Colloidal interactions in dilute solution singularly enable selection of well-behaved antibodies during drug discovery.
Abstract
Despite the therapeutic success of monoclonal antibodies (mAbs), early identification of developable mAb drug candidates with optimal manufacturability, stability, and delivery attributes remains elusive. Poor solution behavior, which manifests as high solution viscosity or opalescence, profoundly affects the developability of mAb drugs. Using a diverse dataset of 59 mAbs, including 43 approved products, and an array of molecular descriptors spanning colloidal, conformational, charge-based, hydrodynamic, and hydrophobic properties, we show that poor solution behavior is prevalent (>30%) in mAbs and is singularly predicted (>90%) by the diffusion interaction parameter (kD), a dilute-solution measure of colloidal self-interaction. No other descriptor, individually or in combination, was found to be as effective as kD. We also show that well-behaved mAbs, a substantial subset of which bear high positive charge and pI, present no disadvantages with respect to pharmacokinetics in humans. Here, we provide a systematic framework with quantitative thresholds for selecting well-behaved therapeutic mAbs during drug discovery.
INTRODUCTION
Monoclonal antibodies (mAbs) have emerged as a major therapeutic class with more than 60 approved products for treating a wide spectrum of disease, from arthritis to cancer (1) and even infectious diseases (2). The success of mAbs is attributed to high target affinity, exquisite specificity, superior safety, and long half-lives compared to small-molecule drugs. A continued, deeper understanding of antibody structure-function relationships is now routinely used during selection of lead candidates for modulating affinity, specificity, effector function, immunogenicity, and pharmacokinetics (PK) (3). In addition to optimizing biological function, however, there is equal emphasis on early identification of mAbs with optimal manufacturability, stability, and delivery profiles collectively known as developability attributes. It is now well-appreciated that developability characteristics determine the feasibility of mAb drug development, cost-of-goods, and product outcomes. The emergence of mAb drugs has also caused a fundamental transformation in how drugs are delivered to patients. While traditional small-molecule–based medicine is predominantly self-administered orally, mAbs, given their poor oral bioavailability, must be administered parenterally. Intravenous infusion is a common route of administration for mAbs. However, it is mainly restricted to hospital settings. Subcutaneous injection, on the other hand, is less invasive, is more convenient, and enables patient adherence and compliance (4). This is particularly important in the treatment of chronic diseases where self-injection is possible or in settings with limited medical infrastructure. Patient-centric products suitable for subcutaneous injection are essential to realizing the full therapeutic potential of mAbs. However, administration by this route is predominantly restricted to delivery volumes of up to 2 ml. This, in turn, necessitates the development of high-concentration mAb formulations of greater than 100 mg/ml, given that the average dose estimated from currently marketed mAb drugs is ~500 mg.
Here, we focus on predicting two problematic mAb solution behaviors, high solution viscosity and opalescence, which are commonly encountered during the development of high-concentration (> 100 mg/ml), subcutaneously administered mAb products. Both viscosity and opalescence impact mAb developability broadly, affecting all three aspects, manufacturability, stability, and delivery. High solution viscosities [>30 centipoise (cP)] cause limiting back-pressures in ultrafiltration/diafiltration during the mAb concentration unit operation (5). Similarly, viscous mAb solutions also result in forbidding injection forces when administering a subcutaneous injection, particularly via patient friendly autoinjectors (4). In effect, solution viscosity becomes the determining factor for the maximum mAb dose possible via a single subcutaneous injection. Solution opalescence in therapeutic mAbs can be equally, if not more, problematic as it can indicate predisposition for liquid-liquid phase separation, precipitation, or aggregation (6). Given that therapeutic mAbs are routinely exposed to a wide range of solvent streams and greater than a 100°C temperature range (approximately −80°C to 40°C) from production until use, any observation of solution opalescence warrants careful, exhaustive study during development to mitigate potentially catastrophic consequences to product stability and safety.
That molecular properties of mAbs profoundly affect developability is now well recognized; however, selection of mAb candidates with desirable developability profiles has remained difficult. Particularly, predicting mAb solution viscosity and/or opalescence early during candidate screening has been a challenge because definitive molecular determinants of the phenomena have been elusive. Myriad molecular attributes such as molecular charge, domain charge, charge anisotropy, charge patches, and hydrophobicity have been implicated to underlie high viscosity and opalescence (7, 8). Furthermore, any supporting evidence has been with limited mAb datasets (9, 10), closely related mAbs varying as point mutants (10), or with mAbs sharing framework sequences (7, 10). Another practical hurdle is that undesirable solution behavior usually manifests only at higher mAb concentrations, at ~100 mg/ml, and producing necessary quantities of many candidates during early screening is not feasible. In this context, the salient questions that we aimed to address here are what features do well-behaved mAbs have in common? And, could we exploit these features to enable mAb candidate selection early in the discovery process?
To determine general predictors of antibody solution behavior, we analyzed a large and diverse set of 59 therapeutic mAbs, 43 of which are approved products by the U.S. Food and Drug Administration (FDA). Through systematic analysis of 23 measured and calculated molecular descriptors spanning colloidal, conformational, charge-based, hydrophobic, and hydrodynamic properties, we show that mAb solution behavior in terms of viscosity and opalescence can be singularly predicted by measurement of self-interactions in dilute solution. While predicated by colloidal attractive interactions in dilute solution, we show viscosity and opalescence to manifest in a mutually exclusive manner, suggestive of pathway-dependent mechanisms. Colloidally repulsive mAbs are always well behaved, i.e., form inviscid and clear solutions, with a substantial subset bearing high isoelectric points (pI) and positive molecular charge. Because these molecular attributes have been previously implicated in adversely affecting PK, we performed a systematic analysis with human PK data of approved mAbs (n = 41) and their molecular properties. We find no correlation between well-behaved mAbs and their PK behavior in humans. Through this work, we provide a systematic framework for the study of antibody solution behavior with accompanying quantitative thresholds, which can be used to select well-behaved antibodies of therapeutic interest.
RESULTS
Poor solution behavior: Viscosity and opalescence
We sought to develop a systematic dataset of mAb solution behavior in terms of viscosity and opalescence that could be interrogated to identify underlying molecular attributes that predisposed the solution behavior. We measured the viscosity and opalescence of a large set of mAbs (n = 59) at 150 mg/ml, a concentration at which problems related to high viscosity or opalescence generally manifest (Fig. 1). Measurements were made in a 10 mM histidine·HCl (pH 6.0) buffer system, which has emerged as a consensus pH/buffer system for mAb-based products (fig. S1). This is because at pH 6.0 chemical degradation of proteins is minimized, and given adequate solution stability and behavior, liquid formulations become feasible.
Fig. 1. Poor solution behavior is prevalent in mAbs.
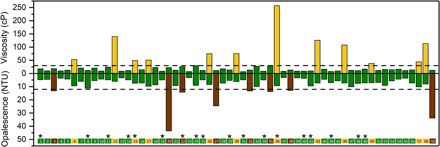
Viscosity and opalescence of 59 mAbs at 150 mg/ml in 10 mM histidine·HCl (pH 6.0). Dashed lines indicate threshold values of 30 cP and 12 NTU for viscosity and opalescence, respectively. Antibodies with viscosity ≤30 cP and turbidity ≤12 NTU are designated green; those with viscosity >30 cP are designated tan; and those with turbidity >12 NTU are designated brown. In the inset at the bottom, mAbs were categorized overall as inviscid and clear (green), viscous (tan), or opalescent (brown). Products approved for subcutaneous injection are indicated by asterisks.
While viscosity measurements with small sample volumes is possible, measurement of solution opalescence of small samples remains challenging. To overcome this, we developed a novel, sensitive, microscale method for accurate measurement of intrinsic solution opalescence measured in terms of nephelometric turbidity units (NTU) in a 10-μl sample (see Materials and Methods). Confirmation of opalescence being related to mAb phase behavior or being a result of large reversible complexes, and not resulting from the presence of large irreversible aggregates, was based on concentration- and temperature-dependent reversibility of the opalescence (fig. S2). Macroscopic phase separation was not observed in any sample. Furthermore, all mAb solutions were of high purity with >95% monomer content (table S1) as determined by size exclusion chromatography (SEC) and sedimentation velocity analytical ultracentrifugation (SV-AUC).
We categorized mAbs based on their viscosity and opalescence (Fig. 1, top and bottom, respectively) using threshold values of 30 cP for viscosity and 12 NTU for opalescence. As stated earlier, solution viscosities >30 cP cause problems during manufacturing and delivery. A turbidity value of 12 NTU was used as it is the midpoint of the opalescent range of 6 to 18 NTU for parenteral products as classified by the European Pharmacopeia (11). Consequently, solutions with viscosities of >30 cP were categorized as viscous, while those below were deemed inviscid (Fig. 1, top; tan and green bars, respectively). Similarly, solutions of >12 NTU were categorized as opalescent and those below were designated as clear (Fig. 1, bottom; brown and green bars, respectively). In all cases, we observed that viscous mAbs tended to exhibit relatively low opalescence and conversely opalescent mAbs were inviscid. This enabled us to categorize the mAbs in accordance with their solution behaviors as viscous (tan), opalescent (brown), and well behaved (green), as indicated in the inset at the bottom of Fig. 1. The inverse correlation between viscosity and opalescence in poorly behaved mAbs provided the first indication that these solution behaviors may be different phenotypic manifestations resulting from colloidal self-association.
The approved therapeutic mAbs in our dataset (n = 43) have been developed for diverse targets using different discovery and manufacturing platforms. Confirmation of mAbs within our dataset representing a diverse sampling of properties was sought by comparison to the larger clinical stage mAb landscape assembled by Raybould et al. (8) (n = 236 variable domain sequences). The calculated physicochemical properties of mAbs within our dataset spanning pI, charge, and hydrophobicity are similar to those in the larger dataset based on average, variance, and span of values (fig. S3).
The viscosity and opalescence data reveal that over a third of the mAbs (37%) became either viscous (19%) or opalescent (18%), suggesting that these undesirable mAb qualities are common, even among manufacturable mAbs. Of the 18 mAbs that have been developed for subcutaneous injection within our dataset (Fig. 1, asterisks), 15 were categorized as well behaved, supporting our categorization thresholds. Given that our dataset is enriched in manufacturable products, development challenges may be encountered in higher incidence in typical early development stages.
Colloidal interactions govern antibody solution behavior
With the systematically assembled solution behavior dataset in hand, we sought to determine the underlying molecular attributes that could serve as predictors. Colloidal interactions have been consistently linked to viscosity as well as opalescence and related phase behaviors in mAb solutions (6, 10, 12). Here, we used the diffusion interaction parameter (kD), which is linearly correlated (fig. S4) to the osmotic second virial coefficient (B2) and is well suited for high-throughput measurement of colloidal interactions (10). The kD measurements of mAbs were in the same 10 mM histidine·HCl (pH 6.0) buffer/pH system (kD,His) as used for viscosity and opalescence measurements. Our results reveal that mAbs, which tend to form inviscid and clear solutions, have high, positive kD values as evident from the clustering of well-behaved mAbs to the left in Fig. 2 (A and B) or the lower left corner in Fig. 2C. In the context of antibody profiling, our data enable us to pose a threshold value of 20 ml/g for kD,His to differentiate well-behaved and poorly behaved mAbs with a remarkable success rate of ~95% (56 of 59 mAbs correctly categorized; Fig. 2, A and D). All 11 immunoglobulin G4s (IgG4s) in our dataset had kD,His values below the threshold (20 ml/g) and displayed disproportionately high propensity for opalescence (7 of 11 IgG4s) compared to IgG1s and IgG2s (Fig. 2E).
Fig. 2. Colloidal interactions in dilute solution underlie solution behavior.
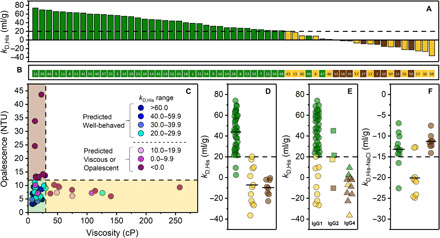
(A) Antibodies arranged in descending order of kD,His values reflecting most repulsive to most attractive. (B) Strip plot of rank-ordered mAbs as arranged in (A). (C) Opalescence versus viscosity of mAbs measured at 150 mg/ml in 10 mM histidine·HCl (pH 6.0). Data points are color-coded according to kD,His ranges as indicated in the figure inset. Dashed lines bordering shaded areas represent threshold values for opalescence (12 NTU; brown) and viscosity (30 cP; tan). Well-behaved mAbs with high kD,His values are clustered in the lower left corner (green). (D) kD,His values for mAbs in each solution property category. A threshold of 20 ml/g is indicated with the dashed line. (E) kD,His values organized by subclass: IgG1 (circles), IgG2 (squares), and IgG4 (triangles). (F) kD,His-NaCl values for mAbs in each solution property category. A threshold of −15 ml/g is indicated with the dashed line. In (A), (B), and (D) to (F), individual mAbs are color-coded according to their solution behavior category: inviscid and clear (green), viscous (tan), or opalescent (brown). In (D) and (F), mean values within each category are indicated by the bold horizontal lines.
Our ability to distinguish between poorly behaving and well-behaving mAbs on the basis of kD,His, measured at low ionic strength [10 mM histidine·HCl (pH 6.0)], suggested that electrostatic interactions are important in governing solution behavior. We then measured kD with 150 mM NaCl added (kD,His-NaCl), i.e., under charge screening conditions, to probe the relative contributions of shorter-range interactions and to potentially differentiate viscous and opalescent mAbs. Eight well-behaved mAbs were randomly selected, as were 17 of the 20 poorly behaved mAbs (due to availability). While kD,His-NaCl values of well-behaved and opalescent mAbs were generally above an arbitrary threshold of −15 ml/g, those for the viscous mAbs were substantially lower (Fig. 2F), indicative of increased self-attraction under charge screening conditions. Differentiation between viscous and opalescent mAbs may thus be possible using a secondary kD,His-NaCl threshold value of −15 ml/g. During early screening, such differentiation may not be essential. However, there will be instances where selection of mAbs with high kD values is not possible, superseded by low-kD mAb candidates with more favorable biological activity. In such cases, it is beneficial to identify the specific liability, whether viscosity or opalescence, early so that focused mitigation strategies can be pursued.
Molecular descriptors of antibody solution behavior
With compelling evidence of attractive colloidal interactions underlying solution viscosity and opalescence, we sought to determine its molecular basis. Our dataset offered us a unique opportunity to test these propositions in the context of measured solution behavior. We assembled a comprehensive array of 23 molecular descriptors for each mAb, categorized as colloidal, electrostatic, conformational, hydrodynamic, and hydrophobic (Fig. 3A). To evaluate the predictive ability of each descriptor, mAbs per their designated color were arranged in ascending or descending order for each descriptor as a strip plot (Fig. 3A). Among all the descriptors, kD,His, as evidenced by the sorting of the well-behaved mAbs to the left of the strip, was singularly effective in identifying viscosity- and opalescence-prone mAbs (Fig. 3A, row 1). No other descriptor was as selective. Moreover, a multivariate approach using partial least squares (PLS) was used to assess the relevance of combining multiple descriptors to predict viscosity. The model (fig. S5) was found optimum using nine descriptors: kD,His, measured pI, calculated pI, sedimentation coefficient (s), effective charge (z*) at pH 6.0, calculated charge at pH 6.0, diffusion coefficient (D), weak cation exchange (WCX) retention time, and Fab/Fc charge separation parameter (Fab/Fc-CSP). The model describes [R2Y(cum)] 61.9% of the variability in viscosity [Q2(cum) = 54.7% by cross-validation]. As a descriptor, kD,His was the highest contributor in the model as per the variable importance to projection plot. Removing kD,His from the model, the remaining eight descriptors only explain 30.1% (R2Y) of the variability in viscosity [Q2(cum) = 20.5%], confirming the significant impact from kD,His to describe this physical characteristic. A similar PLS model was constructed to assess the ability from multiple variables to describe opalescence. The model was found optimum using 11 descriptors that included predominantly electrostatic and hydrodynamic variables. As with viscosity, kD,His was one of the main contributing descriptors in this model.
Fig. 3. Electrostatic repulsion dominates colloidal interactions in dilute solution.
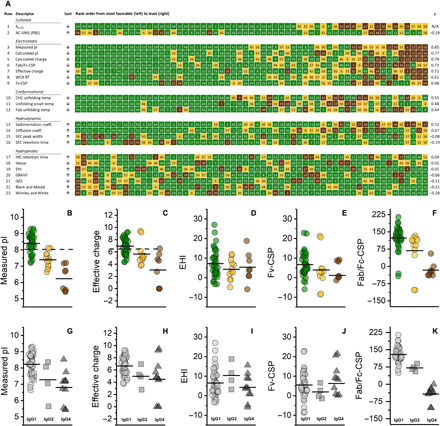
(A) Twenty-three measured and calculated molecular descriptors arranged in order of decreasing favorable value from left to right. Individual mAbs are identified by the numbers in each box and color-coded according to their solution behavior category: inviscid and clear (green), viscous (tan), or opalescent (brown). The Spearman’s rank order correlation (ρ) for each descriptor with respect to kD,His (row 1) is indicated in the rightmost column. (B) to (F) indicate selected molecular descriptors of mAbs [color-coded as in (A)]: (B) measured pI, (C) measured charge, (D) EHI, (E) Fv-CSP, and (F) Fab/Fc-CSP. (G) to (K) indicate the same descriptors organized by subclass as follows: IgG1 (circles), IgG2 (squares), and IgG4 (triangles). Selection thresholds for measured pI and charge (B and C) are indicated with black dashed lines. Solid horizontal lines in (B) to (K) represent mean values of a descriptor in a given category.
We further evaluated the level of discrimination afforded by each of the other descriptors relative to kD,His by calculating the Spearman’s rank order correlation (ρ). Our results show that the measured pI and z* were strongly correlated (ρ = 0.85 and 0.71, respectively; Fig. 3A, rows 3 and 7), confirming the dominant electrostatic component of interactions in dilute solution. Basic mAbs with pI > 8.0 and z* > 6.5 with rare exceptions were always well behaved (Fig. 3, B and C). The Fv charge separation parameter (Fv-CSP), which has been proposed to be important in governing antibody solution viscosity (7), was observed to be the weakest in its correlation relative to the other charge-related parameters (ρ = 0.48; Fig. 3A, row 9). Although a similar parameter representing the calculated charge differential between the Fab and Fc, Fab/Fc-CSP, was strongly correlated (ρ = 0.73; Fig. 3A, row 6). The Fab/Fc-CSP parameter was substantially more negative (greater charge differential at pH 6.0) for the IgG4s compared to the other subtypes within our dataset (Fig. 3K), while the distributions of the other descriptors were comparable (Fig. 3, G to J). The reduced number of basic amino acids in the IgG4 Fc compared with other subtypes leads to a negative charge at pH 6.0, whereas a positive charge is anticipated for IgG1 and IgG2 (table S2). With a generally positively charged Fab at pH 6.0, this backbone-driven charge asymmetry may explain the consistently low kD,His values observed for the IgG4s and hint at a molecular factor underlying opalescence.
For calculated descriptors, the absence of structural information, particularly with respect to charge distribution and hydrophobicity, could cause inaccuracy given contributions from solvent accessibility and local structural environment. To evaluate structural effects on the predictability of calculated descriptors, we analyzed the publicly available sequences in the dataset (n = 43) using the recently published Therapeutic Antibody Profiler (TAP) computational tool (8). The TAP approach uses variable domain structural homology modeling to generate five calculated descriptors based on complementarity-determining region (CDR) length, surface hydrophobicity and charge patches, and charge symmetry. While these factors have been previously linked to undesirable solution behaviors of viscosity, opalescence, and aggregation (8), the TAP analysis did not provide any differentiation between poorly and well-behaved mAbs in terms of viscosity or opalescence (fig. S6).
Properties underlying favorable solution behavior do not negatively affect PK behavior
To evaluate whether our proposed selection criteria are mutually permissive with PK outcomes, we correlated the calculated mAb pI and charge at pH 6.0 of approved products with human subcutaneous bioavailability (F%, n = 14), half-life (t1/2, n = 38), and steady-state clearance (CL, n = 29) data from their respective product package inserts (table S3). Data corresponding to complex elimination behaviors including parallel elimination pathways and concentration-dependent t1/2 were not included. Our analysis (fig. S7) shows no significant correlation between human PK parameters with either calculated mAb charge at pH 6.0 or pI (|r| < 0.2). Similarly, no significant correlations were found with any of the measured molecular descriptors in this study, including with kD,His (r = 0.056 with F%, −0.006 with t1/2, and 0.042 with CL), effective charge measured at pH 6.0 (r = 0.022 with F%, −0.029 with t1/2, and −0.158 with CL), and measured pI (r = 0.058 with F%, 0.074 with t1/2, and −0.113 with CL).
DISCUSSION
Predictors of solution behavior: Viscosity and opalescence
Poor mAb solution behavior, which often manifests as high viscosity and opalescence, presents challenges in the development of mAb drugs. High solution viscosity often defines the highest achievable concentration for a mAb drug product because of the limiting back pressures encountered during production and prohibitive injection forces during delivery via subcutaneous injection. Similarly, high opalescence in mAb solutions is a recognized risk factor for poor phase behavior as recently reviewed [(6) and references therein]. These complications necessitate mitigation strategies, imposing limits on the product manufacturing and design space and consuming development time and resources. Moreover, even following extensive development, the impact of poor solution behavior may not be adequately controlled through customization of the manufacturing process or by developing enabling formulations. Compounding this uncertainty, during preclinical development when there is opportunity to select from a pool of potential candidate mAbs, there is generally insufficient quantity of sample to directly characterize solution behavior, particularly across the potential process and formulation design space. Therefore, we must rely on a risk-based strategy centered on the identification of attributes that reflect a general predisposition to poor behavior. Despite this need, the identification of molecular descriptors that reliably predict the general tendency of mAb solution behavior has remained elusive.
Our results show that the occurrence of high solution viscosity or opalescence is quite common, even among manufacturable mAbs. Through systematic and exhaustive analysis, we also show that both opalescence and viscosity can be most reliably predicted by measurement of weak self-interactions in dilute solution (B2 or kD). While low kD in dilute solution predicts either viscous or opalescent solutions to manifest at higher concentrations, high kD (>+20 ml/g) is a hallmark of well-behaved mAbs. The measurement of kD is made in dilute solution, where the average distance between individual mAb molecules is large relative to the molecular size. In this regime, isotropic charge-charge repulsion is expected to dominate intermolecular interactions due to the shallow distance dependence, receding as exp(−rκ)/r, where r is the intermolecular distance and κ is the inverse Debye length. The high degree of rank order correlation between either pI or z* and kD (Fig. 3A) confirms the predominant electrostatic component of the interaction under these conditions. Of the various intermolecular interactions, only the long-range charge-charge interactions and ultrashort-range excluded volume effects are repulsive, and the rest are attractive (13). Therefore, the high fidelity in the prediction of well-behaved mAbs by kD or B2 may not be readily anticipated given expectation of greater contribution from shorter-range attractive interactions at higher concentrations. Our data point to such shorter-range, attractive interactions cumulatively having a subtle but detectable impact in dilute solution. On the other hand, electrostatic repulsion, while dominant in dilute solution, will also contribute to antibody behavior in concentrated solutions.
As a parameter, kD is agnostic to any specific underlying interaction. While electrostatic repulsion is clearly dominant in governing solution behavior, not all our data can be explained solely based on electrostatics. We recognize that additional shorter-range interactions may be important in governing solution behavior (14). Our screening and high-concentration measurements were made at low ionic strength, where electrostatic repulsions are minimally screened. The kD of opalescent mAbs was least affected by charge screening from sodium chloride (Fig. 2F). Well-behaved mAbs had comparable and negative kD values under these conditions. The viscous mAbs, however, appeared to be particularly sensitive to the addition of sodium chloride, exhibiting more negative values (Fig. 2F). While this enables the practical differentiation of viscous and opalescent mAbs, the underlying mechanistic, molecular relationships between kD, viscosity, and opalescence require further study. In concentrated solutions, it is possible that shorter-range interactions have differentiating impact on solution behavior under these conditions. This hypothesis is supported by the observation that sodium chloride (15), hydrophobic salts (16), and amino acid salts (14, 15) can modulate solution behavior. Arginine salts have been shown to be particularly effective (15). However, we note that, even under these conditions, good correlations between kD and viscosity have been reported in arginine salts (10). However, it stands to reason that well-behaved mAbs are amenable to achieving higher concentrations and enable expedient development. Given that the underlying attractive interactions fully manifest only at sufficiently high mAb concentrations and are likely promiscuous with respect to interaction surface complementarity, it may be difficult to predict solution behavior only based on sequence or structural surface properties. While each of these molecular properties may determine behavior for a given set of mAbs as is supported by literature, reliable generalized prediction would only be possible by understanding overall colloidal interactions in solution, which reflects the individual contributions within the total envelope of interactions.
Computational approaches for developability remain an opportunity to enhance mAb selection and screening. However, our results suggest limited predictability of these tools including Fv charge, hydrophobicity index, Fv-CSP, and the TAP suite of homology-based calculations (Fig. 3 and fig. S6) as general predictors of mAb solution behavior. While the measurement of kD has great utilitarian value in identifying poorly and well-behaved mAbs based on relevant criteria, it may be limited in providing quantitative regressions for viscosity- and opalescence-mAb concentration relationships. In this regard, studies of empirical measurements of solution microstructure at high concentrations coupled with coarse-grain modeling of protein-protein interactions hold promise for providing rich additional insight (17, 18). Deeper study and validation of such approaches would present a powerful, practical approach for predicting antibody solution behavior.
Another key finding of our work is that viscosity and opalescence appear to manifest in a mutually exclusive, nonintersecting manner. Viscous solutions were not opalescent and vice versa. This interrelationship, to the best of our knowledge, has not been previously reported and suggests that opalescence and viscosity may share an underlying mechanism that stems from colloidal self-attraction, but which diverges with respect to molecular orientations or other factors that lead to different solution microstructures. Large molecular clusters or network-like microstructures, which resist deformation under shear, have been linked to solution viscosity (15, 19–21). It is plausible that opalescent solutions also arise from oligomeric clusters, but which are distinct in configuration, scattering properties, or behavior under shear flow (22). Low-viscosity, concentrated nanocluster dispersions of mAbs have been produced by careful modulation of solvent composition (23).
No PK disadvantage for selecting mAbs that display favorable solution behavior
An emerging theme in the literature is that mAbs with high pI or positive charge may have lower subcutaneous bioavailability or be susceptible to rapid clearance. These proposals, which have largely been based on limited mutant sets and preclinical models (24–26), deserve greater scrutiny given that a high proportion of well-behaved mAbs within our dataset bear a high pI and positive charge at pH 6.0. Consideration of PK behavior is a critically important developability attribute and cannot be secondary to manufacturability, stability, or delivery via injection, particularly so because high-concentration liquid formulations are often implicit for subcutaneously administered mAb products. The mAbs within our retrospective analysis are approved products and are therefore likely to have PK properties acceptable for their clinical indication. However, they encompass a broad range of electrostatic properties (calculated pI ranging from 6.4 to 9.1 and charge at pH 6.0 ranging from 7.4 to 37.6 for the approved products). Given this molecular diversity, our analysis shows that there is no PK penalty incurred when selecting mAbs with high pI or charge at pH 6.0 for favorable solution behavior. Conversely, our results do not suggest that electrostatic properties or self-interaction as measured in our screening buffer could identify mAbs with PK-related issues. Previous work in this area has suggested that self-interactions and nonspecific interactions in phosphate-buffered saline or similar buffers may be important (27, 28). Recently, assays based on cellular trafficking processes, facilitated by specific binding and transcytosis through the FcRn receptor, have been shown to be important in determining mAb PK (29). With respect to molecular attributes, PK modeling (30) and specific engineering of IgGs at the FcRn binding interface, between the CH2 and CH3 domains, have yielded definitive evidence of half-life extension or reduced clearance of mAbs in preclinical settings (31, 32). These studies suggest that it may be possible to identify mAbs with favorable PK properties through assays that can be included in a screening paradigm along with the assays that we describe herein for identifying mAbs with favorable solution behavior.
Antibody subclass contributes to solution behavior
Given the sequence and structural homology between IgG subtypes, an implicit assumption in developability studies has been that differences in antibody solution behavior predominantly stem from mAb variable domains. In a recent study, 137 different antibodies were produced using a single IgG1 framework, regardless of natural subtype and framework, to provide a common context for comparing biophysical properties of mAbs (33). The TAP methodology recently described by Raybould et al. (8) also focuses on parameters obtained from structural homology modeling of Fv domains. However, even with a moderate subset of 11 IgG4s, our results suggest that this subclass may be predisposed to different solution behavior at higher concentrations at pH 6.0 compared to IgG1s. In dilute solution, a broad range (positive to negative) of colloidal interactions was observed for IgG1 mAbs, whereas consistently low kD,His values (<20 ml/g) were measured for all 11 IgG4 mAbs (Fig. 2E). Upon concentration, opalescence was observed in 7 of 11 in this cohort. The molecular basis of this divergent behavior is not immediately obvious because many characteristics are shared among the IgG1, IgG2, and IgG4 cohorts. This includes a similar breadth in distribution of pI, solution charge, hydrophobicity indices, and Fv-CSP (Fig. 3, G to J). However, the calculated charge of the IgG4 Fc dimer at pH 6.0 is distinctly different and lower (−1.92) than that of IgG1s (+6.50) and IgG2s (+5.22). The calculated charge of the Fv at pH 6.0 is generally positive. Therefore, IgG4s can be expected to bear a pronounced charge anisotropy between the Fab and Fc domains. A simplistic reflection of this anisotropy can be represented by calculating a Fab/Fc-CSP (Fig. 3, F and K) akin that of the Fv-CSP (7). Given that the Fab/Fc-CSP is most dissimilar for the IgG4s as compared to IgG1 and IgG2s (Fig. 3K), it is possible that the observed opalescence in IgG4s stems from such charge anisotropy. We recognize that while none of the 45 IgG1s in our dataset exhibited opalescence, such behavior has been reported in literature under similar conditions. However, weak colloidal self-interactions as a key underlying factor for opalescence has been a common feature of each of these reports (34, 35).
Thresholds for predicting antibody solution behavior during developability screening
Perhaps the single most important decision contemplated in drug discovery, before initiating clinical development, is the selection of the mAb candidate. Once a candidate is chosen, subsequent development must be conducted with all liabilities inherent to the molecule. Robust tools predictive of developability attributes are prerequisite in preventing the elimination of otherwise biologically functional mAb candidates. Given that identification of functionally optimized mAb candidates is exceedingly challenging, the imposition of additional developability criteria demands a high level of stringency. It is equally important to understand the relative importance of various molecular descriptors, which often provide divergent readouts. Candidate selection is best enabled with a minimal set of predictive descriptors supported by definitive evidence of development risk. We have established and validated a systematic framework for screening viscous and opalescent mAb solution behavior based on measurement of colloidal interactions in dilute solution. Within this framework, kD,His > +20 ml/g (or + ~45 ml/g for 2B2M) in 10 mM histidine·HCl (pH 6.0) has been shown to be singularly predictive of solution viscosity and opalescence with a successful categorization rate of ~95%. Our categorization thresholds are supported by analysis of approved mAb products within our dataset. Of the 18 mAbs developed for subcutaneous injection (Fig. 1, asterisks), 15 were correctly categorized as well behaved. Likewise, parsing the results for approved versus nonapproved products in our dataset further illustrates the greater prevalence of well-behaved mAbs in the approved mAb population (fig. S8). Further differentiation between viscous and opalescent mAbs is possible using a secondary kD,His-NaCl threshold value of −15 ml/g. Effective charge (measured at pH 6.0) > 6.5 and pI > 8.0 are common, but not universal, features of well-behaved mAbs and provide orthogonal supporting criteria. We note that the selection of molecules with high colloidal stability would also be beneficial with respect to reduced aggregation propensity, as is established in literature (36, 37). It is important to note that three mAbs in our dataset with poor solution behavior in the consensus buffer (mAbs 15, 22, and 36) have been developed for subcutaneous injection. This illustrates that high viscosity or opalescence can be overcome through optimization of buffer/pH and excipient content in the drug product, which in these three cases appears to have been achieved with the amino acids arginine or proline. However, selection of well-behaved mAbs permits more expedient development and dosage form flexibility.
Even with the large size and diversity of our dataset, we anticipate the proposed thresholds and guidelines to be continually refined with increasing dataset size, perhaps even leading to subclass-specific thresholds, particularly for IgG4s. Well-established developability guidelines are in place for small molecules (38). Our work provides a systematic framework for formulating one such rule for predicting the solution behavior of mAbs via measurement of colloidal interactions in dilute solution.
MATERIALS AND METHODS
Antibodies
A total of 59 mAbs, consisting of 44 IgG1, 4 IgG2, and 11 IgG4 subclass, were used in this study. Samples were either commercially purchased (Myoderm or BAP Inc.) or produced internally at Sanofi using a CHO (Chinese hamster ovary cell) expression system. Approved mAbs (n = 43) included adalimumab, alemtuzumab, alirocumab, atezolizumab, avelumab, belimumab, beralizumab, bevacizumab, cemiplimab, cetuximab, daratumumab, denosumab, dupilumab, durvalumab, eculizumab, elotuzumab, evolocumab, fremanezumab, galcanezumab, golimumab, guselkumab, infliximab, mepolizumab, necitumumab, nivolumab, obinutuzumab, ocrelizumab, ofatumumab, olaratumab, omalizumab, palivizumab, panitumumab, pembrolizumab, pertuzumab, ramucirumab, rituximab, sarilumab, secukinumab, siltuximab, tocilizumab, trastuzumab, ustekinumab, and vedolizumab. Surfactant was removed from the drug product formulations using DetergentOUT Tween spin columns (G-Biosciences), and the samples were exchanged into either a basis buffer of 10 mM histidine·HCl (pH 6.0) for low-concentration biophysical characterization or a surfactant-free formulation buffer of 10 mM histidine·HCl, 8% (w/v) sucrose (pH 6.0) for high-concentration testing. Exchange was conducted by exhaustive dialysis into the surfactant-free formulation buffer by four successive dialysis cycles, wherein the protein was dialyzed against a total of 8 liters of buffer using 20,000 MWCO Slide-A-Lyzer cassettes (Thermo Fisher Scientific). Following exchange, the samples were concentrated to a target of 220 mg/ml and diluted with surfactant-free formulation buffer to 200, 175, 165, 150, 125, 100, and 50 mg/ml concentrations. Out of these concentrations, 165 mg/ml was further diluted with 10 mM histidine·HCl, 8% (w/v) sucrose, 0.22% (w/v) polysorbate 80 (pH 6.0) so as to achieve a target concentration of 150 mg/ml with 0.02% polysorbate 80.
Viscosity measurement
Samples were passed through 5-μm microcentrifuge filters (Merck Millipore). The mAb concentrations were measured using a SoloVPE attachment (C Technologies) on a Cary 50 spectrophotometer. Solution viscosities were measured at 20°C with a VROC Initium automatic high-throughput viscometer (RheoSense) using shear rate adapted for each solution viscosity such that the pressure drop was in the optimal range for the pressure sensor array of the microfluidic chip. The concentration-viscosity data were reduced by fitting to a simple exponential function (10)
where η is the solution viscosity, η0 is the solvent viscosity, c2 is the mAb concentration, and k is an empirical constant. The best-fit parameters were used to interpolate the viscosity at 150 mg/ml. In 11 cases (mAbs 6, 11, 13, 14, 22, 24, 28, 29, 41, 53, and 57), the sample quantities available did not permit generation of the concentration-viscosity data. Instead, a single viscosity measurement was conducted on the sample at a target concentration of 150 mg/ml. In these cases, the measured concentrations varied from the target (150 mg/ml) by 0.4 to 12.5% (mean 4.9%), leading to corresponding discrepancies in the viscosity value compared to those interpolated from the concentration-viscosity data.
Opalescence measurement
High-concentration samples were passed through 0.22-μm microcentrifuge filters (Millipore) and analyzed by μ-nephelometry using a “de-tuned” static light scattering channel of a Wyatt DynaPro NanoStar laser light scattering instrument. Briefly, samples were loaded in 10-μl quantities into NanoStar disposable cuvettes. The instrument was set to acquire data with the laser power diminished to 1% to reduce sensitivity to excipients, monomeric mAbs, and smaller soluble aggregates. The static scattering detector voltage was recorded and calibrated against that of a set of polymer bead turbidity standards of 1, 4, 10, 20, and 50 NTU (Millipore Sigma).
Measurement of the diffusion interaction parameter by dynamic light scattering (kD-DLS)
Samples dialyzed against 10 mM histidine·HCl (pH 6.0) were diluted to 1.0 to 4.5 mg/ml in 0.5 mg/ml increments and loaded in triplicate into an AcroPrep 384-well 0.45-μm filter plate (Pall Life Sciences) set into a Greiner Bio-One UV-STAR 384-well plate. The plate sandwich was centrifuged briefly, and the apparent diffusion coefficient (Dapp) was measured in each well using a Wyatt DynaPro Plate Reader II dynamic light scattering instrument. Data were fit with the cumulants model, and the fidelity of the fit was verified by filtering the data with an SOS of 0.5. The absorbance of each well at 280 nm was determined using a Molecular Devices i3 plate reader. The mean Dapp (in cm2/s) of each triplicate sample was plotted as a function of the mean concentration (in grams per milliliter) and linearly regressed to determine the diffusion interaction parameter (kD; in milliliters per gram).
Measurement of the diffusion interaction parameter by Taylor dispersion (kD-TD)
Taylor dispersion was conducted on Viscosizer TD using 1.3-m-long capillaries (75 μm inner diameter, 360 mm outer diameter) that were precoated with 2% (w/v) DEAE-dextran in 20 mM sodium citrate (pH 5.0). Samples were dialyzed against 10 mM histidine·HCl, 150 mM sodium chloride (pH 6.0) and adjusted to 25 mg/ml either by ultracentrifugal concentration or by dilution in the same buffer as required. The Viscosizer capillary was filled with dialysis buffer and thermostated at 35°C, and samples were injected at 140 mbar for 2.5 min. Runs were conducted at 140 mbar with a linear velocity of 2.4 mm/s while acquiring absorbance data at 280 nm. Taylorgram curves were analyzed with the vendor software, and a linear regression was applied to the resulting diffusion coefficient versus concentration data. All measurements were conducted in triplicate.
Measurement of effective charge (z*) by capillary zonal electrophoresis
Samples dialyzed against 10 mM histidine·HCl (pH 6.0) were diluted to 0.3 mg/ml and loaded into CE vials with inserts and analyzed using a Sciex PA800Plus capillary electrophoresis instrument. A sample of 0.02% dimethyl sulfoxide (DMSO) to mark electroosmotic flow (EOF) was injected using 0.5 psi forward pressure for 3 s. This was followed immediately by injecting the mAb sample at 0.5 psi for 5 s. Electrophoresis was conducted in an amine-coated capillary using reverse polarity. Data were acquired at each of five different potentials: 6, 8, 10, 12, and 14 kV. The capillary was rinsed with water, cleaned with 1 N NaOH, regenerated, and equilibrated with 10 mM histidine·HCl (pH 6.0) between each measurement. The migration times of the sample and EOF marker were plotted as a function of applied electric field to calculate the electrophoretic mobility for each (μEOF for the EOF marker and appμp for the apparent mobility of the protein). The EOF-corrected electrophoretic mobility of the protein (μp) was calculated as appμp − μEOF. The effective charge (z*) was calculated as
where kB is the Boltzmann constant (1.3806 × 10−23 J/K), T is the temperature in kelvin, e is the elementary charge (1.6022 × 10−19 C), and D is the diffusion coefficient at zero-field and assumed constant for all mAbs (3.855 × 10−7 cm2/s), a value consistent with a hydrodynamic radius in 10 mM histidine·HCl (pH 6.0) of 5.5 nm. The effective charge, a context-dependent empirical parameter, was used directly to screen and compare among different mAbs because all measurements were made in the same buffer. No further modeling was used to relate z* to the molecular charge.
Measurement of isoelectric point by imaged capillary isoelectric focusing
Samples were mixed into a solution containing 0.35% methyl cellulose, a 4% mixture of 3 to 10 and 8 to 10.5 Pharmalyte, pI markers, and the mAb at 0.35 mg/ml and then analyzed using a Protein Simple icE3 imaged capillary isoelectric focusing. Samples were prefocused for 1 min at 1500 V followed by a focusing time of at least 5 min at 3000 V. Data were analyzed with Chromperfect software, and the pI and percentage main peak were recorded.
Chromatographic analyses by SEC, WCX, and hydrophobic interaction
Chromatography was conducted using Agilent 1200 or 1260 systems equipped with binary pumps and variable wavelength detectors. For SEC, 50-μg samples were injected onto a Tosoh TSKgel G3000SWXL column (7.8 × 300 mm, 5-μm particle size) with guard and eluted at 0.5 ml/min with a mobile phase of 40 mM sodium phosphate, 150 mM sodium chloride, 0.05% sodium azide (pH 7.2). WCX was conducted using a pH-based elution system. Samples (20 μg) were injected onto a Thermo ProPac WCX-10 column (4 × 100 mm, 10-μm particle size), preequilibrated in mobile phase A [2.5 mM tris hydrochloride, 1.5 mM imidazole, 11.5 mM piperazine, 20 mM sodium chloride (pH 6.0)], and heated to 40°C. The column was eluted with a linear gradient of mobile phase B [2.5 mM tris hydrochloride, 1.5 mM imidazole, 11.5 mM piperazine, 20 mM NaCl (pH 9.5)] over 7 min at 1.5 ml/min. For hydrophobic interaction, 5-μg samples were injected onto a Tosoh TSKgel butyl-NPR column (4.6 × 35 mm, 2.5-μm particle size) preequilibrated in mobile phase A [1.5 M ammonium sulfate, 25 mM sodium phosphate (pH 7.0)] and thermostated at 25°C. The column was eluted with a linear gradient of mobile phase B [25 mM sodium phosphate (pH 7.0)] over 15 min at 0.5 ml/min. Chromatograms were analyzed using Agilent ChemStation software, and the retention time and percentage main peak were recorded. For SEC, the peak width was also logged.
Sedimentation velocity analytical ultracentrifugation
Samples dialyzed against 10 mM histidine·HCl (pH 6.0) were diluted to 0.3 mg/ml and loaded into AUC cells assembled with charcoal-filled Epon dual-sector centerpieces and quartz windows. Cells were loaded into an AN60-Ti rotor and equilibrated for at least 2 hours under vacuum at 20°C. Analyses were conducted at 40,000 rpm with continuous acquisition of 280-nm absorbance scans using a radial step size of 30 μm. Datasets were analyzed with the continuous c(s) distribution and noninteracting discrete species models implemented in the sedfit analysis platform (39). All data were well modeled as evidenced by Z-scores less than 10 in all cases, and typically less than 5 (table S1). The primary analytic output from SV-AUC was the relative percent monomeric protein, the sedimentation coefficient (s), and the diffusion coefficient (D).
Differential scanning calorimetry
Samples dialyzed against 10 mM histidine·HCl (pH 6.0) were diluted to 0.5 mg/ml and analyzed with a Malvern PEAQ-DSC Automated system. Samples were preequilibrated at 10°C for 15 min and then heated to 110°C at a rate of 60°C per hour with a filtering period of 8 s and passive feedback. The resulting endotherms were modeled using the vendor software, and the unfolding onset (Tonset) and apparent melting temperatures (Tm) of each transition were recorded.
Measurement of sedimentation interaction parameter (ks) and calculation of second virial coefficient (B2)
Samples were loaded into AUC cells assembled with 0.3-cm path-length charcoal-filled Epon dual-sector centerpieces and quartz windows. Analyses were conducted at 42,000 rpm with continuous acquisition of absorbance at either 280 nm (target concentrations of 1.0, 1.5, and 2.0 mg/ml) or 250 nm (target concentrations of 2.5, 3.0, 3.5, 4.0, and 4.5 mg/ml) and a radial step size of 30 μm. The apparent sedimentation coefficient (sapp) for each dataset was determined by the time derivative method as used in the program DCDT+ (40). The inverse of sapp (in s−1) of each sample was plotted as a function of the mean concentration (in grams per milliliter) and linearly regressed to determine the sedimentation interaction parameter (ks; in milliliters per gram). The osmotic second virial coefficient (B2) was approximated as (41)
where the partial specific volume () was assumed as 0.735 ml/g and the molecular weight (M) was assumed as 150,000 Da.
Calculation of properties from amino acid composition
Sequences for approved mAbs were obtained in the public domain. In instances where only the variable domain sequence was available (33), a consensus Fc sequence of matching subtype was assumed. The clinical antibody landscape comparator dataset was assembled with variable domain sequences from Raybould et al. (8) matched with subtype-specific consensus Fc sequences. Of the total compiled sequences (n = 242), four non-IgG sequences and two of uncertain subtype were excluded from analysis (n = 236 final). Charge-based calculations were determined from composition using the analysis platform sednterp (42) and assuming the following amino acid pKa (where Ka is the acid dissociation constant) values: Arg = 12, Asp = 4.5, Glu = 4.6, His = 6.2, Lys = 10.4, and Tyr = 9.7. All sulfhydryl side chains in Cys residues were assumed disulfide-bonded with no contributing pKa. Hydrophobicity was estimated using the methods of Eisenberg et al. [Eisenberg hydrophobicity index (EHI)] (43), Kyte and Doolittle [grand average of hydropathy (GRAVY)] (44), Black and Mould (45), Hessa et al. (46), Wimley and White (47), and Engelman et al. [Goldman/Engelman/Steitz (GES)] (48). The Fv-CSP (7) was calculated as the product of the net charge at pH 6.0 of the VH and VL domains as determined in sednterp (42). The Fab/Fc-CSP was calculated as the product of the net charge at pH 6.0 of the Fab and Fc domains. Publicly available variable domain sequences were analyzed using the TAP (8) web server (http://opig.stats.ox.ac.uk/webapps/sabdab-sabpred/TAP.php).
PLS regression analyses
PLS is a multivariate regression method (49) that models the relationship between two blocks of data: in this work, the descriptors characterizing each mAb (X-block) and the responses (viscosity and opalescence, Y-block). PLS is a supervised regression technique that is based on the decomposition of the dataset into a reduced number of dimensions (latent variables) that maximizes the covariance between the variables in the X-block and response(s) in the Y-block.
Supplementary Material
Acknowledgments
We thank S. Mitragotri, A. Saluja, B. Perez-Ramirez, P. Wils, and I. Kather for critical discussions and review of the manuscript. Funding: This work was supported by Sanofi. Author contributions: J.S.K., J.Z., and Y.R.G. designed research; J.S.K., A.S., S.M.A., L.F., M.M.L., and K.T.H. performed research; J.S.K., C.C.-R., W.S., and C.Y.A. analyzed data; J.S.K. and Y.R.G. wrote the manuscript with all authors providing feedback. Competing interests: All authors are employed by Sanofi, a company that discovers and markets antibody therapies. The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Materials can be provided by Sanofi pending scientific review and a completed material transfer agreement. Requests for materials should be submitted to: Y.R.G. or J.S.K.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eabb0372/DC1
REFERENCES AND NOTES
- 1.Walsh G., Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 36, 1136–1145 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A., Dadachova E., Pirofski L.-a., Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2, 695–703 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Carter P. J., Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Anselmo A. C., Gokarn Y., Mitragotri S., Non-invasive delivery strategies for biologics. Nat. Rev. Drug Discov. 18, 19–40 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Shire S. J., Shahrokh Z., Liu J., Challenges in the development of high protein concentration formulations. J. Pharm. Sci. 93, 1390–1402 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Raut A. S., Kalonia D. S., Pharmaceutical perspective on opalescence and liquid–liquid phase separation in protein solutions. Mol. Pharm. 13, 1431–1444 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Sharma V. K., Patapoff T. W., Kabakoff B., Pai S., Hilario E., Zhang B., Li C., Borisov O., Kelley R. F., Chorny I., Zhou J. Z., Dill K. A., Swartz T. E., In silico selection of therapeutic antibodies for development: Viscosity, clearance, and chemical stability. Proc. Natl. Acad. Sci. U.S.A. 111, 18601–18606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raybould M. I. J., Marks C., Krawczyk K., Taddese B., Nowak J., Lewis A. P., Bujotzek A., Shi J., Deane C. M., Five computational developability guidelines for therapeutic antibody profiling. Proc. Natl. Acad. Sci. U.S.A. 116, 4025–4030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomar D. S., Singh S. K., Li L., Broulidakis M. P., Kumar S., In silico prediction of diffusion interaction parameter (kD), a key indicator of antibody solution behaviors. Pharm. Res. 35, 193 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Connolly B. D., Petry C., Yadav S., Demeule B., Ciaccio N., Moore J. M. R., Shire S. J., Gokarn Y. R., Weak interactions govern the viscosity of concentrated antibody solutions: High-throughput analysis using the diffusion interaction parameter. Biophys. J. 103, 69–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ph. Eur. 2.2.1. Clarity and degree of opalescence of liquids. European Pharmacopoeia 7.0. (2008), pp. 21–22.
- 12.Lewus R. A., Levy N. E., Lenhoff A. M., Sandler S. I., A comparative study of monoclonal antibodies. 1. Phase behavior and protein-protein interactions. Biotechnol. Prog. 31, 268–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laue T., Proximity energies: A framework for understanding concentrated solutions. J. Mol. Recognit. 25, 165–173 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Tilegenova C., Izadi S., Yin J., Huang C. S., Wu J., Ellerman D., Hymowitz S. G., Walters B., Salisbury C., Carter P. J., Dissecting the molecular basis of high viscosity of monospecific and bispecific IgG antibodies. MAbs 12, e1692764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Nguyen M. D., Andya J. D., Shire S. J., Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J. Pharm. Sci. 94, 1928–1940 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Du W., Klibanov A. M., Hydrophobic salts markedly diminish viscosity of concentrated protein solutions. Biotechnol. Bioeng. 108, 632–636 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Ferreira G. M., Calero-Rubio C., Sathish H. A., Remmele R. L. J. Jr., Roberts C. J., Electrostatically mediated protein-protein interactions for monoclonal antibodies: A combined experimental and coarse-grained molecular modeling approach. J. Pharm. Sci. 108, 120–132 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Dear B. J., Bollinger J. A., Chowdhury A., Hung J. J., Wilks L. R., Karouta C. A., Ramachandran K., Shay T. Y., Nieto M. P., Sharma A., Cheung J. K., Nykypanchuk D., Godfrin P. D., Johnston K. P., Truskett T. M., X-ray scattering and Coarse-Grained simulations for clustering and interactions of monoclonal antibodies at high concentrations. J. Phys. Chem. B 123, 5274–5290 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Yearley E. J., Godfrin P. D., Perevozchikova T., Zhang H., Falus P., Porcar L., Nagao M., Curtis J. E., Gawande P., Taing R., Zarraga I. E., Wagner N. J., Liu Y., Observation of small cluster formation in concentrated monoclonal antibody solutions and its implications to solution viscosity. Biophys. J. 106, 1763–1770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung J. J., Dear B. J., Karouta C. A., Chowdhury A. A., Godfrin P. D., Bollinger J. A., Nieto M. P., Wilks L. R., Shay T. Y., Ramachandran K., Sharma A., Cheung J. K., Truskett T. M., Johnston K. P., Protein–protein interactions of highly concentrated monoclonal antibody solutions via static light scattering and influence on the viscosity. J. Phys. Chem. B 123, 739–755 (2019). [DOI] [PubMed] [Google Scholar]
- 21.von Bülow S., Siggel M., Linke M., Hummer G., Dynamic cluster formation determines viscosity and diffusion in dense protein solutions. Proc. Natl. Acad. Sci. U.S.A. 116, 9843–9852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stradner A., Sedgwick H., Cardinaux F., Poon W. C. K., Egelhaaf S. U., Schurtenberger P., Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 432, 492–495 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Johnston K. P., Maynard J. A., Truskett T. M., Borwankar A. U., Miller M. A., Wilson B. K., Dinin A. K., Khan T. A., Kaczorowski K. J., Concentrated dispersions of equilibrium protein nanoclusters that reversibly dissociate into active monomers. ACS Nano 6, 1357–1369 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Igawa T., Tsunoda H., Tachibana T., Maeda A., Mimoto F., Moriyama C., Nanami M., Sekimori Y., Nabuchi Y., Aso Y., Hattori K., Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng. Des. Sel. 23, 385–392 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Li B., Tesar D., Boswell C. A., Cahaya H. S., Wong A., Zhang J., Meng Y. G., Eigenbrot C., Pantua H., Diao J., Kapadia S. B., Deng R., Kelley R. F., Framework selection can influence pharmacokinetics of a humanized therapeutic antibody through differences in molecule charge. MAbs 6, 1255–1264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav D. B., Sharma V. K., Boswell C. A., Hotzel I., Tesar D., Shang Y., Ying Y., Fischer S. K., Grogan J. L., Chiang E. Y., Urban K., Ulufatu S., Khawli L. A., Prabhu S., Joseph S., Kelley R. F., Evaluating the use of antibody variable region (Fv) charge as a risk assessment tool for predicting typical cynomolgus monkey pharmacokinetics. J. Biol. Chem. 290, 29732–29741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hötzel I., Theil F.-P., Bernstein L. J., Prabhu S., Deng R., Quintana L., Lutman J., Sibia R., Chan P., Bumbaca D., Fielder P., Carter P. J., Kelley R. F., A strategy for risk mitigation of antibodies with fast clearance. MAbs 4, 753–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avery L. B., Wade J., Wang M., Tam A., King A., Piche-Nicholas N., Kavosi M. S., Penn S., Cirelli D., Kurz J. C., Zhang M., Cunningham O., Jones R., Fennell B. J., Donnell B. M., Sakorafas P., Apgar J., Finlay W. J., Lin L., Bloom L., O’Hara D. M., Establishing in vitro in vivo correlations to screen monoclonal antibodies for physicochemical properties related to favorable human pharmacokinetics. MAbs 10, 244–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung S., Nguyen V., Lin Y. L., Lafrance-Vanasse J., Scales S. J., Lin K., Deng R., Williams K., Sperinde G., Li J. J., Zheng K., Sukumaran S., Tesar D., Ernst J. A., Fischer S., Lazar G. A., Prabhu S., Song A., An in vitro FcRn-dependent transcytosis assay as a screening tool for predictive assessment of nonspecific clearance of antibody therapeutics in humans. MAbs 11, 942–955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagan L., Pharmacokinetic modeling of the subcutaneous absorption of therapeutic proteins. Drug Metab. Dispos. 42, 1890–1905 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Zalevsky J., Chamberlain A. K., Horton H. M., Karki S., Leung I. W. L., Sproule T. J., Lazar G. A., Roopenian D. C., Desjarlais J. R., Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng R., Meng Y. G., Hoyte K., Lutman J., Lu Y., Iyer S., DeForge L. E., Theil F.-P., Fielder P. J., Prabhu S., Subcutaneous bioavailability of therapeutic antibodies as a function of FcRn binding affinity in mice. MAbs 4, 101–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain T., Sun T., Durand S., Hall A., Houston N. R., Nett J. H., Sharkey B., Bobrowicz B., Caffry I., Yu Y., Cao Y., Lynaugh H., Brown M., Baruah H., Gray L. T., Krauland E. M., Xu Y., Vásquez M., Wittrup K. D., Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. U.S.A. 114, 944–949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukumar M., Doyle B. L., Combs J. L., Pekar A. H., Opalescent appearance of an IgG1 antibody at high concentrations and its relationship to noncovalent association. Pharm. Res. 21, 1087–1094 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Salinas B. A., Sathish H. A., Bishop S. M., Harn N., Carpenter J. F., Randolph T. W., Understanding and modulating opalescence and viscosity in a monoclonal antibody formulation. J. Pharm. Sci. 99, 82–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi E. Y., Krishnan S., Randolph T. W., Carpenter J. F., Physical stability of proteins in aqueous solution: Mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 20, 1325–1335 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Saluja A., Fesinmeyer R. M., Hogan S., Brems D. N., Gokarn Y. R., Diffusion and sedimentation interaction parameters for measuring the second virial coefficient and their utility as predictors of protein aggregation. Biophys. J. 99, 2657–2665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J., Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997). [DOI] [PubMed] [Google Scholar]
- 39.P. Schuck, in Analytical Ultracentrifugation: Techniques and Methods, D. J. Scott, S. E. Harding, A. J. Rowe, Eds. (The Royal Society of Chemistry, 2005), chap. 2, pp. 26–49. [Google Scholar]
- 40.Philo J. S., A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279, 151–163 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Harding S. E., Johnson P., The concentration-dependence of macromolecular parameters. Biochem. J. 231, 543–547 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T. M. Laue, B. D. Shah, T. M. Ridgeway, S. L. Pelletier, in Analytical Ultracentrifugation in Biochemistry and Polymer Science, S. E. Harding, A. J. Rowe, J. C. Horton, Eds. (Royal Society of Chemistry, 1991), chap. 7, pp. 90–124. [Google Scholar]
- 43.Eisenberg D., Schwarz E., Komaromy M., Wall R., Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 (1984). [DOI] [PubMed] [Google Scholar]
- 44.Kyte J., Doolittle R. F., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
- 45.Black S. D., Mould D. R., Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 193, 72–82 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G., Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Wimley W. C., White S. H., Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Engelman D. M., Steitz T. A., Goldman A., Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15, 321–353 (1986). [DOI] [PubMed] [Google Scholar]
- 49.Wold S., Sjöström M., Eriksson L., PLS-regression: A basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 58, 109–130 (2001). [Google Scholar]
- 50.Lehermayr C., Mahler H.-C., Mäder K., Fischer S., Assessment of net charge and protein–protein interactions of different monoclonal antibodies. J. Pharm. Sci. 100, 2551–2562 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Saito S., Hasegawa J., Kobayashi N., Kishi N., Uchiyama S., Fukui K., Behavior of monoclonal antibodies: Relation between the second virial coefficient (B2) at low concentrations and aggregation propensity and viscosity at high concentrations. Pharm. Res. 29, 397–410 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eabb0372/DC1


