Abstract
Mitochondrial damage is involved in many pathophysiological processes, such as tumor development, metabolism, and neurodegenerative diseases. The mitochondrial unfolded protein response (mtUPR) is the first stress-protective response initiated by mitochondrial damage, and it repairs or clears misfolded proteins to alleviate this damage. Studies have confirmed that the sirtuin family is essential for the mitochondrial stress response; in particular, SIRT1, SIRT3, and SIRT7 participate in the mtUPR in different axes. This article summarizes the associations of sirtuins with the mtUPR as well as specific molecular targets related to the mtUPR in different disease models, which will provide new inspiration for studies on mitochondrial stress, mitochondrial function protection, and mitochondria-related diseases, such as neurodegenerative diseases.
Keywords: SIRT, mitochondrial unfolded protein response, mitochondrial stress
1. Introduction
Mitochondria are essential cellular elements that generate energy. In addition to being critical energy-generating organelles, mitochondria are also involved in cell differentiation, cellular information transmission, and cell growth. The complex associations between mitochondria and other components require coordination between the nuclear genome and the mitochondrial genome. Emerging evidence has confirmed that the functional and structural integrity of mitochondria plays a vital role in tumor development, aging, metabolism, and neurodegenerative disease [1]. The key mechanism by which cells maintain their normal state is called mitochondrial protein quality control (PQC), which determines mitochondrial fate. The early-onset mitochondrial unfolded protein response (mtUPR), an emerging adaptive stress response that is the first line of defense for mitochondrial PQC, ensures the optimal quality and function of the mitochondrial proteome as a unique process that mitigates mitochondrial damage [2]. Subtle adjustments in the level of mtUPR activation in order to stop the onset and development of diseases at the initial stage are critical for the early intervention and treatment of irreversible diseases, such as metabolic diseases, cancer, and neurodegenerative diseases.
2. Mitochondrial unfolded protein response
The mtUPR is a PQC mechanism [2] that can individually perceive the increased expression of chaperones such as mitochondrial heat shock protein 70, heat shock protein 60 (HSP60), and heat shock protein 60 (HSP10). Some proteases related to the mitochondrial PQC system are encoded by nuclear genes and maintain mitochondrial protein homeostasis, such as the Clp protease proteolytic subunit and the ATP-dependent metalloprotease YME1L1 [3, 4]. Chaperone proteins help restore misfolded proteins to their regular conformations and promote the correct folding of newly synthesized proteins, while proteases degrade unneeded proteins. If protein homeostasis is not maintained correctly, reactive oxygen species (ROS) levels can increase due to the dysfunction of the electron transport chain. When mitochondrial damage is severe, the stress response cannot mitigate the damage, and the process of injury is accelerated. The mtUPR monitors not only the degradation of aberrant proteins in mitochondria but also the balance between mitochondrial protein entry and exit, which serves as an indirect indicator of mitochondrial adaptation and is primarily related to the transcription of PQC components [5]. The mtUPR has been investigated in yeast, worms, flies, and mammalian cells [6]. Current related studies suggest that the mechanism of the mtUPR can be divided into three axes with an additional mtUPR translational axis. Importantly, one review article has proposed the existence of a sirtuin (SIRT) axis in the mtUPR and emphasized the role of this axis in the mtUPR [7]. Here, we have summarized the functions of the mtUPR axes shown in Fig. 1.
Fig. (1).
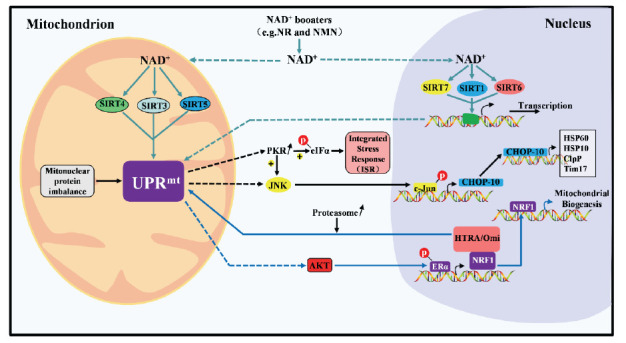
Overview of the Molecular mechanism in the mitochondrial unfolded protein response, including the classical axis (marked in black), UPRIMS-ERα axis (marked in blue), and mtUPR Sirtuin axis. (marked in green). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.1. Classic mtUPR Axis
Activating transcription factor 5 (ATF5), a homolog of nematode activating transcription factor associated with stress-1, shuttles between the mitochondria and nuclei and induces the mtUPR to promote cell proliferation and mitochondrial functional recovery under mitochondrial stress. In addition, in mammalian cells overexpressing misfolded deposition of massive amounts of unfolded proteins in the mitochondrial matrix activates Jun N‐terminal kinase 2, which in turn triggers the binding of c-Jun to activating protein-1 and the subsequent upregulation of the C/EBP homologous protein (CHOP) and C/EBPβ promoters. Dimers of the latter two proteins can act as transcription factors that bind to the promoters of mtUPR-related genes, thereby inducing the expression of mitochondrial heat shock proteins and proteases [8]. Furthermore, phosphorylation of eukaryotic initiation factor 2α (eIF2α) kinase mediated by protein kinase R leads to the increased translation of transcription factor 4, which activates many genes, including CHOP [9].
2.2. Unfolded Protein Response in the Intermembrane Space (IMS) of Mitochondria (UPRIMS)-Estrogen
Receptor Alpha (ERΑ) Axis
The accumulation of misfolded proteins in mitochondrial membranes leads to ROS-dependent AKT kinase phosphorylation, which activates nuclear ERα and induces the nuclear respiratory factor 1 (NRF1) gene to promote mitochondrial biosynthesis. At the same time, the expression of the membrane clearance protein high-temperature-required protein A2 increases, which restores mitochondrial function [10, 11].
2.3. mtUPR Translational Axis
Wade Harper et al. [12, 13] used two inhibitors of mitochondrial heat shock protein 90 and Lon protease homolog 1 (LONP1) to rapidly induce the mtUPR in order to examine the mtUPR translational axis. This axis is activated only when mitochondrial damage is mild and functions to regulate protein homeostasis rapidly without affecting other cells. In short, it is a local reaction that is mostly independent of transcriptional effects in the nucleus.
2.4. mtUPR SIRT Axis
SIRTs, which belong to the class III histone deacetylase enzyme family, are highly conserved enzyme homologs of the yeast Sir2 protein with nicotine adenine dinucleotide-dependent protein deacetylase and single ADP ribosyltransferase activity. The SIRT family consists mainly of seven proteins (SIRT1-SIRT7) and is involved in a variety of cellular processes, including metabolism, DNA repair, stress responses, apoptosis, tumorigenesis, and aging [14, 15]. Among SIRTs, SIRT1, SIRT6, and SIRT7 are found mainly in the nucleus; SIRT3, SIRT4, and SIRT5 are located mainly in mitochondria; and SIRT2 exists mainly in the cytoplasm. In particular, SIRT3 may shuttle from mitochondria to the nucleus under conditions of cellular stress. In addition, SIRT1 and SIRT2 can shuttle between the nucleus and cytoplasm [14].
3. SIRT axis in the MTUPR
SIRTs can be used to produce therapeutic agents capable of slowing down aging [16], and are hot topics in current research. The specific mechanism by which the mtUPR protects mitochondrial function is not yet precisely known. A growing number of related studies have confirmed that SIRT proteins participate in the mtUPR by targeting different molecules, and research has shown that the mtUPR may be an essential component of SIRT-mediated longevity [17]. The activation of SIRTs stimulates the mtUPR; such stimulation most commonly occurs after SIRT1 activation in mice and sir-2.1 activation in worms [6, 18]. SIRT1 activation is associated with increased mitochondrial autophagy, the mtUPR, and the maintenance of mitochondrial protein balance [19]. In a mouse model of nonalcoholic fatty liver disease (NAFLD), nicotinamide riboside (NR) has been found to prevent and alleviate NAFLD by inducing SIRT1- and SIRT3-dependent mtUPRs [20]. Concerning metabolism-related clinical conditions such as obesity, the mtUPR downregulates SIRT1, SIRT3, SIRT5 and NAD-dependent biosynthesis-related genes in the subcutaneous adipose tissue of larger twins [21]. In breast cancer cells, the mtUPR activates SIRT3, CHOP, and estrogen receptor alpha (ERα) via the accumulation of misfolded proteins in mitochondria. By coordinating antioxidant mechanisms and mitochondrial autophagy in a manner that is independent of CHOP and ERα, SIRT3 helps overcome toxic protein stress and mitochondrial stress, and the mechanisms may be important for tumor adaptability [22]. In muscle stem cells from aged mice, supplementation with NR activates SIRT1-dependent mtUPR signaling to improve mitochondrial function and delay aging [23]. Furthermore, in the MV389 worms, the knockdown of sir-2.1 blocks the expression of HSP-6 in the mtUPR. The effect of sir-2.1 overexpression is also wholly dependent on the mtUPR, and the lifespan of the MV389 strain is almost wholly restored to wild-type levels upon sir-2.1 overexpression [24]. Notably, interference with or blockade of the mtUPR pathway prevents changes in longevity induced by sir-2.1 overexpression or NR supplementation in worms [25]. Similar to the case in sir-2.1 transgenic worms, the ratio between ATP5 (encoded by nuclear [nDNA]) and mitochondrial cytochrome c oxidase subunit 1 (MTCO1), which is associated with mtUPR induction, is significantly reduced in SIRT1-overexpressing primary hepatocytes compared with wild-type hepatocytes, while the ratio of CLPP to HSP60 is significantly increased, showing the occurrence of the mitochondrial stress response and the existence of mitochondrial clearance protein imbalance [6]. Related articles have also reported that SIRT3 activates a branch of the mtUPR, regulates the mtUPR and induces mitochondrial autophagy and antioxidant responses in human cells [26]. SIRT7 may be the key to mitochondrial protein homeostasis, and its absence can induce the mtUPR [27]. Danica Chen and colleagues have further demonstrated that increased expression of the SIRT7 gene inhibits NRF1-induced multidrug resistance proteins(MRPs) in an essential mechanism by which the mtUPR attenuates mitochondrial protein folding stress (PFSmt) in mice [25, 28, 29]. Below, we will explain the relationships between the different SIRTs (SIRT1-SIRT7) and the mtUPR, which are demonstrated in Figs. 2 and 3.
3.1. SIRT1 AND THE MTUPR
SIRT1 is the most studied functional member of the SIRT family. SIRT1 can deacetylate many different substrate proteins after activation, including peroxisome proliferation activating receptor coactivating factor 1α (PGC-1α), peroxisome proliferator-activated receptor γ (PPARγ), Ku70, hypoxia-inducible factor 2α, transducer of regulated CREB protein 2, nuclear factor kappa B (NF-κB), acetyl-CoA synthetase (AceCS1), myocyte enhancer factor-2, and p53, thus playing essential roles in aging, tumor development, stress responses and metabolic regulation [5, 30, 31].
Mitochondrial ribosomal protein S5 (MRPS5), a component of the 28S small subunit of mitochondrial ribosomes, is closely related to the function of mitochondrial complex I. Its primary role is to synthesize proteins of the mitochondrial respiratory chain, thus regulating lifespan in nematodes and mice, and its functions are closely related to aging, energy metabolism, tumor susceptibility, cell senescence, etc. [32]. In addition, it is widely distributed in almost all tissues and organs. Hepatoma cell lines exhibit both cytoplasmic and nuclear distribution of MRPS5. Moreover, bioinformatics analyses have revealed that both a nuclear localization sequence and a mitochondrial targeting domain exist in the protein sequence of MRPS5 [33]. The combined results of mouse population genetics and nematode RNAi studies have confirmed that Mrps5 and other mitochondrial ribosomal proteins (MRPs) are metabolism and longevity regulators. Knockdown of MRPs, which triggers ribonucleoprotein imbalance, reduces mitochondrial respiration and activates the mtUPR [21]. In a recent study [33], MRPS5 was found to be abundant in liver tumor stem cells and to decrease in abundance during differentiation. In experiments in which MRPS5 was coexpressed with SIRT1 and SIRT2, only SIRT1 reduced the acetylation degree of MRPS5; SIRT2 did not. Furthermore, cells treated with SIRT1 shRNA or the inhibitor EX527 showed increased acetylation of MRPS5, confirming that SIRT1 directly regulates the acetylation status of MRPS5.
In conclusion, both in vivo and in vitro experiments have confirmed the interaction between SIRT1 and MRPS5 and the critical role of SIRT1 in the regulation of MRPS5 function. In addition, it has been confirmed that the expression of SIRT1 is increased in liver cancer stem cells (CSCs); these cells must induce the mtUPR to cope with oxidative stress by regulating the deacetylation of MRPS5. In general, the
existence of the mtUPR-SIRT1/MRPS5 axis has been confirmed in liver tumor stem cells, but this axis has rarely been studied in other disease models and needs to be further explored.
Poly-(ADP-ribose) polymerases (PARPs) are a group of enzymes that are responsible for detecting and repairing DNA damage and are primarily involved in cellular stress responses, acting as both sensors of cell injury and active participants in the stress response [34]. Seventeen different genes encode PARP-related proteins, but PARP1 and PARP2 account for the majority of PARP activity in cells. PARPs are activated by HSP70 during heat shock stress to alter the nucleosome structure and tryptophan tRNA synthase. SIRT1 and PARP1 associate with each other to affect gene transcription and mitochondrial function, and SIRT1 can be acetylated to respond to mechanical stresses. In addition, SIRT1 is capable of negatively regulating the PARP1 promoter, while the SIRT1 promoter is affected by PARP2 [35]. PARP1 is involved in mechanisms related to mitochondrial function maintenance, including the regulation of reverse mitochondrial signal transduction to the nucleus [36]. A PARP inhibitor (Parib) has been found to trigger the mtUPR in a SIRT1-dependent manner to increase mitochondrial respiratory capacity in worms [7]. In addition, findings of increased levels of CLPP and HSP60 in muscles treated with MRL-45696, another PARP inhibitor, have indicated that MRL-45696 strongly affects mitochondrial translation and ultimately induces the mtUPR. Nevertheless, it is not clear how Paribs and MRL-45696 can cause nuclear and cytoplasmic imbalances in mammalian cells to activate the mtUPR [33].
Decreases in nicotinamide adenine dinucleotide (NAD+) levels lead to the formation of complexes between the NAD+-binding protein Deleted in Breast Cancer 1 and PARP1 and inhibit the catalytic activity of PARP. Similarly, decreases in NAD+ levels can decrease SIRT1 function and limit the mtUPR, but these effects can be overcome by NR supplementation [37]. SIRT1 activation is indispensable for the NR-induced mtUPR [38]. In an alcohol-induced liver injury cell model, increased levels of NR were confirmed to reverse liver injury by activating the mtUPR and regulating lipid metabolism; at the same time, the activity of SIRT1 and PGC-1α was enhanced [39] in the SIRT1/PGC-1α axis of the mtUPR. Increased SIRT activity activates transcriptional regulators, such as PGC-1α, which increases mitochondrial content and metabolism as well as upregulating the mtUPR [40]. PGC-1α is a transcriptional regulator known for its ability to drive oxidative gene expression in various tissues and cells. A lack of PGC-1α leads to increased oxidative stress and makes mitochondria prone to apoptosis [41]. PGC-1α controls mitochondrial biogenesis by controlling the expression of NRF1 and nuclear respiratory factor 2 (NRF2). NRF1 and NRF2 themselves control the expression of mitochondrial transcription factors. siRNAs targeting estrogen-related receptor alpha (ERRα) can inhibit the expression of PGC-1α, while ERRα overexpression can induce PGC-1α overexpression, which indicates that PGC-1α is closely related to ERRα and that ERRα is a key participant in the mitochondrial IMS in the context of the mtUPR [42]. Notably, one of the mechanisms by which resveratrol (RSV), an agonist of SIRT1, prolongs life span relies on mtUPR activation. RSV improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α [43]. Although PGC-1α is coexpressed with SIRT1, RSV is not involved in cardiac mitochondrial biogenesis. Similar changes may be due to the existence of cell-specific associations between PGC-1α and other transcription factors or cofactors. Overactivation of PARP1, a DNA damage sensor, can also induce the NAD+-SIRT1-PGC-1α pathway [44]. PARP1 regulates PGC-1α by mediating the activity of SIRT1, and inhibition of PARP2 can induce SIRT1 to acetylate PGC-1α. PARP1 regulates forkhead box O (FOXO) 1 by affecting SIRT1 activity, and inhibition of PARP2 induces SIRT1 to deacetylate FOXO1, thereby further regulating mitochondrial activity [45]. The regulation of FOXO activity by SIRT1 can affect mitochondrial function and retrograde signal transduction from mitochondria to the nucleus by affecting the expression of nuclear genes, including FOXO1, FOXO3A, and FOXO4. This communication forms the SIRT1/FOXO axis of the mtUPR [30, 46].
Adenosine 5-monophosphate (AMP)-activated protein kinase (AMPK) is the primary cellular energy sensor. AMPK activates related transcription procedures, enhances oxidative metabolism, and recharges cell energy reserves [35]. PARP1 activation leads to increases in AMPK activity. AMPK can also trigger the mtDNA quality control system. AMPK may activate SIRT1 by indirectly increasing the levels of NAD+ and SIRT1. In addition, SIRT1 can deacetylate the AMPK gene, which may increase SIRT1 activity more than other stimuli [47]. Under mtUPR conditions induced by a medium dose of RSV, SIRT1 can stimulate AMPK, which plays an essential role in improving mitochondrial function in vivo and in vitro. All the major participants in the NAD+-SIRT1-hypoxia-inducible factor 1 alpha (HIF-1α)-oxidative phosphorylation axis exist in lower eukaryotes; this axis coordinates nuclear and mitochondrial synchronization in response to changes in energy supply and oxygen levels. With respect to these changes, the regulation of HIF-1α may have different effects on life expectancy in Caenorhabditis elegans depending on the animal diet and whether the mtUPR is activated [48]. Although the mtUPR has not yet been detected in skeletal muscle, the possibility that the mtUPR works in other tissues or under different conditions cannot be ruled out [19].
3.2. SIRT2 AND THE MTUPR
SIRT2 is mainly distributed in the cytoplasm but also exists in the nucleus, especially in tissues and organs with active metabolism, including the brain, kidneys, pancreas, testes, liver, and adipose tissue in mice [49, 50]. SIRT2 interacts with many histone and nonhistone protein substrates, including tubulin and histone H4. It participates in cell growth, differentiation, energy metabolism, and other cell functions and is associated with cancer, neurodegeneration, and metabolic diseases. Liu G et al. [51] demonstrated for the first time that SIRT2 regulates mitochondrial metabolism. SIRT2 is the principal regulatory factor of ROS production by mitochondria [49, 52]. At present, the relationship between SIRT2 and the mtUPR has not been confirmed in the relevant literature, and the specific molecular mechanism is not clear. However, findings of increases in Parkin and Parkin 1 (PINK1) in SIRT2 knockout mice indicate that SIRT2 is associated with mitochondrial PQC [51] . In addition, studies have shown that SIRT2 is involved in mitochondrial stress; for example, SIRT2 can regulate PGC-1α, which can protect cells from oxidative stress-induced apoptosis by inhibiting mitochondrial autophagy [50, 52]. SIRT2 can also bind to FOXO3A and deacetylate it, which increases the transcriptional activity of FOXO3A and ultimately upregulates the expression of target genes such as manganese-dependent superoxide dismutase (MnSOD), Bcl-2-interacting mediator of cell death (Bim) and Cyclin-dependent kinase inhibitor 1B. These changes in turn reduce ROS production [49, 53]. One way to inhibit SIRT2 is by regulating mitochondrial biogenesis and kinetics via downregulation of transcription factor A (TFAM) and mitofusin-2 (Mfn2) and upregulation of dynamin-related protein 1 (Drp1) [54]. This evidence indicates that SIRT2 may play an important role in the mtUPR and can provide new ideas for further exploring the relationship between SIRT2 and the mtUPR.
3.3. SIRT3 AND THE MTUPR
SIRT3 is the main deacetylase in mitochondria and is abundant in the liver, heart, brain, and brown adipose tissue. It not only regulates the activity of metabolic enzymes but also promotes mitochondrial function by regulating mitochondrial autophagy, the mtUPR, and optic atrophy 1 (OPA1)-dependent mitochondrial dynamics [26, 55]. Acetylation of HSP10, a molecular chaperone that controls the folding of mitochondrial proteins, is increased in SIRT3 knockout mice. Therefore, SIRT3 agonists [56] can reduce the degree of acetylation. Compared with wild-type mice, SIRT3 knockout mice have 35% lower misfolded protein levels [57]. Protein cytotoxic stress can induce SIRT3 expression. Deletion of SIRT3 decreases mitochondrial membrane potential and cytotoxic stress activity and increases mitochondrial protein aggregation [57, 58]. In breast cancer, the expression of SIRT3 is decreased significantly, increasing ROS levels. Increased ROS activates the mtUPR by upregulating mitochondrial chaperone proteins [57, 58]. In aged hematopoietic stem cells (HSCs), the mitigating effects of the mitochondrial protection mechanism are related to decreased SIRT3 expression, which may be related to dysfunction of the SIRT3-dependent mtUPR pathway [22]. These findings indicate that SIRT3 is the key coordinator of the mtUPR induced by mitochondrial protein cytotoxic stress. Normally, the accumulation of ROS in the matrix is controlled by two important proteins, superoxide dismutase (SOD) and SIRT3 [38]. SIRT3 activated by the toxic stress of mitochondrial proteins can deacetylate FOXO3A, leading to its translocation to the nucleus. The activation of FOXO3A increases the expression of target genes such as PGC-1α, SOD2, MnSOD, and catalase and reduces the production of ROS. In general, this process participates in the antioxidant mechanism and the expression of mitochondrial autophagy genes. Such effects make up the SIRT3/FOXO3A axis of the mtUPR [53, 59], which is found in C. elegans [60]. The direct association between FOXO3A and SIRT3 in mitochondria is controversial. At present, there is little evidence that FOXO3A exists in mitochondria; therefore, the effect of SIRT3 on FOXO3A may be indirect [58]. In primary breast cancer cases, activation of the SIRT3/FOXO3A/SOD2 axis of the mtUPR has demonstrated that NRF1 is necessary for invasion and metastasis [61]. In addition, trans sodium crocetinate alleviates ischemia/reperfusion-induced myocardial oxidative stress and apoptosis via the SIRT3/FOXO3A/SOD2 signaling pathway [62]. Inhibition of SIRT3 does not affect the expression levels of LONP1, HSP10 or HSP60. The CHOP axis and the SIRT3/FOXO3A axis of the mtUPR are independent. They can simultaneously cope with the toxic stress of matrix proteins and control protein quality [11]. The major antioxidant enzyme SOD2 in the mitochondrial matrix is the downstream target gene of SIRT3. Relevant studies have revealed that upregulation of SOD2 expression is consistent with changes in SIRT3. Therefore, SIRT3 directly controls the activity of SOD2 by deacetylation. Inadequate deacetylation of SOD2 by SIRT3 will lead to ROS accumulation and mitochondrial damage, thus inducing the mtUPR [60]. For example, SOD2 deacetylation mediated by SIRT3 can regulate ROS production and mitochondrial function in endothelial progenitor cells, thereby improving vascular endothelial repair capacity in hypertension [63]. The specific induction of SOD2 can eliminate excess mitochondrial superoxide, and deacetylation of Lys68 can increase the expression of SIRT3 and the activity of SOD2. In contrast, knockdown of SOD2 and SIRT3 results in inhibition of osteoblast differentiation. Therefore, the SIRT3/SOD2 axis is necessary for mitochondrial stress regulation [64]. Furthermore, accumulation of SOD1 containing copper and zinc ions in the catalytic core in the IMS can activate the mtUPR. This process works with the mtUPR ERα pathway in IMS to resist mitochondrial damage. Opposite trends in SOD1 and SIRT3 have been observed in MCF10A cells [65], as have been found in both breast cancer cell lines and nonmalignant cell lines. The SIRT3/SOD1 axis in the mtUPR may thus be a new target to control tumors. Notably, AMPK is a key molecule in the regulation of bioenergetic metabolism, and choline attenuates cardiac hypertrophy via SIRT3 upregulation. Downregulation of SIRT3 or AMPK eliminates the ability of choline to activate the mtUPR, which indicates that the SIRT3/AMPK axis in the mtUPR plays a beneficial role in restoring the structure and function of mitochondria. This process represents a unique cellular mechanism by which cells adapt to changes in metabolic needs [66].
3.4. SIRT4, SIRT5, AND SIRT6 AND THE MTUPR
Compared with other SIRT members, SIRT4/5/6 have been rarely studied in the context of the mtUPR. SIRT4 is negatively regulated during the maintenance of normal mitochondrial function [67] and is involved in the regulation of ROS production in mitochondria. SIRT4 inhibits the binding of SOD2 and SIRT3, increasing acetylation and thereby reducing the activity of SOD2. Thus, SIRT4 is a very promising target in the mtUPR [49]. Large amounts of SIRT5 accumulate in the IMSs of mitochondria in the brain, heart, liver, and lymphoblasts. Recently, the estrogen receptor and mitochondrial matrix-localized SIRT5 have also been found to play roles in mtUPR regulation in response to unfolded protein accumulation in the IMS [68]. Since SIRT5 can deacetylate FOXO3A to promote SOD2 expression, similar to SIRT3 in the SIRT3/FOXO3A axis, this antioxidant may play a role in protecting lung epithelial cells from cigarette smoke extract-induced apoptosis [69]. SIRT5 inhibits lung cancer cell growth by affecting SOD1 to mediate reductions in ROS, which may be a SIRT5-mediated mtUPR mechanism. Furthermore, SIRT6 overexpression has been shown to protect cardiomyocytes from ischemia/reperfusion injury through the energy-sensitive AMPK-FOXO3A axis, which is required for oxidative stress resistance [14]. Therefore, SIRT6 may be critical in redox and antioxidant homeostasis and may be involved in the mtUPR. Similarly, SIRT6 has been shown to protect human mesenchymal stem cells (hMSCs) from oxidative stress through coactivation of NRF2 [50].
3.5. SIRT7 AND THE MTUPR
SIRT7 is also a nucleoprotein, and it is located mainly in the nucleolus. SIRT7 has NAD+-dependent deacetylating enzyme activity, and its catalytic activity is related to the His187 residue. SIRT7 can deacetylate histone H3 Lys18 on part of the gene promoter and interact with various nonhistone proteins, including U3 small nuclear riboprotein factor 55K, GA binding protein β1 (GABPβ1), p53, Nucleophosmin 1, DNA damage-binding protein 1, etc. SIRT7 is of great significance in RNA polymerase transcription, ribosomal synthesis, cellular stress and metabolism, genome stability, mitochondrial function maintenance and mitochondria-related disease prevention. The deletion of SIRT7 in mice can cause a wide range of types of mitochondrial dysfunction, resulting in different degrees of injury to multiple organs and tissues [70]. Treating cells with an mtUPR inducer upregulates the expression of SIRT7 in response to stress. SIRT7 has been shown to coordinate the correct stoichiometric ratio of nDNA and mtDNA-encoded mtDNA proteins. When the intracellular protein mass balance is disrupted, mtDNA protein stress initiates the mtUPR [71]. SIRT7 promotes the apparent genetic stability of cancer-related gene expression programs by regulating the reaction of nuclear-encoded mtDNA genes and mtDNA unfolded proteins and modulates the homeostasis of mtDNA [72]. ATF5 homologous substances in Rhizoma japonicum play essential roles in the mtUPR. Luciferase reporter gene assays have revealed that ATF5 can activate transcription driven by the SIRT7 promoter [71]. Mohrin and colleagues first linked SIRT7 to the mtUPR in HSCs. They found that HSCs inhibited mitochondrial biosynthesis and oxidative phosphorylation through the mtUPR signaling pathway to coordinate the metabolism of stem cells. Inhibiting the expression of SIRT7 in HSCs can activate the mtUPR, leading to decreased mitochondrial biogenesis and impaired HSC regeneration, whereas increasing the levels of SIRT7 in aged stem cells reduces mitochondrial protein toxicity and improves differentiation potential, which suggests that SIRT7 is the primary regulator of mitochondrial homeostasis and is involved in mtUPR processing. ChIP-seq analysis has shown that SIRT7 binds not only to the promoter of the ribosomal subunit but also to the promoters of many mitochondrial ribosomal subunits and mitochondrial translation proteins. In addition, immunoprecipitation and mass spectrometry of 293T cells transfected with SIRT7-Flag have shown that NRF1 is one of the proteins pulled down by SIRT7-Flag. NRF1 knockdown experiments performed to determine whether SIRT7 is targeted at the mtDNA promoter through NRF1 have revealed that knockdown significantly reduces the enrichment at the promoter of SIRT7 but does not reduce the enrichment of other SIRT7 targets [73-76]. In particular, NRF1 explicitly mediates the binding of SIRT7 and the mitochondrial promoter [79]. The complexity of the mtUPR is reflected in the toxic stress of the mtUPR, which also induces the expression of NRF1 [77, 78]. Although the loss of mitochondrial protein homeostasis triggers the mtUPR, the interaction between SIRT7 and NRF1 downregulates mitochondrial stress according to the SIRT7/NRF1 axis of the mtUPR.
GABPβ1 is the primary regulator of nuclear coding for mtDNA genes. Recent studies have revealed that SIRT7 is the most closely related to GABP among all transcription factors that regulate mitochondrial biosynthesis and function. A whole-genome ChIP-seq analysis has shown that a complex formed by SIRT7 and GABP acts at the promoters of the nuclear-encoded mitochondrial genes, controlling the expression of multiple MRPs and maintaining the mitochondrial steady state [81]. The effect of SIRT7 on the mitochondrial steady state is mainly mediated by deacetylation of GABPβ1. The GABPα complex forms in the ETS domain and with transcriptional activation of the GABPα/GABPβ heterotetramer complex [79]. Dynamic signal changes in the SIRT7/GABPβ1 axis in the mtUPR are essential for various cell stresses, the maintenance of mitochondrial homeostasis, and the repair of various types of mitochondrial dysfunction. Although the potential role of SIRT7 in mtUPR induction is not very clear, SIRT7 may be essential to mitochondrial protein homeostasis. Because of its primary effects on MRPs, its absence can induce the mtUPR [6].
Conclusion and future directions
In this review, we summarize the relationships between SIRTs and the mtUPR as well as the molecular mechanisms underlying these relationships. This knowledge will not only deepen our understanding of diseases related to mitochondrial dysfunction but also clarify new molecular targets for controlling the occurrence and development of these diseases and for developing related inhibitors and agonists as potential treatments for cancer, metabolic diseases, and neurodegenerative diseases. To date, there are still many uncertain mechanisms of the SIRT-mediated mtUPR, and more complex interactions need to be further explored and verified in additional models. Future research will surely promote progress in the diagnosis and treatment of cancer, metabolic diseases, and neurodegenerative diseases.
Fig. (2).
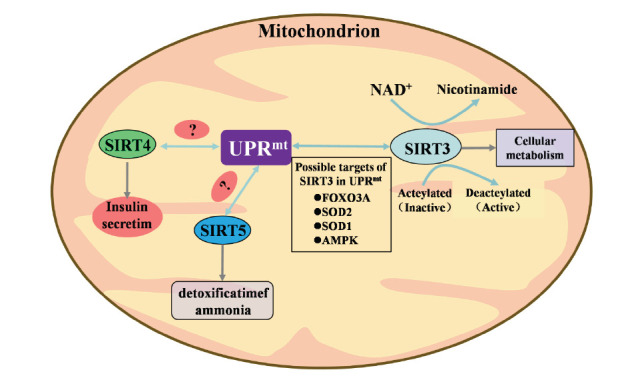
Overview of the correlation between the species, distribution, and function of SIRT3, SIRT4, SIRT5, and mtUPR reflect in mitochondrial. The current targets for SIRT4 and SIRT5 in sirtuin and m tUPR are unclear. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (3).
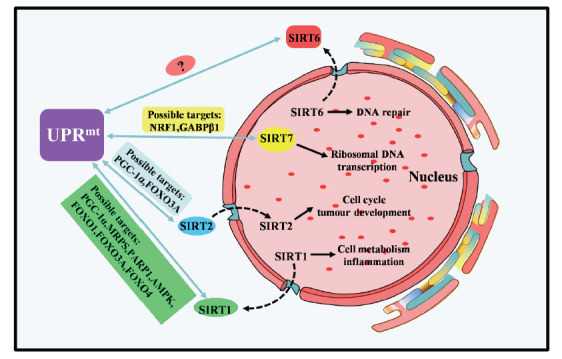
Overview of the species, distribution, and function of sirtuin protein (SIRT1, SIRT2, SIRT6, SIRT7) showed in the nucleus and its correlation with mtUPR.The current targets for SIRT6 in sirtuin and mtUPR are unclear. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Table 1.
Classification, distribution, target, function, and Homologs in C.elegans, yeast, Drosophila melanogaster, which obtained of the sirtuin protein family.
|
Sirtuin Protein
Classification of Mammalian |
Homologs of
Mammalian [ 80 , 81 ] |
Intracellular Location [ 81 ] | Enzymatic Activity [ 81 ] | Substrates/targets [ 82 - 93 ] | Possible Targets of Sirtuin in mtUPR [ 19 , 58 , 70 , 71 ] | Function [ 22 ] |
|---|---|---|---|---|---|---|
| SIRT1 | sir-2.1 in Caenorhabditis elegans SIR2 in yeast (SIR2 homologs in yeast include Hst1, Hst2, Hst3 and Hst4) SIRT1 in Drosophila melanogaster |
Cell nucleus; Cytoplasm |
Deacetylase | P53, H3, H4, H1, P300, PGC-1α, FOXO, HIF-1α, NF-κB, Hi-2α, MYC, PPARγ, PCAF, AceCS1, PGAM-1 | PGC-1α, MRPS, PARP1, AMPK, FOXO1, FOXO3A, FOXO4 |
Cell metabolism, Inflammation |
| SIRT2 | sir-2.1 in Caenorhabditis elegans SIRT2 in Drosophila melanogaster |
Cell nucleus, Cytoplasm |
Deacetylase | FOXO3A, EIF5A, P53, G6PD, MY, Tubulin, FOXO, H4K16 |
PGC-1α, FOXO3A | Cell cycle, Tumor development |
| SIRT3 | sir-2.1 in Caenorhabditis elegans | Mitochondrion | Deacetylase | FOXO3A, SOD2, PDMC1a, IDH2, GOT2, OTC, AceCS1, HMG-COA, Synthase2, LCAD, GDH, SDH | FOXO3A, SOD2, SOD1, AMPK |
Cell metabolism |
| SIRT4 | sir-2.1 in Caenorhabditis elegans SIRT4 in Drosophila melanogaster |
Mitochondrion | ADP-ribosyltransferase | GDH, PDH | Unknown | Insulin secretion |
| SIRT5 | sir-2.1 in Caenorhabditis elegans | Mitochondrion | Deacetylase, Desuccinylase, Demalonylase |
CPS1 | Unknown | Detoxification of ammonia |
| SIRT6 | sir-2.1 in Caenorhabditis elegans sir-2.4 in Caenorhabditis elegans SIRT6 in Drosophila melanogaster |
Cell nucleus | Deacetylase, ADP-ribosyltransferase | H3K9, H3K56, CtlP, PARP1, NF-κB, HIF-1α, DNA-PK, PPARγ | Unknown | DNA repair processes, Cell metabolism, TNF factor secretion |
| SIRT7 | sir-2.1 in Caenorhabditis elegans sir-2.4 in Caenorhabditis elegans SIRT7 in Drosophila melanogaster |
Nucleolus | Deacetylase | HIF-1α, Hi-2α | NRF1, GABPβ1 | Ribosomal DNA transcription |
Acknowledgements
Declared none.
Consent for Publication
Not applicable.
Funding
The author's research was supported by the National Natural Science Foundation of China. (Grant No: 81671265)and the Natural Science Foundation of the Fujian Province, China (Grant No. 2016Y0043).
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Hamon M. P., Bulteau A. L., Friguet B. Mitochondrial proteases and protein quality control in ageing and longevity. 2015. [DOI] [PubMed]
- 2.Liang R., Ghaffari S. Mitochondria and FOXO3 in stem cell homeostasis, a window into hematopoietic stem cell fate determination. J. Bioenerg. Biomembr. 2017;49(4):343–346. doi: 10.1007/s10863-017-9719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naresh N.U., Haynes C.M. Signaling and Regulation of the Mitochondrial Unfolded Protein Response. Cold Spring Harb. Perspect. Biol. 2019;11(6):a033944. doi: 10.1101/cshperspect.a033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreux P.A., Houtkooper R.H., Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 2013;12(6):465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mora A.L., Bueno M., Rojas M. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J. Clin. Invest. 2017;127(2):405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jovaisaite V., Auwerx J. The mitochondrial unfolded protein response—synchronizing genomes. Curr. Opin. Cell Biol. 2015;33:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Münch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018;16(1):81. doi: 10.1186/s12915-018-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldridge J.E., Horibe T., Hoogenraad N.J. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2(9):e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papa L., Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J. Cell Sci. 2011;124(Pt 9):1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny T.C., Germain D. From discovery of the CHOP axis and targeting ClpP to the identification of additional axes of the UPRmt driven by the estrogen receptor and SIRT3. J. Bioenerg. Biomembr. 2017;49(4):297–305. doi: 10.1007/s10863-017-9722-z. [DOI] [PubMed] [Google Scholar]
- 12.Kang B.H., Plescia J., Song H.Y., Meli M., Colombo G., Beebe K., Scroggins B., Neckers L., Altieri D.C. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J. Clin. Invest. 2009;119(3):454–464. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein S.H., Venkatesh S., Li M., Lee J., Lu B., Hilchey S.P., Morse K.M., Metcalfe H.M., Skalska J., Andreeff M., Brookes P.S., Suzuki C.K. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 2012;119(14):3321–3329. doi: 10.1182/blood-2011-02-340075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon J., Kim H.R., Shin M.G. Rejuvenating aged hematopoietic stem cells through improvement of mitochondrial function. Ann. Lab. Med. 2018;38(5):395–401. doi: 10.3343/alm.2018.38.5.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerrer B., Gertler A.A., Cohen H.Y. The complex role of SIRT6 in carcinogenesis. Carcinogenesis. 2016;37(2):108–118. doi: 10.1093/carcin/bgv167. [DOI] [PubMed] [Google Scholar]
- 16.Wątroba M., Dudek I., Skoda M., Stangret A., Rzodkiewicz P., Szukiewicz D. Sirtuins, epigenetics and longevity. Ageing Res. Rev. 2017;40:11–19. doi: 10.1016/j.arr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Rose G., Santoro A., Salvioli S. Mitochondria and mitochondria-induced signalling molecules as longevity determinants. BMC Biol. 2017;16(1):81. doi: 10.1016/j.mad.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Dahlmans D., Houzelle A., Schrauwen P., Hoeks J. Mitochondrial dynamics, quality control and miRNA regulation in skeletal muscle: implications for obesity and related metabolic disease. Clin. Sci. (Lond.) 2016;130(11):843–852. doi: 10.1042/CS20150780. [DOI] [PubMed] [Google Scholar]
- 19.Mendelsohn A.R., Larrick J.W. The NAD+/PARP1/SIRT1 Axis in Aging. Rejuvenation Res. 2017;20(3):244–247. doi: 10.1089/rej.2017.1980. [DOI] [PubMed] [Google Scholar]
- 20.Gariani K., Menzies K.J., Ryu D., Wegner C.J., Wang X., Ropelle E.R., Moullan N., Zhang H., Perino A., Lemos V., Kim B., Park Y.K., Piersigilli A., Pham T.X., Yang Y., Ku C.S., Koo S.I., Fomitchova A., Cantó C., Schoonjans K., Sauve A.A., Lee J.Y., Auwerx J. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2016;63(4):1190–1204. doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jukarainen S., Heinonen S., Rämö J.T., Rinnankoski-Tuikka R., Rappou E., Tummers M., Muniandy M., Hakkarainen A., Lundbom J., Lundbom N., Kaprio J., Rissanen A., Pirinen E., Pietiläinen K.H., Obesity Is Associated With Low N.A.D. Obesity is associated with low nad(+)/sirt pathway expression in adipose tissue of bmi-discordant monozygotic twins. J. Clin. Endocrinol. Metab. 2016;101(1):275–283. doi: 10.1210/jc.2015-3095. [DOI] [PubMed] [Google Scholar]
- 22.O’Callaghan C., Vassilopoulos A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell. 2017;16(6):1208–1218. doi: 10.1111/acel.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Menzies K.J., Auwerx J. The role of mitochondria in stem cell fate and aging. Development. 2018;145(8):dev143420. doi: 10.1242/dev.143420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., Guarente L., Auwerx J. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai S., Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24(8):464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho E.H. SIRT3 as a Regulator of non-alcoholic fatty liver disease. J. Lifestyle Med. 2014;4(2):80–85. doi: 10.15280/jlm.2014.4.2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J.P., Chen R. Stressed SIRT7: facing a crossroad of senescence and immortality. Clin. Exp. Pharmacol. Physiol. 2015;42(6):567–569. doi: 10.1111/1440-1681.12423. [DOI] [PubMed] [Google Scholar]
- 28.Jensen M.B., Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20(2):214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai S.I., Guarente L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. Aging Mech. Dis. 2016;2:16017. doi: 10.1038/npjamd.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calabrese V., Cornelius C., Mancuso C., Pennisi G., Calafato S., Bellia F., Bates T.E., Giuffrida Stella A.M., Schapira T., Dinkova Kostova A.T., Rizzarelli E. Cellular stress response: a novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem. Res. 2008;33(12):2444–2471. doi: 10.1007/s11064-008-9775-9. [DOI] [PubMed] [Google Scholar]
- 31.Pillai V.B., Sundaresan N.R., Jeevanandam V., Gupta M.P. Mitochondrial SIRT3 and heart disease. Cardiovasc. Res. 2010;88(2):250–256. doi: 10.1093/cvr/cvq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirinen E., Cantó C., Jo Y.S., Morato L., Zhang H., Menzies K.J., Williams E.G., Mouchiroud L., Moullan N., Hagberg C., Li W., Timmers S., Imhof R., Verbeek J., Pujol A., van Loon B., Viscomi C., Zeviani M., Schrauwen P., Sauve A.A., Schoonjans K., Auwerx J. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab. 2014;19(6):1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z., Ye J. Inhibition of transcriptional activity of c-JUN by SIRT1. Biochem. Biophys. Res. Commun. 2008;376(4):793–796. doi: 10.1016/j.bbrc.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luna A., Aladjem M.I., Kohn K.W. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome Integr. 2013;4(1):6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikó E., Kovács T., Fodor T., Bai P. Methods to Assess the Role of Poly(ADP-Ribose) Polymerases in Regulating Mitochondrial Oxidation. Methods Mol. Biol. 2017;1608:185–200. doi: 10.1007/978-1-4939-6993-7_13. [DOI] [PubMed] [Google Scholar]
- 37.Cantó C., Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De I., Dogra N., Singh S. The mitochondrial unfolded protein response: role in cellular homeostasis and disease. Curr. Mol. Med. 2017;17(9):587–597. doi: 10.2174/1566524018666180308110130. [DOI] [PubMed] [Google Scholar]
- 39.Ge J., Zhang C., Sun Y-C., Zhang Q., Lv M-W., Guo K., Li J-L. Cadmium exposure triggers mitochondrial dysfunction and oxidative stress in chicken (Gallus gallus) kidney via mitochondrial UPR inhibition and Nrf2-mediated antioxidant defense activation. Sci. Total Environ. 2019;689:1160–1171. doi: 10.1016/j.scitotenv.2019.06.405. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann S., Costa A.C., Celardo I., Loh S.H., Martins L.M. Parp mutations protect against mitochondrial dysfunction and neurodegeneration in a PARKIN model of Parkinson’s disease. Cell Death Dis. 2016;7:e2166. doi: 10.1038/cddis.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan M., Tang C., Zhang Y., Cheng Y., Cai L., Chen X., Gao Y., Deng Y., Pan M. SIRT1/PGC-1α signaling protects hepatocytes against mitochondrial oxidative stress induced by bile acids. Free Radic. Res. 2015;49(8):935–945. doi: 10.3109/10715762.2015.1016020. [DOI] [PubMed] [Google Scholar]
- 42.Wang S., Wan T., Ye M., Qiu Y., Pei L., Jiang R., Pang N., Huang Y., Liang B., Ling W., Lin X., Zhang Z., Yang L. Nicotinamide riboside attenuates alcohol induced liver injuries via activation of SirT1/PGC-1α/mitochondrial biosynthesis pathway. Redox Biol. 2018;17:89–98. doi: 10.1016/j.redox.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Mihaela T., Savu D., Moisoi N. Intracellular and intercellular signalling mechanisms following DNA damage are modulated by pink1. Oxid. Med. Cell. Longev. 2018;2018:1–15. doi: 10.1155/2018/1391387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai P., Nagy L., Fodor T., Liaudet L., Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol. Metab. 2015;26(2):75–83. doi: 10.1016/j.tem.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Tang B.L. Sirt1 and the Mitochondria. Mol. Cells. 2016;39(2):87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price N.L., Gomes A.P., Ling A.J., Duarte F.V., Martin-Montalvo A., North B.J., Agarwal B., Ye L., Ramadori G., Teodoro J.S., Hubbard B.P., Varela A.T., Davis J.G., Varamini B., Hafner A., Moaddel R., Rolo A.P., Coppari R., Palmeira C.M., de Cabo R., Baur J.A., Sinclair D.A. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15(5):675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes A.P., Price N.L., Ling A.J.Y., Moslehi J.J., Montgomery M.K., Rajman L., White J.P., Teodoro J.S., Wrann C.D., Hubbard B.P., Mercken E.M., Palmeira C.M., de Cabo R., Rolo A.P., Turner N., Bell E.L., Sinclair D.A., Declining N.A.D. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155(7):1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitada M., Ogura Y., Monno I., Koya D. Sirtuins and type 2 diabetes: role in inflammation, oxidative stress, and mitochondrial function. Front. Endocrinol. (Lausanne) 2019;10:187–187. doi: 10.3389/fendo.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh C.K., Chhabra G., Ndiaye M.A., Garcia-Peterson L.M., Mack N.J., Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antioxid. Redox Signal. 2018;28(8):643–661. doi: 10.1089/ars.2017.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu G., Park S.H., Imbesi M., Nathan W.J., Zou X., Zhu Y., Jiang H., Parisiadou L., Gius D. Loss of NAD-dependent protein deacetylase sirtuin-2 alters mitochondrial protein acetylation and dysregulates mitophagy. Antioxid. Redox Signal. 2017;26(15):849–863. doi: 10.1089/ars.2016.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu W-N., Yang R-Z., Zheng H-L., Yu W., Zheng X-F., Li B., Jiang S-D., Jiang L-S. PGC-1α acts as an mediator of Sirtuin2 to protect annulus fibrosus from apoptosis induced by oxidative stress through restraining mitophagy. Int. J. Biol. Macromol. 2019;136:1007–1017. doi: 10.1016/j.ijbiomac.2019.06.163. [DOI] [PubMed] [Google Scholar]
- 53.Zhou S., Tang X., Chen H.Z. Sirtuins and insulin resistance. Front. Endocrinol. 2018;9:748. doi: 10.3389/fendo.2018.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D., Wu L., Jiang X., Yang L., Cheng J., Chen H., Hua R., Geng G., Yang L., Li Q. SIRT2 inhibition results in meiotic arrest, mitochondrial dysfunction, and disturbance of redox homeostasis during bovine oocyte maturation. Int. J. Mol. Sci. 2019;20(6):E1365. doi: 10.3390/ijms20061365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pehar M., Harlan B.A., Killoy K.M., Vargas M.R. Nicotinamide adenine dinucleotide metabolism and neurodegeneration. Antioxid. Redox Signal. 2018;28(18):1652–1668. doi: 10.1089/ars.2017.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomes P., Leal H., Mendes A.F., Reis F., Cavadas C. Dichotomous Sirtuins: Implications for drug discovery in neurodegenerative and cardiometabolic diseases. Trends Pharmacol. Sci. 2019;40(12):1021–1039. doi: 10.1016/j.tips.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Lu Z., Chen Y., Aponte A.M., Battaglia V., Gucek M., Sack M.N. Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. J. Biol. Chem. 2015;290(4):2466–2476. doi: 10.1074/jbc.M114.606228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papa L., Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell. Biol. 2014;34(4):699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srivastava S. Emerging therapeutic roles for NAD(+) metabolism in mitochondrial and age-related disorders. Clin. Transl. Med. 2016;5(1):25–25. doi: 10.1186/s40169-016-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenny T.C., Craig A.J., Villanueva A., Germain D. Mitohormesis primes tumor invasion and metastasis. Cell Rep. 2019;27(8):2292–2303.e6. doi: 10.1016/j.celrep.2019.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenny T.C., Hart P., Ragazzi M., Sersinghe M., Chipuk J., Sagar M.A.K., Eliceiri K.W., LaFramboise T., Grandhi S., Santos J., Riar A.K., Papa L., D’Aurello M., Manfredi G., Bonini M.G., Germain D. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis. Oncogene. 2017;36(31):4393–4404. doi: 10.1038/onc.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang G., Chen Y., Zhang H., Zhou W. Trans sodium crocetinate alleviates ischemia/reperfusion-induced myocardial oxidative stress and apoptosis via the SIRT3/FOXO3a/SOD2 signaling pathway. Int. Immunopharmacol. 2019;71:361–371. doi: 10.1016/j.intimp.2019.03.056. [DOI] [PubMed] [Google Scholar]
- 63.He J., Liu X., Su C., Wu F., Sun J., Zhang J., Yang X., Zhang C., Zhou Z., Zhang X., Lin X., Tao J. Inhibition of mitochondrial oxidative damage improves reendothelialization capacity of endothelial progenitor cells via sirt3 (sirtuin 3)-enhanced sod2 (superoxide dismutase 2) deacetylation in hypertension. Arterioscler. Thromb. Vasc. Biol. 2019;39(8):1682–1698. doi: 10.1161/ATVBAHA.119.312613. [DOI] [PubMed] [Google Scholar]
- 64.Gao J., Feng Z., Wang X., Zeng M., Liu J., Han S., Xu J., Chen L., Cao K., Long J., Li Z., Shen W., Liu J. SIRT3/SOD2 maintains osteoblast differentiation and bone formation by regulating mitochondrial stress. Cell Death Differ. 2018;25(2):229–240. doi: 10.1038/cdd.2017.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomez M., Germain D. Cross talk between SOD1 and the mitochondrial UPR in cancer and neurodegeneration. Mol. Cell. Neurosci. 2019;98:12–18. doi: 10.1016/j.mcn.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu M., Xue R.Q., Lu Y., Yong S.Y., Wu Q., Cui Y.L., Zuo X.T., Yu X.J., Zhao M., Zang W.J. Choline ameliorates cardiac hypertrophy by regulating metabolic remodelling and UPRmt through SIRT3-AMPK pathway. Cardiovasc. Res. 2019;115(3):530–545. doi: 10.1093/cvr/cvy217. [DOI] [PubMed] [Google Scholar]
- 67.Lin S., Xing H., Zang T., Ruan X., Wo L., He M. Sirtuins in mitochondrial stress: Indispensable helpers behind the scenes. Ageing Res. Rev. 2018;44:22–32. doi: 10.1016/j.arr.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Bergström S., Danielsson H., Samuelsson B. The enzymatic formation of prostaglandin E2 from arachidonic acid prostaglandins and related factors 32. Biochimica et Biophysica Acta (BBA) -. General Subjects. 1964;90(1):207–210. doi: 10.1016/0304-4165(64)90145-X. [DOI] [PubMed] [Google Scholar]
- 69.Zhou L., Wang F., Sun R., Chen X., Zhang M., Xu Q., Wang Y., Wang S., Xiong Y., Guan K-L., Yang P., Yu H., Ye D. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. 2016;17(6):811–822. doi: 10.15252/embr.201541643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryu D., Jo Y.S., Lo Sasso G., Stein S., Zhang H., Perino A., Lee J.U., Zeviani M., Romand R., Hottiger M.O., Schoonjans K., Auwerx J.A. SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab. 2014;20(5):856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 71.Li L., Shi L., Yang S., Yan R., Zhang D., Yang J., He L., Li W., Yi X., Sun L., Liang J., Cheng Z., Shi L., Shang Y., Yu W. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 2016;7:12235–12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang E.F., Scheibye-Knudsen M., Chua K.F., Mattson M.P., Croteau D.L., Bohr V.A. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 2016;17(5):308–321. doi: 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C.M., Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347(6228):1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohrin M., Chen D. The mitochondrial metabolic checkpoint and aging of hematopoietic stem cells. Curr. Opin. Hematol. 2016;23(4):318–324. doi: 10.1097/MOH.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Y., Cheung H.H., Tu J., Miu K.K., Chan W.Y. New insights into the unfolded protein response in stem cells. Oncotarget. 2016;7(33):54010–54027. doi: 10.18632/oncotarget.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ajami M., Pazoki-Toroudi H., Amani H., Nabavi S.F., Braidy N., Vacca R.A., Atanasov A.G., Mocan A., Nabavi S.M. Therapeutic role of sirtuins in neurodegenerative disease and their modulation by polyphenols. Neurosci. Biobehav. Rev. 2017;73:39–47. doi: 10.1016/j.neubiorev.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 77.Ocampo A., Izpisua Belmonte J.C. Stem cells. Holding your breath for longevity. Science. 2015;347(6228):1319–1320. doi: 10.1126/science.aaa9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He C., Hart P.C., Germain D., Bonini M.G. SOD2 and the mitochondrial upr: partners regulating cellular phenotypic transitions. Trends Biochem. Sci. 2016;41(7):568–577. doi: 10.1016/j.tibs.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang B.L. SIRT7 and hepatic lipid metabolism. Front. Cell Dev. Biol. 2015;3:1–1. doi: 10.3389/fcell.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiang W-C., Tishkoff D.X., Yang B., Wilson-Grady J., Yu X., Mazer T., Eckersdorff M., Gygi S.P., Lombard D.B., Hsu A-L.C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet. 2012;8(9):e1002948–e1002948. doi: 10.1371/journal.pgen.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haigis M.C., Guarente L.P. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 82.Lu S-P., Lin S-J. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim. Biophys. Acta. 2010;1804(8):1567–1575. doi: 10.1016/j.bbapap.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai H., Sinclair D.A., Ellis J.L., Steegborn C. Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol. Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wood J.G., Schwer B., Wickremesinghe P.C., Hartnett D.A., Burhenn L., Garcia M., Li M., Verdin E., Helfand S.L. Sirt4 is a mitochondrial regulator of metabolism and lifespan in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2018;115(7) Suppl. 1:1564–1569. doi: 10.1073/pnas.1720673115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fusco S., Maulucci G., Pani G. Sirt1: def-eating senescence? Cell Cycle. 2012;11(22):4135–4146. doi: 10.4161/cc.22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014;25(3):138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teng Y.B., Jing H., Aramsangtienchai P., He B., Khan S., Hu J., Lin H., Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lombard D.B., Tishkoff D.X., Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb. Exp. Pharmacol. 2011;206:163–188. doi: 10.1007/978-3-642-21631-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Y., Yan Y., Principe D.R., Zou X., Vassilopoulos A., Gius D. SIRT3 and SIRT4 are mitochondrial tumor suppressor proteins that connect mitochondrial metabolism and carcinogenesis. Cancer Metab. 2014;2:15–15. doi: 10.1186/2049-3002-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirschey M.D., Zhao Y. Metabolic Regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteomics. 2015;14(9):2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsai Y-C., Greco T.M., Cristea I.M. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell. Proteomics. 2014;13(1):73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H.C., Hao Q., Lin H. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kupis W., Pałyga J., Tomal E., Niewiadomska E. The role of sirtuins in cellular homeostasis. J. Physiol. Biochem. 2016;72(3):371–380. doi: 10.1007/s13105-016-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


