cause GI and urinary tract infections in HIV-infected children [38]; however, the clinical significance is unclear, because Enterobacteriaceae are found as part of the normal intestinal flora. Burkholderiaceae, also enriched in those with low CD4+ T-cell counts (≤400 cells/mm3), includes species known to cause severe lung infections in patients with cystic fibrosis [39].
Gut-Lung Axis
Recent evidence suggests that gut microbiome is involved in maintaining lung health, and an altered gut microbiome composition is often observed in patients with lung diseases [15, 16]. For example, low gut microbiome diversity during infancy has been linked to asthma at school age [32]. In our study, we did not observe any difference in alpha diversity estimates between participants with and without HIV-associated chronic lung disease, but there were some significant differences in relative abundance of specific taxa. For example, the genus Faecalibacterium was enriched in HIV-infected individuals with chronic lung disease, whereas Prevotella was enriched in HIV-infected individuals without chronic lung disease.
Faecalibacterium have previously been regarded as a protective commensal and is associated with a healthy gut. Depletion of this genus has been linked to the development of inflammatory bowel disease and asthma, and low levels have been shown in patients with cystic fibrosis [16, 40]. Some studies have challenged this, showing increased levels of the species Faecalibacterium prausnitzii in gut microbiome of pediatric patients with untreated Crohn's disease at the time of diagnosis [41]. It is interesting to note that a recent study also showed increased levels of Faecalibacterium in the gut microbiome of patients with active TB [42].
Co-trimoxazole Prophylaxis
Because the majority (89%) of HIV-infected participants in our study received co-trimoxazole prophylaxis, it is not possible to completely tease apart the effect of HIV from that of cotrimoxazole. Although it is known that antibiotics cause substantial changes in the gut microbiota, data regarding the impact of co-trimoxazole prophylaxis on gut microbiota in HIV-infected, ART-treated individuals are limited. However, recent evidence suggests that co-trimoxazole does not affect global gut microbial composition but rather specific inflammatory pathways in HIV-infected individuals [43]. In our study, the negative impact of positive HIV status on richness estimates remained significant after accounting for co-trimoxazole prophylaxis. In addition, no effect of co-trimoxazole administration on alpha diversity in HIV-infected participants was observed. Our results are in line with several other studies where no significant difference in alpha diversity was observed in HIV-infected individuals who took co-trimoxazole and those who did not [44–46].
Study Strengths and Limitations
Our study is one of the few to assess the gut microbiome composition in children and adolescents with perinatally acquired HIV infection. A relatively large sample size and detailed characteristics of study participants allowed us to perform extensive statistical analysis. All participants in our study were from the same region, thus increasing the internal validity of our data.
Our study was cross-sectional and is therefore unable to directly assess relationships over time. The group of HIV-infected participants without chronic lung disease was small and therefore gave limited power to detect differences. Furthermore, we did not assess diet and social factors such as housing or level of education, which may have an impact on gut microbiota. Age imbalance between HIV-infected and -uninfected participants is also a limitation of this study.
CONCLUSIONS
Our study is among the first to assess gut microbial composition of HIV-infected children and adolescents in a very high HIV burden setting. Our results indicate that gut microbiota is altered in HIV-infected children, although diversity improves with increasing duration of ART. Further studies, in which the gut microbiota, markers of microbial translocation, and immunological markers are measured, are warranted to provide better insight to the pathogenesis of HIV and its related complications.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all of the participants for taking part in this study. We also thank to the BREATHE study team members: Carmen Gonzalez-Martinez, Katharina Kranzer, Elizabeth L. Corbett, Hilda Mujuru, Sarah Rowland-Jones, Andrea M. Rehman, Tsitsi Bandason, Ethel Dauya, Edith Majonga, Beauty Makamure, Gugulethu Newton Mapurisa, Brewster Wisdom Moyo, Lucky Gift Ngwira, Jamie Rylance, Victoria Simms, Helen Anne Weiss, Louis-Marie Yindom, and Slee Mbhele.
Author contributions. M. P. N., T. F., J. P. C., S. C.-W., E. S., and T. T. F. conceived and designed the study and participated in data analysis and revision of the manuscript. The BREATHE study team was responsible for sample collection and management. T. T. F. performed the laboratory experiments, analyzed the data, and wrote the first draft of the manuscript. S. C.-W. performed the laboratory experiments, participated in data analysis, and reviewed the final manuscript. E. S. analyzed the data and wrote the first draft of the manuscript. E. H. and K. S. M. analyzed the data. J. Ø O., R. A. F., G. M., M. P. N., J. P. C., and T. F. revised the manuscript. All authors approved the final version.
Financial support. The BREATHE trial was funded by the Global Health and Vaccination Programme of the Medical Research Council of Norway. The analysis of rectal swabs was funded by Northern Norway Regional Health Authority (Helse Nord RHF), RH Grant Number 1448-19. The microbiome platform at the University of Cape Town is supported by National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director, National Institute of Environmental Health Sciences, and National Human Genome Institute of Health of the National Institutes of Health under Award Numbers U54HG009824 and 1U01HG006961.
Potential conflicts of interest. R. A. F. received a grant from Wellcome Trust. T. J. G. received personal fees from Gilead, ABBVIE, MSD, ROCHE, outside the submitted work, related to teaching on hepatitis B virus and hepatitis C virus. M. P. N. received a grant from the Research Council of Norway. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
BREATHE Study Team:
Carmen Gonzalez-Martinez, Katharina Kranzer, Elizabeth L Corbett, Hilda Mujuru, Sarah Rowland-Jones, Andrea M Rehman, Tsitsi Bandason, Ethel Dauya, Edith Majonga, Beauty Makamure, Gugulethu Newton Mapurisa, Brewster Wisdom Moyo, Lucky Gift Ngwira, Jamie Rylance, Victoria Simms, Helen Anne Weiss, Louis-Marie Yindom, and Slee Mbhele
References
- 1. Bhaijee F, Subramony C, Tang SJ, Pepper DJ. Human immunodeficiency virus-associated gastrointestinal disease: common endoscopic biopsy diagnoses. Patholog Res Int 2011; 2011:247923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 2004; 200:749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 2016; 11:182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mwansa J, Mutela K, Zulu I, Amadi B, Kelly P. Antimicrobial sensitivity in enterobacteria from AIDS patients, Zambia. Emerg Infect Dis 2002; 8:92–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 6. Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bandera A, De Benedetto I, Bozzi G, Gori A. Altered gut microbiome composition in HIV infection: causes, effects and potential intervention. Curr Opin HIV AIDS 2018; 13:73–80. [DOI] [PubMed] [Google Scholar]
- 9. Nowak P, Troseid M, Avershina E, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 2015; 29:2409–18. [DOI] [PubMed] [Google Scholar]
- 10. Ji Y, Zhang F, Zhang R, et al. Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count. Emerg Microbes Infect 2018; 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014; 5:562–70. [DOI] [PubMed] [Google Scholar]
- 12. Li SX, Armstrong A, Neff CP, Shaffer M, Lozupone CA, Palmer BE. Complexities of gut microbiome dysbiosis in the context of HIV infection and antiretroviral therapy. Clin Pharmacol Ther 2016; 99:600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Far M, Tremblay CL. Gut microbial diversity in HIV infection post combined antiretroviral therapy: a key target for prevention of cardiovascular disease. Curr Opin HIV AIDS 2018; 13:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med 2011; 62:141–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol 2017; 15:55–63. [DOI] [PubMed] [Google Scholar]
- 16. Anand S, Mande SS. Diet, microbiota and gut-lung connection. Front Microbiol 2018; 9:2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez-Martinez C, Kranzer K, McHugh G, et al. Azithromycin versus placebo for the treatment of HIV-associated chronic lung disease in children and adolescents (BREATHE trial): study protocol for a randomised controlled trial. Trials 2017; 18:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bogaert D, Keijser B, Huse S, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 2011; 6:e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 2011; 108Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claassen-Weitz S, Gardner-Lubbe S, Nicol P, et al. HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a South African birth cohort. Sci Rep 2018; 8:5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Illumina Proprietary. MiSeq sequencing system guide. San Diego, CA: Illumina; 2018. [Google Scholar]
- 22. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat 1984; 11:265–70. [Google Scholar]
- 23. Morris EK, Caruso T, Buscot F, et al. Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol Evol 2014; 4:3514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sorenson T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content. Kongelige Danske Videnskabernes Selskab 1948; 5.1–34:4–7. [Google Scholar]
- 25. Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral Ecol 2001; 26:32–46. [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57:289–300. [Google Scholar]
- 27. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 2017; 45:W180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. World Health Organization. Guidelines on post-exposure prophylaxis for HIV and the use of co-trimoxazole prophylaxis for HIV-related infections among adults, adolescents and children: recommendations for a public health approach - December 2014 supplement to the 2013 consolidated ARV guidelines. Geneva: World Health Organization; 2014. [PubMed] [Google Scholar]
- 30. McHardy IH, Li X, Tong M, et al. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang Y, Cai Y. Altered gut microbiota in HIV infection: future perspective of fecal microbiota transplantation therapy. AIDS Res Hum Retroviruses 2019; 35:229–35. [DOI] [PubMed] [Google Scholar]
- 32. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy 2014; 44:842–50. [DOI] [PubMed] [Google Scholar]
- 33. Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One 2010; 5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nowak RG, Bentzen SM, Ravel J, et al. Rectal microbiota among HIV-uninfected, untreated HIV, and treated HIV-infected in Nigeria. AIDS 2017; 31:857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dinh DM, Volpe GE, Duffalo C, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noguera-Julian M, Rocafort M, Guillén Y, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iyamba JM, Wambale JM, Takaisi-Kikuni NZ. Antimicrobial susceptibility patterns of enterobacteriaceae isolated from HIV-infected patients in Kinshasa. Pan Afr Med J 2014; 17:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zlosnik JE, Zhou G, Brant R, et al. Burkholderia species infections in patients with cystic fibrosis in British Columbia, Canada. 30 years' experience. Ann Am Thorac Soc 2015; 12:70–8. [DOI] [PubMed] [Google Scholar]
- 40. Miquel S, Martín R, Rossi O, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 2013; 16:255–61. [DOI] [PubMed] [Google Scholar]
- 41. Hansen R, Russell RK, Reiff C, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol 2012; 107:1913–22. [DOI] [PubMed] [Google Scholar]
- 42. Maji A, Misra R, Dhakan DB, et al. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol 2018; 20:402–19. [DOI] [PubMed] [Google Scholar]
- 43. Bourke CD, Gough EK, Pimundu G, et al. Cotrimoxazole reduces systemic inflammation in HIV infection by altering the gut microbiome and immune activation. Sci Transl Med 2019; 11:eaav0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS 2014; 28:753–60. [DOI] [PubMed] [Google Scholar]
- 45. Monaco CL, Gootenberg DB, Zhao G, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016; 19:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.