Abstract
Background
Over 100 people die daily from opioid overdose and $78.5B per year is spent on treatment efforts, however, the real societal cost is multifold greater. Alternative strategies to eradicate/manage drug misuse and addiction need consideration. The perception of opioid addiction as a social/criminal problem has evolved to evidence-based considerations of them as clinical disorders with a genetic basis. We present evaluations of the genetics of addiction with ancestry-specific risk profiles for consideration.
Objective
Studies of gene variants associated with predisposition to substance use disorders (SUDs) are monolithic, and exclude many ethnic groups, especially Hispanics and African Americans. We evaluate gene polymorphisms that impact brain reward and predispose individuals to opioid addictions, with a focus on the disparity of research which includes individuals of African and Hispanic descent.
Methodology
PubMed and Google Scholar were searched for: Opioid Use Disorder (OUD), Genome-wide association studies (GWAS); genetic variants; polymorphisms, restriction fragment length polymorphisms (RFLP); genomics, epigenetics, race, ethnic group, ethnicity, ancestry, Caucasian/White, African American/Black, Hispanic, Asian, addictive behaviors, reward deficiency syndrome (RDS), mutation, insertion/deletion, and promotor region.
Results
Many studies exclude non-White individuals. Studies that include diverse populations report ethnicity-specific frequencies of risk genes, with certain polymorphisms specifically associated with Caucasian and not African-American or Hispanic susceptibility to OUD or SUDs, and vice versa.
Conclusion
To adapt precision medicine-based addiction management in a blended society, we propose that ethnicity/ancestry-informed genetic variations must be analyzed to provide real precision-guided therapeutics with the intent to attenuate this uncontrollable fatal epidemic.
Keywords: Dopamine homeostasis, genetics, Precision Addiction Management (PAM), Reward Deficiency Syndrome (RDS), ethnic groups, gene guided therapy
1. INTRODUCTION
It is well documented that individuals with exposure to opioids can develop addictive behavior. This possibility is not just specific for street use of heroin but also when used legally for pain relief. Opioid medications used to alleviate pain are very effective, but chronic use does induce dependence. In America, there has been a steady increase in opioid-related deaths from all walks of life and economic status as a result of overdose from natural opiates or synthetic opioids and their many new isoforms which include deadly fentanyl derivatives. The Center for Disease Control (CDC) reports greater than 400,000 people died from opioid overdose in the US from 1999 to 2017 (https://www.cdc.gov/drugoverdose/epidemic/index.html) and this number continues to rise. Unfortunately, this unwanted opioid epidemic is, in part, the result of the pharmaceutical companies falsely claiming that their opioids have a low liability to induce addiction possibly because of extended release formulae; this has led to the widespread use of opioid drugs to attenuate moderate pain [1-3].
The two main treatment options for opioid replacement therapy (ORT) are buprenorphine and methadone and these have proven efficacious in ameliorating opioid addiction [1, 4] and are arguably the gold standard for treatment. However, patients under ORT are not protected from relapse [5, 6], and long term chronic use of ORTs also lead to respiratory depression and overall behavioral health problems along with reward deficiency [7]. Genetics is an important component in an individual’s susceptibility to addiction and in the efficacy of opioid replacement therapy. Allelic variations can influence how the brain reacts to substances of abuse, especially to opioids. For alcohol, it has been shown that people of different ancestry metabolize alcohol differently [8, 9] and certain groups may be more susceptible to the addictive properties of alcohol [9]. Alcohol, opioids and other substances of abuse act on the reward pathway and modulate neurotransmitters, neuropeptides, their receptors and modulatory enzymes. These SUDs can act at different anatomical and subcellular regions of the neural circuitry of reward and addiction to modulate neurotransmitter release, processing, and metabolism.
Whereas individuals regardless of whether of European, African, Asian, or Hispanic descent can be impacted by the devastating effects of opioid addiction, we show that susceptibility and approach to treatment will vary widely. With a one-size fits all approach to the opioid addiction problem in America, a cycle is perpetuated where medications prescribed to one societal group may be effective whereas in another group it is grossly ineffective, and this ineffectiveness can perpetuate drug relapse due to inadequate therapy. Precision addiction medicine through pharmacogenomics involves accounting for the individual’s genetic profile to determine dosing based on metabolizer phenotype for more effective opioid therapy. In addition, a great many gene polymorphisms influence how the brain process reward and the propensity to develop addiction. We analyze the literature to 1) determine the inclusion of various ethnic groups in important genome-wide studies, and 2) when included we report similarities and differences for expressing allelic SNPs between groups. This approach highlights the necessity to include people of all backgrounds to reduce the disparity of access to innovative treatment in America. Furthermore, genetics can present the opioid addiction problem as a medical disorder to de-stigmatize its impact and encourage others to adopt early prevention measures in individuals.
The reward pathway and the point at which other substances of abuse influence its function have been reviewed extensively elsewhere [10, 11]. In the first part of this review, we discuss the natural reward pathway and the neurobiology and anatomy of opioid addiction. Next, we focus on studies to examine genetic links to opioid use disorder, with a specific focus on disparities in different populations, examining ancestry-specific differences or ancestry-neutral (shared) commonalities in susceptibility to addiction, relapse, and drug metabolism. We focus on specific genes implicated in reward and discuss the single nucleotide polymorphisms (SNP) evaluated in different ethnicities. As society moves toward precision medicine, OUD will require a genetic basis for precision addiction management [12].
2. NEUROBIOLOGY OF OPIOID ADDICTION
We now know how opioids induce their powerful neuropharmacological effects as established by discoveries of scientists in the ’70s who presented evidence for endogenous opioid peptide biology [13-15], characterization of opioid peptides [16-18] and even their role in alcoholism [19, 20]. The three major opioid receptors, mu (MOR), delta (DOR), and kappa (KOR) were cloned in the early nineties [21-24] and further understanding of their neuropharmacology has been derived from knock-outs [25].
In general, psychoactive abusable drugs induce an increase in dopamine in the nucleus accumbens (NAc). Continued chronic stimulation can desensitize or upregulate dopamine receptors, thus increasing the dopamine requirement to achieve the same amount of pleasure [26]. The molecular mechanism underlying opioid addiction is an intricate biological process that involves exposure of opioids/opiates to and activation of opioid receptors in the ventral tegmental area (VTA). Specifically, opioids bind mu opioid receptors (MOR), which belong to the G protein-coupled receptor (GPCR) superfamily of seven transmembrane receptors [27], on the inhibitory neurons in the VTA to initiate a cascade of downstream reactions via the Gα/Gβγ protein pathway [28, 29]. Consequently, adenylyl cyclase and the voltage-gated Ca2+ channels are inhibited by Gα and Gβγ proteins, respectively, while G protein-activated inwardly rectifying K+ channels (GIRKs) and phospholipase Cβ effectors are both stimulated by Gαi [30, 31].
Studies have shown that opioid-mediated adenylyl cyclase inhibits the release of some transmitters including gamma-aminobutyric acid (GABA) [32, 33]. Important mediators of opioid actions involve MOR-mediated [34] downstream signaling with inhibitory effects on neural excitability occasioned by the Gαi interaction with GIRKs and the reduction in Ca2+ currents across the membrane [35]. Overall, the combined effects of opioids on MORs result in the release of dopamine from VTA terminals innervating the nucleus accumbens (NAc), a process reported to be facilitated by concomitant MOR-mediated GABA downregulation in the VTA [36, 37]. Furthermore, the VTA itself is controlled by or communicates with a variety of brainstem structures. The NAc is referred to as the pleasure center of the brain because of its connection with and modulation of other regions that translate the rewarding properties of substances as pleasurable; These substances encompass not just drugs of abuse, but also other substances like food, sex, exercise, and other activities/behaviors that cause dopamine release from VTA terminals onto the NAc [38]. Thus, activation of the NAc results in and from firing through secondary, parallel series of integrated circuits to the reward system comprising the amygdala, hippocampus and prefrontal cortex (Fig. 1), as well as the striatum, which respectively control emotion [39, 40], memories [41], and habit and processing of reward [42]. While the production of dopamine itself does not result in addiction, its unregulated production in the NAc, and sustained firing in other regions of the brain as a result of continuous exposure to opioids in the mesolimbic reward pathway, produces a reinforcement of gratifying sensations. Over time, brain reward systems may readjust via altered regulation of receptors and modulation of transporters, enzymes, reuptake pumps. Various signaling machinery to accommodate these changes will require more opioids/opiates to achieve comparatively the same level of gratification, a condition known as tolerance [43]. Eventually, subjects may become addicted to drugs of abuse as a result of defective or impaired rewiring of the reward system and the prefrontal complex, which controls planning and decision making [10, 44]. Whereas MORs mediate the rewarding effects of opioids, kappa opioid receptors (KORs) have antagonistic actions in terms of reward and addiction processes and mediate the dysphoric and aversive effects of the drug in animals and humans [45, 46]. The role of delta opioid receptor (DOR) is unclear since morphine reward and motivation seems independent of DOR based on knock–out mouse experiments [47, 48].
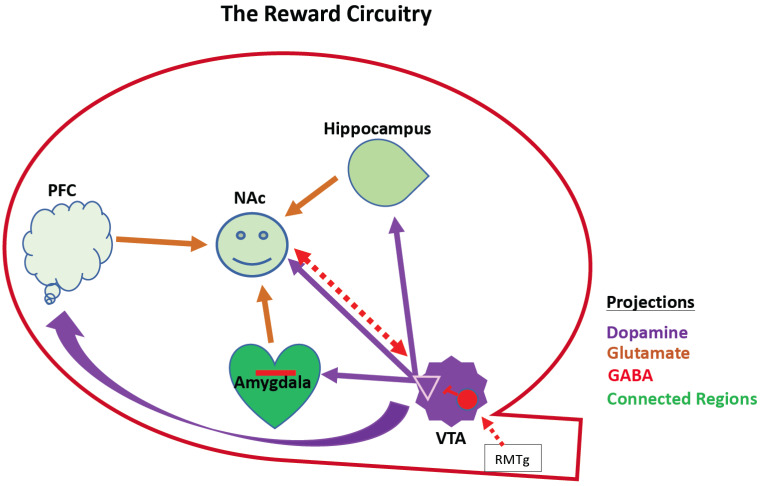
Fig. (1).
Schematic of reward circuitry. This shows interconnected mesocorticolimbic regions that interact to control reward behavior. These include the hippocampus, the amygdala, the VTA, the PFC, and the NAc. This is a VTA-NAc centric cartoon, but there are reciprocal interactions between the PFC, Hippocampus, and amygdala (not shown). Except for the RMTg, downstream components of the Brain Reward Cascade are omitted for simplicity. Dopaminergic neurons emanate from the VTA (purple). The NAc receives its dopaminergic input from the VTA and integrates that with cortical hippocampal and amygdaloid afferents to effect dopamine reward (converging arrows in NAc). NAc-VTA share reciprocal inhibitory projections and the VTA receives inhibitory afferents from the RMTg that modulate its activity. Opioid receptors modulate GABA neuron activity (red). PFC, prefrontal cortex; NAc, nucleus accumbens; VTA, ventral tegmental area; Red interneuron in the VTA represents GABAergic neurons/synapses expressing opioid receptors. NAc, Nucleus Accumbens; PFC, prefrontal cortex; VTA, ventral tegmental area; RMTg; rostromedial tegmental area. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3. OPIOID USE DISORDER, A GLOBAL ISSUE
The brain has a family of endogenous opioids peptides such as endorphins, enkephalins, Substance P, the release of which is controlled by the hypothalamus. These opioids are used as the brain’s endogenous analgesics and also activate the reward system and register pleasure and satisfaction. Research on these endogenous resident peptides and their modulation by and interaction with exogenous opioid drugs as well as alcohol or other illicit substances supports the notion of common neurochemical mechanisms of seemingly diverse addictive drugs [49]. It is strongly supported by clinical and biological data that opioid addiction is a brain disorder that causes neuroadaptive alterations in neural circuits leading to dependence, craving, and relapse or drug reinstatement. Jones [50] has provided evidence that addiction to prescribed opioids is associated with transition to illicit heroin. Moreover, Schiller [51] reported that opioid-related overdoses have risen since the 1990s and these overdoses include not only heroin but fentanyl and oxycodone. Interestingly, the so-called global opioid epidemic seems to have a lower impact in Australia [52] and Europe [53] but these regions require careful scrutiny for methods that work and continued vigilance to prevent an epidemic [54].
4. THE NATURAL REWARD PATHWAY AS WE KNOW IT
As previously discussed, the mesocorticolimbic system regulates the natural reward pathway of the brain [55], and accounts for the motivated adaptive behaviors that condition the brain system to continually seek for beneficially, rewarding events such as food, sex and pleasures [38]. The molecular mechanism underlying this pathway has been unraveled by several studies. The mesolimbic pathway starts at the midbrain’s ventral tegmental area (VTA), where resident dopamine-synthesizing projection neurons innervate brain regions such as the nucleus accumbens (NAc), amygdala, hippocampus, striatum and the prefrontal cortex (PFC) [56]. The amygdala, hippocampus and parts of the ventral striatum constitute the limbic system and are embedded deep to cerebral cortical regions in the basal, inferior aspects of the brain [57, 58]. They, along with the PFC, send glutamatergic input to the NAc.
Specifically, the amygdala mediates negative emotions, [39, 40], motivational components of emotion, and along with the ventral hippocampus, modulates emotional memories and learning [59]. By contrast, the dorsal and ventral striatum contribute to habit forming and the processing of reward, respectively [42]. Together, the limbic system controls important processes such as motivation, emotion, learning and memory and is part of the stress surfeit domain of addiction [26, 60]. The NAc, referred to as the brain pleasure center, is part of the ventral striatum (VS) [42, 61] and is the responsive site where reward (dopamine) is processed through activation of dopaminergic receptors (DRD) and induction of subsequent signaling cascades. The medial prefrontal cortex is crucial to planning, thinking, decision making and for memory storage [62]. Together these contribute to the anticipation/preoccupation phase of addiction and reward seeking. When the brain is exposed to potentially rewarding events, some of which are critical to maintaining life, the VTA neurons become activated [63]. The stimulation of VTA neurons induces the release of dopamine in the synaptic cleft or junctions separating VTA axons and respective resident neurons of the cortex, limbic and NAc regions [56, 62, 64] (Fig. 1). Released dopamine binds to the dopamine receptors on the surface of postsynaptic neurons in the limbic and NAc regions. Unbound dopamine is reabsorbed into the VTA presynaptic neurons through the dopamine reuptake pumps as a mechanism to check uncontrolled stimulation [65].
Understandably, there tends to be a strong focus on the VTA-NAc projections when addressing reward. However, convergence of many systems is needed in order for dopamine to be released from the VTA, be processed into a rewarding stimulus that affects post-NAc systems and be recorded through behavior or sensation as a rewarding stimulus. In the ‘Brain Reward Cascade’ (BRC) as termed by Blum & Kozlowski in 1990 [66], these interactions between neurotransmitters and neuromodulators expand the areas where reward processing may be influenced. They discuss the complex actions of alcohol to model activation of serotonergic, opioidergic, glutamatergic, GABA-ergic and norepinephrine neurons from a network of interconnected brain systems (including the hypothalamus, Raphe nuclei, and other brain stem/limbic regions) that converge onto the VTA as part of the BRC. Genetic, epigenetic or biochemical modulation at any point -- on the enzymes necessary for synthesis and breakdown, on receptors that transduce the signal, or on enzymes and transporters that terminate the signal -- could significantly impact how the reward is processed. The BRC effects in the limbic system and the NAc are translated into emotional manifestations, memorable activities and habitually-worthy events by the amygdala, hippocampus and the striatum, respectively. The overall sensation is stored in the PFC. The continuous stimulation of the VTA dopaminergic pathway through prolonged exposure to these events creates a positive reinforcement of pleasurable feelings worthy of perpetuation.
5. DRUG-DISRUPTIVE REWARD PATHWAY
The natural reward pathway/brain reward cascade can be disrupted by exogenous substances, including natural and synthetic drug molecules. In addition to the long-held and widely accepted hypothesis that dopamine is the catecholamine involved in the mediation of reward by alcohol and heroin [67, 68], it is also fairly widely established that nicotine, cocaine, methamphetamines, ketamine etc. also use dopamine to communicate their rewarding properties [69]. In addition, behaviors such as pornography [70], gambling [71] and video games [72] also illicit dopamine. Prolonged exposure to these substances impacts the reset point of neurotransmitter balance. Although the mechanism of disrupted reward differs slightly among drugs that modulate abuse and addictive behaviors, the resultant impact is on dopamine-mediated reward [64]. Opiates/opioids, including heroin, morphine, methadone, buprenorphine, fentanyl and its many derivatives, have a high affinity for their receptors and disrupt the reward pathway through the continuous stimulation of the dopaminergic pathway in the VTA [73, 74]. Cocaine and Methamphetamines on the other hand block the dopamine reuptake pump in the VTA presynaptic neuron, causing dopamine to be continually available to stimulate the limbic system, creating a positive reinforcement of pleasurable feelings [75, 76] Prolonged exposure to these drugs of abuse increases their baseline requirement for rewarding effects by disrupting the balance of excitatory tone, and leads to abuse, dependence, and addiction [43].
6. BUPRENORPHINE GENETIC CATABOLISM AS A FUNCTION OF ETHNICITY
Differences in the metabolism of buprenorphine, one of the three pharmacological therapies for opioid use disorder (OUD), is a good example of how genetics influences the effective dose for treatment. Buprenorphine, a partial agonist of the mu opioid receptor, is considered by many as a safe and effective therapy for opioid addiction and is heavily relied upon in the current epidemic for the treatment of individuals with OUD. When taken orally, buprenorphine has a delayed peak circulating level (~1hr) and can remain in the bloodstream for up to 22 hours, and thus is ideal for a once a day, long-acting treatment [4, 77, 78]. The cytochrome P450 enzyme CYP3A4 is the metabolizing enzyme for buprenorphine, and allelic variations such as the *1B allele confer a faster metabolism of buprenorphine, clearing it from the system, and this may precipitate relapse in individuals. Since dosing limitations vary widely from state to state, insurance plan limits on buprenorphine dose could adversely, differentially affect certain populations.
This discrepancy of appropriate dosing has been further vetted by Accurso & Rastegar [79] who argue that the optimal dose for office-based buprenorphine is unknown. They found that when insurance companies imposed a buprenorphine dose decrease for patients, this was associated with an increase in aberrant drug tests [80]. Further, patients in a control group with higher buprenorphine doses had greater retention in treatment. These findings suggest that buprenorphine dosages greater than 16 mgs are more effective for some patients and dose limits at 16 mgs or lower are harmful. Therefore, with a cap on the prescription of buprenorphine, patients who have the risk allele for CYP3A4, may be doubly disadvantaged.
To this end, the National Human Genome Center (NHGC) at Howard University, Howard University College of Pharmacy, and Chapman and colleagues [81] pharmacogenetically tested a population of 98% African-Americans for CYP3A4, the major metabolizing enzyme for buprenorphine (+Naloxone). Genotyping 144 patients resulted in a wide polymorphic variation whereby a total of 85% of the population carried the extended metabolizer CYP3A4 *1B promoter variant allele. In fact, it was found that 43% of the population carried *1/*1B; and 42% carried *1B/*1B homozygote, whereas a larger mixed population database indicated that only 26% carried the extended *1B metabolizer [81]. This is further supported by Wandel, C. et al. [80] who found that CYP3A4*1B (-290A>G), has an allelic frequency that was very low in European Americans but high in African American participants. This reveals a possible genetic disparity between Caucasian and African American individuals. This disparate higher expression of *1B may extend to individuals of African descent in general, as revealed by a study of 6 North African countries executed by Fernandez- Santander and colleagues [82]. Individuals from these regions of Africa, in spite of their heterogeneous, genetically diverse populations, had allelic frequencies in the CYP3A4*1B allele that largely exceeded the variation ranges described in European counterparts [82]. Since the CYP450 family is large, it is highly likely, in fact, that different ethnicities metabolize the pharmacological intervention differently, and are susceptible to overdose or even addiction, based on genetic alleles of the CYP450 family. Together, these data suggest that higher dosing of buprenorphine may be necessary to reduce treatment failure in certain populations of our society, e.g. African-Americans [81-83]. These data further indicate that treatment for OUD should be matched to the individual patient’s genetic and epigenetic variations and pharmacological profile in drug metabolism. Additional pharmacogenetic studies of cytochromes P450 such as CYP3A4 for metabolizing buprenorphine and CYP3A4 and CYP2D6 for methadone are needed, as well as studies that lend an understanding of how efflux pumps function. Indeed, inhibiting CYP3A4 with delavirdine, an antiretroviral medication used in HIV- treatment induces an elevation of methadone plasma concentration and drug delayed clearance [84]. From a consensus of the CYP450 system literature, it appears that although some variants of genes are associated with undesired circulating plasma levels of OUD therapeutics, these data have not influenced dose requirements or overall provider behavior [85]. However, if more studies consider the ethnic ancestry in the study cohorts, perhaps a more solid foundation or consensus can be reached, as the results thus far are spurious at best.
7. NEUROGENETICS OF OPIOID USE DISORDER AS A FUNCTION OF ETHNICITY
A major issue concerning the underpinnings of genetics and associated polymorphic risk alleles is the disregard for consideration for the role played by ethnic ancestry, differentially displayed in various nationalities and regions. Also understudied is whether or not the carrying of these risk alleles actual load onto OUD. In this overview, we carefully consider the paucity of this important work required by clinicians/scientists in truly understanding the benefits of genetic testing especially in underserved populations. While the neurogenetics of the brain reward circuitry is a work in progress since the seminal finding of the DRD2 A1 allele association with severe alcoholism [86], more work is needed.
With this brief snapshot of the brain reward cascade and effector brain regions and related neurotransmitters leading to a net release of dopamine at the NAc and subsequent receipt of reward and well-being, the following provides a very brief overview of a number of genes involved in the reward pathway and their associated risk alleles as presented in the literature. Our focus here is to identify these genes and polymorphisms especially related to differential prevalence or resilience among African-Americans and other ethnic groups. While there are hundreds of genes implicated in the brain reward circuitry, for this review we have selected just a few candidate genes and associated polymorphisms and well-known risk alleles for drug related addictive behaviors as an example of the literature to showcase the importance of neurogenetics and ethnicity (Table 1).
Table 1.
Publications on gene variants associated with opioid use disorder (OUD) as related to diversity of study cohort.
| Genes | Variants (SNPs and Haplotypes) | Publication Describing Association with Opioid Dependence | Publications that Consider Caucasians | Publications that Consider Chinese | Publications that Consider Hispanics | Publications that Consider African Ancestry |
|---|---|---|---|---|---|---|
| DRD2 | rs6275; rs6277; rs1076560; rs1799978; rs1800496; rs1801028 | Clarke, T.K. et al. [94]; Hou, Q.F. and Li, S.B. [95]; Vereczkei, A. et al. [96]; Lawford, B.R. et al. [98]. | Clarke, T.K. et al. [94]; Vereczkei, A. et al. [96]; Lawford, B.R. et al. [98]. | Hou, Q.F. and Li, S.B. [95]. | Clarke, T.K. et al. [94]. | |
| DRD3 | rs6280 rs9825563 rs2654754 rs9288993 rs1486009 |
Levran, O. et al. [99]; Kuo, S.C. et al. [100]. | Levran, O. et al. [99]. | Kuo, S.C. et al. [100]. | ||
| DRD4 | rs1800955 | Vereczkei, A. et al. [96]; Szilagyi, A. et al. [102]; Ho. A.M. et al. [103]; Lai, J.H. et al. [104]. | Vereczkei, A. et al. [96]; Szilagyi, A. et al. [102]. | Ho. A.M. et al. [103]; Lai, J.H. et al. [104]. | ||
| OPRM1 | rs1799971 rs1799972 |
Bond, C. et al. [115]; Crowley, J.J. et al. [120]; Szeto, C.Y. et al. [123]; Shi, J. et al. [124]; Hastie, B.A. et al. [118]. | Bond, C. et al. [115]; Crowley, J.J. et al. [120]; Hastie, B.A. et al. [118]. | Szeto, C.Y. et al. [123]; Shi, J. et al. [124]. | Bond, C. et al. [115]; Crowley, J.J. et al. [120]; Hastie, B.A. et al. [118]. | Bond, C. et al. [115]; Crowley, J.J. et al. [120]; Hastie, B.A. et al. [118]. |
| OPRD1 | rs1042114 rs678849 rs10753331 rs529520 rs581111 rs2234918 |
Zhang, H. et al. [129]; Levran, O. et al. [130]; Nelson, E.C. et al. [131]; Levran, O. et al. [132]; Crist, R.C. et al. [122]; Sharafshah, A. et al. [133]; Beer, B. et al. [134]. | Zhang, H. et al. [129]; Crist, R.C. et al. [122];Levran, O. et al. [130]; Nelson, E.C. et al. [131]; Beer, B. et al. [134]. | Levran, O. et al. [132]; Crist, R.C. et al. [122]. | ||
| KCNC1; KCNG2 |
rs60349741 rs62103177 |
Gelernter, J. et al. [146]. | Gelernter, J. et al. [146]. | Gelernter, J. et al. [146]. | ||
| OPRK1 | rs1051660 | Yuferov, V. et al. [135]; Jones, J.D. et al. [136]; Nagaya, D. et al. [137]; Zhang, H. et al. [129]; Albonaim, A. et al. [138]; Gerra, G. et al. [139]; Kumar, D. et al. [140]; Levran, O. et al. [130]. | Yuferov, V. et al. [135]; Jones, J.D. et al. [136]; Zhang, H. et al. [129]; Gerra, G. et al. [139]; Levran, O. et al. [130]. | Yuferov, V. et al. [135]; Jones, J.D. et al. [136]. | Yuferov, V. et al. [135]; Jones, J.D. et al. [136]. | |
| BDNF | rs6265; rs11030104; rs10767664; rs13306221; rs56164415; rs13306221; rs16917204; rs7127507; rs1967554; rs11030118; rs988748; rs2030324; rs11030119 | Jin, T. et al. [152]; Jia, W. et al. [153]; Su, H. et al. [155]; de Cid, R. et al. [156]. | Jin, T. et al. [152]; Jia, W. et al [153]; Su, H. et al. [155]. | |||
| NRXN3 | rs10144398 rs10151731 rs10083466 rs1424850 rs221497 rs221473 |
Panagopoulos, V.N. et al. [159]; Lachman, H.M. et al. [160]. | Panagopoulos, V.N. et al. [159]; Lachman, H.M. et al. [160]. | Lachman, H.M. et al. [160]. | Lachman, H.M. et al .[160]. | |
| COMT | rs4680 | Henker, R.A. et al. [162]; Vereczkei, A. et al. [96]; Li, T. et al. [163]; Rakvåg, T.T. et al. [164]. | Henker, R.A. et al. [162]. | Vereczkei, A. et al. [96]; Li, T. et al. [163]; Rakvåg, T.T. et al. [164]. | ||
| SLC6A4 | rs25531 rs1042173 |
Saiz, P.A. et al. [166]; Gerra, G. et al. [167]; Szilagyi, A. et al. [102]; Iamjan, S.A. et al. [154]. | Gerra, G. et al. [167]; Szilagyi, A. et al. [102]. | Saiz, P.A. et al. [166]; Iamjan, S.A. et al. [154]. |
8. POLYMORPHISM OF REWARD GENES AS A FUNCTION OF ETHNICITY
8.1. DRD2
The DRD2 gene was cloned by Grandy, D. et al. [87] and mapped to chromosome 11 at q22-q23 [88]. It encodes the dopamine receptor type 2 [87, 88] and was first associated with alcohol and cocaine addiction in the early 1990s [86, 89, 90]. DRD2 has also been implicated in learning, memory and disorders like Alzheimer’s disease [91]. In rat brains, DRD2 is expressed highly in the cortex, hippocampus, ventral and dorsal striatum and the claustrum [92]. Additionally, substantial amounts of DRD2 have been found to be expressed in the thalamus, amygdala, and pons [92, 93]. Prominent DRD2 SNPs in humans that have been identified to predispose people to substance (alcohol, nicotine, cocaine and opioids) misuse or addictions include rs6275, rs6277, rs1076560, rs1799978, rs1800497 (Taq1A) and rs1079597 (Taq1B). Recent studies have now shown that rs1800497 is located on ANKK1 gene, downstream of DRD2. Whereas rs1076560, rs1800497 and rs1079597 have all been explicitly linked with opioid addiction, only rs1076560 has been investigated in African-Americans (AA) [94]. Clarke and colleagues [94] established an association between rs1076560 and opioid addictions in both European Americans (EAs) and AAs. To establish this association, 1041 EA and 284 AA
opioid addicts were genotyped for rs1076560. Initial analysis found a significant association between rs1076560 and both ethnic groups at p = 0.02 for EA and p = 0.03 for AA. Final analysis after correction for multiple testing also revealed an association. While Clarke, T. et al. [94] included significant numbers of AA in their studies, other research groups did not. For instance, in their effort to establish an association between rs1800497 and heroin dependence, Hou and Li, 2009 [95] only included Chinese population in their studies while Vereczkei, A. et al. [96] considered subjects of Hungarian descent (Caucasians) in their multivariate analysis involving many DRD2 genes to establish an association between rs1800497 (Taq1A) and rs1079597(Taq1B) and heroin dependence. These findings are in agreement with earlier work from Blum’s group in 1993 showing the association of rs1079597 (Taq1B) and severe alcoholism [89] and with the work of others at the National Institute of Drug Abuse (NIDA) showing that polysubstance users with histories of heavy daily preferential psychostimulant use more often displayed [97] one or two copies of the TaqI A1 (27/62 = 43.5% vs 33/119 = 27.7% for controls), and B1 (20/62 = 32.3% vs 23/119 = 19.8% for controls) markers at the DRD2 locus. However, once again Caucasians were prominent as the cohort [97].
In a follow-up study of an out-patient clinic using methadone for the treatment of OUD, Lawford, B. et al. [98] evaluated only 95 patients, all of whom were of European ancestry. Apart from the high correlation between rs1800497 and opioid dependence, this study revealed a negative treatment response for rs1800497 (A) allele group, thereby presenting the A allele as a risk factor of opioid dependence.
8.2. DRD3
The DRD3 gene, which is found on chromosome 3q13.3, encodes dopamine receptor type 3. The D3 receptors are densely located in the limbic subcortical regions of the brain, specifically the NAc, thalamus, hypothalamus, and the cerebellum. More than 50 SNPs have been identified in DRD3. Of these, few have been implicated in impacting the dopaminergic-mediated reward pathway. rs2654754, rs9288993 and rs1486009 showed significantly high association with heroin addiction in subjects predominantly of European ancestry [99], the only ethnic background considered. In another study evaluating the impacts of DRD3 variants with heroin addiction in Han Chinese subjects, rs6280 and rs9825563 SNPs showed a significant association with the development of early-onset heroin dependence [100]. So far, only a few studies have been done to link DRD3 variants with opioid use disorder. However, to our knowledge, none of these studies considered African Americans or Hispanics.
8.3. DRD4
Dopamine Receptor Type 4 (DRD4) is encoded by the DRD4 gene on chromosome 11, specifically at 11p15.5 [101]. Notable DRD4 SNPs include rs1800955, rs936462 and rs747302. Several studies have reported an association of rs1800955 (-521 C/T) with heroin dependence, with a protective effect of the C allele [96]. To arrive at these results, Vereczkei, A. et al. [96] and Szilagyi, A. et al. [102] have all considered only Caucasian population while Ho, A. et al. [103], and Lai, J. et al. [104] focused their study on subjects of Chinese descent.
In addition to SNPs, variable number of tandem repeats (VNTRs) have been reportedly found in DRD4 exon 3 [105]. These VNTRs exist in 3 different allelic repeats of two, four (48bp; ≤ 4 = short allele) and seven (120bp; ≥ 7 = long allele) [106]. It has been shown that subjects with seven repeats or greater exhibit higher vulnerability to high alcohol consumption [105] including among youths [107, 108]. Shao et al. [109] have reported a high frequency of VNTR long allele in Chinese subjects, while Franke et al. [110] reported a high frequency in heroin addicts of German descent. Multiple authors have considered both long and short alleles of DRD4 in their association studies with heroin in Caucasian and Chinese descent and found no correlation [96, 102, 104, 111]. However, in an earlier study, Kotler et al. [112] had reported that the 7-repeat allele is significantly over-represented in an opioid-dependent Israeli population cohort, with a relative risk of 2.46. Similar to the Kotler et al. study, Li, T. et al. [113] also reported a high vulnerability of heroin addiction for Chinese subjects with high frequency long allele DRD4 exon 3, while Chien, C. et al. reported that long-repeat allelic and 2-repeat allele variants of DRD4 exon 3 might be associated with heroin addiction in Chinese men [114].
8.4. OPRM1
The OPRM1 gene encodes the µ opioid receptor located on chromosome 6q24-q25. Mu opioid receptor activation is a powerful mediator of the rewarding effects of opioids. In-depth study of this gene has revealed several SNPs that have been linked with substance misuse, including alcohol, opioids, cocaine and nicotine. Studies of OPRM1 polymorphisms lack adequate address of African ancestry. Although there are many OPRM1 SNPs, rs1799971 (A/G) and rs1799972 (C/T) remain the most prominent and investigated, with a record of linkage with opioid dependence.
In 1998, Bond, C. et al. [115] described disparate allele frequencies for A118G and C17T. According to this study, allele frequencies among African-Americans, Caucasians and Hispanics in the study population were respectively 0.984, 0.885 and 0.858 for rs1799971 (A), and 0.016, 0.115 and 0.142 for rs1799971(G). Therefore, African-Americans present with the highest allele frequency for rs1799971(A) and lowest for rs1799971(G). In this case, the risk allele associated with opioid addiction is rs1799971(G) and is present at a minimal frequency in AAs. Although these authors did report a higher allele frequency of rs1799971(G) in opioid-dependent compared with non -dependent subjects in Hispanic subgroups, they could not find any significant differences in allele frequencies between opioid-dependent and nondependent subjects in all ethnic groups combined. rs1799971(A) may provide protection against opioid addiction in African-Americans. It is noteworthy that while this may be true, poorly screened controls free of hidden reward deficiency syndrome (RDS) behaviors could have skewed the results [116]. A thorough definition of RDS and the diseases that encompass this syndrome has been published [117]. In another study comprising 81 African Americans, 79 Hispanics, and 87 Caucasians, a lower frequency of the G allele in African Americans was also reported [118]. In Caucasians, the G allele was associated with decreased pain sensitivity, i.e., higher pain threshold. Furthermore, even though the Hispanic cohort possessed the G allele at similar frequencies to the Caucasian group, their pain sensitivity did not change. While the reasons for this observation remains unknown, the authors postulated that the involvement of ethnic differences in haplotypic structure of A118G may be responsible [118].
For rs1799972(C), the major allele frequencies were 0.790, 0.981 and 0.963 for African-American, Caucasian and Hispanic individuals, respectively whereas the respective minor allele frequencies for rs1799972 (T) were 0.210, 0.019 and 0.037 in the overall population. Consequently, the minor allele rs1799972 (T) was found to be predominantly higher in opioid-dependent persons in all the subjects combined, presenting the rs1799972 (T) as a risk factor for opioid dependence. The authors recommended more investigation to validate these results, especially within each ethnic group. Another study reported no association between polymorphic haplotypes in OPRM1 in African-Americans and alcohol and opioid dependence, although significant haplotype frequencies were observed in European-Americans. The SNPs considered were -2044C/A, -1793T/A, -1699insT, -1469T/C, - 1320A/G, -111C/T, +17C/T (Ala6Val), and +118A/G [119]. This finding must be taken seriously because it suggests that even though there is an increased prevalence of a particular SNP or variant in AA it does not always translate to similar addictive behaviors as is observed in other ethnic groups. The potential effect of these various haplotypes in modulating reward is represented in Fig. 2.
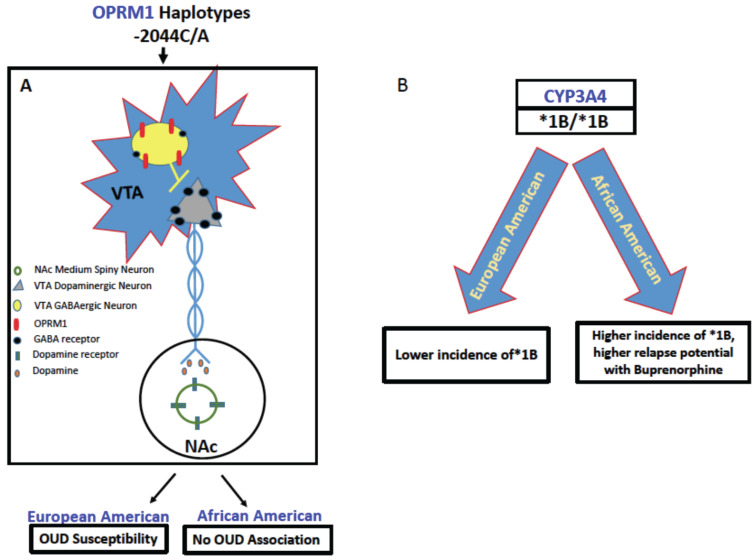
Fig. (2).
Representative differential impact of gene variants on opioid metabolism in European and African Americans. (A) -2044C/A haplotypes modulate the reward pathway to have a net negative effect on OUD, where European Americans with this allele are more susceptible to OUD than African Americans (Luo et al., 2003 [119]). By contrast, (B) a higher frequency of the *1B allele is present in African Americans, and this allele confers the extended metabolizer phenotype (Chapman et al. 2018 [81]; Wandel et al., 2000 [83]; Fernandez-Santander et al., 2016. [82], thus increasing chances of relapse in African Americans undergoing opioid replacement therapy with Buprenorphine/Suboxone. CYP3A4, Cytochrome P450 3A4; GABA, gamma-aminobutyric acid; NAc, Nucleus accumbens; OPRM1, mu opioid receptor; VTA, ventral tegmental area. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
A study by Crowley, J. et al. in 2003 [120] revealed no significant association between 5 SNPs, including rs1799971 and rs1799972, and opioid dependence in neither African nor European ancestry, although significant allele differences were apparent for all the SNPs between African American and European Americans. This study was accomplished through a genome association study in which 213 subjects (89 African-Americans and 124 European-Americans) with severe opioid dependence and 196 “supercontrol” subjects (96 African-Americans, 100 European-Americans) were genotyped and screened for five SNPs residing in the OPRM1 gene. The SNPs include three in the promoter region (T-1793A, -1699T insertion and A-1320G) and two in exon 1 (C+17T and A+118G). Clarke and colleagues investigated rare SNP variants in 1377 European Americans and 1238 African Americans [121]. The group found an association between rs62638690 (not rs17174794) and heroin and cocaine addiction in European Americans. In the 1238 African Americans, the association was with two rare SNPs, rs199971 and rs17174801. A limitation of this study is that the same SNPs were not analyzed across ethnic groups for direct comparison [121, 122].
On the whole, there are contrasting reports regarding the connection between OPRM1 and opioid dependence. Some authors have reported an association between OPRM1 SNPs and opioid dependence, while others have refuted this claim. While Szeto, C. et al. 2001 [123] reported an association between the G allele of rs1799971 and heroin addiction in Chinese heroin addicts, Shi, J. et al. 2002 [124] reported a contrary result also in Chinese heroin addicts. In another study of subjects from the Bulgaria population, no evidence of association was found between rs1799971 and heroin dependence. Again, these discrepant findings may be due to poorly screened controls [117]. Another study suggests that the G allele of rs1799971 is emerging as a potentially protective allele. A Tufts University study of neonatal abstinence syndrome (NAS) infants showed that patients with this G allele had an average shorter stay for treatment in the hospital and these infants are 25% less likely to require treatment for NAS. It is postulated that b-endorphins bind to the mu opioid receptors with such affinity and upon exposure to additional opiates fewer MORs are produced and thus fewer opioid receptors are available to respond to the parent’s intake of opiates while in utero [125]. Ancestry of the infants was not specified.
Additionally, SNPs can have implications for the efficacy of methadone, a full and potent agonist of the MOR, used to treat heroin/opioid dependence. Crist and colleagues show that SNP alleles A/G and G/G but not A/A of rs10485058 located in the 3’ UTR may be associated with relapse and reduced efficacy of methadone. The G allele mRNA presumably binds to miRNAs which inhibits translation of MOR, reducing methadone action, perhaps rendering individuals susceptible to relapse [126]. Therefore, genetic considerations are important for determining the efficacy and choice of various pharmacotherapies. This effectiveness will differ based on ancestry. Several other studies investigated an association between OPRM1 gene variants and opioid dependence but did not consider subjects of African descent [127, 128].
Taken together, with respect to OPRM1, some studies indicate that SNPs associated with OUD in European American subgroups, although these very SNPS may be present in African Americans, do not seem to confer susceptibility to OUD in AAs, and furthermore some SNPs may have neuroprotective characteristics conferring resilience to opioids [120, 121, 125].
8.5. OPRD1
Prominent SNPs in OPRD1 include rs1042114, rs678849, rs10753331, rs529520, rs581111, and rs2234918 and these OPRD1 gene variants have been investigated for a link with opioid addiction [122, 129-134]. Of all these studies, only Levran, O. et al. [132] and Crist, R. et al. [122] considered AAs in their study cohort. In fact, Levran, O. et al. [132] exclusively considered subjects of African descent in which 1350 SNPs of 130 different genes were genotyped in 202 former severe heroin addicts undergoing methadone treatment and 167 healthy controls with no history of drug abuse. In the Levran, O. et al. study, no significant association with OUD was found for SNPs in OPRDs. Of the other genes, there were many associations with OUD, but of interest was the finding that glutamate receptor subunit GRIN2A haplotype G-A-T (rs4587976-rs1071502-rs1366076) had a protective effect on AA heroin addicts.
On the other hand, in their comparative analysis of the effect of OPRD1 variants with respect to Methadone and Buprenorphine treatment in African-Americans and European- Americans, Crist, R. et al. [122] genotyped five SNPs on the OPRD1 locus. While the result of the analysis revealed no association or outcome with methadone and buprenorphine treatment in European- Americans, subjects in the African-American group with the intronic rs678849 (CC) genotype and on buprenorphine treatment were observed to have less opioids in their urine than subjects in the same subgroup with the rs678849 (CT) or rs678849 (TT). Further analysis in the same subgroup revealed a contrasting result as subjects with the rs678849(CC) genotype and on methadone treatment were observed to have more opioids than subjects in the same subgroup with the rs678849 (CT) or rs678849 (TT). This result showed that African-Americans addicted to opioids may show differential outcomes with methadone and buprenorphine depending on the OPRD1 SNP they possess, with rs678849 (CT) presenting as a good predictor of positive response to two common treatments for opioid dependence, buprenorphine and methadone, whereas the CC haplotype showed an inverse relationship for the effectiveness of buprenorphine or methadone.
8.6. OPRK1
Another important gene that has been hypothesized to play a critical role in the reward pathway is OPRK1. Like other opioid receptor genes, some of its variants have been extensively studied to evaluate their influence on the reward pathway vis-à-vis addictions. Again, of all the studies that focused on finding an association between OPRK1 polymorphisms and addictions, only a few considered AAs, Hispanics or comparative ethnicities. While trying to completely unravel the structure of this gene, 12 SNPs were identified in three different ethnic populations, including African Americans, European Americans and Hispanics [135]. Nine of these were novel and 3 had been previously reported. Rs1051660 (36G>T) was found to show a significant association with opioid addiction [135]. In another study, AA was included to investigate the mediation of opioid withdrawal by opioid receptor gene polymorphisms [136]. In this
study, the two OPRK1 SNPs considered, rs6473797 and rs963549, were found to show no association with severity of abstinence-induced withdrawal. Other studies that investigated the impact of OPRK1 polymorphisms on the reward pathway but that did not include AA subjects include: [129, 130, 135, 137-140].
8.7. KCNC1 and KCNG2
KCNC1 belongs to a subgroup of the family of tetrameric voltage-gated K+ channels, subfamily C member 1 expressed in the inhibitory fast-spiking GABAergic interneurons of the neocortex, thalamus, hippocampus, and striatum [141]. In addition to its expression in GABA neurons, KCNC1 is also expressed in T-lymphocytes, oligodendrocyte precursor cells, astrocytes, and neural progenitor cells (NPC) [142-144]. KCNG2 is a member to subfamily G member 2 of the voltage-gated K+ channels and is expressed throughout the brain where it regulates the release of neurotransmitters. KCNC1 has been linked with a variety of neurological disorders, including seizure [145]. A genome-wide association study (GWAS) of two populations of opioid addicts which included both African Americans and European Americans in the United States revealed an association between the novel KCNC1 (rs60349741) and KCNG2 (rs62103177) gene SNPs, and opioid dependence as defined in the Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) [146]. Although the association of these SNPs with opioid dependence was found in both AA and EA study participants. The association was more pronounced in AA. This study was important because it is the first time variants of these genes involved in potassium ion signaling pathways would be linked with opioid addiction.
8.8. BDNF
Brain-derived neurotrophic factor (BDNF), located on chromosome 11, encodes a member of the nerve growth factor family of proteins [147, 148]. Because of alternate splicing, this gene is able to code for multiple protein variants. BDNF-mediated activity enhances the survival of neurons [149, 150]. Studies have implicated BDNF expressions in the etiologies of Alzheimer’s, Parkinson’s and Huntington’s diseases. Thus far, BDNF has been linked with opioid addiction [151]. A notable variant of BDNF is the rs6265 (Val66Met), with an MAF of 0.2013/1008 (1000 Genomes), present across all ethnic groups. In this variant, the original valine amino acid is replaced with methionine. In their study to evaluate the relationship between polymorphisms of BDNF genes and heroin addiction in the Han Chinese population, Jin, T. et al. [152] found that rs6265, rs11030104 and rs10767664 in BDNF may be associated with a decreased risk of heroin addiction. In another study, Jia, W. et al. [153] evaluated the protective association between four BDNF variants including rs13306221, rs6265, rs56164415, and rs16917204 in heroin-dependent subjects in the Chinese population, and found a role of BDNF rs6265 and rs13306221 polymorphisms in heroin addiction. Other studies that focused on characterizing the association and influence of BDNF variants on opioid addiction include Iamjan, S. et al. [154], Su, H. et al. [155] and de Cid, R et al. [156]. While Iamjan, S. et al. [154] considered subjects only from a Thai population in their study to link rs6265 with methamphetamine dependence, Su, H. et al. [155], using fine mapping, identified BDNF SNPs haplotype comprising rs7127507, rs1967554, rs11030118, rs988748, rs2030324 and rs11030119 associated with differential response to Methadone Maintenance Therapy (MMT) in Chinese subjects. These six haplotype blocks of BDNF were found to be associated with a positive response to MMT in Caucasian subjects, even though no direct relationship had been established between each SNP and OUD [156]. Apparently, no study has yet considered AA in association studies linking BDNF variants with opioid dependence or addictions.
8.9. NRXN3
Neurexin 3 (NRXN3) is a cell adhesion molecule associated with maintaining synaptic connections [157, 158]. In Australian subjects with Bipolar Disorder (BPD), NRXN3 variants rs10144398, rs10151731 and rs10083466 was found to be associated with BPD in heroin addicts [159]. Another linkage study analysis comprising Hispanics, AA and EA suggested linkage of opioid dependence to chromosome 14q, which encompasses the NRXN3 gene [160], but no ancestry specific differences were reported.
8.10. COMT
Catechol-O-Methyltransferase (COMT) breaks down catecholamines including epinephrine, norepinephrine and dopamine, reducing the level of neurotransmitters involved in reward, cognition, craving behavior, and motor skills. Since dopamine is a substrate of COMT, if the ability of COMT to effectively catabolize dopamine is impacted, many aspects of reward will be affected. A number of gene variants encoding COMT and shown to impact reward have been identified [161]. In trying to investigate the associations between OPRM1 and COMT genotypes and postoperative pain, opioid use, and opioid-induced sedation in a EA based population, Henker, R. et al. [162] found rs4818 to be modestly associated with the amount of opioids patients consumed in the post-anesthesia care unit during the first 45 minutes. The GG homozygous SNP rs4818 was especially associated with the consumption of less opioids in the same time range. Another study failed to establish an association between rs4818 and heroin dependence in Hungarian patients [97]. It has been reported that the TT allele of COMT rs737866 may be associated with the early onset of heroin use in Chinese subjects [163]. Genetic variability in the COMT gene may influence the efficacy of morphine in cancer patients with pain. Rakvåg, T. et al. [164] showed that cancer patients of Norwegian populations who carried the A allele of COMT rs4680 required lower doses of morphine to alleviate their pain when compared to the G variant. Although rs4680 is the most studied COMT SNP, its influence in opioid catabolism in subjects of African descents is yet to be fully investigated.
8.11. SLC6A4
Solute Carrier Family 6 Member 4 (SLC6A4) is a serotonin transporter encoded by the SLC6A4 gene [165]. There are very few primary genetic studies emphasizing the association between SLC6A4 gene variants and opioid addiction. In trying to investigate a differential role of serotonergic polymorphisms, including A-1438G, 5-HTTLPR, and STin2 VNTR in alcohol and heroin dependence, Saiz, P. et al. [166] showed that there may be an association between these variants and heroin dependence in a study comprising of only Spanish subjects. Before this time, Gerra, G et al. [167] had shown that there may be an association between the SS genotype of the most investigated SLC6A4 gene variants for the serotonin transporter-like polymorphic region, 5-HTTLPR, and heroin addiction in Caucasians subjects, whereas Szilagyi, A. et al. [103] showed that although 5-HTTLPR did not impact heroin dependence in Caucasian subjects, its interaction with DRD4 SNPs, specifically -521 C/T significantly odds ratio (OR) from 2.14 to 4.82, underscores the relevance and significance of the combined effects of the dopaminergic and serotonergic systems in heroin dependence/addiction.
9. DISPARITY IN STUDIES OF GENES VARIANTS INVOLVED IN OUD
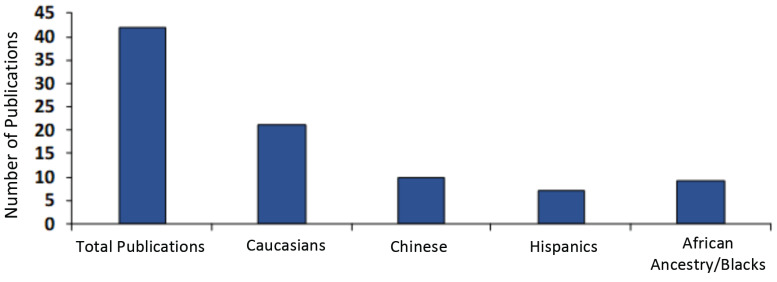
Past and recent studies have shown that OUD may have some genetic basis which may further be complicated by ethnicity and epigenetics. Because of this, it is pertinent that when studying the molecular mechanism underlying the genetic basis of OUD, that ethnicity is considered whenever possible. Obviously, from this review article sum analysis, Hispanics and people of African descent have been overlooked in studies emphasizing the genetic basis of opioid dependence/OUD. The literature on this subject is summarized in Fig. 3. Asians are included in exclusive studies from that region. Ironically in America, African-American people are as highly affected by the opioid crisis as people of European descent (as a percentage of each population). In fact, according to a recent report by the New York Times, drug deaths, including opioid related, among blacks in urban counties rose by 41 percent in 2016, exceeding other ethnic groups and especially whites by 19 percent in similar urban communities. Some genetic testing is available for these genes, especially the genetic addiction risk score where many of the genes discussed are tested on a panel to determine risk based on the number of polymorphisms [12, 168]. However, specialized gene panels that target specific populations will need to be developed as more disparities are identified between the genetic susceptibility of individual groups to addiction. It is well established that African populations or individuals of African descent have greater genetic diversity, and thus may have a diverse threshold and efficacy profiles in response to drugs or stimuli [169]. Thus, precision addiction medicine is critical for the proper treatment of opioid use disorder in diverse populations.
Fig. (3).
Studies of gene variants and their association with opioid use in different ethnic groups. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
10. MITOCHONDRIAL DAMAGE AND OUD
Although the focus of this review is to examine ancestry-specific gene variants within the reward pathway, there are other metabolic pathways that can impact the severity of OUD. Chronic use of psychostimulant drugs correlates with oxidative stress [170], opioid-induced mitochondrial damage/accumulation [171], and increased reactive oxygen species (ROS) production [172, 173]. A study of OUD patients showed decreased mtDNA copy number and elevated mtDNA damage in peripheral blood [174].
One study of Rs1695, an Ile105Val polymorphism of Glutathione S-transferase P1 (GSTP1, OMIM: 134660) involved in catalyzing the detoxification of endogenous and exogenous substances by their conjugation with glutathione (GSH), showed no association with the risk of heroin and opium dependency in 442 heroin-dependent subjects compared to 794 healthy controls [175]. However, for superoxide dismutase-2 (SOD2, OMIM: 147460) involved in the detoxification of superoxide anions during oxidative stress, an Iranian cohort study comprising 442 heroin- dependent subjects and 799 healthy controls showed an association between two SOD2 variants, rs2758339 and rs5746136, and heroin dependence. The AC (OR = 0.72, 95% CI = 0.56-0.93, P = 0.013) and CC (OR = 0.64, 95% CI = 0.45-0.92, P = 0.015) genotypes of the rs2758339 were negatively associated with the risk of HD, while the AA genotype of the rs5746136 increased the risk of HD (OR = 1.46, 95% CI = 1.03-2.07, P = 0.031) [176].
There are very few studies dedicated to showing a correlation between oxidative stress genes variants and opioid consumption or addiction. Of those, none investigated ethnic variations in mitochondrial SNPs. However, this is an important consideration.
CONCLUSION
It is our opinion that the dearth of studies focusing on opioid misuse and abuse including OUD among minorities, especially Blacks, may be due in part to the low funding availabilities for such investigations in African countries. There is also a paucity of neurogenetic based research in non-African countries with significant populations of African ancestry, especially in the United States. Moreover, this identified disproportionate research as discussed in this review could also be due to a lack of understanding of the importance of results tied to different ethnic ancestry in cohorts under study, and lack of prioritization of treatment for all groups. It is important to note that carrying a risk allele does not automatically confer addiction, especially if the individual is never exposed. Furthermore, many common minor alleles and some rare alleles confer protection against OUD, and their identification will be important for genetic-based treatment strategies.
We propose that it will be highly beneficial and greatly advantageous to encourage our scientific community especially the American Society of Addiction Medicine (ASAM), American Board of Addiction Medicine (ABAM), American Psychological Association (APA) and other groups to be more aware of these genetic differences as reported in this review and design studies capturing cohorts of multi-ethnic groups. In addition, we encourage community leaders especially clinicians to more clearly communicate with their AA/Hispanic communities to understand the menace of OUD and the benefits of participating in clinical studies designed to understand the genetic basis of opioid misuse disorder. As scientists, we must do our part to sensitize institutes of the National Institutes of Health (NIH) such as National Institute on Drug Abuse (NIDA), National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute of Neurological Disorders and Stroke (NINDS), National Institute of Mental Health (NIMH) and other NIH agencies to fund larger studies on neurogenetics of all addictive behaviors with special focus on gene polymorphic differences as a function of diversity.
ACKNOWLEDGEMENTS
MGL developed the topic idea and concepts; MGL and TA wrote the first draft of the review. KB contributed ideas. All authors edited, vetted, and approved the final document.
Consent for Publication
Not applicable.
Funding
National Institute of Health (NIH) grants AA021262 to MGL, AI117970 DC CFAR (DC Center for AIDS Research) and #MD007597 RCMI (Research Centers at Minority Institutions) SubProjects to MGL, as well as #MD012318 to MGL and KB.
Conflict of Interest
KB owns stock in some companies holding patents on genetic testing. MGL is a member of the scientific advisory board for a company that conducts genetic testing. TA has no conflict of interest to declare.
REFERENCES
- 1.Blanco C., Volkow N.D. Management of opioid use disorder in the USA: present status and future directions. Lancet. 2019;393(10182):1760–1772. doi: 10.1016/S0140-6736(18)33078-2. [DOI] [PubMed] [Google Scholar]
- 2.Van Zee A. The promotion and marketing of oxycontin: commercial triumph, public health tragedy. Am. J. Public Health. 2009;99(2):221–227. doi: 10.2105/AJPH.2007.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morone N.E., Weiner D.K. Pain as the fifth vital sign: exposing the vital need for pain education. Clin. Ther. 2013;35(11):1728–1732. doi: 10.1016/j.clinthera.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble F., Marie N. Management of Opioid Addiction With Opioid Substitution Treatments: Beyond Methadone and Buprenorphine. Front. Psychiatry. 2019;9:742. doi: 10.3389/fpsyt.2018.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kakko J., Svanborg K.D., Kreek M.J., Heilig M. 1-year retention and social function after buprenorphine-assisted relapse prevention treatment for heroin dependence in Sweden: a randomised, placebo-controlled trial. Lancet. 2003;361(9358):662–668. doi: 10.1016/S0140-6736(03)12600-1. [DOI] [PubMed] [Google Scholar]
- 6.Sees K.L., Delucchi K.L., Masson C., Rosen A., Clark H.W., Robillard H., Banys P., Hall S.M. Methadone maintenance vs 180-day psychosocially enriched detoxification for treatment of opioid dependence: a randomized controlled trial. JAMA. 2000;283(10):1303–1310. doi: 10.1001/jama.283.10.1303. [DOI] [PubMed] [Google Scholar]
- 7.Lo Re M., Chaplin M., Aronow B., Modesto-Lowe V. Buprenorphine Overdose in Young Children: An Underappreciated Risk. Clin. Pediatr. (Phila.) 2019;58(6):613–617. doi: 10.1177/0009922819829038. [DOI] [PubMed] [Google Scholar]
- 8.Edenberg H.J. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 9.Scott D.M., Taylor R.E. Health-related effects of genetic variations of alcohol-metabolizing enzymes in African Americans. Alcohol Res. Health. 2007;30(1):18–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Adinoff B. Neurobiologic processes in drug reward and addiction. Harv. Rev. Psychiatry. 2004;12(6):305–320. doi: 10.1080/10673220490910844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias-Carrión O., Stamelou M., Murillo-Rodríguez E., Menéndez-González M., Pöppel E. Dopaminergic reward system: a short integrative review. Int. Arch. Med. 2010;3:24. doi: 10.1186/1755-7682-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum K., Modestino E.J., Gondre-Lewis M., Chapman E.J., Neary J., Siwicki D., Baron D., Hauser M., Smith D.E., Roy A.K., Thanos P.K., Steinberg B., McLaughlin T., Fried L., Barh D., Dunston G.A., Badgaiyan R.D. 2018.
- 13.Kieffer B.L. Opioids: first lessons from knockout mice. Trends Pharmacol. Sci. 1999;20(1):19–26. doi: 10.1016/S0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 14.Terenius L. Stereospecific interaction between narcotic analgesics and a synaptic plasm a membrane fraction of rat cerebral cortex. Acta Pharmacol. Toxicol. (Copenh.) 1973;32(3):317–320. doi: 10.1111/j.1600-0773.1973.tb01477.x. [DOI] [PubMed] [Google Scholar]
- 15.Dhawan B.N., Cesselin F., Raghubir R., Reisine T., Bradley P.B., Portoghese P.S., Hamon M. International Union of Pharmacology. XII. Classification of opioid receptors. Pharmacol. Rev. 1996;48(4):567–592. [PubMed] [Google Scholar]
- 16.Hughes J., Smith T.W., Kosterlitz H.W., Fothergill L.A., Morgan B.A., Morris H.R. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature. 1975;258(5536):577–580. doi: 10.1038/258577a0. [DOI] [PubMed] [Google Scholar]
- 17.Pert C.B., Snyder S.H. Opiate receptor: demonstration in nervous tissue. Science. 1973;179(4077):1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- 18.Simon E.J., Hiller J.M., Edelman I. Stereospecific binding of the potent narcotic analgesic (3H) Etorphine to rat-brain homogenate. Proc. Natl. Acad. Sci. USA. 1973;70(7):1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blum K., Briggs A.H., DeLallo L. Clonidine enhancement of ethanol withdrawal in mice. Subst. Alcohol Actions Misuse. 1983;4(1):59–63. [PubMed] [Google Scholar]
- 20.Blum K. Alcohol and central nervous system peptides. Subst. Alcohol Actions Misuse. 1983;4(2-3):73–87. [PubMed] [Google Scholar]
- 21.Chen Y., Mestek A., Liu J., Hurley J.A., Yu L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol. Pharmacol. 1993;44(1):8–12. [PubMed] [Google Scholar]
- 22.Evans C.J., Keith D.E., Jr, Morrison H., Magendzo K., Edwards R.H. Cloning of a delta opioid receptor by functional expression. Science. 1992;258(5090):1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- 23.Kieffer B.L., Befort K., Gaveriaux-Ruff C., Hirth C.G. The delta-opioid receptor: isolation of a cDNA by expression cloning and pharmacological characterization. Proc. Natl. Acad. Sci. USA. 1992;89(24):12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasuda K., Raynor K., Kong H., Breder C.D., Takeda J., Reisine T., Bell G.I. Cloning and functional comparison of kappa and delta opioid receptors from mouse brain. Proc. Natl. Acad. Sci. USA. 1993;90(14):6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charbogne P., Kieffer B.L., Befort K. 2014. [DOI] [PMC free article] [PubMed]
- 26.Koob G.F., Volkow N.D. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3(8):760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Mestek A., Liu J., Yu L. Molecular cloning of a rat kappa opioid receptor reveals sequence similarities to the mu and delta opioid receptors. Biochem. J. 1993;295(Pt 3):625–628. doi: 10.1042/bj2950625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Childers S.R. Opioid receptor-coupled second messenger systems. Life Sci. 1991;48(21):1991–2003. doi: 10.1016/0024-3205(91)90154-4. [DOI] [PubMed] [Google Scholar]
- 29.Svingos A.L., Chavkin C., Colago E.E., Pickel V.M. Major coexpression of kappa-opioid receptors and the dopamine transporter in nucleus accumbens axonal profiles. Synapse. 2001;42(3):185–192. doi: 10.1002/syn.10005. [DOI] [PubMed] [Google Scholar]
- 30.Waldhoer M., Bartlett S.E., Whistler J.L. Opioid receptors. Annu. Rev. Biochem. 2004;73:953–990. doi: 10.1146/annurev.biochem.73.011303.073940. [DOI] [PubMed] [Google Scholar]
- 31.Williams J.T., Ingram S.L., Henderson G., Chavkin C., von Zastrow M., Schulz S., Koch T., Evans C.J., Christie M.J. Regulation of μ-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 2013;65(1):223–254. doi: 10.1124/pr.112.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chieng B., Williams J.T. Increased opioid inhibition of GABA release in nucleus accumbens during morphine withdrawal. J. Neurosci. 1998;18(17):7033–7039. doi: 10.1523/JNEUROSCI.18-17-07033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoji Y., Delfs J., Williams J.T. Presynaptic inhibition of GABA(B)-mediated synaptic potentials in the ventral tegmental area during morphine withdrawal. J. Neurosci. 1999;19(6):2347–2355. doi: 10.1523/JNEUROSCI.19-06-02347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capogna M., Gähwiler B.H., Thompson S.M. Mechanism of mu-opioid receptor-mediated presynaptic inhibition in the rat hippocampus in vitro. J. Physiol. 1993;470:539–558. doi: 10.1113/jphysiol.1993.sp019874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Hasani R., Bruchas M.R. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology. 2011;115(6):1363–1381. doi: 10.1097/ALN.0b013e318238bba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Contet C., Kieffer B.L., Befort K. Mu opioid receptor: a gateway to drug addiction. Curr. Opin. Neurobiol. 2004;14(3):370–378. doi: 10.1016/j.conb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Johnson S.W., North R.A. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J. Neurosci. 1992;12(2):483–488. doi: 10.1523/JNEUROSCI.12-02-00483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blum K., Sheridan P.J., Wood R.C., Braverman E.R., Chen T.J., Cull J.G., Comings D.E. The D2 dopamine receptor gene as a determinant of reward deficiency syndrome. J. R. Soc. Med. 1996;89(7):396–400. doi: 10.1177/014107689608900711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalumiere R.T. Optogenetic dissection of amygdala functioning. Front. Behav. Neurosci. 2014;8:107. doi: 10.3389/fnbeh.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieh E.H., Kim S.Y., Namburi P., Tye K.M. Optogenetic dissection of neural circuits underlying emotional valence and motivated behaviors. Brain Res. 2013;1511:73–92. doi: 10.1016/j.brainres.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eichenbaum H., Yonelinas A.P., Ranganath C. The medial temporal lobe and recognition memory. Annu. Rev. Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yager L.M., Garcia A.F., Wunsch A.M., Ferguson S.M. The ins and outs of the striatum: role in drug addiction. Neuroscience. 2015;301:529–541. doi: 10.1016/j.neuroscience.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christie M.J. Cellular neuroadaptations to chronic opioids: tolerance, withdrawal and addiction. Br. J. Pharmacol. 2008;154(2):384–396. doi: 10.1038/bjp.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell T.G. The best of a bad bunch: the ventromedial prefrontal cortex and dorsal anterior cingulate cortex in decision-making. J. Neurosci. 2007;27(3):447–448. doi: 10.1523/JNEUROSCI.4967-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mucha R.F., Herz A. Motivational properties of kappa and mu opioid receptor agonists studied with place and taste preference conditioning. Psychopharmacology (Berl.) 1985;86(3):274–280. doi: 10.1007/BF00432213. [DOI] [PubMed] [Google Scholar]
- 46.Pfeiffer A., Brantl V., Herz A., Emrich H.M. Psychotomimesis mediated by kappa opiate receptors. Science. 1986;233(4765):774–776. doi: 10.1126/science.3016896. [DOI] [PubMed] [Google Scholar]
- 47.Chefer V.I., Shippenberg T.S. Augmentation of morphine-induced sensitization but reduction in morphine tolerance and reward in delta-opioid receptor knockout mice. Neuropsychopharmacology. 2009;34(4):887–898. doi: 10.1038/npp.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Merrer J., Plaza-Zabala A., Del Boca C., Matifas A., Maldonado R., Kieffer B.L. Deletion of the δ opioid receptor gene impairs place conditioning but preserves morphine reinforcement. Biol. Psychiatry. 2011;69(7):700–703. doi: 10.1016/j.biopsych.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 49.Blum K., Gaskill H., DeLallo L., Briggs A.H., Hall W. Methionine enkephalin as a possible neuromodulator of regional cerebral blood flow. Experientia. 1985;41(7):932–933. doi: 10.1007/BF01970019. [DOI] [PubMed] [Google Scholar]
- 50.Jones C.M. Heroin use and heroin use risk behaviors among nonmedical users of prescription opioid pain relievers - United States, 2002-2004 and 2008-2010. Drug Alcohol Depend. 2013;132(1-2):95–100. doi: 10.1016/j.drugalcdep.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Schiller E., Mechanic O. Opioid overdose. In: Mechanic O.J., editor. StatPearls Treasure Island. FL: StatPearls Publishing LLC; 2019. [Google Scholar]
- 52.Shipton E.A., Shipton E.E., Shipton A.J. A Review of the Opioid Epidemic: What Do We Do About It? Pain Ther. 2018;7(1):23–36. doi: 10.1007/s40122-018-0096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Häuser W., Schug S., Furlan A.D. The opioid epidemic and national guidelines for opioid therapy for chronic noncancer pain: a perspective from different continents. Pain Rep. 2017;2(3):e599. doi: 10.1097/PR9.0000000000000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Amsterdam J., van den Brink W. The Misuse of Prescription Opioids: A Threat for Europe? Curr. Drug Abuse Rev. 2015;8(1):3–14. doi: 10.2174/187447370801150611184218. [DOI] [PubMed] [Google Scholar]
- 55.Blum K., Gold M.S. Neuro-chemical activation of brain reward meso-limbic circuitry is associated with relapse prevention and drug hunger: a hypothesis. Med. Hypotheses. 2011;76(4):576–584. doi: 10.1016/j.mehy.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Ikemoto S. Brain reward circuitry beyond the mesolimbic dopamine system: a neurobiological theory. Neurosci. Biobehav. Rev. 2010;35(2):129–150. doi: 10.1016/j.neubiorev.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swenson R. 2016.
- 58.Rajmohan V., Mohandas E. The limbic system. Indian J. Psychiatry. 2007;49(2):132–139. doi: 10.4103/0019-5545.33264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malenka R.C., Nestler E.J., Hyman S.E. 2009.
- 60.Blum K., Thanos P.K., Oscar-Berman M., Febo M., Baron D., Badgaiyan R.D., Gardner E., Demetrovics Z., Fahlke C., Haberstick B.C., Dushaj K., Gold M.S. Dopamine in the Brain: Hypothesizing Surfeit or Deficit Links to Reward and Addiction. J. Reward Defic. Syndr. 2015;1(3):95–104. doi: 10.17756/jrds.2015-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berridge K.C., Kringelbach M.L. Pleasure systems in the brain. Neuron. 2015;86(3):646–664. doi: 10.1016/j.neuron.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobsen C.F. 1936.
- 63.Sher L. The role of endogenous opioids in the placebo effect in post-traumatic stress disorder. Forsch. Komplementarmed. Klass. Naturheilkd. 2004;11(6):354–359. doi: 10.1159/000082817. [DOI] [PubMed] [Google Scholar]
- 64.Everitt B.J., Belin D., Economidou D., Pelloux Y., Dalley J.W., Robbins T.W. Review. Neural mechanisms underlying the vulnerability to develop compulsive drug-seeking habits and addiction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363(1507):3125–3135. doi: 10.1098/rstb.2008.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baik J.H. Dopamine signaling in reward-related behaviors. Front. Neural Circuits. 2013;7:152. doi: 10.3389/fncir.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blum K., Kozlowski G. In: Ethanol and Neuromodulator Interactions: A Cascade Model of Reward.Alcohol and Behavior; Ollat, H.; Parvez, S.; Parvez, H. Press V.S.P., editor. Utrecht: 1990. pp. 131–149. [Google Scholar]
- 67.Corre J., van Zessen R., Loureiro M., Patriarchi T., Tian L., Pascoli V., Lüscher C. Dopamine neurons projecting to medial shell of the nucleus accumbens drive heroin reinforcement. eLife. 2018;7:7. doi: 10.7554/eLife.39945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilpin N.W., Koob G.F. Neurobiology of alcohol dependence: focus on motivational mechanisms. Alcohol Res. Health. 2008;31(3):185–195. [PMC free article] [PubMed] [Google Scholar]
- 69.Salamone J.D., Correa M. The mysterious motivational functions of mesolimbic dopamine. Neuron. 2012;76(3):470–485. doi: 10.1016/j.neuron.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park B.Y., Wilson G., Berger J., Christman M., Reina B., Bishop F., Klam W.P., Doan A.P. Is Internet Pornography Causing Sexual Dysfunctions? A Review with Clinical Reports. Behav. Sci. (Basel) 2016;6(3):E17. doi: 10.3390/bs6030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yau Y.H., Potenza M.N. Gambling disorder and other behavioral addictions: recognition and treatment. Harv. Rev. Psychiatry. 2015;23(2):134–146. doi: 10.1097/HRP.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bavelier D., Green C.S., Han D.H., Renshaw P.F., Merzenich M.M., Gentile D.A. Brains on video games. Nat. Rev. Neurosci. 2011;12(12):763–768. doi: 10.1038/nrn3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chao J., Nestler E.J. Molecular neurobiology of drug addiction. Annu. Rev. Med. 2004;55:113–132. doi: 10.1146/annurev.med.55.091902.103730. [DOI] [PubMed] [Google Scholar]
- 74.Kalivas P.W., Volkow N.D. The neural basis of addiction: a pathology of motivation and choice. Am. J. Psychiatry. 2005;162(8):1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 75.Cameron K., Kolanos R., Vekariya R., De Felice L., Glennon R.A., Glennon R.A. Mephedrone and methylenedioxypyrovalerone (MDPV), major constituents of “bath salts,” produce opposite effects at the human dopamine transporter. Psychopharmacology (Berl.) 2013;227(3):493–499. doi: 10.1007/s00213-013-2967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lacey M.G., Mercuri N.B., North R.A. Actions of cocaine on rat dopaminergic neurones in vitro. Br. J. Pharmacol. 1990;99(4):731–735. doi: 10.1111/j.1476-5381.1990.tb12998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiang C.N., Hawks R.L. Pharmacokinetics of the combination tablet of buprenorphine and naloxone. Drug Alcohol Depend. 2003;70(2) Suppl.:S39–S47. doi: 10.1016/S0376-8716(03)00058-9. [DOI] [PubMed] [Google Scholar]
- 78.Strain E.C., Moody D.E., Stoller K.B., Walsh S.L., Bigelow G.E. Relative bioavailability of different buprenorphine formulations under chronic dosing conditions. Drug Alcohol Depend. 2004;74(1):37–43. doi: 10.1016/j.drugalcdep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 79.Accurso A.J., Rastegar D.A. The Effect of a Payer-Mandated Decrease in Buprenorphine Dose on Aberrant Drug Tests and Treatment Retention Among Patients with Opioid Dependence. J. Subst. Abuse Treat. 2016;61:74–79. doi: 10.1016/j.jsat.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 80.Wandel C., Witte J.S., Hall J.M., Stein C.M., Wood A.J., Wilkinson G.R. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5′-promoter region polymorphism. Clin. Pharmacol. Ther. 2000;68(1):82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 81.Chapman E., Ettienne E., Clarke M., Dunston G. Wide Pharmacogenetic Variations in Dosing of Buprenorphine in the Treatment of Opioid Addiction in African Americans; 2nd International Conference on Addiction Medicine and Reward Deficiency Syndrome; Baltimore, MD. 2017. [Google Scholar]
- 82.Fernández-Santander A., Novillo A., Gaibar M., Romero-Lorca A., Moral P., Sánchez-Cuenca D., Amir N., Chaabani H., Harich N., Esteban M.E. Cytochrome and sulfotransferase gene variation in north African populations. Pharmacogenomics. 2016;17(13):1415–1423. doi: 10.2217/pgs-2016-0016. [DOI] [PubMed] [Google Scholar]
- 83.Wandel C., Kim R.B., Guengerich F.P., Wood A.J. Mibefradil is a P-glycoprotein substrate and a potent inhibitor of both P-glycoprotein and CYP3A in vitro. Drug Metab. Dispos. 2000;28(8):895–898. [PubMed] [Google Scholar]
- 84.McCance-Katz E.F., Rainey P.M., Smith P., Morse G.D., Friedland G., Boyarsky B., Gourevitch M., Jatlow P. Drug interactions between opioids and antiretroviral medications: interaction between methadone, LAAM, and delavirdine. Am. J. Addict. 2006;15(1):23–34. doi: 10.1080/10550490500419029. [DOI] [PubMed] [Google Scholar]
- 85.Fonseca F., Torrens M. Pharmacogenetics of Methadone Response. Mol. Diagn. Ther. 2018;22(1):57–78. doi: 10.1007/s40291-017-0311-y. [DOI] [PubMed] [Google Scholar]
- 86.Blum K., Noble E.P., Sheridan P.J., Montgomery A., Ritchie T., Jagadeeswaran P., Nogami H., Briggs A.H., Cohn J.B. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA. 1990;263(15):2055–2060. doi: 10.1001/jama.1990.03440150063027. [DOI] [PubMed] [Google Scholar]
- 87.Grandy D.K., Marchionni M.A., Makam H., Stofko R.E., Alfano M., Frothingham L., Fischer J.B., Burke-Howie K.J., Bunzow J.R., Server A.C. Cloning of the cDNA and gene for a human D2 dopamine receptor. Proc. Natl. Acad. Sci. USA. 1989;86(24):9762–9766. doi: 10.1073/pnas.86.24.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grandy D.K., Litt M., Allen L., Bunzow J.R., Marchionni M., Makam H., Reed L., Magenis R.E., Civelli O. The human dopamine D2 receptor gene is located on chromosome 11 at q22-q23 and identifies a TaqI RFLP. Am. J. Hum. Genet. 1989;45(5):778–785. [PMC free article] [PubMed] [Google Scholar]
- 89.Blum K., Noble E.P., Sheridan P.J., Montgomery A., Ritchie T., Ozkaragoz T., Fitch R.J., Wood R., Finley O., Sadlack F. Genetic predisposition in alcoholism: association of the D2 dopamine receptor TaqI B1 RFLP with severe alcoholics. Alcohol. 1993;10(1):59–67. doi: 10.1016/0741-8329(93)90054-R. [DOI] [PubMed] [Google Scholar]
- 90.Noble E.P., Blum K. Alcoholism and the D2 dopamine receptor gene. JAMA. 1993;270(13):1547–1548. doi: 10.1001/jama.270.13.1547. [DOI] [PubMed] [Google Scholar]
- 91.Blum K., Badgaiyan R.D., Dunston G.M., Baron D., Modestino E.J., McLaughlin T., Steinberg B., Gold M.S., Gondré-Lewis M.C. The DRD2 Taq1A A1 Allele May Magnify the Risk of Alzheimer’s in Aging African-Americans. Mol. Neurobiol. 2018;55(7):5526–5536. doi: 10.1007/s12035-017-0758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meador-Woodruff J.H., Mansour A., Civelli O., Watson S.J. Distribution of D2 dopamine receptor mRNA in the primate brain. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1991;15(6):885–893. doi: 10.1016/0278-5846(91)90016-T. [DOI] [PubMed] [Google Scholar]
- 93.Lee S.A., Suh Y., Lee S., Jeong J., Kim S.J., Kim S.J., Park S.K. Functional expression of dopamine D2 receptor is regulated by tetraspanin 7-mediated postendocytic trafficking. FASEB J. 2017;31(6):2301–2313. doi: 10.1096/fj.201600755RR. [DOI] [PubMed] [Google Scholar]
- 94.Clarke T.K., Weiss A.R., Ferarro T.N., Kampman K.M., Dackis C.A., Pettinati H.M., O’brien C.P., Oslin D.W., Lohoff F.W., Berrettini W.H. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann. Hum. Genet. 2014;78(1):33–39. doi: 10.1111/ahg.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hou Q.F., Li S.B. Potential association of DRD2 and DAT1 genetic variation with heroin dependence. Neurosci. Lett. 2009;464(2):127–130. doi: 10.1016/j.neulet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 96.Vereczkei A., Demetrovics Z., Szekely A., Sarkozy P., Antal P., Szilagyi A., Sasvari-Szekely M., Barta C. Multivariate analysis of dopaminergic gene variants as risk factors of heroin dependence. PLoS One. 2013;8(6):e66592. doi: 10.1371/journal.pone.0066592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Persico A.M., Bird G., Gabbay F.H., Uhl G.R. D2 dopamine receptor gene TaqI A1 and B1 restriction fragment length polymorphisms: enhanced frequencies in psychostimulant-preferring polysubstance abusers. Biol. Psychiatry. 1996;40(8):776–784. doi: 10.1016/0006-3223(95)00483-1. [DOI] [PubMed] [Google Scholar]
- 98.Lawford B.R., Young R.M., Noble E.P., Sargent J., Rowell J., Shadforth S., Zhang X., Ritchie T. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am. J. Med. Genet. 2000;96(5):592–598. doi: 10.1002/1096-8628(20001009)96:5<592:AID-AJMG3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 99.Levran O., Peles E., Randesi M., Correa da Rosa J., Ott J., Rotrosen J., Adelson M., Kreek M.J. Dopaminergic pathway polymorphisms and heroin addiction: further support for association of CSNK1E variants. Pharmacogenomics. 2014;15(16):2001–2009. doi: 10.2217/pgs.14.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuo S.C., Yeh Y.W., Chen C.Y., Huang C.C., Chang H.A., Yen C.H., Ho P.S., Liang C.S., Chou H.W., Lu R.B., Huang S.Y. DRD3 variation associates with early-onset heroin dependence, but not specific personality traits. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2014;51:1–8. doi: 10.1016/j.pnpbp.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 101.Van Tol H.H., Bunzow J.R., Guan H.C., Sunahara R.K., Seeman P., Niznik H.B., Civelli O. Cloning of the gene for a human dopamine D4 receptor with high affinity for the antipsychotic clozapine. Nature. 1991;350(6319):610–614. doi: 10.1038/350610a0. [DOI] [PubMed] [Google Scholar]
- 102.Szilagyi A., Boór K., Székely A., Gaszner P., Kalász H., Sasvári-Székely M., Barta C. Combined effect of promoter polymorphisms in the dopamine D4 receptor and the serotonin transporter genes in heroin dependence. Neuropsychopharmacol. Hung. 2005;7(1):28–33. [PubMed] [Google Scholar]
- 103.Ho A.M., Tang N.L., Cheung B.K., Stadlin A. Dopamine receptor D4 gene -521C/T polymorphism is associated with opioid dependence through cold-pain responses. Ann. N. Y. Acad. Sci. 2008;1139:20–26. doi: 10.1196/annals.1432.054. [DOI] [PubMed] [Google Scholar]
- 104.Lai J.H., Zhu Y.S., Huo Z.H., Sun R.F., Yu B., Wang Y.P., Chai Z.Q., Li S.B. Association study of polymorphisms in the promoter region of DRD4 with schizophrenia, depression, and heroin addiction. Brain Res. 2010;1359:227–232. doi: 10.1016/j.brainres.2010.08.064. [DOI] [PubMed] [Google Scholar]
- 105.Hutchison K.E., McGeary J., Smolen A., Bryan A., Swift R.M. The DRD4 VNTR polymorphism moderates craving after alcohol consumption. Health Psychol. 2002;21(2):139–146. doi: 10.1037/0278-6133.21.2.139. [DOI] [PubMed] [Google Scholar]
- 106.Van Tol H.H., Wu C.M., Guan H.C., Ohara K., Bunzow J.R., Civelli O., Kennedy J., Seeman P., Niznik H.B., Jovanovic V. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358(6382):149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- 107.Larsen H., van der Zwaluw C.S., Overbeek G., Granic I., Franke B., Engels R.C. A variable-number-of-tandem-repeats polymorphism in the dopamine D4 receptor gene affects social adaptation of alcohol use: investigation of a gene-environment interaction. Psychol. Sci. 2010;21(8):1064–1068. doi: 10.1177/0956797610376654. [DOI] [PubMed] [Google Scholar]
- 108.Creswell K.G., Sayette M.A., Manuck S.B., Ferrell R.E., Hill S.Y., Dimoff J.D. DRD4 polymorphism moderates the effect of alcohol consumption on social bonding. PLoS One. 2012;7(2):e28914. doi: 10.1371/journal.pone.0028914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shao C., Li Y., Jiang K., Zhang D., Xu Y., Lin L., Wang Q., Zhao M., Jin L. Dopamine D4 receptor polymorphism modulates cue-elicited heroin craving in Chinese. Psychopharmacology (Berl.) 2006;186(2):185–190. doi: 10.1007/s00213-006-0375-6. [DOI] [PubMed] [Google Scholar]
- 110.Franke P., Nöthen M.M., Wang T., Knapp M., Lichtermann D., Neidt H., Sander T., Propping P., Maier W. DRD4 exon III VNTR polymorphism-susceptibility factor for heroin dependence? Results of a case-control and a family-based association approach. Mol. Psychiatry. 2000;5(1):101–104. doi: 10.1038/sj.mp.4000583. [DOI] [PubMed] [Google Scholar]
- 111.Li T., Zhu Z.H., Liu X., Hu X., Zhao J., Sham P.C., Collier D.A. Association analysis of polymorphisms in the DRD4 gene and heroin abuse in Chinese subjects. Am. J. Med. Genet. 2000;96(5):616–621. doi: 10.1002/1096-8628(20001009)96:5<616:AID-AJMG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 112.Kotler M., Cohen H., Segman R., Gritsenko I., Nemanov L., Lerer B., Kramer I., Zer-Zion M., Kletz I., Ebstein R.P. Excess dopamine D4 receptor (D4DR) exon III seven repeat allele in opioid-dependent subjects. Mol. Psychiatry. 1997;2(3):251–254. doi: 10.1038/sj.mp.4000248. [DOI] [PubMed] [Google Scholar]
- 113.Li T., Xu K., Deng H., Cai G., Liu J., Liu X., Wang R., Xiang X., Zhao J., Murray R.M., Sham P.C., Collier D.A. Association analysis of the dopamine D4 gene exon III VNTR and heroin abuse in Chinese subjects. Mol. Psychiatry. 1997;2(5):413–416. doi: 10.1038/sj.mp.4000310. [DOI] [PubMed] [Google Scholar]
- 114.Chien C.C., Lin C.H., Chang Y.Y., Lung F.W. Association of VNTR polymorphisms in the MAOA promoter and DRD4 exon 3 with heroin dependence in male Chinese addicts. World J. Biol. Psychiatry. 2010;11(2 Pt 2):409–416. doi: 10.3109/15622970903304459. [DOI] [PubMed] [Google Scholar]
- 115.Bond C., LaForge K.S., Tian M., Melia D., Zhang S., Borg L., Gong J., Schluger J., Strong J.A., Leal S.M., Tischfield J.A., Kreek M.J., Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc. Natl. Acad. Sci. USA. 1998;95(16):9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen T.J., Blum K., Mathews D., Fisher L., Schnautz N., Braverman E.R., Schoolfield J., Downs B.W., Comings D.E. Are dopaminergic genes involved in a predisposition to pathological aggression? Hypothesizing the importance of “super normal controls” in psychiatricgenetic research of complex behavioral disorders. Med. Hypotheses. 2005;65(4):703–707. doi: 10.1016/j.mehy.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 117.Blum K., Gondré-Lewis M.C., Baron D., Thanos P.K., Braverman E.R., Neary J., Elman I., Badgaiyan R.D. Introducing Precision Addiction Management of Reward Deficiency Syndrome, the Construct That Underpins All Addictive Behaviors. Front. Psychiatry. 2018;9:548. doi: 10.3389/fpsyt.2018.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hastie B.A., Riley J.L., III, Kaplan L., Herrera D.G., Campbell C.M., Virtusio K., Mogil J.S., Wallace M.R., Fillingim R.B. Ethnicity interacts with the OPRM1 gene in experimental pain sensitivity. Pain. 2012;153(8):1610–1619. doi: 10.1016/j.pain.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo X., Kranzler H.R., Zhao H., Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2003;120B(1):97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- 120.Crowley J.J., Oslin D.W., Patkar A.A., Gottheil E., DeMaria P.A., Jr, O’Brien C.P., Berrettini W.H., Grice D.E. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatr. Genet. 2003;13(3):169–173. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 121.Clarke T.K., Crist R.C., Kampman K.M., Dackis C.A., Pettinati H.M., O’Brien C.P., Oslin D.W., Ferraro T.N., Lohoff F.W., Berrettini W.H. Low frequency genetic variants in the μ-opioid receptor (OPRM1) affect risk for addiction to heroin and cocaine. Neurosci. Lett. 2013;542:71–75. doi: 10.1016/j.neulet.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Crist R.C., Ambrose-Lanci L.M., Vaswani M., Clarke T.K., Zeng A., Yuan C., Ferraro T.N., Hakonarson H., Kampman K.M., Dackis C.A., Pettinati H.M., O’Brien C.P., Oslin D.W., Doyle G.A., Lohoff F.W., Berrettini W.H. Case-control association analysis of polymorphisms in the δ-opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend. 2013;127(1-3):122–128. doi: 10.1016/j.drugalcdep.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Szeto C.Y., Tang N.L., Lee D.T., Stadlin A. Association between mu opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport. 2001;12(6):1103–1106. doi: 10.1097/00001756-200105080-00011. [DOI] [PubMed] [Google Scholar]
- 124.Shi J., Hui L., Xu Y., Wang F., Huang W., Hu G. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum. Mutat. 2002;19(4):459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 125.Wachman E.M., Hayes M.J., Brown M.S., Paul J., Harvey-Wilkes K., Terrin N., Huggins G.S., Aranda J.V., Davis J.M. Association of OPRM1 and COMT single-nucleotide polymorphisms with hospital length of stay and treatment of neonatal abstinence syndrome. JAMA. 2013;309(17):1821–1827. doi: 10.1001/jama.2013.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Crist R.C., Doyle G.A., Nelson E.C., Degenhardt L., Martin N.G., Montgomery G.W., Saxon A.J., Ling W., Berrettini W.H. A polymorphism in the OPRM1 3′-untranslated region is associated with methadone efficacy in treating opioid dependence. Pharmacogenomics J. 2018;18(1):173–179. doi: 10.1038/tpj.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.2013.
- 128.Haerian B.S., Haerian M.S. OPRM1 rs1799971 polymorphism and opioid dependence: evidence from a meta-analysis. Pharmacogenomics. 2013;14(7):813–824. doi: 10.2217/pgs.13.57. [DOI] [PubMed] [Google Scholar]
- 129.Zhang H., Kranzler H.R., Yang B.Z., Luo X., Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol. Psychiatry. 2008;13(5):531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Levran O., Londono D., O’Hara K., Nielsen D.A., Peles E., Rotrosen J., Casadonte P., Linzy S., Randesi M., Ott J., Adelson M., Kreek M.J. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7(7):720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nelson E.C., Lynskey M.T., Heath A.C., Wray N., Agrawal A., Shand F.L., Henders A.K., Wallace L., Todorov A.A., Schrage A.J., Madden P.A., Degenhardt L., Martin N.G., Montgomery G.W. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict. Biol. 2014;19(1):111–121. doi: 10.1111/j.1369-1600.2012.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Levran O., Londono D., O’Hara K., Randesi M., Rotrosen J., Casadonte P., Linzy S., Ott J., Adelson M., Kreek M.J. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8(5):531–540. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharafshah A., Fazel H., Albonaim A., Omarmeli V., Rezaei S., Mirzajani E., Ajamian F., Keshavarz P. Association of OPRD1 Gene Variants with Opioid Dependence in Addicted Male Individuals Undergoing Methadone Treatment in the North of Iran. J. Psychoactive Drugs. 2017;49(3):242–251. doi: 10.1080/02791072.2017.1290303. [DOI] [PubMed] [Google Scholar]
- 134.Beer B., Erb R., Pavlic M., Ulmer H., Giacomuzzi S., Riemer Y., Oberacher H. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case-control study. PLoS One. 2013;8(9):e75359. doi: 10.1371/journal.pone.0075359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuferov V., Fussell D., LaForge K.S., Nielsen D.A., Gordon D., Ho A., Leal S.M., Ott J., Kreek M.J. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenetics. 2004;14(12):793–804. doi: 10.1097/00008571-200412000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jones J.D., Luba R.R., Vogelman J.L., Comer S.D. Searching for evidence of genetic mediation of opioid withdrawal by opioid receptor gene polymorphisms. Am. J. Addict. 2016;25(1):41–48. doi: 10.1111/ajad.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nagaya D., Zahari Z., Saleem M., Yahaya B.H., Tan S.C., Yusoff N.M. An analysis of genetic association in opioid dependence susceptibility. J. Clin. Pharm. Ther. 2018;43(1):80–86. doi: 10.1111/jcpt.12585. [DOI] [PubMed] [Google Scholar]
- 138.Albonaim A., Fazel H., Sharafshah A., Omarmeli V., Rezaei S., Ajamian F., Keshavarz P. Association of OPRK1 gene polymorphisms with opioid dependence in addicted men undergoing methadone treatment in an Iranian population. J. Addict. Dis. 2017;36(4):227–235. doi: 10.1080/10550887.2017.1361724. [DOI] [PubMed] [Google Scholar]
- 139.Gerra G., Somaini L., Leonardi C., Cortese E., Maremmani I., Manfredini M., Donnini C. Association between gene variants and response to buprenorphine maintenance treatment. Psychiatry Res. 2014;215(1):202–207. doi: 10.1016/j.psychres.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 140.Kumar D., Chakraborty J., Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2012;36(2):225–230. doi: 10.1016/j.pnpbp.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 141.Poirier K., Viot G., Lombardi L., Jauny C., Billuart P., Bienvenu T. Loss of Function of KCNC1 is associated with intellectual disability without seizures. Eur. J. Hum. Genet. 2017;25(5):560–564. doi: 10.1038/ejhg.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deutsch C., Chen L.Q. Heterologous expression of specific K+ channels in T lymphocytes: functional consequences for volume regulation. Proc. Natl. Acad. Sci. USA. 1993;90(21):10036–10040. doi: 10.1073/pnas.90.21.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tiwari-Woodruff S., Beltran-Parrazal L., Charles A., Keck T., Vu T., Bronstein J.K. + channel KV3.1 associates with OSP/claudin-11 and regulates oligodendrocyte development. Am. J. Physiol. Cell Physiol. 2006;291(4):C687–C698. doi: 10.1152/ajpcell.00510.2005. [DOI] [PubMed] [Google Scholar]
- 144.Schaarschmidt G., Wegner F., Schwarz S.C., Schmidt H., Schwarz J. Characterization of voltage-gated potassium channels in human neural progenitor cells. PLoS One. 2009;4(7):e6168. doi: 10.1371/journal.pone.0006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nascimento F.A., Andrade D.M. Myoclonus epilepsy and ataxia due to potassium channel mutation (MEAK) is caused by heterozygous KCNC1 mutations. Epileptic Disord. 2016;18(S2):135–138. doi: 10.1684/epd.2016.0859. [DOI] [PubMed] [Google Scholar]
- 146.Gelernter J., Kranzler H.R., Sherva R., Koesterer R., Almasy L., Zhao H., Farrer L.A. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol. Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jones K.R., Reichardt L.F. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc. Natl. Acad. Sci. USA. 1990;87(20):8060–8064. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Maisonpierre P.C., Le Beau M.M., Espinosa R., III, Ip N.Y., Belluscio L., de la Monte S.M., Squinto S., Furth M.E., Yancopoulos G.D. Human and rat brain-derived neurotrophic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics. 1991;10(3):558–568. doi: 10.1016/0888-7543(91)90436-I. [DOI] [PubMed] [Google Scholar]
- 149.Rutherford L.C., Nelson S.B., Turrigiano G.G. BDNF has opposite effects on the quantal amplitude of pyramidal neuron and interneuron excitatory synapses. Neuron. 1998;21(3):521–530. doi: 10.1016/S0896-6273(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 150.McAllister A.K., Katz L.C., Lo D.C. Neurotrophins and synaptic plasticity. Annu. Rev. Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- 151.Heberlein A., Dürsteler-MacFarland K.M., Lenz B., Frieling H., Grösch M., Bönsch D., Kornhuber J., Wiesbeck G.A., Bleich S., Hillemacher T. Serum levels of BDNF are associated with craving in opiate-dependent patients. J. Psychopharmacol. (Oxford) 2011;25(11):1480–1484. doi: 10.1177/0269881111411332. [DOI] [PubMed] [Google Scholar]
- 152.Jin T., Zhang H., Yang Q., Li L., Ouyang Y., Yang M., Wang F., Wang Z., Zhang J., Yuan D. The relationship between polymorphisms of BDNFOS and BDNF genes and heroin addiction in the Han Chinese population. J. Gene Med. 2016;18(10):288–293. doi: 10.1002/jgm.2927. [DOI] [PubMed] [Google Scholar]
- 153.Jia W., Shi J.G., Wu B., Ao L., Zhang R., Zhu Y.S. Polymorphisms of brain-derived neurotrophic factor associated with heroin dependence. Neurosci. Lett. 2011;495(3):221–224. doi: 10.1016/j.neulet.2011.03.072. [DOI] [PubMed] [Google Scholar]
- 154.Iamjan S.A., Thanoi S., Watiktinkorn P., Nudmamud-Thanoi S., Reynolds G.P. BDNF (Val66Met) genetic polymorphism is associated with vulnerability for methamphetamine dependence. Pharmacogenomics. 2015;16(14):1541–1545. doi: 10.2217/pgs.15.96. [DOI] [PubMed] [Google Scholar]
- 155.Su H., Tao J., Zhang J., Xie Y., Han B., Lu Y., Sun H., Wei Y., Wang Y., Zhang Y., Zou S., Wu W., Zhang J., Xu K., Zhang X., He J. The analysis of BDNF gene polymorphism haplotypes and impulsivity in methamphetamine abusers. Compr. Psychiatry. 2015;59:62–67. doi: 10.1016/j.comppsych.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 156.de Cid R., Fonseca F., Gratacòs M., Gutierrez F., Martín-Santos R., Estivill X., Torrens M. BDNF variability in opioid addicts and response to methadone treatment: preliminary findings. Genes Brain Behav. 2008;7(5):515–522. doi: 10.1111/j.1601-183X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 157.Li C.Y., Liu Q.R., Zhang P.W., Li X.M., Wei L., Uhl G.R. OKCAM: an ontology-based, human-centered knowledgebase for cell adhesion molecules. Nucleic Acids Res. 2009;37(Database issue):D251–D260. doi: 10.1093/nar/gkn568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aoto J., Martinelli D.C., Malenka R.C., Tabuchi K., Südhof T.C. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154(1):75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Panagopoulos V.N., Trull T.J., Glowinski A.L., Lynskey M.T., Heath A.C., Agrawal A., Henders A.K., Wallace L., Todorov A.A., Madden P.A., Moore E., Degenhardt L., Martin N.G., Montgomery G.W., Nelson E.C. Examining the association of NRXN3 SNPs with borderline personality disorder phenotypes in heroin dependent cases and socio-economically disadvantaged controls. Drug Alcohol Depend. 2013;128(3):187–193. doi: 10.1016/j.drugalcdep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lachman H.M., Fann C.S., Bartzis M., Evgrafov O.V., Rosenthal R.N., Nunes E.V., Miner C., Santana M., Gaffney J., Riddick A., Hsu C.L., Knowles J.A. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum. Mol. Genet. 2007;16(11):1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- 161.Blum K., Oscar-Berman M., Demetrovics Z., Barh D., Gold M.S. Genetic Addiction Risk Score (GARS): molecular neurogenetic evidence for predisposition to Reward Deficiency Syndrome (RDS). Mol. Neurobiol. 2014;50(3):765–796. doi: 10.1007/s12035-014-8726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Henker R.A., Lewis A., Dai F., Lariviere W.R., Meng L., Gruen G.S., Sereika S.M., Pape H., Tarkin I.S., Gowda I., Conley Y.P. The associations between OPRM 1 and COMT genotypes and postoperative pain, opioid use, and opioid-induced sedation. Biol. Res. Nurs. 2013;15(3):309–317. doi: 10.1177/1099800411436171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li T., Du J., Yu S., Jiang H., Fu Y., Wang D., Sun H., Chen H., Zhao M. Pathways to age of onset of heroin use: a structural model approach exploring the relationship of the COMT gene, impulsivity and childhood trauma. PLoS One. 2012;7(11):e48735. doi: 10.1371/journal.pone.0048735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rakvåg T.T., Klepstad P., Baar C., Kvam T.M., Dale O., Kaasa S., Krokan H.E., Skorpen F. The Val158Met polymorphism of the human catechol-O-methyltransferase (COMT) gene may influence morphine requirements in cancer pain patients. Pain. 2005;116(1-2):73–78. doi: 10.1016/j.pain.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 165.Wang J.Y., Fan Q.Y., He J.H., Zhu S.G., Huang C.P., Zhang X., Zhu J.H. SLC6A4 Repeat and Single-Nucleotide Polymorphisms Are Associated With Depression and Rest Tremor in Parkinson’s Disease: An Exploratory Study. Front. Neurol. 2019;10:333. doi: 10.3389/fneur.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Saiz P.A., Garcia-Portilla M.P., Florez G., Arango C., Corcoran P., Morales B., Bascaran M.T., Alvarez C., San Narciso G., Carreño E., Alvarez V., Coto E., Bobes J. Differential role of serotonergic polymorphisms in alcohol and heroin dependence. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2009;33(4):695–700. doi: 10.1016/j.pnpbp.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 167.Gerra G., Garofano L., Santoro G., Bosari S., Pellegrini C., Zaimovic A., Moi G., Bussandri M., Moi A., Brambilla F., Donnini C. Association between low-activity serotonin transporter genotype and heroin dependence: behavioral and personality correlates. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;126B(1):37–42. doi: 10.1002/ajmg.b.20111. [DOI] [PubMed] [Google Scholar]
- 168.Blum K., Chen A.L.C., Thanos P.K., Febo M., Demetrovics Z., Dushaj K., Kovoor A., Baron D., Smith D.E., Roy A.K., III, Fried L., Chen T.J.H., Chapman E., Sr, Modestino E.J., Steinberg B., Badgaiyan R.D. Genetic addiction risk score (GARS) ™, a predictor of vulnerability to opioid dependence. Front. Biosci. (Elite Ed.) 2018;10:175–196. doi: 10.2741/e816. [DOI] [PubMed] [Google Scholar]
- 169.Rajman I., Knapp L., Morgan T., Masimirembwa C. African Genetic Diversity: Implications for Cytochrome P450-mediated Drug Metabolism and Drug Development. EBioMedicine. 2017;17:67–74. doi: 10.1016/j.ebiom.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Walker J., Winhusen T., Storkson J.M., Lewis D., Pariza M.W., Somoza E., Somoza V. Total antioxidant capacity is significantly lower in cocaine-dependent and methamphetamine-dependent patients relative to normal controls: results from a preliminary study. Hum. Psychopharmacol. 2014;29(6):537–543. doi: 10.1002/hup.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kong H., Jiang C.Y., Hu L., Teng P., Zhang Y., Pan X.X., Sun X.D., Liu W.T. Morphine induces dysfunction of PINK1/Parkin-mediated mitophagy in spinal cord neurons implying involvement in antinociceptive tolerance. J. Mol. Cell Biol. 2019;11(12):1056–1068. doi: 10.1093/jmcb/mjz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sharma H.S., Sjöquist P.O., Ali S.F. Drugs of abuse-induced hyperthermia, blood-brain barrier dysfunction and neurotoxicity: neuroprotective effects of a new antioxidant compound H-290/51. Curr. Pharm. Des. 2007;13(18):1903–1923. doi: 10.2174/138161207780858375. [DOI] [PubMed] [Google Scholar]
- 173.Abdel-Zaher A.O., Mostafa M.G., Farghaly H.S., Hamdy M.M., Abdel-Hady R.H. Role of oxidative stress and inducible nitric oxide synthase in morphine-induced tolerance and dependence in mice. Effect of alpha-lipoic acid. Behav. Brain Res. 2013;247:17–26. doi: 10.1016/j.bbr.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 174.Feng Y.M., Jia Y.F., Su L.Y., Wang D., Lv L., Xu L., Yao Y.G. Decreased mitochondrial DNA copy number in the hippocampus and peripheral blood during opiate addiction is mediated by autophagy and can be salvaged by melatonin. Autophagy. 2013;9(9):1395–1406. doi: 10.4161/auto.25468. [DOI] [PubMed] [Google Scholar]
- 175.Rezaei M., Saadat M. Association Between GSTP1 Ile105Val Genetic Polymorphism and Dependency to Heroin and Opium. Biochem. Genet. 2019;57(2):214–221. doi: 10.1007/s10528-018-9885-2. [DOI] [PubMed] [Google Scholar]
- 176.Boroumand F., Mahmoudinasab H., Saadat M. Association of the SOD2 (rs2758339 and rs5746136) polymorphisms with the risk of heroin dependency and the SOD2 expression levels. Gene. 2018;649:27–31. doi: 10.1016/j.gene.2018.01.074. [DOI] [PubMed] [Google Scholar]