Abstract
Objectives
To assess the methodologies used in the estimation of diagnostic accuracy of SARS-CoV-2 real-time reverse transcription polymerase chain reaction (rRT-PCR) and other nucleic acid amplification tests (NAATs) and to evaluate the quality and reliability of the studies employing those methods.
Methods
We conducted a systematic search of English-language articles published December 31, 2019-June 19, 2020. Studies of any design that performed tests on ≥10 patients and reported or inferred correlative statistics were included. Studies were evaluated using elements of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) guidelines.
Results
We conducted a narrative and tabular synthesis of studies organized by their reference standard strategy or comparative agreement method, resulting in six categorizations. Critical study details were frequently unreported, including the mechanism for patient/sample selection and researcher blinding to results, which lead to concern for bias.
Conclusions
Current studies estimating test performance characteristics have imperfect study design and statistical methods for the estimation of test performance characteristics of SARS-CoV-2 tests. The included studies employ heterogeneous methods and overall have an increased risk of bias. Employing standardized guidelines for study designs and statistical methods will improve the process for developing and validating rRT-PCR and NAAT for the diagnosis of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Diagnostic accuracy, Sensitivity, Specificity, QUADAS-2
Introduction
After its emergence in December 2019, the virus now known as SARS-CoV-2 was identified and sequenced in early January 2020,1 allowing for the rapid development of diagnostic testing based on the detection of viral nucleic acid (i.e., real-time reverse transcription polymerase chain reaction [rRT-PCR]).2 Because infected patients can present with non-specific symptoms or be asymptomatic, the development of accurate diagnostic tests for both clinical and epidemiological purposes was a crucial step in the response to the COVID-19 pandemic.3
In the United States, the spread of SARS-CoV-2 rapidly outpaced the capacity to test for it, resulting in the Food and Drug Administration (FDA) relaxing regulatory requirements to increase testing availability. The FDA granted the first Emergency Use Authorization (EUA) for a SARS-CoV-2 rRT-PCR diagnostic test on February 4, 2020. Consequently, hundreds of tests for SARS-CoV-2, among them rRT-PCRs, other types of nucleic acid amplification tests (NAATs), and automated and/or multiplex methods based on proprietary platforms, obtained FDA Emergency Use Authorization (EUA). As of August 4th, 2020, the FDA has granted EUAs to 203 diagnostic tests, including 166 molecular tests, 35 antibody assays, and 2 antigen tests. Although the FDA began requiring the submission of validation methods and results as part of EUA application for SARS-CoV-2 diagnostic tests, these tests were not initially required to undergo the rigorous assessment that would normally be part of the FDA approval process. Researchers also began developing alternative nucleic-acid based methodologies to detect SARS-CoV-2, including reverse-transcription loop-mediated isothermal amplification (RT-LAMP), and others.
Concurrently with rapid test production, publications emerged reporting clinical diagnostic test performance characteristics, such as “sensitivity” and “specificity”, though some lacked the rigorous methodologies usually required to formally estimate diagnostic accuracy. Here we present a scoping review of the literature with two main objectives: 1) to assess the methodologies used in the estimation of diagnostic accuracy of SARS-CoV-2 tests and 2) to evaluate the quality and reliability of the studies employing those methods.
Methods
Data sources and searches
Searches were performed through MEDLINE (Ovid), EMBASE (Elsevier), Scopus, Web of Science, CINAHL, and PubMed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines 4 between December 1, 2019 and June 19, 2020. The following search string was used: (2019-nCOV or SARS-CoV-2 or SARS-CoV2 or COVID-19 or COVID19 or COVID) and (“positive agreement” or “negative agreement” or “overall agreement” or “diagnostic accuracy” or “positive rate” or “positivity rate” or “test performance” or “reference standard” or “gold standard” or sensitivity or specificity or “percent agreement” or “concordance” or “test agreement” or “predictive value” or "false negative" or “false positive”) and (“polymerase chain reaction” or PCR or “reverse transcriptase” or “nucleic acid amplification test” or NAAT or isothermal or “RT-LAMP” or “RT-PCR” or “molecular test”). The literature hub LitCovid's “Diagnosis” section was screened in its entirety once and then daily for relevant titles.
Study selection
We liberally screened articles by title and abstract for further evaluation. Articles were included if they met the following criteria on screening: 1) Peer-reviewed publication, 2) Study evaluated diagnostic test accuracy of NAAT, 3) Diagnostic test performed on ≥10 patients, 4) Diagnostic/Clinical sensitivity, specificity, other correlative statistics, or test positive rate were either identified by name or were included in the publication as a numerical value and we could reproduce the calculations. Exclusion criteria included: 1) Pre-print status, 2) Guidelines, consensus, review, opinion, and other summary articles 3) Entirely pregnant or pediatric populations, 4) Overlap of study population with another included publication.
Data extraction and quality assessment
Four authors independently extracted data and two authors reviewed data for accuracy. For study characteristics, we extracted: first author name, country, study design, patient population, total number of patients or samples included in test performance calculations, and number of cases according to rRT-PCR (Table 1, Table 2, Table 3, Table 4, Table 5 ) or total number of cases based on positive result of any platform tested (Table 6 ). For patient characteristics, we extracted age and sex. For index test and reference standard characteristics, we extracted: test type (NAAT) or definition (clinical diagnosis, composite reference standards), specimen (NAAT), specimen dry/collection liquid status (for studies evaluating Abbott ID NOW), proprietary automated and/or multiplex systems – henceforth called “platforms” (NAAT), and target genes of primers (NAAT). For outcomes, we extracted the values of test performance characteristics with their designation according to the original authors, without our interpretation. For this reason, we indicate these outcomes as “reported” (r): reported sensitivity (rSN), specificity (rSP), positive predictive value (rPPV), negative predictive value (rNPV), accuracy (rAcc), positive percent agreement (rPPA), negative percent agreement (rNPA), overall agreement (rOA), and Kappa coefficient. Additionally, we extracted “positive rate,” a non-standard term used by the included studies to refer to the number of positive NAATs in a population of patients suspected to have COVID-19 (Table 1), or to the number of positive samples in a total population of positive samples after repeat testing (Table 2). We constructed 2 × 2 contingency tables and reproduced test performance characteristic calculations to demonstrate the methods of how the original authors obtained the values (Supplementary Table 1). We report additional pertinent study data in Supplementary Table 2: enrollment dates, number of sites of enrollment, symptomatic status, and chest radiology status. No articles were excluded on the basis of quality in order to present the most comprehensive summary of the currently available evidence.
Table 1.
Studies reporting the “positive rate” of rRT-PCR testing within a population of patients suspected to have COVID-19.
| Authors | Country | Study Type | No. Patients | Demographics† | Index Test | Reference Standard: Case Definition/Clinical Diagnosis | Study Findings | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | rRT-PCR positive* | Age (y)‡ | % Male | Type Specimen (No.) | Primers | rRT-PCR Kit Company | PR§ (95% CI) | ||||
| Ai et al. 7 | China | Cross Sectional|| | 1014 | 601 | 51 ± 15 | 46.0% | rRT-PCR Throat Swab (1014) | ORF1ab, N | Shanghai Huirui Biotech, Shanghai BioGerm | “patients…who were suspected of novel coronavirus infection” (p. 5) | 59% (56 - 62) |
| Liu et al. 8 | China | Cross Sectional|| | 4880 | 1875 | 50 (IQR 27) | 46.13% | rRT-PCR Nasal swab, pharyngeal swab, BAL, sputum | ORF1ab, N | Shanghai Huirui Biotech | “All the cases were suspected of SARS-CoV-2 infection because of, (1) typical respiratory infection symptoms such as fever, cough and dyspnea, or (2) close contact with a COVID-19 patient.” (p.172) | 38.42% |
| Xie et al. 9 | China | Cross Sectional|| | 19 | 9 | 33 (8 - 62) | 42.1% | rRT-PCR OP swab (19) | ORF1ab, N | GeneoDx, Maccura, Life-river | “…suspected cases…” (p. 264) | 47.4% |
Reported instead of “cases according to reference standard” as present in other tables.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD.
PR: positive rate, which is the number of rRT-PCR patients out of the number of patients suspected to have COVID-19 (i.e. the reference standard). Patient population.
Hospitalized patients. Abbreviations- BAL: bronchoalveolar lavage, CI: confidence interval, IQR: Interquartile range, N: nucleocapsid, OP: oropharyngeal, ORF1ab: open reading frame 1ab, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, y: years.
Table 2.
Studies reporting test performance characteristics of initial rRT-PCR result compared to result after repeated tests of rRT-PCR as reference standard.
| Authors | Country | Study Type | Demographics† | Specimen Type (No.) | Primers/ Platform No. samples (%) | No. pts with 1st rRT-PCR positive | Total No. of pts with rRT-PCR ever positive | Total No. of pts in calculations* | Interval (d) between each re-test | No. of tests performed per pt until positive | Interval (d) between initial and positive (final) rRT-PCR | Study Findings | PR | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (y)‡ | % Male | Correlative Statistics (95% CI) | ||||||||||||||
| rSN | rSP | rAcc | ||||||||||||||
| Bernheim et al. 10 | China | Cases only|| | 45.3 ± 15.6 | 50.4% | NPS, OPS, Trach Asp, BAL (nr) | ORF1ab, N rRT-PCR |
90 | 102 | 102 | nr | 1 test: 90 >1 test: 12 |
nr | 88% | |||
| Fang et al. 11 | China | Cases only|| | 45 (IQR 39–55) |
56.9% | Throat swab (45), sputum (6) | ORF1ab, N rRT-PCR |
36 | 51 | 51 | ≥1 | 1 test: 36 2 tests: 12 3 tests: 2 4 tests: 1 |
nr | 71% (56–83) | |||
| Green et al. 12 | USA | Cohort|| | 53.1 ± 22.3 | 45.5% | NPS, OPS (nr) | RdRp, E Roche cobas 19,195 (70.1%) |
10,070 | 17,405⁎⁎ | 22,061 | median 8 range (1 – 49) |
1 test: 10,070 >1 test: 7335⁎⁎ |
1 - 49 | Lower bound estimate:⁎⁎ 57.9% (55.2–60.5) |
|||
| N2, E Cepheid Xpert 6219 (22.7%) |
||||||||||||||||
| N1, N2 rRT-PCR 1884 (6.9%) |
||||||||||||||||
| 10,643⁎⁎ | 22,061 | median 8 range (1 – 49) |
1 test: 10,070 >1 test: 573⁎⁎ |
1 - 49 | Upper bound estimate:⁎⁎ 94.6% (94.2–95.0) |
|||||||||||
| RdRp Abbott ID NOW 53 (0.2%) |
||||||||||||||||
| ORF1ab (x2) Hologic Panther 26 (0.1%) |
||||||||||||||||
| He et al. 13 | Hong Kong | Case-Control|| | 52 (8 - 74) | 50% | NPS, OPS, Trach Asp, BAL (nr) | RdRp, S rRT-PCR |
27 | 34 | 82 | nr | 1 test: 27 >1 test: 7 |
1–14 | 79% (66–93) | 100% (100) | 92% (91–92) | |
| Lee et al. 14 | Singapore | Cases only|| | nr | nr | NPS (70) | ORF1ab, N rRT-PCR |
62 | 70 | 70 | 1st-2nd: 1 2nd-Xth: 1–2 |
1 test: 62 2 tests: 5 3 tests: 1 5 tests: 1 6 tests: 1 |
1st-2nd: 1 2nd-Final: 2,4,7 |
88.6% | |||
| Long et al. 15 | China | Cases only|| | 44.8 ± 18.2 | 55.6% | OPS, NPS (nr) | ORF1ab, N rRT-PCR |
30 | 36 | 36 | nr | 1 test: 30 2 tests: 3 3 tests: 3 |
2 - 8 | 83.3% | |||
| Wong et al. 16 | Hong Kong | Cases only|| | 56 ± 19 | 40.6% | NPS, throat swab (nr) | RdRp/Hel rRT-PCR |
58 | 64 | 64 | nr, “not uniform” | 1 test: 58 >1 test: 6 |
nr | 91% (83–97) | |||
| Wu et al. 17 | China | Cases only|| | 46.1 ± 15.4 | 48.8% | Nose swab, throat swab (nr) | ORF1ab, N rRT-PCR |
41 | 80 | 80 | 1 | 1 test: 41 2 tests: 30 3 tests: 9 |
1 - 2 | 51.25% | |||
Pts included in test performance calculations.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD. Patient Population.
Hospitalized patients. Abbreviations- BAL: Bronchoalveolar lavage, CI: confidence interval, d: days, E: envelope, Hel: helicase, IQR: Interquartile range, N: nucleocapsid, No.: number, NPS: nasopharyngeal swab, nr: not reported, OPS: oropharyngeal swab, ORF1ab: open reading frame 1ab, PR: positive rate, pts: patients, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, rAcc: reported accuracy, RdRp: RNA-dependent RNA polymerase, rSN: reported sensitivity, rSP: reported specificity, S: spike, Trach Asp: Tracheal Aspirate, y: years.
Sensitivity estimates for the first test conducted on patients were calculated based on different assumptions about true negatives. The estimate of the upper bound estimate assumes that any negative test results (whether negative on a single test or consistently negative across multiple, repeated tests) was a true negative (aka, false negative rate=0%). The estimate of the lower bound uses the proportion of repeatedly tested cases who initially tested negative but then tested positive in repeated tests to calculate a false negative rate (16.8%) and apply that rate to the patients who only received a single test to calculate an assumed number of false negative cases. Additional details are provided in Suppl. Fig. 1 and Green et al.
Table 3.
Studies that calculate test performance characteristics of rRT-PCR or automated rRT-PCR platforms compared to composite reference standards.
| Authors | Country | Study Type | No. Patients | Demographics† | Specimen | Index Test | Composite Reference Standard Definition | Study Findings (95% CI) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Cases* | Age (y)‡ | % M | Type | Primers | Platform | rSN | rSP | rPPV | rNPV | rAcc | rPPA | rNPA | Cohen's κ | |||||
| Cradic et al. 18 | USA | Cohort||¶ | 184 | 33 | nr | nr | NPS in VTM (184) | Automated Multiplex rRT-PCR | ORF1ab, S | Diasorin Simplexa | Result obtained from at least 2 of the 3 assays is consensus result. | 100% (90–100) | 100% (98–100) | ||||||
| Automated Isothermal NAAT | RdRp | Abbott ID NOW |
91% (79–97) | 100% (98–100) | |||||||||||||||
| Automated Multiplex rRT-PCR | ORF1, E | Roche cobas 6800 | 100% (90–100) | 100% (98–100) | |||||||||||||||
| Suo et al. 19,a | China | Cohort⁎⁎ | 58 | 52 | nr | nr | Throat swab (58) | Initial rRT-PCR (China CDC protocol) | ORF1ab, N | N/A | Positive result of repeated rRT-PCR, or serology is considered a positive result. | 40% (27–55) | 100% (54–100) | 100% (N/A) | 16% (13–19) | 47% (33–60) | |||
| Zhen & Mangi et al. 20 | USA | Case Control†† | 104 | 51 | nr | nr | NPS (104) | rRT-PCR (US CDC protocol) | N1, N2 | N/A | Result obtained by 3 out of 4 assays tested is consensus result | 100% (93–100) | 98% (89–99) | 0.98 (0.94–1) | |||||
| Automated multiplex rRT-PCR | ORF1ab, S | Diasorin Simplexa |
100% (93–100) | 100% (93–100) | 1.0 (0.99–1) | ||||||||||||||
| Automated rRT-PCR w/sensor | N | GenMark ePlex | 96% (87–99) | 100% (93–100) | 0.96 (0.91–1) | ||||||||||||||
| Automated multiplex rRT-PCR | ORF1ab (2 targets) |
Hologic Panther | 100% (93–100) | 96% (87–99) | 0.96 (0.91–1) | ||||||||||||||
Cases according to composite reference standard.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD. Patient Population.
Hospitalized patients.
Emergency Room patients.
Outpatients, some of whom were later hospitalized.
not reported.
Suo et al. data is also present in Table 4. Abbreviations- E: envelope, IQR: Interquartile range, κ: kappa coefficient, M: male, N/A: not applicable, N: nucleocapsid, NPS: nasopharyngeal swab, nr: not reported, ORF1ab: open reading frame 1ab, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, rAcc: reported accuracy, RdRp: RNA-dependent RNA polymerase, rNPA: reported negative percent agreement, rNPV: reported negative predictive value, rPPA: reported positive percent agreement, rPPV: reported positive predicted value, rSN: reported sensitivity, rSP: reported specificity, S: spike, VTM: viral transport media, y:years.
Table 4.
Studies reporting test performance characteristics of other nucleic acid amplification test methods compared to rRT-PCR.
| Authors | Country | Study Type | No. Patients | Demographics† | Specimen (No.) | Index Test | Ref Stnd: rRT-PCR |
Study Findings (95% CI or p-value) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Cases* | Age (y)‡ | % Male | Type | Primers | Platform | ||||||||||||
| Primers | rSN | rSP | rPPV | rNPV | rAcc | rOA | Cohen's κ | |||||||||||
| Baek et al. 21 | Korea | Case Control|| | 154 | 14 | nr | nr | Nasal swab (154) | RT-LAMP | N | nr | ORF1ab, S | 100% | 98.7% | 0.826 | ||||
| Kitagawa et al. 22 | Japan | Cohort|| | 76 | 30 | nr | nr | NPS (76) | RT-LAMP | nr | LA-200 turbidimeter | N | 100% | 95.6% | 97.4% | ||||
| Lau et al. 23 | Malaysia | Case Control†† | 89‡‡ | 47‡‡ | nr | nr | NPS (89) | RT-LAMP | N | LA-320 turbidimeter | RdRp,E | 100% | 100% | |||||
| Lu et al. 24 | China | Case Control|| | 56 | 36 | nr | nr | Throat swab (56) | RT-LAMP | N | nr | ORF1ab, N | 92.9% | ||||||
| Yan et al. 25 | China | Cohort†† | 130 | 58 | nr | nr | Throat swab, BAL (nr) | RT-LAMP | ORF1ab, S | nr | ORF1ab, N | 100% (92.3–100) | 100% (93.7–100) | |||||
| Wang, Cai, & He et al. 26 | China | Cohort†† | 947 | 338 | 44 ± 17.1 | 60% | OPS (834), sputum (82), NPS (16), nasal swab (8), BAL (4), stool (2), blood (1) | RT-RAA | ORF1ab | RAA-F1620 fluorescent detector | ORF1ab &N, or ORF1ab | 97.6% (95.2–98.9) |
97.8% (96.2–98.8) |
96.2% (93.4–97.8) |
98.6% (97.3–99.3) |
0.952 p<0.05 |
||
| Xue et al. 27 | China | Cohort†† | 120‡‡ | 22‡‡ | nr | nr | NPS, sputum (nr) | RT-RAA | ORF1ab | RAA-1620 fluorescent detector | ORF1ab, S | 100% | 100% | 1.0 p < 0.001 |
||||
| Perchetti et al. 28 | USA | Case Control†† | 356 | 186 | nr | nr | NPS (356) | Triplex rRT-PCR | N1, N2 | n/a | N1, N2 | 98.4% | 100% | 99.2% | ||||
| Waggoner et al. 29 | USA | Cohort|| | 27 | 11 | nr | nr | NPS, OPS (nr) | Triplex rRT-PCR | N2, E | n/a | N2, E | 100% | ||||||
| Li et al. 30 | China | Cohort†† | 303‡‡ | 126‡‡ | nr | nr | throat swab (267), sputum (22), nose swab (8), BAL (3), blood (3) | AIGS | ORF1ab, N, S | LifeReady 1000 | ORF1ab, N | 97.62% (93.2–99.5) |
100% | |||||
| Suo et al. 19,a | China | Cohort⁎⁎ | 58 | 52 | nr | nr | Throat swab (58) | ddPCR | ORF1ab, N | QX200 System | ORF1ab, N | 94% (83–99) |
100% (48–100) |
100% (NA) |
63% (36–83) |
95% (84–99) |
||
| Bulterys et al. 31 | USA | Cohort†† | 80 | 30 | nr | nr | NPS (80) | Isothermal amplification | ORF1ab, N | Atila iAMP kit | E | 82.8% (65.0–92.9) |
0.86 (0.74–0.98) |
|||||
| Wang , Cai, & Zhang et al. 32 | China | Cohort†† | 181‡‡ | 25‡‡ | nr | nr | Throat swab (181) | OSN-qRT-PCR | ORF1ab, N | Life Tech. 480 | ORF1ab, N | 0.737 | ||||||
Cases according to reference standard.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD. Patient Population.
Hospitalized patients
¶Emergency Room/Immediate Care Center patients.
Outpatients, who were later hospitalized.
not reported.
Number of samples (when number of patients not reported).
Suo et al. data is also present in Table 2. Abbreviations- AIGS: Automatic integrated gene detection system, BAL: Bronchoalveolar lavage, CI: confidence interval, ddPCR: digital droplet polymerase chain reaction, E: envelope, iAMP: isothermal amplification, IQR: Interquartile range, κ: kappa statistic, n/a: not applicable, N: nucleocapsid, No.: number, NPS: nasopharyngeal swab, nr: not reported, OPS: oropharyngeal swab, ORF1ab: open reading frame 1ab, OSN-qRT-PCR: one-step single-tube nested quantitative real-time polymerase chain reaction, rAcc: reported accuracy, RdRp: RNA-dependent RNA polymerase, Ref Stnd: reference standard, rNPA: reported negative percent agreement, rNPV: reported negative predictive value, rOA: reported overall agreement, rPPA: reported positive percent agreement, rPPV: reported positive predictive value, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, rSN: reported sensitivity, rSP: reported specificity, RT-LAMP: reverse transcription loop-mediated isothermal amplification, RT-RAA: reverse-transcription recombinase-aided amplification, S: spike, y: years.
Table 5.
Studies estimating NAAT platform test performance characteristics compared to rRT-PCR as the reference standard.
| Country | Study Type | No. Patients | Demographics† | Specimen | Index Test | Ref Stnd: rRT-PCR | Study Findings (95% CI) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Cases* | Age (y)‡ | % M | Type | Primers | Platform | Primers | rSN | rSP | rPPV | rNPV | rPPA | rNPA | rOA | Cohen's κ | ||||
| Mitchell et al. 33 | USA | Case Control†† | 61‡‡ | 46‡‡ | nr | nr | NPS in VTM (61) | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
N1, N2 | 71.70% | 100% | 78.70% | |||||
| Rhoads et al. 35,b | US | Cases only|| | 96‡‡ | 96‡‡ | nr | nr | NPS (85), nasal swab (11) in NS or UTM | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
N1, N2 | 94% (87–98) |
|||||||
| Moore et al. 34,a | USA | Cohort||¶,⁎⁎ | 200 | 119 | 50 ± 17 | 46% | NPS in VTM (200) |
Automated Isothermal NAAT | RdRp | Abbott ID NOW |
80.3% (71.9–87.1) | 100% (95.4–100) |
|||||||
| Automated Multiplex RT-PCR | RdRp, N |
Abbott RealTime | N1, N2 | 100% (96.9–100) |
92.4% (84.2–97.2) | ||||||||||||||
| DegliAngeli et al. 36 | USA | Case Control†† | 60‡‡ | 30‡‡ | nr | nr | Nasal swab, NPS (nr) | Automated Multiplex RT-PCR | RdRp, N |
Abbott RealTime | N1, N2 | 93% | 100% | ||||||
| Hou et al. 37 | China | Cohort||,⁎⁎ | 285 | 153 | <65 y: 77.2% | 55.8% | OPS (285) |
Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | ORF1ab, N |
96.1% (91.3–98.4) |
96.2% (90.9–98.6) |
96.1%§ | 0.92 (0.88–0.97) |
||||
| Lieberman et al. 38 | USA | Cohort†† | 26‡‡ | 13‡‡ | nr | nr | NPS (26) |
Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | N1, N2 | 100%§ | |||||||
| Loeffelholz et al. 39,a | USA, UK, FR, IT | Cohort†† (enriched for positive cases) | 88‡‡ | 13‡‡ | nr | nr | NPS (339), NPS+OPS (97), Trach Asp (30), OPS (15) | Automated Multiplex RT-PCR | N2, E, RdRp | Cepheid Xpert Xpress | N1, N3 | 99.5% (97.5–99.9) | 95.8% (92.6–97.6) | ||||||
| 129‡‡ | 60‡‡ | S, E | 100% (94.0–100) |
100% (94.7–100) |
|||||||||||||||
| 99‡‡ | 74‡‡ | N1, N2 | 100% (94.2–100) |
92.0% (75.0–97.8) | |||||||||||||||
| 65‡‡ | 30‡‡ | RdRP | 100% (88.7–100) |
74.3% (57.9–85.8) | |||||||||||||||
| 79‡‡ | 35‡‡ | RdRp, E, N |
100% (67.6–100) |
100% (92.0–100) |
|||||||||||||||
| Bordi et al. 40 | Italy | Cohort + Controls†† | 278 + 20 | 99 | nr | nr | Nasal swab, NPS (nr) | Multiplex RT-PCR | ORF1ab, S | Diasorin Simplexa |
RdRp,E | 100% | 100% | 0.938 (0.89–0.98) |
|||||
| Rhoads et al35,b | US | Cases only|| | 96‡‡ | 96‡‡ | nr | nr | NPS (85), nasal swab (11) | Multiplex RT-PCR | ORF1ab, S | Diasorin Simplexa | N1, N2 | 96% (90–99) |
|||||||
| Poljak et al. 41 | Slovenia | Cohort†† | 501 | 63 | nr | nr | NPS (489), NPS+OPS (12) | Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | RdRP, E | 100% (92.8–100) |
99.5% (98.2–99.9) | 99.6% (98.4–99.9) | 0.98 (0.96–1.0) |
||||
| Pujadas et al. 42 | USA | Cohort†† | 963‡‡ | 640‡‡ | nr | nr | NPS (963) |
Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | N1, N2, N3 | 94.2% (92.2–95.9) | 99.6% (98.1–99.9) |
95.8% (94.4–97.0) | 0.904 (0.87–0.93) |
||||
| Rahman et al. 43 | Australia | Cohort†† | 52‡‡ | 5‡‡ | 31.5 (0–84) | 58% | NPS+OPS (30), NPS (16), N Asp (5), sputum (1) | Multiplex RT-PCR | ORF1 | Aus Diagnostics |
RdRp, E | 100% | 92.16% | 55.56% | 100% | ||||
| Hogan et al. JCV, 4–24 44 |
USA | Cohort†† | 180‡‡ | 77‡‡ | nr | nr | NPS (184) |
Automated Multiplex RT-PCR | ORF1ab | ||||||||||
| (2) | Hologic Panther | E | 98.7% (93.0–100) | 98.1% (93.1–99.8) | 98.3% (95.2–99.7) | 0.97 (0.93–1.0) |
|||||||||||||
| Chen et al. 45 | Hong Kong | Cohort|| | 214 | 91 | 51 (IQR 31–69) |
nr | NPS (214) |
Multiplex RT-PCR | ORF1ab, E, N | Luminex NxTAG CoV |
RdRp/Hel, E |
97.8% (92.2–99.7) |
100% (97.1–100) |
100% (95.9–100) |
98.4% (94.3–99.8) | 0.98 (0.95–1.0) |
|||
| Hogan et al. JCM 46 | USA | Case Control || | 100 | 50 | nr | nr | NPS (100) |
Automated PCR with LFA | ORF1ab (2) | Mesa BioTech Accula | E | 68.0% (53.3–80.5) | 100% (92.9–100) |
84.0% (75.3–90.6) | 0.74 (0.61–0.87) |
||||
| Visseaux et al. 47 | France | Case Control|| | 69 | 40 | nr | nr | NPS (66), BAL(1), Trach Asp (2) | Automated Multiplex RT-PCR | ORF1, E | QIAstat-Dx | RdRp, E | 100% | 93% | 97% | |||||
Cases according to reference standard.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD. Patient Population.
Hospitalized patients.
Emergency Room/Immediate Care Center patients.
Outpatients.
not reported.
Number of samples (when number of patients not reported).
Reported as concordance.
Loeffelholz et al. and Moore et al. also appear in Table 6.
Rhoads et al. appears twice in Table 5 for ease of comparison of studies of the same platform. Abbreviations- BAL: Bronchoalveolar lavage, CI: confidence interval, E: envelope, Hel: helicase, IQR: Interquartile range, κ: kappa statistic, LFA: lateral flow assay, M: male, n/a: not applicable, N: nucleocapsid, NAAT: nucleic acid amplification test, N Asp: nasopharyngeal aspirate, No.: number, NPS: nasopharyngeal swab, nr: not reported, NS: normal saline, OPS: oropharyngeal swab, ORF1ab: open reading frame 1ab, RdRp: RNA-dependent RNA polymerase, Ref Stnd: reference standard, rNPA: reported negative percent agreement, rNPV: reported negative predictive value, rOA: reported overall agreement, rPPA: reported positive percent agreement, rPPV: reported positive predictive value, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, rSN: reported sensitivity, rSP: reported specificity, S: spike, Trach Asp: Tracheal Aspirate, UTM: Universal transport medium, VTM: viral transport medium, y: years.
Table 6.
Studies assessing agreement between NAAT platforms.
| Authors | Country | Study Type | No. Patients | Demographics† | Specimen | Platform #1 | Platform #2 | Study Findings (95% CI) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Cases* | Age (y)‡ | % M | Type | Primers | Platform | Type | Primers | Platform | rPPA | rNPA | rOA | rPPV | rNPV | Cohen's κ | ||||
| Harrington et al. 48 | USA | Cohort¶ | 524 | 188 | nr | nr | Paired NPS in VTM (RealTime) & foam nasal swab (ID NOW) (524 pairs) | Automated Multiplex RT-PCR | RdRp, N | Abbott RealTime | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
75% (67.7–80.6) |
99% (97.6–99.8) |
||||
| Moore et al34,a | USA | Cohort||,¶,⁎⁎ | 200 | 125 | 50 ± 17 | 46% | NPS in VTM (200) | Automated Multiplex RT-PCR | RdRp, N | Abbott RealTime | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
75.2% (66.7–82.5) |
100% (95.4–100) |
||||
| Basu et al. 49 | USA | Cohort¶ | 101 | 32 | (28 - 90) | nr | NPS dry (101) | Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress |
Automated Isothermal NAAT | RdRp | Abbott ID NOW |
54.8% (37.8–70.8) |
98.6% (92.3–99.7) |
85.1% (76.9–90.8) |
94.4% (74.3–99) |
83.1% (73–89.7) |
|
| Cases only¶ | 15 | 15 | nr | nr | NPS in VTM (15) | 66.7% | |||||||||||||
| Hogan et al. JCV, 5–1 50 | USA | Case Control†† | 100‡‡ | 53‡‡ | nr | nr | NPSc (100) | Automated Multiplex RT-PCR | ORF1ab (2) | Hologic Panther | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
80.4% (66.9–90.2) | 95.9% (86.0–99.5) | ||||
| Zhen, Smith et al51,b | USA | Cohort†† | 108 | 58 | nr | nr | NPS in VTM (108) | Automated Multiplex RT-PCR | ORF1ab (2) | Hologic Panther | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
87.7% (76–95) |
100% (93–100) |
0.87 (0.78–0.96) |
|||
| Smithgall et al. 52 | USA | Case Control||¶ | 113 | 90 | 65 (0–101) |
60.2% | NPS in VTM or UTM (113) | Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | Automated Isothermal NAAT | RdRp | Abbott ID NOW |
73.9% (63.2–82.3) |
100% (83.4–100) |
||||
| Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | 98.9% (92.9–100) |
92% (72.4–98.6) |
||||||||||||
| Moran et al. 53 | USA | Cohort||,⁎⁎ | 103‡‡ | 42‡‡ | nr | nr | NPS (95), nasal swab (8) | Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | 99% | |||||
| Craney et al. 54 | USA | Cohort†† | 389 | 147 | nr | nr | NPS (389) | Automated Multiplex RT-PCR | ORF1ab (2) | Hologic Panther | Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | 96.4% | 0.922 | ||||
| Zhen, Smith et al51,b | USA | Cohort†† | 108 | 58 | nr | nr | NPS in VTM (108) | Automated Multiplex RT-PCR | ORF1ab (2) | Hologic Panther | Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | 98.3% (91–100) |
100% (93–100) |
0.98 (0.95–1.0) |
|||
| Automated Multiplex RT-PCR | ORF1ab (2) | Hologic Panther | Automated RT-PCR w/sensor | N | GenMark ePlex |
91.4% (81–97) |
100% (93–100) |
0.91 (0.83–0.99) |
|||||||||||
| Loeffelholz et al39,a | USA, UK, FR, IT | Cohort†† (enriched for positive cases) | 18‡‡ | 8‡‡ | nr | nr | NPS, OPS, NPS+OPS, Trach Asp (nr) | Automated Multiplex RT-PCR | RdRp, N | Abbott RealTime | Automated Multiplex RT-PCR | N2, E | Cepheid Xpert Xpress | 100% (67.6–100) |
100% (77.2–100) |
||||
| Norz et al. 55 | Germany | Case Control†† | 165‡‡ | 36‡‡ | nr | nr | NPS, OPS (nr) | Automated Multiplex RT-PCR | ORF1, E | Roche cobas 6800 | Automated RT-PCR |
E | NeuMoDx 96 | 100% | 99.2% | ||||
Case estimated as a positive result of any evaluated platform.
Of cohort or cases.
Format: median (range), median(IQR), or mean±SD. Patient Population.
Hospitalized patients.
Emergency Room/Immediate Care Center patients.
Outpatient.
not reported.
Number of samples (when number of patients not reported).
Loeffelholz et al. and Moore et al. also appear in Table 5.
Zhen, Smith et al. appears twice in Table 6 for ease of comparison of studies of the same platform.
Dry or VTM status not reported. Rhoads et al. appears twice in Table 5 for ease of comparison of studies of the same platform Abbreviations- CI: confidence interval, E: envelope, IQR: Interquartile range, κ: kappa statistic, M: male, N: nucleocapsid, NAAT: nucleic acid amplification test, No.: number, NPS: nasopharyngeal swab, nr: not reported, OPS: oropharyngeal swab, ORF1ab: open reading frame 1ab, RdRp: RNA-dependent RNA polymerase, rNPA: reported negative percent agreement, rNPV: reported negative predictive value, rOA: reported overall agreement, rPPA: reported positive percent agreement, rPPV: reported positive predictive value, rRT-PCR: real-time Reverse Transcription Polymerase Chain Reaction, S: spike, Trach Asp: Tracheal Aspirate, UTM: universal transport medium, VTM: viral transport medium, y: years.
Data synthesis and analysis
We presented the extracted data in tabular form mirrored by a descriptive synthesis 5 in two broad categories: diagnostic accuracy studies for rRT-PCR (Table 1, Table 2, Table 3), and diagnostic accuracy or comparative agreement studies of two NAATs (Table 4, Table 5, Table 6). Tables are thematically divided based on the reference standard strategy, or approach to obtaining comparative agreement measures. Diagnostic accuracy studies for rRT-PCR were arranged alphabetically in tables by first author last name (Table 1, Table 2, Table 3). Diagnostic accuracy and comparative agreement studies for two NAATs were arranged by decreasing order of studies per methodology, then alphabetically by methodology or platform (Table 4, Table 5, Table 6) for easy comparison. Due to significant diversity in methods and reporting of results, we reported grouped summary data for study characteristics, patient characteristics, and outcomes.
We used the framework of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2)6 to evaluate our selected articles (Supplementary Table 3). We collected data, or noted their absence, for a narrative description of risk of bias and concerns of applicability based on the QUADAS domains. For assessment of bias in patient selection, we evaluated author conflicts of interest, study design type, inclusion/exclusion criteria, method of patient enrollment, and reporting of patient demographics and characteristics (i.e. symptomatic status). For assessment of bias in reference standard and index test, we evaluated the accuracy of the reference standard, the description of duration of symptoms at time of testing, whether the threshold to determine a positive test was prespecified, and researcher blinding to reference standard and index test results. For assessment of bias in flow and timing, we evaluated whether the reference standard was the same for all patients, the sequence and timing of the performance of the reference standard and index test, whether test performance characteristics were calculated based on sample numbers or patient numbers, and whether indeterminate or invalid results were included in test performance calculations.
Results
Study selection
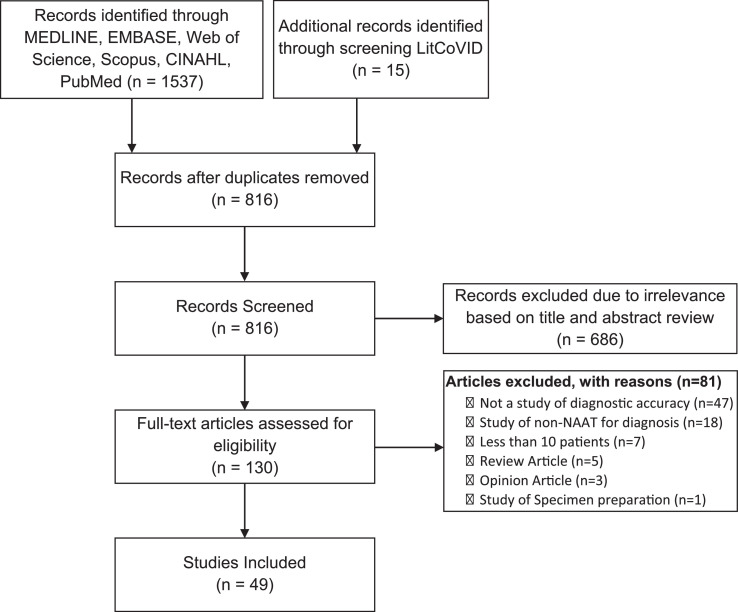
Our search yielded 1537 articles, with 816 unique articles after deduplication. After screening title and abstract, 130 articles underwent full text evaluation. Ultimately, 49 articles were included in our review (Fig. 1 ).
Fig. 1.
PRISMA Flow diagram of studies included in the review.
The performance of rRT-PCR compared to case definitions or clinical diagnoses
Three studies, with 19 to 1014 patients, report a “positive rate” as the number of positive rRT-PCR out of the number of suspected cases of COVID-19, with a range of 38.42% to 59% (Table 1).7, 8, 9 The studies do not report these values as “sensitivity” directly, however these values were interpreted as reflective of the accuracy of rRT-PCR. Ai et al. compared the positive rate of rRT-PCR (59%) with the positive rate of Chest CT in order to draw conclusions about the accuracy of Chest CT for the diagnosis of COVID-19,7 and Liu et al. and Xie et al. expressed concern that their low calculated positive rates (38.42% and 47.4%, respectively) were indicative of a failure of rRT-PCR to diagnose COVID-19.8 , 9
In terms of quality assessment, the studies lack specific details as to how patients were classified as having suspected COVID-19 infection. The accuracy of clinical diagnosis based on case definitions is unclear but is likely not ideal for diagnosis. Additionally, duration of symptoms at the time of clinical diagnosis or rRT-PCR testing was not provided (Supplementary Table 3).
The performance of rRT-PCR compared to end result after multiple repetitions of rRT-PCR
Eight studies, with a range of 36 to 22,061 patients per study, attempted to determine the accuracy of rRT-PCR by comparing the initial rRT-PCR result to the result after multiple repeated samples from the patient submitted for rRT-PCR testing, which was called the reference standard (Table 2).10, 11, 12, 13, 14, 15, 16, 17 Three studies reported this value as a “positive rate,” ranging from 51.25% to 88%,10 , 14 , 17 and five reported sensitivity, with a range of 57.9% to 94.6%.11, 12, 13 , 15 , 16 Of these studies, only He et al. included an rSp of 100%, calculated from patients who remained negative for SARS-CoV-2 after repeated sample testing (Supplementary Table 1).13
Green et al. included patients in their study regardless of whether they were tested once or multiple times, using data from these subsets of patients to make assumptions for estimating clinical test characteristics. In addition, this study also conducted multiple different NAATs and rRT-PCRs on patients, whereas other studies employing this strategy used only one type of NAAT. Additionally, the authors do not clarify whether patients who had repeat SARS-CoV-2 test were consistently tested with the same NAAT/rRT-PCR test or a different one. They also calculated test performance characteristics differently from other studies: two estimates of sensitivity were calculated, one in which the rate of false negatives for single-tested patients was 0%, and one in which the “false negative” rate was the same as in repeat-tested patients in their study of approximately 16.8%.12 However, the details of how they calculated test characteristics were not presented. To clarify the two assumptions made in the calculations, we reconstructed the calculation in Supplementary Figure 1 which demonstrated the range of rSN with an estimate of the lower bound of 57.9% (55.2%−60.5%) and an estimate of the upper bound of 94.6% (94.2%−95.0%).
In terms of quality assessment, most of the studies were performed with non-cohort design, and six consisted of only patients who were determined to have COVID-19 by rRT-PCR, i.e. cases only (Table 2).10 , 11 , 14, 15, 16 Five of the studies had inclusion criteria which caused pertinent patients to be excluded by necessitating patients to have had a well-performed CT Chest or X-Ray (Supplementary Table 3). This excluded several patients who would otherwise have been pertinent to the study of test diagnostic accuracy.10 , 11 , 13 , 16 , 17 The studies involved repeating rRT-PCR several times for a reference standard, but each patient received a different number of repeat tests over a different time period, resulting in each patient receiving a different reference standard. One study tracked negative-to-positive conversion over 1 to 49 days,12 and another tracked over 1 to 14 days,13 leading to concern that potentially a patient could have been infected in the time between the initial test and the final test and confounding results. One study counted invalid results as negative results and indeterminate results as positive results when calculating test performance characteristics,12 otherwise the rationale and ways invalid and indeterminate results were handled were not reported in these studies.
The performance of rRT-PCR compared to various composite reference standards
Three studies determined the accuracy or agreement of rRT-PCR or automated rRT-PCR platforms/instruments compared to a reference standard based on the results of several tests as a “composite reference standard” (Table 3).18, 19, 20 There were between 58 and 184 patients per study. Suo et al. considered a positive result of either repeated measurements of rRT-PCR or serology to indicate a positive test according to the reference standard; reported sensitivity of initial rRT-PCR result was 40%, rSp 100%, rPPV 100%, and rNPV 16%.19 Zhen et al. compared rRT-PCR performed according to the US CDC protocol to a composite reference standard in which the consensus result of 3 or more out of 4 molecular assays was considered the correct result. The rRT-PCR had an rPPA of 100%, an rNPA of 98%, and Cohen's kappa coefficient of 0.98.20 Cradic et al. did not study rRT-PCR but studied three automated molecular assays and used a composite reference standard of the consensus result of two or more of the three assays. While Abbott ID NOW had a rPPA of 91%, the Roche cobas 6800 and Diasorin Simplexa assays had a rPPA of 100%.18
These studies either did not report how samples were selected for evaluation (Supplementary Table 3),19 , 20 or reported that only samples which had sufficient residual volume and had been properly stored were selected.18 Cradic et al. and Zhen & Mangi et al. had initially tested samples with one platform, and some or all samples were frozen, and subsequently thawed and tested with other platforms, leading to confounding factors in the reference standard and test performance calculations involving the various platform results.18 , 20 Suo et al. used repeat rRT-PCR testing as part of the reference standard, with repeat tests performed 2–10 days after the initial test, after the patient had been discharged from the hospital, leading to potential exposure for initial infection or reinfection.19
The performance other nucleic acid amplification test methods compared to standard rRT-PCR
Fourteen studies compared other nucleic acid amplification test methods to detect SARS-CoV-2 to rRT-PCR (Table 4), with between 27 and 356 patients per study. Five of the studies evaluated reverse transcription loop mediated isothermal amplification (RT-LAMP),21, 22, 23, 24, 25 four reported sensitivity of 100% and specificity of 95.6 to 100%,21, 22, 23 , 25 and one study reported accuracy of 92.9%.24 Two studies, Wang, Cai, & He et al. and Xue et al., evaluated reverse-transcription recombinase-aided amplification (RT-RAA) with Cohen's kappa of 0.952 and 1.0.26 , 27 Two studies, Perchetti et al. and Waggoner et al., evaluated triplex rRT-PCR, reporting overall agreement as 99.2 and 100%.28 , 29 Li et al. evaluated an automatic integrated gene detection system (AIGS) with rSN of 97.2% and rSp 98.5%.30 Suo et al. evaluated digital droplet polymerase chain reaction (ddPCR), with rSN 94%, rSp 100%, rPPV 100%, rNPV of 63%, and rAcc 95%.19 Bulterys et al. study evaluated an isothermal amplification method with rSN 82.8% and Cohen's kappa 0.86.31 Wang, Cai, and Zhang et al. evaluated one-step single-tube nested quantitative polymerase chain reaction (OSN-qRT-PCR) with Cohen's kappa of 0.737.32
Regarding evaluation of quality (Supplementary Table 3), the majority of studies did not report how patient samples were selected for evaluation.21 , 22 , 24 , 25 , 27, 28, 29, 30 , 32 In the study conducted by Bulterys et al., sample selection was a convenience selection of samples with residual volume that had been stored correctly.31 Most studies did not report symptomatic status of the patient 21, 22, 23, 24, 25, 26, 27, 28 , 30, 31, 32 or patient demographics.21, 22, 23, 24, 25 , 27, 28, 29, 30, 31, 32 Problematically, many of the studies did not report when the reference standard was conducted on the patient samples compared to the index test, or whether actions that could potentially alter test results (such as freeze/thaw cycles) occurred between reference standard or index test.21, 22, 23, 24 , 27 , 28 , 31 Four studies calculated test performance characteristics based on number of samples rather than number of patients.23 , 27 , 30 , 32 The management of indeterminate and invalid test results went largely unreported.21, 22, 23, 24, 25 , 27 , 30
The performance of NAAT platforms compared to rRT-PCR as the reference standard
Fifteen studies compared automated NAAT platforms to various rRT-PCR assays to determine test performance characteristics (Table 5), with between 26 and 963 patients or samples per study. Three studies evaluated Abbott ID NOW, an isothermal NAAT platform, with rPPA or rSN of 71.7% to 94%, and rNPA or rSP of 100%.33, 34, 35 Two studies evaluated Abbott RealTime with rSN or rPPA of 93% to 100%, and rSP or rNPA of 92.4% to 100%.34 , 36 Three studies evaluated Cepheid Xpert Xpress, with rPPA 96.1% to 100%, rNPA 74.3% to 100%, rOA 96.1% to 100%, and Cohen's Kappa of 0.92.37, 38, 39 Two studies evaluated Diasorin Simplexa with rSN or rPPA of 96% to 100%, and rSN of 100%.35 , 40 Two studies evaluated Roche cobas 6800 with rPPA 94.2% to 100%, rNPA 99.5% to 99.6%, and Cohen's Kappa of 0.904 to 0.98.41 , 42 Other studies evaluated AusDiagnostics (rSN 100%, rSP 92.16%),43 Hologic Panther Fusion (rPPA 98.7%, rNPA 98.1%),44 Luminex NxTAG (rSN 97.8%, rSP 100%),45 Mesa BioTech Accula (rPPA 68.0%, rNPA 100%),46 or QIAstat-Dx (rSN 100%, rSP 93%) 47 compared to rRT-PCR.
With regards to quality evaluation (Supplementary Table 3), most studies did not report method of sample collection/patient recruitment,33 , 35 , 37 , 41, 42, 43, 44, 45, 46, 47 and four studies conducted a convenience selection of samples, including enrichment for positive samples.34, 35, 36 , 39 Eight studies conducted test performance calculations on sample numbers instead of patient numbers.33 , 35 , 36 , 38 , 39 , 42, 43, 44 Four studies conducted calculation of test performance characteristics with indeterminate or inconclusive results as “positive,” 35 , 38 , 39 , 42 and the management of indeterminate/inconclusive as well as invalid results went unreported in an additional three studies.33 , 37 , 47 No study reported the blinding of researchers to the reference standard or index test results.
The agreement of NAAT platforms compared to other NAAT platforms
Ten studies, containing between 15 and 524 patients per study, evaluated the agreement between two different types of NAAT platforms (Table 6), typically under the circumstances where one platform was the standard of care at the institution, and another was introduced. The Abbott ID NOW platform, using isothermal amplification, was the most frequently studied test, with an rPPA of 75–75.2% compared to Abbott Real Time,34 , 48 54.8% compared to Cepheid Xpert Xpress,49 80.4–87.7% compared to Hologic Panther Fusion,50 , 51 and 73.9% compared to Roche cobas 6800.52 Two studies evaluated Cepheid Xpert Xpress compared to Roche cobas 6800, with rPPA 98.9% and rNPA 92% in one,52 and overall agreement of 99% in another.53 Several platforms were compared to Hologic Panther Fusion, including Roche cobas 6800 with rOA 96.4%,54 Cepheid Xpert Xpress with rPPA 98.3% and rNPA 100%, and GenMark ePlex with rPPA 91.4% and rNPA 100%.51
In the studies, some platforms were identified as the “comparator” or “reference” platforms, including Cepheid Xpert Xpress,49 Abbott RealTime,34 , 48 Hologic Panther Fusion,50 , 51 and Roche cobas 6800,52 , 55 and these were listed as “Platform #1″ in Table 6. Three studies did not identify any studied platform as the “comparator” or “reference standard,” and instead only reported general, non-directional measures of agreement such as overall agreement, Cohen's Kappa, or alternatively, the calculations of PPA and NPA were identical no matter their method of calculation (Supplementary Table 1).39 , 53 , 54
Regarding quality evaluation (Supplementary Table 3), the samples used for calculating test performance characteristics were reported to be selected for enrichment of positive samples,34 , 39 for diversity of viral load,52 , 54 otherwise curated,50 or the method of selecting samples was unreported.49 , 51 , 53 , 55 Symptomatic status of the patients was largely unreported.39 , 49, 50, 51, 52, 53, 54, 55 Five studies included samples where one test was conducted, then interim freezing, cooling, or other storage, before performance of the second test.39 , 50, 51, 52 , 54 Two studies did not report the sequence of testing of the two platforms or interim handling or storage of the samples.49 , 53 The status of researcher blinding to either platform result was not reported in any study.
Discussion
In our scoping review of 49 articles concerning test performance characteristics of rRT-PCR and other NAAT used for the diagnosis of COVID-19, we were able to observe several overarching themes. Clinical diagnosis by the case definition for COVID-19 used in the early period of the pandemic does not correlate well with positive rates of COVID-19 rRT-PCR (Table 1). The result of the initial rRT-PCR performed on a patient, if negative, may not be reflective of the result after multiple repeated rRT-PCRs for that patient (Table 2). Several alternative NAAT methods, many of which are easier or faster to perform, may be comparable to standard rRT-PCR (Table 4). Proprietary multiplex, automated, and/or point-of-care methods are comparable to in accuracy to rRT-PCR (Table 5) and to each other (Table 6), although the Abbott ID NOW SARS-CoV-2 test appears to have lower comparative agreement to other platforms.34 , 48, 49, 50, 51, 52
These findings should be viewed cautiously as the SARS-CoV-2 tests in these studies have not undergone rigorous evaluation necessary for FDA approval due to the emergency state generated by the COVID-19 pandemic. In addition, during our scoping review, we found substantial heterogeneity among available studies in terms of test types, reference standards, metrics, and details of study design and methodology.
We categorized the included studies by four different reference standard strategies: clinical diagnosis/case definitions (Table 1), repeated index testing (Table 2), composite reference standard (Table 3), and rRT-PCR (Table 4 and 5). Additionally, we identified a fifth category, where instead of using a reference standard, comparative agreement between two NAAT platforms was calculated (Table 5 and 6).
The main limitation of the first group of studies (Table 1) was the use of a “case definition” as the reference standard to report a “positive rate” of rRT-PCR. During novel disease outbreaks, standard case definitions are often developed to assist clinicians in case identification before a diagnostic test is available. Unfortunately, the studies included in this group were unable to use a clear case definition; instead they refer to a population of “suspected cases,” for which the definition is not reported.7, 8, 9 Because this group enrolled patients prior to February 15, 2020 in China, during the time in which the Chinese National Guideline for Diagnosis and Treatment of COVID-19 (NGDTC) published five different versions of the COVID-19 case definition, the case definitions in use at the time of these studies varied.56 A recent study estimated that if a single guideline (specifically, version 5 of the NGDTC) had been used to identify cases from the beginning of the outbreak to February 20, 2020, there would have been more than three times as many identified cases in Hubei province.56 This is relevant to our review because the two largest studies that evaluated the rRT-PCR positive rate of patients with a clinical diagnosis of COVID-19 took place in Wuhan, Hubei province, and included patients evaluated before February 14, 2020 7 , 8 (Supplementary Table 2). This increased case estimate due to diagnosis of COVID-19 based on case definition complicates the legitimacy and reported accuracy of the “positive rate” of rRT-PCR referred to in these studies.
The second group assessed rRT-PCR test performance characteristics via repeated index rRT-PCR testing (Table 2). Most studies in this group reported “sensitivity” by dividing the number of participants with positive baseline rRT-PCRs by the total number of participants who eventually had a positive rRT-PCR after repeated measurements. While such an approach may have some advantages over the use of a case definition alone as a reference standard, this strategy is, nonetheless, an imperfect solution with its own set of inherent limitations. SARS-CoV-2 infection is transient and the associated viral loads are time-varying because of the natural pathophysiology of the infection. Therefore, the time interval between each repeated test becomes crucially important, and even relatively small time differences (and/or lack of uniformly used intervals) could complicate the interpretation of re-test results and their quality as reference standards. Furthermore, repeated use of the same test as a reference standard for itself does not eliminate the inaccuracies or limitations of the test. Such comparisons ultimately reflect the reliability of the test (assuming a short, uniform time interval between tests), rather than providing a true view of test accuracy.
The third group of three studies calculated test performance characteristics of rRT-PCR according to a composite reference standard (Table 3). Using arbitrary rules to combine multiple different and imperfect tests inevitably creates a reference standard with some degree of bias.57 Furthermore, all three studies in this group included the test under evaluation as part of the composite reference standard, which leads to additional bias, described below.58 Use of a biased composite standard is likely to lead to reduced sensitivity, among other errors affecting true test performance characteristics.59
The fourth group of studies evaluated SARS-CoV-2 diagnostic tests that are under development as well as proprietary testing platforms (most of which are based on standard rRT-PCR methods). These studies used traditional rRT-PCR as a reference standard; results are summarized in Tables 4 and 5, respectively. Importantly, while these studies were not designed to estimate the accuracy of rRT-PCR, their results indicate that the index tests did not identify significantly more positive samples than rRT-PCR.
Finally, the last group of studies compared SARS-CoV-2 NAAT platforms (Table 6). These comparative accuracy studies examined the agreement between two non-reference standard tests. Although most of the testing platforms evaluated in these studies were based on standard rRT-PCR, the agreement between two non-reference standard tests is not equivalent to test accuracy, as mentioned previously.
This scoping review is limited by the lack of reporting of several key study features in the majority of the articles evaluated, which is an important indicator of quality and potential bias. Based on the QUADAS-2 criteria, most of the included studies had concern for bias (Supplementary Table 3). The most prominent concerns were unclear inclusion/exclusion criteria, unclear method of enrollment/selection of patients and samples, and unclear handling of indeterminate/inconclusive and invalid results. Additionally, many of the studies were conducted in a so-called “two gate” (case-control) design, in which cases and controls were known and selected ahead of time, rather than performing the test on a group of patients or samples with suspected COVID-19. These factors likely incorporate bias that significantly confounds the results of the studies, thus, the accuracy of the tests in other settings with different prevalences (such as asymptomatic screening, other age groups) may not be truly generalizable. Furthermore, few studies were able to evaluate both the index and reference tests simultaneously or within a short period of time, which is key to avoiding biases caused by changes in the patient's true disease status; this bias can also affect the diagnostic accuracy of the index test.
The best approach to determining diagnostic test performance characteristics in the absence of a “gold” standard is an open question in diagnostic accuracy methodology. While many methods have been described, there are only a few well-defined statistical approaches that use a reference standard in lieu of a gold standard, reviewed elsewhere.60 Latent class analysis is one commonly used approach in situations in which neither the true error rates of the reference standard nor the true prevalence of the disease are known. This approach uses the results of a set of imperfect tests to estimate parameters related to sensitivity, specificity, and prevalence often using maximum likelihood methods. However, this is not the only method available and every method has its own strengths and limitations.57 Therefore, careful interpretation by studies that attempt to estimate test characteristics is warranted to account for and clarify the inherent limitations of assessing accuracy-related metrics when a gold standard is unavailable.
Evaluation of the performance characteristics of SARS-CoV-2 diagnostic tests is vital to control of the ongoing COVID-19 pandemic. While more than 200 SARS-CoV-2 molecular diagnostic tests have received FDA EUAs, we have described in this scoping review that the performance of few of these tests has been assessed appropriately. The lack of robust test performance that we noted in many studies published to date is undoubtably due in part to the critical need for tests, which resulted in accelerated test development. However, our scoping review also uncovered imperfect methods for estimating diagnostic test performance in the absence of a gold standard and demonstrate that the accuracy of these tests should be interpreted with caution. Future studies would benefit from employing statistical methods such as latent class analysis and other methods referenced above to accurately analyze their data. Indeed, instituting national requirements for test performance analysis and reporting, perhaps based on the existing FDA guidelines on diagnostic tests,61 would advance the goal of standardizing the evaluation SARS-CoV-2 diagnostic test performance. Such an initiative would lead to statistically robust conclusions regarding the accuracy of the index test, which will in turn support hospitals and clinicians as they determine the optimal test to use for COVID-19 diagnosis.
Declaration of Competing Interest
None
Acknowledgments
Acknowledgments
None.
Funding
This work was supported by the National Institutes of Health [R01CA232890 to E.Y.C.].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.08.043.
Appendix. Supplementary materials
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei W.E., Li Z., Chiew C.J., Yong S.E., Toh M.P., Lee V.J. Presymptomatic Transmission of SARS-CoV-2 - Singapore. MMWR Morb Mortal Wkly Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. January 23-March 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. W64. [DOI] [PubMed] [Google Scholar]
- 5.Campbell M., McKenzie J.E., Sowden A., Katikireddi S.V., Brennan S.E., Ellis S., et al. Synthesis without meta-analysis (SWiM) in systematic reviews: reporting guideline. BMJ. 2020;368:l6890. doi: 10.1136/bmj.l6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 7.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H., et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green D.A., Zucker J., Westblade L.F., Whittier S., Rennert H., Velu P., et al. Clinical performance of SARS-CoV-2 molecular testing. J Clin Microbiol. 2020:e00995–20. doi: 10.1128/JCM.00995-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He J.L., Luo L., Luo Z.D., Lyu J.X., Ng M.Y., Shen X.P., et al. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir Med. 2020;168 doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee T.H., Lin R.J., Lin R.T.P., Barkham T., Rao P., Leo Y.S., et al. Testing for SARS-CoV-2: can We Stop at Two. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., et al. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT. Eur J Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong H.Y.F., Lam H.Y.S., Fong A.H., Leung S.T., Chin T.W., Lo C.S.Y., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2019 doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J., Liu J., Li S., Peng Z., Xiao Z., Wang X., et al. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cradic K., Lockhart M., Ozbolt P., Fatica L., Landon L., Lieber M., et al. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;28:28. doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020;27:27. doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek Y.H., Um J., Antigua K.J.C., Park J.H., Kim Y., Oh S., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa Y., Orihara Y., Kawamura R., Imai K., Sakai J., Tarumoto N., et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau Y.L., Ismail I., Mustapa N.I., Lai M.Y., Tuan Soh T.S., Hassan A., et al. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2. PeerJ. 2020;8:e9278. doi: 10.7717/peerj.9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int. 2020;21:18. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Cai K., He X., Shen X., Wang J., Liu J., et al. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin Microbiol Infect. 2020;15:15. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue G., Li S., Zhang W., Du B., Cui J., Yan C., et al. A reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2) Anal Chem. 2020;22:22. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- 28.Perchetti G.A., Nalla A.K., Huang M.L., Jerome K.R., Greninger A.L. Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A., et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Li J., Zhang Y., Dai L., Li L., Liu J., et al. Development of an automatic integrated gene detection system for novel Severe acute respiratory syndrome-related coronavirus (SARS-CoV 2) Emerg. 2020:1–24. doi: 10.1080/22221751.2020.1782774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulterys P.L., Garamani N., Stevens B., Sahoo M.K., Huang C., Hogan C.A., et al. Comparison of a laboratory-developed test targeting the envelope gene with three nucleic acid amplification tests for detection of SARS-CoV-2. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Cai K., Zhang R., He X., Shen X., Liu J., et al. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal Chem. 2020;15:15. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell S.L., George K.S. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M.K. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV RT-PCR assay for the detection of SARS-CoV-2. J Clin Microbiol. 2020;27:27. doi: 10.1128/JCM.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020:e00760–20. doi: 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degli-Angeli E., Dragavon J., Huang M.L., Lucic D., Cloherty G., Jerome K.R., et al. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou H., Chen J., Wang Y., Lu Y., Zhu Y., Zhang B., et al. Multi-center evaluation of the Cepheid Xpert Xpress SARS-CoV-2 assay for the detection of SARS-CoV-2 in oropharyngeal swab specimens. J Clin Microbiol. 2020:e01288–20. doi: 10.1128/JCM.01288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020;29:29. doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A., et al. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J Clin Microbiol. 2020;04:04. doi: 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordi L., Piralla A., Lalle E., Giardina F., Colavita F., Tallarita M., et al. Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa TM COVID-19 direct assay. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poljak M., Korva M., Knap Gasper N., Fujs Komlos K., Sagadin M., Ursic T., et al. Clinical Evaluation of the cobas SARS-CoV-2 Test and a Diagnostic Platform Switch during 48 Hours in the Midst of the COVID-19 Pandemic. J Clin Microbiol. 2020;58:26. doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pujadas E., Ibeh N., Hernandez M.M., Waluszko A., Sidorenko T., Flores V., et al. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020;08:08. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman H., Carter I., Basile K., Donovan L., Kumar S., Tran T., et al. Interpret with caution: an evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogan C.A., Sahoo M.K., Huang C., Garamani N., Stevens B., Zehnder J., et al. Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J.H., Yip C.C., Chan J.F., Poon R.W., To K.K., Chan K.H., et al. Clinical performance of the Luminex NxTAG CoV Extended Panel for SARS-CoV-2 detection in nasopharyngeal specimens of COVID-19 patients in Hong Kong. J Clin Microbiol. 2020;01:01. doi: 10.1128/JCM.00936-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hogan C.A., Garamani N., Lee A.S., Tung J.K., Sahoo M.K., Huang C., et al. Comparison of the Accula SARS-CoV-2 Test with a Laboratory-Developed Assay for Detection of SARS-CoV-2 RNA in Clinical Nasopharyngeal Specimens. J Clin Microbiol. 2020:e01072–20. doi: 10.1128/JCM.01072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., et al. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020;27:27. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P., et al. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020:e00798–20. doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basu A., Zinger T., Inglima K., Woo K.M., Atie O., Yurasits L., et al. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. J Clin Microbiol. 2020:e01136–20. doi: 10.1128/JCM.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogan C.A., Sahoo M.K., Huang C., Garamani N., Stevens B., Zehnder J., et al. Five-minute point-of-care testing for SARS-CoV-2: not there yet. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., et al. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 Assays. J Clin Microbiol. 2020:e00772–20. doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Craney A.R., Velu P., Satlin M.J., Fauntleroy K.A., Callan K., Robertson A., et al. Comparison of two high-throughput reverse transcription-polymerase chain reaction systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol. 2020;07:07. doi: 10.1128/JCM.00890-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norz D., Fischer N., Schultze A., Kluge S., Mayer-Runge U., Aepfelbacher M., et al. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsang T.K., Wu P., Lin Y., Lau E.H.Y., Leung G.M., Cowling B.J. Effect of changing case definitions for COVID-19 on the epidemic curve and transmission parameters in mainland China: a modelling study. Lancet Public Health. 2020;5:e289–ee96. doi: 10.1016/S2468-2667(20)30089-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rutjes A.W., Reitsma J.B., Coomarasamy A., Khan K.S., Bossuyt P.M. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol Assess. 2007;11 doi: 10.3310/hta11500. iii, ix-51. [DOI] [PubMed] [Google Scholar]
- 58.Alonzo T.A., Pepe M.S. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat Med. 1999;18:2987–3003. doi: 10.1002/(sici)1097-0258(19991130)18:22<2987::aid-sim205>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 59.Naaktgeboren C.A., Bertens L.C., van Smeden M., de Groot J.A., Moons K.G., Reitsma J.B. Value of composite reference standards in diagnostic research. BMJ. 2013;347:f5605. doi: 10.1136/bmj.f5605. [DOI] [PubMed] [Google Scholar]
- 60.Umemneku Chikere C.M., Wilson K., Graziadio S., Vale L., Allen A.J. Diagnostic test evaluation methodology: a systematic review of methods employed to evaluate diagnostic tests in the absence of gold standard - An update. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0223832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.U.S. Food and Drug Administration CfDaRH. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. https://www.fda.gov/media/71147/download: [Accessed May 15, 2020]; 2007.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.