Abstract
Background
Chlamydia trachomatis and Chlamydia muridarum are intracellular bacterial pathogens of mucosal epithelial cells. CD4 T cells and major histocompatibility complex (MHC) class II molecules are essential for protective immunity against them. Antigens presented by dendritic cells (DCs) expand naive pathogen-specific T cells (inductive phase), whereas antigens presented by epithelial cells identify infected epithelial cells as targets during the effector phase. We previously showed that DCs infected by C trachomatis or C muridarum present epitopes from a limited spectrum of chlamydial proteins recognized by Chlamydia-specific CD4 T cells from immune mice.
Methods
We hypothesized that Chlamydia-infected DCs and epithelial cells present overlapping sets of Chlamydia-MHC class II epitopes to link inductive and effector phases to generate protective immunity. We tested that hypothesis by infecting an oviductal epithelial cell line with C muridarum, followed by immunoaffinity isolation and sequencing of MHC class I- and II-bound peptides.
Results
We identified 26 class I-bound and 4 class II-bound Chlamydia-derived peptides from infected epithelial cells. We were surprised to find that none of the epithelial cell class I- and class II-bound chlamydial peptides overlapped with peptides presented by DCs.
Conclusions
We suggest the discordance between the DC and epithelial cell immunoproteomes has implications for delayed clearance of Chlamydia and design of a Chlamydia vaccine.
Keywords: Bm1.11, Chlamydia, MHC class II, T cell, vaccine
Optimally, DCs expand antigen-specific naive T cells that recognize infected cells. We tested this postulate by determining the Chlamydia-epithelial immunoproteome. Chlamydia antigens presented to CD4 T cells by DC and epithelial cells are discordant, with major implications for vaccine development.
Chlamydia trachomatis genital tract infections are the most prevalent sexually transmitted bacterial infection worldwide, an important cause of pelvic inflammatory disease (PID) and its sequelae, and a focus of public health efforts. Although case finding and treatment reduced PID rates, infection rates are increasing worldwide [1]. Natural history studies show a C trachomatis spontaneous clearance rate of ~50% at 1 year, demonstrating that de novo establishment of sterilizing immunity is a ponderous process [2–4]. It is frustrating to note that early antibiotic therapy arrests development of protective immunity [5], and infection control measures are likely increasing herd susceptibility [6]. The optimal strategy to control C trachomatis is a vaccine.
Progress has been made elucidating the nature of protective immunity and developing a molecular Chlamydia vaccine [7]. Protective Chlamydia immunity is CD4 T cell-mediated requiring major histocompatibility complex (MHC) class II-presented antigens [8]. Immunoproteomics using tandem mass spectrometry to identify microbial peptides complexed to infected host cell MHC molecules is useful for designing vaccines against intracellular pathogens. We have used this technology to identify T-cell antigens for Chlamydia [9–11]. The underlying assumption is that MHC-bound peptides identify microbial proteins that enter antigen-processing pathways (epitope source proteins), and that those proteins are useful vaccine candidates. Our previous studies characterized the MHC class I- and II-bound immunoproteome of murine dendritic cells (DCs) infected with Chlamydia muridarum and C trachomatis [9, 10]. Those studies demonstrated that many class II-bound peptides originate from outer membrane proteins and were substantially protective against C muridarum infections in vivo [12, 13].
Direct (contact-dependent) and indirect (diffusion-dependent) mechanisms of interferon gamma (IFN-γ)-mediated immunity to Chlamydia exist. Interferon gamma indirectly activates indoleamine 2,3-dioxygenase, depleting intracellular tryptophan to starve C trachomatis of an essential nutrient [14]. The vaginal microbiome supplies indole used by C trachomatis tryptophan synthase to synthesize tryptophan and escape nutrient deprivation [15]. Because passive transfer of the immune antibody is ineffective during primary infection [16], and Chlamydia replicates almost exclusively in the reproductive tract epithelium, the critical event in protective immunity is likely CD4 T-cell interaction with infected epithelial cells. The immunoproteome of Chlamydia-infected reproductive tract epithelial cells has not been investigated. Among the Chlamydia T-cell antigens identified from the infected DC immunoproteome, polymorphic membrane protein (PmpG), a source protein for the MHC class II-presented epitope PmpG303-311, provided the greatest vaccine-induced protection against C muridarum genital tract challenge [17]. We derived a PmpG303-311-specific CD4 T-cell clone PmpG1.1 from an immune mouse [18]. PmpG1.1 recognized infected epithelial cells and terminated bacterial replication in them, suggesting an overlap between immunoproteomes of infected DCs and epithelial cells. We hypothesized that Chlamydia-infected epithelial cells and DCs present overlapping epitopes, linking inductive and effector phases to generate protective immunity. To test this hypothesis, we determined the immunoproteome of a C muridarum-infected epithelial cell line. Available to us were epithelial cell lines derived from C57BL/6 (C57epi.1) [19], bm12 (Bm12.4) [20], and bm1 (Bm1.11) [21] mice. Compared with DCs, epithelial cells grow at low density with low levels of MHC class II expression. It was unclear whether infected epithelial cells had sufficient MHC class II-peptide complexes to determine an immunoproteome with existing technology. Therefore, even though our DC immunoproteome is from C57BL/6 mice, we chose Bm1.11 cells matched at MHC class II IAb and class I Db, but not class I K allele, because Bm1.11 cells had the highest inducible level of MHC class II, minimizing a potentially prohibitive technological barrier.
MATERIALS AND METHODS
Chlamydia
Chlamydia muridarum (Nigg strain) was grown in HeLa 229 cells. Discontinuous density gradients of Renografin-76 was used to purify the chlamydial elementary bodies (EBs) [22].
Mice
Female C57BL/6 (H2-Kb, Db, and IAb) mice (6–8 weeks) from Charles River were housed at the Jack Bell Research Center. Animal experiments were conducted in accordance with University of British Columbia guidelines for animal care and use.
Cell Lines
Bm1.11 is a murine oviduct epithelial cell line derived from a B6.C-H2bm1/ByJ mouse [21]. B6.C-H2bm1/ByJ (bm1) and C57BL/6 (H-2b) mice differ in 3 amino acids of the MHC class I K allele, potentially altering the repertoire of class I K but not D presented peptides. Bm1.11 cells were grown in supplemented Dulbecco’s modified Eagle’s medium:F12K media as previously described [21]. Bone marrow-derived DCs were generated from femur marrow of C57BL/6 mice cultured in Iscove’s modified Dulbecco’s media supplemented with granulocyte-macrophage colony-stimulating factor and interleukin-4, harvesting and characterizing the nonadherent fraction as previously described [9].
Optimization of Chlamydia muridarum Infection of Bm1.11
Bm1.11 cells were treated with IFN-γ (0, 5, 10, 20, and 40 ng/mL) for 18 hours, and the optimal concentration (10 ng/mL) was determined based on expression of MHC class I and II. The Bm1.11 cells pretreated with 10 ng/mL IFN-γ for 6 hours were infected with C muridarum at a different multiplicity of infection ([MOI] 0, 1.25, 2.5, 5, 10, and 20) for different intervals (12, 14, 16, 18, and 20 hours). Chlamydial inclusions were stained using anti-EB mouse polyclonal antibody from mice recovered from C muridrum infection, followed by peroxidase-antimouse immunoglobulin G (Jackson ImmunoResearch) and a 3,3′-diaminobenzidine substrate kit (Thermo Fisher Scientific). Cell viability was determined by trypan blue staining. Major histocompatibility complex class I and II expression and mean fluorescence intensities (MFIs) were determined with anti-MHC I-fluorescein isothiocyanate (FITC) (clone AF6-88.5.3) and anti-MHC II-phycoerythrin (clone M5/114.15.2); costimulatory molecules CD80, CD86, CD40, and CD54 (intercellular adhesion molecule 1 [ICAM-1]) expression was evaluated with anti-CD80-APC (clone 16-10A1), anti-CD86-allophycocyanin (APC) (clone GL1), anti-CD40-APC (clone 3/23), and anti-CD54-FITC (clone 3E2) using flow cytometry.
Purification of Major Histocompatibility Complex-Bound Peptides
Bm1.11 cells in 150-mm dishes were infected with C muridarum at the optimal condition. Infected cells were harvested in 5 mL phosphate-buffered saline containing 0.2 mM phenylmethyl sulfonyl fluoride (PMSF) and cocktail protease inhibitors (1:200) using a cell scraper. Cells from 10 dishes were pelleted and stored at −80°C. Batches of infected cells (5 × 109 in total) were pooled and solubilized in cold lysis buffer (1% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate [Sigma-Aldrich], 150 mM NaCl, 20 mM Tris-HCl, pH 8, 0.04% sodium azide, protease inhibitors (5 µg/mL aprotinin, 10 µg/mL leupeptin, 10 µg/mL pepstatin A, 0.04% Na azide, and 1 mM PMSF) by rocking at 4°C for 2–3 hours. Lysates were centrifuged at 110 000 ×g for 1 hour at 4°C; pellets were discarded and supernatant containing MHC-bound peptides were centrifuged at 110 000 ×g for 30 minutes at 4°C. Supernatant was transferred to a 50-mL tube with prewashed rPAS beads (rProtein A Sepharose Fast Flow; GE Healthcare) rotated at 4°C for 2–3 hours to preclear supernatant. rPAS beads were spun down, and supernatant were transferred into a 50-mL tube containing monoclonal antibody (mAb)-bound rPAS and rotated at 4°C overnight with mAb HB11 to isolate MHC class I H-2Db molecules; HB158 for class I H-2Kb molecules; HB183 for class II I-Ab molecules. Beads were transferred to a 15-mL tube and washed twice with 10 mL lysis buffer, centrifuging at 160 ×g for 1 minute, and then transferred to a Bio-Rad Poly-prep chromatography column, washing with 5 mL 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, then with 5 mL of the following buffers: 1 time with 20 mM Tris-HCl, pH 8.0, 150 mM NaCl; 2 times with 20 mM Tris-HCl, pH 8.0, 1 M NaCl; and 3 times with 20 mM Tris-HCl, pH 8.0. Major histocompatibility complex molecules were eluted from rPAS with 4 bed volumes (~4 mL) of 0.2 N acetic acid, then 380 μL acetic acid was added to the elute peptides from MHC molecules. For isolation of peptides, eluate was transferred to a prewetted filter unit with 10 000 dalton cutoff and centrifuged at 1000 ×g.
Identification of Major Histocompatibility Complex-Bound Peptides
Isolated peptides were purified, concentrated, filtered then desalted (STop And Go Extraction tips), and analyzed by liquid chromatography-tandem mass spectrometry using a Q-Exactive mass spectrometer online coupled to a EasyLC 1000 UHPLC. Analytical columns were 30-cm long × 75-μm inner diameter-fused silica columns packed with 2.1-μm diameter ReproSil Pur C18 AQ beads, joint with 2 cm-long, 100 μm-inner diameter-fused silica trap column with 5 μm-diameter Aqua C-18 beads and a 20-μm-inner diameter-fused silica gold-coated spray tip with 6-μm diameter opening. Liquid chromatography buffer A consisted of 0.5% acetic acid, and buffer B consisted of 0.5% acetic acid and 80% acetonitrile. Gradients were run in buffer B from 10% to 32% over 51 minutes, then from 32% to 40% in the next 5 minutes, then increased to 100% over a 2-minute period, and held at 100% for 2.5 minutes. The Q-Exactive acquired a full-range scan at 60 000 event resolution; the 10 most intense multiply-charged ions per cycle were isolated for fragmentation. The search was performed with Andromeda search engine [23] in the MaxQuant (version 1.5.1.12) software against a database comprising protein sequences from mouse and Chlamydia proteomes; an identity score of ≥25 had a false-discovery rate below 2%. Prediction of Db and Kb T-cell epitopes based on peptide sequences was performed using CBS prediction server NetMHC 4.0 Server (http://www.cbs.dtu.dk/services/NetMHC/) [24, 25].
Enzyme-Linked Immunospot Assay
Interferon gamma enzyme-linked immunospot (ELISPOT) assay using splenocytes from mice recovered from C muridarum infection was performed to screen antigenicity of class II-bound Chlamydia peptides from the epithelial immunoproteome. Four Chlamydia peptides (score >25) were synthesized (Thermo Fisher Scientific). MultiScreen-HA filter 96-well plates (Millipore) were coated overnight at 4°C with 2 μg/mL antimouse IFN-γ antibody (clone R4-6A2; BD PharMingen). Splenocytes from C muridarum-immune mice were seeded at 106 per well in presence of individual Chlamydia peptides at 2 μg/mL. One irrelevant ovalbumin peptide and medium alone were used as negative controls. The immunodominant PmpG303-311 peptide previously identified by DC immunoproteomics and heat-killed C muridarum EBs (HK-Cm EB) were used as positive controls. After 20 hours incubation at 37°C and 5% CO2, plates were washed then incubated with biotinylated antimouse IFN-γ antibodies (Clone XMG1.2; BD PharMingen) at 2 μg/mL for 2 hours at room temperature. This was followed by incubation with streptavidin-alkaline phosphatase (BD PharMingen) at a 1:1000 dilution for 1 hour at room temperature; spots visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (NCIP/NBT tablet; Sigma-Aldrich).
RESULTS
Chlamydia muridarum Infection of Bm1.11
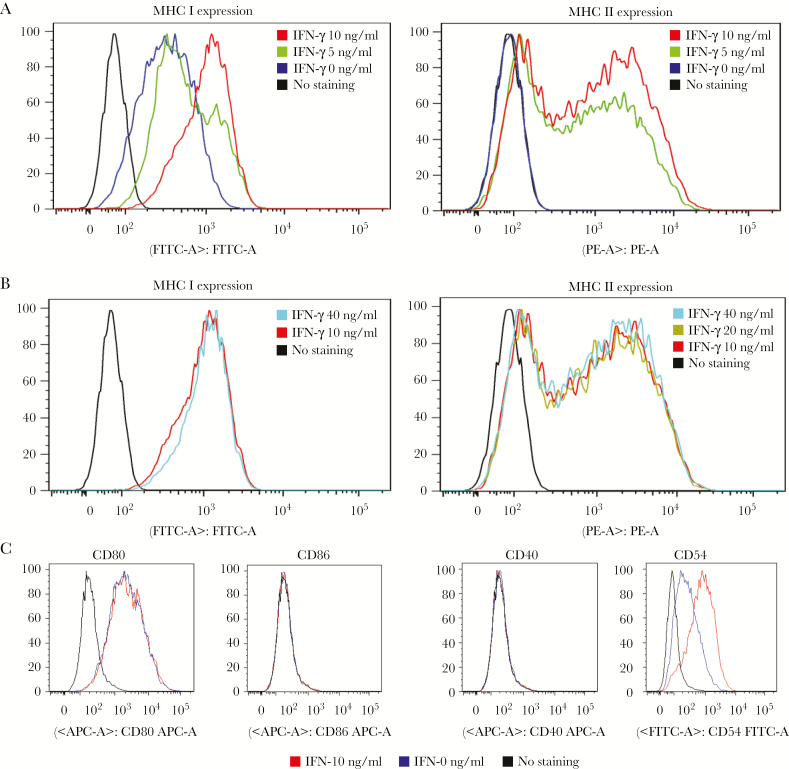
Unlike professional antigen-presenting cells, semiprofessional oviductal epithelial cells such as Bm1.11 do not constitutively express MHC class II and have a limited repertoire of costimulatory molecules. Major histocompatibility complex II expression is controlled by the IFN-inducible CIITA IV promoter, and IFN-γ is known to enhance antigen presentation in multiple epithelial cell types. We tested IFN-γ regulation of MHC II expression on Bm1.11 cells (Figure 1). Major histocompatibility complex class I and II expression on Bm1.11 peaked with 10 ng/mL at 18 hours (Figure 1A), and higher concentrations of IFN-γ had no further effect (Figure 1B). We measured CD80, CD86, CD40, and CD54 (ICAM-1) because costimulation is crucial for T-cell activation. CD80 was constitutively expressed by Bm1.11 cells and unaffected by IFN-γ, whereas CD54, minimally expressed on nonactivated Bm1.11 cells, was substantially increased with IFN-γ treatment. CD86 and CD40 were not expressed on Bm1.11 cells with or without IFN-γ treatment (Figure 1C).
Figure 1.
Interferon gamma (IFN-γ) increased the expression of major histocompatibility complex (MHC) class I and class II on Bm1.11 cells. (A) Expression of MHC class I and II on Bm1.11 cells after the treatment with the doses of 0, 5, and 10 ng/mL IFN-γ. (B) Expression of MHC class I and II on Bm1.11 cells after the treatment with the doses of 10, 20, and 40 ng/mL IFN-γ. The MHC class I and II expression on Bm1.11 cells reached the peak after the treatment with 10 ng/mL IFN-γ. (C) Expression of costimulatory molecules CD80, CD86, CD40, and CD54 on Bm1.11 cells treated with or without 10 ng/mL IFN-γ. The data are representative of 2 similar experiments.
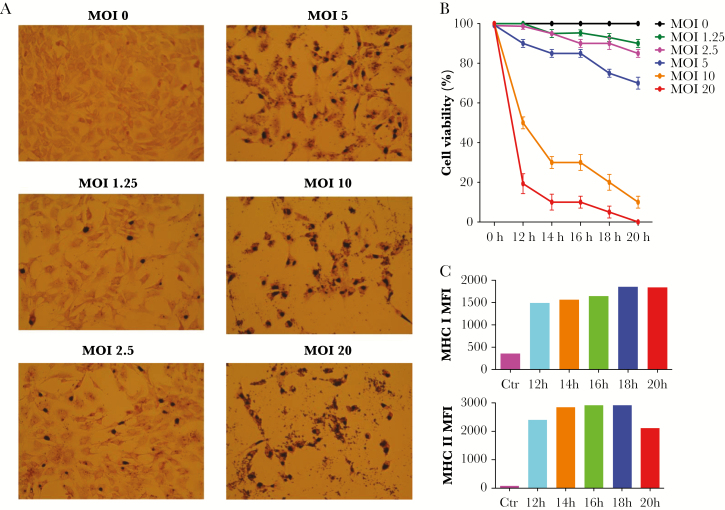
To determine the optimal chlamydial antigen presentation conditions, Bm1.11 cells were infected with C muridarum at a different MOI for different intervals. Chlamydia muridarum infectivity, Bm1.11 viability, and MHC class I and II expression were determined (Figure 2). Although an MOI of 10 and 20 resulted in 100% infectivity (Figure 2A), viability was low at those MOIs (Figure 2B). An MOI of 5 was selected for further evaluation. At an MOI of 5, MHC class I expression peaked 18 hours postinfection and class II expression peaked at 16 hours; extending incubation did not increase MHC class II expression (Figure 2C). The optimal combination of IFN-γ pretreatment, MOI, and infection interval were determined to be 10 ng/mL IFN-γ for 6 hours, followed by 17 hours infection with 5 infectious units/cell.
Figure 2.
Cell viability and major histocompatibility complex (MHC) expression of Bm1.11 cells infected with Chlamydia muridarum at different multiplicity of infection (MOI). (A) Bm1.11 cells were pretreated with 10 ng/mL interferon gamma for 6 hours and infected with C muridarum at different MOI. The inclusions after 18-hour infection were stained using anti-elementary body polyclonal antibodies. (B) Cell viability at different infection time points and MOI determined by trypan blue staining. The results represent the average of 3 replicate samples and are expressed as the means ± standard deviation. (C) Major histocompatibility complex class I and class II mean fluorescence intensity (MFI). Bm1.11 cells were pretreated with 10 ng/mL interferon gamma for 6 hours and infected with C muridarum at an MOI of 5 for 12, 14, 16, 18, or 20 hours. The cells were stained with anti-MHC I-fluorescein isothiocyanate and anti-MHC II-phycoerythrin. The cells not treated with interferon gamma and noninfected cells were set up as control. The data in C are representative of 2 similar experiments.
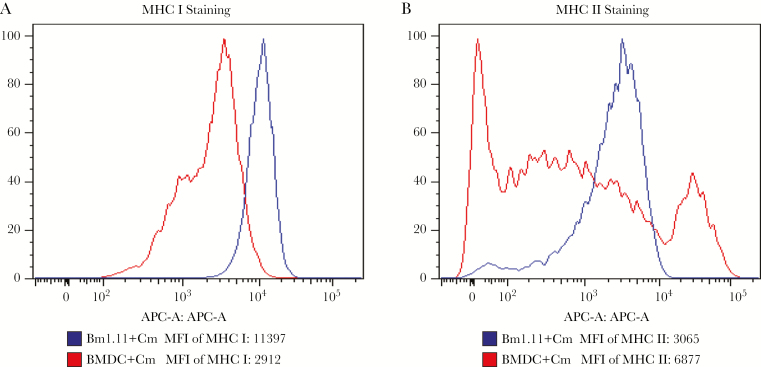
We performed flow cytometry to compare MHC class I and II expression of C muridrum-infected Bm1.11 cells and DCs (Figure 3). The results showed that Bm1.11 epithelial cells expressed higher levels of MHC class I but lower levels of class II compared with DCs. The MFI values for MHC class I on Bm1.11 cells versus DCs is 1696 versus 1114, and the MFI of class II on Bm1.11 versus DC is 7155 versus 15661.
Figure 3.
Comparison of major histocompatibility complex (MHC) class I and class II expression in Bm1.11 cells and bone marrow-derived dendritic cells (BMDCs). Bm1.11 cells were pretreated with 10 ng/mL interferon gamma and infected with Chlamydia muridarum at a multiplicity of infection (MOI) of 4 for 18 hours. The BMDCs from C57BL/6 mice were infected with C muridarum at an MOI of 4 for 18 hours. The cells were stained with anti-MHC I-fluorescein isothiocyanate (A) or anti-MHC II-phycoerythrin (B). Chlamydia muridarum-infected Bm1.11 cells expressed a higher level of MHC I (A) but lower level of MHC II (B) compared with C muridarum-infected BMDCs. The data are representative of 3 similar experiments.
Identification of Chlamydia-Derived Major Histocompatibility Complex-Bound Peptides
Using the optimized protocol, we isolated epithelial MHC class I and II molecules, then we eluted and identified their resident peptides by tandem mass spectrometry. We identified 1569 peptides in total, 1138 of which were MHC class I-bound and 431 peptides that were class II-bound. Among the 1138 MHC class I-bound peptides, 26 peptides were derived from C muridarum and the rest were derived from the mouse proteome. Because the class I K allele of Bm1.11 cells and C57BL/6 DCs differ by 3 amino acids, comparable class I data are limited to 7 chlamydial peptides predicted to be class I Db bound highlighted in Table 1. Among the 431 MHC class II-bound peptides, 4 peptides were derived from C muridarum proteins (TsaD, MnmG, DnaG, and TC0014) and the rest were derived from the mouse proteome (Table 1, Chlamydia and Supplementary Table S1, mouse).
Table 1.
MHC Class I and Class II-Bound Chlamydia Peptides Identified Using Immunoproteomics From Infected Oviductal Epithelial Cells Bm1.11
| Peptide | UniProt Number—Protein Name | Score |
|---|---|---|
| Class I | ||
| VNISSIVGL | Q9PKF7—3-oxoacyl-[acyl-carrier-protein] reductase (FabG) | 45.51 |
| QGLKLSMFS | Q9PL27—Cytochrome D ubiquinol oxidase, subunit I (CydA) | 39.95 |
| PWAEMGVAVTIIA | Q9PL57—Hypothetical protein (TC0251) | 38.74 |
| SAATFPRL | Q9PLP1—Hypothetical protein (TC0058) | 37.44 |
| SAPEQLLKM | Q9PKX4—Hypothetical protein (TC0336) | 35 |
| LEEGSVEIVK | Q9PKT9—Transcription termination/antitermination protein (NusA) | 34.07 |
| EETLLAVL | Q9PJD0—RNA polymerase sigma factor, sigma-54 family(TC0899) | 34.03 |
| LPILESAIG | Q9PKM6—Adherence factor (TC0439) | 33.53 |
| RGPDYGLQLL | Q9PLG7—UDP-N-acetylmuramoyl-tripeptideD-alanyl-d-alanine ligase | 33.31 |
| PNILISLTSA | Q9PK55—Hypothetical protein (TC0617) | 32.8 |
| ASLALSYRL | P75024—Major outer membrane protein (MOMP) | 32.58 |
| FSSHKLYIF | Q9PJE7—Succinate dehydrogenase, cytochrome b558 subunit | 32.57 |
| YGVKNVGSL | Q9PJQ0—Transcription termination factor Rho | 30.94 |
| VVVEGINV | Q9PJM5—50S ribosomal protein (L24) | 30.54 |
| AVAILSLQN | Q9PKQ5—Hypothetical protein (TC0410) | 30.23 |
| MTLLVDR | Q9PLC9—DNA primase (DnaG) | 29.86 |
| SMGSVLSL | Q9PJW1—ATP-dependent Clp protease proteolytic subunit 1 | 29.7 |
| VSPASTTDLL | P56961—Shikimate biosynthesis protein (AroDE) | 28.93 |
| SIALSLETENKHF- | Q9PK55—Hypothetical protein (TC0617) | 28.9 |
| -YQDLPCSLVSS | ||
| ISGIKDFLP | Q59322—60 kDa chaperonin Hsp60 (GroEL1) | 27.41 |
| EAVLKMIP | Q9PJC2—Hypothetical protein (TC0909) | 27.17 |
| DQVFKALI | Q9PLU1—Transcription elongation factor (GreA) | 26.76 |
| SVSVLTSAIQI | Q9PK60—UvrABC system protein A (UvrA) | 26.4 |
| MKIMTRMG | Q9PLL6—Phosphoenolpyruvate carboxykinase (PckG) | 26.31 |
| AIVDWDKQAQ | Q9PJT7—UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 25.16 |
| GAGGYDLTTQGKISINLK | Q9PKP6—Hypothetical protein (TC0419) | 25 |
| Class II | ||
| APFPEV | Q9PKJ5—tRNA N6-adenosine threonylcarbamoyltransferase (TsaD) | 29.88 |
| VIVIGAGHAGCEAAYCA | Q9PJP3—tRNA uridine 5-carboxymethylaminomethyl modification enzyme (MnmG) | 29.42 |
| NTELMRDI | Q9PLC9—DNA primase (DnaG) | 26.33 |
| CLGTIPFVILKIVRFI | Q9PLT1—Hypothetical protein (TC0014) | 25.61 |
Peptides in bold face are predicted to be Db-bound.
Abbreviations: DNA, deoxyribonucleic acid; MHC, major histocompatibility complex; RNA, ribonucleic acid.
Properties of Major Histocompatibility Complex-Bound Peptides
In the C muridarum model, MHC class I and CD8 T cells are nonessential and do not make significant contributions to protective immunity [8]. In our previous study, the C57BL/6 mouse DC immunoproteome had a single class I-bound Chlamydia peptide, SSLFLVKL, predicted to have high affinity for Kb. There were no Db-associated Chlamydia peptides in our published DC immunoproteome. Because Bm1.11 cells differ from C57BL/6 at the MHC class I K allele, we are limited to the observation that infected Bm1.11 epithelial cells present class I Db-bound Chlamydia peptides, whereas infected DC do not.
Bm1.11 cells, matched with C57BL/6 DCs at the MHC class II IAb locus, were chosen to optimize investigation of the IAb immunoproteome because CD4 T cells and MHC class II are critical to protective immunity [8]. We compared the 4 epithelial class II-bound Chlamydia peptides identified in this study (TsaD, MnmG, DnaG, TC0014) with the Chlamydia peptides previously identified in DCs. To our surprise, none of the epithelial class II-bound Chlamydia peptides or their source proteins overlapped with class II-bound Chlamydia peptides and source proteins identified from DCs. Chlamydia source proteins for 3 of the epithelial MHC class II-bound peptides are enzymes involved in replication and translation; TC0014 is a hypothetical protein that may be membrane bound.
Identification of Immunodominant Chlamydia Peptides
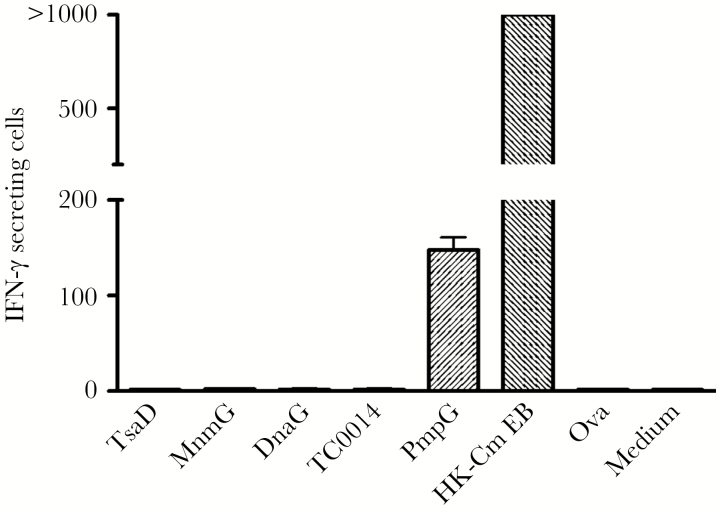
We performed IFN-γ ELISPOT assays using splenocytes from immune C57BL/6 mice that resolved C muridarum genital tract infections to test whether epithelial class II-bound Chlamydia peptides (TsaD, MnmG, DnaG, TC0014) were immunodominant. Splenocytes were stimulated for 20 hours with individual peptides and controls, respectively. The results (Figure 4) showed that immune splenocytes did not recognize any of the epithelial cell-derived Chlamydia peptides, whereas immune splenocytes exposed to heat-killed EBs had very high numbers of IFN-γ-secreting cells including a prominent PmpG303-311-specific subset. Although it was plausible that epithelial cells act as supplemental APCs to present epithelial Chlamydia peptides, addition of Bm1.11 epithelial cells to the ELISPOT system had no effect (data not shown). These results indicate that the primary adaptive response to a C muridarum genital tract infection does not include T cells specific for the 4 dominant MHC class II-bound Chlamydia peptides presented by infected epithelial cells.
Figure 4.
Recognition of individual major histocompatibility complex (MHC) class II-bound Chlamydia peptides eluted from the Bm1.11 epithelial cell line in C57BL/6 mice recovered from live Chlamydia muridarum infection identified by interferon gamma (IFN-γ)—enzyme-linked immunospot assay. Splenocytes from C muridarum immune mice (n = 6) were stimulated in vitro for 20 hours with 4 individual epithelial-derived Chlamydia MHC class II peptides, respectively, 1 irrelevant peptide Ova and medium alone were used as negative controls, and peptide PmpG and heat-killed C muridarum elementary bodies (HK-Cm EB) were used as positive controls. The results represent the average of duplicate wells and are expressed as the means ± standard error of the means of Chlamydia antigen-induced IFN-γ-secreting cells per 106 splenocytes for 2 independent experiments.
DISCUSSION
We found that an infected murine oviductal epithelial cell line processed and loaded Chlamydia peptides onto MHC class I and II molecules, and that the repertoire of class I- and II-bound Chlamydia peptides in infected epithelial cells did not overlap those of infected DCs. Furthermore, unlike infected DC immunoproteome peptides (eg, PmpG303-311), mice that self-resolved C muridarum genital tract infections did not have a detectable T-cell response against the dominant MHC class II-bound Chlamydia peptides presented by infected epithelial cells.
Epithelial cell studies outside the Chlamydia field have focused on intestinal epithelial cells and lung alveolar epithelial cells. Intestinal epithelial cells from humans, mice, and rats constitutively express a low level of MHC class II that becomes elevated during mucosal inflammation [26]. Intestinal epithelial cells contain abundant cathepsin proteases that function in MHC class II antigen presentation [27]. Activation of intestinal epithelial cells by IFN-γ induced the expression of the antigen-processing machinery including invariant chain and H2-DM [27, 28]. Multiple studies demonstrated that intestinal epithelial cells could stimulate CD4 T-cell hybridomas or clones [29, 30]. Based on limited expression of costimulatory molecules on epithelial cells including Bm1.11 in this study, epithelial cells likely serve as targets of effector T-cell responses rather than participating in activation and expansion of naive antigen-specific T cells, the latter function well documented to be performed by DCs. This basic immunobiology semiprofessional (epithelial cell) versus professional antigen-presenting cell (DC) dichotomy has the potential for mismatches between antigen specificity of DC-expanded T cells and microbial peptide epitopes presented by infected epithelial cell targets.
In 1994, Igietseme et al [31, 32] began studies of lymphocyte-epithelial cell interactions in Chlamydia immunobiology, demonstrating that MHC class II-restricted CD4 T-cell lines and clones inhibited the growth of C muridarum in an MHC identical Serotoli cell-derived epithelial cell line. Growth inhibition required contact between the epithelial cell and the T cell and was dependent on ICAM-1 expression. They also showed that CD4 T clones and lines that secreted IFN-γ inhibited C muridarum growth in vitro and adoptively transferred protection in vivo [33]. A decade later, Johnson [21] and others cloned epithelial cell lines from the oviducts of B6.C-H2 bm1 and bm12 mice and characterized their responses to C muridarum infection [20, including the Bm1.11 cell line used in this study. They subsequently demonstrated that MHC class II-restricted CD4 T-cell clones recognized infected oviductal epithelial cells in an IFN-dependent fashion [19], and that CD4 T cells controlled C muridarum replication in epithelial cells by redundant mechanisms, the most potent of which required T-cell degranulation [34]. T-cell expression of Plac8 was identified as being associated with the degranulation-dependent mechanism [35].
To clear Chlamydia-infected reproductive tract epithelium, presumably, an optimal inductive phase response would expand antigen-specific naive T cells capable of recognizing infected epithelial cells during the effector phase. Linking inductive and effector phases requires overlapping presentation of Chlamydia epitopes by DCs and epithelial cells. We were surprised that there was no overlap in the class I Db- or class II IAb-bound Chlamydia peptides or epitope source proteins in the infected DC and epithelial immunoproteomes. This discordance likely reflects differences in antigen processing and permissiveness for Chlamydia replication between the 2 cell types. Although we are limited in analyzing the MHC class I immunoproteome due to differences in the MHC class I K allele in our system, the MHC class II immunoproteome shows significantly more class II-bound Chlamydia peptides in DCs than in Bm1.11 epithelial cells (13 among 344 class II-bound peptides in DCs versus 4 among 431 in epithelial cells, P = .005), supporting differences in the class II presentation pathways between these 2 cell types. That conjecture is further supported by differences in the self-peptide repertoire presented by MHC class II in DCs and Bm1.11 epithelial cells. Only 27 (13%) of 272 MHC class II-presented self-peptides by DCs were shared among the 427 self-peptides presented by epithelial cell class II molecules. The overlap in class II self-peptide presentation was much higher (63%) comparing DCs infected with C muridarum or Salmonella enterica [11] (data not shown). Beyond likely differences in intrinsic antigen-processing pathways, epithelial cells and DCs experience Chlamydia infection differently. Epithelial cells are productively infected, going through the full developmental cycle with expression of ~900 genes [36], whereas murine and human DCs that take up viable Chlamydia appear to be persistently infected [37, 38], a state in which a limited spectrum of Chlamydia proteins are expressed and available for antigen-processing presentation [39].
In our previous DC immunoproteomics studies, we found that the MHC class II-bound Chlamydia peptides were recognized by CD4 T cells from C muridarum immune mice [6, 10], and that adoptive transfer of DCs pulsed ex vivo with those peptides accelerated clearance of intranasal and genital tract Chlamydia infections [6]. In this study, class II-bound Chlamydia peptides identified in infected epithelial cells were not recognized by T cells from C muridarum immune mice. During primary infection, there was no expansion of naive T cells recognizing 4 dominant epithelial MHC class II-bound Chlamydia peptides that apparently go unused during clearance of bacteria from reproductive tract epithelium. This discordance in MHC class II peptide presentation between infected DCs and epithelial cells may explain (1) why DC immunoproteome-based class II antigens used as vaccines only induce partial protection against genital tract infections, (2) the inefficient self-clearance of Chlamydia infections generally, and (3) the failure of sexually transmitted infection vaccines against other epitheliotropic pathogens including herpes simplex virus [40], where the target tissue during primary infection is reproductive tract epithelium [41].
CONCLUSIONS
The ability of individuals to self-clear infection is generally regarded as proof-in-principle of vaccine feasibility, and that vaccines work by giving vaccinees a head-start toward “self-clearance” protective immunity. Likewise, microbial proteins recognized by immune individuals are considered immunodominant antigens important for consideration in subunit vaccines. However, this general paradigm may not be suitable for infections in which natural clearance of primary infections is inefficient. It was recently shown that a varicella-zoster vaccine is effective because it expands a T-cell response to glycoprotein E that is minimal or absent in individuals who have recovered from chickenpox (self-cleared), even when that response is boosted with the attenuated varicella-zoster virus vaccine strain [42]. For infections like Chlamydia, designing vaccines to mimic marginally effective immune states induced by self-resolution of untreated infections may be a suboptimal vaccine strategy. For epitheliotropic pathogens, it may be that vaccines based on antigens presented by infected epithelial cells would best limit intracellular pathogen replication and prevent both transmission and immunopathology.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was funded by a grant from the National Institutes of Health (Grant No. R01AI113103). The mass spectrometry infrastructure was supported by Genome Canada and Genome BC funding (214PRO; to L. J. F.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Rekart ML, Gilbert M, Meza R, et al. Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ectopic pregnancy. J Infect Dis 2013; 207:30–8. [DOI] [PubMed] [Google Scholar]
- 2. Molano M, Meijer CJ, Weiderpass E, et al. The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J Infect Dis 2005; 191:907–16. [DOI] [PubMed] [Google Scholar]
- 3. Morré SA, van den Brule AJ, Rozendaal L, et al. The natural course of asymptomatic Chlamydia trachomatis infections: 45% clearance and no development of clinical PID after one-year follow-up. Int J STD AIDS 2002; 13(Suppl 2):12–8. [DOI] [PubMed] [Google Scholar]
- 4. van den Brule AJ, Munk C, Winther JF, et al. Prevalence and persistence of asymptomatic Chlamydia trachomatis infections in urine specimens from Danish male military recruits. Int J STD AIDS 2002; 13(Suppl 2):19–22. [DOI] [PubMed] [Google Scholar]
- 5. Su H, Morrison R, Messer R, Whitmire W, Hughes S, Caldwell HD. The effect of doxycycline treatment on the development of protective immunity in a murine model of chlamydial genital infection. J Infect Dis 1999; 180:1252–8. [DOI] [PubMed] [Google Scholar]
- 6. Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J Infect Dis 2005; 192:1836–44. [DOI] [PubMed] [Google Scholar]
- 7. Brunham RC. IMMUNOLOGY. A Chlamydia vaccine on the horizon. Science 2015; 348:1322–3. [DOI] [PubMed] [Google Scholar]
- 8. Morrison RP, Feilzer K, Tumas DB. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 1995; 63:4661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karunakaran KP, Rey-Ladino J, Stoynov N, et al. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol 2008; 180:2459–65. [DOI] [PubMed] [Google Scholar]
- 10. Karunakaran KP, Yu H, Jiang X, et al. Outer membrane proteins preferentially load MHC class II peptides: implications for a Chlamydia trachomatis T cell vaccine. Vaccine 2015; 33:2159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karunakaran KP, Yu H, Jiang X, et al. Identification of MHC-bound peptides from dendritic cells infected with Salmonella enterica strain SL1344: implications for a nontyphoidal salmonella vaccine. J Proteome Res 2017; 16:298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu H, Karunakaran KP, Jiang X, Brunham RC. Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice. Vaccine 2014; 32:4672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol 2009; 182:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roshick C, Wood H, Caldwell HD, McClarty G. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect Immun 2006; 74:225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McClarty G, Caldwell HD, Nelson DE. Chlamydial interferon gamma immune evasion influences infection tropism. Curr Opin Microbiol 2007; 10:47–51. [DOI] [PubMed] [Google Scholar]
- 16. Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol 2005; 175:7536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu H, Karunakaran KP, Jiang X, Shen C, Andersen P, Brunham RC. Chlamydia muridarum T cell antigens and adjuvants that induce protective immunity in mice. Infect Immun 2012; 80:1510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson RM, Yu H, Kerr MS, Slaven JE, Karunakaran KP, Brunham RC. PmpG303-311, a protective vaccine epitope that elicits persistent cellular immune responses in Chlamydia muridarum-immune mice. Infect Immun 2012; 80:2204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jayarapu K, Kerr MS, Katschke A, Johnson RM. Chlamydia muridarum-specific CD4 T-cell clones recognize infected reproductive tract epithelial cells in an interferon-dependent fashion. Infect Immun 2009; 77:4469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derbigny WA, Kerr MS, Johnson RM. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J Immunol 2005; 175:6065–75. [DOI] [PubMed] [Google Scholar]
- 21. Johnson RM. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect Immun 2004; 72:3951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 1981; 31:1161–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794–805. [DOI] [PubMed] [Google Scholar]
- 24. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016; 32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 2003; 12:1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mayer L, Eisenhardt D, Salomon P, Bauer W, Plous R, Piccinini L. Expression of class II molecules on intestinal epithelial cells in humans. Differences between normal and inflammatory bowel disease. Gastroenterology 1991; 100:3–12. [DOI] [PubMed] [Google Scholar]
- 27. Hershberg RM, Framson PE, Cho DH, et al. Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J Clin Invest 1997; 100:204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, Justinich CJ. Antigen presentation and MHC class II expression by human esophageal epithelial cells: role in eosinophilic esophagitis. Am J Pathol 2011; 178:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaiserlian D, Vidal K, Revillard JP. Murine enterocytes can present soluble antigen to specific class II-restricted CD4+ T cells. Eur J Immunol 1989; 19:1513–6. [DOI] [PubMed] [Google Scholar]
- 30. Hershberg RM, Cho DH, Youakim A, et al. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J Clin Invest 1998; 102:792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Igietseme JU, Wyrick PB, Goyeau D, Rank RG. An in vitro model for immune control of chlamydial growth in polarized epithelial cells. Infect Immun 1994; 62:3528–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Igietseme JU, Uriri IM, Hawkins R, Rank RG. Integrin-mediated epithelial-T cell interaction enhances nitric oxide production and increased intracellular inhibition of Chlamydia. J Leukoc Biol 1996; 59:656–62. [DOI] [PubMed] [Google Scholar]
- 33. Igietseme JU, Ramsey KH, Magee DM, Williams DM, Kincy TJ, Rank RG. Resolution of murine chlamydial genital infection by the adoptive transfer of a biovar-specific, Th1 lymphocyte clone. Reg Immunol 1993; 5:317–24. [PubMed] [Google Scholar]
- 34. Jayarapu K, Kerr M, Ofner S, Johnson RM. Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms. J Immunol 2010; 185:6911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson RM, Kerr MS, Slaven JE. Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J Immunol 2012; 188:1896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Belland RJ, Zhong G, Crane DD, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 2003; 100:8478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rey-Ladino J, Jiang X, Gabel BR, Shen C, Brunham RC. Survival of Chlamydia muridarum within dendritic cells. Infect Immun 2007; 75:3707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Datta B, Njau F, Thalmann J, Haller H, Wagner AD. Differential infection outcome of Chlamydia trachomatis in human blood monocytes and monocyte-derived dendritic cells. BMC Microbiol 2014; 14:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Belland RJ, Nelson DE, Virok D, et al. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci U S A 2003; 100:15971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cohen J. Immunology. Painful failure of promising genital herpes vaccine. Science 2010; 330:304. [DOI] [PubMed] [Google Scholar]
- 41. Taylor JM, Lin E, Susmarski N, et al. Alternative entry receptors for herpes simplex virus and their roles in disease. Cell Host Microbe 2007; 2:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levin MJ, Kroehl ME, Johnson MJ, et al. Th1 memory differentiates recombinant from live herpes zoster vaccines. J Clin Invest 2018; 128:4429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.