Abstract
We evaluated the effects of a chemical additive on the microbial communities, fermentation profile, and aerobic stability of whole-plant corn silage with or without air stress during storage. Whole-plant corn was either untreated or treated with a chemical additive containing sodium benzoate, potassium sorbate, and sodium nitrite at 2 or 3 liters/t of fresh forage weight. Ten individually treated and replicated silos (7.5 liters) were made for each treatment. Half of the silos remained sealed throughout a 63-d storage period, and the other half was subjected to air stress for 2 h/wk. The composition of the bacterial and fungal communities of fresh forage and silages untreated or treated with 2 liters/t of fresh forage weight was analyzed by Illumina Miseq sequencing. Treated silage had greater (P < 0.05) aerobic stability than untreated, even when subjected to air stress during storage, but the numbers of yeasts culturable on selective agar were not affected. However, the additive reduced the relative abundance (RA) of the lactating-assimilating yeast Candida tropicalis (P < 0.01). In air-stressed silages, untreated silage had a greater (P < 0.05) RA of Pichia kudriavzevii (also a lactate assimilator) than treated silage, whereas treated silage was dominated by Candida humilis, which is usually unable to assimilate lactate or assimilates it slowly. The additive improved the aerobic stability by specifically preventing the dominance of yeast species that can consume lactate and initiate aerobic spoilage. To the best of our knowledge, this is the first work that identifies the specific action of this additive on shifting the microbial communities in corn silage.
Keywords: aerobic stability, corn silage, potassium sorbate
Introduction
Corn silage is the predominant forage fed to lactating dairy cows, but many factors can affect its quality. For example, harvesting losses, respiration and fermentation losses, effluent production, and exposure to air account for most of the dry matter (DM) losses during silage production (Borreani et al., 2018). Preventing air from entering the silo is essential to producing high-quality silage, but exposure to air occurs to varying degrees during storage and is unavoidable at feed-out. Under aerobic conditions, lactate-assimilating yeasts are primarily responsible for initiating aerobic spoilage in silages because they oxidize sugars and lactic acid, producing CO2, H2O, and heat (Pahlow et al., 2003). (In some instances, acetic acid bacteria have been reported to initiate the aerobic deterioration of corn silage; Spoelstra et al., 1988; Dolci et al., 2011). Metabolism of lactic acid subsequently results in an increased silage pH, which allows for the growth of aerobic bacteria and molds that cause further spoilage (Oude Elferink et al., 2000).
Chemical additives can be used in silage production to attenuate the risks brought by specific crop characteristics (e.g., legumes have a high buffering capacity), challenging environmental conditions, and (or) poor management practices, by inhibiting the growth of microorganisms that cause undesirable fermentations or spoilage, and consequently, reducing the DM losses and maintaining the nutritional quality of silages (Muck et al., 2018). Compounds with antifungal properties have been evaluated for their ability to inhibit yeasts that initiate aerobic spoilage. Specifically, sodium benzoate, potassium sorbate, and sodium nitrite have been used in combination to inhibit various undesirable microorganisms (Stanojevic et al., 2009). Knicky and Spörndly (2011, 2015) reported that together, these compounds improved the fermentation and aerobic stability of silages. However, it is unclear how this additive specifically accomplishes this because improvements in aerobic stability have been observed sometimes even when the additive did not reduce total numbers of yeasts culturable on selective agar (Da Silva et al., 2015; Kung et al., 2018a). Additionally, there is a need for more studies evaluating the effectiveness of this additive on improving the aerobic stability of corn silage under challenging management issues that can occur in the field, such as the infiltration of air into the silo during storage (Borreani et al., 2018).
McAllister et al. (2018) reported that next-generation sequencing (NGS) technology is a useful tool to help the understanding of silage fermentation and aerobic deterioration. For example, Romero et al. (2018) reported that treatment with a bacterial inoculant resulted in a distinct microbial community dominated by Lactobacillaceae and Debaryomycetaceae, and Zhang et al. (2020) showed that a mixture of potassium sorbate and sodium benzoate increased the relative abundance (RA) of Lactobacillus paralimentarius and Pediococcus spp. and reduced the RA of Lactobacillus reuteri, Lactobacillus coryniformis, and Klebsiella, in corn silage. Therefore, the analysis of the composition of the bacterial and fungal communities of corn silage treated with sodium benzoate, potassium sorbate, and sodium nitrite by NGS might help in explaining how this additive improves the aerobic stability.
The objective of this experiment was to evaluate the effects of an additive containing sodium benzoate, potassium sorbate, and sodium nitrite on the composition of the microbial community of whole-plant corn silage (stored without or with air stress) and its relationship to silage fermentation and aerobic stability. The hypothesis was that the additive would inhibit microorganisms that cause undesirable fermentation and spoilage, modifying the composition of the microbial community and improving the fermentation profile and aerobic stability of corn silage subjected to air stress.
Materials and Methods
Crops and silos
Whole corn plant (P1449AMX, DuPont Pioneer, Johnston, USA) was harvested at approximately 39% DM from the University of Delaware Farm (Newark, DE, USA) and chopped to a theoretical length of 19 mm using a pull-type chopper (3975, John Deere, Moline, IL) equipped with kernel processer (roller gap set to 1.4 mm). We prepared five individually treated and replicated forage piles for each of the following treatments: 1) control (CTRL), untreated, 2) treatment with Safesil (SF, active ingredients: 20% sodium benzoate, 10% potassium sorbate, and 5% sodium nitrite, Salinity, Göteborg, Sweden) at 2 liters/t of fresh forage weight (SF2), or 3) treatment with SF at 3 liters/t of fresh forage weight (SF3). A total of 200 mL of water or additive diluted in water were applied into each of the five individually replicated forage piles (32 kg each) per treatment, using a hand sprayer. Forage from each treatment and pile was packed into two 7.5-liters laboratory silos (total of 30 silos) using a custom-made gas-driven hydraulic press, modified from a log splitter, at a packing density of approximately 224 kg of DM/m3 and sealed with plastic lids with O-ring seals. Half of the silos for each treatment remained sealed during ensiling (no air stress, NS), and the other half was subjected to air stress (AS). This resulted in the following treatment combinations: NS-CTRL, AS-CTRL, NS-SF2, AS-SF2, NS-SF3, and AS-SF3. Silos subjected to air stress had three holes of 1.60 cm diameter each, which were plugged with butyl rubber stoppers and sealed with silicone glue. Two of these holes were located 5 cm above the bottom of the silo, 180° from each other, and the third hole was on the lid of the silo. Silos were stored in the dark at a temperature of 22 ± 2 °C. During air stress, the holes in each silo were opened for 2 h per wk, then resealed with the plugs and silicone glue, over a 9-wk period.
Aerobic stability measurements
Upon silo opening after 63 d of ensiling, approximately 2 kg of silages from each silo was returned to clean laboratory silos. Silos were covered with two layers of cheesecloth and constantly exposed to air in the laboratory at 22 ± 2 °C. Thermocouple wires were positioned in the geometric center of each forage mass, and temperatures were recorded every 15 min using a data logger DataTaker DT85 (Thermo Fisher Scientific, Australia Pty, Scoresby, VIC, Australia). We calculated the aerobic stability of silage as the number of hours before the temperature of the silage rose 2 °C above the baseline temperature of its mass.
Analytical procedures
We measured the DM content and pH of fresh forages from four CTRL piles at day 0 and of five samples from each silo after ensiling. The DM content was determined after 48 h of incubation in a 60 °C forced-air oven. The weight of the forage mass in the silo and its DM content at day 0 and at day 63 were used to calculate the DM recovery (DMR). Representative 25-g portions of fresh forage and silage were mixed with 225 mL of sterile quarter-strength Ringers solution (Oxoid BR0052G, Oxoid, Unipath Ltd., Basingstoke, UK) and homogenized for 1 min in a Proctor-Silex 57171 blender (Hamilton Beach⁄Proctor-Silex Inc., Washington, DC, USA). The homogenate was filtered through four layers of cheesecloth. The pH was determined on homogenized samples using an Oakton pH700 Meter (Oakton Instruments, Vernon Hills, USA).
Portions of the water extracts previously filtered through four layers of cheesecloth were filtered through Whatman 54 filter paper (Whatman Inc., Clifton, USA). The extract was acidified with H2SO4 50% vol/vol and frozen. Later, the extract was analyzed for the concentrations of lactic, acetic, and ethanol (Muck and Dickerson, 1988) by high-performance liquid chromatography using a Shimadzu LC-20AD pump (Shimadzu Scientific Instruments, Columbia, MD). The concentration of NH3-N was determined on the water extracts by the phenol-hypochlorite method of Weatherburn (1967), and water-soluble carbohydrates (WSC) by the colorimetric procedure of Nelson (1944).
The nutrient analysis of fresh forage described below was performed by Cumberland Valley Analytical Services (Waynesboro, USA). A portion of each dried sample was ground using a Udy Cyclone Sample Mill (Udy Corp., Fort Collins, USA) to pass through a 1-mm screen. Total N was measured by combustion of the sample following the AOAC method 990.03 (AOAC, 2010) using a Leco CNS2000 Analyzer (Leco Corporation, St. Joseph, MI), and the concentration of crude protein (CP) was calculated by multiplying the resulting total N concentration by 6.25. Soluble protein (SP) was determined by the method of Krishnamoorthy et al. (1982) and expressed as % of CP. The concentration of acid detergent fiber (ADF) was quantified on dried ground samples using the AOAC method 973.18 (AOAC, 2010), with the modification of using Whatman 934-AH glass microfiber filters with 1.5-µm particle retention instead of fritted glass crucibles. The concentration of neutral detergent fiber (NDF) was analyzed on dried samples according to the methodology of Van Soest et al. (1991), with sodium sulfite and amylase, and not corrected for ash content. Another portion of the dry sample was ground to pass a 3-mm screen and analyzed for starch by the methodology of Hall (2008).
Enumeration of microorganisms on agar plates
Representative 25-g portions of fresh forages and silages were mixed with 225 mL of sterile quarter-strength Ringers solution (Oxoid BR0052G, Oxoid, Unipath Ltd., Basingstoke, UK) and homogenized for 1 min in a Proctor-Silex 57171 blender (Hamilton Beach/Proctor-Silex Inc., Washington, USA). The homogenate was filtered through four layers of cheesecloth. The filtrate was serially diluted in Ringers solution, and the numbers of lactic acid bacteria (LAB) were determined by pour-plating in de Man, Rogosa, and Sharpe (MRS) agar (CM3651, Oxoid, Unipath Ltd., Basingstoke, UK). Plates were incubated anaerobically at 35 °C in sealed plastic containers with anaerobic packs (Mitsubishi Gas Chemical Co., Tokyo, Japan) and an anaerobic indicator (BR0055, Oxoid, Unipath Ltd., Basingstoke, UK). Containers were vacuum-sealed in nylon-polyethylene bags (3.5-mil embossed pouches, 15.2 × 30.5 cm; Doug Care Equipment Inc., Springville, USA) using a Best Vac vacuum machine (distributed by Doug Care Equipment Inc.). Plates with a number of colonies between 30 and 300 were counted after 48 and 72 h to obtain the number of colony-forming units. The numbers of yeasts and molds were determined on malt extract agar (MEA, CM0059, Oxoid, Unipath Ltd., Basingstoke, UK) acidified after autoclaving with lactic acid (85%) at a rate of 0.5% vol/vol. The plates were incubated aerobically at 32 °C and enumerated after 48 and 72 h. Plates with a number of colonies between 30 and 300 were counted to obtain the number of colony-forming units.
Analysis of the composition of the microbial communities by NGS
Samples from four replicates of fresh forage and silages (CTRL-NS, CTRL-AS, SF2-NS, and SF2-AS) were analyzed for the composition of their microbial community by NGS. Representative 25-g portions of the samples were mixed with 225 mL of autoclaved Ringers solution (Oxoid BR0052G, Oxoid, Unipath Ltd., Basingstoke, UK) for 2 min using a Colworth 400 stomacher (Seward, London, UK). The homogenates were filtered through four layers of cheesecloth. Next, 2 mL of the filtrate was centrifuged for 3 min at 21,130.2 × g using a Centrifuge 5424 R (Eppendorf AG, Hamburg, Germany). The supernatant was discarded, and 100 µL of autoclaved Ringers solution was used to resuspend the pellet. Samples were kept at −80 °C for further analysis.
Extraction of DNA, amplification of the 16S rRNA and internal transcribed spacer 1 (ITS1), and Illumina MiSeq-based sequencing were performed by the Research and Testing Laboratory (Lubbock, USA). DNA was extracted using the MoBio PowerMag Soil kit (MoBio Laboratories Inc., CA, USA), according to the manufacturer’s instructions. For bacterial analysis, the primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGTCAATTCMTTTRAGTTT-3′) (Baker et al., 2003) were used to amplify the V4 and V5 hypervariable regions of the 16S rRNA gene. For fungal analysis, the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2aR (5′-GCTGCGTTCTTCATCGATGC-3′) (White et al., 1990; Gardes and Bruns, 1993) were used to amplify the ITS1. Sequencing was performed on an Illumina MiSeq (San Diego, CA, USA) platform using the 2 × 250 bp paired-end method.
Paired-end reads were merged and converted into FASTA files using FLASH (version 1.2.11; Magoč and Salzberg, 2011). The quality profile of the reads was checked on FastQC (Andrews, 2010). Next, reads were filtered for quality, and the primers were trimmed using QIIME 1.9.1 (Caporaso et al., 2010). Operational taxonomic units (OTU) picking was done using open reference clustering at 97% sequence similarity against Greengenes (version 13.8) (DeSantis et al., 2006) for bacteria or UNITE (version 12.11) (Nilsson et al., 2019) for fungi. To obtain the final OTU taxonomic classification table, singles and double reads were filtered.
Alpha-diversity estimates (numbers of observed OTU, Chao1, Shannon, and Simpson) and beta-diversity evaluation, based on principal coordinate analysis (PCoA), were performed using the Phyloseq and Vegan packages on R (McMurdie and Holmes, 2013; Oksanen et al., 2013; R Core Team, 2018). A targeted loci BLAST search was performed against the sequences of the 10 most abundant fungal OTU. (Altschul et al., 1990). The sequences with the greatest alignment score (Max score) were used to build a phylogenetic tree in MEGA 7 (Kumar et al., 2016) using the neighbor-joining method (Saitou and Nei, 1987) with the maximum composite likelihood substitution model after 1,000 bootstrap replications.
Statistical analysis
Agar counts of LAB and yeasts and molds were transformed to log10 before statistical analysis and are presented on a fresh weight basis. Data were analyzed using JMP version 12 (SAS Institute, Cary, NC). Data on the chemical, microbial, and physical parameters, as well as of the RA of bacterial families and fungal species of silages at silo opening, were analyzed as a 3 × 2 factorial arrangement of treatments in a completely randomized design. The model included the fixed effects of the additive treatment, air stress, and their interaction. The alpha diversity indexes data were analyzed separately as a completely randomized design. Differences were considered significant when P < 0.05 and trends toward significance were considered when P < 0.10. Pairwise mean comparisons were performed using Tukey’s test at P < 0.05 (Snedecor and Cochran, 1980).
We used the weighted UniFrac dissimilarity, for bacteria, and Bray–Curtis dissimilarity, for fungi, to calculate the distance matrices to build the PCoA plot using the phyloseq (McMurdie and Holmes, 2013) package in R (R Core Team, 2018). Statistical analysis of the distance matrices was conducted using the PERMANOVA (permutational multivariate analysis of variance) method of the Adonis function on Vegan (Oksanen et al., 2013). Pearson correlation coefficient matrix was built using the corrplot package (Wei and Simko, 2017) in R (R Core Team, 2018)
Results and Discussion
Nutrient composition and numbers of culturable microorganisms in fresh forage
Freshly chopped whole corn plant was 36.30% DM and had a pH of 5.60. The numbers of LAB and yeasts were 7.59 and 5.63 log10 cfu/g of fresh forage, respectively. Regarding the nutrient composition of the fresh forage, the concentrations of CP, NH3-N, ADF, NDF, starch, and WSC were respectively: 8.80%, 0.03%, 20.03%, 35.40%, 35.08%, and 12.48% of DM. The concentration of SP was 32.95% of CP. The nutrient composition and numbers of culturable microorganisms of the fresh whole-corn plants used in this study were typical for this feedstuff (Kleinschmit et al., 2005; Hafner et al., 2015).
Nutrient composition, fermentation profile, and numbers of culturable microorganisms in silages
We found no interactions between air stress and treatment with the additive for the nutrient composition of silages shown in Table 1. Air stress had no direct effects on the concentrations of CP, SP, ADF, NDF, starch, or WSC, but silages subjected to air stress did have a greater (P < 0.01) DM content and lower concentration of NH3-N than NS silages. There were numerous effects of the additive on the nutrient composition of silage which were somewhat unexpected. For example, we found that compared with CTRL, treatment with SF2 and SF3 unexpectedly reduced (P < 0.05) the concentration of CP regardless of air stress during storage. This effect was not observed in previous experiments using the same additive (Kung et al., 2018a) or additives containing sorbate or benzoate (Kleinschmidt et al., 2005; Hafner et al., 2015). A possible explanation for the additive effect on the CP content is that a higher CP content is expected with a decrease in DMR, which occurred in CTRL silages but not in treated silages, which had a higher DM recovery and maintained a CP content similar to the levels found in the fresh corn plant. Silages treated with the two additive levels in the current study also had lower (P < 0.05) concentrations of SP and NH3-N compared with CTRL, a finding also not observed in the experiments of Kung et al. (2018a). However, in high moisture corn, Da Silva et al. (2015) showed that the additive lowered the concentrations of SP and NH3-N compared with the untreated control. Knicky and Spörndly (2015) also reported that SF (at 5 liters/t of fresh forage weight) reduced the concentration of NH3-N when applied to a variety of crops packed at a low density and subjected air stress during the storage period. Because no butyric acid, indicative of the presence of clostridia, was detected in their silages, they attributed the lower concentration of NH3-N to the inhibition of enterobacteria by SF.
Table 1.
The DM content (%) and chemical composition (% of DM, unless stated otherwise) of whole-plant corn silage untreated or treated with two concentrations of an additive containing sodium benzoate, potassium sorbate, and sodium nitrite subjected or not to air stress, ensiled for 63 d
| Item1 | DM | CP | SP, % of CP | NH3-N | ADF | NDF | Starch | WSC |
|---|---|---|---|---|---|---|---|---|
| Air stress × additive | ||||||||
| NS | ||||||||
| CTRL | 31.42 | 9.38 | 50.53 | 0.09 | 22.16 | 37.92 | 31.94 | 0.00 |
| SF2 | 33.53 | 8.84 | 46.15 | 0.06 | 19.62 | 33.64 | 35.78 | 0.12 |
| SF3 | 34.18 | 9.02 | 46.34 | 0.06 | 20.72 | 35.38 | 31.64 | 0.20 |
| AS2 | ||||||||
| CTRL | 32.74 | 9.32 | 48.07 | 0.08 | 21.86 | 36.94 | 33.34 | 0.00 |
| SF2 | 33.94 | 8.92 | 45.07 | 0.05 | 20.72 | 35.80 | 33.64 | 0.11 |
| SF3 | 35.76 | 8.94 | 44.97 | 0.04 | 21.06 | 35.46 | 32.10 | 0.29 |
| Air stress | ||||||||
| NS | 33.04b | 9.08 | 47.69 | 0.07a | 20.83 | 35.65 | 33.12 | 0.11 |
| AS | 34.15a | 9.06 | 46.03 | 0.06b | 21.21 | 36.07 | 33.03 | 0.13 |
| Additive | ||||||||
| CTRL | 32.08c | 9.35a | 49.30a | 0.09a | 22.01a | 37.43a | 32.64 | 0.00c |
| SF2 | 33.74b | 8.98b | 45.66b | 0.06b | 20.17b | 34.72b | 34.72 | 0.12b |
| SF3 | 34.97a | 8.88b | 45.61b | 0.05b | 20.89ab | 35.42ab | 31.87 | 0.25a |
| SEM | 0.45 | 0.08 | 1.05 | <0.01 | 0.61 | 0.90 | 1.48 | 0.03 |
| Significance | ---------------------------------------------- P–value3 ------------------------------------------------------------ | |||||||
| Additive | <0.01 | <0.01 | <0.01 | <0.01 | 0.02 | 0.02 | 0.16 | <0.01 |
| Air stress | <0.01 | 0.75 | 0.08 | <0.01 | 0.45 | 0.57 | 0.94 | 0.17 |
| Air stress × Additive | 0.42 | 0.53 | 0.66 | 0.78 | 0.52 | 0.23 | 0.48 | 0.15 |
1NS, no air stress; CTRL, control; SF2, Safesil at 2 L/t of fresh forage weight; SF3, Safesil at 3 L/t of fresh forage weight.
2AS, 2 h/wk air stress.
3Means in columns within a section with unlike superscripts differ (P < 0.05).
Only the application of SF2 resulted in silage with statistically lower (P < 0.05) ADF and NDF compared with CTRL. Reduced concentrations of ADF and NDF in SF2 compared with CTRL were not observed in our previous experiments (Da Silva et al., 2015; Kung et al., 2018a). However, similarly to what probably occurred with the CP content, the DMR might also have affected the concentrations of ADF and NDF. An increase in fiber content is expected when the DMR decreases, which occurred in CTRL silages but not in treated silages, which had a higher DMR and maintained concentrations of ADF and NDF similar to the ones of the fresh corn plant. The additive did not affect the concentrations of starch among treatments as expected.
The end products of fermentation, numbers of agar-culturable microorganisms, DMR, and aerobic stability of silages are presented in Table 2. Treated silages (P < 0.05) had a lower pH than CTRL. Treatment with SF also resulted in silages with less (P < 0.05) acetic acid. Air stress resulted in silage with a greater (P < 0.01) pH than NS, which was expected because of competition between aerobic and anaerobic microorganisms. Specifically, we found that AS caused a decrease (P < 0.05) in the numbers of LAB culturable on MRS agar plates in CTRL and SF2 treatments, a decrease (P < 0.05) in the concentration of lactic acid in CTRL and SF2 treatments, and a numerical decrease in lactic acid for SF3. The numbers of LAB in silages treated with the high dose of the additive were below the lowest dilution (10–4) evaluated in this study. Kim and Adesogan (2006) also reported a decrease in the production of lactic acid due to air stress.
Table 2.
The pH, concentrations of lactic and acetic acids and ethanol (% of DM), numbers of LAB and yeasts (log10 cfu/g of fresh weight), DM recovery (%), and aerobic stability (h) of whole-plant corn silage untreated or treated with two concentrations of an additive containing sodium benzoate, potassium sorbate, and sodium nitrite subjected or not to air stress, ensiled for 63 d
| Item1 | pH | Lactic acid | Acetic acid | Ethanol | LAB | Yeasts | DMR | AERS2 |
|---|---|---|---|---|---|---|---|---|
| Air stress × additive | ||||||||
| NS | ||||||||
| CTRL | 3.69 | 6.81a | 1.35 | 4.78a | 6.98a | 2.74b | 85.04 | 131.8b |
| SF2 | 3.56 | 5.79ab | 1.03 | 1.44c | 6.61a | 1.86bc | 91.53 | >260.0a |
| SF3 | 3.53 | 5.17bc | 0.97 | 0.86d | <4.00c | <1.00c | 93.45 | >260.0a |
| AS3 | ||||||||
| CTRL | 3.75 | 4.86bc | 1.31 | 3.86b | 5.30b | 4.91a | 88.59 | 41.3c |
| SF2 | 3.60 | 4.64c | 1.01 | 1.50c | 5.22b | 4.71a | 92.48 | 97.6b |
| SF3 | 3.59 | 4.94bc | 1.13 | 1.02d | <4.00c | 4.54a | 97.16 | 239.2a |
| Additive | ||||||||
| CTRL | 3.72a | 5.84 | 1.33a | 4.32 | 6.14 | 3.83 | 86.81c | 86.6 |
| SF2 | 3.58b | 5.22 | 1.02b | 1.47 | 5.92 | 3.29 | 92.00b | 178.8 |
| SF3 | 3.56b | 5.06 | 1.05b | 0.94 | <4.00 | 2.77 | 95.31a | 249.6 |
| Air stress | ||||||||
| NS | 3.60b | 5.92 | 1.12 | 2.36 | 5.86 | 1.87 | 90.00b | 217.3 |
| AS | 3.64a | 4.81 | 1.15 | 2.13 | 4.84 | 4.72 | 92.74a | 126.0 |
| SEM | 0.01 | 0.24 | 0.06 | 0.09 | 6.98 | 0.22 | 1.19 | 9.5 |
| Significance | ------------------------------------------------------ P–value4 ----------------------------------------------------- | |||||||
| Additive | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Air stress | <0.01 | <0.01 | 0.44 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Air stress × Additive | 0.20 | <0.01 | 0.21 | <0.01 | <0.01 | 0.02 | 0.43 | <0.01 |
1NS, no air stress; CTRL, control; SF2, Safesil at 2 L/t of fresh forage weight; SF3, Safesil at 3 L/t of fresh forage weight.
2AS, aerobic stability.
3AERS, 2 h/wk air stress.
4Means in columns within sections with unlike superscripts differ (P < 0.05).
Although many different microorganisms (yeasts, enterobacteria, and heterofermentative LAB) have the potential to produce ethanol during ensiling, high levels of ethanol in silages are most likely due to yeasts that can ferment WSC to ethanol (Kung et al., 2018b). Treatment with SF3 produced silage with the greatest, SF2 intermediate, and CTRL the lowest concentration of residual WSC (P < 0.05; Table 1). SF2 and SF3 had lower (P < 0.05) ethanol content than CTRL in both NS and AS silages (Table 2). In a series of experiments with corn silage, Kung et al. (2018a) reported that the most consistent effect on the end products of fermentation in silage treated with SF was a reduction in the production of ethanol, and this effect was observed in the current study and documented in past studies with the same additive (Knicky and Spörndly, 2011; Da Silva et al., 2015). Because the conversion of sugars to ethanol by yeasts also produces large quantities of CO2, DMR is poor in this type of fermentation (McDonald et al., 1991). Thus, it was not surprising to find greater (P < 0.05) DMR in SF silages compared with CTRL silage. Unexpectedly, AS silages had a greater (P < 0.01) DMR than NS silages.
Air stress markedly increased (P < 0.01) the numbers of yeasts regardless of additive treatment. Only the treatment with the greatest dose of the additive had fewer (P < 0.05) yeasts cultured on MEA agar when compared with CTRL in NS silages. The numbers of molds were below the detection limit of 1 log cfu/g fresh forage weight (data not shown).
The addition of SF2 and SF3 consistently improved (P < 0.05) the aerobic stability of corn silage compared with CTRL when exposed to air after silo opening. Air stress caused a decrease (P < 0.05) in aerobic stability for CTRL and SF2, but SF2 was still stable for more than two times longer than CTRL. Air stress did not negatively affect the aerobic stability of SF3. The numbers of yeasts culturable on MEA did not fully explain the improvements in aerobic stability afforded by SF and are similar to the findings reported by Da Silva et al. (2015) and Kung et al. (2018a). Kung et al. (2018a) pointed out that MEA agar supports the growth of a greater variety of yeast species than just the lactate-assimilating yeasts that are the primary microorganisms initiating aerobic spoilage in many silages. They also pointed out that the salts of organic acids in SF may not be fungicidal in low concentrations, or after moderate lengths of exposure, but they still could be negatively affecting the metabolism of yeasts and improving stability, while not affecting their numbers in MEA agar.
Analysis of the composition of the bacterial and fungal communities by NGS
From a total of 704,298 reads for bacteria and of 1,052,064 for fungi found in fresh forages and silages, 270,347 bacterial sequences and 415,477 fungal sequences were classified (data not shown). Sequencing depth was enough to describe the diversity of the microbial population as the rarefaction curves reached a plateau (Supplementary Figures S1 and S2). Fresh forage had a more diverse bacterial population than silage as indicated by a greater (P < 0.05) Shanon index than silages (Supplementary Table S1). The greater Shanon index indicated that the ensiling process reduced the richness and evenness of the bacterial community. A decrease in bacterial diversity during the ensiling process has been previously reported and was not unexpected (Dunière et al., 2017; Romero et al., 2017) as fermentation and end-product inhibition result in a more select group of organisms. Regarding the fungal community diversity, silages treated with the additive and exposed to air during storage had a lower (P < 0.05) Chao1 index than fresh forage or silages from other treatments, indicating that the additive in combination with air stress reduced the fungal diversity.
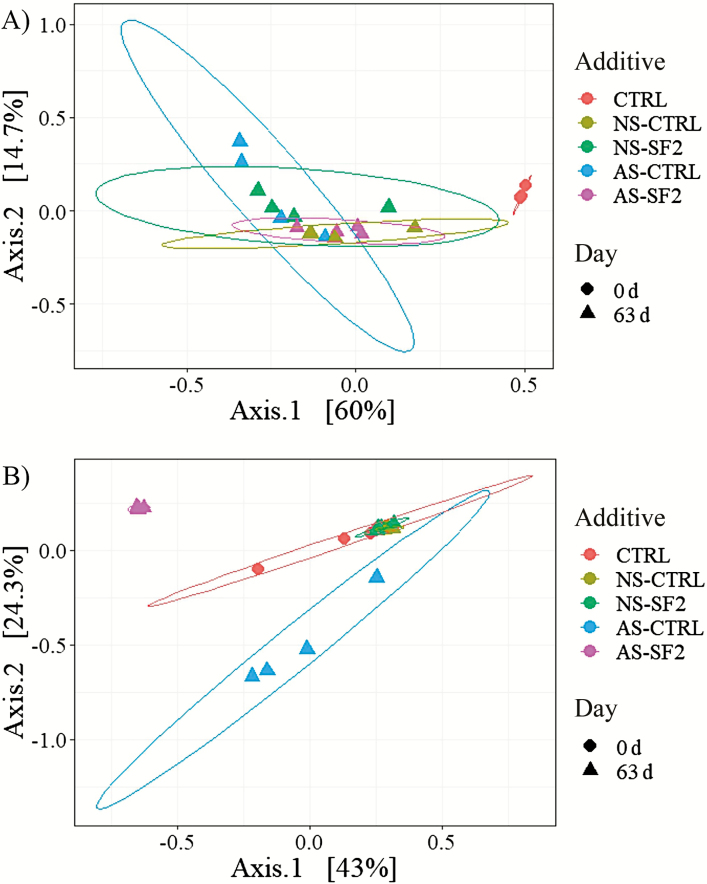
The PCoA plots for the composition of the bacterial and fungal communities of fresh forage and silages after 63 d of ensiling are shown in Figure 1. Figure 1A shows the PCoA plot for the composition of the bacterial community of fresh forage and silage. A significant permutational multivariate analysis of variance (Adonis) test (P < 0.01) proved that the different treatments did not have the same centroid, and a nonsignificant betadisper test (P = 0.27) showed that differences in group dispersions did not cause the results observed. The bacterial community of fresh forage samples formed a cluster separated from all the silage samples, which clustered together themselves. By analyzing the pattern of samples clustering on the PCoA plot and the statistical results of the dissimilarity matrices, we found that ensiling affected the composition of the bacterial community more than did air stress and treatment with SF.
Figure 1.
Principal coordinate analysis plots of the microbial community composition. Principal coordinate analysis (PCoA) plots with (A) weighted UniFrac dissimilarity of the composition of the bacterial community, as analyzed by the sequencing of the V4-V5 region of the 16S rRNA and with (B) Bray–Curtis dissimilarity of the composition of the fungal community, as analyzed by the sequencing of the ITS1 of four replicates per treatment using Illumina Miseq. Treatments consisted of fresh forage (0 d) and corn silage (63 d) untreated (CTRL) or treated with Safesil 2 L (active ingredients of potassium sorbate, sodium benzoate, and sodium nitrite; Salinity, Göteborg, Sweden)/t of fresh matter (SF2), and not subjected (NS) or subjected (AS) to a 2 h/wk air stress.
Figure 1B shows the PCoA plot for the composition of the fungal community of fresh forage and silage. A significant permutational multivariate analysis of variance (Adonis) test (P < 0.01) proved that the different treatments did not have the same centroid, and a nonsignificant betadisper test (P = 0.20) showed that differences in group dispersions were not responsible for the results. Fresh forage and NS silages samples formed a cluster, whereas AS-CTRL and AS-SF2 formed exclusively different clusters. Both treated and untreated silages that were not subjected to air stress had a fungal community similar to the one found in fresh forages.
Table 3 shows the 10 most abundant bacterial families and fungal genera on fresh forages and silages. A total of 9 out of the 10 main bacterial families found in fresh forage were members of the phylum Proteobacteria, which comprises gram-negative bacteria (Kersters et al., 2006). Ni et al. (2017) also found a high RA of Proteobacteria in fresh corn plants, and the main species of Proteobacteria identified by them were Streptomyces spp., Klebsiella spp., Agrobacterium spp., and Pseudomonas spp.
Table 3.
RA (%) of the 10 most abundant bacterial families and fungal genera in fresh forage and corn silage untreated or treated with an additive containing sodium benzoate, potassium sorbate, and sodium nitrite subjected or not to a 2 h/wk air stress ensiled for 63 d, as analyzed by the sequencing of the V4-V5 region of the 16S rRNA, for bacteria, and ITS1, for fungi, using the Illumina MiSeq platform
| Bacteria | |||||
|---|---|---|---|---|---|
| Fresh forage | Silages | ||||
| RA, % | Means | SD | RA, % | Means | SD |
| Xanthomonadaceae | 18.64 | 3.50 | Lactobacillaceae | 72.41 | 14.31 |
| Sphingobacteriaceae | 17.16 | 5.85 | Leuconostocaceae | 5.09 | 3.74 |
| Alcaligenaceae | 11.55 | 4.61 | Enterobacteriaceae | 4.47 | 2.85 |
| Enterobacteriaceae | 11.52 | 4.87 | Xanthomonadaceae | 3.01 | 2.43 |
| Comamonadaceae | 9.27 | 1.15 | Acetobacteraceae | 2.59 | 6.22 |
| Sphingomonadaceae | 6.53 | 1.39 | Sphingobacteriaceae | 1.71 | 1.50 |
| Rhizobiaceae | 4.33 | 0.74 | Sphingomonadaceae | 1.27 | 1.02 |
| Burkholderiaceae | 3.03 | 1.72 | Alcaligenaceae | 1.19 | 1.43 |
| Brucellaceae | 2.51 | 1.30 | Comamonadaceae | 1.18 | 1.45 |
| Weeksellaceae | 2.09 | 0.47 | Paenibacillaceae | 1.15 | 2.18 |
| Fungi | |||||
| Fresh forage | Silages | ||||
| RA, % | Means | SD | RA, % | Means | SD |
| Acremonium | 27.53 | 21.63 | Candida | 62.44 | 33.08 |
| Candida | 23.21 | 16.55 | Pichia | 18.44 | 34.85 |
| Kodamaea | 6.81 | 9.12 | Kodamaea | 2.05 | 2.49 |
| Pichia | 4.03 | 2.66 | Saccharomyces | 0.92 | 2.81 |
| Epicoccum | 0.77 | 0.96 | Acremonium | 0.58 | 1.06 |
| Gibberella | 0.60 | 0.89 | Cryptococcus | 0.24 | 0.40 |
| Occultifur | 0.59 | 0.64 | Fusarium | 0.22 | 0.44 |
| Nigrospora | 0.59 | 0.93 | Occultifur | 0.21 | 0.37 |
| Fusarium | 0.54 | 0.55 | Cercospora | 0.16 | 0.34 |
| Cercospora | 0.13 | 0.22 | Bullera | 0.15 | 0.29 |
Table 4 shows the effect of air stress and additive treatment on the four most abundant bacterial families in the corn silages (Lactobacillaceae, Leuconostocaceae, Enterobacteriaceae, and Acetobacteraceae), except for Xanthomonadaceae, which did not show significative differences among treatments. There was an interaction between air stress and additive treatment for the RA of Lactobacillaceae (P < 0.01), Enterobacteriaceae (P = 0.02), and Leuconostocaceae (P < 0.01) in silages. The addition of SF2 did not affect the numbers of culturable LAB on MRS when compared with CTRL in NS or AS silage, but it increased (P < 0.05) the RA of Lactobacillaceae in NS (although not in AS) compared with CTRL. In contrast, SF3 caused a marked decrease in culturable LAB. Treatment with SF2 reduced (P < 0.05) the RA of Leuconostocaceae and Enterobacteriaceae in NS (but not in AS), and this could account for the reduction in the concentration of acetic acid brought about by SF2 and SF3. We (Da Silva et al., 2016) previously reported that when SF was applied to alfalfa at 4 liters/t, it reduced the numbers of culturable enterobacteria during the early stages of ensiling. In support of that finding, treatment with SF2 in our current study reduced (P < 0.05) the RA of Enterobacteriaceae in NS (but not in AS), which could potentially explain the lower concentrations of SP and NH3-N due to the additive.
Table 4.
RA (%) of bacterial families and fungal species in corn silages untreated or treated with an additive containing sodium benzoate, potassium sorbate, and sodium nitrite subjected or not to air stress ensiled for 63 d, as analyzed by the sequencing of the V4-V5 region of the 16S rRNA, for bacteria, and ITS1, for fungi, using the Illumina MiSeq platform
| Bacterial families | Fungal species | ||||||
|---|---|---|---|---|---|---|---|
| Item1 | Lactobacillaceae | Enterobacteriaceae | Leuconostocaceae | Acetobacteraceae | C. tropicalis | C. humilis | P. kudriavzevii |
| Air stress × additive | |||||||
| NS | |||||||
| CTRL | 62.03c | 7.30a | 10.26a | 0.02 | 77.01 | 0.08b | 1.02b |
| SF2 | 79.76ab | 1.87b | 2.58b | 0.00 | 49.82 | 0.05b | 0.55b |
| AS2 | |||||||
| CTRL | 84.28a | 4.02ab | 2.95b | 1.36 | 21.91 | 0.04b | 71.51a |
| SF2 | 63.56bc | 4.69ab | 4.56b | 8.99 | 0.49 | 98.09a | 0.00b |
| Additive means | |||||||
| CTRL | 73.16 | 5.66 | 6.61 | 0.69 | 49.46a | 0.06 | 36.27 |
| SF2 | 71.66 | 3.28 | 3.57 | 4.50 | 25.16b | 49.07 | 0.27 |
| Air stress means | |||||||
| NS | 70.90 | 4.58 | 6.42 | 0.01 | 63.42a | 0.07 | 0.78 |
| AS | 73.92 | 4.36 | 3.75 | 5.17 | 11.20b | 49.07 | 35.76 |
| SEM | 5.68 | 1.14 | 1.10 | 2.73 | 8.25 | 0.33 | 5.76 |
| Significance | ----------------------------------------------------------- P–value3 ----------------------------------------------------------------- | ||||||
| Air stress | 0.60 | 0.85 | 0.03 | 0.08 | <0.01 | <0.01 | <0.01 |
| Additive | 0.80 | 0.06 | 0.02 | 0.19 | 0.01 | <0.01 | <0.01 |
| Air stress × additive | <0.01 | 0.02 | <0.01 | 0.19 | 0.73 | <0.01 | <0.01 |
1NS, no air stress; CTRL, control; SF2, Safesil at 2 L/t of fresh forage weight.
2AS, 2 h/wk air stress.
3Means in columns within sections with unlike superscripts differ (P < 0.05).
Air stress during ensiling also resulted in a trend (P = 0.08) for increased RA of Acetobacteraceae, which is comprised of species of bacteria that can consume the organic acids of corn silage and sometimes initiate aerobic spoilage (Spoelstra et al., 1988). Because Acetobacter are aerobic bacteria, stimulation of growth by oxygen was expected. Dolci et al. (2011) observed the presence of Acetobacter on corn silage covered with polyethylene film after 2 d of exposure to air but in silage covered with an oxygen barrier film, these bacteria occurred only after 7 d of exposure to air.
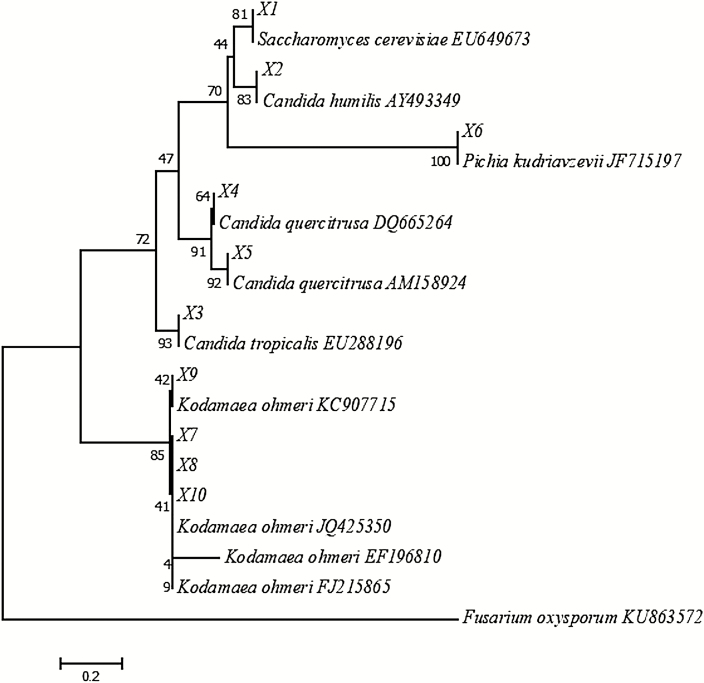
Figure 2 shows the phylogenetic position relative to the top-scoring BLAST hits of the 10 most frequent fungal OTU found in the NS-CTRL, AS-CTRL, NS-SF2, and AS-SF2 silages. OTU X1 was assigned to Saccharomyces cerevisiae; X2 to Candida humilis; X3 to Candida tropicalis; X4 and X5 to Candida quercitrusa; X6 to Pichia kudriavzevii; and X7, X8, X9, and X10 to Kodamaea ohmeri. The three most abundant fungal species found in the silages ensiled for 63 d were C. tropicalis (37.31%), C. humilis (24.57%), and P. kudriavzevii (18.27%; data not shown). The presence of these yeasts has been previously documented in corn silage and total mixed ration silage (Santos et al., 2017, Wang et al., 2018, Jiang et al., 2020). Jiang et al. (2020) observed that a high RA of P. kudriavzevii was associated with the aerobic deterioration of total mixed ration silage containing sweet potato residue.
Figure 2.
Phylogenetic tree of yeast species. Phylogenetic tree constructed by the neighbor-joining method showing the position of yeast species based on ITS1 region sequences. The evolutionary distances were computed using the maximum composite likelihood method. Bootstraps value for 1,000 replicates is shown at the nodes. X1 to X10 represent OTU at the species level. Fusarium oxysporum was used as an outgroup. Scale bar indicates an evolutionary distance of 0.2 nucleotide substitution.
We found that in NS silage, C. tropicalis was the dominant species of yeast independent of additive treatment (Table 4). The RA of both C. humilis and P. kudriavzevii were low in NS and not affected by the additive. Air stress during ensiling caused a marked decrease (P < 0.01) in the RA of C. tropicalis and increased (P < 0.05) the RA of C. humilis in SF2 but P. kudriavzevii in CTRL. Dolci et al. (2011) compared the fungal communities, during aerobic exposure, of corn silage stored under polyethylene film or oxygen barrier film. The authors observed that P. kudriavzevii was found only in silage covered with the polyethylene film, after 7 d of aerobic exposure, but not in silage covered with the oxygen barrier film. Such finding suggests that P. kudriavzevii was stimulated by air infiltration in the silo, similar to what was observed in the present study. A possible explanation for the decline in C. tropicalis is that it might be less tolerant of aerobic conditions than these other species. In AS silages, the reasons for different yeast species to be selected in treated and untreated silages might be due to different degrees of resistance to the additive itself or due to a response to changes in silage characteristics caused by the additive. In AS silages, CTRL had a greater (P < 0.05) concentration of ethanol and a numerically greater concentration of acetic acid than SF2. C. humilis is less tolerant to ethanol than P. kudriavzevii (De Melo Pereira et al., 2012) and is inhibited by acetic acid (Neysens and Vuyst, 2005), and this can explain why this species did not develop well in CTRL silages. In fact, we observed strong negative correlations (P < 0.0001) between the RA of C. humilis and the concentrations of acetic acid (−0.62) and ethanol (−0.69) in the silage (Supplementary Figure S3).
We observed that the greater the RA of C. tropicalis the greater was the pH of silages (0.58, P < 0.0001). Opposingly, the greater the RA of C. humilis the lower was the pH (−0.74, P < 0.0001; Supplementary Figure S3). This fact can be explained by the capacity of certain yeast species to assimilate lactate and initiate aerobic deterioration. It is known that C. tropicalis and P. kudriavzevii are efficient lactate-assimilators (Meroth et al., 2003; Wang et al., 2018), whereas C. humilis is usually unable to assimilate lactate or assimilates this compound slowly (Gori et al., 2011; Vigentini et al., 2014). The additive markedly improved (P < 0.05) the aerobic stability of NS silages by reducing the RA of C. tropicalis, which is known to assimilate lactate (Wang et al., 2018). In AS silages, SF2 had greater (P < 0.05) aerobic stability than CTRL as the additive treatment prevented the dominance of the lactate-assimilating yeasts P. kudriavzevii. We also observed in AS silages that, during aerobic exposure, SF2 had a slow increase in temperature (0.2 °C/h until reaching the maximum temperature) and reached a maximum temperature of 29.5 °C, whereas CTRL had a heating speed of 0.7 °C/h and reached a maximum temperature of 40.8 °C (data not shown). Because a slower heating speed and a more moderate heating curve are indicatives of deterioration by acetic acid bacteria (Muck and Pitt, 1994), such findings suggest that the aerobic deterioration of AS silage treated with SF2 was most likely initiated by acetic acid bacteria rather than by yeasts. This is corroborated by the fact that Acetobacteraceae was detected in treated AS silages, as previously discussed, and that selective inhibition of yeasts by additives can indirectly stimulate the acetic acid bacteria population (Dolci et al., 2011).
Conclusions
NGS was a useful tool to help elucidate the mode of action of SF. Treatment with SF improved the aerobic stability of corn silage. As reported in several previous studies from our lab, we found that the numbers of yeasts culturable on selective agar could not completely explain this finding. However, changes in the RA of the main populations of yeasts better explained the improvements in aerobic stability by the additive. This is the first study showing that aerobic stability can be improved by SF by changing the RA of varying types of yeasts.
Supplementary Material
Acknowledgments
This study was partially supported by Salinity, Göteborg, Sweden, and the Coordination for the Improvement of Higher Education Personnel (CAPES) of the Brazilian Ministry of Education, Brazil, grant (99999.013556/2013-04).
Glossary
Abbreviations
- ADF
acid detergent fiber
- CP
crude protein
- DM
dry matter
- DMR
dry matter recovery
- ITS1
internal transcribed spacer 1
- LAB
lactic acid bacteria
- MEA
malt extract agar
- MRS
Man, Rogosa, and Sharpe
- NDF
neutral detergent fiber
- NGS
next-generation sequencing
- OTU
operational taxonomic unit
- PCoA
principal coordinate analysis
- RA
relative abundance
- SP
soluble protein
- WSC
water-soluble carbohydrate
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Literature Cited
- Altschul S F, Gish W, Miller W, Myers E W, and Lipman D J. . 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andrews S. FASTQC. A quality control tool for high throughput sequence data, 2010. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- AOAC 2010. Official methods of analysis. 18th ed. Gaithersburg (MD): Association of Official Analytical Chemists. [Google Scholar]
- Baker G C, Smith J J, and Cowan D A. . 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Borreani G, Tabacco E, Schmidt R J, Holmes B J, and Muck R E. . 2018. Silage review: factors affecting dry matter and quality losses in silages. J. Dairy Sci. 101:3952–3979. doi: 10.3168/jds.2017-13837 [DOI] [PubMed] [Google Scholar]
- Caporaso J G, Kuczynski J, Stombaugh J, Bittinger K, Bushman F D, Costello E K, Fierer N, Peña A G, Goodrich J K, Gordon J I, . et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva E B, Savage R M, Smith M L, Polukis S A, Laubach A E, Pacer K M, and Kung L Jr.. 2016. Effects of chemical additives on fermentation characteristics of high-moisture alfalfa silage. J. Anim. Sci. 94:327–327. doi: 10.2527/jam2016-068526812338 [DOI] [Google Scholar]
- Da Silva T C, Smith M L, Barnard A M, and Kung L Jr. 2015. The effect of a chemical additive on the fermentation and aerobic stability of high-moisture corn. J. Dairy Sci. 98:8904–8912. doi: 10.3168/jds.2015-9640 [DOI] [PubMed] [Google Scholar]
- De Melo Pereira G V, Miguel M G D C P, Ramos C L, and Schwan R F. . 2012. Microbiological and physicochemical characterization of small-scale cocoa fermentations and screening of yeast and bacterial strains to develop a defined starter culture. Appl. Environ. Microbiol. 78:5395–5405. doi: 10.1128/AEM.01144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T Z, Hugenholtz P, Larsen N, Rojas M, Brodie E L, Keller K, Huber T, Dalevi D, Hu P, and Andersen G L. . 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci P, Tabacco E, Cocolin L, and Borreani G. . 2011. Microbial dynamics during aerobic exposure of corn silage stored under oxygen barrier or polyethylene films. Appl. Environ. Microbiol. 77:7499–7507. doi: 10.1128/AEM.05050-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunière L, Xu S, Long J, Elekwachi C, Wang Y, Turkington K, Forster R, and McAllister T A. . 2017. Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. doi: 10.1186/s12866-017-0947-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, and Bruns T D. . 1993. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Gori K, Bjørklund M K, Canibe N, Pedersen A Ø, and Jespersen L. . 2011. Occurrence and identification of yeast species in fermented liquid feed for piglets. Microb. Ecol. 61:146–153. doi: 10.1007/s00248-010-9706-6 [DOI] [PubMed] [Google Scholar]
- Hafner S D, Windle M, Merrill C, Smith M L, Franco R B, and Kung L Jr. 2015. Effects of potassium sorbate and Lactobacillus plantarum MTD1 on production of ethanol and other volatile organic compounds in corn silage. Anim. Feed Sci. Technol. 208:79–85. doi: 10.1016/j.anifeedsci.2015.07.007 [DOI] [Google Scholar]
- Hall M B. 2008. Determination of starch, including maltooligosaccharides, in animal feeds: comparison of methods and a method recommended for AOAC collaborative study. J. AOAC Int. 92:42–49. [PubMed] [Google Scholar]
- Jiang D, Niu D, Zuo S, Tian P, Zheng M, and Xu C. . 2020. Yeast population dynamics on air exposure in total mixed ration silage with sweet potato residue. Anim. Sci. J. 91:e13397. doi: 10.1111/asj.13397 [DOI] [PubMed] [Google Scholar]
- Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, and Stackebrandt E. . 2006. Introduction to the proteobacteria. In: Dworkin M., Falkow S, Rosenberg E, Schleifer K H, and Stackebrandt E, editors. The prokaryotes. Proteobacteria: alpha and beta subclasses. New York (NY):Springer; p 3–37. [Google Scholar]
- Kim S C, and Adesogan A T. . 2006. Influence of ensiling temperature, simulated rainfall, and delayed sealing on fermentation characteristics and aerobic stability of corn silage. J. Dairy Sci. 89:3122–3132. doi: 10.3168/jds.S0022-0302(06)72586-3 [DOI] [PubMed] [Google Scholar]
- Kleinschmit D H, Schmidt R J, and Kung L Jr. 2005. The effects of various antifungal additives on the fermentation and aerobic stability of corn silage. J. Dairy Sci. 88:2130–2139. doi: 10.3168/jds.S0022-0302(05)72889-7 [DOI] [PubMed] [Google Scholar]
- Knicky M, and Spörndly R. . 2011. The ensiling capability of a mixture of sodium benzoate, potassium sorbate, and sodium nitrite. J. Dairy Sci. 94:824–831. doi: 10.3168/jds.2010-3364 [DOI] [PubMed] [Google Scholar]
- Knicky M, and Spörndly R. . 2015. Short Communication: Use of a mixture of sodium nitrite, sodium benzoate, and potassium sorbate in aerobically challenged silages. J. Dairy Sci. 98:5729–5734. doi: 10.3168/jds.2015-9332 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy U, Muscato T V, Sniffen C J, and Van Soest P J. . 1982. Nitrogen fractions in selected feedstuffs. J. Dairy Sci. 65:217–225. doi: 10.3168/jds.S0022-0302(82)82180-2 [DOI] [Google Scholar]
- Kumar S, Stecher G, and Tamura K. . 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung L Jr, Shaver R D, Grant R J, and Schmidt R J. . 2018b. Silage review: interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 101:4020–4033. doi: 10.3168/jds.2017-13909 [DOI] [PubMed] [Google Scholar]
- Kung L Jr., Smith M L, da Silva E B, Windle M C, da Silva T C, and Polukis S A. . 2018a. An evaluation of the effectiveness of a chemical additive based on sodium benzoate, potassium sorbate, and sodium nitrite on the fermentation and aerobic stability of corn silage. J. Dairy Sci. 101:5949–5960. doi: 10.3168/jds.2017-14006 [DOI] [PubMed] [Google Scholar]
- Magoč T, and Salzberg S L. . 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister T A, Dunière L, Drouin P, Xu S, Wang Y, Munns K, and Zaheer R. . 2018. Silage review: using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 101:4060–4074. doi: 10.3168/jds.2017-13704 [DOI] [PubMed] [Google Scholar]
- McDonald P, Henderson A R, and Heron S J E. . 1991. The biochemistry of silage. In: McDonald P, Henderson A R, and Heron S J E, editors. Marlow (UK): Chalcombe Publications. [Google Scholar]
- McMurdie P J, and Holmes S. . 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8:e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroth C B, Hammes W P, and Hertel C. . 2003. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:7453–7461. doi: 10.1128/aem.69.12.7453-7461.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muck R E, and Dickerson J T. . 1988. Storage temperature effects on proteolysis in alfalfa silage. Trans. ASAE. 31:1005–1009. doi: 10.13031/2013.30813 [DOI] [Google Scholar]
- Muck R E, Nadeau E M G, McAllister T A, Contreras-Govea F E, Santos M C, and Kung L Jr. 2018. Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101:3980–4000. doi: 10.3168/jds.2017-13839 [DOI] [PubMed] [Google Scholar]
- Muck R E, and Pitt R E. . 1994. Aerobic deterioration in corn silage relative to the silo face. Trans. ASAE. 37:735–743. doi: 10.13031/2013.28134 [DOI] [Google Scholar]
- Nelson N. 1944. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153:375–380. [Google Scholar]
- Neysens P, and De Vuyst L. . 2005. Kinetics and modelling of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:95–103. doi: 10.1016/j.jpgs.2004.02.016 [DOI] [Google Scholar]
- Ni K, Minh T T, Tu T T, Tsuruta T, Pang H, and Nishino N. . 2017. Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl. Microbiol. Biotechnol. 101:1385–1394. doi: 10.1007/s00253-016-7900-2 [DOI] [PubMed] [Google Scholar]
- Nilsson R H, Larsson K H, Taylor A F S, Bengtsson-Palme J, Jeppesen T S, Schigel D, Kennedy P, Picard K, Glöckner F O, Tedersoo L, . et al. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47:D259–D264. doi: 10.1093/nar/gky1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F G, Kindt R, Legendre P, Minchin P R, O’hara R B, Simpson G L, Solymos P, Henry M, Stevens H, . et al. 2013. Package ‘vegan’. Community ecology package, version 2. Available from https://CRAN.R-project.org/package=vegan [Google Scholar]
- Oude Elferink S J W H, Driehuis F, Gottschal J C, and Spoelstra S F. . 2000. Silage fermentation processes and their manipulation. In: ‘t Mannetje L., editor. FAO Electronic Conference on Tropical Silage: Silage making in the tropics with particular emphasis on smallholders Rome (Italy): Food and Agriculture Organization; p. 17–30. [Google Scholar]
- Pahlow G, Muck R E, Driehuis F, Oude Elferink S J W H, and Spoelstra S F. . 2003. Microbiology of ensiling. In: Buxton D R, Muck R E, and Harrison H J, editors. Silage science and technology (agronomy series no. 42). Madison (WI): American Society of Agronomy; p. 31–93. [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria):R Foundation for Statistical Computing; Available from https://www.R-project.org/ [Google Scholar]
- Romero J J, Joo Y, Park J, Tiezzi F, Gutierrez-Rodriguez E, and Castillo M S. . 2018. Bacterial and fungal communities, fermentation, and aerobic stability of conventional hybrids and brown midrib hybrids ensiled at low moisture with or without a homo- and heterofermentative inoculant. J. Dairy Sci. 101:3057–3076. doi: 10.3168/jds.2017-13754 [DOI] [PubMed] [Google Scholar]
- Romero J J, Zhao Y, Balseca-Paredes M A, Tiezzi F, Gutierrez-Rodriguez E, and Castillo M S. . 2017. Laboratory silo type and inoculation effects on nutritional composition, fermentation, and bacterial and fungal communities of oat silage. J. Dairy Sci. 100:1812–1828. doi: 10.3168/jds.2016-11642 [DOI] [PubMed] [Google Scholar]
- Saitou N, and Nei M. . 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Santos M C, Golt C, Joerger R D, Mechor G D, Mourão G B, and Kung L Jr. 2017. Identification of the major yeasts isolated from high moisture corn and corn silages in the United States using genetic and biochemical methods. J. Dairy Sci. 100:1151–1160. doi: 10.3168/jds.2016-11450 [DOI] [PubMed] [Google Scholar]
- Snedecor G W, and Cochran W G. . 1980. Statistical methods. 7th ed. Ames (IA): The Iowa State University Press. [Google Scholar]
- Spoelstra S F, Courtin M G, and Van Beers J A C. . 1988. Acetic acid bacteria can initiate aerobic deterioration of whole crop maize silage. J. Agri. Sci. 111:127–132. doi: 10.1017/S0021859600082915 [DOI] [Google Scholar]
- Stanojevic D, Comic L, Stefanovic O, and Solujic-Sukdolak S. . 2009. Antimicrobial effects of sodium benzoate, sodium nitrite and potassium sorbate and their synergistic action in vitro. Bulg. J. Agric. Sci. J. 15:307–311. Available from https://www.agrojournal.org/15/04-05.htm [Google Scholar]
- Van Soest P J, Robertson J B, and Lewis B A. . 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Vigentini I, Antoniani D, Roscini L, Comasio A, Galafassi S, Picozzi C, Corte L, Compagno C, Dal Bello F, Cardinali G, . et al. 2014. Candida milleri species reveals intraspecific genetic and metabolic polymorphisms. Food Microbiol. 42:72–81. doi: 10.1016/j.fm.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Wang H, Hao W, Ning T, Zheng M, and Xu C. . 2018. Characterization of culturable yeast species associating with whole crop corn and total mixed ration silage. Asian-Australas. J. Anim. Sci. 31:198–207. doi: 10.5713/ajas.17.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherburn D. 1967. The preparation and properties of some quaternary arsonium compounds. Aust. J. Chem. 20:2771–2776. doi: 10.1071/CH9672771 [DOI] [Google Scholar]
- Wei T, and Simko V. 2017. R package “corrplot”: visualization of a Correlation Matrix (Version 0.84). Available from https://github.com/taiyun/corrplot. [Google Scholar]
- White T J, Bruns T, Lee S J W T, and Taylor J L. . 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A., Gelfand D H, Sninsky J J, and White T J, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press Inc.; p. 315–322. [Google Scholar]
- Zhang Y, Liu Y, Meng Q, Zhou Z, and Wu H. . 2020. A mixture of potassium sorbate and sodium benzoate improved fermentation quality of whole-plant corn silage by shifting bacterial communities. J. Appl. Microbiol. 128:1312–1323. doi: 10.1111/jam.14571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.