Abstract
Salivary amylase, encoded by the AMY1 gene, is responsible for the digestion of carbohydrates. We investigated associations of AMY1 genetic variations with general and central adiposity changes considering dietary carbohydrate intake among 32,054 adults from four prospective cohort studies. A genetic risk score (GRS) was calculated based on nine AMY1 single-nucleotide polymorphisms, with higher AMY1-GRS indicating higher activity of salivary amylase. We meta-analyzed interactions between AMY1-GRS and dietary intake for changes in general and central adiposity over 5.5–10 years. We found that carbohydrate food intake significantly altered associations of AMY1-GRS with changes in BMI (Pinteraction = 0.001) and waist circumference (Pinteraction < 0.001). Results were consistent and significant in female cohorts rather than in male cohorts. Among women, higher AMY1-GRS was associated with more increases in adiposity if dietary carbohydrate food intake was high, while higher AMY1-GRS was associated with less gains in adiposity when the dietary intake was low. Also, in a 2-year randomized dietary intervention trial, associations of AMY1-GRS with changes in weight (Pinteraction = 0.023) and waist circumference (Pinteraction = 0.037) were significantly modified by carbohydrate intake. Our results suggest the importance of precision nutrition strategies considering participants’ genetic adaptation to carbohydrate-rich diets in regulating general and central adiposity.
Introduction
Carbohydrates are the primary dietary source of energy for humans and play a major role in determining energy balance and regulating the degree of adiposity (1). Dietary carbohydrate quantity and quality have been associated with weight gain, as well as risks of complications of obesity, such as type 2 diabetes (2,3).
The digestion of polysaccharide carbohydrates begins in the mouth by the action of salivary α-amylase, and individual differences in salivary amylase amount and activity are, in part, determined by copy number variations in the salivary α-amylase (AMY1) gene (4–8). Higher AMY1 copy number is related to the improved digestion of starch (6,8). The AMY1 copy variation is likely due to human genetic adaptation to starch-rich diets (4,9), which occurred as consequences of positive selection on the human genome under environmental changes (10). The genetic adaptation to improve the digestion of starchy foods, reflected by the AMY1 gene, may also affect profiles of the oral and gut microbiome (11,12). Gut microbiota alterations have been related to obesity and related diseases in the host, via interactions with dietary factors (13).
Previous studies have shown that the AMY1 copy number variation and single-nucleotide polymorphisms (SNPs) are associated with the risk of obesity (12,14–19) and adiposity changes after a dietary intervention (20), while conflicting results also exist (8,21–23). A recent cross-sectional study of Europeans suggested that associations of AMY1 copy number with adiposity may be modified by dietary starch intake (24). However, no large-scale prospective analysis has addressed the potential modification effects of starch and carbohydrate intakes on associations of AMY1 genotypes with long-term changes in adiposity.
In the present analysis of four prospective cohort studies, we investigated the associations of copy number–related AMY1 genetic variants (assessed by an AMY1 genetic risk score [AMY1-GRS]) with longitudinal changes in general and central adiposity, and we tested whether intakes of starch and carbohydrates might modify the associations. Further, in a 2-year randomized weight-loss dietary intervention trial of overweight and obese adults (25), we investigated whether associations of the AMY1-GRS with changes in general and central adiposity were significantly modified by weight-loss diets varying in carbohydrate intakes.
Research Design and Methods
Study Participants in Prospective Cohort Studies
The current study included a total of 32,054 adults (17,171 women and 14,883 men) from four large-scale prospective cohort studies: the Nurses’ Health Study (NHS), the Health Professionals Follow-Up Study (HPFS), the Atherosclerosis Risk in Communities (ARIC) Study, and the UK Biobank. In all cohorts, participants with cardiovascular disease (CVD) (myocardial infarction or stroke) or cancer at baseline were excluded. We included participants with a complete data set on AMY1 genotypes, dietary intake at baseline, and adiposity measurements (i.e., BMI and waist circumference [WC]) at baseline and follow-up.
The NHS is a cohort of 121,701 female registered nurses aged 30–55 years at enrollment in 1976, and the HPFS is a cohort of 51,529 male health professionals aged 40–75 years at enrollment in 1986. The study protocol was approved by the institutional review boards of Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health. In both the NHS and HPFS cohorts, participants were asked to report data on demographic factors, diet, lifestyle factors, and disease status at study inception. Updated information was collected through follow-up questionnaires. The baseline year of the present analysis in the NHS and the HPFS was 1986 when detailed data on dietary intake and WC were available. In the HPFS, WC was assessed in 1987. A total of 11,339 women in the NHS and 6,851 men in the HPFS had available genetic data and were eligible for the present analysis. We excluded participants with a history of CVD or cancer at baseline, with missing data on dietary intake or obesity measurements at baseline, with implausible energy intake at baseline, or with missing data on outcome measurements at a follow-up time (1996). Consequently, a total of 6,107 women in the NHS and 4,621 men in the HPFS were included in the current study. Details on the selection of participants in the NHS and HPFS are described in Supplementary Figs. 1 and 2.
The ARIC study is a prospective study conducted in four U.S. communities (Forsyth County, NC; Jackson, MS; the northwest suburbs of Minneapolis, MN; and Washington County, MD). We retrieved genotype, diet, and clinical data from the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGaP) server. The institutional review boards at Tulane University approved the study protocol using the dbGaP data. A total of 15,792 participants received a baseline examination (1987–1989), including assessments of medical, lifestyle/diet, and demographic factors. The participants were re-examined approximately every 3 years after the initial evaluation. We used the first examination occurring in 1987–1989 as a baseline, and the third examination in 1993–1995 as a follow-up time point. A total of 8,365 white individuals of European descent with available AMY1 genotype data were eligible in this study. After similar exclusions, a total of 6,515 individuals were included in the ARIC study cohort (Supplementary Fig. 3).
In the UK Biobank, participants provided various information on health and diseases, and a total of 197,080 participants completed at least one dietary assessment (2009–2011). A total of 154,921 white British participants had had available AMY1 genotype data and were eligible in this study. Participants with complete data on diet, genotype, and adiposity measurements at follow-up time (2012–2014) were included in the present analysis. After similar exclusions, the final analytic cohort of UK Biobank included 14,811 individuals (Supplementary Fig. 4). The UK Biobank has approval from the North West Multi-Center Research Ethics Committee that covers the U.K. and the Community Health Index Advisory Group that covers Scotland. This study was covered by the general ethical approval for UK Biobank studies from the National Health Service, National Research Ethics Service. The study participants provided written informed consent to participate in UK Biobank.
Dietary Assessment in Cohort Studies
In the NHS, HPFS, and the ARIC, semiquantitative food frequency questionnaires (FFQs) were used for the assessment of dietary intake. In the NHS and HPFS, validated 131-item FFQs were used, and participants reported how often on average they consumed a specified standard portion size or serving size of specific foods. Nutrient intakes were calculated by multiplying the frequency of consumption by the nutrient content of the specified portion size of each food. Nutrient values were derived from the U.S. Department of Agriculture sources supplemented with information from food manufacturers and published research. Previous studies have provided the reproducibility and validity of the FFQs and reported good correlations between nutrients assessed by the FFQs and multiple food records (26,27). In the ARIC study, dietary data were collected using a validated 66-item semiquantitative FFQ, which was modified from the Harvard FFQ by Willet et al. (28). Nutrient intakes were calculated using the Harvard Nutrient Database. The reproducibility of dietary intakes was validated in the subset of the ARIC study participants (29).
In UK Biobank, participants completed a web-based dietary assessment, the Oxford WebQ, during the period 2009–2011. The Oxford WebQ asked about consumption of >200 types of foods and >30 types of drinks during the previous 24 h using standard categories to indicate the amount consumed. A standard amount/portion size was given for each food item. Nutrient intakes were calculated to provide information comparable to a traditional interviewer-administrated 24-h dietary recall with moderate-to-strong correlations for the majority of nutrients (30).
In these four cohorts, we assessed baseline intake of starch (g/day) and carbohydrate foods (including grains, potatoes, sugar-sweetened beverages, and sweets/desserts) (servings/day). A summary of the food groups in each cohort is described in Supplementary Table 1. We also calculated a total intake of food groups that are major sources of dietary resistant starch (such as bread, cooked cereals/pasta, starchy vegetables other than legumes, and banana) based on estimated data from the U.S. National Health and Nutrition Examination Survey (31) (Supplementary Table 2).
Assessments of Changes in BMI and WC in Cohort Studies
Changes in BMI and WC from baseline to follow-up time were calculated (10-year changes for the NHS and HPFS; 6-year changes for the ARIC; and 5.5-year changes for the UK Biobank). BMI (kg/m2) was calculated as weight in kilograms divided by height in meters squared in all cohorts. In the NHS, height was reported at original study entry in 1976 and was combined with weight reported on the 1986 and 1996 questionnaires. In the HPFS, height and weight were collected at baseline and follow-up questionnaires (in 1986 and 1996). Correlations between self-reported measures of height or weight with direct measurements have been high (r > 0.9 in the NHS and HPFS) (32). WC was self-reported via questionnaires. The Pearson correlations between self-reported WC values and an average of two technician-measured WC values were 0.95 for men and 0.89 for women (32). In the ARIC study and UK Biobank, weight and height were measured by the technicians, and WC was measured at the level of the umbilicus.
Analyses in a Weight-Loss Dietary Intervention Trial
To imply the causality of associations for whether consuming weight-loss diets with different amounts of carbohydrates may have modified the associations of AMY1 genotypes with changes in adiposity, we also analyzed the data of a 2-year randomized weight-loss dietary intervention, the Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trial (25). The trial was conducted from October 2004 through December 2007 at two sites: Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital, and the Pennington Biomedical Research Center of Louisiana State University System. The POUNDS Lost trial was approved by the human subjects committee at each institution and by a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute. All participants in the trial gave written informed consent.
Major exclusion criteria were the presence of diabetes or unstable CVD, the use of medications that affect body weight, and insufficient motivation (25). A total of 811 overweight and obese individuals were randomly assigned to one of four energy-reduced diets varying in macronutrient composition to compare their effects on weight change over 2 years (ClinicalTrials.gov, NCT00072995). Macronutrient intake goals for the four diet groups were 1) 20% fat, 15% protein, and 65% carbohydrate; 2) 20% fat, 25% protein, and 55% carbohydrate; 3) 40% fat, 15% protein, and 45% carbohydrate; 4) 40% fat, 25% protein, and 35% carbohydrate. We recategorized carbohydrate intake into three groups (35% [diet group 4], 45–55% [diet groups 3 and 2], or 65% [diet group 1] of total energy) to indicate low, moderate, and high carbohydrate intake. Height, weight, and WC were measured. A total of 506 participants had available data on genotyping and outcome measurements. Regains of adiposity were used as the study outcomes because most of the participants lost weight and WC in the early phase of the intervention (such as at 6 months) and tended to regain after 6 months to the end of the intervention. Changes in weight (kg) and WC (cm) from 6 months to 2 years in response to the dietary intervention were calculated.
Calculation of GRS
We calculated AMY1-GRS using nine SNPs associated with copy number variation in AMY1 gene loci based on a previous publication of European data (21). The correlations between AMY1 copy number and their SNPs were statistically significant, and the correlations were replicated in two independent cohorts of individuals with European ancestry sampled in the U.S. and Europe in the previous study (21). The detailed information on genotyping and imputation in each study was described previously (20,33–39). Each SNP was recoded as 0, 1, or 2 according to the number of AMY1 copy number–increasing alleles. We calculated a weighted GRS using the following equation: AMY1-GRS = (β1 × SNP1 + β2 × SNP2 + … + βn × SNPn) × (n/sum of the β coefficients), where β is the β coefficient of each SNP for change in AMY1 copy number, and n was 9. We also calculated an unweighted version of AMY1-GRS for conducting the sensitivity analyses. Details of SNPs, β coefficient of each SNP, and distribution of the AMY1-GRS are presented in Supplementary Table 3 and Supplementary Fig. 5.
Statistical Analysis
General linear models were used to analyze the associations of AMY1-GRS with changes in BMI and WC. We assessed multiplicative interactions between AMY1-GRS (as a continuous variable, per 1 unit) and a dietary factor of interest (as a continuous variable, per 1 SD) for the outcomes by adding a cross-product term into a model. We used the intake of carbohydrate food (servings/day) as a primary dietary exposure, and starch (g/day) as a confirmative exposure since the intake of starch is reflected by overall carbohydrate food intake. We also performed sensitivity analyses using the intakes of foods high in resistant starch as dietary exposures to examine whether there were similar associations. Covariates of the multivariate-adjusted model included age, sex (in the ARIC and UK Biobank), source of genotyping data (in the NHS and HPFS), five ancestry principal components (in the UK Biobank), Townsend Social Deprivation Index (in the UK Biobank), education (except for the HPFS due to unavailability of the variable), smoking, physical activity, total energy intake, total fat intake, alcohol consumption, and baseline value of the respective outcome. Participants in the NHS, HPFS, and ARIC had complete data on the covariates. In the UK Biobank, a few participants with missing data on physical activity and the Townsend Social Deprivation Index were included using missing indicators. We analyzed data of the four cohorts separately, and then performed meta-analyses based on β coefficients and SE values for outcomes. Heterogeneity was assessed by the Cochran Q test and I2 statistic. We used a fixed effects model when there was no significant heterogeneity. As the sensitivity analyses, a random effects model was also performed when significant or considerable heterogeneity was observed. We estimated changes in adiposity per 5-point increment in AMY1-GRS according to tertiles of the dietary exposure to examine trends of the gene–diet interactions. To examine potential sex differences in the interaction effects, we analyzed interaction effects among men and women separately using sex-specific SD values and tertiles of the dietary intake.
As for the analysis of data of the POUND Lost trial, we similarly analyzed gene–diet interactions using the general linear model and tested multiplicative interactions between AMY1-GRS and carbohydrate amount on changes in adiposity. The multivariate-adjusted model included covariates of age, sex, ethnicity, and the initial value (at 6 months) of weight or WC in the analysis of total participants. A sex-stratified analysis was also performed. Statistical analyses were performed with the SAS version 9.4 (SAS Institute Inc.) and STATA SE 14.0 (StataCorp).
Data and Resource Availability
Data used in this study are available from the corresponding authors on reasonable request for purposes of reproducing the results or replicating the procedure. No applicable resources were generated or analyzed in the present epidemiological study. The data sets used for the analyses of the ARIC study (dbGaP accession numbers: phs000280 and phs000090) were obtained from the dbGaP at https://www.ncbi.nlm.nih.gov/gap/.
Results
The mean age ranged 54.1–56.3 years, and the median AMY1-GRS (weighted version) ranged 8.5–8.6 points (Supplementary Table 4). Mean values of BMI (25–27 kg/m2) at baseline were comparable across studies, and the mean baseline WC in female and male cohorts ranged from 79 cm (in NHS) to 100 cm (in male participants of ARIC). Mean carbohydrate food intake (a total of grains, potatoes, sugar-sweetened beverages, and sweets/desserts) was also comparable across the cohorts, with a range of 5–6 servings/day. Overall, men (n = 14,883) had higher intakes of total energy (mean [SD], 2,109 [653] kcal/day, when all data were combined) and carbohydrate foods (6.1 [3.0] servings/day), as compared with women (n = 17,171; total energy: 1,801 [547] kcal/day; carbohydrate foods: 5.0 [2.4] servings/day). Energy- and study-place–adjusted intake of carbohydrate food was also comparable in men (mean [SE] 5.6 [0.02] servings/day) and women (5.5 [0.02] servings/day). There was no significant genetic association of AMY1-GRS for differences in the consumption of starch or carbohydrate food at baseline (Supplementary Table 5).
In results of genetic associations of AMY1-GRS with changes in BMI and WC without considering dietary starch/carbohydrate intake, we did not find significant associations of AMY1-GRS with the outcomes (Supplementary Tables 6 and 7 and Supplementary Figs. 6–8). Associations of carbohydrate and starch intakes with these outcomes varied across the studies with significant heterogeneity, and meta-analyzed summary effects were not significant (Supplementary Figs. 9 and 10). On the other hand, except in the male-only HPFS cohort, higher AMY1-GRS was associated with less increase in BMI if dietary carbohydrate food intake was low (in the lowest tertile category, tertile 3) (Pinteraction [between AMY1-GRS and carbohydrate food] = 0.13 in NHS; Pinteraction = 0.019 in ARIC; Pinteraction = 0.004 in UK Biobank) (Table 1). Differences in carbohydrate food intake also modified associations of AMY1-GRS with changes in WC (Pinteraction = 0.04 in NHS; Pinteraction = 0.056 in the ARIC; Pinteraction = 0.005 in UK Biobank). In results of meta-analysis on interactions of all cohorts (Table 1 and Supplementary Fig. 11), we found significant interactions between AMY1-GRS and carbohydrate foods on BMI changes (summary Pinteraction = 0.001; I2 = 53%; heterogeneity test: P = 0.095) and WC changes (summary Pinteraction < 0.001; I2 = 0%; heterogeneity test: P = 0.548). Also, we observed significant interactions between AMY1-GRS and starch intake (g/day) on BMI changes (Pinteraction = 0.031; I2 = 0%; heterogeneity test, P = 0.585) and WC changes (Pinteraction < 0.001; I2 = 32.9%; heterogeneity test, P = 0.215) (Supplementary Fig. 12).
Table 1.
Changes in BMI and WC by increases in AMY1-GRS across tertile categories of carbohydrate food intake
| Outcomes and cohorts | Tertiles of carbohydrate food intake | Pinteraction | |||||
|---|---|---|---|---|---|---|---|
| Tertile 1 | Tertile 2 | Tertile 3 | |||||
| β (SE) | P | β (SE) | P | β (SE) | P | ||
| BMI changes | |||||||
| NHS (n = 6,107) | −0.09 (0.16) | 0.6 | −0.20 (0.13) | 0.13 | 0.08 (0.14) | 0.57 | 0.13 |
| HPFS (n = 4,621) | 0.17 (0.12) | 0.15 | 0.03 (0.13) | 0.84 | −0.09 (0.12) | 0.44 | 0.26 |
| ARIC (n = 6,515) | −0.31 (0.11) | 0.005 | −0.08 (0.11) | 0.48 | 0.07 (0.1) | 0.47 | 0.019 |
| UK Biobank (n = 14,811) | −0.11 (0.06) | 0.06 | −0.03 (0.06) | 0.65 | 0.08 (0.06) | 0.17 | 0.004 |
| Total* | −0.1 (0.05) | 0.025 | −0.05 (0.04) | 0.28 | 0.05 (0.04) | 0.21 | 0.001 |
| WC changes | |||||||
| NHS (n = 6,107) | −0.69 (0.64) | 0.28 | −0.06 (0.57) | 0.91 | 1.17 (0.58) | 0.045 | 0.04 |
| HPFS (n = 4,621) | 0.47 (0.47) | 0.32 | −0.19 (0.45) | 0.67 | −0.09 (0.42) | 0.82 | 0.79 |
| ARIC (n = 6,515) | −0.74 (0.36) | 0.038 | −0.26 (0.35) | 0.46 | 0.2 (0.32) | 0.53 | 0.056 |
| UK Biobank (n = 14,811) | −0.57 (0.22) | 0.01 | 0.21 (0.22) | 0.34 | 0.15 (0.21) | 0.47 | 0.005 |
| Total* | −0.48 (0.17) | 0.004 | 0.03 (0.16) | 0.85 | 0.2 (0.16) | 0.2 | <0.001 |
β (SE) per 5-point increase in AMY1-GRS for the respective outcome using general linear model after adjusting for age, sex (in ARIC and UK Biobank), source of genotyping data (in NHS and HPFS), five ancestry principal components (in UK Biobank), Townsend Social Deprivation Index (in UK Biobank), education, smoking habit (never, former, or current), physical activity, total energy intake, total fat intake, alcohol consumption, and baseline value of the respective outcome.
Meta-analyzed data.
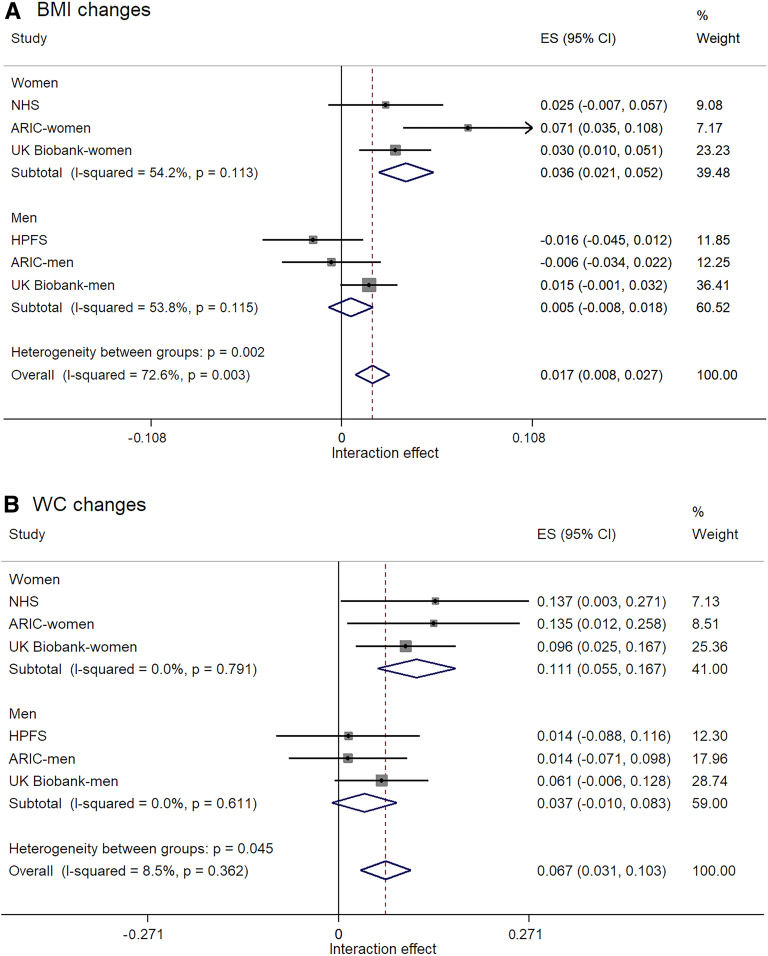
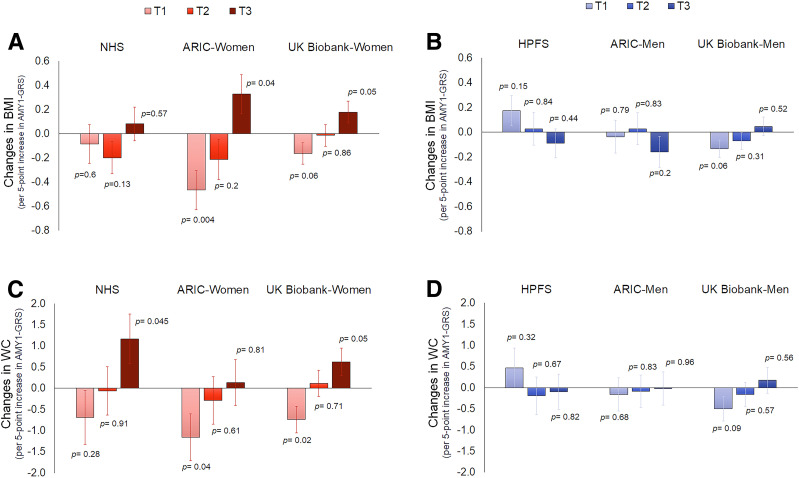
Since the results of the male-only HPFS cohort were different from other cohorts including women, we further analyzed results in men and women separately (Fig. 1). The interactions of AMY1-GRS and carbohydrates were consistently found in women than in men (P for heterogeneity between groups: P = 0.002 for BMI changes) (Fig. 1A). The summary interaction effect for BMI changes was significant in women (Pinteraction < 0.001) but not in men (Pinteraction = 0.44), although there was a moderate heterogeneity within the subgroups (I2 = 54.2% in women; I2 = 53.8% in men). Sensitivity analyses using the random effects model yield similar sex differences for BMI changes. Results were significant in women (Pinteraction = 0.002) (Supplementary Fig. 13). For changes in WC (Fig. 1B), the overall test for heterogeneity between women and men was significant (P = 0.045) without the evidence of heterogeneity within the subgroups (I2 = 0% in both groups). Again, the interaction effect for WC changes was significant in women (Pinteraction < 0.001) but not in men (Pinteraction = 0.12). Figure 2 shows changes in BMI and WC per 5-point increment of AMY1-GRS according to the sex-specific tertile categories of carbohydrate food intake in women and men separately. Supplementary Figure 14 also shows the magnitude of these effects as changes in weight. In female cohorts, an increasing number of AMY1-GRS tended to be related to increases in adiposity when the dietary intake was high (in the tertile 3 group). On the other hand, higher GRS was associated with less gains in adiposity among women who had a low intake of carbohydrate foods (in the lowest tertile, tertile 1 group). There was no consistent trend across the male cohorts. Supplementary Figure 15 shows changes in BMI and WC per 1 SD increment of dietary carbohydrate food intake according to tertile categories of AMY1-GRS when we viewed the data differently.
Figure 1.
A: Sex differences in interaction effects of AMY1-GRS and dietary carbohydrate food intake for BMI changes with the test of effect size (ES): Pinteraction < 0.001 (women); Pinteraction = 0.44 (men); Pinteraction < 0.001 (overall). B: Sex differences in interaction effects of AMY1-GRS and dietary carbohydrate food intake for WC changes with the test of ES: Pinteraction < 0.001 (women); Pinteraction = 0.12 (men); Pinteraction < 0.001 (overall). The ES (95% CI) indicates the effect of interactions between AMY1-GRS (per 1 unit) and carbohydrate foods (per 1 SD) for changes in BMI or changes in WC after adjusting for the same covariates listed in Table 1 (except for sex).
Figure 2.
Changes in BMI and changes in WC per 5-point increment in AMY1-GRS according to tertile categories of carbohydrate food intake in female and male cohorts. Data after adjusting for the same covariates listed in Table 1 (except for sex). A: Effect of AMY1-GRS across tertile 1 (T1) to tertile 3 (T3) of carbohydrate food intake in each cohort (NHS, ARIC-women, and UK Biobank-women) for changes in BMI. B: Effect of AMY1-GRS across T1 to T3 of carbohydrate food intake in each cohort (HPFS, ARIC-men, and UK Biobank-men) for changes in BMI. C: Effect of AMY1-GRS across T1 to T3 of carbohydrate food intake in each cohort (NHS, ARIC-women, and UK Biobank-women) for changes in WC. D: Effect of AMY1-GRS across T1 to T3 of carbohydrate food intake in each cohort (HPFS, ARIC-men, and UK Biobank-men) for changes in WC.
We performed several sensitivity analyses. In results of testing interactions between AMY1-GRS and individual carbohydrate food groups for WC changes in women, each food group showed a similar interaction (Pinteraction = 0.046 for grains, Pinteraction = 0.021 for potatoes, Pinteraction = 0.017 for sugar-sweetened beverages, and Pinteraction = 0.009 for sweets) (Supplementary Fig. 16). When we used food sources of dietary registrant starch as exposures, there were significant interactions between AMY1-GRS and intakes of foods high in dietary resistant starch on changes in BMI (Pinteraction = 0.042) and WC (Pinteraction = 0.006) (Supplementary Fig. 17), also showing significant interactions on WC changes in women (Supplementary Fig. 18). Results using an unweighted version of GRS were similar to the main results (detailed data available upon request). Results using individual SNPs showed no heterogeneity across SNPs except for two SNPs (rs1566154 and rs1930212) in the gene–diet interactions for WC changes among women. Four SNPs (rs4244372, rs11577390, rs11185098, and rs1999478) showed significant interactions without heterogeneity (Supplementary Fig. 19).
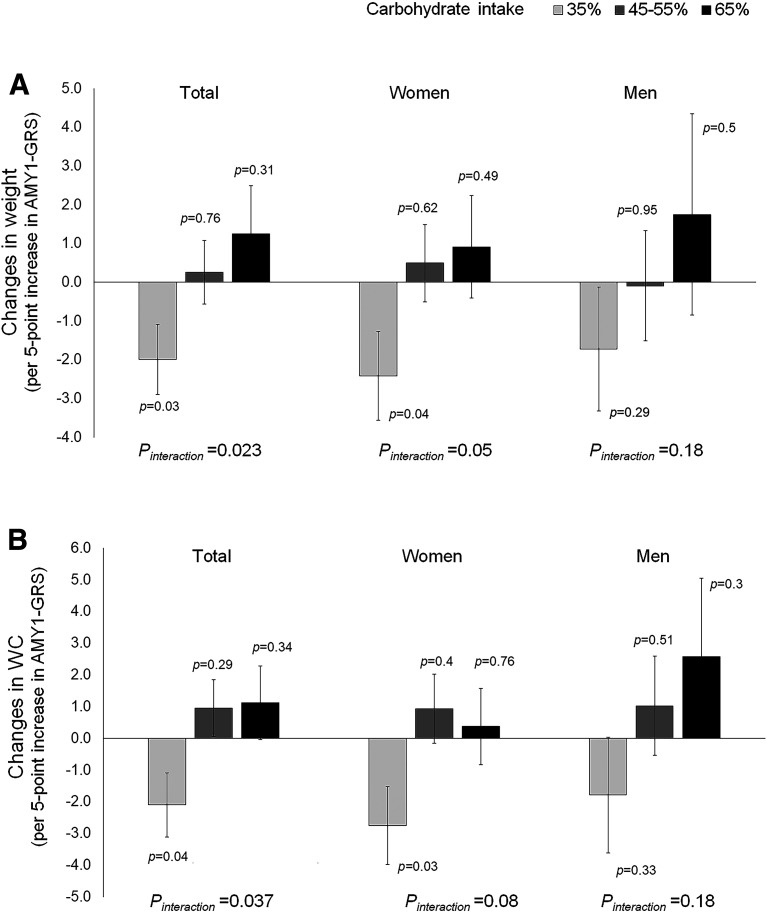
Lastly, we investigated data of the 2-year dietary intervention, POUNDS Lost trial (n = 506). The characteristics of the study participants are shown in Supplementary Table 8. In the POUNDS Lost trial, we found similar patterns of interactions for weight changes (Pinteraction = 0.023) (Fig. 3A) and WC changes (Pinteraction = 0.037) (Fig. 3B). In response to a low-carbohydrate diet (35% of total energy intake), higher AMY1-GRS was associated with greater decreases in weight and WC. Conversely, higher AMY1-GRS tended to be associated with increases in weight and WC in response to a high-carbohydrate diet (65% of total energy intake). Similarly, in participants with high AMY1-GRS scores (based on median), higher carbohydrate intake was related to greater gains in weight (Ptrend = 0.037) and WC (Ptrend = 0.069); consuming a high-carbohydrate diet was related to greater increases in weight (mean [SE], 3.3 [0.7] kg) and WC (1.8 [0.76] cm), as compared with consuming a low-carbohydrate diet (weight: 1.5 [0.7] kg, WC: 0.1 [0.7] cm) (Supplementary Fig. 20).
Figure 3.
A: Effect of a per 5-unit increase in AMY1-GRS for changes in weight. B: Effect of a per 5-unit increase in AMY1-GRS for changes in WC. The effect is in response to low-calorie weight-loss diets varying in carbohydrate intake (35%, 45–55%, and 65% of total energy intake) in the POUNDS Lost trial. Bars indicate β and SE values for changes in weight or WC from 6 months to 2 years after dietary intervention per 5-unit increase in AMY1-GRS after adjustment for age, sex (in total participants), ethnicity, the initial value (at 6 months) of weight or WC. The number of total participants was 506, including 301 women and 205 men.
Discussion
We found that dietary starch and carbohydrate intakes significantly modified associations of the AMY1 genetic variants with long-term changes in general and central adiposity. The genetically determined higher amylase activity was related to less increases in general and central adiposity when the dietary carbohydrate intake was low, while the trend was opposite when the dietary intake was high, particularly among female cohorts. Further, we replicated similar interactions between AMY1-GRS and diets with different carbohydrate amounts in long-term maintenance of adiposity among obese adults who participated in the POUNDS Lost trial.
Our findings on the gene–diet interactions are in line with the results from a recent cross-sectional study (24), in which it was found that having a higher AMY1 copy number was related to lower BMI among people with lower starch intake, while the trend was opposite among people with higher starch intake (24). In the POUNDS Lost trial, we found that starch digestion–related genetic variations were associated with determining individual variations in long-term maintenance of adiposity after consuming weight-loss diets varying in carbohydrate intakes. Such results suggest the causality of changes in carbohydrate intake in modifying the effect of human genetic adaptation to starch-rich diets on the regulation of general and central adiposity, and they also highlight the importance of precision dietary intervention strategies for obese patients. Also, our findings may at least partly explain the inconsistent findings between the AMY1 and obesity in previous studies (8,21–23). We did not observe significant main genetic associations of AMY1-GRS with changes in adiposity when differences in dietary intake of starch or carbohydrate foods were not considered. We speculate that the null association may be due to mixed associations of genetically determined amylase activity in regulating adiposity in populations with varying amounts of starch/carbohydrate intakes. These results collectively suggest that favorable amounts of starch and carbohydrates for determining individual variations of changes in general and central adiposity may be varied across participants based on the AMY1 genetic variations.
Although detailed mechanisms underlying the observed interaction associations need further investigations, a situation where people with higher AMY1-GRS (i.e., people with higher activity and amount of salivary amylase) consume high amounts of starch/carbohydrate foods can be interpreted as a metabolic status where the digestion of carbohydrates occurs faster to raise postprandial blood glucose levels (8). Such a concept is similar to the glycemic index (GI) (40). Low GI foods are more slowly digested and cause slower increases in blood glucose and insulin levels, as compared with carbohydrate foods with a high GI (40). A meta-analysis of randomized controlled trials shows diets achieving a substantial decrease in GI were moderately effective in lowering weight (41). Also, the copy number of AMY1 may be correlated to that of pancreatic amylase gene (42), and there might be indirect effects of pancreatic amylase gene variations on adiposity changes (17). We also speculated that differential effects of carbohydrate amount or AMY1 copy number variation on energy metabolism (43) and insulin resistance (3,44,45) might partly be an underlying issue. In a previous metabolomics study, AMY1 copy number variation was associated with metabolomics markers of energy production, carbohydrate metabolism, as well as lipid metabolism (43). We speculated that associations of AMY1 gene with energy metabolism and weight change might be regulated complex pathways. Further research is warranted to quantify whether the magnitude of AMY1 effects on caloric absorption is consistent with the amount of weight changes explained by the effect, with detailed assessments of metabolomicmarkers, nutrients and total energy intake, and total energy expenditure.
Interestingly, we observed that AMY1-GRS and carbohydrate interactions for adiposity changes were consistent, particularly in women, implicating the substantial heterogeneity in men, which requires more research. Many of gene loci determining body fat show significant sexual dimorphism, with a stronger effect in women (46), and hormonal regulations, sex chromosomes, and other factors are thought to be the major contributors to sex differences in the regulation of obesity and energy homeostasis (46). A recent experimental study suggested potential sex differences in intestinal carbohydrate metabolism (48). Studies have also shown that serum amylase levels were associated with visceral adipose tissues particularly in women (49) and that associations of AMY1 gene copies with obesity may be different in men and women (16,50), although directions of sex-specific AMY1 associations have been inconsistent in the previous studies (16,50). Although further investigations are necessary to understand the complex gene–sex–diet interactions in the development of obesity, our study suggests a potential necessity of sex-specific nutrition strategies regarding carbohydrate amount for controlling adiposity.
Our study has several strengths. We have performed the cross-validation from independent prospective cohorts with well-validated measures of dietary factors and longitudinal assessments of changes in general and central adiposity. The large sample size allowed us to examine potential sex differences in the observation. Secondly, we replicated findings in one of the largest and longest randomized dietary intervention trials, which minimized a potential bias in observational studies and warranted the robustness of our findings. Our study also has several potential limitations. Firstly, our study mainly included white individuals of European descent. The consumption of carbohydrate food would be substantially different in regions, and there might be diversity in the distribution of AMY1 copy number across different populations (4,51). Although the assessment of dietary intake was validated, self-reported data on dietary intake was also a limitation. We did not directly measure copy numbers using the droplet digital PCR or salivary amylase concentrations. Further investigations are warranted to explore potential underlying mechanisms. Even so, our study should motivate future large-scale gene–diet interaction studies focusing on structural variations in the human genome to understand the development of metabolic diseases.
In conclusion, dietary intakes of starch and carbohydrate foods significantly modified associations of starch digestion–related genetic variations with general and central adiposity changes, particularly among women. Our study suggests the importance of precision nutrition strategies considering participants’ genetic adaptation to carbohydrate-rich diets in regulating adiposity.
Article Information
Acknowledgments. The authors thank the participants and staff of the NHS and HPFS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors thank Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School for their assistance. The authors thank participants and staff of the ARIC study and UK Biobank for their valuable contributions. The ARIC study data sets used for the analyses were obtained from the dbGaP.
Funding. This study is supported by National Institutes of Health grants from the National Cancer Institute (UM1-CA-186107, UM1-CA-167552, R01-CA-137178, and P01-CA-87969, R01-CA-49449), the National Heart, Lung, and Blood Institute (R01-HL-034594, R01-HL-35464, R01-HL-088521, HL-071981, and HL-126024), the National Institute of Diabetes and Digestive and Kidney Diseases (DK-091718, DK-100383, DK-078616, DK-098311, and DK-115679), the Boston Obesity Nutrition Research Center (DK-46200), and United States–Israel Binational Science Foundation (2011036). The ARIC study has been supported in whole or in part by the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I). Funding for the Gene Environment Association Studies initiative (GENEVA) was provided by National Human Genome Research Institute grant U01HG004402. L.Q. was supported by the American Heart Association (Scientist Development Award 0730094N). Y.H. was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research and JSPS Overseas Research Fellowship). Y.H. is supported by the American Heart Association (Postdoctoral Fellowship 19POST34380035).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.H. contributed to the study concept and design, statistical analysis and interpretation of the data, drafting and revision of the manuscript, and study supervision. T.Z., C.Y., and T.H. contributed to the statistical analysis and interpretation of data and revision of the manuscript. W.C.W., F.B.H., G.A.B., and F.M.S. contributed to acquisition of data, interpretation of data, revision of the manuscript, and funding. L.Q. contributed to the study concept and design, acquisition of data, analysis and interpretation of the data, drafting and revision of the manuscript, funding, and study supervision. All authors contributed to the interpretation of results and critical revision of the manuscript for important intellectual content, and they approved the final version of the manuscript. Y.H. and L.Q. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Clinical trial reg. no. NCT00072995, clinicaltrials.gov
This article contains supplementary material online at https://doi.org/10.2337/figshare.12389399.
References
- 1.Bray GA, Heisel WE, Afshin A, et al. The science of obesity management: an endocrine society scientific statement. Endocr Rev 2018;39:79–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Hao T, Rimm EB, Willett WC, Hu FB. Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med 2011;364:2392–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig DS, Hu FB, Tappy L, Brand-Miller J. Dietary carbohydrates: role of quality and quantity in chronic disease. BMJ 2018;361:k2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007;39:1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA. Individual differences in AMY1 gene copy number, salivary α-amylase levels, and the perception of oral starch. PLoS One 2010;5:e13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandel AL, Breslin PA. High endogenous salivary amylase activity is associated with improved glycemic homeostasis following starch ingestion in adults. J Nutr 2012;142:853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang ZM, Lin J, Chen LH, Zhang M, Chen WW, Yang XR. The roles of AMY1 copies and protein expression in human salivary α-amylase activity. Physiol Behav 2015;138:173–178 [DOI] [PubMed] [Google Scholar]
- 8.Atkinson FS, Hancock D, Petocz P, Brand-Miller JC. The physiologic and phenotypic significance of variation in human amylase gene copy number. Am J Clin Nutr 2018;108:737–748 [DOI] [PubMed] [Google Scholar]
- 9.Perry GH, Kistler L, Kelaita MA, Sams AJ. Insights into hominin phenotypic and dietary evolution from ancient DNA sequence data. J Hum Evol 2015;79:55–63 [DOI] [PubMed] [Google Scholar]
- 10.Mathieson I, Lazaridis I, Rohland N, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015;528:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole AC, Goodrich JK, Youngblut ND, et al. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 2019;25:553–564.e7 [DOI] [PubMed] [Google Scholar]
- 12.León-Mimila P, Villamil-Ramírez H, López-Contreras BE, et al. Low salivary amylase gene (AMY1) copy number is associated with obesity and gut Prevotella abundance in Mexican children and adults. Nutrients 2018;10:E1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falchi M, El-Sayed Moustafa JS, Takousis P, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet 2014;46:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mejía-Benítez MA, Bonnefond A, Yengo L, et al. Beneficial effect of a high number of copies of salivary amylase AMY1 gene on obesity risk in Mexican children. Diabetologia 2015;58:290–294 [DOI] [PubMed] [Google Scholar]
- 16.Marcovecchio ML, Florio R, Verginelli F, et al. Low AMY1 gene copy number is associated with increased body mass index in prepubertal boys. PLoS One 2016;11:e0154961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnefond A, Yengo L, Dechaume A, et al. Relationship between salivary/pancreatic amylase and body mass index: a systems biology approach. BMC Med 2017;15:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatapoorna CMK, Ayine P, Parra EP, et al. Association of salivary amylase (AMY1) gene copy number with obesity in Alabama elementary school children. Nutrients 2019;11:E1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquina C, Mousa A, Belski R, Banaharis H, Naderpoor N, de Courten B. Increased inflammation and cardiometabolic risk in individuals with low AMY1 copy numbers. J Clin Med 2019;8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heianza Y, Sun D, Wang T, et al. Starch digestion-related amylase genetic variant affects 2-year changes in adiposity in response to weight-loss diets: the POUNDS lost trial. Diabetes 2017;66:2416–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usher CL, Handsaker RE, Esko T, et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet 2015;47:921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shwan NAA, Armour JAL. No evidence for association of BMI with salivary amylase gene copy number in the UK 1958 birth cohort. Obesity (Silver Spring) 2019;27:1533–1538 [DOI] [PubMed] [Google Scholar]
- 23.Valsesia A, Kulkarni SS, Marquis J, et al. Salivary α-amylase copy number is not associated with weight trajectories and glycemic improvements following clinical weight loss: results from a 2-phase dietary intervention study. Am J Clin Nutr 2019;109:1029–1037 [DOI] [PubMed] [Google Scholar]
- 24.Rukh G, Ericson U, Andersson-Assarsson J, Orho-Melander M, Sonestedt E. Dietary starch intake modifies the relation between copy number variation in the salivary amylase gene and BMI. Am J Clin Nutr 2017;106:256–262 [DOI] [PubMed] [Google Scholar]
- 25.Sacks FM, Bray GA, Carey VJ, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feskanich D, Rimm EB, Giovannucci EL, et al. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. J Am Diet Assoc 1993;93:790–796 [DOI] [PubMed] [Google Scholar]
- 27.Salvini S, Hunter DJ, Sampson L, et al. Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int J Epidemiol 1989;18:858–867 [DOI] [PubMed] [Google Scholar]
- 28.Willett WC, Sampson L, Stampfer MJ, et al. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol 1985;122:51–65 [DOI] [PubMed] [Google Scholar]
- 29.Savoca MR, Steffen LM, Bertoni AG, Wagenknecht LE. From neighborhood to genome: three decades of nutrition-related research from the Atherosclerosis Risk in Communities Study. J Acad Nutr Diet 2017;117:1881–1886.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Young H, Crowe FL, et al. Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr 2011;14:1998–2005 [DOI] [PubMed] [Google Scholar]
- 31.Murphy MM, Douglass JS, Birkett A. Resistant starch intakes in the United States. J Am Diet Assoc 2008;108:67–78 [DOI] [PubMed] [Google Scholar]
- 32.Rimm EB, Stampfer MJ, Colditz GA, Chute CG, Litin LB, Willett WC. Validity of self-reported waist and hip circumferences in men and women. Epidemiology 1990;1:466–473 [DOI] [PubMed] [Google Scholar]
- 33.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007;39:870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L, Cornelis MC, Kraft P, et al.; Meta-Analysis of Glucose and Insulin-related traits Consortium (MAGIC); Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet 2010;19:2706–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornelis MC, Monda KL, Yu K, et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet 2011;7:e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiggs JL, Kang JH, Yaspan BL, et al.; GENEVA Consortium . Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 2011;20:4707–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen MK, Pers TH, Dworzynski P, Girman CJ, Brunak S, Rimm EB. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ Cardiovasc Genet 2011;4:549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GENEVA: The Atherosclerosis Risk in Communities (ARIC) Study Accessed 23 April 2020. Available from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000090.v3.p1.
- 39.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willett W, Manson J, Liu S. Glycemic index, glycemic load, and risk of type 2 diabetes. Am J Clin Nutr 2002;76:274S–280S [DOI] [PubMed] [Google Scholar]
- 41.Zafar MI, Mills KE, Zheng J, Peng MM, Ye X, Chen LL. Low glycaemic index diets as an intervention for obesity: a systematic review and meta-analysis. Obes Rev 2019;20:290–315 [DOI] [PubMed] [Google Scholar]
- 42.Carpenter D, Dhar S, Mitchell LM, et al. Obesity, starch digestion and amylase: association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum Mol Genet 2015;24:3472–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arredouani A, Stocchero M, Culeddu N, et al.; D.E.S.I.R. Study Group . Metabolomic profile of low-copy number carriers at the salivary α-amylase gene suggests a metabolic shift toward lipid-based energy production. Diabetes 2016;65:3362–3368 [DOI] [PubMed] [Google Scholar]
- 44.Ebbeling CB, Feldman HA, Klein GL, et al. Effects of a low carbohydrate diet on energy expenditure during weight loss maintenance: randomized trial. BMJ 2018;363:k4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi YJ, Nam YS, Yun JM, et al. Association between salivary amylase (AMY1) gene copy numbers and insulin resistance in asymptomatic Korean men. Diabet Med 2015;32:1588–1595 [DOI] [PubMed] [Google Scholar]
- 46.Shungin D, Winkler TW, Croteau-Chonka DC, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium . New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015;518:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lovejoy JC, Sainsbury A; Stock Conference 2008 Working Group . Sex differences in obesity and the regulation of energy homeostasis. Obes Rev 2009;10:154–167 [DOI] [PubMed] [Google Scholar]
- 48.Hudry B, de Goeij E, Mineo A, et al. Sex differences in intestinal carbohydrate metabolism promote food intake and sperm maturation. Cell 2019;178:901–918.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias JP, Schrack JA, Shardell MD, Egan JM, Studenski S. Association of abdominal fat with serum amylase in an older cohort: the Baltimore Longitudinal Study of Aging. Diabetes Res Clin Pract 2016;116:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viljakainen H, Andersson-Assarsson JC, Armenio M, et al. Low copy number of the AMY1 locus is associated with early-onset female obesity in Finland. PLoS One 2015;10:e0131883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inchley CE, Larbey CD, Shwan NA, et al. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci Rep 2016;6:37198. [DOI] [PMC free article] [PubMed] [Google Scholar]