Abstract
Destructive impacts of COVID-19 pandemic worldwide necessitates taking more appropriate measures for mitigating virus spread and development of the effective theranostic agents. In general, high heterogeneity of viruses is a major challenging issue towards the development of effective antiviral agents. Regarding the coronavirus, its high mutation rates can negatively affect virus detection process or the efficiency of drugs and vaccines in development or induce drug resistance. Bioengineered nanomaterials with suitable physicochemical characteristics for site-specific therapeutic delivery, highly-sensitive nanobiosensors for detection of very low virus concentration, and real-time protections using the nanorobots can provide roadmaps towards the imminent breakthroughs in theranostics of a variety of diseases including the COVID-19. Besides revolutionizing the classical disinfection procedures, state-of-the-art nanotechnology-based approaches enable providing the analytical tools for accelerated monitoring of coronavirus and associated biomarkers or drug delivery towards the pulmonary system or other affected organs. Multivalent nanomaterials capable of interaction with multivalent pathogens including the viruses could be suitable candidates for viral detection and prevention of further infections. Besides the inactivation or destruction of the virus, functionalized nanoparticles capable of modulating patient’s immune response might be of great significance for attenuating the exaggerated inflammatory reactions or development of the effective nanovaccines and medications against the virus pandemics including the COVID-19.
Keywords: COVID-19, Coronavirus, Nanotechnology, Nanoparticle
Graphical abstract
1. Introduction
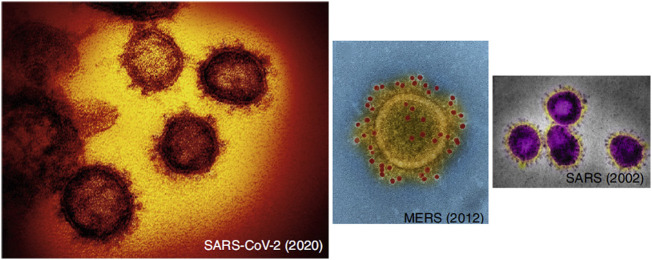
Viral infections have been considered as a major cause of mortality and morbidity worldwide [1]. Coronaviruses with the largest genome size among the RNA viruses and spike peplomers (Fig. 1 ) are able to cause fatal diseases in both mammals and birds [1,2].
Fig. 1.
The micrographs of coronavirus structures. Adapted from Ref. [249].
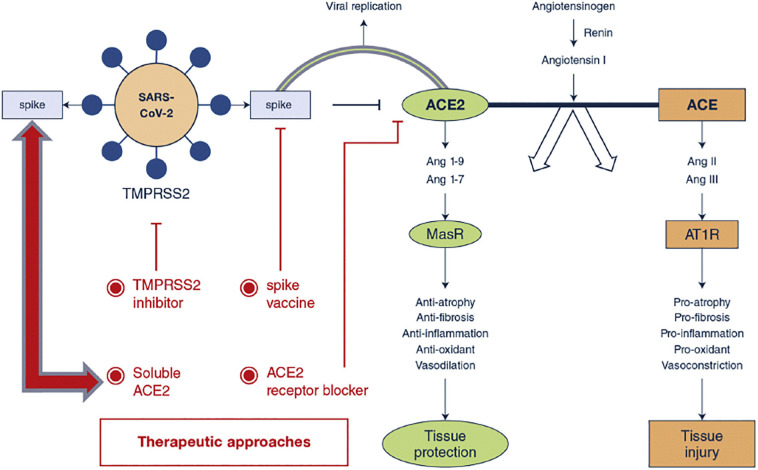
Following detecting high-degree plasticity in the coronaviruses (CoVs) genomes, it has been found that the large genomes of CoVs could be associated with increased probability of mutations. Furthermore, the virus can easily enter to the cells or attach them through various receptor types [[3], [4], [5], [6], [7]]. Spike glycoprotein attachment to the receptor is usually followed by the protease cleavage, spike protein activation, virus entrance into the cell by endocytosis or fusion of the virus lipid envelop with the membrane of cell, virus un-coating, and RNA transcription and replication [[8], [9], [10] ]. Angiotensin converting enzyme II (ACE-2) has been found to be a major virus target which facilitates its cellular uptake [10]. This has provoked a growing interest for designing ACE-2-based therapeutics [9,10], (Fig. 2 ).
Fig. 2.
The role of Angiotensin-converting enzyme-2 in the pathophysiology of COVID-19 and its receptor blockade for disease treatment. Adapted from Ref. [250].
Virus isolation in humans has been carried out in 1960s followed by developing new cultivating methods in 1965 which facilitated isolated virus inoculation into the volunteers, and identifying various coronaviruses in animals and humans (2003-2019) [[11], [12], [13], [14], [15] ]. This type of viruses are capable of long-term coexisting with humans, targeting the epithelial cells, transmission by several routes, inducing fever, cough, respiratory tract infections such as pneumonia and bronchitis, mild to severe gastrointestinal symptoms, or other less severe symptoms. Damage to the organs including the kidney, eye, brain, and heart have also been reported [[16], [17], [18], [19]]. Noteworthy, occurrence of the symptoms may be due to the patient’s immune response activation [20]. This necessitates controlling the inflammation or hyperactive responses and potentiating host’s immune system.
2. The present anti-COVID-19 strategies and their potential limitations
Limitations associated with the antibody tests such as technical problems in protein (antigen) production and identifying the suitable one for producing antibody and false positive or negative results regarding the screening of coronavirus has prompted FDA warnings [21]. Real-time RT-PCR with high sensitivity and specificity, capability of early stage infection diagnosis, and reduced contamination or error risk appears more appropriate for detecting the genetic material of SARS-Cov2 [22,23]. This method may also be associated with several pitfalls and any inappropriate virus loads or collecting procedures could enhance false negative rates [24,25], hence, it could not be considered as the most appropriate test in the clinical settings.
Development of vaccines including the viral or peptide ones against a variety of disorders such as the infectious diseases has been a medical breakthrough [[26], [27], [28]]. An in silico approach has been applied for designing the epitope-based peptide vaccine against the spike protein of coronavirus and triggering suitable immune response [29], meanwhile, the results have remained for further validations. In order to overcome limitations of the peptide-based vaccines, peptide NPs may be suitable alternatives [30]. Regarding the hypervariable viruses, determination of the immunogenic domains of the virus proteins is of critical significance for obtaining an appropriate immune response [31]. Application of the monoclonal antibodies (mAbs) which are able to target widely-shared protein motifs, provides the possibilities to characterize peptides for epitope mimicking and stimulate suitable immune response [32]. Post-immunization assessment and mAbs cloning appears as a promising approach for recognizing the new epitopes with protective characteristics [33]. In general, mAbs can play an important role in designing immunogens and antiviral therapies, however, various limitations such as technical problems could negatively affect the process of characterizing the protective epitopes of the mAbs [34]. In this sense, epitope-based predictive algorithms have been developed in order to overcome some limitations associated with designing the epitope-based vaccines [35]. In silico approaches may also be applied for empirical data analysis and immunogen prediction for development of epitope-based vaccine [36]. Noteworthy, there are a variety of challenging issues towards designing safe and effective vaccines such as pathogenic diversity, high mutation rates of virus, and host-related failures including the inappropriate immune response or problems in immune response prediction [30,[37], [38], [39], [40]]. Regarding SARS-CoV-2, more than 148 mutation sites have been identified across its genome [41]. This might provide major obstacles against the development of the effective vaccines. Various techniques such as the nucleotide sequence alteration or optimization and mRNA modification have been suggested for the development of COVID-19 vaccines [[43], [42]]. This necessitates evaluation of the efficiency and safety of vaccine in animals via application of several viral strains and assessment of their immune responses. Noteworthy, the safety of mRNA platforms for delivery of COVID-19 vaccine in humans has not been well-established [44]. Furthermore, high virus pathogenicity may negatively affect the phases of clinical trials. Virus-like particles (VLPs) which are able to mimic the organization and conformation of the native viruses, could be used for assessment of the viral infection mechanism(s) or the efficiency of therapeutics, vaccines, or drug delivery [45]. They may also be used for identifying the antibodies induced by coronavirus vaccine and evaluating the efficiency of vaccine [46]. Accelerating the vaccine development against the coronavirus necessitates application of suitable adjuvants for immune response enhancement [47,48]. Selection of the optimal adjuvants is usually based on the process of vaccine development. For enhancing the efficiency of immune response, combination of several adjuvants could be useful [49]. In general, experimental vaccines or therapeutics against the COVID-19 may be associated with safety concerns [50]. Even S protein, an antigen candidate for vaccine development could induce serious liver or lung damages and enhanced infection risk [51,52]. In this sense, structure or function of S protein should be strictly evaluated. Regarding the proposed anti-COVID-19 medications, chloroquine and hydroxychloroquine have shown disappointing results including their toxic effects against the body organs [53,54]. Remdesivir has also been presented to prevent coronavirus replication. It has been previously used against MERS-CoV (in non-human primates) and SARS-CoV-2 (in vitro) [55]. In some of the patients affected by COVID-19, the drug has provided relatively accelerated recovery rate and prolonged survival time [56,57], however, well-designed and multicenter clinical trials should confirm the observational data. Furthermore, remdesivir may be associated with adverse effects including the hepatotoxicity [57] that may negatively affect its efficiency. In patients with severe conditions, application of lopinavir-ritonavir has also led to disappointing findings [58]. The efficiency of favipiravir (RNA polymerase inhibitor) which has been presented for rapid coronavirus clearance awaits further confirmation by health authorities [59]. Host-directed therapeutic agents capable of targeting virus-host interaction may be promising candidates against the virus infections [60]. In general, therapeutics capable of direct virus targeting appear more beneficial. The potential benefits or harms of ACE inhibitors or angiotensin receptor antagonists in COVID-19 patients have not been well-established [61,62]. In addition, this type of medications can increase the expression of ACE-2 and patient’s susceptibility to the virus entrance and propagation into host cells [63]. Since the receptor binding domain of surface protein in SARS-CoV-2 binds to either ACE-2 or heparin [62], it could be a suitable target in the antiviral design projects. Treatment with recombinant ACE-2 for competitive binding inhibition through the S1 protein of SARS-CoV-2 has been suggested for preventing the virus uptake and further infections [63]. The efficiency of the entry/fusion inhibitors, HR2- and EK1-derived peptides, against SARS-CoV-2 infections has also been suggested [64]. Using the clustered regularly interspaced short palindromic repeats (CRISPR) system to target and cleave SARS-CoV-2 genome or inhibition of the coronavirus replication by poly(ADP-ribose) polymerases [65,66] are other proposed anti-COVID-19 strategies that require further confirmation. According to the receptor-mediated coronavirus endocytosis, targeting endocytosis process may be another option against SARS-CoV-2 [67]. Because of its inhibitory effect on AP-2-associated protein kinase-1 activity, baricitinib has been proposed as an anti-COVID-19 drug [68]. Since p21-activated protein kinase-1 is implicated in the viral replication and entry, its inhibitors have also been suggested as therapeutics against the disease [69]. Because of the poor solubility and cell penetrability of older inhibitors such as ketorolac or caffeic acid, newer inhibitors like frondoside-A and minnelide have been designed with improved potency and solubility [70,71] that needs further assessment and approval. For reduction of SARS-CoV-2-induced inflammation, prevention of Fc receptor activation has been suggested to be useful [72]. Furthermore, blockage of granulocyte-macrophage colony stimulating factor or interleukin-6 receptors could attenuate the immunopathologic conditions induced by SARS-CoV-2 [73]. In the absence of a specific treatment approach, convalescent plasma (CP) obtained from a recovered COVID-19 patient may be considered as a treatment strategy, however, the efficiency or safety of CP therapy is a challenging issue that may be due to its non-specific mechanism of action [74,75]. In National Health Commission of the People's Republic of China (NHC) guidelines, CP therapy has been considered only for patients in the critical conditions and rapid disease progression [76].
In order to neutralize or directly attack the virus, prevent host cell infection, or block spike proteins, application of the mAbs appears promising [77]. In China and Italy, tocilizumab has been recently used as an immunosuppressive drug in COVID-19 patients in severe conditions [78]. Meanwhile, clinical trials in more countries should be performed in order to obtain more precise data. In the recovered COVID-19 patients, functional copy engineering or reproduction could provide antibodies to enhance or mimic the immune system attack against the virus [79]. Application of the corticosteroids for hyper-inflammatory response suppression in COVID-19 patients has remained challenging and needs further evaluations. Some clinical evidence do not support corticosteroid therapy for COVID-19-associated lung injury [80]. Meanwhile, beneficial effects of corticosteroids (in low doses) in patients with critical conditions has been reported [81]. Dexamethasone due to its high potency could be applied to suppress the immunologic or hyperinflammatory responses in severe conditions of disease, however, after appropriate control of infection. Noteworthy, dexamethasone may act as a double-edged sword; despite attenuating the inflammatory or immune reactions, it may worsen the present infection or reactivate the previously managed one, intensify the negative impacts of COVID-19 on various body organs such as the heart that may result in the increased risks of arrhythmia (interested reader is referred to dexamethasone-related precautions and warnings). In this respect, exaggerated optimisms or propaganda regarding the currently available therapeutics or newly-emerged ones may be associated with life-threatening outcomes.
Drug repurposing has also been suggested against the viral infections [82]. Noteworthy, the efficiency and safety of the proposed treatment strategies against COVID-19 have not been fully supported by the health authorities. Developing efficient antiviral strategies (treatment or preventative) with minimal safety concerns necessitates focusing on the unique characteristics of the virus, selecting suitable sample size and sharing datasets during the experiments, and identification of the study restrictions.
2.1. The role of tissue engineering
Tissue engineering (TE) methods which could be used for replacement or repairing damaged organs, immunomodulation, and improvement of the efficiency conventional treatment approaches [83,84], may also be applied against the outbreaks including the coronavirus through viral model designing or facilitating the development of platforms for delivery of drugs or vaccines [85]. TE-based lung models could be used to assess the pathological conditions of the organ during the viral infections [86]. Meanwhile, various limitations may be associated with TE process that necessitates using novel technologies (e.g., 3D printing) and computational modeling in order to obtain biocompatible materials with minimal safety concerns [87].
2.2. The significance of artificial intelligence
In recent years, advanced techniques and strategies have been increasingly applied in theranostics design projects. Technologies of the artificial intelligence (AI) have enabled multivariate data analysis in large scale, making accurate decisions, solving complicated problems, providing new insights towards the pathophysiology of various diseases, accelerated designing of more efficient therapeutics or drug carriers, and identification of new compounds such as biomarkers and prediction of their targets, bioactivities, and interactions [88]. AI-based platforms could be applied for matching the patients with appropriate clinical trials [88] that might significantly reduce error rates and costs.
3. Nanomaterials against the COVID-19
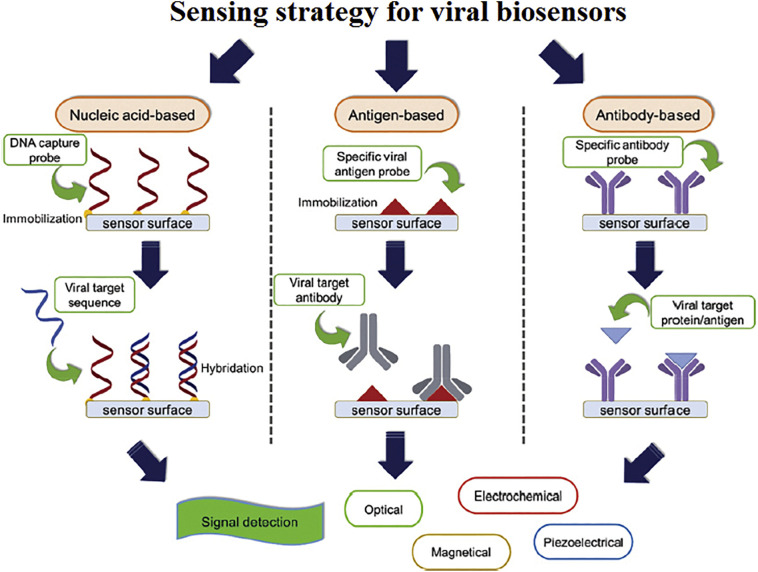
Over the last decades, the emergence of nanotechnology has revolutionized theranostic approaches. Engineering of the functional systems at molecular scales, nanovaccines with efficient immunization and minimal safety concerns, or nanovectors capable of carrying therapeutic or imaging agents for early disease detection, targeted treatment, and monitoring treatment outcomes [89,90] might exert enormous impacts on the biomedical approaches. Nanobiosensors with improved stability and biocompatibility in a variety of media have shown great potentials for detecting bacteria and viruses at too low concentrations or disease bio-signature [91] that might be of great significance in pandemic outbreaks. Reverse transcription loop-mediated isothermal amplification assay coupled with nanobiosensors could be applied for diagnose of SARS-CoV-2 within 30-40 min [91]. Using nanopore target sequencing method enables simultaneous detection of SARS-CoV-2 and other viruses implicated in the respiratory tract infections within 6–10 h [90]. Photonic crystals, Metallic NPs, grapheme, and carbon nanotubes (CNTs) with suitable surface chemistry for bio-conjugation and capable of signal amplification are promising candidates for viral detection [91,92], (Fig. 3 ).
Fig. 3.
Schematic illustration of various biosensor types for detecting of MERS-CoV and SARS-CoV-1. Biosensors are classified based on the biomolecules (antibodies, nucleic acids, or antigens) which are immobilized onto the surface of sensors. Interaction between the target and immobilized probe results in a physicochemical alteration which is transducible to the quantifiable signals. Adapted from Ref. [92].
Hybrid nanobiosystems in which virus-derived biomolecules have been entrapped, could be applied as specific probes for detecting the viruses. These nanoplatforms have shown more stability, specificity, and sensitivity than traditional devices or techniques [93]. Besides simultaneous detection of multiplex respiratory viruses, development of the surface plasmon resonance-based biosensor enables immobilizing SARS-CoV-specific oligonucleotide on a chip and targeting the virus [94]. Identification of several single-stranded DNA aptamers capable of biding with high affinity to SARS-CoV nucleocapsid N protein has led to effective detection of the protein [95]. Photonic crystals functionalized with carboxyl and amine groups which could attach to S protein, enables detection of SARS-CoV-1 antibodies [96]. Using CNTs, field-effect biosensors with high reliability and sensitivity have been designed for selective detecting of SARS-CoV nucleocapsid protein in the physiological conditions [97], (Fig. 4 ).
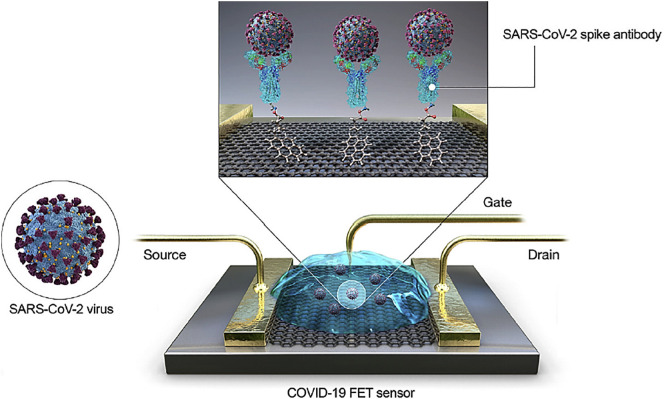
Fig. 4.
Field-effect transistor-based sensor and the process for rapid diagnosis of COVID-19. (FET: field-effect transistor). Adapted from Ref. [105].
Multi-target, arch-shaped sensors enable rapid recognizing of target nucleic acids from miscellaneous pathogens with enhanced sensitivity of detection [98]. Following RNA strand addition into the device channel, a microfluidic platform has provided the possibility of optical fluorescence or visual virus detection in 20 min with significantly improved limit of quantification [99]. Designing of a microfluidic platform with enhanced surface area for rolling circle amplification reaction has reduced time of virus detection to 15 min [100]. Multiplex immunoassay based on the electrospun polystyrene microfibers has been designed for accelerated detecting of specific antibodies of MERS-CoV which can be prepared within 5 min [101].
Gold NPs (Au-NPs) due to their distinctive catalytic, photonic, and electric characteristics and capability of interactions with a variety of biomolecules could be applied in the viral detection settings [100]. Using Au-NPs for detecting coronaviruses is based on the development of specific and rapid detection of molecules via colorimetric and electrochemical assays. These assays particularly the colorimetric one put an end to the requirement of skilled personnel or complex instrumentation and provide negative or positive results only in the liquid phase which can be easily identified (within 5 min) by the unveiled eye. Using the technique enables detecting target SARS virus nucleic acids with the sensitivity limit of about 100 fM [100,101] indicating the appropriateness of the technique for early virus diagnosis. Au-NP-modified carbon electrodes array has been demonstrated as a promising immunosensor for detection of coronavirus in 20 min using the spike protein as biomarker [102]. The immunosensor with high selectivity has shown limit of detection (LOD) of 1 pg/ml for protein of MERS-CoV [102]. Highly sensitive and simple Au-NPs-based assays have represented Au-NPs as promising nanoplatforms for detecting of other virus types [101,102]. Meanwhile, special attention should be paid to the application of advanced and rigorous methods for synthesis of the stable and uniform Au-NPs with several surface modification possibilities, appropriate conjugation of the specific targeting biomolecule to the NP surface, application of the negative and positive controls and reference tests, and in-field assessment of the novel Au-NPs-based techniques for confirming their specificity and sensitivity. In recent years, a highly selective colorimetric analysis has been performed for lysine assay using the molecular-driven Au nanorods. As known, lysine is capable of viral growth [98,100]. Application of the multifunctional Staphylococcus aureus-based nanobioprobes is another interesting strategy for rapid detecting the viral antibodies [103]. NPs are biosynthesized within the S. aureus cells where the viral nucleoproteins could be conjugated using cell wall-binding domain from the bacteriophage lysine (PlyV12). Using S. aureus-based nanobioparticles for performing agglutination test enables detecting IgG antibodies of MERS or EBOV nucleoprotein in 20 min [103]. For highly-accurate detecting SARS-CoV-2 nucleic acid, dual-functional plasmonic photothermal biosensors have been recently applied [104]. Integration of device onto a chip by 2D Au nano-islands enables local plasmonic photothermal heat generation leading to the sensitive and rapid nucleic acid detection by improvement of the fully-matched strands hybridization kinetics. For system validation, hybridization detecting has been carried out on various genome sequences of SARS-CoV-1 and SARS-CoV-2. Findings revealed high sensitivity of dual-functional biosensor towards the selected sequences of SARS-CoV-2 (LOD ~ 0.22 pM), [104]. Field effect transistor-based biosensor has been recently designed for accelerated SARS-CoV-2 detection [105]. Graphene sheets of the transistor were coated with specific antibody of SARS-CoV-2 and biosensor LOD in phosphate-buffered saline, universal transport media, and SARS-CoV-2 culture medium were 1,100 fg/ml, and 1.6 × 101 pfu/ml, respectively. Furthermore, the sensor is capable of discriminating between the non-infected and infected people [105].
The promising antiviral activities of nanoplatforms has prompted application of graphene oxide (GO) and its derivatives against the viral infections. Based on findings, charge and structure of composites significantly affect the antiviral effects [106]. It has also been shown that GO inhibits viral infections via virus inactivation prior its entrance into the cells. Moreover, some parts of envelope and spikes are destructed following incubation of virus with GO [106]. Interaction of the silver NPs with cell surface receptors and inhibition of the virus entrance into the host cells have been well-documented [100,101]. Application of the silver-graphene nanocomposites against the non-enveloped and enveloped viruses (feline coronavirus) has led to the inhibition of coronavirus in a concentration-dependent manner[107]. This type of nanocomposite inhibited the infectivity of both non-enveloped and enveloped viruses and provided higher coronavirus inhibition as compared to GO (24.8% vs. 16.3%) [107]. Silver NPs (30 nm) on the magnetic hybrid colloid (containing amine-functionalized SiO2-Fe3O4 particles) represent a promising nanosystem for viral inactivation [108]. The systems is capable of interaction with virus proteins via binding between the silver ions and thiol groups. Furthermore, silver ions may generate reactive oxygen species (ROS) for inactivation of the viruses [108]. Using silver NPs coated with various materials for investigating the interaction of silver with HIV-1 virus has revealed the impacts of NP size and thiol groups on virus-NP interaction [109]. Glutathione-capped silver-based nanoclusters have also been developed for inhibition of coronavirus proliferation [110]. These nanoclusters are recognized as quantum dots with suitable stability and optical characteristics. They have inhibited virus proliferation via blocking virus budding and RNA synthesis [110]. Quantum dots as modifiable semiconductor particles are capable of emitting the photons with specific wavelengths and providing highly-sensitive and robust fluorescence perspective for point-of-care viral detection [109]. Functional carbon quantum dots have also been prepared as therapeutic agents against the human coronavirus. After successful cell internalization and interaction with S protein, the nanosystems inhibited virus activity in a concentration-dependent manner (EC50 ~ 52 μg/mL) [111]. In recent years, curcumin-based cationic carbon dots (~ 1.7 nm) have been synthesized (using the hydrothermal procedure) against the enteric coronavirus [112]. The positively-charged dots could bind to the cell membranes via the electrostatic interaction and competition between the virus and dots for cell membrane binding was observed. Dots were capable of inhibiting the viral entrance, RNA (negative-stranded) synthesis, virus budding, and virus-induced ROS generation [112]. Didodecyldimethylammonium bromide-coated silica NPs or those including boronic acid moiety have shown promising antiviral activities via reducing virus entrance [113,114].
A nanoclay modified by surfactant has demonstrated antiviral activity with broad spectrum and high potency [115]. Nanoclay systems appear to exhibit their inhibitory effects at the early stage of infection or life cycle of virus via the electrostatic binding. They have provided no protection at 24 h postinfection [116]. Moreover, polyethylene glycol-functionalized carbon nanohorns for eliminating T7 phage via photothermal effect (Fig. 5 ), [117], or protoporphyrin-modified multi-walled CNTs for reducing Influenza A virus infectivity [118] all represent the promising antiviral activity of nanoplatforms.
Fig. 5.
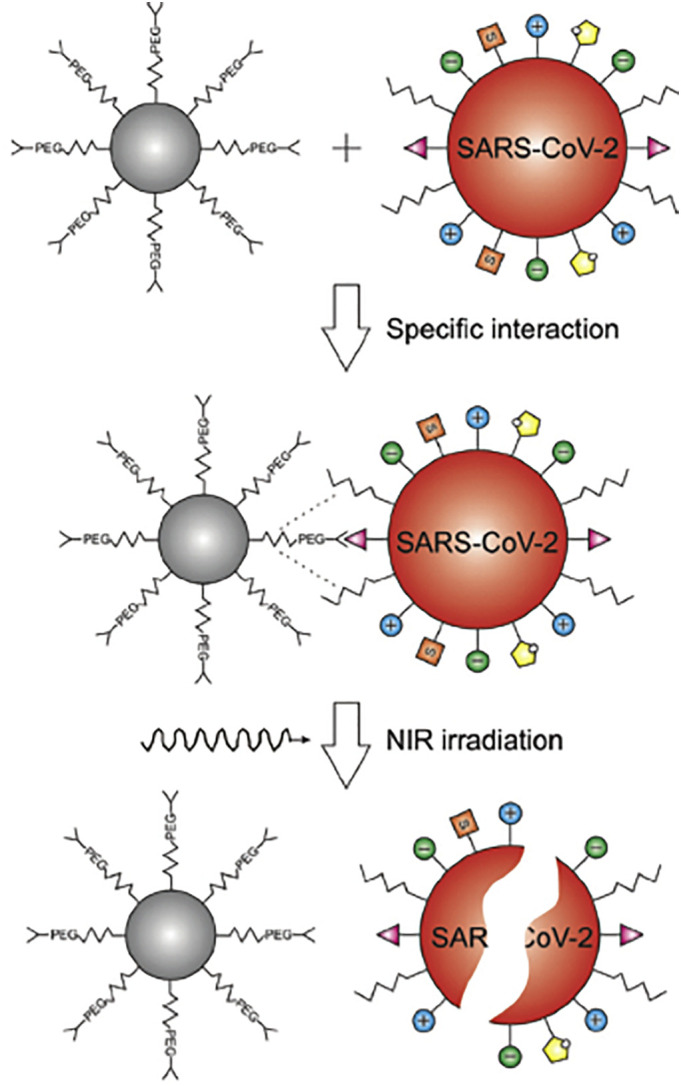
Specific interaction of SARS-CoV-2 virus and targeted nano-system. Photothermal heating and viral inactivation has been performed under NIR laser irradiation (1064 nm). Adapted from Ref. [117].
After SARS-CoV-2 entrance into the human body, deaminase enzymes including ADARs and APOBECs are able to edit virus RNA and affect its replication and fate [119] this might be of key importance against COVID-19. RNA sequencing data analysis in COVID-19 patients has shown lower levels of mutations and nucleotide alterations that may be due to RNA editing [120]. A variety of nanoplatforms such as mesoporous silica or gold NPs and nanoblades could be applied for viral genome editing [121]. CNTs with extensive potentials for targeted delivery of various theranostics, could be used as nanocarriers of the antiviral agents and increase their efficiency [[122], [123], [124], [125], [126]]. Chemical linkage of isoprinosine or ribavirin on the single-walled CNT surface has provided improved efficiency of drugs [127,128]. Moreover, CNT-based nanosystems have been shown promising for editing the viral genomes and attenuating their activity [129]. Lipid NPs which are able to deliver a wide variety of therapeutics [[130], [131], [132], [133], [134]], have provided efficient vaccination against the influenza and Zika viruses [135]. mRNA-lipid NPs enable durable and highly-efficient editing or silencing the target genes [136,137]. siRNA ability for prophylaxis or therapy of the coronavirus-related infections has been previously evaluated [138]. siRNA preexisting in the host cells is capable of inhibiting the replication of SARS-CoV and further infections because of disrupting virus RNA and inactivating the replication machinery of the virus [139]. siRNA duplexes have been assessed for anti-SARS-CoV effects in the primate cells and active duplexes and showed prolonged inhibitory effects on the replication of SARS-CoV and further infection. Combination of the active sequences provided increased antiviral potency and reduced viral escape because of the mutation in siRNA target. In this sense, integrating siRNA duplexes has been shown as a promising approach for development of the antiviral therapeutics [140]. Noteworthy, application of siRNA duplexes for treating patients infected by CoV necessitates efficient delivery of this type siRNAs to the lungs. In the Rhesus macaque affected by SARS coronavirus, siRNA has reduced viral load and replication and protected lungs against the diffuse alveolar damage. It has been represented as a safe biological agent with enormous potentials in targeted therapy or prophylactic antiviral regimens [141]. Meanwhile, further clinical evaluations are required for supporting such findings. M protein of SARS-CoV is critically involved in the viral integration and infections [142]. Two siRNAs have been created which targeted well-conserved and unexploited regions in M (membrane) gene. Both siRNAs specifically and effectively inhibited the expression of SARS-CoV membrane gene at new targeting sites [143]. siRNA-loaded lipid NPs have been successfully applied against a variety of viruses and counteracting the lethal symptoms [144,145]. Liver is one of the most vulnerable organs to the virus attacks. Patients affected by the liver diseases may be more vulnerable to the negative outcomes of COVID-19 [146]. Based on the reports, dendrimer-based lipid NPs have effectively delivered miRNAs/siRNAs for normalizing the functions of liver [147]. mRNA-encapsulated lipid NPs can be applied for production of the therapeutic proteins and gene-editing complexes for correcting disease-induced mutations in the hepatocytes [148]. Synthetic NP vectors composing of the nucleic acids poly(β-amino esters) have been shown promising for delivery of nucleic acids against a variety of diseases [149]. For expanding this advantage for systemic mRNA delivery, hybrid lipid-polymer NPs have been developed to deliver mRNA into the lungs [150]. Co-formulation of poly(β-amino esters) with polyethylene glycol-lipid has led to the development of mRNA formulations with enhanced stability and potency and capable of mRNA delivery to the lungs in mice following intravenous injection [151] indicating the effectiveness of degradable lipid-polymer NPs for systemic mRNA administration. Molar ratios and components of lipid NPs which usually consist of the cholesterol, phospholipids, polyethylene glycol lipids, and cationic lipids, may be optimized for ensuring efficient delivery of the nucleic acids to the tissue and providing potent silencing of genes [152,153]. Increasing the molar components of lipid NPs with additional molecules for tuning NP internal charge can facilitate the delivery of therapeutics in an organ-specific fashion and affect the cellular fate of NPs [154]. Based on the ideality of nanomaterials for delivery of antigens, RNA vaccines could be packaged within the lipid NPs as vectors [155]. This type of vaccines are currently under development for fighting against the coronavirus pandemic. mRNA vaccines could be suitable substitutes to the traditional vaccine technology that may be due to their high potency and short cycles of production [156]. mRNA vaccine entrapped in the lipid NPs can generate vigorous immune responses and it is possible to improve the tolerability of the vaccine without influencing its potency [157,158]. Besides the liposomes, dendrimers, nanoemulsions, and gold NPs for mRNA-based vaccine delivery, VLPs have attracted a growing interest for development of nanovaccines against SARS-CoV-2. VLPs with suitable safety, modularity, scalability, and immunogenicity have enhanced production of IL-4 and IFN-γ [[159], [160], [161]]. Chimeric VLPs have provided complete protection for mice immunized by the intranasal or intramuscular SARS protein (0.8 μg) [162]. VLPs from the insect, plant, and mammalian viruses have been applied for immunotherapy and vaccine applications [163,164], (Fig. 6 ).
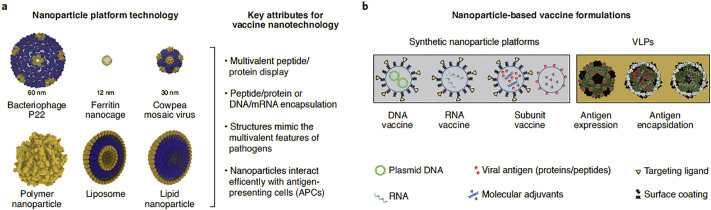
Fig. 6.
Nanotechnology-based technologies for vaccine development. a: Various size of nanoplatforms (10–1000 nm). Protein nanoparticles have been designed by Chimera software. b: nanovaccine components. Adapted from Ref. [164].
Novel technologies including the nanotech-based appear to exert significant impacts on designing next-generation modern vaccines. These approaches enable vaccine trafficking to proper subcellular or cellular locations [164].
Based on the zinc activity against a variety of viruses such as the influenza, rhinoviruses, and SARS coronavirus, zinc-based NPs have been suggested promising against the COVID-19 via inhibiting the mucosal binding of virus, suppressing the virus replication, interferon gamma or alpha generation, activating the enzymes implicated in various cellular functions, attenuating the inflammatory response, or boosting the immune system of host [165,166]. Even impregnation of the surface of masks with zinc-based NPs has provided protective effects against the pathogens [167]. As aforementioned, nanostructures could be applied for boosting the immune system and specific directing the immune response against the antigens. Rational designing of the immune-targeted nanocarriers such as the lipid- or polymeric-based NPs could result in the amplification of the hosts’ immune responses [168,169]. Besides the lipid-based NPs carrying siRNA or mRNA, polymer-entrapped antigen is capable of triggering appropriate immune response depending on the polymer type [170,171]. Indeed, nanostructures as promising systems for presenting antigens might be of great importance against the viral infections. Based on the probabilities of random mutations of viruses leading to the shape alterations of antigens, application of the appropriately-functionalized nanoplatforms capable of targeting the viruses or their specific motifs could enhance nanovaccine efficiency and inhibit virus-induced infections. At present, the efficiency of the viral vector, attenuated, inactivated, and recombinant-protein-based vaccine candidates are being investigated against SARS-CoV-2 [172,173]. Noteworthy, the safety of the inactivated and recombinant-protein-based vaccines is higher than other vaccine types, however, adjuvants should be applied for immunogenicity enhancement. Regarding SARS-CoV-2, adjuvants elevate the vaccine efficiency particularly in the elderly and patients with dysfunctions in the immune systems. Indeed, COVID-19 pandemic has provided opportunities for application of the nanotechnology-based approaches to design vaccine adjuvants. This necessitates performing coherent experiments both in vitro and in vivo for selecting vaccine adjuvant candidates [174] and testing them for clinical approvals. Studies regarding the vaccine adjuvants based on the nanomaterials have shown their immunomodulatory effects on the immune signaling. For instance, GO has activated macrophages and stimulated inflammatory reactions in animals or cells [175,176] indicating its usefulness for being used as an adjuvant and inducing cytokine release.
Recently, nanoimmunity-by-design concept based on the rational designing of functionalized nanomaterials with definite physicochemical characteristics has been proposed in order to finely tune the effects of nanomaterials on the immune system [177]. In this context, application of the nanostructures appears as a promising approach for modulating (suppressing or stimulating) the immune reactions. This might be of preventive or therapeutic significance against the viral infections including those induced by SARS-CoV-2. Based on their functionalization, nanomaterials including the CNTs, graphene, polystyrene particles, and nanodiamonds have shown the intrinsic capacity of the immune system activation [[178], [179], [180]]. Amino group-functionalized GO has induced interferon (STAT1/IRF1 ) signaling activation in T cells and monocytes leading to T cell chemo-attractant production with minimal toxicity [181]. Development of T helper 1-related response has been shown critical for infection control in COVID-19 [182]. Cytokine storm triggering or the syndrome of cytokine release within the body has been known as a major aspect of COVID-19 [168] that might be due to an exaggerated immune reaction. Cytokine inflammatory storm could be associated with severe respiratory distress syndrome and failure of other organs. In this context, increasing efforts have been attracted towards the suppression of this storm such as development of IL-6 blockers [183]. It is worth mentioning that prolonged and unbalanced immune reactions could induce sever adverse effects. Besides increasing drug solubility and capability of high drug loading, designing the nanoplatforms for immune response adjustment, inhibiting the release of inflammatory cytokines, or targeted delivery of immunosuppressive agents towards the immune cells might reduce non-specific distribution of drug, drug dosage or frequency, and potential harmful effects leading a more efficient immunosuppressive therapy [184]. In COVID-19 patients, Fas upregulation and implication of macrophages in the hyperinflammation, virus spread, and lymphocytic apoptosis as well as enhanced IL-6 release in the infected lymph nodes and spleen has been reported [185]. Histological evaluations have shown enhanced alveolar exudate because of the prolonged monocyte and neutrophil infiltrations into the capillaries of lung that may result in gas exchange problems [172]. Using the appropriate nanomedical approaches enables targeting the immune cells and limiting the aforementioned complications [178,181]. Functionalized nanodiamonds capable of adsorbing the anti-inflammatory drugs have provided pro-regenerative and anti-inflammatory effects in the macrophages of human in vitro and reduction of the infiltration of macrophages and proinflammatory mediators (TNF-α and iNOS ) expression in mice [186]. This suggests the immunomodulatory potentials of the nanodiamond-based platforms. Inflammatory cytokine removal from the blood plasma might significantly enhance patients’ survival rates in the initial stages of sepsis. Porous carbon-based nanomaterials capable of the effective cytokine (e.g., TNF-α or IL-6) adsorption and graphene-based nanomaterials with tunable surface chemistry and pore size have been shown promising for rapid inflammatory cytokine removal and reducing the mortality rates induced by the uncontrolled inflammation [187,188]. This provides opportunities for activation of the human defense mechanisms against the viral attacks and further infections. Over the last decades, photodynamic therapy has been applied against the viral diseases [189]. This approach has also been suggested for inactivating SARS-CoV-2 via attacking the target cells followed by ROS generation and cell death [190]. A variety of nanomaterials such as graphene and fullerene appear as suitable candidates for inactivating viruses via improving the efficiency of phototherapy [190].
In recent years, the significance of multivalency in development of the effective anti-infective therapeutics has attracted a growing interest [180,181,191]. In general, high heterogeneity and parasitic characteristics of viruses are challenging issues against development of the antiviral agents capable of inhibiting the replication of viruses. Targeting the initial steps of the infections such as virus particle entrance into the host cells might result in more efficient viral inhibition [192]. Because of the multivalent interactions between a cell and virus, application of the monovalent medications for inhibiting the entry and spread of viruses has not been a successful approach [191]. In the biological systems, multivalent interactions are critically involved in the adhesion, recognition, and signaling events [193]. Acquiring a deeper knowledge about this type of interactions at molecular levels might be of key significance for designing the optimal multivalent ligands for acquiring suitable biological effects. Multivalent ligands by efficient crosslinking the membrane receptors could regulate the signaling events [191]. Multivalent interactions at cell-pathogen interfaces could be inhibited competitively leading to the prevention of the cellular adhesion of pathogens during the early infection stages [194]. Pathogen (bacteria, fungi, or virus) adhesion to host cells has been considered as an initial step in the process of infection [191]. Viral adherence to the receptors on cell surface occurs via multivalent interactions followed by the cellular uptake, delivery of the genetic materials, and making novel infectious particle copies [195]. Using the monovalent medications against the fungal, bacterial, or viral infections could be associated with drug resistance [196,197]. This necessitates designing the antiviral therapeutics based on the multivalent interactions for an efficient shielding the particles of virus and inhibiting cell-virus interactions, virus entrance, and further infection. In general, using the multivalent inhibitors with optimal characteristics facilitates binding to the pathogen receptors, shielding them, and inhibiting cell surface adhesion of pathogens (Fig. 7 ), [198,199].
Fig. 7.

Application of the multivalent inhibitors for binding to various receptors on the pathogens for inhibiting their adhesion to the surface of cell. Comparison of the multivalent binding of virus to the surface of cell (a) and monovalent inhibition by a conventional drug (b). Treatment with monovalent therapeutics (in red) even at high doses could not prevent cellular binding of virus. c: Ligand-decorated multivalent inhibitor binds to the surface of virus with access to the limited numbers of receptors because of its rigidity. d: Highly-adaptive star-like and dendritic polymers which can more efficiently prevent virus adhesion as compared to the monovalent ligands. e: Ligand-decorated coiled linear polymer capable of stretching and obtaining various conformations for accessing higher numbers of receptors and shielding some of them from inappropriate interactions. f: Ligand-decorated multivalent scaffold capable of interaction with a virus and shielding it efficiently. Adapted from Ref. [193].
Based on the significance of multivalency in designing the effective therapeutics against the infectious diseases including the viral ones [191], multivalent NPs capable of blocking the virus binding to the host cells have been presented as promising antiviral agents [200]. Linear and dendritic polymers, Au-NPs, Ag-NPs, and other functionalized nanomaterials including CNTs, carbon dots, graphene oxide, or nanodiamonds have been applied as the multivalent inhibitors of the entry of various viruses [[200], [201], [202]]. Mercaptoethanesulfonate-capped Au-NPs have been shown as efficient HSV-1 inhibitors due to their ability of mimicking cell surface receptor (heparan sulfate) and binding to the virus competitively [200]. Polyvalent Au-NPs could inhibit binding of virus to the host cells in a size-dependent manner [203]. Multivalent sialic acid-decorated Au-NPs have efficiently inhibited influenza virus binding to the target cell and prevented further infection [204]. Besides Au-NPs, other antiviral NPs including those with Fe2O3 core with flexible and long linkers capable of mimicking the heparan sulfate and binding and inactivating viruses have provided permanent viral malformation in a model of lung infection. In this context, functionalized and biocompatible NPs have been represented as broad-spectrum and nontoxic antiviral agents [205].
According to the multivalency significance in reliable blocking of the host-virus interactions, nanocarriers have been shown promising for improving the stability and delivery of the entrapped therapeutics and significant enhancement of the strength of binding [206]. Multivalent glycoarchitectures including the sialic acid-coupled polyglycerol-based NPs (1–100 nm) have successfully inhibited the cellular binding or fusion of the influenza A virus and prevented its infectivity. Larger particles (50–100 nm) demonstrated more efficient inhibitory effects against the viral infection (~ 80%). Besides the NP size, ligand density has been shown as a major determinant of the efficiency of viral activity inhibition [207].
Considering the complexity of the virus entrance procedure into the host cells and multivalent interactions with various receptors on the surface of cell, designing the multivalent interaction-based antiviral therapeutics for shielding viral particles and blocking the preliminary interactions with receptors have been the focus of intense research. In this context, multivalent nanogels with various flexibility degrees have been designed which demonstrated broad spectrum antiviral effects via inhibiting the entry of virus [208]. Nontoxic nanogels could exert multivalent interaction with the glycoproteins of virus, shield the surface of virus, and block efficiently the infectivity of virus. Visualizing the interactions between the nanogels and virus and cellular uptake of nanogels via clathrin-dependent endocytosis represented the flexible nanogels as strong virus inhibitors [208].
Based on the high potential of hybrid polyvalent nanoarchitectures for binding and inhibiting various microorganisms including the viruses, several polyvalent carbon-based nanostructures capable of binding and neutralizing the animal and human herpesviruses, EHV-1 and HSV-1, have been developed which efficiently inhibited the infections [202]. Furthermore, 2D multivalent sulfated dendritic polyglycerol-functionalized nanosystems have been synthesized to inhibit orthopoxvirus particles [209]. The flexible carbon-based hybrid architecture with large surface area demonstrated high levels of binding and infection inhibition efficiency due to the negatively-charged sulfate groups which facilitated interactions with the membrane proteins of virus. This multifunctional nanoarchitecture has been presented as a promising candidate against the viruses with heparan sulfate-based mechanism of cell entrance [209].
Polysulfated nanostructures capable of mimicking the extracellular cell matrix have shown great antiviral potentials [210]. Since controlling over the functional group density which is important for effective interactions, have remained challenging, an efficient and facile method has been developed for controlling the attachment of heparin sulfate on the graphene surface for achieving highly-effective 2D nanoplatforms for interactions with pathogen. Scanning force and electron microscopies and bioassays revealed highly-efficient interactions between the stomatitis virus and sulfated polymer-covered graphene sheets for binding and neutralizing the pathogens and modulating the cellular response [210]. 2D nanomaterial consisting of the sulfated polyglycerol (94%) and graphene (6%) significantly reduced the viral titer as compared to the hydroxyl group-functionalized analog [210]. Meanwhile, because of some safety concerns associated with the application of carbon-based nanomaterials that may be due to the ROS generation and oxidative stress in target cells [211], nanogels have been recently represented as promising alternatives for development of the inhibitors of virus entrance [207]. Following degradation to the small fragments, polyglycerol nanogels have been easily removed by the renal clearance [212]. In this sense, bioactive ligand-functionalized polymeric nanogels could play an important role against the viral infections. Recently, the inhibitory effects of nanosponges against the infectivity of SARS-CoV-2 has been reported [213], (Fig. 8 ).
Fig. 8.
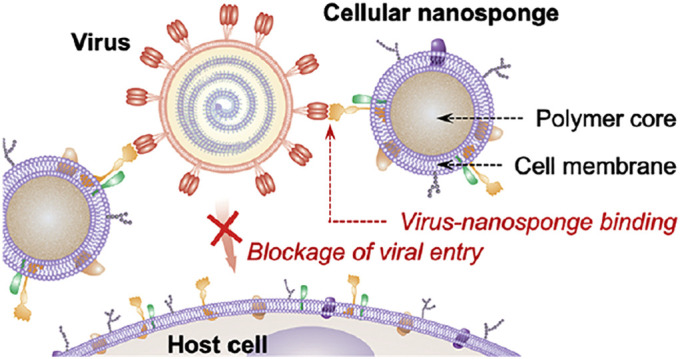
Schematic illustration of the mechanism of SARS-CoV-2 infectivity inhibition by the cellular nanosponges. Nanosponges have been prepared via wrapping the cores of polymeric NPs with the membranes of target cells including the macrophages and epithelial cells of the lungs. They inherited the antigen profiles of their source cells and served as decoys for binding to SARS-CoV-2 leading to the inhibition of virus entrance and infectivity. Adapted from Ref. [213].
They have been made using the plasma membranes of epithelial (type II) cells or macrophages of the human and exhibited similar receptors necessary for cellular entrance of SARS-CoV-2. Incubating with nanosponges resulted in the neutralization of virus and prevented its infectivity. Furthermore, the prepared broad acting nanoplatform has been shown agnostic to the viral mutations [213].
Based on the key significance of the nanostructures and biological system interactions and surface recognizing in successful delivery of therapeutics, engineering of the bio-inspired virus- and cell-like NPs for delivery of therapeutics has been the focus of intense research projects which have been performed according to the bio-inspiration and bio-mimicry principles [214]. Inorganic nanoplatforms are capable of functioning as the globular proteins due to their close physicochemical characteristics including their ability of chemical functionalization which resembles the proteins [215]. This type of common features may be applied for controlling the interactions of NPs with cell surface receptors or virus-related pathways [215]. Development of the virus-like nanocarriers capable of circulating in the blood, overcoming the biological barriers, and efficient delivery of their payloads has been suggested as a useful approach to discover the mechanisms of viral adaptations [216].
4. The importance of in silico approaches
In silico methods which are usually applied for drug repurposing, could be useful for assessment and rational designing of anti-COVID-19 nanotherapeutics. Combination of large drug-related databases and in silico techniques enables selection of the best drug candidates among a huge number of compounds. Several drug-repurposing strategies have been recently evaluated against the COVID-19 pandemic for evaluating the ability of previously-approved medications for interacting with host cell receptors or virus proteins [217,218]. In recent years, computational models have been increasingly applied to identify the mechanisms of nanomaterial interactions with the biosystems and predict their pharmacological profiles in order to improve their efficiencies. Indeed, computational modeling is of critical importance in creation of the functional systems for nanotherapeutic delivery and clinical translation [219,220]. Obtaining deeper understanding about the mechanisms of bio-events or nano-bio interactions and prediction of the effects of formulation factors on delivery and distribution of the encapsulated agents or dose-response are of key significance for development of nanotherapeutics with improved efficiency of targeting and minimal safety concerns. Patient-specific models capable of providing special opportunities can be applied for personalized theranostics [220,221]. In recent years, advancements in the molecular and structural biology and computational genomics have provided a better understanding about the protein target structures in the host and virus that might facilitate designing more effective antiviral medications [222]. Computer simulations through mutational analyses and application of the structural information enable rational designing of novel antiviral therapeutics such as the nucleotide inhibitors [223]. Advancements in the pharmacogenomics, structural biology, translational bioinformatics, and virtual designing of ligands facilitate fighting against the highly-mutating viruses and designing new vaccines or a variety of antiviral agents with high specificity such as those capable of inhibition of the proteins of virus [224,225]. Inverse computational fluid dynamics modeling enables identification of the contaminants and their spread including the coronavirus spread [226]. Regarding the COVID-19, coarse-grained molecular dynamics simulation enables evaluating the internalization of Au-based nanostructures into the mammalian cells and mechanism of targeting and delivery, and predicting the pharmacological profiles of NPs [217,227]. This might provide useful guidance towards the development of novel nanoformulations against SARS-CoV-2.
A project has been recently designed using the computational methods in order to construct COVID-19 disease map for obtaining a deeper knowledge about the mechanisms of interactions between the host and SARS-CoV-2 virus and high-quality model development capable of linking to data repositories [228]. The map could serve as an appropriate platform for visual evaluations and analysis of the molecular procedures implicated in the entrance and replication of virus and its interactions with host, cell recovery, immune response, and mechanisms of repair. Furthermore, it would be possible to obtain a deeper knowledge about the disease pathomechanisms and nature of the infection, host susceptibility characteristics including the age and gender, progression of disease, mechanisms of defense, and treatment response, and facilitate the design process of more efficient theranostics [228]. Several pathways have been included in the map such as the replication cycle of virus and mechanisms of transcription, the effect of SARS-CoV-2 on the pulmonary blood pressure, interferon-2 signaling, apoptosis, and heme catabolism. COVID-19 diagram collections and metabolic model of the alveolar macrophages in patients affected by the virus are also incorporated in the map [228]. Combination of the illustrative representations of the mechanisms of COVID-19 with underlying models in the map provides a suitable plan for the virologists, immunologists, and clinicians for collaboration with the computational biologists and data analyzers in order to build rigorous models and accurate interpretation of data. Moreover, application of the map with other disease maps enables assessment of the comorbidities [228,229].
Over the last decade, remarkable advancements in AI-based techniques have facilitated designing intelligent mechanical nanoplatforms powered by various sources of chemical energy or bio-molecular motors for accelerating the process of nanofabrication and solving the associated problems, developing smarter and hybrid technologies, producing the nanoarchitectures with enhanced power of computation, and evaluating the impact of nanostructures on the biosystems [230,231]. These highly-durable nanomachines enable monitoring the activities and internal chemistry of organs and accessing the malfunctioned regions [232]. Development of the biologically-inspired nanorobots with powerful engines capable of processing of information, sensing, signaling, actuation, entering the cells, combating a variety of diseases, or cutting out the defective genes, is indeed an eminent breakthrough in medicine. Nanoswimmers which are able to maneuver through the physiological fluids for targeted-therapy, 3D DNA nanomachines, and remote-controlled nanorockets capable of targeted delivery of therapeutics [233,234] can revolutionize the traditional theranostic interventions. Besides predicting and preventing the hazards of chemicals, nanorobots can cross over the body for cell repair, assisting a mal-functioning organ, repairing tissues, targeted gene or drug delivery, improvement of drug efficiency, or monitoring the patient. Nanobots along with the wireless transmitters provide the possibility of modifying the treatment protocols [232,235]. Besides detecting or destroying the toxic agents, stimuli-responsive nanorobots can be used against a variety of disorders including the viral infections [236]. MRI-guided nanomachines enable simultaneous tracking and actuating NPs or precise magnetic particle localizations [237]. These nanostructures could be programmed for performing the biological functions at cellular level and attacking the viruses [235]. Folate substances may be attached onto the nanorobot body which are powered by the flagella motors [238]. This approach could be taken for drug delivery against the viral infections in humans [239]. In general, nanorobot payload is released at distinct points via manipulation of the physiological parameters. Targeted release may be triggered by the alteration of pH or temperature [240]. Based on the unpredicted alterations of the physiological parameters, development of the externally-triggered nanomachines might result in more appropriate payload delivery [241]. Nucleic acid-based nanorobots capable of biosensing such as sensing the flow rate, delivery of the cellular-compatible message and biological activators, assessment the intra- or inter- molecular forces, manipulating the NPs or molecules for fabrication of more advanced nanoplatforms, controlling the chemical reactions, and apoptosis triggering [242,243] might be the major components of the modern theranostic settings. These smart machines with new generations of nanomotors can move through the fluid environments and execute specific missions [244]. Advanced simulation techniques enable acquiring a deeper knowledge about the mechanisms of nanorobot interactions within the living organisms [245]. Regarding the virus pandemics, application of the programmed nanorobots provides the possibility of detecting various levels of specific proteins in the bloodstream [245] that might facilitate characterizing of a specific virus. Cell invasion by the influenza virus and secretion of α-N-acetyl-galactosaminidase (α-NAGA) protein has resulted in the virus spread throughout the body and immunosuppression [246]. α-NAGA overexpression in bloodstream triggers the prognostic behavior of nanorobot and electromagnetic signals could be transmitted to a mobile phone and satellites followed by identification of the contaminated person position. Programmable nanorobot are capable of sensing and detecting α-NAGA concentration in bloodstream [247]. For positioning of nanorobots in vivo, radio frequency identification device and complementary metal oxide semiconductor transponder system are applied in their architectures [248]. Furthermore, embedment of nanobiosensors in the structure of nanorobot facilitates α-NAGA monitoring. Detecting the overexpression of protein indicates time of viral contamination [247]. For SARS-CoV-2 inhibition, ACE-2-based peptides have been recently designed using the classical MD simulations [206]. Conformational matching of the virus and ACE-2-extracted peptides enables improvement of the binding affinities and designing more appropriate inhibitors capable of selective binding. It would be possible to increase the binding affinity via multivalent binding of several peptides which are attached to the surfaces of NPs, clusters, or dendrimers [206]. In an analogy to the broad spectrum antiviral NPs with virucidal inhibitory mechanism, it would be possible to attach the sulphonated ligand capable of mimicking heparane sulfate to α1 helix and provide the inhibitors which bind to the positively-charged residues at receptor binding domain and can be applied as inhalation for prevention of viral activation in the lungs [205,206].
5. Bench-to-bedside knowledge transfer
Following the COVID-19 healthcare crisis, clinicians and researchers worldwide have been working hard for identifying the most appropriate preventive or treatment strategies. Collecting and analysis of the laboratory data sets from the hospitals as well as improving the diagnosis speed and capacity of detection in the laboratories might facilitate providing more appropriate preventive or treatment strategies. Biochemical and hematological parameters have provided useful information regarding COVID-19. Laboratory abnormalities (specially the hematological alterations) enable checking the conditions of infection induced by SARS-CoV-2 [251]. Based on the recent laboratory findings, some researchers have suggested the ratio of neutrophil-to-lymphocyte as the independent risk factor for the severity of disease and the ratio of > 3.13 was represented as a threshold for progressing towards the critical conditions [252]. Leukocytosis, thrombocytopenia, and lymphopenia are associated with higher severity or fatality in patient with COVID-19 [251]. In this sense, monitoring of the hematological parameters might be of key significance for evaluating the prognosis or progression of disease. Sever conditions may also be associated with enhancement of the serum procalcitonin [253]. The inflammatory marker, C-reactive protein, may be applied for tracking the disease severity. Other inflammatory markers such as tumor necrosis factor-α or interleukins 4 and 6 have been significantly increased in some critically-ill patients [253]. Recently, mesenchymal stem cells have been shown promising in COVID-19 patients with critical conditions [254], however, these cells are still in the initial stages of developing anti-COVID-19 therapies and further clinical trial-based data are required for confirming of their efficiencies. Regarding the application of nano-based tools for inactivation of SARS-CoV-2, incubation with copper-containing surfaces has led to the fragmentation of the viral genome and irreversibile virus inactivation [255].
6. Concluding remarks
COVID-19 outbreak could impose serious negative impacts on the infrastructures of societies including the healthcare systems. Considering the unique characteristics of the pathogens and avoiding the simplistic views on the prophylactic or therapeutic approaches including the one-size-fits-all ones or presenting multiple medications that may be associated with synergistic toxicities rather than enhanced efficiencies might pave the way towards the development of more appropriate treatment strategies with reduced safety concerns. Over the last decades, remarkable progress in nanotechnology has provided unique sets of advanced tools and methods for obtaining deeper knowledge of pathophysiology of various diseases including the viral ones and designing more effective theranostic platforms. Nanocarriers because of their appropriate physicochemical properties which provide facilitated penetration across the cell membranes, increased bioavailability of the encapsulated therapeutics, and reduced drug resistance or side effects could be suitable carriers for antiviral agents. A variety of nanocomposites, polymeric, silver, or lipid NPs, nanosuspensions, liposomal formulations, dendrimers, or nanoemulsions have been developed against the viral infections. Multivalent nanostructures with suitable surface areas for interacting with multivalent pathogen could be considered as promising theranostics against the viral diseases including the recent pandemic. Development of the scalable NPs with improved tissue penetration and proof-of-concept nanorobots capable of early detection of pathogens and destroying them, editing the genomes, and smart delivery of therapeutics could be of great significance against the disease. Meanwhile, effective management or treatment of COVID-19 necessitates performing well-designed basic and clinical investigations. Focusing on the intracellular trafficking machinery and inter-patient variables might provide smarter insights into the disease pathomechanism and treatment response. High-performance multi-scale modeling and simulation methods with ever-increasing predictability and power and conclusive impacts on nanomedicine provide a deeper knowledge about the bio-interactions and disease pathomechanism that might result in the development of more efficient therapeutics with improved outcomes. Even after development of the preventive or therapeutic agents including the membrane-modifiers and inhibitors of the virus entrance or membrane anchoring, they should undergo long-term safety, efficiency, and immunogenicity assessments. In this respect, exaggerated optimisms or propaganda regarding the currently available or newly-developed therapeutics may be associated with serious negative outcomes.
Role of the funding source
This work did not receive any specific grant from the funding agencies in public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
Author declares no conflicts of interest.
Acknowledgements
Not applicable
References
- 1.Almeida J.D., Berry D.M., Cunningham C.H., Hamre D., Hofstad M.S., Mallucci L., McIntosh K., Tyrrell D.A. Virology: Coronaviruses. Nature. 1968;220(5168):650. [Google Scholar]
- 2.Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam T.T.Y., Ho-Hin Shum M., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020 doi: 10.1101/2020.02.13.945485. [DOI] [Google Scholar]
- 4.Wang N., Rosen O., Wang L., Turner H.L., Stevens L.J., Kizzmekia S., et al. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S., Zhou P., Wang P., Li Y., Jiang L., Jia W., et al. Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein. Cell Rep. 2018;24:441–452. doi: 10.1016/j.celrep.2018.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun C., Chen L., Yang L., Luo C., Zhang Y., Li J., Yang J., et al. SARS-CoV-2 and SARS-CoV spike-RBD structure and receptor binding comparison and potential implications on neutralizing antibody and vaccine development. bioRxiv. 2020 doi: 10.1101/2020.02.16.951723. [DOI] [Google Scholar]
- 7.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong P.B., Lin L.Y., Tran T.H. Coronaviruses pandemics: Can neutralizing antibodies help? Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagan W.A., Bruner D.W., Gillespie J.H., Timoney J.F., Scott F.W., Barlough J.E. Cornell University Press; 1988. Hagan and Bruner’s Microbiology and Infectious Diseases of Domestic Animals: With Reference to Etiology, Epizootiology, Pathogenesis, Immunity, Diagnosis, and Antimicrobial Susceptibility; p. 440. ISBN 978-0-8014-1896-9. [Google Scholar]
- 11.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. The Pediatric Infectious Disease Journal. 2005;24(11 Suppl):S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. discussion S226. [DOI] [PubMed] [Google Scholar]
- 12.Kendall E.J., Bynoe M.L., Tyrrell D.A. Virus isolations from common colds occurring in a residential school. Br Med J. 1962;2(5297):82–86. doi: 10.1136/bmj.2.5297.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., et al. In: Ninth Report of the International Committee on Taxonomy of Viruses. King AM, Lefkowitz E, Adams MJ, Carstens EB, International Committee on Taxonomy of Viruses, International Union of Microbiological Societies. Virology Division, editor. 806–828. Elsevier; Oxford: 2011. Family Coronaviridae. ISBN 978-0-12-384684-6. [Google Scholar]
- 14.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trend Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New Eng J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decaro N., Tidona C., Alphacoronavirus Darai G. Springer; 2011. The Springer Index of Viruses; pp. 371–383. [Google Scholar]
- 17.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui D.S., Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infectious Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neher RA, Dyrdak R, Druelle V, Hodcroft EB, Albert J. Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. MedRxiv 2020. [DOI] [PubMed]
- 20.Tay Matthew Zirui, Poh Chek Meng, Rénia Laurent, MacAry Paul A., Ng Lisa F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn Stephen M. Food and Drug Administration; April 07, 2020. Coronavirus (COVID-19) update: Serological tests. [Google Scholar]
- 22.Carter Linda J., Garner Linda V., Smoot Jeffrey W., Li Yingzhu, Zhou Qiongqiong, Saveson Catherine J., et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A., Yao J.D.C., et al. Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clin Microbiol Rev. 2006;19(1):165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustin Stephen A., Nolan Tania. Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. J Biomol Tech. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- 25.Tahamtan Alireza, Ardebili Abdollah. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chianese-Bullock K.A., Irvin W.P., Petroni G.R., Murphy C., Smolkin M., Olson W.C., Coleman E., et al. A Multipeptide vaccine is safe and elicits T-cell responses in participants with advanced stage ovarian cancer. J Immunother. 2008;31:420–430. doi: 10.1097/CJI.0b013e31816dad10. [DOI] [PubMed] [Google Scholar]
- 27.Koellhoffer J.F., Higgins C.D., Lai J.R. Protein Engineering Strategies for the Development of Viral Vaccines and Immunotherapeutics. FEBS Lett. 2014;588:298–307. doi: 10.1016/j.febslet.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skwarczynski Mariusz, Toth Istvan. Peptide-based synthetic vaccines. Chem Sci. 2016;7(2):842–854. doi: 10.1039/c5sc03892h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oany Arafat Rahman, Emran Abdullah-Al, Jyoti Tahmina Pervin. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Design Dev Ther. 2014;(8):1139–1149. doi: 10.2147/DDDT.S67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malonis Ryan J., Lai Jonathan R., Vergnolle Olivia. Peptide-based vaccines: Current progress and future challenges. Chem. Rev. 2020;120(6):3210–3229. doi: 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelli Matteo, Cappelletti Francesca, Diotti Roberta Antonia, Sautto Giuseppe, Criscuolo Elena, Dal Peraro Matteo, et al. Peptide-Based Vaccinology: Experimental and Computational Approaches to Target Hypervariable Viruses through the Fine Characterization of Protective Epitopes Recognized by Monoclonal Antibodies and the Identification of T-Cell-Activating Peptides. Clin Dev Immunol. 2013;2013 doi: 10.1155/2013/521231. 1-12. Article ID: 521231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Yedidia T., Arnon R. Epitope-based vaccine against influenza. Expert Rev Vaccines. 2007;6:939–948. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]
- 33.Lee T.H., Song B.H., Yun S.I., et al. A cross-protective mAb recognizes a novel epitope within the flavivirus NS1 protein. J General Virol. 2012;93:20–26. doi: 10.1099/vir.0.036640-0. [DOI] [PubMed] [Google Scholar]
- 34.Mancini N., Solforosi L., Clementi N., De Marco D., Clementi M., Burioni R. A potential role for monoclonal antibodies in prophylactic and therapeutic treatment of influenza. Antiviral Res. 2011;92(1):15–26. doi: 10.1016/j.antiviral.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Gershoni J.M., Roitburd-Berman A., Siman-Tov D.D., Freund N.T., Weiss Y. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs. 2007;21(3):145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seyed N., Zahedifard F., Safaiyan S., et al. In silico analysis of six known leishmania major antigens and in vitro evaluation of specific epitopes eliciting HLA-A2 restricted CD8 T cell response. PLoS Negl Trop Dis. 2011;5(9) doi: 10.1371/journal.pntd.0001295. Article ID e1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed Nabil, Gottschalk Stephen. How to design effective vaccines: lessons from an old success story. Expert Rev Vaccines. 2009 May;8(5):543–546. doi: 10.1586/erv.09.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller E., Beverley P.C.L., Salisbury D.M. Vaccine programmes and policies. Br Med Bull. 2002;62(1):201–211. doi: 10.1093/bmb/62.1.201. [DOI] [PubMed] [Google Scholar]
- 39.Wiedermann Ursula, Garner-Spitzer Erika, Wagner Angelika. Primary vaccine failure to routine vaccines: Why and what to do? Hum. Vaccines Immunotherapeutics. January 2016;12(1):239–243. doi: 10.1080/21645515.2015.1093263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frei J.C., Wirchnianski A.S., Govero J., Vergnolle O., Dowd K.A., Pierson T.C., Kielian M., et al. Engineered Dengue Virus Domain III Proteins Elicit Cross- Neutralizing Antibody Responses in Mice. J. Virol. 2018;92 doi: 10.1128/JVI.01023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uddin M., Mustafa F., Rizvi T.A., Loney T., Suwaidi H.A., Hassan Al-Marzouqi A.H., et al. SARS-CoV-2/COVID-19: Viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12(5):526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8(2):153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA Vaccines for Infectious Diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong H., Guo H.C., Sun S.Q. Virus-like particles in picornavirus vaccine development. Appl. Microbiol. Biotechnol. 2014;98:4321–4329. doi: 10.1007/s00253-014-5639-1. [DOI] [PubMed] [Google Scholar]
- 46.Temperton N.J., Chan P.K., Simmons G., Zambon M.C., Tedder R.S., Takeuchi Y., Weiss R.A. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 2005;11:411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogenesch H. Mechanism of immunopotentiation and safety of aluminum adjuvants. Front. Immunol. 2012;3:406. doi: 10.3389/fimmu.2012.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Didierlaurent A.M., Laupeze B., Di Pasquale A., Hergli N., Collignon C., Garcon N. Adjuvant system AS01: Helping to overcome the challenges of modern vaccines. Expert Rev. Vaccines. 2017;16:55–63. doi: 10.1080/14760584.2016.1213632. [DOI] [PubMed] [Google Scholar]
- 49.Petrovsky N. Comparative Safety of Vaccine Adjuvants: A Summary of Current Evidence and Future Needs. Drug Saf. 2015;38:1059–1074. doi: 10.1007/s40264-015-0350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang S. Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature. 2020;579:321. doi: 10.1038/d41586-020-00751-9. [DOI] [PubMed] [Google Scholar]
- 51.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh Awadhesh Kumar, Singh Akriti, Shaikh Altamash, Singh Ritu, Misra Anoop. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr. 2020 May-June;14(3):241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browning David J. Hydroxychloroquine and Chloroquine Retinopathy. Vol. 4. 2014. Pharmacology of Chloroquine and Hydroxychloroquine; pp. 35–63. [Google Scholar]
- 55.Wang Y., Zhang D., Du G., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grein J., Ohmagari N., Shin D., et al. Compassionate use of remdesivir for patients with severe COVID-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon Calvin J., Tchesnokov Egor P., Woolner Emma, Perry Jason K., Feng Joy Y., Porter Danielle P., Gotte Matthias. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao B., Wang Y., Wen D., et al. A trial of lopinavir–ritonavir in adults hospitalized with severe COVID-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saiz J.C., Oya N.J., Blázquez A.B., Escribano-Romero E., Martín- Acebes M.A. Host-Directed Antivirals: A Realistic Alternative to Fight Zika Virus. Viruses. 2018;10(9):E453. doi: 10.3390/v10090453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuster G.M., Pfister O., Burkard T., Zhou Q., Twerenbold R., Haaf P., et al. SARS-CoV2: should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. 2020;41(19):1801–1803. doi: 10.1093/eurheartj/ehaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang P., Zhu L., Cai J., et al. Association of Inpatient Use of Angiotensin Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers with Mortality Among Patients With Hypertension Hospitalized With COVID-19. Circulation Res. 2020;126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel Ankit B., Verma Ashish. COVID-19 and Angiotensin-Converting Enzyme Inhibitors and Angiotensin Receptor Blockers: What Is the Evidence? JAMA. 2020;323(18):1769–1770. doi: 10.1001/jama.2020.4812. [DOI] [PubMed] [Google Scholar]
- 64.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen T.M., Zhang Y., Pandolfi P.P. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grunewald Matthew E., Chen Yating, Chad KunyI, Maejima Takashi, Lease Robert, Ferraris Dana, et al. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019;15(5) doi: 10.1371/journal.ppat.1007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van den Broeke C., Radu M., Chernoff J., Favoreel H.W. An emerging role for p21-activated kinases (Paks) in viral infections. Trends Cell Biol. 2010;20:160–169. doi: 10.1016/j.tcb.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen B.C.Q., Takahashi H., Uto Y., Shahinozzaman M.D., Tawata S., Maruta H. 1,2,3-Triazolyl ester of Ketorolac: A “Click Chemistry”-based highly potent PAK1-blocking cancer-killer. Eur. J. Med. Chem. 2017;126:270–276. doi: 10.1016/j.ejmech.2016.11.038. [DOI] [PubMed] [Google Scholar]
- 71.Nguyen B.C.Q., Yoshimura K., Kumazawa S., Tawata S., Maruta H. Frondoside A from sea cucumber and nymphaeols from Okinawa propolis: Natural anti-cancer agents that selectively inhibit PAK1 in vitro. Drug Discov. Ther. 2017;11:110–114. doi: 10.5582/ddt.2017.01011. [DOI] [PubMed] [Google Scholar]
- 72.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: From mechanisms to potential therapeutic tools. Virol Sin. 2020;35:266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y.F.B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv. 2020 doi: 10.1101/2020.02.12.945576. [DOI] [Google Scholar]
- 74.Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garraud O., Heshmati F., Pozzetto B., Lefrere F., Girot R., Saillol A., Laperche S. Plasma therapy against infectious pathogens, as of yesterday, today andtomorrow. Transfus Clin Biol. 2016;23:39–44. doi: 10.1016/j.tracli.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.National Health Commission of the People's Republic of China Notice on printing and distributing the convalescent plasma treatment for novel coronavirus pneumonia. 2020; trial version 2: 5a2.shtml. http://www.nhc.gov.cn/yzygj/s7658/202003/61d608a7e8bf49fca418a6074cbf
- 77.Gooa Junghyun, Jeonga Yuji, Parka Young-Shin, Yanga Eunji, Junga Dae-Im, Rho Semi. Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Research. 2020;278:1–10. doi: 10.1016/j.virusres.2020.197863. 197863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.González-Gay Miguel A., Mayo José, Castañeda Santos, Cifrián José M., Hernández-Rodríguez José. Tocilizumab: from the rheumatology practice to the fight against COVID-19, a virus infection with multiple faces. Expert Opin. Biol. Ther. 2020;20(7):717–723. doi: 10.1080/14712598.2020.1770222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 80.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou W., Liu Y., Tian D., 369 Wang C., Wang S., Cheng J., Hu M., Fang M., Gao Y. Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduct Target Ther. 2020;5 doi: 10.1038/s41392-020-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Méndez-Lucio O., Tran J., Medina-Franco J.L., Meurice N., Muller M. Toward drug repurposing in epigenetics: olsalazine as a hypomethylating compound active in a cellular context. Chem Med Chem. 2014;9:560–565. doi: 10.1002/cmdc.201300555. [DOI] [PubMed] [Google Scholar]
- 83.Khademhosseini A., Langer R. A decade of progress in tissue engineering. Nat Protoc. 2016;11:1775. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- 84.Hassanzadeh P. Tissue engineering and growth factors: updated evidence. Biomed Rev. 2012;23:19–35. [Google Scholar]
- 85.Tatara Alexander M. Vol. 26. 2020. Role of tissue engineering in COVID-19 and future viral outbreaks; pp. 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller A.J., Spence J.R. In vitro models to study human lung development, disease and homeostasis. Physiology (Bethesda) 2017;32:246. doi: 10.1152/physiol.00041.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. Tissue engineering: Still facing a long way ahead. J. Controlled Release. 2018;279:181–197. doi: 10.1016/j.jconrel.2018.04.024. [DOI] [PubMed] [Google Scholar]
- 88.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. The significance of artificial intelligence in drug delivery system design. Adv. Drug Deliv. Rev. 2019;151–152:169–190. doi: 10.1016/j.addr.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Kubik T., Bogunia-Kubik K., Sugisaka M. Nanotechnology on duty in medical applications. Curr Pharm Biotechnol. 2005;6:17–33. doi: 10.2174/1389201053167248. [DOI] [PubMed] [Google Scholar]
- 90.Hassanzadeh P. Nanopharmaceuticals: Innovative theranostics for the neurological disorders. Biomed Rev. 2014;25:25–34. [Google Scholar]
- 91.Hassanzadeh P. New perspectives in biosensor technology. Gastroenterol. Hepatol. Bed Bench. 2010;3(3):105–107. [Google Scholar]
- 92.Palestino G., Garcia-Silva I., Gonzalez-Ortega O., Rosales-Mendoza S. Can nanotechnology help in the fight against COVID-19? Expert Rev Anti-infective Therapy. 2020;1:1–17. doi: 10.1080/14787210.2020.1776115. [DOI] [PubMed] [Google Scholar]
- 93.Wang X., Liu L.-H., Ramstroem O., Yan M. Engineering Nanomaterial Surfaces for Biomedical Applications. Exp Biol Med. 2009;234:1128–1139. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi L., Sun Q., He J., et al. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Biomed Mater Eng. 2015;26:S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 95.Cho S.J., Woo H.M., Kim K.S., et al. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J Biosci Bioeng. 2011;112(6):535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park T.J., Lee S.K., Yoo S.M., et al. Development of reflective biosensor using fabrication of functionalized photonic nanocrystals. J Nanosci Nanotechnol. 2011;11(1):632–637. doi: 10.1166/jnn.2011.3269. [DOI] [PubMed] [Google Scholar]
- 97.Ishikawa F.N., Curreli M., Olson C.A., et al. Importance of controlling nanotube density for highly sensitive and reliable biosensors functional in physiological conditions. ACS Nano. 2010;4(11):6914–6922. doi: 10.1021/nn101198u. [DOI] [PubMed] [Google Scholar]
- 98.Wang J., Zhang P., Li C.M., Li Y.F., Huang C.Z. A highly selective and colorimetric assay of lysine by molecular-driven gold nanorods assembly. Biosensors Bioelectronics. 2012;34:197–201. doi: 10.1016/j.bios.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 99.Jung I.Y., You J.B., Choi B.R., et al. Highly sensitive molecular detection platform for robust and facile diagnosis of middle east respiratory Syndrome (MERS) corona virus. Adv Healthc Mater. 2016;5(17):2168–2173. doi: 10.1002/adhm.201600334. [DOI] [PubMed] [Google Scholar]
- 100.Ting Zhao V.X., Wong T.I., Zheng X.T., Tan Y.N., Zho X. Colorimetric biosensors for point-of-care virus detections. Mater Sci Energy Technol. 2020;3:237–249. doi: 10.1016/j.mset.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoy C.F., Kushiro K., Yamaoka Y., et al. Rapid multiplex microfiber-based immunoassay for anti-MERS-CoV antibody detection. Sens Biosensing Res. 2019;26 doi: 10.1016/j.sbsr.2019.100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Mikrochim Acta. 2019;186(4) doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qiao J., Li Y., Wei C., et al. Rapid detection of viral antibodies based on multifunctional Staphylococcus aureus nanobioprobes. Enzyme Microb Technol. 2016;95:94–99. doi: 10.1016/j.enzmictec.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu G., Gai Z., Tao Y., et al. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 105.Seo G., Lee G., Kim M.J., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 106.Ye S., Shao K., Li Z., et al. Antiviral activity of graphene oxide: how sharp edged structure and charge matter. ACS App Mater. 2015;7(38):21571–21579. doi: 10.1021/acsami.5b06876. [DOI] [PubMed] [Google Scholar]
- 107.Chen Y.N., Hsueh Y.H., Hsieh C.T., et al. Antiviral activity of graphene– silver nanocomposites against non-enveloped and enveloped viruses. Int J Environ Res Public Health. 2016;13(4):430. doi: 10.3390/ijerph13040430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park S., Park H.H., Kim S.Y., et al. Antiviral properties of silver nanoparticles on a magnetic hybrid colloid. Appl Environ Microbiol. 2014;80(8):2343–2350. doi: 10.1128/AEM.03427-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elechiguerra J.L., Burt J.L., Morones J.R., et al. Interaction of silver nanoparticles with HIV-1. J Nanobiotechnology. 2005;3(1):6. doi: 10.1186/1477-3155-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Du T., Liang J., Dong N., et al. Glutathione-capped Ag2S nanoclusters inhibit coronavirus proliferation through blockage of viral RNA synthesis and budding. ACS Appl Mater. 2018;10(5):4369–4378. doi: 10.1021/acsami.7b13811. [DOI] [PubMed] [Google Scholar]
- 111.Loczechin A., Seron K., Barras A., et al. Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl Mater. 2019;11(46):42964–42974. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ting D., Dong N., Fang L., et al. Multisite inhibitors for enteric coronavirus: antiviral cationic carbon dots based on curcumin. ACS Appl Nano Mater. 2018;1(10):5451–5459. doi: 10.1021/acsanm.0c00970. [DOI] [PubMed] [Google Scholar]
- 113.Botequim D., Maia J., Lino M.M.F., et al. Nanoparticles and surfaces presenting antifungal, antibacterial and antiviral properties. Langmuir. 2012;28(20):7646–7656. doi: 10.1021/la300948n. [DOI] [PubMed] [Google Scholar]
- 114.Lee J.H., Ozorowski G., Ward A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351(6277):1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liang J.J., Wei J.C., Lee Y.L., et al. Surfactant-modified nanoclay exhibits an antiviral activity with high potency and broad spectrum. J Virol. 2014;88(8):4218–4228. doi: 10.1128/JVI.03256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang M.C., Lin J.J., Tseng H.J., et al. Characterization, antimicrobial activities, and biocompatibility of organically modified clays and their nanocomposites with polyurethane. ACS Appl Mater. 2012;4(1):338–350. doi: 10.1021/am2014103. [DOI] [PubMed] [Google Scholar]
- 117.Miyako E., Nagata H., Hirano K., et al. Photoinduced antiviral carbon nanohorns. Nanotechnology. 2008;19(7) doi: 10.1088/0957-4484/19/7/075106. [DOI] [PubMed] [Google Scholar]
- 118.Banerjee I., Douaisi M.P., Mondal D., et al. Light-activated nanotube–porphyrin conjugates as effective antiviral agents. Nanotechnology. 2012;23(10) doi: 10.1088/0957-4484/23/10/105101. [DOI] [PubMed] [Google Scholar]
- 119.Di Giorgio S., et al. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv. 2020;6(25) doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Di Giorgio S., et al. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 2020 doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Esvelt K.M., Wang H.H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 2013;9(1):641. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hassanzadeh Parichehr, Arbabi Elham, Rostami Fatemeh, Atyabi Fatemeh, Dinarvand Rassoul. Carbon nanotubes prolong the regulatory action of nerve growth factor on the endocannabinoid signaling. Physiol Pharmacol. 2015;19:167–176. [Google Scholar]
- 123.Hassanzadeh Parichehr, ElhamArbabi Fatemeh Atyabi, Dinarvand Rassoul. Application of carbon nanotubes as the carriers of the cannabinoid, 2-arachidonoylglycerol: Towards a novel treatment strategy in colitis. Life Sciences. 2017;179:66–72. doi: 10.1016/j.lfs.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 124.Hassanzadeh Parichehr, Arbabi Elham, Atyabi Fatemeh, Dinarvand Rassoul. Nerve growth factor-carbon nanotube complex exerts prolonged protective effects in an in vitro model of ischemic stroke. Life Sciences. 2017;179:15–22. doi: 10.1016/j.lfs.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 125.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. Application of Carbon Nanotubes for Controlled Release of Growth Factors or Endocannabinoids: A Breakthrough in Biomedicine. Biomed. Rev. 2017;27:41. doi: 10.14748/bmr.v27.2105. [DOI] [Google Scholar]
- 126.Hassanzadeh Parichehr, Arbabi Elham, Atyabi Fatemeh, Dinarvand Rassoul. Carbon nanotubes provide longer lasting gastroprotective effects for anandamide in stressinduced gastric ulcer in rat. Physiol Pharmacol. 2018;22:38–47. [Google Scholar]
- 127.Zhu S., Li J., Huang A.-G., Huang J.-Q., Huang Y.-Q., Wang G.-X. Anti-betanodavirus activity of isoprinosine and improved efficacy using carbon nanotubes based drug delivery system. Aquaculture. 2019;512 [Google Scholar]
- 128.Zhu B., Liu G.-L., Ling F., Wang G.-X. Carbon nanotube-based nanocarrier loaded with ribavirin against grass carp reovirus. Antivir. Res. 2015;118:29–38. doi: 10.1016/j.antiviral.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 129.Demirer G.S., et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019;14:456–464. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul, Dehpour Ahmad-Reza, Azhdarzadeh Morteza, Dinarvand Meshkat. Application of nanostructured lipid carriers: the prolonged protective effects for sesamol in in vitro and in vivo models of ischemic stroke via activation of PI3K signalling pathway. DARU J. Pharm. Sci. 2017;25 doi: 10.1186/s40199-017-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hassanzadeh Parichehr, Arbabi Elham, Atyabi Fatemeh, Dinarvand Rassoul. Ferulic acid-loaded nanostructured lipid carriers: A promising nanoformulation against the ischemic neural injuries. Life Sciences. 2018;193:64–76. doi: 10.1016/j.lfs.2017.11.046. [DOI] [PubMed] [Google Scholar]
- 132.Hassanzadeh P., Arbabi E., Rostami F., Atyabi F., Dinarvand R. Aerosol delivery of ferulic acid-loaded nanostructured lipid carriers: A promising treatment approach against the respiratory disorders. Physiol Pharmacol. 2017;21:331–342. [Google Scholar]
- 133.Hassanzadeh P., Atyabi F., Dinarvand R. Nanoencapsulation: A promising strategy for biomedical applications of ferulic acid. Biomed. Rev. 2017;28:22–30. [Google Scholar]
- 134.Hassanzadeh P. Nanopharmaceuticals: Innovative theranostics for the neurological disorders. Biomed. Rev. 2014;25:25–34. [Google Scholar]
- 135.Pardi N., et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8 doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Conway A., et al. Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Akinc Akin, Maier Martin A., Manoharan Muthiah, Fitzgerald Kevin, Jayaraman Muthusamy, Barros Scott, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 138.Wu Chang-Jer, Chan Yi-Lin. Antiviral applications of RNAi for Coronavirus. Expert Opin. Investig. Drugs. 2006;15:89–97. doi: 10.1517/13543784.15.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng B.-J., Guan Y., Tang Q., et al. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir Ther. 2004;9:365–374. [PubMed] [Google Scholar]
- 140.Leonard J.N., Schaffer D.V. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li B.-j., Tang Q., Cheng D., et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Frieman M., Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008;72:672–685. doi: 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Ying, Cao Ying-Li, Yang Fan, Zhang Yun, Wang Shu-Hui, Liu Li. Small Interfering RNA Effectively Inhibits the Expression of SARS Coronavirus Membrane Gene at Two Novel Targeting Sites. Molecules. 2010;15:7197–7207. doi: 10.3390/molecules15107197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Watanabe T., Umehara T., Yasui F., et al. Liver target delivery of small interfering RNA to the HCV gene by lactosylated cationic liposome. J. Hepatol. 2007;47(6):744–750. doi: 10.1016/j.jhep.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 145.Torrecilla Josune, Rodríguez-Gascón Alicia, Solinís María Ángeles, del Pozo-Rodríguez Ana. Lipid Nanoparticles as Carriers for RNAi against Viral Infections: Current Status and Future Perspectives. BioMed Res. Int. 2014;2014:1–17. doi: 10.1155/2014/161794. Article ID 161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Spengler Ulrich. 2020. Liver Disease Associated with Non-Hepatitis Viruses. Encyclopedia of Gastroenterology; pp. 363–376. [Google Scholar]
- 147.Cheng Qiang, Wei Tuo, Jia Yuemeng, Farbiak Lukas, Zhou Kejin, Zhang Shuyuan, et al. Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH mRNA to Normalize Liver Function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I. Adv. Mater. 2018;1805308:1–10. doi: 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- 148.van der Meel Roy. Nanotechnology for organ-tunable gene editing. Nat Nanotechnol. 2020;15:253–255. doi: 10.1038/s41565-020-0666-9. [DOI] [PubMed] [Google Scholar]
- 149.van der Meel Roy, Sulheim Einar, Shi Yang, Kiessling Fabian, Mulder Willem J.M., Lammers Twan. Smart cancer nanomedicine. Strategic directions to improve translation and exploitation. Nat Nanotechnol. 2019 November;14(11):1007–1017. doi: 10.1038/s41565-019-0567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kaczmarek James C., Patel Asha K., Kauffman Kevin J., Fenton Owen S., Webber Matthew J., Heartlein Michael W., et al. Polymer–Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. 2016;128:1–6. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Vol. 52. 2019. Acc. Chem. Res; pp. 2435–2444. [DOI] [PubMed] [Google Scholar]
- 153.Cheng Qiang, Wei Tuo, Farbiak Lukas, Johnson Lindsay T., Dilliard Sean A., Siegwart Daniel J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khan I., Saeed K., Khan I. Nanoparticles: Properties, applications and toxicities. Arab J Chem. 2019;12:908–931. [Google Scholar]
- 155.Andreas Reichmuth, Matthias Oberli, Ana Jeklenec, Robert S Langer. mRNA vaccine delivery using lipid nanoparticles. Therapeutic Delivery 7(5):319-334. [DOI] [PMC free article] [PubMed]
- 156.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11:885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 157.John S., Yuzhakov O., Woods A., Deterling J., Hassett K., Shaw C.A., Ciaramella G. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine. 2018;36:1689–1699. doi: 10.1016/j.vaccine.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 158.Hassett Kimberly J., Benenato Kerry E., Jacquinet Eric, Lee Aisha, Woods Angela, Yuzhakov Olga, et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. 2019;15(1):1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ohno S., Kohyama S., Taneichi M., et al. Synthetic peptides coupled to the surface of liposomes effectively induce SARS coronavirusspecific cytotoxic T lymphocytes and viral clearance in HLA-A* 0201 transgenic mice. Vaccine. 2009;27(29):3912–3920. doi: 10.1016/j.vaccine.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sekimukai H., Iwata-Yoshikawa N., Fukushi S., et al. Gold nanoparticle-adjuvanted S protein induces a strong antigen-specific IgG response against severe acute respiratory syndrome-related coronavirus infection, but fails to induce protective antibodies and limit eosinophilic infiltration in lungs. Microbiol Immunol. 2020;64(1):33–51. doi: 10.1111/1348-0421.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lu X., Chen Y., Bai B., et al. Immune responses against severe acute respiratory syndrome coronavirus induced by virus-like particles in mice. Immunology. 2007;122(4):496–502. doi: 10.1111/j.1365-2567.2007.02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Y.V., Massare M.J., Barnard D.L., et al. Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV. Vaccine. 2011;29(38):6606–6613. doi: 10.1016/j.vaccine.2011.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Plummer E.M., Manchester M. Viral nanoparticles and virus-like particles: platforms for contemporary vaccine design. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011;3:174–196. doi: 10.1002/wnan.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shin Matthew D., Shukla Sourabh, Chung Young Hun, Beiss Veronique, Chan Soo Khim, Ortega-Rivera Oscar A., et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 165.Read S.A., Obeid S., Ahlenstiel C., Ahlenstiel G. The role of zinc in antiviral immunity. Adv Nutr. 2019;10:696–710. doi: 10.1093/advances/nmz013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discoveries Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 167.Wimalawansa S.J. Global epidemic of coronavirus—covid-19: what can we do to minimize risks. Eur. J Biomed. 2020;7:432–438. [Google Scholar]
- 168.Hirano T., Murakami M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity. 2020;52:731. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu M., Wu J., Shi J., Farokhzad O.C. Nanotechnology for Protein Delivery: Overview and Perspectives. J. Controlled Release. 2016;240:24–37. doi: 10.1016/j.jconrel.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li B., Zhang X., Dong Y. Nanoscale Platforms for Messenger RNA Delivery. WIREs Nanomed. Nanobiotechnol. 2019;11 doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van der Zee R., Jiskoot W., van Eden W., Broere F. PLGA, PLGA-TMC and TMC-TPP Nanoparticles Differentially Modulate the Outcome of Nasal Vaccination by Inducing Tolerance or Enhancing Humoral Immunity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., et al. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. No. eabc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knudsen N.P., Olsen A., Buonsanti C., Follmann F., Zhang Y., Coler R.N., et al. Different Human Vaccine Adjuvants Promote Distinct Antigen-Independent Immunological Signatures Tailored to Different Pathogens. Sci. Rep. 2016;6 doi: 10.1038/srep19570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sun B., Xia T. Nanomaterial-Based Vaccine Adjuvants. J. Mater. Chem. B. 2016;4:5496–5509. doi: 10.1039/C6TB01131D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ma J., Liu R., Wang X., Liu Q., Chen Y., Valle R.P., Zuo Y.Y., Xia T., Liu S. Crucial Role of Lateral Size for Graphene Oxide in Activating Macrophages and Stimulating Pro-Inflammatory Responses in Cells and Animals. ACS Nano. 2015;9:10498–10515. doi: 10.1021/acsnano.5b04751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gazzi A., Fusco L., Orecchioni M., Ferrari S., Franzoni G., Yan J.S., et al. Graphene, Other Carbon Nanomaterials and the Immune System: Toward Nanoimmunity-by-Design. J Phys Mater. 2020;3 [Google Scholar]
- 178.Orecchioni M., Bedognetti D., Sgarrella F., Marincola F.M., Bianco A., Delogu L.G. Impact of Carbon Nanotubes and Graphene on Immune Cells. J. Transl. Med. 2014;12:138. doi: 10.1186/1479-5876-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fuchs A.K., Syrovets T., Haas K.A., Loos C., Musyanovych A., Mailänder V., Landfester K., Simmet T. Carboxyl- and Amino-Functionalized Polystyrene Nanoparticles Differentially Affect the Polarization Profile of M1 and M2 Macrophage Subsets. Biomaterials. 2016;85:78–87. doi: 10.1016/j.biomaterials.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 180.Fusco L., Avitabile E., Armuzza V., Orecchioni M., Istif A., Bedognetti D., Da Ros T., Delogu L.G. Impact of the Surface Functionalization on Nanodiamond Biocompatibility: A Comprehensive View on Human Blood Immune Cells. Carbon. 2020;160:390–404. [Google Scholar]
- 181.Orecchioni M., Bedognetti D., Newman L., Fuoco C., Spada F., Hendrickx W., et al. Single-Cell Mass Cytometry and Transcriptome Profiling Reveal the Impact of Graphene on Human Immune Cells. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Prompetchara E., Ketloy C., Palaga T. Immune Responses in COVID-19 and Potential Vaccines: Lessons Learned from SARS and MERS Epidemic. Asian Pac. J. Allergy Immunol. 2020;(38):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 183.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Al-Lawati H., Aliabadi H.M., Makhmalzadeh B.S., Lavasanifar A. Nanomedicine for Immunosuppressive Therapy: Achievements in Pre-Clinical and Clinical Research. Expert Opin. Drug Delivery. 2018;15:397–418. doi: 10.1080/17425247.2018.1420053. [DOI] [PubMed] [Google Scholar]
- 185.Chen, Y.; Feng, Z.; Diao, B.; Wang, R.; Wang, G.; Wang, C.; et al. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV- 2) Directly Decimates Human Spleens and Lymph Nodes. medRxiv 2020, DOI: 10.1101/2020.03.27.20045427. [DOI]
- 186.Pentecost A., Kim M.J., Jeon S., Ko Y.J., Kwon I.C., Gogotsi Y., Kim K., Spiller K.L. Immunomodulatory Nanodiamond Aggregate-Based Platform for the Treatment of Rheumatoid Arthritis. Regen. Biomater. 2019;6:163–174. doi: 10.1093/rb/rbz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Presser V., Yeon S.H., Vakifahmetoglu C., Howell C.A., Sandeman S.R., Colombo P., Mikhalovsky S., Gogotsi Y. Hierarchical Porous Carbide-Derived Carbons for the Removal of Cytokines from Blood Plasma. Adv. Healthcare Mater. 2012;(1):796–800. doi: 10.1002/adhm.201200044. [DOI] [PubMed] [Google Scholar]
- 188.Seredych M., Haines B., Sokolova V., Cheung P., Meng F., Stone L., et al. Graphene-Based Materials for the Fast Removal of Cytokines from Blood Plasma. ACS Appl. Bio Mater. 2018;1:436–443. doi: 10.1021/acsabm.8b00151. [DOI] [PubMed] [Google Scholar]
- 189.Wainwright M. Local Treatment of Viral Disease Using Photodynamic Therapy. Int. J. Antimicrob. Agents. 2003;21:510–520. doi: 10.1016/s0924-8579(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 190.Wiehe A., O’Brien J.M., Senge M.O. Trends and Targets in Antiviral Phototherapy. Photochem. Photobiol. Sci. 2019;18:2565–2612. doi: 10.1039/c9pp00211a. [DOI] [PubMed] [Google Scholar]
- 191.Bhatia S., Camacho L.C., Haag R. Pathogen Inhibition by Multivalent Ligand Architectures. J. Am. Chem. Soc. 2016;138:8654–8666. doi: 10.1021/jacs.5b12950. [DOI] [PubMed] [Google Scholar]
- 192.De Clercq E. Antiviral Drugs in Current Clinical Use. J. Clin. Virol. 2004;30:115–133. doi: 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 193.Fasting C., Schalley C.A., Weber M., Seitz O., Hecht S., Koksch B., Dernedde J., Graf C., Knapp E.-W., Haag R. Multivalency as a chemical organization and action principle. Angew Chem Int Ed Engl. 2012;51(42):10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 194.Bhatia S., Dimde M., Haag R. Multivalent glycoconjugates as vaccines and potential drug candidates. MedChemComm. 2014;5:862. [Google Scholar]
- 195.Bhella D. The role of cellular adhesion molecules in virus attachment and entry. Philos. Trans. R. Soc., B. 2015;370 doi: 10.1098/rstb.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Moscona A. Annu. Medical Management of Influenza Infectio. Rev. Med. 2008;59:397. doi: 10.1146/annurev.med.59.061506.213121. [DOI] [PubMed] [Google Scholar]
- 197.Schirmer P., Holodniy M. Oseltamivir for treatment and prophylaxis of influenza infection. Expert Opin. Drug Saf. 2009;8:357. doi: 10.1517/14740330902840519. [DOI] [PubMed] [Google Scholar]
- 198.Papp I., Sieben C., Sisson A.L., Kostka J., Bottcher C., Ludwig K., Herrmann A., Haag R. Inhibition of influenza virus activity by multivalent glycoarchitectures with matched sizes. ChemBioChem. 2011;12:887. doi: 10.1002/cbic.201000776. [DOI] [PubMed] [Google Scholar]
- 199.Qi Z., Bharate P., Lai C.H., Ziem B., Bottcher C., Schulz A., Beckert F., Hatting B., Mulhaupt R., Seeberger P.H., Haag R. Multivalency at Interfaces: Supramolecular Carbohydrate-Functionalized Graphene Derivatives for Bacterial Capture, Release, and Disinfection. Nano Lett. 2015;15:6051. doi: 10.1021/acs.nanolett.5b02256. [DOI] [PubMed] [Google Scholar]
- 200.Baram-Pinto D., Shukla S., Gedanken A., Sarid R. Inhibition of HSV-1 Attachment, Entry, and Cell-to-Cell Spread by Functionalized Multivalent Gold Nanoparticles. Small. 2010;6:1044–1050. doi: 10.1002/smll.200902384. [DOI] [PubMed] [Google Scholar]
- 201.Hu R.L., Li S.R., Kong F.J., Hou R.J., Guan X.L., Guo F. Inhibition Effect of Silver Nanoparticles on Herpes Simplex Virus 2. GMR. Genet. Mol. Res. 2014;13 doi: 10.4238/2014.March.19.2. [DOI] [PubMed] [Google Scholar]
- 202.Ziem B., Azab W., Gholami M.F., Rabe J.P., Osterrieder N., Haag R. Size-Dependent Inhibition of Herpesvirus Cellular Entry by Polyvalent Nanoarchitectures. Nanoscale. 2017;9:3774–3783. doi: 10.1039/c7nr00611j. [DOI] [PubMed] [Google Scholar]
- 203.Vonnemann J., Sieben C., Wolff C., Ludwig K., Böttcher C., Herrmann A., Haag R. Virus Inhibition Induced by Polyvalent Nanoparticles of Different Sizes. Nanoscale. 2014;6:2353–2360. doi: 10.1039/c3nr04449a. [DOI] [PubMed] [Google Scholar]
- 204.Papp I., Sieben C., Ludwig K., Roskamp M., Böttcher C., Schlecht S., Herrmann A., Haag R. Inhibition of Influenza Virus Infection by Multivalent Sialic-Acid-Functionalized Gold Nanoparticles. Small. 2010;6:2900–2906. doi: 10.1002/smll.201001349. [DOI] [PubMed] [Google Scholar]
- 205.Cagno V., Andreozzi P., D’Alicarnasso M., Jacob Silva P., Mueller M., Galloux M., et al. Broad-Spectrum Non-Toxic Antiviral Nanoparticles with a Virucidal Inhibition Mechanism. Nat. Mater. 2018;17:195–203. doi: 10.1038/nmat5053. [DOI] [PubMed] [Google Scholar]
- 206.Han Y., Král P. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Papp Ilona, Sieben Christian, Sisson Adam L., Kostka Johanna, Bçttcher Christoph, Ludwig Kai, et al. Inhibition of Influenza Virus Activity by Multivalent Glycoarchitectures with Matched Sizes. ChemBioChem. 2011;12:887–895. doi: 10.1002/cbic.201000776. [DOI] [PubMed] [Google Scholar]
- 208.Dey Pradip, Bergmann Tobias, Cuellar-Camacho Jose Luis, Ehrmann Svenja, Chowdhury Mohammad Suman, Zhang Minze, et al. Multivalent Flexible Nanogels Exhibit Broad-Spectrum Antiviral Activity by Blocking Virus Entry. ACS Nano. 2018;12:6429–6442. doi: 10.1021/acsnano.8b01616. [DOI] [PubMed] [Google Scholar]
- 209.Ziem Benjamin, Thien Hendrik, Achazi Katharina, Yue Constanze, Stern Daniel, Silberreis Kim, et al. Highly Efficient Multivalent 2D Nanosystems for Inhibition of Orthopoxvirus Particles. Adv. Healthcare Mater. 2016;5:2922–2930. doi: 10.1002/adhm.201600812. [DOI] [PubMed] [Google Scholar]
- 210.Mohammad Fardin Gholami M.F., Lauster Daniel, Ludwig Kai, Storm Julian, Ziem Benjamin, Severin Nikolai, et al. Functionalized Graphene as Extracellular Matrix Mimics: Toward Well-Defined 2D Nanomaterials for Multivalent Virus Interactions. Adv. Funct. Mater. 2017;1606477:1–12. 1606477. [Google Scholar]
- 211.Jastrzębska A.M., Kurtycz P., Olszyna A.R. Recent Advances in Graphene Family Materials Toxicity Investigations. J. Nanopart. Res. 2012;14:1320–1341. doi: 10.1007/s11051-012-1320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Steinhilber D., Sisson A.L., Mangoldt D., Welker P., Licha K., Haag R. Synthesis, Reductive Cleavage, and Cellular Interaction Studies of Biodegradable, Polyglycerol Nanogels. Adv. Funct. Mater. 2010;20:4133–4138. [Google Scholar]
- 213.Zhang Qiangzhe, Honko Anna, Zhou Jiarong, Gong Hua, Downs Sierra N., Vasquez Jhonatan Henao, et al. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nanoletters. 2020;20(7):5570–5574. doi: 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Parodi A., Molinaro R., Sushnitha M., Evangelopoulos M., Martinez J.O., Arrighetti N., Corbo C., Tasciotti E. Bio-Inspired Engineering of Cell- and Virus-Like Nanoparticles for Drug Delivery. Biomaterials. 2017;147:155–168. doi: 10.1016/j.biomaterials.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 215.Kotov N.A. Inorganic Nanoparticles as Protein Mimics. Science. 2010;330:188. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- 216.Czapar A.E., Zheng Y.-R., Riddell I.A., Shukla S., Awuah S.G., Lippard S.J., Steinmetz N.F. Tobacco Mosaic Virus Delivery of Phenanthriplatin for Cancer Therapy. ACS Nano. 2016;10:4119–4126. doi: 10.1021/acsnano.5b07360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mullard A. Flooded by the Torrent: The COVID-19 Drug Pipeline. Lancet. 2020;395:1245–1246. doi: 10.1016/S0140-6736(20)30894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Senanayake S.L. Drug Repurposing Strategies for COVID-19. Future Drug Discovery. 2020;2(2):FDD40. [Google Scholar]
- 219.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. Ignoring the modeling approaches: Towards the shadowy paths in nanomedicine. J. Controlled Release. 2018;280:58–75. doi: 10.1016/j.jconrel.2018.04.042. [DOI] [PubMed] [Google Scholar]
- 220.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. Application of modelling and nanotechnology-based approaches: The emergence of breakthroughs in theranostics of central nervous system disorders. Life Sciences. 2017;182:93–103. doi: 10.1016/j.lfs.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 221.Hassanzadeh P. Computational modelling: moonlighting on the neuroscience and medicine. Biomed Rev. 2013;24:25–31. [Google Scholar]
- 222.Ferreira L.G., Dos Santos R.N., Oliva G., Andricopulo A.D. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Šebera J., Dubankova A., Sychrovsky V., Ruzek D., Boura E., Nencka R. The structural model of Zika virus RNA-dependent RNA polymerase in complex with RNA for rational design of novel nucleotide inhibitors. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-29459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shameer K, Johnson KW, Readhead B. Rapid Therapeutic Recommendations in the Context of a Global Public Health Crisis using Translational Bioinformatics Approaches: A proof-of-concept study using Nipah Virus Infection. bioRxiv; 333021 , 10.1101/333021. [DOI]
- 225.Zhang T.F., Chen Q. Identification of contaminant sources in enclosed environments by inverse CFD modeling. Indoor Air. 2007;17:167–177. doi: 10.1111/j.1600-0668.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- 226.Mazumdar S., Poussou S.B., Lin C.-H., Isukapalli S.S., Plesniak M.W., Chen Q. Impact of scaling and body movement on contaminant transport in airliner cabins. Atmos. Environ. 2011;45:6019–6028. [Google Scholar]
- 227.Lunnoo T., Assawakhajornsak J., Puangmali T. In Silico Study of Gold Nanoparticle Uptake into a Mammalian Cell: Interplay of Size, Shape, Surface Charge, and Aggregation. J. Phys. Chem. C. 2019;123:3801–3810. [Google Scholar]
- 228.Ostaszewski Marek, Mazein Alexander, Gillespie Marc E., Kuperstein Inna, Niarakis Anna, Hermjakob Henning, et al. COVID-19 Disease Map, building a computational repository of SARS-CoV-2 virus-host interaction Mechanisms. Scientific Data. 2020;7:136. doi: 10.1038/s41597-020-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ostaszewski M., et al. Community-driven roadmap for integrated disease maps. Brief. Bioinform. 2019;20:659–670. doi: 10.1093/bib/bby024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sacha G.M., Varona P. Artificial intelligence in nanotechnology. Nanotechnology. 2013;24:1–13. doi: 10.1088/0957-4484/24/45/45200. 452002. [DOI] [PubMed] [Google Scholar]
- 231.Cavalcanti A., Shirinzadeh B., Freitas R.A., Jr., Kretly L.C. Medical nanorobot architecture based on nano bioelectronics. Recent Pat Nanotechnol. 2007;1:1–10. doi: 10.2174/187221007779814745. [DOI] [PubMed] [Google Scholar]
- 232.Yamaan Saadeh B.S., Vyas Dinesh. Nanorobotic Applications in Medicine: Current Proposals and Designs. Am J Robot Surg. 2014;1(1):4–11. doi: 10.1166/ajrs.2014.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Murphy D., Challacombe B., Nedas T., Elhage O., Althoefer K., Seneviratne L., et al. Equipment and technology in robotics. Arch Esp Urol. 2007;60:349–354. doi: 10.4321/s0004-06142007000400004. [DOI] [PubMed] [Google Scholar]
- 234.Hassanzadeh Parichehr, Atyabi Fatemeh, Dinarvand Rassoul. Creation of nanorobots: Both state-of-the science and state-of-the-art. Biomed. Rev. 2016;27:37–44. [Google Scholar]
- 235.Patel G.M., Patel G.C., Patel R.B., Patel J.K., Patel M. Nanorobot: A versatile tool in nanomedicine. J Drug Target. 2006;14:63–67. doi: 10.1080/10611860600612862. [DOI] [PubMed] [Google Scholar]
- 236.Lee Y.S., Bae J.Y., Koo H.Y., Lee Y.B., Choi W.S. A remote-controlled generation of gold@polydopamine (core@ shell) nanoparticles via physical-chemical stimuli of polydopamine/gold composites. Sci Rep. 2016;6 doi: 10.1038/srep22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Martel S., Mathieu J.B., Felfoul O., Chanu A., Aboussouan E., Tamaz S., et al. A computer-assisted protocol for endovascular target interventions using a clinical MRI system for controlling untethered microdevices and future nanorobots. Comput Aided Surg. 2008;13:340–352. doi: 10.3109/10929080802551274. [DOI] [PubMed] [Google Scholar]
- 238.Balasubramanian S., Kagan D., Jack H.C.M., Campuzano S., Lobo-Castañon M.J., Lim N., et al. Micromachine-enabled capture and isolation of cancer cells in complex media. Angew Chem Int Ed Engl. 2011;50:4161–4164. doi: 10.1002/anie.201100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Patel R.R. Nanorobotics ideas in nanomedicine. Asian J Pharm Sci Res. 2013;3:15–22. [Google Scholar]
- 240.Martel S., Felfoul O., Mohammadi M., Mathieu J.B. Interventional procedure based on nanorobots propelled and steered by flagellated magnetotactic bacteria for direct targeting of tumors in the human body. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:2497–2500. doi: 10.1109/IEMBS.2008.4649707. [DOI] [PubMed] [Google Scholar]
- 241.Somanna M.B. Nanobots: The future of medical treatments. Int J Sci Tech Res. 2015;4:276–278. [Google Scholar]
- 242.Torelli E., Marini M., Palmano S., Piantanida L., Polano C., Scarpellini A., et al. A DNA origami nanorobot controlled by nucleic acid hybridization. Small. 2014;10:2918–2926. doi: 10.1002/smll.201400245. [DOI] [PubMed] [Google Scholar]
- 243.Douglas S.M., Bachelet I., Church G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 244.Behkam B., Sitti M. Bacterial flagella-based propulsion and on/off motion control of microscale objects. Appl Phys Lett. 2007;90:1–3. [Google Scholar]
- 245.Dubey A., Sharma G., Mavroidis C., Tomassone M.S., Nikitczuk K., Yarmushc M.L. Computational studies of viral protein nano-actuators. J Comp Theor Nanosci. 2004;1:18–28. [Google Scholar]
- 246.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 247.Ahuja S.P., Myers J.R. A survey on wireless grid computing. J. Supercomput. 2006;37(1):3–21. [Google Scholar]
- 248.Ricciardi L., Pitz I., Al-Sarawi S.F., Varadan V., Abbott D. Investigation into the future of RFID in biomedical applications. Proc. SPIE - Int. Soc. Opt. Eng. 2003;5119:199–209. [Google Scholar]
- 249.Kostarelos Kostas. Nanoscale nights of COVID-19. Nat. Nanotechnol. 2020;15:343–344. doi: 10.1038/s41565-020-0687-4. [DOI] [PubMed] [Google Scholar]
- 250.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.de Oliveira Toledo S.L., Nogueira L.S., das Graças Carvalho M., Alves Rios D.R., de Barros Pinheiro M. COVID-19: Review and hematologic impact. Clinica Chimica Acta. 2020;510:170–176. doi: 10.1016/j.cca.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Singh A., Shaikh A., Singh R., Sing A.K. COVID-19: From bench to bed side. Diabetes Metabolic Syndrome. 2020;14:277e281. doi: 10.1016/j.dsx.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tanindi A., Cemri M. Troponin elevation in conditions other than acute coronary syndromes. Vasc Health Risk Manag. 2011;7:597e603. doi: 10.2147/VHRM.S24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sahu K.K., Siddiqui A.D., Cerny J. Mesenchymal Stem Cells in COVID-19: A Journey from Bench to Bedside. Lab Med. 2020;1:1–12. doi: 10.1093/labmed/lmaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Warnes S.L., Little Z.R., Keevil C.W. Human Coronavirus 229E Remains Infectious on Common Touch Surface Materials. mBio. 2015;6 doi: 10.1128/mBio.01697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]