Summary
SARS-CoV-2 has recently been detected in feces, which indicates that wastewater may be used to monitor viral prevalence in the community. Here, we use RT-qPCR to monitor wastewater for SARS-CoV-2 RNA over a 74-day time course. We show that changes in SARS-CoV-2 RNA concentrations follow symptom onset gathered by retrospective interview of patients but precedes clinical test results. In addition, we determine a nearly complete (98.5%) SARS-CoV-2 genome sequence from wastewater and use phylogenetic analysis to infer viral ancestry. Collectively, this work demonstrates how wastewater can be used as a proxy to monitor viral prevalence in the community and how genome sequencing can be used for genotyping viral strains circulating in a community.
Keywords: SARS-CoV-2, COVID-19, wastewater-based epidemiology, genome sequencing
Graphical Abstract

Highlights
SARS-CoV-2 RNA concentrations in wastewater correlate with COVID-19 epidemiology
SARS-CoV-2 RNA levels in wastewater follow symptom onset by 5–8 days
SARS-CoV-2 RNA levels in wastewater precede clinical PCR test results by 2–4 days
SARS-CoV-2 genome from wastewater can trace phylogenetic origin
Nemudryi et al. demonstrate that wastewater can be used to monitor the progression and abatement of SARS-CoV-2 spread at the community level. The authors show a correlation between epidemiological indicators and viral concentrations measured in wastewater. In addition, they infer viral ancestry using a phylogenetic analysis of sequenced SARS-CoV-2 genome(s) from wastewater.
Introduction
In late December 2019, authorities from the People’s Republic of China (PRC) announced an epidemic of pneumonia.1 A novel coronavirus (severe acute respiratory syndrome-coronavirus-2 [SARS-CoV-2]) was identified as the etiologic agent and the disease was named coronavirus disease 2019 (COVID-19). The virus spread rapidly, first to Thailand, Japan, Korea, and Europe, and now to >188 countries across all of the continents except Antarctica. The global total of infected individuals now exceeds 20 million (https://coronavirus.jhu.edu/).
Public health professionals around the world are working to limit the spread of SARS-CoV-2, and “flatten the curve,” which requires a reduction in cases from one day to the next. However, SARS-CoV-2 containment has been outpaced by viral spread and limited resources for testing. Moreover, mounting evidence suggests that the virus is not only spread by aerosols but may also be transmitted via feces. Both viral RNA and infectious virus have been detected in the stool of COVID-19 patients.2, 3, 4, 5, 6, 7, 8 This has important implications for the spread of the virus and suggests that wastewater may be used to monitor progression or abatement of viral spread at the community level.9, 10, 11, 12, 13, 14, 15, 16, 17
Results
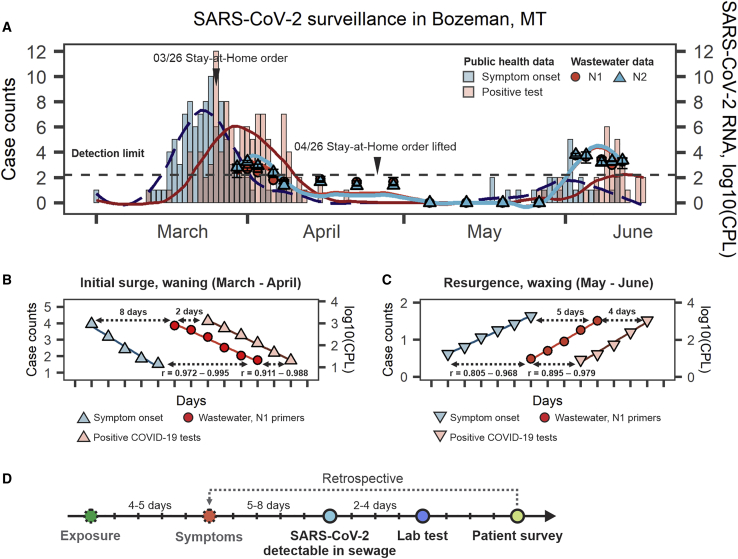
To test whether wastewater could be used for SARS-CoV-2 surveillance, we collected samples from the municipal wastewater treatment plant in Bozeman, Montana (USA). Untreated wastewater samples were collected on 17 different days over the course of a 74-day period using an autosampler that collects a volume proportional to flow for 24 h (Table S1). This composite sample reflects the average characteristics of wastewater over the previous day. The samples were filtered and concentrated before RNA extraction. To concentrate SARS-CoV-2, we used ultrafiltration with spin concentrators that efficiently recover viruses from wastewater.11,18 Extracted RNA was used as a template for one-step quantitative reverse-transcriptase-polymerase chain reaction (qRT-PCR), performed according to Centers for Disease Control and Prevention (CDC) guidelines (https://www.fda.gov/media/134922/download). Each qRT-PCR reaction was performed using 2 primer pairs (N1 and N2), which target distinct regions of the nucleocapsid (N) gene from SARS-CoV-2 (Figures 1A and S1). Composite samples collected in late March and early April 2020 tested positive for SARS-CoV-2, although concentrations of viral RNA steadily declined and then dropped below the limit of detection. After 1 month of undetectable levels of SARS-CoV-2, the wastewater began testing positive again in late May, which coincided with an increase in COVID-19 cases in the community (Figure 1A).
Figure 1.
Detection and Quantification of SARS-CoV-2 in Wastewater and in the Community
(A) Temporal dynamics of SARS-CoV-2 RNA in the municipal wastewater is superimposed on the epidemiological data. Symptom onset data (teal bars) were collected by retrospective interviews of COVID-19 patients who previously tested positive for SARS-CoV-2 (coral bars). The red circles and blue triangles show SARS-CoV-2 RNA concentration in municipal wastewater (means ± SDs) measured with qRT-PCR using the N1 and N2 primer pairs, respectively (see Method Details). The lines show curves fitted to qRT-PCR and epidemiological data using local polynomial regression (LOESS, locally estimated scatterplot smoothing).
(B and C) Linear regions of the epidemiological and wastewater curves. Curves were displaced relative to each other and Pearson correlation coefficients (r) were calculated. The 95% confidence intervals for the highest Pearson’s r values and respective offsets are shown. Data for initial surge (March–April) and resurgence (May) were analyzed separately. Surge boundaries were defined as the earliest reported symptom onset (left boundary) and date with last reported positive test (right boundary). The interval between surges with zero reported cases/symptoms (mid-April–mid-May) was dropped from the analysis.
(D) Timeline of the indicators used in the study. Symptom onset is the earliest available estimate of the viral spread. However, these data are collected retrospectively, which preclude its use for real-time tracking of the outbreak. Wastewater correlates with symptom onset and could be used to track progressing outbreak.
The current methods for tracking the COVID-19 pandemic primarily rely on clinical test results, but this process involves intrinsic delays that preclude real-time tracking of the outbreak. On average, a person develops symptoms 4–5 days after initial exposure, and it is predicted that only 32% of symptomatic individuals are tested.19,20 Test results are typically available 3–9 days after illness onset.14,21 We hypothesized that wastewater levels of SARS-CoV-2 RNA correlate with COVID-19 incidence rates and that these data could be used as an epidemiological indicator to track the outbreak in real time. To test this hypothesis, we compared our wastewater surveillance data to the frequencies of reported lab-confirmed cases and symptom onset dates that were collected by retrospective interviews. In the initial outbreak (mid-March 2020), the SARS-CoV-2 RNA concentration in the wastewater lagged behind symptom onset data by 8 days (Pearson’s r = 0.989; Figure 1B), and preceded laboratory test results for individuals by 2 days (r = 0.969; Figure 1B). When cases resurged in May, wastewater detection trailed symptom onset by 5 days (r = 0.92) and foreshadowed the increase in positive tests by 4 days (r = 0.953; Figure 1C). While wastewater detection trailed symptom onset by 5 days, it is important to note that the retrospective interviews used to collect symptom onset information are available only∼10 days after exposure. Our analysis demonstrates that wastewater surveillance is the earliest real-time measure of SARS-CoV-2 prevalence (Figure 1D).
To verify that the qRT-PCR results reflect bona fide detection of SARS-CoV-2 rather than priming from an unintended template, we repeated the PCR using 10 primer pairs that tile across the SARS-CoV-2 genome.22 These primers were designed to target conserved regions of the genome that flank polymorphic sites that have been used to trace viral ancestry and geographic origins22,23 (Figure S2A). RNA isolated from the Bozeman waste stream on March 27, 2020 was used as a template for these RT-PCR reactions, and all 10 primer pairs produced PCR products of the expected sizes (Figure S2B; Table S2). PCR products were sequenced using the Sanger method and the reads were aligned to the reference genome using MUSCLE.24,25 We observed no sequence heterogeneity in redundant reads derived from each location of the genome (Figure S2C). The same RNA sample was further used to determine a nearly complete (98.5%) SARS-CoV-2 genome sequence using a long-read sequencing platform.22,26
Mutations that do not confer a fitness defect are preserved in viral progeny, and thus serve as genetic landmarks that can be used to trace viral ancestry. Efforts to understand the origins and evolution of SARS-CoV-2 have resulted in ∼82,000 genome sequences from 91 countries as of August 13, 2020 (https://www.gisaid.org/). Phylogenetic analyses of these sequences have enabled molecular tracking of viral spread.27, 28, 29, 30, 31, 32 To determine the ancestry of SARS-CoV-2 strains circulating in Bozeman’s wastewater on March 27, we determined the genome sequence using Oxford Nanopore. Approximately 2,000 copies of the viral RNA (estimated with qPCR) were used to generate an amplicon library.26,33 Nanopore sequencing on the MinION platform resulted in ∼700,000 reads. Quality control and base calling were performed with MinKNOW version 19.06.8 in High Accuracy mode (Oxford Nanopore Technologies), and the sequences were assembled using the bioinformatic pipeline from ARTIC Network (https://artic.network/ncov-2019). This approach resulted in a single viral contig with an average sequencing depth of 6,875× that covered 98.5% of the SARS-CoV-2 reference genome (GenBank: MN908947.3). Unsequenced regions of the genome include the 5′ and 3′ ends and a stretch of 170 bases (22,346–22,515 in the Wuhan-Hu-1 reference genome), which likely had too few reads for basecalling due to PCR bias.34
In total, we found 11 single-nucleotide variants (SNVs) in the assembled genome that distinguish the Bozeman wastewater SARS-CoV-2 sequence from the Wuhan-Hu-1/2019 reference sequence (Figure S2D). To verify the authenticity of these SNVs, we examined raw sequencing data, which is available on Mendeley (Mendeley Data: https://dx.doi.org/10.17632/nfsfvy6xkf.1). During this analysis, we noticed that one of the variants (A23122T) was introduced by incomplete trimming of a sequencing adaptor (5′-CGTATTGCT) that is partially homologous (underlined) to the reference genome (5′-TACATGCA). This variant has been reported in other genomes sequenced using this protocol,35 and similar issues have been identified in other regions of the SARS-CoV-2 genome.36 In contrast to A23122T, we found no other evidence for trimming artifacts for the 10 remaining SNVs. It is possible that the consensus genome presented here is derived from a chimeric assembly of distinct genotypes, but >90% of the reads contain the same variant at each of these 10 positions.
The genome was aligned to 14,970 SARS-CoV-2 genomes from 74 different countries (Global Initiative on Sharing All Influenza Data, https://www.gisaid.org/). The resulting alignment was used to build a phylogenetic tree (Figure 2A), which indicates that the SARS-CoV-2 genome in Bozeman’s wastewater is most closely related to genomes from California and Victoria, Australia). The three mutations that define the Wuhan WA1 linage (C8782T, C18060T, T28144C) are not present in the Bozeman wastewater (WW) genome, while all 10 mutations in the Bozeman wastewater SARS-CoV-2 sequence co-occur in sequences from California; 9 of these 10 mutations are also present in an isolate from Victoria, Australia (Figures 2B, S2C, and S2D).23,37,38 To determine how these sequence variations may have accumulated over space and time, we mapped each mutation onto the phylogenetic tree of SARS-CoV-2 sequences (Figures 2A and S2E). This analysis shows that the A28851T mutation has been acquired most recently and confirms that the assembled genome from the Bozeman wastewater is most closely related to a strain circulating in California. While this sequencing approach reveals the genetic history, it does not measure the fitness of this or any of the mutations associated with distinct geographic locations. We anticipate that temporal genome sequencing from the wastewater will help identify viral strains circulating in a specific community over time.
Figure 2.
Phylogenetic Analysis of SARS-CoV-2 Sequence Isolated from Wastewater
(A) Maximum-likelihood phylogeny of the SARS-CoV-2-related lineage (n = 14,971 sequences). The phylogenetic history of SARS-CoV-2 strain sequenced from Bozeman’s wastewater (WW) is shown in crimson. The outer ring is colored according to regions of the world where the samples were isolated. The tree is rooted relative to the RaTG13 genome (a bat coronavirus with 96% sequence similarity to SARS-CoV-2; GenBank: MN996532.1). Mutations that occurred over space and time are shown in red.
(B) Sequences isolated from Bozeman WW clade with sequences of US and Australian origin (left). The sequences are named according to the geographic origin and the viral isolation date. A comparison of mutations in sequences is shown in the inset (right). The Wuhan reference sequence for each of the positions where mutations occur is shown across the top. The mutated positions and bases present in Bozeman WW sequence are shown in red, the bases matching Wuhan reference sequence are shown in white, and the mutations not present in the Bozeman WW sequence are shown in blue.
Discussion
The results presented here demonstrate that wastewater monitoring for SARS-CoV-2 RNA by qRT-PCR provides a real-time measure of viral prevalence in the community (Figure 1). Clinical testing for COVID-19 typically occurs 3–9 days after symptom onset and may vary, depending on the availability of tests, care-seeking behavior, workloads in testing facilities, and current testing strategy.21,39,40 In our study, wastewater surveillance for SARS-CoV-2 foreshadowed new case reports by 2–4 days.
The statistics of lab-confirmed COVID-19 cases not only lag behind viral spread but they also underestimate the true scale of the pandemic. A recent analysis of outpatient surveillance data estimates that only 32% of SARS-CoV-2-infected individuals in the United States have sought medical care.40 These challenges are overcome with wastewater testing, which captures input from all of the individuals in the local community and thus has the potential for estimating the true prevalence of COVID-19 using computational models that account for the median viral load in stool, virus degradation rates, travel time to the treatment facility, and water use per capita.14,41 Furthermore, wastewater may capture mild and asymptomatic infections that may be used to alert public health officials about emerging undetected transmission events.6
Monitoring wastewater for SARS-CoV-2 provides a useful epidemiological metric that could help track the outbreak and inform policy. The study presented here complements the rapidly emerging body of work by providing an important link between wastewater surveillance, COVID-19 epidemiology, and tracing SARS-CoV-2 spread patterns with genome sequencing.
Limitations of Study
Nanopore sequencing has an error rate of ∼10%–15%, which precludes the reliable detection of rare genotypes.33,42 Therefore, the protocol used here is limited to the detection of genotypes with >10%–15% representation. Due to this limitation of the sequencing approach, we cannot exclude the possibility of chimeric genome assembly. Additional sequencing efforts are required to correlate results from wastewater to clinical isolates and determine how ratios of SARS-CoV-2 variants in wastewater translate to ratios in the population.
An additional limitation of this study is the use of a single method for the concentration of SARS-CoV-2 from wastewater. While others have shown that centrifugal ultrafiltration is effective, comparative analyses of alternative concentration protocols, RNA extraction methods, and diagnostic primer sets are required to correlate results from different groups and build a standard approach for wastewater-based SARS-CoV-2 surveillance.9, 10, 11, 12, 13, 14, 15, 16, 17, 18
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Wastewater sample | Bozeman Water Reclamation Facility, MT, USA | N/A |
| Critical Commercial Assays | ||
| RNeasy Mini kit | QIAGEN | 74104 |
| 2019-nCoV CDC EUA Kit | IDT | 10006606 |
| Positive template control (PTC) plasmid | IDT | 10006625 |
| TaqPath 1-Step RT-qPCR Master Mix | Thermo Fisher Scientific | A15300 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | 18080093 |
| R9.4.1 flow cells | Nanopore Technologies | FLO-MIN106 |
| AMX, LNB, SFB, EB and SQB | Nanopore Technologies | SQK-LSK109 |
| Flow Cell Priming Kit | Nanopore Technologies | EXP-FLP002 |
| NEBNext Ultra II End-prep | New England Biolabs | E7546S |
| NEBNext Quick Ligation Module | New England Biolabs | E6056S |
| Q5 High-Fidelity DNA Polymerase | New England Biolabs | M0491S |
| DNA Clean & Concentrator kit | Zymo Research | D4005 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Q32851 |
| Deposited Data | ||
| SARs-CoV-2 Genome Sequence | GISAID | EPI_ISL_437434 |
| Sequencing reads and phylogenetic materials | Mendeley Data | https://dx.doi.org/10.17632/nfsfvy6xkf.2 |
| Oligonucleotides | ||
| The oligonucleotides used in this study were listed in Table S2 | IDT | N/A |
| Software and Algorithms | ||
| SDS software v1.4 | Applied Biosystems | 4379633 |
| RStudio v1.2.1335 | The R project | RRID: SCR_000432, https://www.r-project.org/ |
| ggplot2 | Tidyverse | RRID: SCR_014601, https://ggplot2.tidyverse.org/ |
| stats | R Core Team | https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html |
| astsa | CRAN | https://cran.r-project.org/web/packages/astsa/index.html |
| spatialEco | CRAN | https://cran.r-project.org/web/packages/spatialEco/index.html |
| MinKNOW software | Oxford Nanopore Technologies | https://community.nanoporetech.com/sso/login?next_url+%2Fdownloads |
| artic-ncov2019 | ARTIC network | https://artic.network/ncov-2019 |
| minimap2 | GitHub | RRID: SCR_018550, https://github.com/lh3/minimap2 |
| MAFFT v7.429 | N/A | RRID: SCR_011811, https://mafft.cbrc.jp/alignment/software/index.html |
| trimAl v1.2rev59 | N/A | RRID: SCR_017334, http://trimal.cgenomics.org/use_of_the_command_line_trimal_v1.2 |
| IQTree | Nextstrain | https://github.com/nextstrain/augur |
| Augur | Nextstrain | https://github.com/nextstrain/augur |
| APE v5.3 | CRAN | RRID: SCR_017343, https://cran.r-project.org/web/packages/ape/index.html |
| ggtree v3.10 | Bioconductor | RRID: SCR_018560, https://bioconductor.org/packages/release/bioc/html/ggtree.html |
| FigTree v1.4.4 | GitHub | RRID: SCR_008515, https://github.com/rambaut/figtree/releases |
| BioStrings | Bioconductor | RRID: SCR_016949, https://bioconductor.org/packages/release/bioc/html/Biostrings.html |
| SnapGene software | GSL Biotech LLC | RRID: SCR_015053, https://snapgene.com:443 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Artem Nemudryi (artem.nemudryi@gmail.com).
Materials Availability
This study has not generated new unique reagents.
Data and Code Availability
The accession number for the SARS-CoV-2 genome sequence reported in this paper is GISAID: EPI_ISL_437434. Sequencing data and phylogenetics materials used in Figure 2 have been deposited to Mendeley Data: https://dx.doi.org/10.17632/nfsfvy6xkf.1. Source data for Figure 1 (SARS-CoV-2 concentrations) is available in Table S1. Clinical tests and symptom onset data are available upon request from the Lead Contact (artem.nemudryi@gmail.com).
Experimental Model and Subject Details
Wastewater samples
Wastewater samples were collected at the Bozeman Water Reclamation Facility (BWRF) that receives and treats domestic, commercial, and industrial wastewater from the City of Bozeman, Montana (USA). Wastewater is sourced from the city limits (∼60 km2, 49’831 population) with an average flow rate of ∼2.31 × 104 m3 / d. Composite samples were collected from raw influent with automatic flow proportional sampler Liquistation CSF34 (Endress+Hauser) located at the entrance to the facility downstream of a rock trap. Autosampler was set to collect 150 mL of influent per 150’000 gal of flow (∼5.68 × 105 L) 7 AM to 7 AM. During collection temperature was kept +2 to +6°C, and samples were stored at +4°C before processing (2-3 h). The composite sample was subsampled in three 500 mL aliquots. No permissions were required for collection of the wastewater.
Symptom onset data and clinical test results
Suspect cases of COVID-19 were tested in a CLIA lab and instructed to self-quarantine until notified of the RT-qPCR test results. All laboratory confirmed positive cases of COVID-19 were contacted via telephone by local public health nurses to complete contact tracing. During this interview, the nurses collected recorded symptoms, symptom onset date, travel history, contact with other known laboratory confirmed cases, close contacts and activities on the two days before symptom onset up until notification of a positive test. Data collection was conducted as part of a public health response. Information on COVID-19 patients COVID-19 (gender, age, disease severity, etc) is not available. The study was reviewed by the Montana State University Institutional Review Board (IRB) For the Protection of Human Subjects (FWA 00000165) and was exempt from IRB oversight in accordance with Code of Federal regulations, Part 46, section 101. All necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived.
Method Details
Wastewater sample processing and RNA extraction
Each wastewater sample (500 mL) was sequentially filtered through 20 μM, 5 μM (Sartorius Biolab Products) and 0.45 μM (Pall Corporation) membrane filters and concentrated down to 150-200 μL using Corning Spin-X UF concentrators with 100 kDa molecular weight cut-off. Total RNA from concentrated samples was extracted with RNeasy Mini Kit (QIAGEN) and eluted with 40 μL of RNase free buffer. This RNA was used as a template for RT-qPCR.
Reverse Transcription quantitative PCR (RT-qPCR)
RT-qPCR was performed using two primers pairs (N1 and N2) and probes from 2019-nCoV CDC EUA Kit (IDT#10006606). SARS-CoV-2 in wastewater was detected and quantified using one-step RT-qPCR in ABI 7500 Fast Real-Time PCR System according to CDC guidelines and protocols (https://www.fda.gov/media/134922/download). 20 μL reactions included 8.5 μL of Nuclease-free Water, 1.5 μL of Primer and Probe mix, 5 μL of TaqPath 1-Step RT-qPCR Master Mix (ThermoFisher, A15299) and 5 μL of the template. Nuclease-free water was used as negative template control (NTC).
Amplification was performed as follows: 25°C for 2 min, 50°C for 15 min, 95°C for 2 min followed by 45 cycles of 95°C for 3 s and 55°C for 30 s. To quantify viral genome copy numbers in the samples, standard curves for N1 and N2 were generated using a dilution series of a positive template control (PTC) plasmid (IDT#10006625) with concentrations ranging from 10 to 10,000 copies per reaction. Three technical replicates were performed at each dilution. The limit of detection was 10 copies of the control plasmid. The NTC showed no amplification over the 40 cycles of qPCR.
Run data was analyzed in SDS software v1.4 (Applied Biosystems). Threshold cycle (Ct) values were determined by manually adjusting the threshold to fall within exponential phase of the fluorescence curves and above any background signal. Ct values of PTC dilutions were plotted against log10(copy number) to generate standard curves. Linear regression analysis was performed in RStudio v1.2.1335 and the trend line equation (Ct = [slope] × [log10(copy number)] + b) was used to calculate copy numbers from mean Ct values of technical replicates for each biological replicate. Primer efficiencies calculated as E = (10(−1/[slope]) – 1) × 100% were 150.36 ± 12.13% for N1 and 129.45 ± 25.5% for N2 (n = 7 runs, mean ± sd).
RT-PCR and SARS-CoV-2 genome sequencing
Reverse transcription was performed with 10 μL of RNA from SARS-CoV-2 positive wastewater sample using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) with 10X RT Random Primers (Applied Biosystems) according to the supplier’s protocol. Approximately 2000 viral RNA copies were used as an input for reverse transcription (estimated with qPCR).
The amplicon library for SARS-CoV-2 whole genome sequencing on Oxford Nanopore was generated as described in protocol developed by ARTIC Network (https://artic.network/ncov-2019).22,26 Briefly, V3 primer pools containing 110 and 108 primers were used for the multiplex PCR (https://artic.network/ncov-2019). PCR reactions were performed using Q5 High-Fidelity DNA Polymerase (New England Biolabs) with the following thermocycling conditions: 98°C for 2min, 35 cycles of 98°C for 15 s and 65°C for 5 min, 35 cycles. Two resulting amplicon pools were combined and used for library preparation pipeline that included end preparation and Nanopore adaptors ligation. 20 ng of final library DNA was loaded onto the MinION flowcell for sequencing. A total of 304.77 Mb of raw sequencing data was collected.
PCR products used for Sanger sequencing were generated with a subset of primers from ARTIC V3 pools (Table S2). PCR reactions were performed as described above. PCR products were analyzed on 1% agarose gels stained with SYBR Safe (Thermo Fisher Scientific), remaining DNA was purified using DNA Clean & Concentrator kit (Zymo Research) and sent to Psomagen for Sanger sequencing. Each PCR product was sequenced with both forward and reverse primers used for PCR.
SARS-CoV-2 genome assembly
Nanopore raw reads (304.77 Mb) were basecalled with MinKNOW software in high-accuracy mode. Successfully basecalled reads (273.8 Mb) were further analyzed using the ARTIC bioinformatic pipeline for COVID-19 (https://artic.network/ncov-2019). Consensus sequence was generated with minimap2 and single nucleotide variants were called with nanopolish (both integrated in the pipeline) relative to Wuhan-Hu-1/2019 reference genome (GenBank: MN908947.3).25,43,44 The resulting assembly had nearly complete genome coverage (98.51%) with 6,875X average sequencing depth. Regions of the genome that were not captured by this sequencing method include 5′ and 3′ ends of the genome and a stretch of 170 nucleotides (22,346 – 22,515 nucleotide positions in reference genome), presumably due to amplicon drop-out. Consensus genome sequence was deposited to GISAID: EPI_ISL_437434.https://www.gisaid.org/
Phylogenetic Analysis
Phylogenetic analysis was performed by aligning the consensus sequence to 14,970 SARS-CoV-2 genomes retrieved from GISAID on 5/5/2020, 8:25:22 AM (https://www.gisaid.org/), using the FFT-NS-2 setting in MAFFT v7.42942,43. Columns composed of more than 70% gaps were removed with trimAl v1.2rev5944. A maximum-likelihood phylogenetic tree was constructed from this alignment using IQTree in the Augur utility of Nextstrain 45,46. The APE v5.3 package in R was used to re-root the tree relative to RaTG13 bat coronavirus genome sequence 47, and the tree was plotted using ggtree v3.10 package in R48. The subtree, visualized in Figure 2B, was rendered in FigTree v1.4.4 49.
Position-specific Mutation Analysis
Position specific mutation analysis was conducted in R using the BioStrings package,45 and chromatograms of Sanger sequencing reads were rendered in SnapGene (GSL Biotech; available at snapgene.com).
Quantification and Statistical Analysis
All statistical analyses were performed in RStudio v1.2.1335. Data in figures are shown as mean of three biological replicates (each with two technical replicates) ± standard deviation (sd). Estimated copy numbers in RT-qPCR reactions were used to calculate titers per liter of wastewater for each biological replicate. Viral RNA concentrations in the composite samples were normalized ([SARS-CoV-2 concentration]Normalized = [SARS-CoV-2 concentration] × (Daily flow / Average flow)).
Correlation analysis was performed in RStudio using stats, astsa and spatialEco R packages. Symptom onset, positive COVID-19 tests and SARS-CoV-2 RNA concentration in wastewater measured with N1 or N2 primers were fit to a local polynomial regression (LOESS method) using the poly.regression wrapper function from spatialEco package. Resulting models were used to impute missing values (day w/o reported cases, periods between wastewater sampling days). Interpolated data were used for correlation analysis that was separately performed for the surge and resurgence of SARS-Co-V-2. Surge boundaries were determined as earliest reported symptom onset (left boundary) and date with last reported positive test (right boundary). The interval (mid-April – mid-May), when SARS-CoV-2 RNA was not detectable and no reported cases/symptoms were reported, was not included in the analysis. LOESS curves for wastewater data were displaced relative to epidemiological curves in one day increments from −14 to +14 days and Pearson correlation coefficient was calculated for each shift using corr function from stats R package.
Acknowledgments
Research in the Wiedenheft lab is supported by the National Institutes of Health (1R35GM134867), the Montana State University (MSU) Agricultural Experimental Station, the M.J. Murdock Charitable Trust, the Gianforte Family Foundation, and the MSU Office of the Vice President for Research. Wastewater data were collected as part of a SARS-CoV-2 surveillance effort supported by the City of Bozeman. We are grateful to Josh French, Justin Roberts, and the other dedicated wastewater technicians who made this work possible. We thank the GISAID EpiFluDatabase and depositing laboratories (Table S3). The phylogenetic analysis in this article would not have been possible without their willingness to share data. We thank the reviewers for helping identify a mutation that was introduced by incomplete trimming of an adaptor.
Author Contributions
Conceptualization, B.W., A. Nemudryi, and A. Nemudraia; Methodology, B.W., A. Nemudryi, and A. Nemudraia; Investigation & Data Collection, A. Nemudryi, A. Nemudraia, R.W., and K.V.; Genomics & Bioinformatic Analysis, T.W., A. Nemudryi, K.S., M.B., and C.C.; Writing – Original Draft, B.W., A. Nemudryi, A. Nemudraia, and T.W.; Writing – Review & Editing, B.W., A. Nemudryi, A. Nemudraia, T.W., K.S., and M.B.
Declaration of Interests
B.W. is the founder of SurGene and VIRIS Detection Systems, and is an inventor on patent applications related to CRISPR-Cas systems and applications thereof.
Published: September 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100098.
Contributor Information
Artem Nemudryi, Email: artem.nemudryi@gmail.com.
Blake Wiedenheft, Email: bwiedenheft@gmail.com.
Supplemental Information
References
- 1.World Health Organization . 2020. Novel Coronavirus (2019-nCoV) Situation Report - 1.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [Google Scholar]
- 2.Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindson J. COVID-19: faecal-oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of Novel Coronavirus by RT-PCR in Stool Specimen from Asymptomatic Child, China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- 10.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 12.Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MedRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 doi: 10.1101/2020.05.19.20105999. [DOI] [Google Scholar]
- 18.Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 19.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R., Prill M., Chai S.J., Kirley P.D., Alden N.B. Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019 - COVID-NET, 14 States, March 1-30, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quick J. 2020. nCoV-2019 sequencing protocol V.1.https://www.protocols.io/view/ncov-2019-sequencing-protocol-bbmuik6w?version_warning=no [Google Scholar]
- 23.Bedford T., Greninger A.L., Roychoudhury P., Starita L.M., Famulare M., Huang M.-L., Nalla A., Pepper G., Reinhardt A., Xie H. Cryptic transmission of SARS-CoV-2 in Washington State. medRxiv. 2020 doi: 10.1101/2020.04.02.20051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T.F., Beutler N.A. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., Ciferri B., Alshammary H., Obla A., Fabre S., Kleiner G., Polanco J., Khan Z. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020;369:297–301. doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castillo A.E., Parra B., Tapia P., Acevedo A., Lagos J., Andrade W., Arata L., Leal G., Barra G., Tambley C. Phylogenetic analysis of the first four SARS-CoV-2 cases in Chile. J. Med. Virol. 2020 doi: 10.1002/jmv.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefanelli P., Faggioni G., Lo Presti A., Fiore S., Marchi A., Benedetti E., Fabiani C., Anselmo A., Ciammaruconi A., Fortunato A., On Behalf Of Iss Covid-Study Group Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25:2000305. doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zehender G., Lai A., Bergna A., Meroni L., Riva A., Balotta C., Tarkowski M., Gabrieli A., Bernacchia D., Rusconi S. Genomic characterization and phylogenetic analysis of SARS-COV-2 in Italy. J. Med. Virol. 2020;92:1637–1640. doi: 10.1002/jmv.25794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., Vogels C.B.F., Brito A.F., Alpert T., Muyombwe A. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell. 2020;181:990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itokawa K., Sekizuka T., Hashino M., Tanaka R., Kuroda M. A proposal of alternative primers for the ARTIC Network’s multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing. bioRxiv. 2020 doi: 10.1101/2020.03.10.985150. [DOI] [Google Scholar]
- 35.Lopez-Alvarez D., Parra B., Cuellar W.J. Genome Sequence of SARS-CoV-2 Isolate Cali-01, from Colombia, Obtained Using Oxford Nanopore MinION Sequencing. Microbiol. Resour. Announc. 2020;9:e00573-20. doi: 10.1128/MRA.00573-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X., Gu W., Federman S., du Plessis L., Pybus O.G., Faria N.R., Wang C., Yu G., Bushnell B., Pan C.-Y. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biggerstaff M., Jhung M.A., Reed C., Fry A.M., Balluz L., Finelli L. Influenza-like illness, the time to seek healthcare, and influenza antiviral receipt during the 2010-2011 influenza season-United States. J. Infect. Dis. 2014;210:535–544. doi: 10.1093/infdis/jiu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman J.D., Hupert N., Washburne A.D. Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States. Sci. Transl. Med. 2020;12:eabc1126. doi: 10.1126/scitranslmed.abc1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagès H., Aboyoun P., Gentleman R., DebRoy S. 2019. Biostrings: efficient manipulation of biological strings.https://bioconductor.org/packages/release/bioc/html/Biostrings.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the SARS-CoV-2 genome sequence reported in this paper is GISAID: EPI_ISL_437434. Sequencing data and phylogenetics materials used in Figure 2 have been deposited to Mendeley Data: https://dx.doi.org/10.17632/nfsfvy6xkf.1. Source data for Figure 1 (SARS-CoV-2 concentrations) is available in Table S1. Clinical tests and symptom onset data are available upon request from the Lead Contact (artem.nemudryi@gmail.com).