Abstract
Francisella tularensis subsp. holarctica is a select agent causing life-threatening tularemia. It has been isolated from humans and animals, mainly lagomorphs and rodents, rarely other wild carnivore species. Increasing numbers of human tularemia cases have been reported during the last 5 years in Switzerland. Here we report the first isolation of Francisella tularensis subsp. holarctica from a domestic cat in Europe and compare its genome sequence with other Swiss isolates. The cat isolate shows a close phylogenetic relationship with a contemporary hare isolate from close geographic proximity, indicating a possible epidemiological link.
Keywords: bacteriuria, cat, feline, Switzerland, tularemia, zoonosis
Introduction, methods and results
Tularemia is a zoonotic disease caused by Francisella tularensis, which comprises the four subspecies tularensis, holarctica, novicida and mediasiatica. The most virulent subspecies are tularensis and holarctica, which have been reported from North America and the whole Northern hemisphere, respectively [1]. The bacterium has a broad host range including different vertebrate groups as well as invertebrates [2]. Transmission occurs via inhalation of infected droplets, contact with infected animals, bites of arthropod vectors or oral uptake [3]. The isolation of the tularemia agent from European carnivores is restricted to isolated cases in free ranging wildlife. Reported cases include a stone marten in Switzerland (2012) [4], a raccoon dog (2012) and a red fox (2008) in Germany [5]. Regarding domestic carnivores, tularemia sporadically occurs in cats in North America where it is almost exclusively caused by subspecies tularensis [6]. In North America, disease occurrence in cats is considered an important factor for humans to contract tularemia [6]. Here we describe the unexpected isolation of F. tularensis subsp. holarctica from the urine of a domestic cat in Switzerland and compare the isolate to contemporary isolates from Swiss hares as well as previously published Swiss isolates [7] from wildlife, humans and ticks.
In March 2019, a 9 year-old male neutered outdoor cat was presented to the Small Animal Clinic of the University of Bern (SAC) for the routine 3-monthly maintenance flush of his bilateral subcutaneous ureteral bypass (SUB) placed 4 years ago. All cultures performed on previous rechecks had been negative. The owners reported no abnormalities and the clinical exam was unremarkable except for a reduced body condition score of 3/9 and a weight loss of 430 g over 3 months (actual body weight 3.47 kg). The cat had concomitant chronic kidney disease International Renal Interest Society (IRIS) stage 3 with a blood creatinine of 298 µmol/L (reference range: 52–138) and urea 26 mmol/L (reference range: 6.5–12.2).
Both sites of the SUB ports -were prepared aseptically and a urine sample was taken under sterile conditions by puncture of the ports chambers with a Huber point needle (Norfolk Vet Products, Skokie, USA). Urine analysis was performed and revealed a specific gravity of 1.012, pH 5.1, hematuria, pyuria, and no visible bacteria on the sediment examination. The rest of the sample was submitted for routine culture at 37 °C on Trypticase Soy Agar II with 5% Sheep Blood (BD, Heidelberg, Germany) to detect bacteria able to cause cystitis. No bacterial colonies had grown after 24 h of incubation, however confluent growth was observed after 96 h and the colonies were identified as F. tularensis by MALDI-TOF (Bruker, Bremen Germany) using an in-house database. Since this was a highly unusual finding, the treating veterinarian was asked to submit a second sample, which was subjected to culture and Real-time PCR [8], both of which were again positive for F. tularensis.
The cat was treated with doxycycline 50 mg once daily for 3 weeks and urine samples for PCR were again collected by puncture of the SUB ports in June, August and September 2019 and again in June 2020. Starting from June 2019, culture results were negative, however PCR remained positive until September 2019 albeit with lower Ct-values (Table 1). Treatment with doxycycline had been restarted mid August 2019 for a total of 2 months due to persistent positive PCR and concern for possible zoonotic risk for the owners. A serum sample was obtained from the cat on 20-Jun-19 (6 weeks after initial urine sampling) and sent to the FLI Friedrich-Löffler-Institut (Jena, Germany) for a microagglutination Test, which resulted in a low positive titer (Table 1).
Table 1.
Diagnosis of tularemia based on urine samples and serum from a domestic cat.
| Sampling date | Culture | qPCR (Ct value) | Serology |
|---|---|---|---|
| May 9, 2019 | Positive | ND | ND |
| May 14, 2019 | Positive | Positive (28) | ND |
| June 20, 2019 | Negative | Positive (28) | 1:20 |
| August 8, 2019 | Negative | Positive (38) | ND |
| September 11, 2019 | Negative | Positive (36) | ND |
| June 2, 2020 | Negative | Negative (> 40) | ND |
To investigate a possible epidemiological relationship with wildlife, the cat isolate and hare isolates derived from cases necropsied in 2019 (Table 2) were subjected to whole genome sequencing and SNP analysis. Illumina 150 bp paired-read sequencing was performed by Eurofins Genomics Germany GmbH (Ebersberg, Germany). On top of the Illumina sequencing PacBio sequencing was performed by the Lausanne Genomics Technologies Facility (GTF) (Lausanne, Switzerland) aiming to make the analyses more accurate. Ambiguous sites were resolved by Sanger sequencing (Microsynth AG, Balgach Switzerland) (primers specified in Additional file 1). The sequence data was submitted to the Sequence Read Archive: PRJNA645814.
Table 2.
Swiss Francisella tularensis subsp. holarctica isolates included in the analyses.
| Sampling date | Isolate ID | Specimen | Host | CanSNP clade |
|---|---|---|---|---|
| March 20, 2019 | 19OD0470 | Liver | Hare | B.45 |
| April 4, 2019 | 19OD0551 | Liver | Hare | B.92 |
| April 11, 2019 | 19OD0587 | Liver | Hare | B.47 |
| April 16, 2019 | 19OD0665 | Liver | Hare | B.61 |
| April 17, 2019 | 19OD0695 | Liver | Hare | B.61 |
| April 18, 2019 | 19OD0700 | Liver | Hare | B.47 |
| April 26, 2019 | 19OD0758 | Spleen | Hare | B.45 |
| May 7, 2019 | 19OD0847 | Liver | Hare | B.61 |
| May 9, 2019 | 19KM1151 | Urine | Cat | B.53 |
| May 14, 2019 | 19KM1164 | Urine | Cat | B.53 |
| May 14, 2019 | 19OD0886 | Liver | Hare | B.45 |
| May 15, 2019 | 19OD0902 | Liver | Hare | B.45 |
| May 29, 2019 | 19OD0988 | Liver | Hare | B.53 |
| June 13, 2019 | 19OD1266 | Liver | Hare | B.46 |
The cat isolates as well as the hare isolate of the same CanSNP clade are marked in italic.
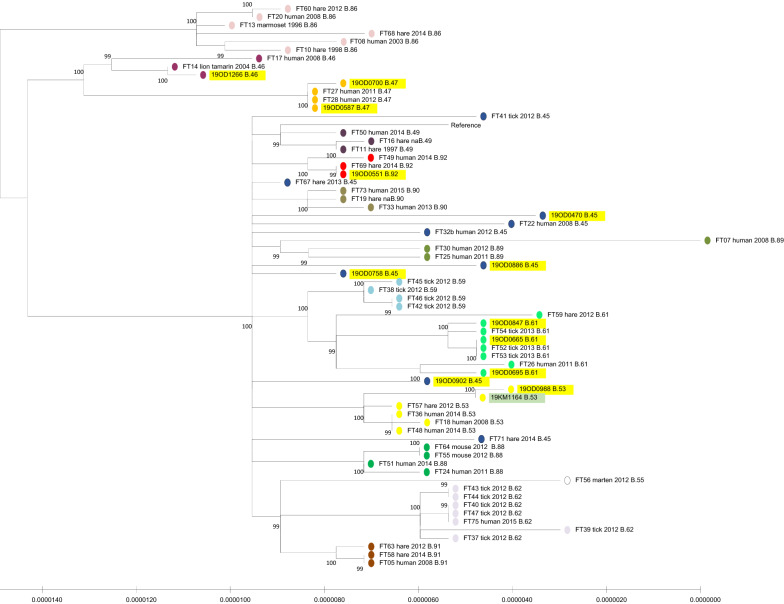
Hybrid assemblies with Illumina and PacBio reads were generated using unicycler 0.4.4 [9]. For SNP analyses the snippy 4.4.5 pipline [10] was applied using F. tularensis subsp. holarctica FTNF002-00 (NC_009749.1) as reference. Furthermore, assemblies of strains from Swiss wildlife, humans and ticks described earlier [7] were downloaded from Genbank and included for comparison. In order to generate phylogenetic trees, at first the best evolutionary model was determined with the program ModelTest-NG 0.1.3 [11], which was followed by tree creation with PhyML 3.3.20180214 [12]. In order to classify isolates in the canonical SNP framework, the program CanSNper 1.0.8 [13] was used. The cat strain was shown to belong to the CanSNP B.53 clade, which is part of the B.11 cluster, the most prevalent in Switzerland [7]. It was very closely related to 19OD0988 isolated in the same month from a brown hare in a neighboring geographical region (Figures 1 and 2).
Figure 1.
Maximum likelihood tree estimated using PhyML 3.3.20180214. Hare strains from the current study are marked in yellow and the cat strain is shown in green. Strains from Wittwer et al. [7] are included for comparison. CanSNP clade B.33 was used as an outgroup (not shown). Only the B.11 cluster is shown, subclades are marked with different colors and indicated for each strain.
Figure 2.
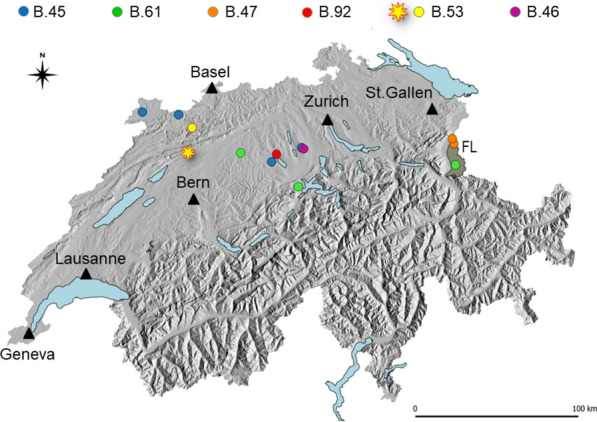
Map of Switzerland depicting the geographical origin of the investigated animals (hares as circles, cat as a star), with landscape relief (shades of grey), main lakes (pale blue), main cities (black triangles) and the neighboring Principality of Liechtenstein (FL, homogenous grey area). Different colors attributed to the animals correspond to the identified CanSNP clusters.
Discussion
Tularemia is a life-threatening zoonotic disease if untreated, which has been increasingly reported in humans in several European countries [3]. In Switzerland, the incidence of tularemia in humans has risen from 0.18/100 000 in 2010 to 1.72 in 2019 [14]. A large outbreak was observed in Sweden in 2019 [15] and in Germany tularemia is also considered a re-emerging disease [16]. A higher frequency of tularemia cases can be related to more sensitive diagnostic procedures, increasing surveillance or spread of the pathogen. Over the last years, Francisella, has also been increasingly isolated from small rodents and hares in Switzerland [17] even though the trend is not as clear as for humans. A higher prevalence in wildlife might also favor spillover events to domestic animals. Since small rodents are part of the diet of domestic cats, oral infection is possible. This is a plausible hypothesis given the fact that the here described cat was reported to be an active hunter. Another possibility would be transmission via ticks, which have been shown to harbor the same isolates as hares and humans [7].
The cat isolate showed a close phylogenetic relationship to a contemporary isolate from a hare that was found in geographical vicinity. This fact supports an epidemiological link between these cases, either directly (contact with small rodents, arthropod vectors) or indirectly (environmental contamination).
Even though, to the best of our knowledge, feline tularemia has not been previously reported in Europe, there are reports of human tularemia following cat bites [18, 19]. This might indicate a transmission via saliva or an undiagnosed infection of the cat. The case described here is also unique in its unexpected clinical presentation as subclinical bacteriuria. However, urine of cats is not routinely analyzed for Francisella and thus previous cases may have been missed due to the fastidious nature and slow growth of the organisms. It remains unclear as to whether infection contributed to the weight loss observed in this cat in the absence of further disease signs. Tularemia in cats caused by subspecies tularensis has been described to manifest as a systemic illness with fever, apathy, lymphadenopathy, hepato-splenomegaly or as oral ulcers after ingestion of infected rodents [20, 21]. While tularemia can cause lesions in the renal pelvis as part of a systemic infection in hares [22], it is not known to occur solely as a urinary tract infection or colonization.
The cat quickly became culture negative after treatment with doxycycline even though increased minimal inhibitory concentrations and treatment failures have been described for Francisella [23]. The positive PCR results over a long time may be due to persistence of dead organisms or DNA in the SUB system.
In 2014, the first case of tularemia caused by subspecies holarctica in a European gun dog was reported from Norway [24]. The dog and his owner had been exposed to an infected mountain hare during hunting and subsequently both dog and owner developed clinical tularemia [24]. A 2018 study in Austrian hunting dogs found a seroprevalence of 6% with most dogs showing no signs of tularemia [25]. In the present cat case, fortunately, none of the exposed humans including owner and veterinary staff were clinically affected, however, no testing was performed. Nevertheless, shedding of Francisella in urine poses a considerable risk for owners, veterinary staff and other people through the contamination of the environment. Most veterinary diagnostic laboratories are not prepared to isolate and identify F. tularensis from urine. Due to its classification as a select agent, it is not present in the standard MALDI-TOF databases used by many veterinary laboratories. The inclusion of F. tularensis in the standard MALDI-TOF database would foster detection of infections in unusual hosts and overall detection of the pathogen, resulting in a better picture of its geographical distribution and host range.
In conclusion, this is the first report of tularemia in a domestic cat in Europe and of a presentation as subclinical bacteriuria. Future studies in regions with increased tularemia should include domestic carnivores as potential source of infection for humans. Furthermore, urine should be considered a possible source of F. tularensis, even when originating from asymptomatic animals.
Supplementary information
Additional file 1. List of primers used for Sanger sequencing.
Acknowledgements
We thank Hatice Akarsu Egger for helpful discussions concerning bioinformatics analyses. We want to acknowledge Stefanie Ackermann and Stefanie Müller for performing the bacteriological cultures on the cat urine samples and Simon Feyer for performing the PCRs. We would also like to thank Estelle Gohl for culturing the hare samples. We thank PD Dr. Herbert Tomaso, head of the NRL for tularemia at the FLI, for the serology. Calculations were performed on UBELIX (https://www.id.unibe.ch/hpc), the HPC cluster at the University of Bern.
Abbreviations
- IRIS
International Renal Interest Society
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- SUB
subcutaneous ureteral bypass
Authors’ contributions
JJ and SK designed the study and drafted the manuscript. SK analysed the data. All authors contributed substantially to the content and reviewed or edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the University of Bern and the Swiss Federal Food Safety and Veterinary Office. Wildlife health surveillance in Switzerland is supported by the Federal Office for the Environment and the Federal Food Safety and Veterinary Office.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the Genbank repository under Bioproject accession number PRJNA645814.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sonja Kittl, Email: sonja.kittl@vetsuisse.unibe.ch.
Thierry Francey, Email: thierry.francey@vetsuisse.unibe.ch.
Isabelle Brodard, Email: isabelle.brodard@vetsuisse.unibe.ch.
Francesco C. Origgi, Email: francesco.origgi@vetsuisse.unibe.ch
Stéphanie Borel, Email: stephanie.borel@vetsuisse.unibe.ch.
Marie-Pierre Ryser-Degiorgis, Email: marie-pierre.ryser@vetsuisse.unibe.ch.
Ariane Schweighauser, Email: ariane.schweighauser@vetsuisse.unibe.ch.
Joerg Jores, Email: joerg.jores@vetsuisse.unibe.ch.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13567-020-00834-5.
References
- 1.Larson MA, Sayood K, Bartling AM, Meyer JR, Starr C, Baldwin J, Dempsey MP. Differentiation of Francisella tularensis subspecies and subtypes. J Clin Microbiol. 2020 doi: 10.1128/JCM.01495-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilo P. Phylogenetic lineages of Francisella tularensis in animals. Front Cell Infect Microbiol. 2018;8:258. doi: 10.3389/fcimb.2018.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hestvik G, Warns-Petit E, Smith LA, Fox NJ, Uhlhorn H, Artois M, Hannant D, Hutchings MR, Mattsson R, Yon L, Gavier-Widen D. The status of tularemia in Europe in a one-health context: a review. Epidemiol Infect. 2015;143(10):2137–2160. doi: 10.1017/S0950268814002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Origgi FC, Wu N, Pilo P. Francisella tularensis infection in a stone marten (Martes foina) without classic pathological lesions consistent with tularemia. J Vet Diagn Invest. 2013;25(4):519–521. doi: 10.1177/1040638713489124. [DOI] [PubMed] [Google Scholar]
- 5.Schulze C, Heuner K, Myrtennas K, Karlsson E, Jacob D, Kutzer P, Große K, Forsman M, Grunow R. High and novel genetic diversity of Francisella tularensis in Germany and indication of environmental persistence. Epidemiol Infect. 2016;144(14):3025–3036. doi: 10.1017/S0950268816001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson MA, Fey PD, Hinrichs SH, Iwen PC. Francisella tularensis bacteria associated with feline tularemia in the United States. Emerg Infect Dis. 2014;20(12):2068–2071. doi: 10.3201/eid2012.131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittwer M, Altpeter E, Pilo P, Gygli SM, Beuret C, Foucault F, Ackermann-Gaumann R, Karrer U, Jacob D, Grunow R, Schurch N. Population genomics of Francisella tularensis subsp. holarctica and its implication on the eco-epidemiology of tularemia in Switzerland. Front Cell Infect Microbiol. 2018;8:89. doi: 10.3389/fcimb.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wicki R, Sauter P, Mettler C, Natsch A, Enzler T, Pusterla N, Kuhnert P, Egli G, Bernasconi M, Lienhard R, Lutz H, Leutenegger CM. Swiss Army Survey in Switzerland to determine the prevalence of Francisella tularensis, members of the Ehrlichia phagocytophila genogroup, Borrelia burgdorferi sensu lato, and tick-borne encephalitis virus in ticks. Eur J Clin Microbiol Infect Dis. 2000;19(6):427–432. doi: 10.1007/s100960000283. [DOI] [PubMed] [Google Scholar]
- 9.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snippy. 2020. https://github.com/tseemann/snippy. Accessed 24 Jun 2020
- 11.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37(1):291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 13.Larkeryd A, Myrtennas K, Karlsson E, Dwibedi CK, Forsman M, Larsson P, Johansson A, Sjodin A. CanSNPer: a hierarchical genotype classifier of clonal pathogens. Bioinformatics. 2014;30(12):1762–1764. doi: 10.1093/bioinformatics/btu113. [DOI] [PubMed] [Google Scholar]
- 14.Federal Office of Public Health. 2020. https://www.bag.admin.ch/bag/de/home/zahlen-und-statistiken/zahlen-zu-infektionskrankheiten.exturl.html/. Accessed 28 May 2020
- 15.Dryselius R, Hjertqvist M, Makitalo S, Lindblom A, Lilja T, Eklof D, Lindstrom A. Large outbreak of tularaemia, central Sweden, July to September 2019. Euro Surveill. 2019 doi: 10.2807/1560-7917.ES.2019.24.42.1900603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber M, Heuner K, Jacob D, Grunow R. Tularemia in Germany-A re-emerging zoonosis. Front Cell Infect Microbiol. 2018;8:40. doi: 10.3389/fcimb.2018.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federal Food Safety and Veterinary Office. 2020. https://www.blv.admin.ch/blv/de/home/tiere/tiergesundheit/ueberwachung/ueberwachung-von-zoonosen.html. Accessed 24 Jun 2020
- 18.Petersson E, Athlin S. Cat-bite-induced Francisella tularensis infection with a false-positive serological reaction for Bartonella quintana. JMM Case Rep. 2017;4(2):e005071. doi: 10.1099/jmmcr.0.005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaqub S, Bjornholt JV, Enger AE. Tularemia from a cat bite. Tidsskr Nor Laegeforen. 2004;124(24):3197–3198. [PubMed] [Google Scholar]
- 20.Foley JE, Nieto NC. Tularemia. Vet Microbiol. 2010;140(3–4):332–338. doi: 10.1016/j.vetmic.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Greene CE. Infectious diseases of the dog and cat. 4. St. Louis: Elsevier/Saunders; 2012. [Google Scholar]
- 22.Hestvik G, Uhlhorn H, Sodersten F, Akerstrom S, Karlsson E, Westergren E, Gavier-Widen D. Tularaemia in European Brown Hares (Lepus europaeus) and mountain Hares (Lepus timidus) characterized by histopathology and immunohistochemistry: organ lesions and suggestions of routes of infection and shedding. J Comp Pathol. 2017;157(2–3):103–114. doi: 10.1016/j.jcpa.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Origgi FC, Pilo P. Francisella Tularensis clades B.FTN002–00 and B.13 are associated with distinct pathology in the European Brown Hare (Lepus europaeus) Vet Pathol. 2016;53(6):1220–1232. doi: 10.1177/0300985816629718. [DOI] [PubMed] [Google Scholar]
- 24.Nordstoga A, Handeland K, Johansen TB, Iversen L, Gavier-Widen D, Mattsson R, Wik-Larssen K, Afset JE, Naeverdal R, Lund A. Tularaemia in Norwegian dogs. Vet Microbiol. 2014;173(3–4):318–322. doi: 10.1016/j.vetmic.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 25.Posautz A, Gyuranecz M, Denes B, Knauer F, Dier H, Walzer C. Seroprevalence of Francisella tularensis in Austrian hunting dogs. Vector Borne Zoonotic Dis. 2018;18(2):117–119. doi: 10.1089/vbz.2017.2193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. List of primers used for Sanger sequencing.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Genbank repository under Bioproject accession number PRJNA645814.