Abstract
Background
To investigate the clinical significance of ubiquitin-specific peptidase 19 (USP19) expression in gastric cancer (GC) compared with that in normal tissues and gastric cell lines.
Methods
USP19 protein expression was analyzed in 212 paired GC samples using immunohistochemical staining. Quantitative real-time PCR (qRT-PCR) and Western blotting were used to detect the level of USP19 in gastric cell lines. The biological functions of USP19 were investigated by MTT assay, colony-forming assay, wound healing assay and gelatin zymography, and apoptotic cells were detected by immunohistochemistry assays in SGC7901 xenograft models.
Results
USP19 expression was significantly increased in GC cells and tissues, and the high level of USP19 expression was positively correlated with the Lauren’s classification and poor prognosis. Moreover, USP19 was identified as a novel independent prognostic biomarker in GC patients. Enhanced USP19 expression promoted GC cell proliferation and metastasis through reduced cleaved caspase-3 and increased MMP2/MMP9 expression and promoted enzyme activity in the study, and verified the results through The Cancer Genome Atlas (TCGA) and bioinformatic websites from the Kaplan–Meier plotter (http://kmplot.com) and GEPIA (http://gepia2.cancer-pku.cn.).
Conclusion
Our study suggests that USP19 promoted tumor progression by inducing MMP2/MMP9 expression and related enzyme activity. Hence, USP19 may be a valuable prognostic predictor for GC, and the USP19-MMP2/MMP9 axis could serve as a therapeutic target.
Keywords: USP19, MMP2/MMP9, gastric cancer, prognosis, invasion, metastasis, TCGA
Introduction
Gastric cancer (GC) is the 5th main cause of cancer-related death worldwide with high mortality and high incidence in China.1,2 Based on the remaining outstanding rate of recurrence and a very low 5-year survival rate (<10% at advanced stages), a molecular understanding of the genetic factors involved in GC progression and advanced-stage disease may contribute to identifying novel biomarkers and highlight potential avenues for targeted therapies. In most patients, the cancer is diagnosed at an advanced stage and may have metastasized, which is one of the main reasons for its relatively poor prognosis. Therefore, it is important to identify novel, robust biomarker candidates for the prognostic assessment of GC patients.
Epithelial–mesenchymal transition (EMT) is a developmental program associated with cancer progression and metastasis. In particular, cancer cells typically spread by secreting various molecules that degrade the extracellular matrix (ECM), invading blood vessels, and migrating to distant sites. Matrix metalloproteinases (MMPs) are a major group of enzymes that regulate ECM composition during normal development and pathological responses.3 MMPs are important in regulating cell migration and invasion through digesting the ECM. Elevated MMP2 and MMP9 expression has been observed in invasive and metastatic cases of GC patients.4,5
Recently, deubiquitinases (DUBs) have been identified as pro- or anti-cancer factors owing to their regulatory effects on key oncogenes or other proteins involved in tumor progression. Of these DUBs, the ubiquitin-specific protease (USP) family plays a critical role in the induction of EMT and in properties of mammary epithelial stem cells. Many USPs have been implicated in human tumorigenesis through regulation of protein localization or activation. For example, USP34 induces mammary gland regeneration or bone formation by induction of EMT and stemness.6,7 In metastatic breast cancer, USP15 is required for cell migration through transforming growth factor-β signaling and p53 stability.8,9 Elevated levels of USPs have been reported in cancer samples compared with those in adjacent normal tissues (ANTs), and this finding has received a great deal of attention.10,11 The ubiquitin-proteasome system is the major pathway that plays an important role in a variety of cellular responses, including cell division, proliferation, metastasis, and apoptosis.12 Moreover, USPs are attractive targets for cancer drug discovery purposes in many cancer-associated pathways.13,14 It has been shown that USPs play important roles in the regulation of unfolded protein response and misfolding-associated protein secretion.15,16 USP6 was shown to induce transcription of MMP9 through the activation of nuclear factor-κB (NF-κB) in a USP-dependent manner.17 USP6 also induced the formation of tumors, which suggested that targeting USP6 with specific inhibitors might be an effective anticancer approach.18–20 Interaction of USP7 with FOXO4 led to FOXO4 inactivation by nuclear export and upregulation of the oncogenic PI3K/Akt pathway.21 The activity or expression of other USPs is thought to have increased in cancer cells, which could serve as prime targets for therapeutic intervention.22,23
USP19 is known to be involved in the following: maintenance of cell proliferation; modulation of DNA damage repair by the targeting of different substrates, such as p27, HIF1α, and cIAP, for de-ubiquitination and inhibition of degradation in a DUB-independent manner;24–26 and inhibition of tumor necrosis factor-α-induced cell apoptosis.11 Pathologically, USP19 is involved in hypoxia and muscle wasting.27 Hence, exploring the underlying molecular mechanism of USP19 may be helpful for developing treatment strategies as well as monitoring the prognosis of GC under hypoxic microenvironment.
To date, little is known about the regulation of USP19 in tumors. It is still unknown whether USP19 is involved in the regulation of growth and metastasis of GC. Identifying the downstream targets of USP19 would help elucidate its role in tumor progression. In this study, we investigated the relationship between USP19 expression and the prognosis of GC patients, and explored the potential effects of USP19 on MMP2/MMP9 expression and enzyme activity in GC. We first identified that USP19 was a positive regulator of metastasis and an independent predictor of clinical outcome in GC patients. Overexpression of USP19 enhanced MMP2/MMP9 protein expression and enzyme activity. Our findings demonstrated that USP19 acted as an enhancer and promising prognostic signature in the metastatic pathway by regulating the MMP2/MMP9 axis in GC.
Materials and Methods
Patients and Tissue Samples
This retrospective study enrolled a total of 212 patients with GC who underwent gastrectomy between January 2012 and May 2018 at Liaoning Province Cancer Hospital & Institute (Liaoning, China). No preoperative radiotherapy or chemotherapy was performed before surgery. All patients had a sufficient number of paraffin-embedded tumor specimens stored. All tissue samples were fixed in 10% neutral formalin and embedded in paraffin. A tissue array was constructed and cut into 4-μm sections. Patients were monitored every 3 to 6 months. Tumor staging conformed to the 8th edition of the American Joint Committee on Cancer/Union International Control Center (AJCC/UICC) TNM staging manual (2017). Median age at surgery was 63.5 years (range 26–81 years); 168 (79.2%) were male, and 44 (20.8%) were female. The clinicopathological parameters of patients included age, sex, Lauren’s classification, TNM stage, T category, N category, and distant metastasis (Table 1). This study was approved by the Liaoning Province Cancer Hospital and Institute Institutional Review Board. Informed consent for specimen preservation was obtained from all patients before their surgery at Liaoning Province Cancer Hospital and Institute.
Table 1.
Univariate Cox Model for the Association Between Survival and Clinicopathological Features in GC
| Clinical Features | No. Cases | P-value | |
|---|---|---|---|
| Sex | |||
| Male | 168 | ||
| Female | 44 | 0.547 | |
| Age | |||
| ≥60 | 131 | ||
| <60 | 81 | 0.243 | |
| TNM stage | |||
| I | 18 | ||
| II | 66 | II vs I | 0.051 |
| III | 118 | III vs I | <0.001 |
| IV | 10 | IV vs I | <0.001 |
| Tumor invasive depth (T) | |||
| 1 | 10 | ||
| 2 | 19 | 2 vs 1 | 0.084 |
| 3 | 115 | 3 vs 1 | 0.002 |
| 4 | 68 | 4 vs 1 | <0.001 |
| Lymph node metastasis (N) | |||
| 0 | 60 | ||
| 1 | 31 | 0.003 | |
| 2 | 53 | 0.003 | |
| 3 | 68 | <0.001 | |
| Distant metastasis (M) | |||
| Yes | 10 | ||
| No | 202 | 0.001 | |
| Lauren’s classification | |||
| Intestinal type | 132 | ||
| Diffused type | 80 | <0.001 | |
| USP19 expression | |||
| High | 141 | ||
| Low | 71 | <0.001 |
Immunohistochemical Staining
The tissue microarray section was dewaxed with xylene, rehydrated, pretreated with 3% H2O2, and then subjected to antigen retrieval. The slides were incubated at 4°C overnight with mouse anti-human USP19 (dilution 1:200; Proteintech, Chicago, IL) for human samples, and with USP19 antibody (dilution 1:200), rabbit anti-human MMP2 and MMP9, cleaved caspase-3 (dilution 1:100, respectively; Cell Signaling Technology, Danvers, MA) for xenograft tissues. The horseradish peroxidase (HRP)-labeled secondary antibody was added for 30 mins at room temperature. At last, the sections were visualized with 3,3ʹ-diaminobenzidine (DAB; Zhongshan Gold Bridge Corporation, China). Slides were then counterstained with hematoxylin, dehydrated with alcohol and xylene, and mounted on coverslips. Immunoreactivity was visualized using an Envision detection system (DAKO, Carpinteria, CA), and the nuclei were counterstained with hematoxylin.
The staining percentage and staining intensity of USP19 were independently graded and calculated for the final staining score by three senior pathologists. The final results were obtained after evaluating the staining intensity of the positively stained cells. This value was calculated as the mean percentage of USP19-positive tumor cells elevated in five areas at 200× magnification.
IHC staining for USP19 was scored based on a semi-quantitative scale (– to +++) system. Cells positively stained for USP19 showed variable degrees of cytoplasmic staining in the tissues. The scoring was based on the staining intensity (0=none; 1=mild; 2=moderate; and 3=intense) and percentage of positive cells (0=0%; 1=1%-25%; 2=26%-50%; 3=51%-75%; 4=76–100%). The final score of each sample (0–12) was obtained by multiplying the staining intensity with the percentage of positive cells, and tissues were ultimately defined as negative (-), score 0; weak expression (+), score 1–4; moderate expression (++), score 5–8; and strong expression (+++), scored ≥9. Tissue samples scoring (+) to (+++) were considered “high expression”, and samples scoring (–) were defined as “low expression”. The median score was used as the expression cutoff point (USP19 median score=4). Tissue samples scores>4 were considered “high expression”, and samples scoring (≤4) were defined as “low expression”.
Cell Lines
The human GC cell lines (BGC823, MGC803, SGC7901, MKN45, AGS, and N87) and gastric immortalized epithelial cell line (GES1) were purchased from the Shanghai Institute of Cell Biology. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (FBS) containing 100 units/mL penicillin and 100 mg/mL streptomycin (P/S, Gibco) at 37°C in a 5% CO2 incubator.
Antibodies
The antibodies used for Western blotting (WB) and immunohistochemistry (IHC) included: mouse anti-human USP19 monoclonal antibody (Proteintech, Chicago, IL), rabbit anti-human MMP2, MMP9, and cleaved caspase-3 antibodies (Cell Signaling Technology), and rabbit anti-human β-actin monoclonal antibody (Sigma-Aldrich, St. Louis, MO) as the internal control.
Fluorescence Microscopy
Gastric cell lines (BGC823, SGC7901, and GES1) were cultured on a glass-bottomed dish and directly observed. For examination by immunofluorescence microscopy, cells grown on gelatin-coated coverslips were fixed with 4% paraformaldehyde, permeabilized using 50μg/mL digitonin, and then stained with specific antibodies. Colocalization images were obtained under a Zeiss LSM 700 confocal microscope equipped with a 100× (NA 1.4) oil-immersion objective, and the LC3 puncta images were captured using Leica DMI3000 B with a 100× oil-immersion objective.
Cell Invasion and Wound Healing Assay
Invasion assays were performed using Boyden chambers (Corning Inc., Corning, NY) with a membrane (8 μm pore size) coated with Matrigel (1 mg/mL; BD Biosciences, Franklin Lakes, NJ) according to the manufacturer’s protocol. Briefly, 1×105 cells in 200μL of FBS-free medium were seeded into the upper chambers. The bottom wells in the system were filled with 600 μL of 10% FBS medium. After the assay had been running for 24 h at 37°C, non-migrated or non-invaded cells were removed from the upper surface of the filter. Cells on the lower surface of the membrane were fixed with ice-cold methanol and stained with crystal violet. Cells were counted under an optical microscope. Each experiment was repeated five times.
Cell migration was measured using a scratch (wounding healing) assay. Cells were plated in 6-well plates to create a confluent monolayer. Then, the monolayer was scraped in a straight line to create a scratch or wound with a p200 pipette tip. The cells were washed once with growth medium and incubated in DMEM containing 2% FBS. The remaining cells were treated with 10% FBS DMEM solution and imaged using a phase-contrast microscope at 0, 12, 24, and 36 h. The wound area was quantified using NIH Image-Pro Plus software. The data are expressed as the mean of three independent experiments ± standard deviation (SD) for three times.
MTT Assay
GC cells transfected with different vectors were seeded in 96-well plates. Viable cells were quantified with the modified 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in six replicates per dilution.
Real-Time PCR
Real-time PCR was performed with an ABI Sep Plus One sequence detection system (Applied Biosystems, Foster City, CA). Primers were designed (https://www.genscript.com/tools/real-time-pcr-tagman-primer- design-tool) and are listed in Table S1. β-actin was used as a loading control. The threshold was set above the non-template control background and within the linear phase of target gene amplification to calculate the cycle number at which the transcript was detected.
Transfection and Generation of Stable Cell Lines
The specific ectopic expression USP19 plasmid (RC214190, USP19 [Myc-DDK-tagged]-Human USP19, NM_006677, transcript variant 4) with the vector control (pCMV6-Entry, CAT#: PS10,0001) were purchased from a commercial company (OriGene Ltd., Rockville, MD), and these plasmids were transfected into GES1 cells with 400μg/mL neomycin (Gibco) addition to form stable clones. The specific kit for knockdown of USP19 was ordered from the commercial company (OriGene, Ltd.; USP19 Human shRNA Plasmid Kit [Locus ID 10,869], CAT#: TR308463, ordered from https://www. origene.com/catalog/rnai/shrna-plasmids/tr308463/usp19-human-shrna- plasmid-kit- locus- id-10,869). These shRNA constructs were designed against multiple splice variants at this gene locus, as well as the negative control-empty vector (pRS, CAT#: TR20003). The ShUSP19 and empty vector plasmids were transfected into SGC7901 cells with 400 μg/mL neomycin (Gibco) for stable transfection. The relative efficacy of transfection was evaluated by Western Blotting analysis.
Determination of MMP2/MMP9 Activity by Gelatin Zymography Analysis
The activities of MMP2/MMP9 in conditioned medium after stable transfection with different plasmids were measured by gelatin zymography protease assay. Briefly, collected media of an appropriate volume were prepared with sodium dodecyl sulfate (SDS) sample buffer without boiling or reduction and subjected to 0.1% gelatin-7% SDS-polyacrylamide gel electrophoresis (PAGE). After electrophoresis, gels were washed with 2.5% Triton X-100 and then incubated in reaction buffer (40mM Tris-HCl, pH 8.0, 10 mM CaCl2, and 0.01% NaN3) for 12 h at 37°C. Then, gel was stained with Coomassie Brilliant Blue R-250. All experiments were repeated for three times.
Protein Preparation and Western Blot Analysis
For total cell lysate preparation, cells were rinsed with phosphate buffered saline (PBS) twice and scraped with 0.2 mL of cold RIPA buffer containing protease inhibitors cocktail, and then vortexed at 4°C for 10 min. Cell lysates were subjected to centrifugation at 10,000 rpm for 10 min at 4°C, and the insoluble pellet was discarded. The protein concentration of the total cell lysate was determined by Bradford assay and then analyzed by WB analysis. The band intensity was analyzed with ImageJ software. Equal amounts of protein were separated by 12% SDS-PAGE and transferred to microporous polyvinylidene difluoride (PVDF) membranes (Roche, Basel, Switzerland). PVDF membranes were incubated with an antibody against USP19 (1:1000; Proteintech, Chicago, IL), MMP2, MMP9, cleaved caspase-3 (Cell Signaling Technology,) or β-actin (internal control; 1:10000; Sigma-Aldrich). Blots were developed using a Luminata Crescendo Western HRP Substrate (Millipore, Billerica, MA). Then, signals were detected using an enhanced chemiluminescence (ECL) commercial kit (Amersham Biosciences, Little Chalfont, UK) and relative photographic density was quantitated by scanning the photographic negatives on a gel documentation and analysis system (AlphaImager 2000; Alpha Innotech Corporation, San Leandro, CA). All experiments were repeated three times.
Bioinformatic Analysis of TCGA Data
We downloaded the relevant data of USP19 expression in Gastric Cancers from TCGA database (https://cancergenome.nih.gov/). The expression and prognostic values of USP19 were analyzed using two online datasets, Gene Expression Profiling Interactive Analysis (GEPIA) and Kaplan-Meier-plotter dataset (http://kmplot.com/analysis/) in GC. GEPIA was an interactive web server for estimating the mRNA expression data from 9736 tumors and 8587 normal samples in the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) dataset projects. The |Log2FC| cutoff of the expression of proposed biomarker was 1. All boxplot analysis used log2 (TPM+1) for log-scale. The Correlation analysis function from GEPIA performs pairwise gene correlation analysis for any given sets of TCGA and/or GTEx expression data using the Spearman correlation statistics. In this study, we examine the simple correlation coefficient between USP19 and MMP2/MMP9 gene pair or check the correlation analysis result between USP19/GAPDH and MMP2/GAPDH or MMP9/GAPDH relative ratios.
Kaplan–Meier-plotter database can analyze prognostic values of gene mRNA expression in breast, gastric, lung and ovarian cancer patients, and also miRNA expression in liver and breast cancer patients. Samples with gene expression data and prognosis information used in Kaplan–Meier-plotter dataset are downloaded from TCGA, EGA and GEO (Affymetrix microarrays only). The database is handled by a PostgreSQL server, which integrates gene expression and clinical data simultaneously. So far, a number of genes have been identified and validated by KM plotter in these four types of cancer. In this online dataset, clinical data from 876 gastric cancer patients including Lauren classification, clinical stage, differentiation, HER2 status and lymph node status were collected. The Affymetrix IDs of USP19 were entering into the web (http://kmplot.com/analysis/index.php?p=serviceandcancer=gastric), and then data were compared through a Kaplan–Meier survival plot. The patient samples were split into two groups by median. The hazard ratio (HR) with 95% confidence intervals (95% CIs) and p-values were calculated by a PostgreSQL server. P<0.05 was considered to be a significant difference.
Mice and Treatment
For tumor-bearing experiments, 2×107 stably infected SGC7901 cells (Vector) or shRNA-USP19 (ShUSP19) were resuspended in sterile PBS (200 µL) and injected subcutaneously into one flank of 4-week-old female BALB/c-nude mice (n=5, per group). Nude mice were obtained from the Animal Center of the Chinese Academy of Science and all animal experiments were approved by the Ethical Committee of Liaoning Cancer Hospital. All protocols were performed under the guidelines followed for the welfare of the laboratory animals. After SGC7901 cell inoculation, tumor sizes in the flanks were measured using vernier calipers twice weekly, and tumor volume was calculated using the formula: V=(L×W2)/2. After 24 days, all surgeries were performed under sodium pentobarbital anesthesia with minimized suffering, and tumor xenografts were harvested for IHC staining.
Statistical Analysis
All data were analyzed with SPSS21.0 statistical software (SPSS Inc. Chicago, IL). All data were expressed as Mean±SD, and statistical analyses were performed using the Student’s t-test. Univariate Cox proportional hazards models were applied to assess the effect of covariates on survival outcomes. All tests were 2-sided. The Pearson Chi-square test and Fisher’s exact test were used to evaluate and compare the clinicopathological characteristics. Multivariate analyses were performed using the Cox proportional hazards regression model. Kaplan–Meier survival plots and log-rank statistics were used to compare the survival rates of GC patients with overall survival data. P-values <0.05 were considered statistically significant.
Results
Elevated USP19 Was Associated with Poor Prognosis in GC
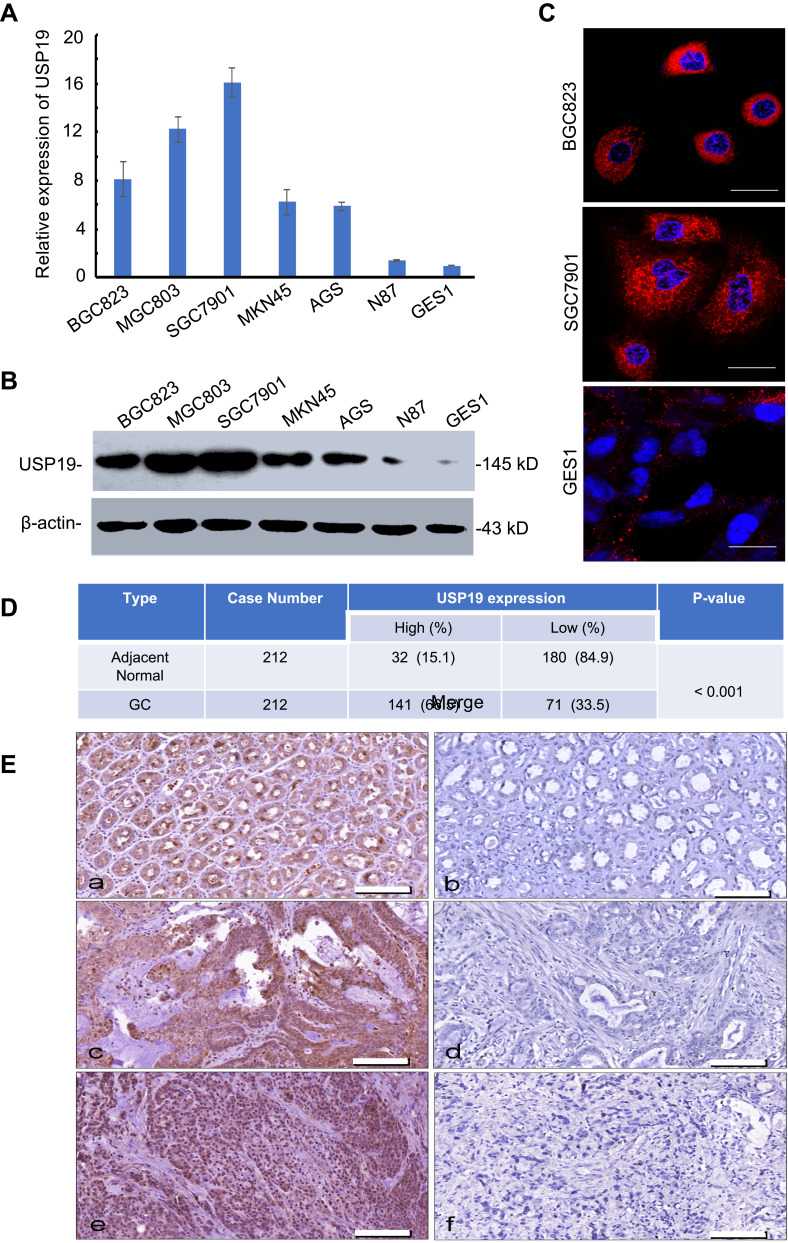
To investigate the expression of USP19 in GC, we first detected the expression of USP19 in GC cell lines (BGC823, MGC803, SGC7901, MKN45, AGS, N87) with an immortal gastric epithelium cell line (GES1) at the mRNA and protein levels with real-time PCR and IHC staining analyses. As shown in Figure 1A and B, most GC cell lines expressed higher levels of USP19 than GES1, and USP19 was found to be localized in the cell plasma in GC cells and tissues by the immunostaining analysis (Figure 1C and E). Furthermore, in order to detect the protein levels of USP19 expression in GC and adjacent normal tissues (NATs), we performed 212 paired GC specimens by IHC staining. The rate of high level of USP19 expression was 66.5% (141/212) in GC samples compared with ANTs (15.1%, 32/212, P<0.001, Table 2 and Figure 1D). Moreover, we investigated the roles of USP19 expression in the outcome of GC patients and other clinicopathological features. As shown in Figure 2A, high level of USP19 expression was associated with apparently poor overall survival (OS) (P<0.001). TNM stage was also a promising indicator of the prognosis of GC patients., advanced GC (including TNM stage III + IV) patients with worse outcome than those with TNM stage I and II (Figure 2B, P<0.001). Moreover, patients with intestinal-type GC had apparently better overall survival than those with diffuse-type GC (Figure 2C, P<0.001). By univariate Cox proportional hazards analysis, the level of USP19 expression, TNM stage, tumor invasive depth (T stage), lymph node metastasis (N stage), distant metastasis (M stage), and Lauren’s classification were the meaningful prognostic factors for the OS of GC patients (Table 2), and further multivariate Cox proportional hazards analysis indicated that USP19, tumor invasive depth (T stage) and lymph node metastasis (N stage), were the independent prognostic factors for OS in GC patients (Table 3).
Figure 1.
Relative expression of USP19 in GC cell lines and GES1. (A) Differential expression of USP19 mRNA in gastric cell lines by real-time PCR assay. (B) The differential expression of USP19 protein in gastric cell lines by WB analysis. (C) Differential expression and distribution of USP19 protein in gastric cell lines by confocal analysis, Scale bar=20 μm. (D) Relative expression of USP19 in paired 212 GC samples and ANTs by IHC staining and analysis by Chi-square test. (E) USP19 protein levels in GC tissues and ANTs were analyzed by IHC staining (200× magnification). High or low expression of USP19 protein in ANTs (a and b); high or low expression of USP19 protein in intestinal-type GC (c and d); high or low expression of USP19 protein in diffuse-type GC (e and f), Scale bar=200 μm.
Table 2.
The Relationship Between USP19 Expression and Other Clinicopathological Parameters in GC
| Clinical Features | No. Cases | USP19 Expression | P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age | ||||
| ≥60 | 131 | 88 | 43 | |
| <60 | 81 | 53 | 28 | 0.794 |
| Sex | ||||
| Male | 168 | 115 | 53 | |
| Female | 44 | 26 | 18 | 0.241 |
| TNM stage | ||||
| I | 18 | 8 | 10 | |
| II | 66 | 43 | 23 | |
| III | 118 | 82 | 36 | |
| IV | 10 | 8 | 2 | 0.168 |
| Tumor invasive depth (T) | ||||
| 1 | 10 | 4 | 6 | |
| 2 | 19 | 9 | 10 | |
| 3 | 115 | 78 | 37 | |
| 4 | 68 | 50 | 18 | 0.051 |
| Lymph node metastasis (N) | ||||
| 0 | 60 | 36 | 24 | |
| 1 | 31 | 19 | 12 | |
| 2 | 53 | 39 | 14 | |
| 3 | 68 | 47 | 21 | 0.404 |
| Distant metastasis (M) | ||||
| Yes | 10 | 8 | 2 | |
| No | 202 | 133 | 69 | 0.501 |
| Lauren’s classification | ||||
| Intestinal type | 132 | 78 | 54 | |
| Diffuse type | 80 | 63 | 17 | 0.003 |
Figure 2.
Kaplan–Meier survival curves of GC patients according to the clinical features and expression of USP19. (A) Patients with high-level USP19 expression showed poor survival. (B) TNM stage was a promising indicator of the prognosis of GC patients. (C) Patients with intestinal-type GC had better survival than those with diffuse-type GC. (D) Low-level USP19 expression predicted better survival in patients with TNM stage III. (E) Low-level USP19 expression predicted better survival in patients with diffuse-type GC. (F) Low-level USP19 expression predicted better survival in patients with intestinal-type GC.
Table 3.
Multivariate Cox Model for the Association Between Survival and Clinicopathological Factors in GC
| Clinical Features | Exp(B) | 95% CI for Exp(B) | P-value | |
|---|---|---|---|---|
| Lower | Upper | |||
| Age | 1.477 | 0.995 | 2.193 | 0.053 |
| Sex | 0.831 | 0.517 | 1.337 | 0.446 |
| TNM stage | 0.814 | 0.404 | 1.639 | 0.565 |
| Tumor invasive depth (T) | 2.226 | 1.520 | 3.261 | <0.001 |
| Lymph node metastasis (N) | 1.634 | 1.206 | 2.214 | 0.002 |
| Distant metastasis (M) | 1.613 | 0.616 | 4.220 | 0.330 |
| Lauren’s classification USP19 expression |
0.709 2.459 |
0.472 1.542 |
1.066 3.921 |
0.099 <0.001 |
USP19 Provided More Accurate Prognostic Prediction
We also performed stratified analyses of patients with diffuse-type GC from our cohort. Patients with high-level USP19 expression showed decreased OS compared with those with low-level USP19 expression (P=0.019, Figure 2E). Similar results were obtained in patients with TNM stage III or intestinal-type GC (P<0.001, Figure 2D; and P=0.002, Figure 2F, respectively). These data indicated that USP19 could be a promising prognostic factor in GC.
We further investigated the association between USP19 expression and other prognostic clinicopathological features. The data are shown in Table 2. No correlation was found between USP19 expression and age (P=0.794), sex (P=0.241), TNM stage (P=0.168), lymph node metastasis (N stage, P=0.414), or distant metastasis (M stage, P=0.501); however, USP19 expression showed a slight positive correlation with tumor invasive depth (T stage, P=0.051) and an inverse correlation with Lauren’s classification (Pearson’s R=–0.202, P= 0.003), which indicated that high-level USP19 expression was observed more frequently in patients with diffuse-type GC than that in patients with intestinal-type GC, suggesting that combined detection of USP19 and TNM stage or Lauren’s classification will provide the novel strategy for precision medicine research.
USP19 Promoted GC Cell Proliferation and Metastasis
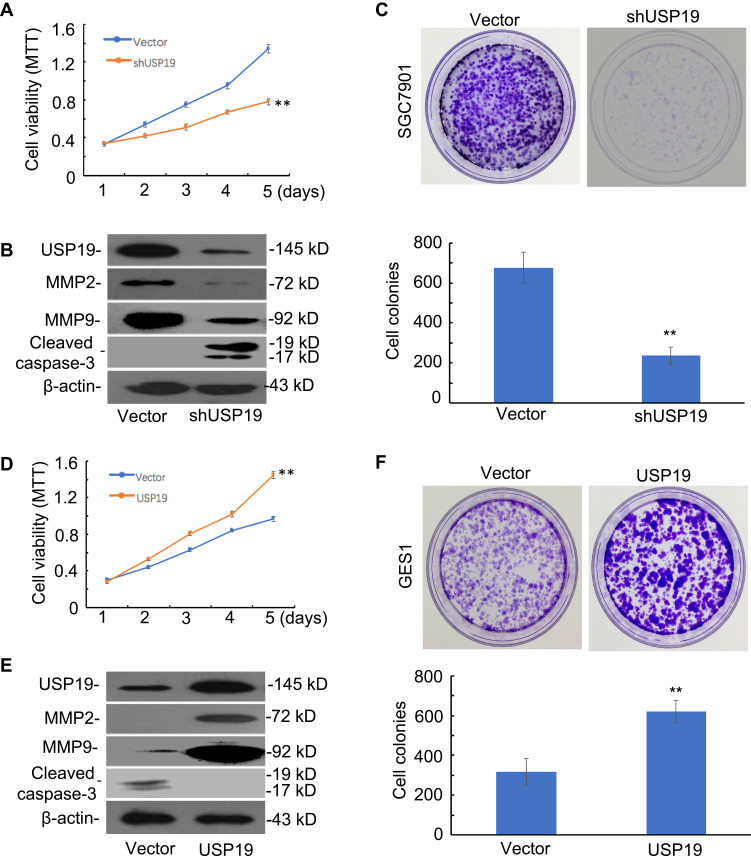
To investigate the potential mechanism of USP19 in the tumorigenesis of gastric mucosa, we first compared the baseline levels of USP19 expression in six GC cell lines with the immortal gastric epithelium cell line, GES1. The highest level of USP19 expression was detected in SGC7901 cells and lowest expression in GES1 cells (Figure 1A and 1B). The SGC7901 GC cell line was selected for knockdown of USP19 in SGC7901 cells, and the GES1 cell line for overexpressing USP19. The transfected efficiencies of knockdown or overexpression were confirmed by WB analyses (Figure 3B and 3E). Next, we investigated the potential effect of USP19 expression on GC in the study. The results showed that knockdown of USP19 suppressed cell proliferation in SGC7901 cells while overexpressing USP19 accelerated cell growth in GES1 cells, as determined by MTT assay (P<0.01, Figure 3A and 3D). A greater number of cell colonies were observed in the USP19-overexpression group, as shown by colony-forming assay (P<0.01, Figure 3C and 3F), and reduced expression of cleaved caspase-3 was observed in USP19 knockdown group, as demonstrated by WB analysis (Figure 4E). In the SGC7901 xenograft model, a greater number of apoptotic cells were detected by Cleaved Caspase-3 staining in the ShUSP19 group in tumor-bearing models (Figure 5A). These results indicated that USP19 enhanced cell proliferation and exhibited anti-apoptotic properties.
Figure 3.
Functional roles of USP19 in cell models. (A) Knockdown of USP19 inhibited SGC7901 cell growth, as determined by MTT assay. Bars: SD; **P<0.01. (B) Knockdown of USP19 decreased the level of MMP2 and MMP9 and increased the level of the apoptotic-related protein, cleaved caspase-3, in SGC7901 cells. (C) Knockdown of USP19 inhibited colony formation in SGC7901 cells. The data are shown as Mean±SD (n=3). **P<0.01. (D) Ectopic expression of USP19 promoted GES1 cell growth, as shown by MTT assay. Bars: SD; **P<0.01. (E) Overexpression of USP19 increased the protein level of MMP2 and MMP9 and decreased the level of the apoptotic-related protein, cleaved caspase-3. (F) Ectopic expression of USP19 increased colony formation in GES1 cells. The data are shown as Mean±SD (n=3). **P<0.01. All WB experiments were repeated three times.
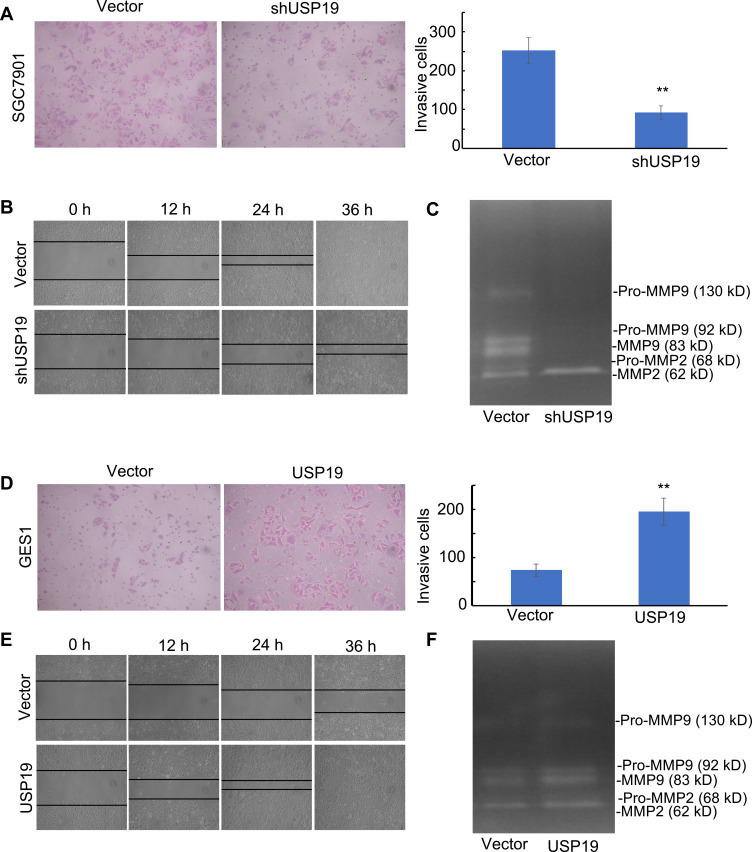
Figure 4.
Effect of USP19 on cell invasion and migration and MMP2/MMP9 enzyme activity. (A) Knockdown of USP19 reduced SGC7901 cell invasion. Images were acquired at the indicated time (200× magnification). Data are shown as Mean±SD (n=5). **P<0.01. (B) Knockdown of USP19 reduced the migration of SGC7901 cells, as determined by wound healing assay at 0, 12, 24, and 36 h. Images were acquired at the indicated time (100× magnification). Data are shown as Mean±SD (n=3). **P<0.01. (C) SGC7901 cells were transfected with ShUSP19 or vector for 36 h and then subjected to gelatin zymography to analyze the activity of MMP2 and MMP9. (D) Ectopic expression of USP19 enhanced invasion of GES1 cells. Images were acquired at the indicated time (200× magnification). Data are shown as Mean±SD (n=5). **P<0.01. (E) Ectopic expression of USP19 increased the migration of GES1 cells, as determined by wound healing assay at 0, 12, 24, and 36 h. Images were acquired at the indicated time (100× magnification). Data are shown as Mean±SD (n=3). **P<0.01. (F) GES1 cells were transfected with USP19-overexpressing plasmid or control vector for 36 h and then subjected to gelatin zymography to analyze the activity of MMP2 and MMP9.
Figure 5.
Knockdown of USP19 reduced gastric carcinogenesis in animal models. Knockdown of USP19 inhibited tumor growth. Five nude mice were injected subcutaneously with 1 × 107 cells/mouse for each of the indicated stable cell lines of SGC7901-ShUSP19 or SGC7901-Vector, respectively. Results were presented as tumor volume at the different time points (C) and isolated xenografts at day 24 (A) with no change in body weight for each group (B). **P<0.01. (D) Tumors were isolated, fixed, and subjected to IHC assay (400× magnification).
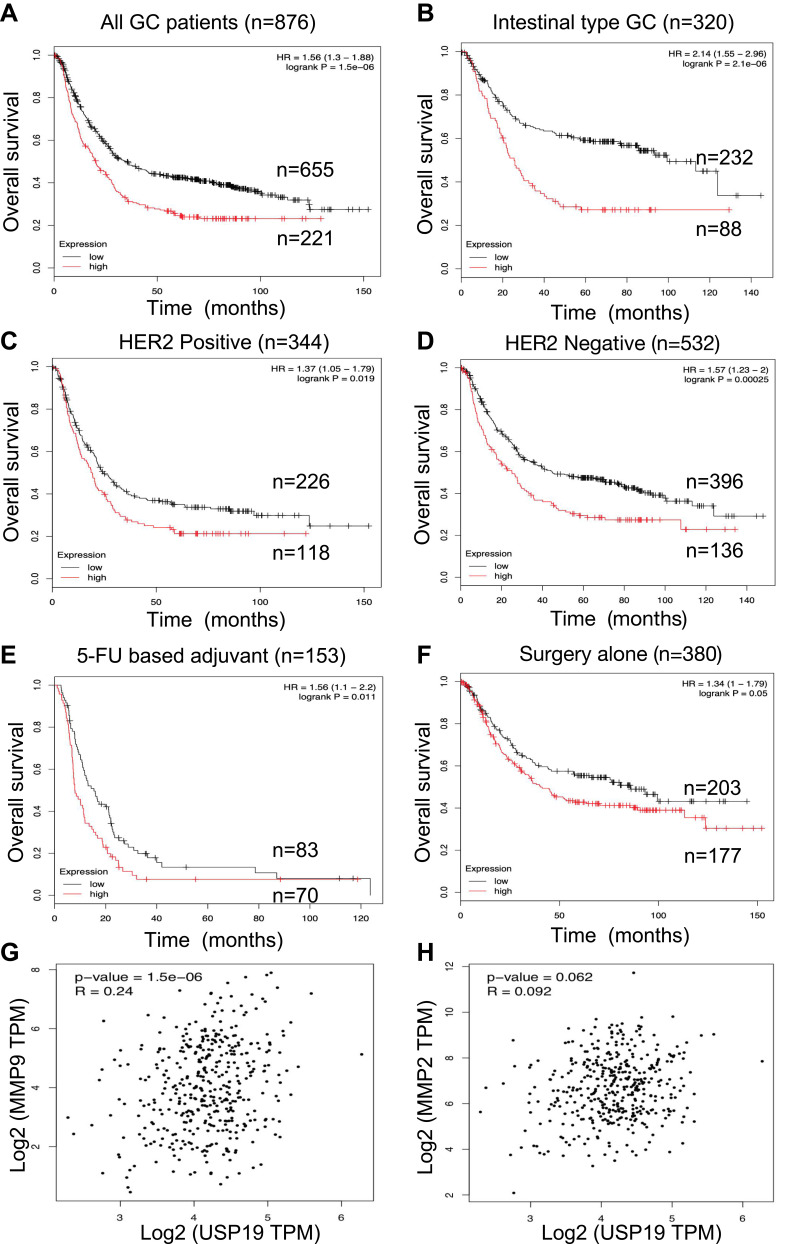
Although rapid progress has been made in basic and translational research on GC-related biomarkers in the past decade, identifying the precise prognostic biomarker for GC by appropriate experimental methods remains a challenge in the field of GC research. In this study, we found that positive USP19 expression was associated with poor survival in GC patients. High-level of USP19 expression was associated with MMP2/MMP9 enzyme activity and translational expression in this study. Furthermore, we used GEPIA website to evaluate the potential positive correlation with USP19 and MMP2/MMP9 in the mRNA levels (Figure 6G and H). The mRNA level of USP19 expression was positive association with MMP2/MMP9 expression (P=0.062, P<0.001, respectively). Especially in Figure 6G, showed a moderate correlation between USP19 and MMP9 (Spearman’s R=0.24, P<0.001).
Figure 6.
Prognostic roles of USP19 was determined in www.kmplot.com and correlation between mRNA levels of USP19 and MMP2/MMP9 in GC dataset from GEPIA Website (http://gepia2.cancer-pku.cn). Overall survival curves were plotted for all GC patients (n=876) with (A) different levels of USP19 expression; (B) Lauren’s classification; (C and D) HER2 status, (E) Different treatments, 5-FU based chemical therapy, (F) Surgery alone. From these data, low level of USP19 expression was correlated with better OS. The mRNA level of USP19 expression was positive association with MMP2/MMP9 expression. (G)P<0.001 correlation between MMP9 and USP19; (H) P=0.062, correlation between MMP2 and USP19. Especially, the moderate correlation was performed between USP19 and MMP9 expression (Spearman’s R=0.24).
Moreover, knockdown of USP19 strongly inhibited the flattening and spreading of SGC7901 cells, whereas ectopic expression of USP19 enhanced the flattening and spreading of GES1 cells using wound healing assay (P<0.01; Figure 4B and E, respectively). Transwell assay showed that knockdown of USP19 decreased invasiveness of SGC7901 cells, whereas overexpression of USP19 enhanced GES1 cell invasion (P<0.01; Figure 4A and 4D, respectively). Taken together, these results indicate that USP19 contributes to the metastatic behavior of GC cells.
USP19 Elevated MMP2/MMP9 Expression and Enzyme Activity
Increased USP19 level was positively associated with metastasis. Next, we explored the regulatory effects of USP19 on the MMP2/MMP9 axis. Elevated MMP2 and MMP9 expression has been previously observed in invasive and metastatic cases of human GC. Our results showed that USP19 positively regulated GC cell migration through the up-regulation of MMP2 and MMP9 expression and the decrease of cleaved caspase-3 activity in cell models (Figure 3B and 3E). Additionally, IHC analysis of xenografts showed that a lower expression of MMP2 and MMP9 was observed in the USP19 knockdown group compared with the negative control. We first found overexpressing USP19 increased MMP2 and MMP9 protein levels. Moreover, the ability of USP19 to enhance the migratory and invasive abilities of GC cells by increasing MMP2 and MMP9 expression was evaluated using gelatin zymography. As shown in Figure 4C, the enzyme activity of MMP2 and MMP9 was suppressed by knockdown of USP19 in SGC7901 cells but was elevated by ectopic expression of USP19 in GES1 cells (Figure 4F). Hence, we suggest that the anti-metastatic effect of knockdown of USP19 expression is at least partially dependent on the inhibition of MMP2/MMP9 expression and related enzyme activity.
Bioinformatic Analysis in GC Dataset
Prognostic roles of USP19 in GC were performed from the website (www.kmplot.com). The valid Affymetrix ID is 214674_at (USP19). Figure 6A represents a survival curve plotted for all GC patients (n=876). From these data, low mRNA expression of USP19 was correlated with better OS in all GC samples [HR=1.56, 95% CIs: (1.3–1.88), P<0.001]. Furthermore, we estimated prognostic values of USP19 in GC patients with different Lauren’s classification, including intestinal, diffuse and mixed type. As shown in Figure 6B, low level of USP19 expression was associated with better OS in intestinal-type gastric cancer patients [HR=2.14, 95% CIs: (1.55–2.96), P<0.001]. Prognostic values of USP19 in GC patients with different HER2 status are shown in Figure 6C and 6D. High mRNA level of USP19 [HR=1.37, 95% CIs: (1.05–1.79), P=0.019] was correlated to poor survival in HER2 positive GC patients (n=344) and a similar result was shown in HER2 negative GC patients [n=532, HR=1.57, 95% CI: (1.23–2), P<0.001]. In 5-FU based chemical therapy, GC patients with high mRNA level of USP19 had worse OS [n=153, HR=1.56, 95% CIs: (1.1–2.2), P<0.011, Figure 6E] than those with a low level of USP19 expression. In the surgery alone subset (Figure 6F), GC patients with high mRNA level of USP19 had also poor outcome [n=380, HR=1.34, 95% CIs: (1–1.79), P=0.05].
Knockdown of USP19 Depressed Tumor Growth in Tumor-bearing Experiments
To further confirm the biological function of USP19 in GC, we performed tumorigenicity assay. SGC7901 cells stably transfected with shUSP19 vector or empty vector were injected into the right flank of nude mice (n=5, each group), respectively. There was no difference in body weight from each group (Figure 5B). Tumor size was significantly smaller in shUSP19 group than that in control group (P<0.001, Figure 5C). Besides, the average tumor mass in shUSP19 group was significantly decreased compared with the control group for the general appearance at the end point (P<0.001, Figure 5A). These data demonstrated that USP19 functions as an oncogene in GC in xenograft models. Knockdown of USP19 in tumor tissues from nude mice was confirmed by IHC staining (Figure 5D). Cell invasion and apoptotic biomarkers in the tumor sections from nude mice were evaluated by MMP2, MMP9 and Cleaved Caspase-3 immunostaining assay, and the shUSP19 group displayed decreased expression of MMP2/MMP9 and elevated expression of Cleaved Caspase-3 compared with the negative control (Figure 5D). Collectively, these findings indicate that silenced USP19 suppressed the tumor growth in GC.
Discussion
DUBs are the novel families of drug targets. However, the physiological function of only few DUBs has been characterized. In this study, we identified USP19 as a promising oncogene in GC. We examined USP19 expression in gastric cell lines and paired GC tissues, as well as its correlation with clinicopathological features and prognosis. These results revealed that USP19 expression was significantly higher in GC tissues compared with that in ANTs. We found that USP19 was a novel independent prognostic factor for GC patients. Reduced expression of USP19 might contribute to the effective prognostic outcome and suppressing gastric tumorigenesis.
Metastasis, which causes approximately 90% of cancer deaths, is the process by which cancer cells spread from the original tumor site to distant organs. Degradation of ECM components and the basement membrane is a critical step in metastasis. There are multiple types of proteases that control ECM degradation and remodeling. MMP2 and MMP9 are the most extensively studied because of their close association with cancer migration and invasion. In our study, the cell invasion and migration induced by USP19 was correlated with MMP2/MMP9 expression and enzyme activity. Our results first indicated that USP19 enhanced MMP2/MMP9 protein expression as well as enzyme activity. The EMT properties of USP19 are similar to those of other USP members. Previous studies have shown that downregulation of USP14 leads to increased expression of E-cadherin and decreased expression of N-cadherin and vimentin in esophageal squamous cell carcinoma (ESCC) cells, as well as suppresses the migration/invasion of ESCC cells through the Wnt/β-catenin signaling pathway.28 USP6 is sufficient to induce the expression of MMP9 and MMP10. Especially, USP6 induces the transcription of MMP9 through activation of NF-κB.17 Downregulation of USP22 has been shown to suppress osteosarcoma growth and metastasis through PI3K/Akt pathway.29
In our study, we first uncovered the potential effect of USP19 on the induction of MMP2/MMP9 axis and enzyme activity. Our results show that USP19 plays an important role in the progression and metastasis of GC, indicating that USP19 may serve as a promising therapeutic target for GC treatment. However, more research is required to study the mechanism underlying the elevation of USP19 levels in GC and its role in the regulation of MMP2/MMP9 axis, which might further our understanding of gastric tumorigenesis and help in the development of precision medicine for GC treatment.
Acknowledgments
We also thank Yidu Cloud (Beijing) Technology Co., Ltd. for support in statistical consultation, data extraction, and processing.
Funding Statement
This study was supported by Liaoning S and T Project, No. 2015020269 and Doctor Fund of Liaoning Province Cancer Hospital and Institute, No. Z1410.
Consent for Publication
The corresponding author gives consent for photograph and other clinical data to be published in the Journal. We have seen and read the material to be published.
Abbreviations
ANTs, Adjacent normal tissues; DAB, 3, 3ʹ-diaminobenzidine; DMEM, Dulbecco’s Modified Eagle Medium; DUBs, De-ubiquitinases; ECM, Extracellular matrix; ECL, Enhanced chemiluminescence; EMT, Epithelial–mesenchymal transition; ESCC, Esophageal squamous cell carcinoma; FBS, Fetal bovine serum; GC: Gastric cancer; HRP, Horseradish peroxidase; MMP, Matrix metalloproteinase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide PAGE, Polyacrylamide gel electrophoresis; PVDF, Polyvinylidene difluoride; qRT-PCR, Quantitative real-time PCR; SDS, Sodium dodecyl sulfate; TNM, Tumor, Node, Metastasis.
Ethics Approval and Consent to Participate
The study was reviewed and approved by the Faculty of Science Ethics Committee at Liaoning Cancer Hospital & Institute (Cancer Hospital of China Medical University) (20,150,308-2) and the animal model experiments were followed the guidelines of animal welfare in China Medical University. All participants provided written informed consent prior to their treatments and study enrollment.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. Zhe Dong and Shuai Guo performed the majority of the experiments and drafted the manuscript; Yan Zhao and Jun Zhang designed the research; Liucun Yan, Kejia Liu, Zhe Sun and Li Song helped with data extraction and processing; Shuai Guo, Yue Wang, and Haojie Luo conducted the IHC assays and assisted in writing the manuscript; Xiangyu Meng, Guoliang Zheng, and Dong Yang collected and analyzed the data; Jianjun Zhang, Tao Zhang, and Zhichao Zheng provided critical revision of the manuscript for important intellectual content.
Disclosure
The authors declare that there are no conflicts of interest related to this study.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30(1):1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang L, Hao X, Tang Y, Wei X, Gong Y. RABEX-5 overexpression in gastric cancer is correlated with elevated MMP-9 level. Am J Transl Res. 2016;8(5):2365–2374. [PMC free article] [PubMed] [Google Scholar]
- 4.Lu S, Zhang Z, Chen M, Li C, Liu L, Li Y. Silibinin inhibits the migration and invasion of human gastric cancer SGC7901 cells by downregulating MMP-2 and MMP-9 expression via the p38MAPK signaling pathway. Oncol Lett. 2017;14(6):7577–7582. doi: 10.3892/ol.2017.7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang X, Xu X, Xue X, et al. NDRG1 controls gastric cancer migration and invasion through regulating MMP-9. Pathol Oncol Res. 2016;22(4):789–796. doi: 10.1007/s12253-016-0071-8 [DOI] [PubMed] [Google Scholar]
- 6.Guo YC, Wang MY, Zhang SW, et al. Ubiquitin-specific protease USP34 controls osteogenic differentiation and bone formation by regulating BMP2 signaling. EMBO J. 2018;37(20):pii:e99398. doi: 10.15252/embj.201899398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lui TT, Lacroix C, Ahmed SM, et al. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/beta-catenin signaling. Mol Cell Biol. 2011;31(10):2053–2065. doi: 10.1128/MCB.01094-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu WT, Huang KY, Lu MC, et al. TGF-beta upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene. 2017;36(19):2715–2723. doi: 10.1038/onc.2016.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inui M, Manfrin A, Mamidi A, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13(11):1368–1375. doi: 10.1038/ncb2346 [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Qiu G, Jiang L, et al. Overexpression of ubiquitin specific proteases 44 promotes the malignancy of glioma by stabilizing tumor-promoter securin. Oncotarget. 2017;8(35):58231–58246. doi: 10.18632/oncotarget.16447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Bedard N, Chevalier S, Wing SS, Polymenis M. Identification of distinctive patterns of USP19-mediated growth regulation in normal and malignant cells. PLoS One. 2011;6(1):e15936. doi: 10.1371/journal.pone.0015936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal A, Donato NJ. Ubiquitin-specific proteases as therapeutic targets for the treatment of breast cancer. Breast Cancer Res. 2014;16(5):461–467. doi: 10.1186/s13058-014-0461-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sippl W, Collura V, Colland F. Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 2011;7(5):619–632. doi: 10.2217/fon.11.39 [DOI] [PubMed] [Google Scholar]
- 14.Pal A, Young MA, Donato NJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014;74(18):4955–4966. doi: 10.1158/0008-5472.CAN-14-1211 [DOI] [PubMed] [Google Scholar]
- 15.Harada K, Kato M, Nakamura N. USP19-mediated deubiquitination facilitates the stabilization of HRD1 ubiquitin ligase. Int J Mol Sci. 2016;17(11):pii:E1829. doi: 10.3390/ijms17111829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne ES, Bedard N, Wykes L, et al. Knockout of USP19 deubiquitinating enzyme prevents muscle wasting by modulating insulin and glucocorticoid signaling. Endocrinology. 2018;159(8):2966–2977. doi: 10.1210/en.2018-00290 [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Pringle LM, Lau AW, et al. TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-kappaB. Oncogene. 2010;29(25):3619–3629. doi: 10.1038/onc.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira AM, Chou MM. USP6-induced neoplasms: the biologic spectrum of aneurysmal bone cyst and nodular fasciitis. Hum Pathol. 2014;45(1):1–11. doi: 10.1016/j.humpath.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 19.Pringle LM, Young R, Quick L, et al. Atypical mechanism of NF-kappaB activation by TRE17/ubiquitin-specific protease 6 (USP6) oncogene and its requirement in tumorigenesis. Oncogene. 2012;31(30):3525–3535. doi: 10.1038/onc.2011.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rueckert C, Haucke V. The oncogenic TBC domain protein USP6/TRE17 regulates cell migration and cytokinesis. Biol Cell. 2012;104(1):22–33. doi: 10.1111/boc.201100108 [DOI] [PubMed] [Google Scholar]
- 21.van der Horst A, de Vries-smits AM, Brenkman AB, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8(10):1064–1073. doi: 10.1038/ncb1469 [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Wen P, Kong R, et al. USP33 mediates Slit-Robo signaling in inhibiting colorectal cancer cell migration. Int J Cancer. 2015;136(8):1792–1802. doi: 10.1002/ijc.29226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia M, Guo Y, Lu X. USP33 is a biomarker of disease recurrence in papillary thyroid carcinoma. Cell Physiol Biochem. 2018;45(5):2044–2053. doi: 10.1159/000488041 [DOI] [PubMed] [Google Scholar]
- 24.Altun M, Zhao B, Velasco K, et al. Ubiquitin-specific protease 19 (USP19) regulates hypoxia-inducible factor 1alpha (HIF-1alpha) during hypoxia. J Biol Chem. 2012;287(3):1962–1969. doi: 10.1074/jbc.M111.305615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Adegoke OA, Nepveu A, et al. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol Cell Biol. 2009;29(2):547–558. doi: 10.1128/MCB.00329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Tian S, Chen Y, et al. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016;35(8):866–880. doi: 10.15252/embj.201593596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedard N, Jammoul S, Moore T, et al. Inactivation of the ubiquitin-specific protease 19 deubiquitinating enzyme protects against muscle wasting. FASEB J. 2015;29(9):3889–3898. doi: 10.1096/fj.15-270579 [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Zhang D, Sun L. Knockdown of ubiquitin-specific protease 14 (USP14) inhibits the proliferation and tumorigenesis in esophageal squamous cell carcinoma cells. Oncol Res. 2017;25(2):249–257. doi: 10.3727/096504016X693164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D, Jiang F, Wang X, Li G. Downregulation of ubiquitin-specific protease 22 inhibits proliferation, invasion, and epithelial-mesenchymal transition in osteosarcoma cells. Oncol Res. 2017;25(5):743–751. doi: 10.3727/096504016X14772395226335 [DOI] [PMC free article] [PubMed] [Google Scholar]