Abstract
Background
Breast cancer (BCa) is an overwhelming malignant tumor mainly in women globally. Circular RNAs (circRNAs) are a special type of noncoding RNAs involved in competing endogenous RNA (ceRNA) network, a classic molecular mechanism of the tumorigenesis of human cancers, including BCa. Here, we intended to explore the role and mechanism of hsa_circ_0000520 (circ_0000520) in BCa cells.
Methods
Expression of circ_0000520, miRNA-1296-5p (miR-1296) and specificity protein 1 (SP1) was measured by real time-quantitative PCR and Western blotting. Cell growth was measured by cell counting kit-8, colony formation assay and flow cytometry method. Cell migration and invasion were assessed by transwell assays and Western blotting. Tumor growth was determined by xenograft models. The direct interaction among circ_0000520, miR-1296 and SP1 was confirmed by dual-luciferase reporter assay and RNA pull-down assay.
Results
circ_0000520 was upregulated in BCa tumors and cell lines (T47D, MCF7, MDA-MB-231, BT549, and SKBR3), and circ_0000520 high expression was associated with poor overall survival. Blocking circ_0000520 suppressed cell viability, colony formation, migration and invasion, but promoted cell cycle arrest and apoptosis rate in MDA-MB-231 and MCF7 cells. circ_0000520 could directly regulate miR-1296 expression, and SP1 was a novel target for miR-1296. Moreover, the anti-tumor role of circ_0000520 silencing was abrogated by miR-1296 downregulation or SP1 restoration. Notably, tumor growth of MDA-MB-231 cells in mice was restrained by circ_0000520 deletion.
Conclusion
circ_0000520 knockdown could suppress cell growth, migration and invasion both in vitro and in vivo through regulating miR-1296/SP1 pathway.
Keywords: circ_0000520, breast cancer, BCa, miR-1296, SP1
Introduction
Breast cancer (BCa) is a common cancer overwhelmingly affecting women globally, and is the leading cause of cancer death.1 It is reported that the incidence of BCa has been rising in the last decade; while, the mortality remains the highest in female patients as well,2 even though advances have been obtained on surgery, endocrine therapy and targeted therapy. Moreover, relapse and metastasis are always occurred in BCa patients, and metastatic breast cancer is refractory to conventional therapies.3 As it’s documented, it has not pinpointed the Achilles heel of BCa as the high heterogeneity at both morphological and molecular phenotypes.4 Genetic changes are involved in tumorigenesis of BCa.5,6 Therefore, novel genetic biomarkers are warranted to be discovered for the early detection and the understanding of the molecular pathways involved in BCa cell growth and metastasis.
The competing endogenous RNA (ceRNA) regulatory mechanism provides a promising look at the hallmarks of human malignant tumors, including BCa.4 Essentially, ceRNA pathway consists of microRNAs (miRNAs), message RNAs (mRNAs), as well as other noncoding RNAs that possess competing shared sequences of miRNAs. Circular RNAs (circRNAs) are a peculiar class of endogenous long noncoding RNAs that are covalently closed, single-stranded transcripts with hundreds to thousands of nucleotides.7 Unlike linear RNAs, circRNAs are resistant to the digestion of exonucleases or RNase R, due to the absence of 5ʹ cap and 3ʹ tail. Thus, circRNAs are more stable in cells and circulating environments, endowing themselves as novel, promising biomarkers for the diagnosis, treatment and prognosis of cancers.8,9 As miRNA sponges, multiple circRNAs have been found to be functionally involved in BCa cells.2 Herein, we wondered the role and mechanism of hsa_circ_0000520 (circ_0000520) in BCa, whose expression is deregulated in BCa tumor tissues according to Gene Expression Omnibus database (GEO; GSE101123 dataset).
MiRNA-1296-5p (miR-1296) is a newly identified metastasis-associated miRNA in several cancers including BCa.10 Moreover, very recently, its expression has also been claimed to be responsible for the prognosis of non-small cell lung cancer.11 In BCa, miR-1296 serves as a tumor suppressor.12–14 However, miR-1296-centered ceRNA network remains largely uncovered in BCa.
Here, we intended to explore the biologic role of circ_0000520 in cell growth and metastasis of BCa, and to figure out the interaction of circ_0000520, miR-1296 and specificity protein 1 (SP1), a basal transcriptional factor that widely regulates genes to contribute to the hallmarks of cancers including BCa.15,16
Materials and Methods
Tissue Samples
A total of 60 patients with primary BCa were recruited from China-Japan Union Hospital of Jilin University after receiving the approval of the Clinical Research Ethics Committee of this hospital. Then, 60 paired BCa tumor tissues and adjacent normal tissues were collected during radical surgery. The tissue collection was after the written informed consent was obtained from every participant, and this study was conducted in accordance with the Declaration of Helsinki. The tissue samples were put into liquid nitrogen immediately. After surgery, all the patients were received standard anti-BCa treatment, and were tracked for 60 months via telephone. The clinical characteristics of these 60 BCa patients are presented in Table 1.
Table 1.
Correlation Between circ_0000520 Expression and Clinicopathological Parameters of Breast Cancer Patients
| Characteristics | Number | circ_0000520 Expression | P | |
|---|---|---|---|---|
| High | Low | |||
| 30 | 30 | |||
| Age (years) | 0.796 | |||
| <50 | 31 | 16 | 15 | |
| ≥50 | 29 | 14 | 15 | |
| Menopause | 0.426 | |||
| No | 37 | 20 | 17 | |
| Yes | 23 | 10 | 13 | |
| Tumor size (cm) | 0.301 | |||
| ≤2 | 28 | 12 | 16 | |
| >2 | 32 | 18 | 14 | |
| Lymph node metastasis | 0.020* | |||
| Yes | 31 | 20 | 11 | |
| No | 29 | 10 | 19 | |
| TNM stage | 0.004* | |||
| I, II | 29 | 9 | 20 | |
| III, IV | 31 | 21 | 10 | |
Note: *Statistically significant.
Abbreviation: TNM, tumor nodes metastasis.
Cell Culture and Cell Transfection
A series of human BCa cell lines including T47D (HTB-133), MCF7 (HTB-22), MDA-MB-231 (HTB-26), BT549 (HTB-122), and SKBR3 (HTB-30), and one normal breast epithelial cell line MCF10A (CRL-10,317) were purchased from American Type Culture Collection (Manassas, VA, USA). All the cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone) in a humidified environment of sterile air (5% CO2, 21% O2 and 74% N2) at 37°C.
For cell transfection, shRNAs targeting circ_0000520 (sh-circ#1: sense 5ʹCGCTATGTGTTCTGGGAAA3ʹ and antisense 5ʹTTTCCCAGAACACATAGCG3ʹ; sh-circ#2: sense 5ʹGAGGTGAGTTCCCAGAGAA3ʹ and antisense 5ʹTTCTCTGGGAACTCACCTC3ʹ), miR-1296 mimic (5ʹUUAGGGCCCUGGCUCCAUCUCC3ʹ), and anti-miRNA against miR-1296 (anti-miR-1296; 5ʹGGAGAUGGAGCCAGGGCCCUAA3ʹ) were synthesized by GenePharma (Shanghai, China), as well as the corresponding negative controls including sh-NC (sense 5ʹCATTAAGCATGATGTCAACCAGACA3ʹ and antisense 5ʹTGTCTGGTTGACATCATGCTTAATG3ʹ), miR-NC mimic (5ʹ UUCUCCGAACGUGUCACGUTT3ʹ), and anti-NC (5ʹCAGUACUUUUGUGUAGUACAA3ʹ). The pIRES2-EGFP vector (Clontech, Mountain View, CA, USA) was selected to construct SP1 overexpression vector (pIRES2-EGFP-SP1). MDA-MB-231 and MCF7 cells were detached by 0.25% trypsin, and passaged at 40% confluency in 6-well plate for 24 h. The transfection was conducted following the manufacturer’s protocol. Briefly, the cells were changed with fresh DMEM without FBS, and incubated with a mixture of Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) and nucleotides (50 nM oligonucleotides or 2 μg vectors) for 6 h. The DMEM medium was refreshed with complete medium for another 24 h for further analysis.
Real Time-Quantitative PCR (RT-qPCR)
Total RNA in tissues and cells was isolated by RNAsimple total RNA kit (TIANGEN, Beijing, China). A total of 500 ng RNA was then used to synthesize cDNA using PrimeScript RT reagent Kit (Takara, Dalian, China), and the cDNA was further amplified using special primers and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (2×) (Takara). The miRNA expression was detected using Mir-X miRNA First-Strand Synthesis Kit (Takara) and Mir-X miRNA qRT-PCR TB Green® Kit (Takara). The primers including circ_0000520 (forward 5ʹGTCTGAGACTAGGGCCAGAGGC3ʹ and reverse 5ʹGACATGGGAGTGGAGTGACAGG3ʹ), miR-1296 (forward 5ʹTTGTTAGGGCCCTGGCTC3ʹ and reverse 5ʹGTGCAGGGTCCGAGGT3ʹ), SP1 (forward 5ʹAATTTGCCTGCCCTGAGTGC3ʹ and reverse 5ʹTTGGACCCATGCTACCTTGC3ʹ), glyceraldehyde-phosphate dehydrogenase (GAPDH; forward 5ʹTCACCAGGGCTGCTTTTAAC3ʹ and reverse 5ʹTGACGGTGCCATGGAATTTG3ʹ), and small nuclear U6 (U6; 5ʹ ATACAGAGAAAGTTAGCACGG3ʹ and reverse 5ʹGGAATGCTTCAAAGAGTTGTG3ʹ). The amplification curve and melting curve were analyzed on ABI 7700 (Foster City, CA, USA), and comparative cycle threshold (Ct) method (2−ΔΔCt) was used to calculate gene expression. The reaction system was conducted in four replicate wells for each sample.
Cell Counting Kit-8 (CCK-8) and Colony Formation Assay
After transfection, the MDA-MB-231 and MCF7 cells were immediately transferred onto 96-well plate at density of 104 cells per well for another 48 h. Afterwards, the cells were stained with 100 μL of CCK‐8 solution (Dojindo, Kumamoto, Japan) for 2 h at 37°C. Next, the absorbance at 450 nm was measured using a microplate reader. The assay was conducted in six replicate wells for each sample, and cells transfected with negative control were considered as 100% cell viability.
After transfection, the MDA-MB-231 and MCF7 cells were passaged in a new 6-well plate at 1000 cells per well. Then, the cells were replaced with fresh complete medium every three days, and cultivated for another 15 days. After that, the cells were washed with cold phosphate buffered saline (PBS), dried in the air, fixed with methanol for 15 min, and stained with 0.25% crystal violet for 20 min. The number of colony (more than 50 cells) was counted under a microscope.
Flow Cytometry (FCM)
After transfection for 24 h, the MDA-MB-231 and MCF7 cells were collected together with the floating cells. For cell apoptosis rate analysis, the cells were washed with cold PBS, re-suspended in the binding buffer, and then stained with 5 μL of Annexin V-fluorescein isothiocyanate (FITC; BD Biosciences, Franklin Lakes, NJ, USA) and 5 μL of propidium iodide (PI; BD Biosciences) for 30 min in the dark. For cell cycle analysis, the cell suspension was fixed with cold 75% ethanol at 4°C for 16 h prior to staining with 5 μL of PI (BD Biosciences) supplemented with 200 μg/mL of RNase A. Finally, the labelled cells were determined on FACScan flow cytometer (BD Biosciences) equipped with Cell Quest software (BD Biosciences). Apoptosis rate (%) was calculated with the percentage of cells in Annexin V+/PI+ and Annexin V+/PI-quadrants. Cell rate (%) in different cell cycle phases was calculated with the percentage of cells in G0/G1, S and G2/M phases, respectively.
Transwell Assays
Transfected cells were harvested to measure abilities of cell invasion and migration by transwell chambers (Corning, Corning, NY, USA) coated with matrigel (BD Biosciences) or not. For invasion assay, matrigel membrane was formed after being diluted in 1:10 serum-free DMEM and then placed at 37°C for 4–6 h. For migration and invasion assays, 5×105 transfected cells were re-suspended in 200 μL of DMEM without FBS, then loaded in the upper layer of the chamber. The 400 μL of DMEM containing 10% FBS was transferred into the lower of the chamber. This transwell migration or invasion system was maintained for 48 h in normal cell culture condition. The cells on the bottom of chamber were fixed with methanol for 15 min and dyed with 0.25% crystal violet for 20 min. A total of 5 randomly fields (100×) were captured under microscope, and the numbers of migrated cells and invaded cells were counted.
Western Blotting
Western blotting was performed according to the standard methods as previously described. The primary antibodies including E-cadherin (E-cad; 20,874-1-AP, 1:50,000), N-cadherin (N-cad; 22,018-1-AP, 1:10,000), SP1 (21,962-1-AP, 1:2500), and GAPDH (10,494-1-AP, 1:40,000) were from Proteintech (Deansgate, Manchester, UK). The secondary antibody goat-anti-rabbit (ab205718, 1:50,000) was from Abcam (Cambridge, UK). GAPDH was the internal control. The densitometry of Western blotting blots was measured using Image J software.
Dual-Luciferase Reporter Assay and RNA Pull-Down Assay
The synthetic circ_0000520 and 3ʹ untranslated region of SP1 (SP1 3ʹUTR) were inserted into pGL4 (Promega, Madison, WI, USA), respectively. The mutant of potential binding sites of miR-1296 on circ_0000520 and SP1 3ʹUTR were mutated into the complementary bases, and then inserted into pGL4 (Promega) as well. The recombinant vector pGL4 carrying circ_0000520 wild type fragment (circ_0000520-WT), circ_0000520 mutant fragment (circ_0000520-MUT), SP1 3ʹUTR wild type fragment (SP1 3ʹUTR-WT), and SP1 3ʹUTR mutant fragment (SP1 3ʹUTR-MUT) were co-transfected into MDA-MB-231 and MCF7 cells (in 24-well plate) together with miR-1296 mimic or miR-NC mimic. After transfection for 48 h, the cells were lysed with commercial lysis buffer (Ambion, Austin, TX, USA), and the luciferase activity was measured on Dual-luciferase Reporter Assay System (Promega). Every transfection group was repeated in three wells.
The biotin labelled miR-1296 (Bio-miR-1296) and miR-NC (Bio-miR-NC) were from Sangon (Shanghai, China). MDA-MB-231 and MCF7 cells in 6-well plate were transfected with 50 nM of Bio-miR-1296/NC for 24 h. After that, the transfected cells were collected, washed with cold PBS, and lysed with lysis buffer (Ambion). Then, the cell lysate was incubated with M-280 streptavidin-coated magnetic beads (Sigma-Aldrich, St Louis, MO, USA) for 4 h at 6°C. By the way, the beads were pre-coated with RNase-free BSA and yeast tRNA. After washing, the RNAs pulled down were detected by RT-qPCR method.
Animal Experiment
MDA-MB-231 cells transfected with sh-circ#1 or sh-NC were harvested at a concentration of 2×107 cells/mL. A total of 8 female BALB/c mice (6-week-old) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China), and randomly divided into sh-NC group (n=4) and sh-circ#1 group (n=4). Then, the mice were subcutaneously injected with 0.2 mL of MDA-MB-231 cells on both sides of posterior flanks prior to another 35 days feeding. During the feeding, the tumor size was measured every 7 days on the length and width, and the tumor weight was examined on the last day after the mice were euthanatized. The tumor volume was calculated by the product of 0.5×length×width2. The animal experiment was approved by the Use and Care of Animals Committee of China-Japan Union Hospital of Jilin University, and was in accordance with the Laboratory Animals-Guideline of welfare and ethics published by “National Institutes of Health”.
Statistical Analysis
Data were presented as the mean ± standard deviation from three independent experiments, and analyzed on GraphPad Prism (GraphPad Software, La Jolla, CA, USA). The difference in groups was performed using paired sample Student’s t-test and one-way analysis of variance. P<0.05 was considered as significant difference, and signed as *<0.01 was very significant difference, and signed as **and <0.001 was extremely significant difference, and signed as ***.
Results
circ_0000520 Was Upregulated in BCa Tumor Tissues and Cells
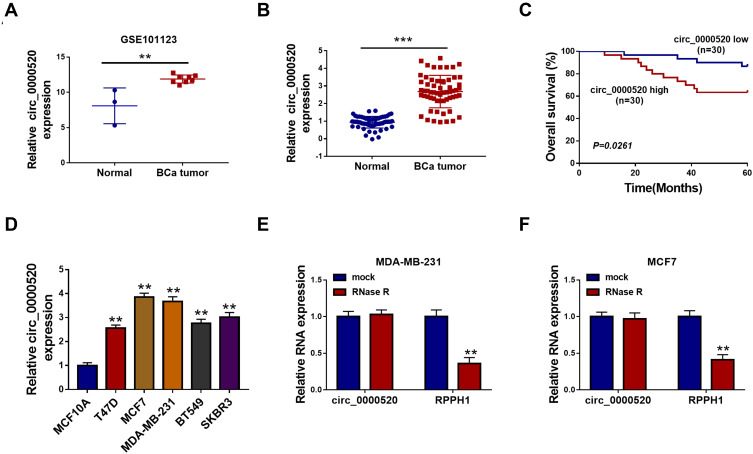
In order to explore the dysregulation of circRNAs in BCa, we grabbled in silicon data in GEO database. According to GSE101123 dataset, microarray-seq data showed that circ_0000520 was comparatively abundant, and was highly expressed in BCa tumor tissues (Figure 1A). Thus, we identified circ_0000520 expression status in recruited 60 BCa patients using RT-qPCR, and found that circ_0000520 level in BCa tumors was 2.68-fold of that in paired normal controls (Figure 1B). Additionally, the relationship between circ_0000520 expression and clinicopathological parameters of this cohort of BCa patients was further analyzed. As a result, high expression of circ_0000520 was associated with lymph node metastasis and TNM stage (Table 1). Moreover, 5-year overall survival rate of patients in circ_0000520 low group (n=30) was 86.7%, paralleled with 63.3% in circ_0000520 high group (n=30) (Figure 1C). Similarly, expression of circ_0000520 was overall upregulated in human BCa cell lines (T47D, MCF7, MDA-MB-231, BT549, and SKBR3) than normal MCF10A cells (Figure 1D). circ_0000520 was located in gene RPPH1 on chromosome 14:20,811,436–20,811,559, with a spliced sequence length of 123 nucleotides. Moreover, circ_0000520 was resistant, while RPPH1 was sensitive to RNase R digestion in MDA-MB-231 and MCF7 cells (Figure 1E and F). These data suggested that circ_0000520 was upregulated in BCa tumors and cells.
Figure 1.
Expression of circular RNA hsa_circ_0000520 (circ_0000520) in breast cancer (BCa) tissues and cells. (A) circRNAs chip determined the expression of circ_0000520 in 8 BCa tumor tissues (BCa tumor) and 3 non-tumor breast tissues (Normal) according to GSE101123 dataset. (B) RT-qPCR measured relative circ_0000520 expression in 60 paired BCa tumor and Normal groups. (C) Kaplan-Meier survival plots analyzed the overall survival (%) of these BCa cases with circ_0000520 high (≥Median, n=30) or low (<Median, n=30). (D) RT-qPCR tested circ_0000520 expression in human BCa cell lines (T47D, MCF7, MDA-MB-231, BT549, and SKBR3) and normal breast epithelial cell line (MCF10A). (E and F) RT-qPCR validated relative RNA expression of circ_0000520 and its parent gene RPPH1 with or without RNase R treatment in MDA-MB-231 and MCF7 cells. **Represented P<0.01 and ***Represented P<0.001.
Blocking circ_0000520 Suppressed Cell Growth, Migration and Invasion of BCa Cells in vitro
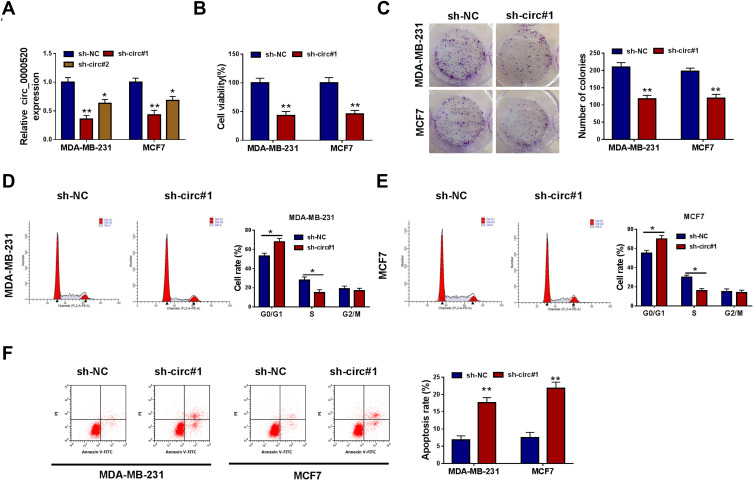
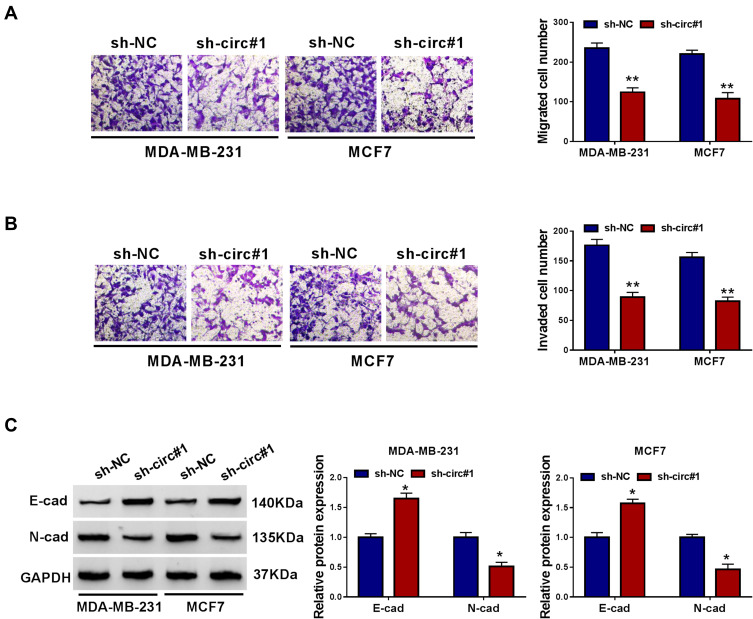
Loss-of-functional experiments in vitro were carried out to investigate the biologic role of circ_0000520 in BCa cells. First of all, circ_0000520 was knocked down using shRNA transfection, and RT-qPCR data depicted a significant reduction of circ_0000520 level in MDA-MB-231 and MCF7 cells due to sh-circ#1 and #2 transfection (Figure 2A); moreover, sh-circ#1 resulted in more than 55% silencing efficiency in both cells, thus sh-circ#1 was selected for the further functional analysis. We observed that cell viability and colony formation ability were attenuated in circ_0000520-silenced MDA-MB-231 and MCF7 cells, as examined by CCK-8 and colony formation assays (Figure 2B and C). FCM method was used to measure cell cycle and apoptosis, and indicated an increase of G0/G1-phase cells and apoptosis rate in MDA-MB-231 and MCF7 cells administrated with sh-circ#1 than sh-NC, as well as reduced S-phase cells (Figure 2D–F). Transwell assays and Western blotting evaluated cell migration and invasion abilities. Introducing sh-circ#1 decreased migrated cell number and invaded cell number in MDA-MB-231 and MCF7 cells, compared to sh-NC introduction (Figure 3A and B). Besides, E-cad expression was promoted, and N-cad expression was inhibited by circ_0000520 silencing (Figure 3C). Taken above results together, blocking circ_0000520 via transfection suppressed cell viability, colony formation, migration and invasion, but promoted cell cycle arrest and apoptosis in MDA-MB-231 and MCF7 cells, suggesting a suppressive role of circ_0000520 knockdown in BCa cell growth, migration and invasion in vitro.
Figure 2.
Blocking circ_0000520 suppressed cell growth of BCa cells in vitro. MDA-MB-231 and MCF7 cells were administrated with shRNAs against circ_0000520 (sh-circ#1 and #2) or scrambled shRNA (sh-NC). (A) RT-qPCR measured relative circ_0000520 expression on 24 h. (B) CCK-8 assay detected cell viability (%) on 48 h, and (C) colony formation assay measured colony numbers on 15 d. (D and E) Flow cytometry (FCM) method examined cell rate (%) in different cell cycle phases on 24 h. (F) FCM method evaluated apoptosis rate (%) on 24 h. *Represented P<0.05 and **Represented P<0.01.
Abbreviations: Annexin V-FITC, fluorescein isothiocyanate-coated annexin V; PI, propidium iodide.
Figure 3.
Blocking circ_0000520 inhibited cell migration and invasion of BCa cells in vitro. MDA-MB-231 and MCF7 cells were administrated with sh-circ#1 or sh-NC. (A and B) Transwell assay measured migrated cell number and invaded cell number on 48 h. The most representative images were presented. (C) Western blotting evaluated E-cadherin (E-cad) and N-cadherin (N-cad) protein expression on 24 h. The most representative images were presented. *Represented P<0.05 and **Represented P<0.01.
miR-1296 Was Targeted by circ_0000520 in BCa Cells
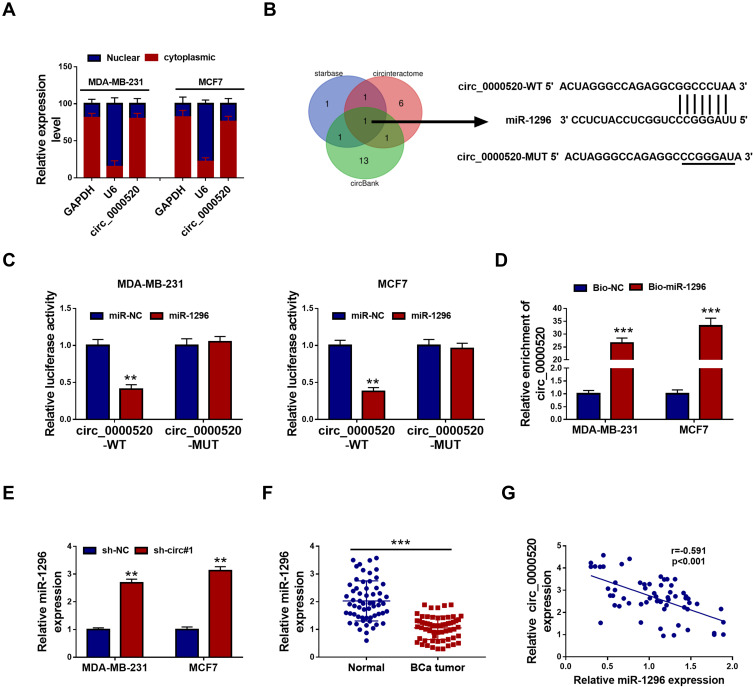
We confirmed a tumor-suppressive role of circ_0000520 knockdown in BCa, and its molecular mechanism was further identified. With separation of nuclear and cytoplasmic fractions, we found that circ_0000520 was enriched in cytoplasmic fractions, similar to GAPDH (Figure 4A), suggesting circ_0000520 was probably mainly distributed in the cytoplasm. Thus, we assumed that circ_0000520 could associate with miRNAs sponging. Furthermore, circinteractome, circBank and starbase were employed to predict the potential target miRNAs of circ_0000520. As shown in Figure 4B, miR-1296 was the common one among these three websites. Then, miR-1296 was the only candidate miRNA for further identification. Dual-luciferase reporter assay showed a decrease of circ_0000520-WT vector in miR-1296 mimic-transfected MDA-MB-231 and MCF7 cells (Figure 4C); RNA pull-down assay illuminated an enrichment of circ_0000520 in Bio-miR-1296 pull-down contents in MDA-MB-231 and MCF7 cells (Figure 4D); in addition, miR-1296 expression could be upregulated in the presence of sh-circ#1 (Figure 4E). Expression status of miR-1296 was lower in BCa tumor tissues in circ_0000520-dependent manner (Figure 4F and G). Thereby, we validated circ_0000520 as a sponge for miR-1296.
Figure 4.
MiRNA-1296-5p (miR-1296) was targeted by circ_0000520 in BCa cells. (A) RT-qPCR analyzed nuclear and cytoplasmic RNA expression levels of glyceraldehyde-phosphate dehydrogenase (GAPDH), small nuclear U6 (U6) and circ_0000520. (B) Bioinformatics algorithms predicted potential target miRNAs of circ_0000520. (C) Dual-luciferase reporter assay identified the direct interaction between circ_0000520 and miR-1296 in MDA-MB-231 and MCF7 cells. (D) RNA pull-down assay validated the enrichment of circ_0000520 by biotin labelled miR-1296 (Bio-miR-1296) or miR-NC (Bio-NC) in MDA-MB-231 and MCF7 cells. (E and F) RT-qPCR tested miR-1296 expression in MDA-MB-231 and MCF7 cells transfected with sh-circ#1 or sh-NC, and in paired BCa tumor and Normal groups (n=60). (G) Pearson correlation coefficient (r) analysis determined the relationship between circ_0000520 and miR-1296 expression in these BCa patients (n=60). **Represented P<0.01 and ***Represented P<0.001.
Abbreviations: miR-1296, miR-1296 mimic; miR-NC, miR-NC mimic.
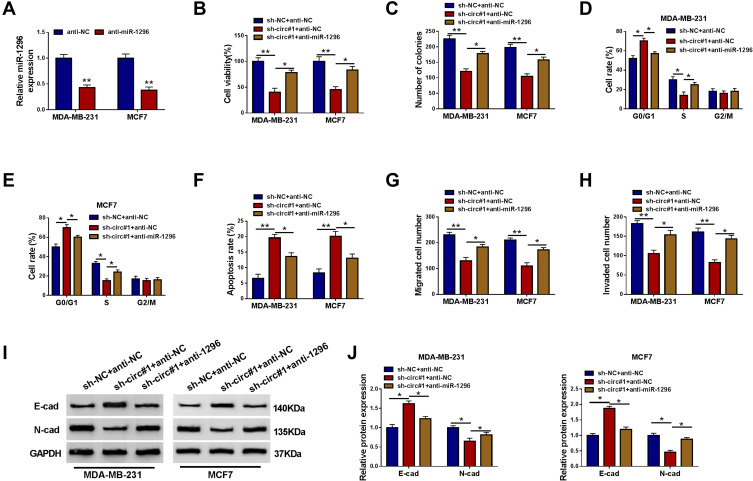
Silencing miR-1296 Countermanded the Suppressive Role of circ_0000520 Knockdown in BCa Cells in vitro
Circ_0000520 knockdown was accompanied with upregulation of miR-1296, thus we examined the effect of miR-1296 inhibition on the anti-tumor role of circ_0000520 knockdown in BCa cells. The exogenous administration of anti-miR-1296 could lead to miR-1296 inhibition in MDA-MB-231 and MCF7 cells (Figure 5A). Rescue experiments demonstrated that the low cell viability and colony formation ability in circ_0000520-silenced MDA-MB-231 and MCF7 cells were elevated by anti-miR-1296 transfection (Figure 5B and C); the sh-circ#1-mediated high rates of G0/G1-phase cells and apoptotic cells were descended by miR-1296 deletion (Figure 5D–F); cell migration and invasion capacities were facilitated by circ_0000520 downregulation, and this promoting activity was partially reversed in the presence of anti-miR-1296, as evidenced by increased migrated cells and invaded cells (Figure 5G and H), as well as higher E-cad and lower N-cad expression (Figure 5I and J). These before-mentioned outcomes indicated that silencing miR-1296 could countermand the suppressive role of circ_0000520 knockdown in BCa cell growth, migration and invasion in vitro.
Figure 5.
The effect of miR-1296 downregulation on the role of circ_0000520 knockdown in BCa cells in vitro. (A) RT-qPCR measured miR-1296 expression in MDA-MB-231 and MCF7 cells introduced with anti-miRNA against miR-1296 (anti-miR-1296) or its negative control anti-NC. (B–J) MDA-MB-231 and MCF7 cells were co-transfected with sh-NC and anti-miR-NC, sh-circ#1 and anti-NC, sh-circ#1 and anti-miR-1296. (B) CCK-8 assay detected cell viability (%) on 48 h, and (C) colony formation assay measured colony numbers on 15 d. (D and E) FCM method examined cell rate (%) in different cell cycle phases on 24 h. (F) FCM method evaluated apoptosis rate (%) on 24 h. (G and H) Transwell assay measured migrated cell number and invaded cell number on 48 h. (I and J) Western blotting evaluated E-cad and N-cad protein expression on 24 h. *Represented P<0.05 and **Represented P<0.01.
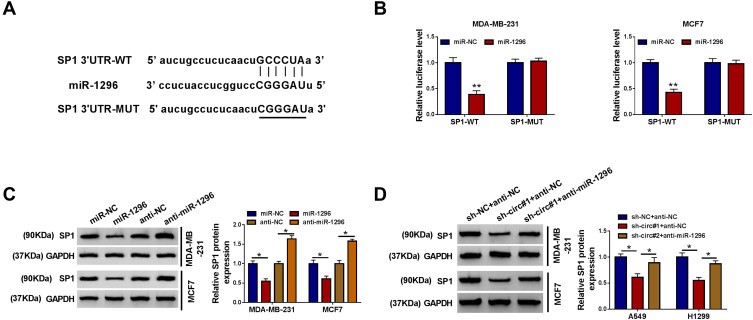
Ectopic Expression of SP1 Counteracted the Suppressive Role of circ_0000520 Knockdown in BCa Cells in vitro Through Directly Regulated by miR-1296
The arithmetic of starbase also predicted a potential binding sequence of miR-1296 on SP1 3ʹUTR (Figure 6A). Luciferase activity of SP1 3ʹUTR-WT was also decreased in MDA-MB-231 and MCF7 cells introduced with miR-1296 mimic (Figure 6B). Besides, SP1 protein expression was inhibited by miR-1296 mimic transfection, and promoted by anti-miR-1296 transfection (Figure 6C). These data supported a direct interaction between miR-1296 and SP1 through target binding.
Figure 6.
Specificity protein 1 (SP1) was a direct target gene for miR-1296. (A) The starbase website predicted a potential binding site of miR-1296 on 3ʹ untranslated region of SP1 (SP1 3ʹUTR). (B) Dual-luciferase reporter assay identified the direct interaction between SP1 3ʹUTR and miR-1296 in MDA-MB-231 and MCF7 cells. (C and D) Western blotting examined SP1 protein expression in MDA-MB-231 and MCF7 cells transfected with miR-1296, miR-NC, anti-miR-1296, or anti-NC, and co-transfected with sh-NC and anti-NC, sh-circ#1 and anti-NC, or sh-circ#1 and anti-miR-1296. *Represented P<0.05 and **Represented P<0.01.
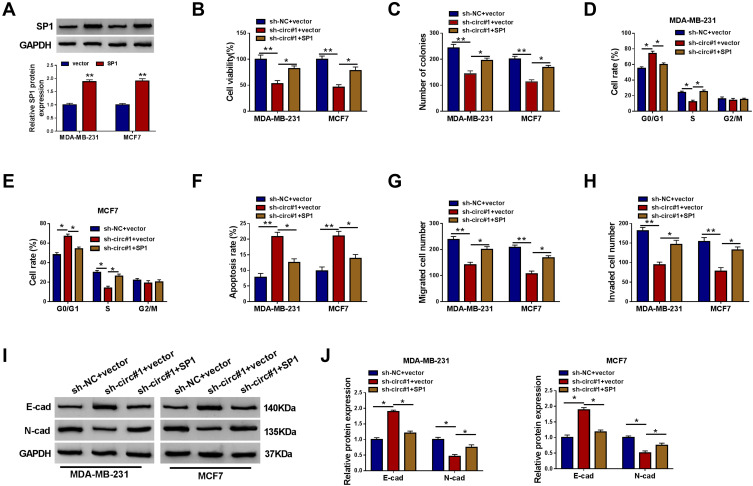
The presence of sh-circ#1 led to lower level of SP1 in MDA-MB-231 and MCF7 cells, which could be improved by anti-miR-1296 (Figure 6D). Moreover, SP1 overexpression vector transfection caused SP1 upregulation (Figure 7A). Restoration of SP1 could rescue cell growth of circ_0000520-silenced MDA-MB-231 and MCF7 cells in vitro, as evidenced by increased cell viability and colony formation ability (Figure 7B and C), as well as S-phase cell rate (Figure 7D and E). Cell apoptosis rate was enhanced by sh-circ#1, and this effect was abolished by SP1 overexpression via transfection (Figure 7F). Cell migration and invasion of MDA-MB-231 and MCF7 cells were facilitated when circ_0000520 was knocked down, which could be further counteracted due to SP1 upregulation (Figure 7G–J). Collectively, we proposed the anti-tumor role of circ_0000520 downregulation was dependent on SP1 inhibition via miR-1296.
Figure 7.
The effect of SP1 restoration on the role of circ_0000520 knockdown in BCa cells in vitro. (A) Western blotting measured SP1 protein expression in MDA-MB-231 and MCF7 cells introduced with pIRES2-EGFP-SP1 vector (SP1) or pIRES2-EGFP vector (vector). (B–J) MDA-MB-231 and MCF7 cells were co-transfected with sh-NC and vector, or sh-circ#1 and vector, sh-circ#1 and SP1. (B) CCK-8 assay detected cell viability (%) on 48 h, and (C) colony formation assay measured colony numbers on 15 d. (D and E) FCM method examined cell rate (%) in different cell cycle phases on 24 h. (F) FCM method evaluated apoptosis rate (%) on 24 h. (G and H) Transwell assay measured migrated cell number and invaded cell number on 48 h. (I and J) Western blotting evaluated E-cad and N-cad protein expression on 24 h. *Represented P<0.05 and **Represented P<0.01.
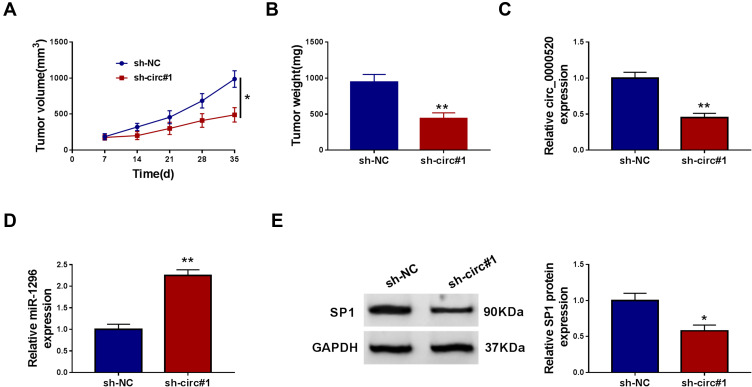
Silencing circ_0000520 Restrained Tumorigenesis of BCa Cells in vivo
We already defined circ_0000520 as an oncogene in BCa cells in vitro, the role of circ_0000520 in tumorigenesis of BCa cells was studied in depth in vivo. Subcutaneous xenograft tumor models displayed a restrained tumor growth in sh-circ#1 group, as supported by reduced tumor volume and tumor weight (Figure 8A and B). Notably, the gene expression in neoplasm tissues was further testified. RT-qPCR analysis demonstrated a low expression of circ_0000520 and SP1, and Western blotting data declared a high expression of miR-1296 in mice neoplasms in circ#1 group (Figure 8C–E). These results preliminarily revealed a tumor-suppressive role of circ_0000520 silencing in tumorigenesis of BCa cells in vivo through regulating miR-1296 and SP1.
Figure 8.
Silencing circ_0000520 restrained tumorigenesis of BCa cells in vivo. MDA-MB-231 cells transfected with sh-circ#1 or sh-NC were subcutaneously transplanted into both sides of posterior flanks in nude mice (n=4). (A) Tumor volume was calculated every 7 days after implantation. (B) Tumor weight was measured after implantation for 35 days. (C and D) RT-qPCR measured circ_0000520 and miR-1296 expression, and (E) Western blotting detected SP1 protein expression in xenograft neoplasm tissues. *Represented P<0.05 and **Represented P<0.01.
Discussion
Very recently, circRNAs had been as emerging new biomarkers for BCa, and potential role of several circRNAs were uncovered. For example, circ-TFF1 was indicated to serve as an enhancer in cell proliferation, migration, invasion and epithelial-mesenchymal transition of BCa cells through absorbing miR-326,17 and circ_002178 was a tumor suppressor in BCa cell viability, energy metabolism and tube formation ability by sponging miR-328-3p.18 Furthermore, circRNA-miRNA networks were proposed to be responsible for the development, prognosis and therapy of BCa.19 However, the link between circRNAs and BCa progression had not been well disclosed. Herewith, we researched the differently expressed circRNAs in GEO database (GSE101123 dataset), and found one interested circRNA, that was circ_0000520. Subsequently, we aimed to identify the expression of circ_0000520 in a group of BCa patients, and to figure out the biological role of circ_0000520 in BCa cells, as well as the underlying circ_0000520-miRNA-mRNA pathway.
Together with RT-qPCR data in BCa tumor tissues, we considered that circ_0000520 was upregulated in BCa. The high expression of circ_0000520 seemed to predict a shorter 5-year overall survival in this cohort of BCa patients. Functional experiments displayed circ_0000520 could be an oncogene in BCa cell growth, migration and invasion both in vitro and in vivo. Therefore, we suggested circ_0000520 as a potential potent biomarker in the prediction and prognosis of BCa, due to its unique structure property. Thus, it could be better to further investigate the expression of circ_0000520 in the plasma of patients, together with the relationship between plasma circ_0000520 and either clinical features or BCa cell processes.20,21 According to previous studies, circ_0000520 was abnormally expressed in hepatocellular carcinoma (HCC) and gastric cancer (GCa).21,22 In HCC, circ_0000520 existed in hepatic tissue, and was highly expressed in HCC tissues (n=3) than non-tumor tissues depending on circRNA microarray scanning.22 In GCa, circ_0000520 was downregulated in tumor tissues, plasma and GCa cell lines from circRNA microarray (n=5) and RT-qPCR (n=56);21 moreover, plasma circ_0000520 showed a remarkable high diagnostic value in GCa patients. Taken our data and above two publications together, it might conclude that circ_0000520 existed in tissues in a content-dependent manner and could be a biomarker in cancers.
The circinteractome database provided a circ_0000520-miRNA-mRNA interaction network, including miR-556-5p, miR-224, miR-146b-3p, and miR-1296.21 Here, we found that miR-1296 was the only one common potential target miRNA according to circinteractome, circBank and starbase. Thus, we took miR-1296 as a candidate target for circ_0000520, and further testified this hypothesis using dual-luciferase reporter assay and RNA pull-down assay. MiR-1296 had also been declared to exhibit tumor suppressive role in BCa. For example, Phan et al14 probably gave birth to the first evidence of the anti-tumor activity of miR-1296 through the decrease of cell proliferation and S-phase cells in triple-negative BCa cells (MDA-MB-231 and MDA-MB-436) by directly regulating cyclinD1. Chen et al12 noticed that miR-1296 overexpression inhibited clonogenic capacity and drug-resistance of human epidermal growth factor receptor 2 (HER2)-positive BCa cells (SK-BR-3 and BT-474) by targeting HER2. In addition, a number of miRNAs including miR-1296 were associated with chemoresistance and self-renewal capability of MCF7 (estrogen receptor (ER)-positive) cells.13 Here, we suggested a similar inhibitory role of miR-1296 in BCa cells MDA-MB-231 and MCF7 cells. The higher level of miR-1296 was underlay the suppressive effect of circ_0000520 silencing on cell viability, colony formation, migration and invasion of BCa cells, as well as the promoting effect on cell apoptosis and cell cycle arrest. The target of miR-1296 was also validated, and SP1 was a novel downstream target of this miRNA.
SP1, belonging to the SP/Kruppel like factor family, was well-documented to be overexpressed in different cancers.15 In BCa, SP1 was 71.67% positive in tumor tissues (n=60), and was correlated with the cell proliferation, invasion and metastasis.23 Mechanically, on one hand, a regulatory axis of miRNAs and SP1 had been established. For example, miR-200b, miR-539 and miR-411 targeted SP1 to suppress cell proliferation, cell cycle progression, cisplatin resistance, epithelial-mesenchymal transition, migration and invasion of BCa cells.24–26 Furthermore, an auto-regulatory loop of miR-22-SP1-c-Myc-miR-22 was also observed to participate in BCa invasion and metastasis.27 On the other hand, an indirect interaction between miRNAs and SP1 had also been discovered. For instance, miR-106b could depress the expression of ZBTB4, a suppressor of SPs including SP1, to regulate E2H2 activity, thus affecting tumor growth and colonization of BCa cells in vivo.28 Besides, Mertens-Talcott et al29 early pointed out miR-27a-ZBTB10 axis to modulate G2/M checkpoint in MDA-MB-231 cells, and ZBTB10 was also a SPs repressor. Here, our data identified that SP1 was one target of miR-1296 in BCa, and could be regulated by circ_0000520. Functionally, restoration of SP1 could improve cell viability, S-phase cells, colony formation ability, and migratory and invasive capacities, but decrease cell apoptosis rate in MDA-MB-231 and MCF7 cells. Lower expression of SP1 was hidden in the suppression of circ_0000520 knockdown on cell proliferation, invasion and tumor growth, which was in line with the previous data in SKBR3 and MDA-MBA-231 cells.30 The pity of this study was that the relevant signaling pathway underlying had not further explored, such as Smad3 signaling, Erk, NF-κB signaling pathways.31,32
In conclusion, this present study showed an upregulation of circ_0000520 in BCa tumor tissues and cell lines, and an anti-tumor role of circ_0000520 inhibition on cell viability, cell cycle progression, apoptosis, migration and invasion in MDA-MB-231 and MCF7 cells, as well as on tumor growth in vivo. The underlying molecular mechanism of circ_0000520 in BCa cells was through regulating miR-1296-SP1 axis. Thereby, we suggested circ_0000520-miR-1296-SP1 pathway as a potential target for the treatment and prognosis of BCa.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Chen Z, Hu G, Jiang Y. Roles of circular RNA in breast cancer: present and future. Am J Transl Res. 2019;11(7):3945–3954. [PMC free article] [PubMed] [Google Scholar]
- 3.Hattori M, Iwata H. Advances in treatment and care in metastatic breast cancer (MBC): are there MBC patients who are curable? Chin Clin Oncol. 2018;7(3):23. doi: 10.21037/cco.2018.05.01 [DOI] [PubMed] [Google Scholar]
- 4.Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi: 10.1002/jcp.27941 [DOI] [PubMed] [Google Scholar]
- 5.Hu X, Stern HM, Ge L, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7(4):511–522. doi: 10.1158/1541-7786.MCR-08-0107 [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Wang Z, Li S, et al. Combinatorial epigenetic regulation of non-coding RNAs has profound effects on oncogenic pathways in breast cancer subtypes. Brief Bioinform. 2018;19(1):52–64. doi: 10.1093/bib/bbw099 [DOI] [PubMed] [Google Scholar]
- 7.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. doi: 10.1080/15476286.2018.1486659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 9.Jahani S, Nazeri E, Majidzadeh AK, Jahani M, Esmaeili R. Circular RNA; a new biomarker for breast cancer: a systematic review. J Cell Physiol. 2020;235(7–8):5501–5510. doi: 10.1002/jcp.29558 [DOI] [PubMed] [Google Scholar]
- 10.Lee SC, Quinn A, Nguyen T, Venkatesh S, Quinn TP. A cross-cancer metastasis signature in the microRNA-mRNA axis of paired tissue samples. Mol Biol Rep. 2019;46(6):5919–5930. doi: 10.1007/s11033-019-05025-w [DOI] [PubMed] [Google Scholar]
- 11.Deng H, Xie C, Ye Y, Du Z. MicroRNA-1296 expression is associated with prognosis and inhibits cell proliferation and invasion by Wnt signaling in non-small cell lung cancer. Oncol Lett. 2020;19(1):623–630. doi: 10.3892/ol.2019.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen G, He M, Yin Y, et al. miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell Int. 2017;17:95. doi: 10.1186/s12935-017-0466-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boo L, Ho WY, Ali NM, et al. MiRNA transcriptome profiling of spheroid-enriched cells with cancer stem cell properties in human breast MCF-7 cell line. Int J Biol Sci. 2016;12(4):427–445. doi: 10.7150/ijbs.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan B, Majid S, Ursu S, et al. Tumor suppressor role of microRNA-1296 in triple-negative breast cancer. Oncotarget. 2016;7(15):19519–19530. doi: 10.18632/oncotarget.6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282(2):224–258. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Li K, Mei Y, et al. Sp1 suppresses miR-3178 to promote the metastasis invasion cascade via upregulation of TRIOBP. Mol Ther Nucleic Acids. 2018;12:1–11. doi: 10.1016/j.omtn.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720. doi: 10.1111/cpr.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24(3):2189–2201. doi: 10.1111/jcmm.14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang SJ, Wang DD, Zhou SY, et al. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12(2):101–125. doi: 10.2217/epi-2019-0058 [DOI] [PubMed] [Google Scholar]
- 20.Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi: 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. doi: 10.3233/CBM-170379 [DOI] [PubMed] [Google Scholar]
- 22.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore). 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XB, Peng WQ, Yi ZJ, Zhu SL, Gan QH. [Expression and prognostic value of transcriptional factor sp1 in breast cancer]. Ai Zheng. 2007;26(9):996–1000. Chinese. [PubMed] [Google Scholar]
- 24.Yao Y, Hu J, Shen Z, et al. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015;19(4):760–769. doi: 10.1111/jcmm.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai F, Chen L, Sun Y, He C, Fu D, Tang J. MiR-539 inhibited the malignant behaviors of breast cancer cells by targeting SP1. Biochem Cell Biol. 2020;98(3):426–433. doi: 10.1139/bcb-2019-0111 [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Yuan J, Xie N, et al. miRNA-411 acts as a potential tumor suppressor miRNA via the downregulation of specificity protein 1 in breast cancer. Mol Med Rep. 2016;14(4):2975–2982. doi: 10.3892/mmr.2016.5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74(14):3764–3778. doi: 10.1158/0008-5472.CAN-13-3555 [DOI] [PubMed] [Google Scholar]
- 28.Yang WS, Chadalapaka G, Cho SG, et al. The transcriptional repressor ZBTB4 regulates EZH2 through a microRNA-ZBTB4-specificity protein signaling axis. Neoplasia. 2014;16(12):1059–1069. doi: 10.1016/j.neo.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67(22):11001–11011. doi: 10.1158/0008-5472.CAN-07-2416 [DOI] [PubMed] [Google Scholar]
- 30.Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget. 2016;7(16):22245–22256. doi: 10.18632/oncotarget.7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Ma J, Fan Y, et al. TGF-beta transactivates EGFR and facilitates breast cancer migration and invasion through canonical Smad3 and ERK/Sp1 signaling pathways. Mol Oncol. 2018;12(3):305–321. doi: 10.1002/1878-0261.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JY, Jung HH, Ahn S, et al. The relationship between nuclear factor (NF)-kappaB family gene expression and prognosis in triple-negative breast cancer (TNBC) patients receiving adjuvant doxorubicin treatment. Sci Rep. 2016;6:31804. doi: 10.1038/srep31804 [DOI] [PMC free article] [PubMed] [Google Scholar]