Abstract
Background
Encephalitis is an inflammatory condition of the brain associated with long-term neurologic sequelae and even death in children. Although viruses are often implicated, an etiology is not identified in the majority of cases. Metagenomics-based next-generation sequencing (mNGS) is a high-throughput sequencing technique that can enhance the detection of novel or low-frequency pathogens.
Methods
Hospitalized immunocompetent children aged 6 months to 18 years with encephalitis of unidentified etiology were eligible for enrollment. Demographic, historical, and clinical information was obtained, and residual blood and cerebrospinal fluid (CSF) samples were subjected to mNGS. Pathogens were identified by querying the sequence data against the NCBI GenBank database.
Results
Twenty children were enrolled prospectively between 2013 and 2017. mNGS of CSF identified 7 nonhuman nucleic acid sequences of significant frequency in 6 patients, including that of Mycoplasma bovis, parvovirus B19, Neisseria meningitidis, and Balamuthia mandrillaris. mNGS also detected Cladophialophora species, tobacco mosaic virus, and human bocavirus, which were presumed to be contaminants or nonpathogenic organisms. One patient was found to have positive serology results for California encephalitis virus, but mNGS did not detect it. Patients for whom mNGS identified a diagnosis had a significantly higher CSF white blood cell count, a higher CSF protein concentration, and a lower CSF glucose level than patients for whom mNGS did not identify a diagnosis.
Conclusion
We describe here the results of a prospective cohort analysis to evaluate mNGS as a diagnostic tool for children with unexplained encephalitis. Although mNGS detected multiple nonpathogenic organisms, it also identified multiple pathogens successfully and was most useful in patients with a CSF abnormality.
Keywords: encephalitis, metagenomics, next-generation sequencing
We describe here the use of next-generation sequencing in 20 children with encephalitis of unknown etiology. A presumptive pathogen (Neisseria meningitidis, Balamuthia mandrillaris, Mycoplasma bovis, or parvovirus B19) was identified in the cerebrospinal fluid and/or blood of 4 of these patients.
Acute encephalitis manifests as altered mental status, fever, and headache and commonly results in long-term neurologic sequelae and even death [1–3]. The incidence of encephalitis is approximately 7.5 cases per 100 000 patient years; the highest incidence occurs in children younger than 1 year [4]. In patients with an identified etiology, viruses are the most common cause, accounting for 40% to 48% of cases [4, 5]. Herpes simplex virus is the most commonly identified virus, followed by arthropod-borne viruses. After infectious etiologies, antibody-mediated processes (specifically anti-N-methyl-d-aspartate receptor encephalitis) are the next most common [6, 7]. However, an etiology is identified in only approximately 50% of hospitalized patients with encephalitis, which leaves clinicians with limited guidance for treatment or prognosis [5, 8].
The substantial disease burden and severity of acute encephalitis necessitates the development of new diagnostics and therapeutics. Many diagnostic assays are based on polymerase chain reaction (PCR), which relies on sequence-specific primers. The main limitation of targeted PCR is the level to which assays can be multiplexed, which constrains the number of targets that can be assessed per reaction. In contrast, metagenomics-based next-generation sequencing (mNGS) is a high-throughput approach that can interrogate all genetic material in a biologic sample simultaneously [9]. mNGS has been used in cases of acute encephalitis to identify Leptospira sp, astrovirus, and Balamuthia mandrillaris in cerebrospinal fluid (CSF) samples in addition to measles and herpes simplex viruses in multiple postmortem brain samples [10–15]. mNGS has been used also to analyze brain and spinal cord specimens in a prospective analysis of 10 patients, which identified Epstein–Barr virus, JC polyomavirus, and Mycobacterium tuberculosis with a high degree of confidence in 3 patients [16].
In this study, we performed a prospective cohort analysis of hospitalized children with encephalitis of unknown etiology using mNGS as a diagnostic modality.
METHODS
Children aged 6 months to 18 years who presented between April 2013 and December 2017 with encephalitis of unknown etiology were recruited and enrolled. Encephalitis was defined as encephalopathy (identified as depressed or altered level of consciousness, lethargy, or change in personality in the past ≥24 hours) plus at least 1 additional finding of fever (temperature, ≥38°C), seizure, focal neurologic finding, CSF pleocytosis (≥5 white blood cells [WBCs]/µL), abnormal electroencephalography (EEG) results, or abnormal neuroimaging results [8, 17]. Children were excluded if they were infected with human immunodeficiency virus, had received an organ transplant, or had a known etiology of encephalitis at the time of enrollment or if informed consent was not obtained. Diagnostic evaluations for encephalitis beyond routine CSF studies (cell counts, protein and glucose measurements, Gram staining, and culture) were performed at the discretion of the clinical team. Of the 20 enrolled patients, 19 underwent a CSF enterovirus PCR assay, 18 underwent a CSF herpes simplex virus type 1/2 PCR assay, 16 underwent head computed tomography, 17 underwent brain magnetic resonance imaging (MRI), and 16 underwent EEG. Although the BioFire FilmArray meningitis/encephalitis panel received clearance from the US Food and Drug Administration during the course of this study, the test was not performed for any of the enrolled patients. A history was obtained by an investigator through an in-person questionnaire with the patient and his or her family, and the medical record was reviewed. Residual serum or plasma and CSF were sent to the Center for Genome Sciences at the US Army Medical Research Institute of Infectious Diseases in Frederick, Maryland, for mNGS testing. Institutional review board approval was obtained before study initiation. mNGS data were considered investigational and were not used to directly influence patient care. All patient families were called 6 to 12 months after enrollment to assess the patient’s clinical outcome.
Sample RNA was extracted using TRIzol, DNase-treated, and amplified using an unbiased sequence-independent single-primer amplification (SISPA) technique as described previously [18]. Illumina sequencing libraries were prepared using the PrepX Complete ILMN 32i DNA library kit (Takara, Mountain View, California) on an Apollo 324 NGS library prep system. Any samples with remaining volume were extracted for DNA sequencing using the QIAamp cador pathogen minikit (Qiagen, Germantown, Maryland). DNA sequencing libraries were prepared using the Nextera XT DNA library prep kit (Illumina, San Diego, California). Sequencing was conducted on either an Illumina MiSeq or NextSeq 500 desktop sequencing platform. Raw sequencing data were filtered by the index reads removing any reads associated with indexes that had a quality score of less than Q20 to help eliminate index bleed-through. Reads were trimmed and quality filtered using Trimmomatic 0.38 [19] to remove Illumina adapter and low-quality bases, Cutadapt [20] to remove the SISPA primer sequence, and Prinseq-lite 0.20.4 [21] to remove duplicate reads. Reads that aligned to the human genome and transcriptome using Bowtie 2 [22] were excluded. The remaining reads were de novo assembled using Ray 2.2 [23] to form contigs, which then were compared to the nr/nt database using Blast 2.2.28+ (National Center for Biotechnology Information). In-house scripts generated a report that detailed the number of reads, size of contigs, taxonomy, and identity metrics, which were used to assess confidence in the result. Reads identified as pathogen specific were aligned to a reference genome and percent coverage calculated.
Statistical analysis was performed to compare the patients with a diagnosis identified by mNGS with patients for whom no pathogen was identified by mNGS. Continuous variables were documented as medians with 25th to 75th percentiles and minimum and maximum values. Categorical variables were expressed as the absolute number of subjects and relative frequencies. Comparisons between groups were made using the Student t or Fischer exact tests, and size effects were calculated.
RESULTS
Baseline Patient Characteristics
Twenty children were enrolled between 2013 and 2017 (Table 1). All these patients had altered mental status, as required by the case definition, and the next most common symptoms of encephalitis included fever and seizure. Nearly all the patients had CSF pleocytosis, and the majority of them had abnormal neuroimaging and/or EEG results.
Table 1.
Baseline Characteristics of Enrolled Children
| Patient Characteristic | Data (N = 20) |
|---|---|
| Age (mean [range]) (years) | 7.5 (0.5–17) |
| Sex, male (n [%]) | 10 (50) |
| Race (n [%]) | |
| White or Caucasian | 10 (50) |
| Black or African American | 9 (45) |
| Other | 1 (5) |
| Encephalitis criteria (n [%]) | |
| Altered mental status | 20 (100) |
| Fever | 18 (90) |
| Seizure | 11 (55) |
| Focal neurologic finding | 8 (40) |
| Pleocytosis (CSF WBC count > 5/µL) | 19 (95) |
| Abnormal neuroimaging result | 15 (75) |
| Abnormal EEG result (N = 16) | 10 (50) |
Abbreviations: CSF, cerebrospinal fluid; EEG, electroencephalography; WBC, white blood cell.
Sequencing Data
Presumptive microbiologic diagnoses were identified in 5 of the 20 patients by mNGS, routine diagnostic testing, or both. Seven nonhuman sequences of significant frequency were identified in 6 patients (Table 2). The suspected pathogens included Neisseria meningitidis, B alamuthia mandrillaris, and parvovirus B19 in CSF and Mycoplasma bovis in both serum and CSF. Presumed contaminants and/or nonpathogenic organisms included Cladophialophora species in CSF, tobacco mosaic virus in CSF and plasma, and human bocavirus in plasma. Routine clinical diagnostic testing identified an infectious etiology in 3 patients. These etiologies included California encephalitis virus, determined by immunoglobulin M (IgM) in the serum, and N meningitidis and B mandrillaris, determined by gene-specific PCR assays of CSF. When mNGS was initially performed by using the RNA extraction methodology, sequences for N meningitidis and B mandrillaris were detected at a low frequency in the respective patients and, thus, were not identified until the data were analyzed retrospectively after a diagnosis was made using conventional assays. When mNGS was subsequently performed using DNA extraction, sequences for both organisms were readily identified at a higher frequency and with a larger percentage of genome coverage.
Table 2.
Findings in 6 Children With an Organism Detected by Next-Generation Sequencinga
| Patient No. | Patient Age/Sex | Specimen Type | Sequencing Type | No. of Raw Reads | No. of Nonhuman Reads | No. of Microorganism- Specific Reads | Microorganism Detected | Genome Coverage (No. [%] of bases) | Additional Testing |
|---|---|---|---|---|---|---|---|---|---|
| 3 | 6 m/M | CSF | RNA | 3 028 | 43 612 | 72 | N meningitidis | 4899 (0.22) |
N meningitidis detected by gene-specific PCR in CSF |
| DNA | 3 339 963 | 15 698 | 1149 | N meningitidis | 199 747 (8.89) | N meningitidis detected by gene-specific PCR in CSF | |||
| 4 | 6 y/M | CSF | DNA | 5 124 949 | 612 431 | 14 | Cladophialophora sp | —b | |
| 5 | 11 y/F | CSF | RNA | 1 993 580 | 28 109 | 1751 | M bovis | 7763 (0.75) | |
| Serum | RNA | 2 379 243 | 726 621 | 3389 | M bovis | 9022 (0.87) | |||
| Hemolyzed serum | RNA | 1 138 614 | 71 163 | 1769 | M bovis | 8795 (0.85) | |||
| 14 | 6 y/M | CSF | RNA | 525 182 | 486 686 | 14 | B mandrillaris | 672 (1.68) | B mandrillaris detected by gene-specific PCR in CSF |
| DNA | 3 661 833 | 35 886 | 119 | B mandrillaris | 18 163 (45.41) | B mandrillaris detected by gene-specific PCR in CSF | |||
| 15 | 13 y/M | CSF | RNA | 2 559 732 | 1 549 662 | 24 586 | Tobacco mosaic virus | 1218 (19.05) | |
| Plasma | RNA | 680 576 | 401 880 | 16 551 | Tobacco mosaic virus | 1444 (22.58) | |||
| 19 | 15 y/M | CSF | RNA | 343 333 | 17 888 | 81 | Human Parvovirus B19 | 457 (8.17) | Parvovirus B19 detected by gene-specific PCR in serum only |
| Plasma | RNA | 316 562 | 20 038 | 2 | Human bocavirus | 149 (2.81) |
Abbreviations: CSF, cerebrospinal fluid; F, female; M, male; PCR, polymerase chain reaction.
aPatients are numbered consecutively on the basis of the time of enrollment. Shown are the sequencing extraction methods, numbers of raw, nonhuman, and microorganism-specific reads, genome coverage, and the microorganism detected. For cases in which multiple specimens of the same type were obtained, the specimen with the highest number of microorganism-specific reads is shown.
bSpecies could not be confirmed because of the lack of a good reference strain.
Compared with patients for whom no pathogen was identified by mNGS, those with a pathogenic diagnosis had a significantly higher CSF WBC count, higher CSF protein concentration, and a lower CSF glucose level (Table 3). Those with an organism detected by mNGS were also noted to have a higher alanine aminotransferase (ALT) level than those without an organism detected my mNGS, although the median ALT level remained in the reference range for both groups. Patients with a diagnosis identified by mNGS had a higher frequency of exposures to animals (100%) and mosquitoes (75%) than those for whom no pathogen was identified by mNGS, although these differences were not statistically significant. We found no other significant differences among the 2 groups in terms of clinical presentation or laboratory parameters.
Table 3.
Comparison of Children for Whom a Microbiologic Diagnosis/Pathogen was Identified and Those With no Identified Microbiologic Diagnosis (According to mNGS)
| Characteristic | Overall (N = 20) | Diagnosis Established by mNGS (N = 4) | No Diagnosis Established by mNGS (N = 16) | P | Effect Size |
|---|---|---|---|---|---|
| Demographics | |||||
| Age (years) | |||||
| Median (25th–75th percentiles) | 7.0 (1.5–12.0) | 8.5 (3.3–13) | 7 (1.5–12) | 1.000 | 0.02 |
| min, max | 0.5, 17.0 | 0.5, 15 | 0.8, 17.0 | ||
| Sex-Male (n [%]) | 10 (50) | 3 (75) | 7 (44) | .582 | 0.67 |
| Race/ethnicity (n [%]) | .648 | 0.72 | |||
| Black | 9 (45) | 1 (25) | 8 (50) | ||
| White | 10 (50) | 3 (75) | 7 (44) | ||
| Other | 1 (5) | 0 (0) | 1 (6) | ||
| Hispanic | 3 (15) | 1 (25) | 2 (13) | .531 | 0.32 |
| Exposures (n [%]) | |||||
| Mosquitoes | 8 (40) | 3 (75) | 5 (31) | .255 | 0.98 |
| Ticks | 1 (5) | 0 (0) | 1 (6) | 1.00 | -0.37 |
| Animals | 15 (75) | 4 (100) | 11 (69) | .530 | 0.95 |
| Clinical manifestations (n [%]) | |||||
| Fever | 18 (90) | 4 (100) | 14 (88) | 1.000 | 0.53 |
| Lethargy | 14 (70) | 3 (75) | 11 (69) | 1.000 | 0.14 |
| Irritability | 6 (30) | 2 (50) | 4 (25) | .549 | 0.53 |
| Seizure | 11 (55) | 1 (25) | 10 (63) | .285 | −0.82 |
| Test results | |||||
| WBCs | |||||
| Median (25th–75th percentiles) | 10.6 (9.2–12.7) | 11.9 (8.1–12.6) | 10.4 (9.2–12.9) | .852 | 0.13 |
| min, max | 4.5, 35.4 | 4.8, 12.7 | 4.5, 35.4 | ||
| CRP | |||||
| Median (25th–75th percentiles) | 0.9(0.3–2.0) | 0.8 (0.5–15.3) | 1 (0.3–2.0) | 1.000 | −0.03 |
| min, max | 0.3, 29.6 | 0.3, 29.6 | 0.3, 15.2 | ||
| Creatinine | |||||
| Median (25th–75th percentiles) | 0.5 (0.3–0.65) | 0.7 (0.4–0.8) | 0.5 (0.3–0.6) | .510 | 0.37 |
| min, max | 0.2, 0.9 | 0.2, 0.9 | 0.2, 0.9 | ||
| ALT | |||||
| Median (25th–75th percentiles) | 21 (14–27.5) | 32 (26–70) | 16.5 (13.5–25.5) | .042b | 1.63 |
| min, max | 8, 102 | 25, 102 | 8, 48 | ||
| CSF test results | |||||
| WBC | |||||
| Median (25th–75th percentiles) | 73 (13–241) | 578 (196–1947) | 45 (12–144) | .032b | 1.74 |
| min, max | 4, 3025 | 105, 3025 | 4, 933 | ||
| Protein | |||||
| Median (25th–75th percentiles) | 44.5 (33–51.5) | 116 (57–173) | 41 (32–48) | .021b | 1.97 |
| min, max | 15, 179 | 49, 179 | 15, 111 | ||
| Glucose | |||||
| Median (25th–75th percentiles) | 59.5 (47–65.6) | 39 (29–49) | 61 (53.5–70) | .032b | −1.84 |
| min, max | 22, 88 | 24, 53 | 22, 88 |
Abbreviations: ALT, alanine aminotransferase; CSF, cerebrospinal fluid; max, maximum; min, minimum; mNGS, metagenomics-based next-generation sequencing; WBC, white blood cell.
aN = 19.
bStatistically Significant result.
Case Descriptions
Patient 3
A 6-month-old previously healthy Caucasian boy was admitted with a 1-day history of fever, lethargy, meningismus, and emesis in the setting of 2 weeks of rhinorrhea and cough. Initial laboratory evaluation revealed an elevated C-reactive protein (CRP) level (29.6 mg/dL), and a lumbar puncture performed on day 2 of hospitalization after the administration of broad-spectrum antibiotics revealed CSF pleocytosis (3025 WBC cells/µL), a protein concentration of 49 mg/dL, and a glucose level of 53 mg/dL. Results of the head computed tomography were concerning for increased intracranial pressure. At the time of enrollment, conventional microbiologic testing (including CSF culture) results were negative. However, after enrollment, CSF from the patient was sent to the Centers for Disease Control and Prevention Bacterial Meningitis Laboratory for a gene-specific PCR assay for N meningitidis, the results of which were positive. The patient was treated with a 7-day course of ceftriaxone, and his symptoms were clinically resolved. N meningitidis was detected by mNGS in both DNA and RNA extracts.
Patient 5
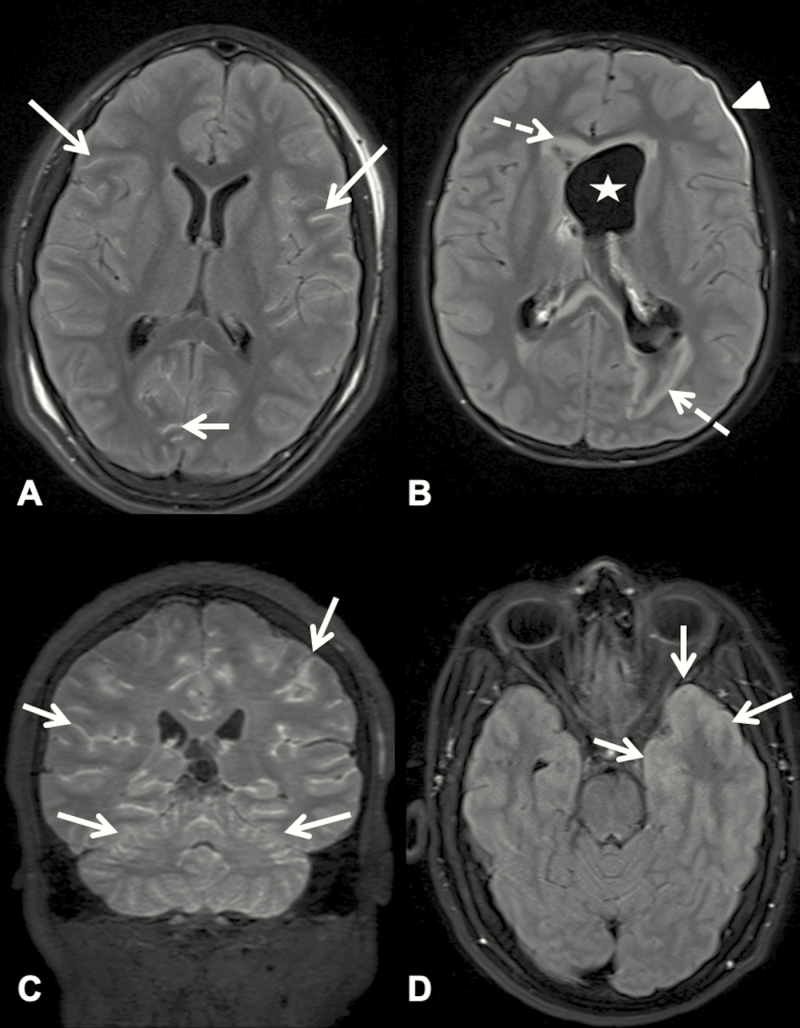
An 11-year-old African American girl presented to the hospital with a 1-week history of fever, headache, emesis, confusion, diplopia, and amnesia. She developed ataxia and unilateral upper- and lower-extremity weakness after admission and was found to be encephalopathic. She had notable animal exposures (to cows, goats, sheep, and pigs at a local fair) in the weeks before admission. She underwent extensive laboratory testing, including 3 lumbar punctures, each of which revealed CSF pleocytosis (80–157 WBCs/µL), an elevated protein concentration (166–600 mg/dL), and a low glucose level (33–45 mg/dL), but no cause of the encephalitis was identified. Brain MRI revealed diffuse leptomeningeal enhancement (Figure 1A). She was treated empirically for tuberculous meningitis with rifampin, isoniazid, pyrazinamide, and ethambutol for 2 months followed by rifampin and isoniazid alone for 10 additional months because of positive tuberculin skin testing results despite the inability to recover acid-fact bacilli from her CSF and the lack of a known tuberculosis exposure. She also received a course of doxycycline (100 mg twice daily for 5 days) in addition to vancomycin, ceftriaxone, and acyclovir. M ycoplasma bovis was identified in her CSF and serum by mNGS. On the basis of their known mechanisms of action, some of the antimicrobial agents she received, particularly doxycycline, might have had activity against the organism, although in vitro antibiotic susceptibilities were not determined. The patient returned to her cognitive baseline 2 to 3 months after discharge from inpatient rehabilitation and regained baseline motor functionality approximately 7 months after discharge. Results of repeat brain MRI 9 months after diagnosis were unremarkable.
Figure 1.
Postcontrast T2-weighted fluid-attenuated inversion recovery magnetic resonance images of patients 5, 14, 19, and 8 (A–D, respectively). (A) Axial image showing abnormal leptomeningeal hyperintensity (arrows) throughout both cerebral hemispheres, consistent with meningitis. (B) Axial image showing dilated left ventricle (star), abnormal signal in periventricular white matter (dotted arrows), and left subdural enhancement (arrowhead), sequelae caused by meningitis. (C) Coronal image showing diffuse abnormal hyperintensity in the leptomeninges of the cerebellar and cerebral hemispheres (arrows). (D) Axial image showing abnormal hyperintensity (arrows) of the left temporal lobe meninges and cortex, consistent with meningoencephalitis.
Patient 14
A 6-year-old Caucasian boy was admitted with a 1-month history of fever and headache after being treated with courses of ceftriaxone and cefprozil for presumed sinusitis. He had been swimming in a backyard pool filled with well water before the onset of illness. During his hospitalization, he developed lethargy, hallucinations, ataxia, diplopia, nystagmus, and cranial nerve palsy. Several lumbar punctures revealed CSF pleocytosis (286–366 WBCs/µL), an elevated protein concentration (65–180 mg/dL), and a low glucose level (24–53 mg/dL), but the results of infectious disease evaluation were negative. Multiple neuroimaging studies revealed progressively worsening cerebral edema, hydrocephalus, and, ultimately, cerebral herniation (Figure 1B). CSF from the third lumbar puncture was sent to the Centers for Disease Control and Prevention for a gene-specific PCR assay for amoebic pathogens, and the results were positive for B mandrillaris. These results were not available at enrollment, but mNGS confirmed this finding by identifying 45% of the B mandrillaris genome by DNA extraction. mNGS post-RNA extraction did not identify the organism initially because of the low number of microorganism-specific reads and low genome coverage percentage. However, the organism was identified in retrospective analysis of the RNA-sequencing data. This patient unfortunately did not survive.
Patient 19
A 15-year-old Hispanic boy was admitted with acute onset of headache, emesis, dizziness, weakness, and fever. He had no remarkable exposures or sick contacts before illness onset. His altered mental status progressed to include confusion, irritability, and hallucinations, and he developed seizures 1 week into his hospitalization. Initial MRI revealed leptomeningeal enhancement, and follow-up MRI results were concerning for decreased cerebral perfusion (Figure 1C). Laboratory evaluation did not reveal hematologic abnormalities, but the results were significant for CSF pleocytosis (869 WBCs/µL), a protein concentration of 179 mg/dL, and a glucose level of 44 mg/dL. His pleocytosis improved on a lumbar puncture 5 days later (457 WBCs/µL), at which time his protein concentration was 186 mg/dL and glucose level was 40 mg/dL. Residual CSF was sent from both samples for mNGS. Parvovirus B19–specific reads were detected in the second CSF sample but were confirmed only by a gene-specific (NS1 and VP) [24] PCR assay of the serum samples. Residual serum was sent for acute-phase parvovirus B19 serology, and no antibodies were detected. Convalescent antibody testing was not performed because of family preference. No parvovirus B19–specific reads were found by mNGS of the plasma sample, but reads for human bocavirus were detected. It was presumed to be a clinically insignificant copathogen, although it is important to note that human bocavirus has been implicated in children with encephalitis [25]. The patient returned to his cognitive baseline over the course of several months after discharge.
Patient 8
A 16-year-old Caucasian boy was admitted with a 2-day history of headache, fever, emesis, urinary incontinence, and aggressive behavior, and he subsequently had 2 generalized seizures. He was an avid outdoorsman and regularly hunted and consumed wild birds, squirrels, frogs, and fish in rural Georgia. He had been bitten by multiple mosquitoes and frequently swam in ponds in wooded areas. CSF evaluation revealed mild CSF pleocytosis (135 WBCs/µL), a protein concentration of 46 mg/dL, and a glucose level of 59 mg/dL, and brain MRI revealed an asymmetric signal in the left hippocampus, amygdala, temporal pole, and cingulate gyrus thought to be consistent with viral encephalitis (Figure 1D). Results of an infectious disease evaluation was notable only for serum-positive California encephalitis virus IgM by indirect immunofluorescence assay, with a titer of 1:256. mNGS did not detect microorganisms in the CSF, plasma, or serum. Given the patient’s substantial risk factors in a location with known endemic transmission of La Crosse virus, this California encephalitis virus serogroup was thought to be the most likely etiology of his symptoms [26]. His symptoms resolved before discharge; however, the patient was lost to long-term follow-up.
Discussion
The findings in this study highlight important clinical scenarios in which mNGS might provide advantages over conventional diagnostic testing for unexplained pediatric encephalitis, such as when pathogen-specific PCR testing is not available or when a rare or emerging pathogen is implicated. For example, we made a presumptive diagnosis of parvovirus B19 acute encephalitis in a previously healthy teenaged male. Parvovirus B19 has been associated with neurologic findings, including encephalitis, meningitis, neuropathy, and stroke [27]. Because it is not commonly implicated in cases of acute encephalitis, it is often not considered in the initial evaluation but is recommended as part of a second-tier evaluation if initial testing results are negative [28]. Although our patient had clinical symptoms consistent with encephalitis, in the absence of serologic confirmation, the mNGS findings could have represented latent infection or reactivation.
The case of M bovis infection also highlights the potential utility of mNGS for uncommonly encountered pathogens. Multiple species of this genus, including Mycoplasma pneumoniae, have been implicated as a causative etiology of encephalitis, but only 1 case of M bovis infection in humans has ever been described to date [29–31]. This case, described in 1979, presented as lobar pneumonia with several extrapulmonary manifestations, including psychosis [30]. M bovis infection is associated with arthritis, pneumonia, brain abscesses, and meningitis in the bovine population [29]. Our patient with M bovis infection detected by mNGS had interacted with livestock at a petting zoo before becoming ill, which might have facilitated exposure to this organism. However, without a confirmatory serologic or culture-based diagnostic test, it is not possible to confirm the diagnosis. These 2 cases emphasize the potential for mNGS to diagnose organisms that might not be considered in the initial differential diagnosis.
Our study also highlights scenarios in which mNGS is not the optimal diagnostic modality. The patient who had detectable California encephalitis virus IgM represents an example of encephalitis that could have been attributable to postinfectious or immune-mediated phenomena or one in which the pathogen was no longer present in the CSF at the time of sampling. In addition, in cases in which the pathogen localizes to brain parenchyma exclusively, alternative diagnostic modalities, such as antibody testing, might be of higher yield than mNGS of the CSF.
The results of this study reveal the challenge of data interpretation that often accompanies mNGS. A Cladophialophora sp was identified in the CSF of 1 patient. In the rare cases in which this organism causes central nervous system disease, the clinical presentation is severe, and the mortality rate is 65% [32, 33]. Given the clinical improvement of the patient in our study without the administration of antifungal therapy, we presumed that the Cladophialophora sp was an environmental contaminant. The issue of contamination deserves special attention, because contaminant DNA could be introduced during manipulation of samples within the clinical or research laboratory or from the sample tubes themselves. The sensitivity of mNGS can create difficulty in distinguishing pathogenic organisms from contaminants or nonpathogenic coinfecting organisms. This was likely the case for our detection of tobacco mosaic virus, which does not infect humans and is commonly found in research laboratories, and human bocavirus, which is typically a coinfecting organism in patients with respiratory disease [34, 35].
Since its advent in 2004, the cost of mNGS has decreased by several orders of magnitude, and its implementation has dispersed, which makes it a more attractive and feasible option to use for diagnosing unexplained encephalitis [36]. Although this study was not designed to provide clinically actionable data, in an ideal situation, laboratory turnaround time of a single assay in our system was estimated at 3 business days, and the cost of reagents was less than $200. Although sequencing based on 16S rRNA for prokaryotic organisms or internal spacer sequences of 28S rRNA for eukaryotes might be cheaper, these analyses are also limited to the detection of nucleic acids from specific classes of pathogens.
We were unable to detect a plausible pathogen in 15 (75%) of the enrollees, which is a frequency similar to that described in the California Encephalitis Study, which used targeted PCR assays for 13 commonly implicated pathogens in unexplained encephalitis [37]. Therefore, although mNGS might not be a panacea for diagnosing acute encephalitis, we have identified clinical scenarios in which it can be beneficial. These results also provide clinically relevant guidance to physicians, because they suggest that mNGS of CSF has the most utility in patients with significant CSF abnormalities, including pleocytosis, an elevated protein concentration, and hypoglycorrhachia. Although the sensitivity of mNGS can theoretically decrease with higher numbers of CSF WBCs because of interfering host nucleic acid sequences, we did not observe this phenomenon in our study. In addition, no pathogens were identified by mNGS of serum or plasma samples that were not also identified in the CSF samples. These results indicate that mNGS of the serum or plasma provides little or no added benefit over that of CSF for diagnosing encephalitis.
In conclusion, although mNGS has limitations, the decreasing costs, faster turnaround times, and improved understanding of its clinical utility make mNGS a potentially useful tool in the arsenal of the physician caring for patients with encephalitis of an unclear etiology.
Notes
Acknowledgments. We thank all the patients and families who participated in this study, and we thank the Children’s Healthcare of Atlanta and Emory University Pediatric Biostatistics Core and the Children’s Healthcare of Atlanta Microbiology Laboratory for their assistance with specimen handling and shipping. We also thank our clinical research coordinators Laila Hussaini and Kathy Stephens.
Financial support. This study was funded in part by a Children’s Healthcare of Atlanta Friend’s grant (to A. L. S. and A.K. M.). C. A. R. is supported by the Pichichero Family Foundation Award from the Pediatric Infectious Diseases Society and by funds from the Children’s Pediatric Research Trust. During the conduct of this work, A. K. M. was supported by the Atlanta Pediatric Scholars Program (grant NIH K12 HD072245) and a start-up award from the Children’s Pediatric Research Trust.
Potential conflicts of interest. C. A. R. has received research funding to her institution from BioFire Diagnostics, LLC, Micron Biomedical, Inc, Sanofi S.A., MedImmune, Pfizer, Janssen Pharmaceuticals, and PaxVax, Inc. All other authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Rismanchi N, Gold JJ, Sattar S, et al. . Neurological outcomes after presumed childhood encephalitis. Pediatr Neurol 2015; 53:200–6. [DOI] [PubMed] [Google Scholar]
- 2. Messacar K, Fischer M, Dominguez SR, et al. . Encephalitis in US children. Infect Dis Clin North Am 2018; 32:145–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mailles A, De Broucker T, Costanzo P, et al. ; Steering Committee and Investigators Group Long-term outcome of patients presenting with acute infectious encephalitis of various causes in France. Clin Infect Dis 2012; 54:1455–64. [DOI] [PubMed] [Google Scholar]
- 4. George BP, Schneider EB, Venkatesan A. Encephalitis hospitalization rates and inpatient mortality in the United States, 2000–2010. PLoS One 2014; 9:e104169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vora NM, Holman RC, Mehal JM, et al. . Burden of encephalitis-associated hospitalizations in the United States, 1998–2010. Neurology 2014; 82:443–51. [DOI] [PubMed] [Google Scholar]
- 6. Dalmau J, Graus F. Antibody-mediated encephalitis. N Engl J Med 2018; 378:840–51. [DOI] [PubMed] [Google Scholar]
- 7. Gable MS, Sheriff H, Dalmau J, et al. . The frequency of autoimmune N-methyl- d -aspartate receptor encephalitis surpasses that of individual viral etiologies in young individuals enrolled in the California Encephalitis Project. Clin Infect Dis 2012; 54:899–904. [Google Scholar]
- 8. Bloch KC, Glaser CA. Encephalitis surveillance through the emerging infections program, 1997–2010. Emerg Infect Dis 2015; 21:1562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schubert RD, Wilson MR. A tale of two approaches: how metagenomics and proteomics are shaping the future of encephalitis diagnostics. Curr Opin Neurol 2015; 28:283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson MR, Naccache SN, Samayoa E, et al. . Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014; 370:2408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naccache SN, Peggs KS, Mattes FM, et al. . Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 2015; 60:919–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frémond ML, Pérot P, Muth E, et al. . f-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc 2015; 4:e53–7. [DOI] [PubMed] [Google Scholar]
- 13. Sato M, Kuroda M, Kasai M, et al. . Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol 2016; 78:66–70. [DOI] [PubMed] [Google Scholar]
- 14. Chan BK, Wilson T, Fischer KF, Kriesel JD. Deep sequencing to identify the causes of viral encephalitis. PLoS One 2014; 9:e93993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greninger AL, Messacar K, Dunnebacke T, et al. . Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med 2015; 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salzberg SL, Breitwieser FP, Kumar A, et al. . Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol Neuroimmunol Neuroinflamm 2016; 3:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glaser CA, Gilliam S, Schnurr D, et al. ; California Encephalitis Project, 1998–2000 In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998–2000. Clin Infect Dis 2003; 36:731–42. [DOI] [PubMed] [Google Scholar]
- 18. Kugelman JR, Wiley MR, Nagle ER, et al. . Error baseline rates of five sample preparation methods used to characterize RNA virus populations. PLoS One 2017; 12:e0171333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17.1:10–2. [Google Scholar]
- 21. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 2010; 17:1519–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Servant A, Laperche S, Lallemand F, et al. . Genetic diversity within human erythroviruses: identification of three genotypes. J Virol 2002; 76:9124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitui MT, Tabib SM, Matsumoto T, et al. . Detection of human bocavirus in the cerebrospinal fluid of children with encephalitis. Clin Infect Dis 2012; 54:964–7. [DOI] [PubMed] [Google Scholar]
- 26. Centers for Disease Control and Prevention. La Crosse encephalitis: epidemiology and geographic distribution Available at: https://www.cdc.gov/lac/tech/epi.html. Accessed 9 May 2019.
- 27. Barah F, Whiteside S, Batista S, Morris J. Neurological aspects of human parvovirus B19 infection: a systematic review. Rev Med Virol 2014; 24:154–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tyler KL. Acute viral encephalitis. N Engl J Med 2018; 379:557–66. [DOI] [PubMed] [Google Scholar]
- 29. Rosales RS, Puleio R, Loria GR, et al. . Mycoplasmas: brain invaders? Res Vet Sci 2017; 113:56–61. [DOI] [PubMed] [Google Scholar]
- 30. Madoff S, Pixley BQ, DelGiudice RA, Moellering RC Jr. Isolation of Mycoplasma bovis from a patient with systemic illness. J Clin Microbiol 1979; 9:709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christie LJ, Honarmand S, Talkington DF, et al. . Pediatric encephalitis: what is the role of Mycoplasma pneumoniae? Pediatrics 2007; 120:305–13. [DOI] [PubMed] [Google Scholar]
- 32. Chakrabarti A, Kaur H, Rudramurthy SM, et al. . Brain abscess due to Cladophialophora bantiana: a review of 124 cases. Med Mycol 2016; 54:111–9. [DOI] [PubMed] [Google Scholar]
- 33. Kantarcioglu AS, Guarro J, De Hoog S, et al. . An updated comprehensive systematic review of Cladophialophora bantiana and analysis of epidemiology, clinical characteristics, and outcome of cerebral cases. Med Mycol 2017; 55:579–604. [DOI] [PubMed] [Google Scholar]
- 34. Peltola V, Söderlund-Venermo M, Jartti T. Human bocavirus infections. Pediatr Infect Dis J 2013; 32:178–9. [DOI] [PubMed] [Google Scholar]
- 35. Harrison BD, Wilson TM. Milestones in the research on tobacco mosaic virus. Philos Trans R Soc Lond B Biol Sci 1999; 354:521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 2019; 14:319–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glaser CA, Honarmand S, Anderson LJ, et al. . Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis 2006; 43:1565–77. [DOI] [PubMed] [Google Scholar]