Abstract
Purpose
To investigate the presence of SARS-CoV-2 RNA in tears of patients with moderate to severe coronavirus disease 2019 (COVID-19).
Design
Cross-sectional study.
Participants
Patients with laboratory-proven moderate to severe COVID-19.
Methods
Tears were collected within 48 hours of laboratory confirmation using 3 methods: conjunctival swab plus Schirmer’s test strips (group 1), conjunctival swab (group 2), and Schirmer’s test strips (group 3). Samples from both the eyes of each patient were transported in a single viral transport media for real-time RT-PCR. Detailed demographic profiles, systemic symptoms, comorbidities, and ocular manifestations were noted.
Main Outcome Measures
Viral load of a sample was determined using cycle threshold (Ct) value of E gene. A specimen was considered to show positive results if the amplification curve for the E gene crossed the threshold line within 35 cycles and if it showed positive results on an RNA-dependent RNA polymerase or open reading frame 1b gene assay.
Results
Of the 78 patients enrolled in the study, samples from 3 patients were found to be inadequate for analysis. Thirty-six patients (48%) had moderate disease, whereas 39 patients (52%) had severe disease, with no ocular involvement in any patient. In the 75 patients, RT-PCR analysis of tears showed positive results in 18 patients (24%), and 29 of 225 samples (12.9%) showed positive results. Positive results were found in 11 (14.7%), 11 (14.7%), and 7 (9.3%) patients in groups 1, 2, and 3, respectively (P = 0.3105). Mean Ct values in groups 1, 2, and 3 were 28.36 ± 6.15, 29.00 ± 5.58, and 27.86 ± 6.46 (P = 0.92), respectively. Five patients showed positive RT-PCR results by all 3 methods (mean Ct value, 25.24 ± 6.33), and 12 patients showed positive results by any of the 3 methods (mean Ct value, 32.16 ± 1.94), the difference in Ct values being statistically significant (P = 0.029). The median value of symptomatology in patients with positive RT-PCR results from tears was 5 days (range, 4–9 days).
Conclusions
SARS-CoV-2 RNA was detected in tears of 24% of patients with laboratory-proven moderate to severe COVID-19. Conjunctival swab remains the gold standard of tear collection for RT-PCR assay. A significantly higher possibility of viral transmission exists through tears in patients with moderate to severe COVID-19.
Keywords: Conjunctival swab, COVID-19, Nasopharyngeal swab, Reverse-transcriptase polymerase chain reaction, SARS-CoV-2
Abbreviations and Acronyms: COVID-19, coronavirus disease 2019; Ct, cycle threshold; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VTM, viral transport media
SARS CoV-2 viral RNA was detected in tears of 24% of laboratory proven moderate to severe COVID 19 patients, in early course of the disease, posing risk of viral transmission through tears.
Coronavirus disease 2019 (COVID 19)-related morbidity, mortality, and economic contraction has taken the world by storm since December 2019. Despite the herculean efforts directed toward curbing its transmission, the disease has continued to spread like a wildfire. Coronaviruses are zoonotic pathogens that can infect human beings by undergoing mutations.1 Airborne respiratory droplet transmission is well recognized; however, alternative methods, such as ocular secretions and oral–fecal transmission, although being held responsible for the spread in many studies, have yet to be proven conclusively.2, 3, 4
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike proteins bind with the host cellular receptor, human angiotensin-converting enzyme 2, and gain entry into the cell in the presence of transmembrane serine protease 2, a cell surface-associated protease.5, 6, 7, 8 Angiotensin-converting enzyme 2 is known to be expressed on epithelial cells in lungs, intestines, and kidney.9 Recent reports indicate that both angiotensin-converting enzyme 2 and transmembrane serine protease 2 are present in human conjunctival and corneal cells, making the ocular surface cells a potential entry point and reservoir for transmission of the virus.10 , 11 The mucous membrane of the ocular surface is continuous from puncta via the nasolacrimal duct to the nasopharynx, resulting in viral transfer in either direction, even to the gastrointestinal tract, if swallowed. Blood-borne infection of the lacrimal gland also has been proposed.12
Despite the above evidence, the reported prevalence of viral RNA detection in tears varies from 0% to 7%, with higher positivity rates in patients with severe COVID-19. This has been attributed to low sensitivity of real-time reverse-transcriptase polymerase chain reaction (RT-PCR) in picking up small quantities of SARS-CoV-2 RNA, missing of window of viral shedding at the time of sample collection and a small sample size. The shedding of viral RNA in tears has been observed in both the presence and the absence of ocular manifestations.13, 14, 15, 16, 17, 18, 19
Inconsistency in study populations exists across different reports with regard to COVID-19 disease severity and laboratory confirmation. Also, variability exists in the method of tear sample collection, that is, conjunctival swabs or Schirmer paper strips, with separate viral transport media (VTM) for each eye. To the best of our knowledge, different techniques for tear collection for SARS-CoV-2 detection have not been compared.
In the present study, to increase the yield of viral RNA in preocular tear film, sampling was performed in laboratory-confirmed hospitalized patients with moderate and severe COVID-19 using different methods within 48 hours of collection of naso-oropharyngeal swabs. Samples from both the eyes were transported in a single VTM. The objectives of this study were to investigate the presence of SARS-CoV-2 RNA in tears of patients with moderate to severe COVID-19 and to ascertain the best method of tear sample collection by assessing the cycle threshold value of E gene.
Methods
A cross-sectional study was conducted from May 22 through June 4, 2020, in Lok Nayak Hospital, New Delhi, India, one of the largest, tertiary, COVID-19–dedicated hospitals in North India. After obtaining the Maulana Azad Medical College institutional ethics committee clearance, the trial was registered (identifier, CTRI/2020/05/025291). Adult patients with moderate and severe laboratory-confirmed COVID-19 with positive results from naso-oropharyngeal RT-PCR analysis, performed 1 day before, who were admitted in the medical block and willing to participate were enrolled in the study consecutively. Exclusion criteria included patients with asymptomatic, mild, and critical COVID-19. In accordance with the Declaration of Helsinki, written informed consent was obtained from all patients. A brief history was obtained and an ocular examination was performed. Special attention was paid to any associated systemic comorbidity.
Moderate disease was diagnosed as those having clinical signs of pneumonia (fever, cough, dyspnea, fast breathing), but no signs of severe pneumonia, including oxygen saturation measured by pulse oximetry of less than 94% (range, 90%–94%) on room air and respiratory rate of 24 breaths/minute or more. Diagnostic criteria for severe disease included clinical signs of pneumonia plus one of the following: respiratory rate of more than 30 breaths/minute, severe respiratory distress, or oxygen saturation of less than 90% on room air.20
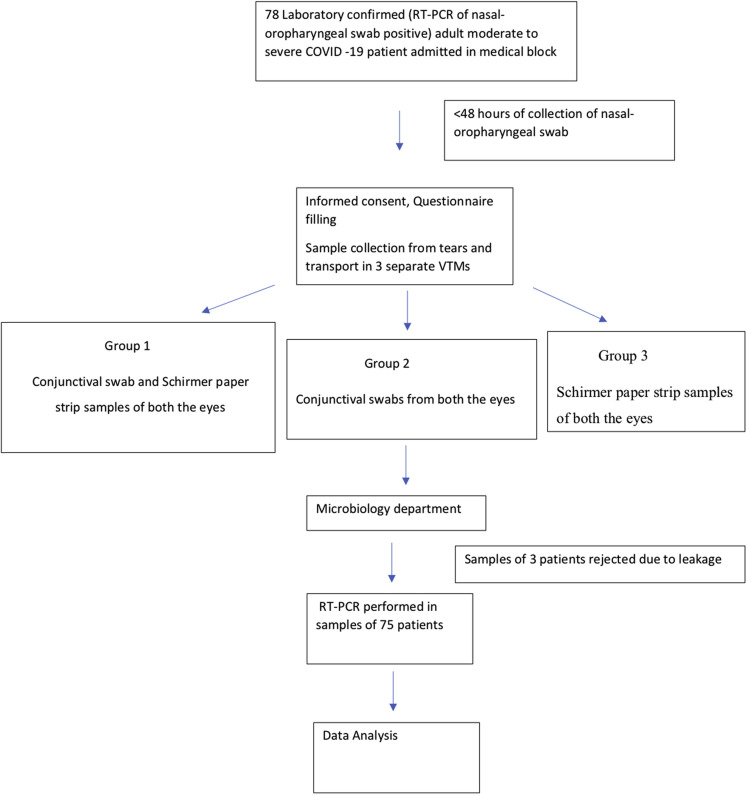
The ocular samples were taken wearing full personal protective equipment within 48 hours of collection of naso-oropharyngeal sample from both the eyes of each patient by ophthalmologists without topical anaesthesia. Tear samples were collected using conjunctival swabs and Schirmer paper strips (Fig 1 ). To obtain the conjunctival swab, the lower eye lid was retracted, the inferior fornix of the eye was swept with a sterile nylon swab for 10 seconds, and a similar procedure was repeated in the other eye.
Figure 1.
Flowchart depicting the study design. COVID-19 = coronavirus disease 2019; RT-PCR = reverse-transcriptase polymerase chain reaction; VTM = viral transport media.
When using the Schirmer paper strip (no. 41 Whatman filter paper, 5 × 35 mm), each strip was folded from one end and inserted at the junction of the middle and outer third of the lower lid of both the eyes. The patient was asked to keep the eyes open and blink normally, and after 3 minutes, the strips were removed. In group 1, the conjunctival swabs from both eyes and Schirmer strips from both the eyes were placed in a single VTM. In group 2, the conjunctival swabs from both eyes were placed in a single VTM. In group 3, the Schirmer strips from both the eyes were placed in a single VTM. A 2-minute gap was observed between each sampling. Further, each sealed and labelled VTM was placed in a falcon tube. The falcon tube was sealed and placed in a zip lock bag and transported to the microbiology department.
Microbiological Methods
All the samples were transported to the laboratory as soon as possible to maintain the cold chain. In case of delay, the sample was stored at 4° C, not beyond 3 days. After reaching the laboratory, the samples were processed immediately or stored at –20 °C until processing.21 , 22
The real-time RT-PCR assay used the TaqMan fluorogenic probe-based chemistry that used the 5′ nuclease activity of Taq DNA polymerase and enabled the detection of a specific RT-PCR product as it is accumulated during RT-PCR cycles. Coronaviruses under the subgenus Sarbecovirus, which includes the 2019 novel coronavirus, severe acute respiratory syndrome coronavirus, and bat severe acute respiratory syndrome-like coronaviruses, were used to generate a nonredundant alignment. Confirmatory assays were based on their matching to the Wuhan virus per inspection of the sequence alignment. Suspected tear samples were tested first for E gene assay and then for confirmation RNA-dependent RNA polymerase (RdRp), and the open reading frame 1b gene assay was used.
RNA Extraction
Viral RNA was extracted from all the samples (150 μl each) using the QIAamp RNA Minikit (Qiagen, Delhi, India) according to the appropriate protocols in the manufacturer’s instructions, with a final elution volume of 60 μl. Extracted RNA was stored at –20°C until required for RT-PCR analysis.
Real-Time Reverse-Transcriptase Polymerase Chain Reaction Analysis
RNA extracted from all the samples was tested for the presence of SARS-CoV-2 by real-time RT-PCR. Quant Studio Dx (Applied Biosystems, Foster City, CA) was used for performing the assay. The viral RNA extract and controls were amplified with primers and probes for screening for E gene and ribonuclease P as an internal control. Negative control (nuclease-free water was used as a template), positive control (Kits control), and MOCK (human source cell line) were included in each test run. Additionally, a specimen with known positive results was included as an internal control during the run. Cycle conditions and RT-PCR reagent preparation were carried out according to the Indian Council of Medical Research protocol. A specimen was considered presumptively positive for the 2019 novel coronavirus if the amplification curve for the E gene crossed the threshold line within 35 cycles. The viral load was assessed in terms of the cycle threshold (Ct) value of the E gene. Only those samples that showed positive results for E gene were tested for confirmatory RT-PCR by using Wuhan strain-specific primers of the viral RdRp gene and HKU-ORF sequence. A specimen was considered to have confirmed positive results for the 2019 novel coronavirus if the reaction growth curves crossed the threshold line within 35 cycles for E gene and both the RdRp gene and the ORF gene or either the RdRp gene or the ORF gene.
Statistical Methods
Paired chi-square tests were used to compare the proportion of patients with positive RT-PCR results in tears in patients with laboratory-confirmed moderate to severe COVID-19. The average Ct value of E gene among the 3 groups was compared using the analysis of variance, and the independent samples median test was used to compare the medians of the Ct values of E gene in tear samples with positive results in all 3 groups versus those with positive results in a single group. The analysis was performed using IBM SPSS statistical software version 25.0 (IBM, Inc, Chicago, IL).
Results
Of the 78 patients enrolled in the study, only 75 were considered for analysis because samples from 3 patients were found to be inadequate for analysis (Table 1 ). Thirty-six of the patients (48%) had moderate disease, whereas 39 patients (52%) had severe COVID-19. The cohort comprised 41 men (54.7%) and 34 women (45.3%) ranging in age from 18 to 81 years. Fifty-six patients (74.7%) had an associated systemic comorbidity. The various comorbidities included hypertension (42%), diabetes mellitus (41%), coronary artery disease (18%), acute or chronic kidney disease (14%), chronic lung disease or asthma (10%), hepatitis (2.6%), B-cell lymphoma (2.6%), anemia or thrombocytopenia (1.3%), rheumatic heart disease (1.3%), epilepsy (1.3%), pancreatitis (1.3%), and multiorgan dysfunction (1.3%). None of the patients showed ocular signs or symptoms.
Table 1.
Demographic Details, Clinical Characteristics, and Reverse-Transcriptase Polymerase Chain Reaction Results of Patients
| Patient No. | Age (yrs) | Gender | Severity | Systemic Symptoms | Duration (Days)∗ | Group 1 |
Group 2 |
Group 3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conjunctival Swab plus Schirmer’s Test Strips |
Conjunctival Swab |
Schirmer’s Test Strips |
|||||||||||||||
| Tear Results | Cyclic Threshold E Gene | Cyclic Threshold ORF1b | Cyclic Threshold RdRP | Tear Results | Cyclic Threshold E Gene | Cyclic Threshold ORF1b | Cyclic Threshold RdRP | Tear Results | Cyclic Threshold E Gene | Cyclic Threshold ORF1b | Cyclic Threshold RdRP | ||||||
| 1 | 73 | M | Moderate | Cough, fever, SOB, DM, HTN | 22 | Negative | Negative | Negative | |||||||||
| 2 | 78 | F | Moderate | Chest pain, cough, CAD, HTN, DM | 5 | Negative | Negative | Negative | |||||||||
| 3 | 68 | F | Severe | Fever, SOB, DM, AKI | 9 | Positive | 30 | 30 | ND | Negative | Negative | ||||||
| 4 | 78 | M | Severe | SOB, cough, COPD, HTN | 7 | Negative | Positive | 29 | 32 | ND | Negative | ||||||
| 5 | 63 | M | Moderate | Fever, sore throat, DM, HTN, CAD | 5 | Negative | Negative | Negative | |||||||||
| 5 | 60 | F | Moderate | Fever, cough, CLD | 6 | Negative | Negative | Negative | |||||||||
| 7 | 27 | M | Moderate | Fever, diarrhea | 8 | Negative | Negative | Negative | |||||||||
| 8 | 48 | F | Moderate | Sore throat | 5 | Negative | Negative | Negative | |||||||||
| 9 | 60 | F | Moderate | Chest pain, DM, HTN | 6 | Negative | Negative | Negative | |||||||||
| 10 | 60 | F | Moderate | Fever, headache, HTN, CKD | 5 | Negative | Negative | Negative | |||||||||
| 11 | 71 | M | Moderate | Generalized weakness, DM, HTN, CAD | 5 | Negative | Negative | Negative | |||||||||
| 12 | 32 | M | Severe | Cough, SOB, fever, asthma | 6 | Negative | Negative | Negative | |||||||||
| 13 | 67 | M | Severe | Cough, SOB, fever, DM, HTN, CAD | 7 | Negative | Negative | Negative | |||||||||
| 14 | 51 | F | Moderate | Diarrhea, vomiting, fever, HTN, depression | 7 | Negative | Negative | Negative | |||||||||
| 15 | 20 | M | Moderate | Fever, vomiting, acute viral hepatitis with pancreatitis | 5 | Negative | Negative | Negative | |||||||||
| 16 | 35 | f | Severe | DM, HTN, RHD (severe MS) | 4 | Negative | Negative | Negative | |||||||||
| 17 | 35 | F | Moderate | Fever, vomiting | 5 | Negative | Positive | 29 | 30 | ND | Negative | ||||||
| 18 | 66 | M | Moderate | Fever, vomiting, seizure disorder | 3 | Negative | Negative | Positive | 33 | 35 | ND | ||||||
| 19 | 30 | M | Severe | Cough, SOB | 5 | Negative | Negative | Negative | |||||||||
| 20 | 55 | M | Moderate | Fever, cough, SOB, DM, HT, active TB | 15 | Negative | Negative | Negative | |||||||||
| 21 | 44 | M | Moderate | Fever | 8 | Negative | Negative | Negative | |||||||||
| 22 | 18 | M | Moderate | Fever, vomiting, anemia, thrombocytopenia | 4 | Negative | Negative | Negative | |||||||||
| 23 | 61 | F | Severe | Fever, SOB, HTN | 5 | Negative | Negative | Negative | |||||||||
| 24 | 64 | M | Moderate | Cough, sore throat, DM, HTN, CAD | 8 | Negative | Negative | Negative | |||||||||
| 25 | 45 | M | Severe | SOB, body aches, dizziness, DM | 4 | Negative | Negative | Negative | |||||||||
| 26 | 60 | M | Severe | SOB, DM | 4 | Negative | Positive | 33 | 32 | ND | Negative | ||||||
| 27 | 66 | F | Severe | Cough, SOB, fever, DM, HTN | 7 | Negative | Negative | Negative | |||||||||
| 28 | 58 | M | Severe | Fever, cough, SOB, DM, HTN, CAD | 11 | Negative | Negative | Negative | |||||||||
| 29 | 73 | F | Severe | SOB, COPD | 4 | Negative | Negative | Negative | |||||||||
| 30 | 74 | M | Severe | SOB, fever, cough | 4 | Negative | Negative | Negative | |||||||||
| 31 | 62 | M | Severe | Fever, cough, SOB, DM | 8 | Positive | 22 | 22 | 23 | Positive | 23 | 23 | 22 | Positive | 28 | 27 | 27 |
| 32 | 21 | F | Severe | Cough | 5 | Negative | Negative | Negative | |||||||||
| 33 | 63 | M | Severe | SOB, fever, DM | 4 | Positive | 34 | 33 | ND | Negative | Negative | ||||||
| 34 | 23 | F | Moderate | SOB, fever, DM, HTN, COPD | 5 | Negative | Positive | 34 | ND | 32 | Negative | ||||||
| 35 | 60 | F | Severe | Cough, SOB, HTN, DM | 5 | Negative | Negative | Negative | |||||||||
| 36 | 52 | M | Moderate | Chest pain, HTN, CAD | 4 | Negative | Negative | Negative | |||||||||
| 37 | 68 | F | Severe | Fever, cough, CKD, DM, HTN | 8 | Positive | 25 | 26 | ND | Positive | 30 | 32 | ND | Positive | 35 | 35 | ND |
| 38 | 66 | M | Moderate | Cough, SOB, DM, HTN | 6 | Negative | Negative | Negative | |||||||||
| 39 | 80 | M | Moderate | SOB, MODS with AKI | 4 | Positive | 26 | 27 | ND | Positive | 29 | 28 | ND | Positive | 28 | 28 | ND |
| 40 | 55 | M | Moderate | Cough, fever, B-cell lymphoma | 5 | Positive | 14 | 16 | 18 | Positive | 15 | 16 | 18 | Positive | 15 | 16 | 18 |
| 41 | 33 | M | Severe | SOB, CKD | 4 | Positive | 32 | 32 | ND | Positive | 31 | 28 | ND | Positive | 26 | 27 | ND |
| 42 | 20 | M | Moderate | SOB, CKD | 4 | Negative | Negative | Negative | |||||||||
| 43 | 60 | F | Severe | Fever, cough, SOB, CKD | 5 | Negative | Negative | Negative | |||||||||
| 44 | 49 | F | Moderate | Fever, SOB | 5 | Positive | 34 | 34 | ND | Negative | Negative | ||||||
| 45 | 51 | F | Severe | Fever, SOB, vomiting, DM, CKD | 8 | Negative | Negative | Negative | |||||||||
| 46 | 34 | M | Moderate | Cough, sore throat, chest pain, DM, HTN, CAD | 4 | Negative | Negative | Negative | |||||||||
| 47 | 53 | F | Moderate | SOB, HTN, CKD | 8 | Negative | Negative | Negative | |||||||||
| 48 | 60 | F | Severe | SOB, cough, fever, asthma | 7 | Negative | Negative | Negative | |||||||||
| 49 | 47 | F | Severe | Diarrhea, HTN, CKD | 5 | Negative | Negative | Negative | |||||||||
| 50 | 78 | F | Moderate | Fever, DM, HTN, CAD | 7 | Negative | Negative | Negative | |||||||||
| 51 | 75 | F | Severe | Cough, SOB | 7 | Negative | Negative | Negative | |||||||||
| 52 | 58 | M | Severe | SOB, DM, CAD | 5 | Positive | 33 | 34 | ND | Negative | Negative | ||||||
| 53 | 40 | M | Moderate | Fever, SOB | 21 | Negative | Negative | Negative | |||||||||
| 54 | 73 | M | Severe | SOB, fever, smoker | 9 | Positive | 31 | 31 | ND | Negative | Negative | ||||||
| 55 | 62 | M | Moderate | SOB, Hodgkin`s lymphoma | 4 | Negative | Negative | Negative | |||||||||
| 56 | 44 | M | Severe | cough, SOB, DM | 6 | Negative | Negative | Negative | |||||||||
| 57 | 81 | F | Moderate | Fever, cough, SOB, DM, HTN, CAD | 14 | Negative | Negative | Negative | |||||||||
| 58 | 63 | F | Moderate | Fever, SOB, cough, DM, HTN | 8 | Negative | Negative | Negative | |||||||||
| 59 | 48 | M | Moderate | Cough, HTN, CKD | 3 | Negative | rejected | Negative | |||||||||
| 60 | 50 | F | Severe | Sore throat, fever, SOB | 5 | Negative | Negative | Negative | |||||||||
| 61 | 73 | F | Severe | Cough, SOB, HTN, CKD | 4 | Negative | Negative | Negative | |||||||||
| 62 | 57 | M | Moderate | Fever, HBs Ag positive | 4 | Negative | Negative | Negative | |||||||||
| 63 | 56 | M | Severe | Fever, cough, SOB, B/L pneumonitis | 11 | Negative | Negative | Negative | |||||||||
| 64 | 47 | M | Severe | SOB, cough, fever, CKD | 9 | Negative | Negative | Negative | |||||||||
| 65 | 60 | F | Severe | Cough, SOB, fever, COPD | 7 | Negative | rejected | Negative | |||||||||
| 66 | 48 | M | Severe | Fever, cough, SOB, DM, HTN, AKI | 8 | Negative | Negative | Negative | |||||||||
| 67 | 60 | M | Moderate | Fever, sore throat, cough, DM, HTN | 8 | Negative | Negative | Negative | |||||||||
| 68 | 62 | F | Severe | Fever, cough, SOB, HTN | 7 | Negative | Negative | Negative | |||||||||
| 69 | 80 | F | Severe | Fever, cough, SOB | 18 | Negative | Positive | 32 | 33 | ND | Negative | ||||||
| 70 | 62 | F | Moderate | Fever, cough, DM | 5 | Negative | Negative | Negative | |||||||||
| 71 | 72 | M | Severe | SOB, HTN, old TB | 7 | Negative | Negative | Negative | |||||||||
| 72 | 72 | F | Severe | Fever, cough, SOB | 8 | Negative | Negative | Negative | |||||||||
| 73 | 43 | M | Severe | Fever, cough, SOB, body aches | 8 | Negative | Negative | Negative | |||||||||
| 74 | 57 | M | Moderate | SOB, fever, sore throat, cough | 21 | rejected | rejected | rejected | |||||||||
| 75 | 57 | F | Severe | Cough, nasal discharge, HTN, CAD | 8 | Negative | Negative | Negative | |||||||||
| 76 | 60 | F | Severe | SOB, cough, fever, chest pain, abdominal pain, HTN | 9 | Positive | 31 | 30 | ND | Negative | Positive | 30 | 30 | ND | |||
| 77 | 60 | M | Moderate | Backache, DM, CAD, pulmonary HTN, CLD | 2 | Negative | Positive | 34 | 32 | ND | Negative | ||||||
| 78 | 35 | M | Moderate | Fever, cough, SOB | 14 | Negative | Negative | Negative | |||||||||
Ag = antigen; AKI = acute kidney injury; B/L = bilateral; CAD = coronary artery disease; CKD = chronic kidney disease; CLD = chronic liver disease; COPB = chronic obstructive pulmonary disease; DM = diabetes mellitus; F = female; HBs = hepatitis; HTN = hypertension; M = male; MS = mitral stenosis; ND = not detectable; RHD = rheumatic heart disease; SOB = shortness of breath; TB = tuberculosis.
Cyclic threshold value of E gene is mentioned only for those patients whose tear film sample showed positive results for severe acute respiratory syndrome coronavirus 2.
Duration from onset of symptoms until collection of sample.
Of 75 patients, RT-PCR showed positive results in the tears of 18 patients (24%). The mean age of patients with positive RT-PCR results in tears was 56.78 ± 16.79 years (P = 0.56). All except 3 of the patients with positive RT-PCR results in tears had associated systemic comorbidities, with 6 having more than 1 comorbidity. Twenty-nine of 225 samples (12.9%) from 75 patients showed positive results: 11 (14.7%) positive samples in group 1 (conjunctival swab plus Schirmer’s paper strip), 11 (14.7%) positive samples in group 2 (conjunctival swab), and 7 (9.3%) positive samples in group 3 (Schirmer’s paper strip; P = 0.3105).
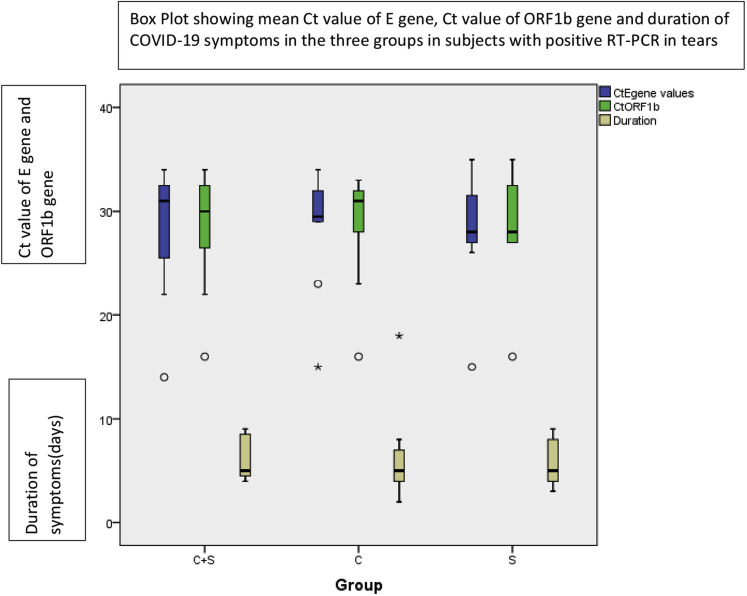
The mean Ct values of E gene in groups 1, 2, and 3 were 28.36 ± 6.15, 29.00 ± 5.58, and 27.86 ± 6.46 (P = 0.92; Fig 2 ). Five patients showed positive RT-PCR results by all the 3 methods. The mean Ct values of E gene of these patients was 25.24 ± 6.33 (Fig S1, available at www.aaojournal.org). A Ct value of E gene of more than 26 was observed in 9 of 11 samples with positive results (P = 0.00; κ = 0.27) in groups 1 and 2 and in 5 of 7 samples with positive results (P = 0.000; κ = 0.42) in group 3. In 12 patients, positive results were obtained by either of the 3 methods (6 patients in group 2, 5 patients in group 1, and 1 patient in group 3), the mean Ct value being 32.16 ± 1.94. The difference in Ct values in patients with positive results in all 3 groups as compared with those with positive results in a single group (conjunctival swab plus Schirmer strip, conjunctival swab, or Schirmer strip) was statistically significant (P = 0.029). In only 1 patient did samples show positive results in 2 groups (conjunctival swab plus Schirmer strip and Schirmer strip; Table 2 ). The median value of the number of days from onset of symptoms to collection of tear samples in patients with positive RT-PCR results in tears was 5 days (range, 4–9 days; Fig 2).
Figure 2.
Boxplot showing the cyclic threshold (Ct) value of E gene, Ct value of ORF1b gene, and duration of coronavirus disease 2019 symptoms in the 3 groups in patients with positive reverse-transcriptase polymerase chain reaction results in tears.
Table 2.
Relationship of Mean Cyclic Threshold Value of E Gene with Positive Results on Reverse-Transcriptase Polymerase Chain Reaction in Tears in Different Groups
| Method Showing Positive Tear Sample Results | No. of patients | Mean Cyclic Threshold Value (E Gene) |
|---|---|---|
| All 3 methods | 5 | 25.24 |
| Single method | 12 | 32 |
| Two methods | 1 | 30.5 |
Discussion
The role of the ocular surface as a possible portal of entry, reservoir for replication, and transmission of SARS-CoV-2 RNA has been explored extensively.9 , 23, 24, 25 Differences in detection of viral RNA in tears have ranged from 0% to 7%, with some researchers claiming minimal viral shedding in ocular secretions.13, 14, 15, 16, 17, 18, 19 Recently, live virus has been demonstrated in ocular fluids by demonstrating cytopathic effect in Vero E6 cells.26 Although initial reports included patients with asymptomatic, mild, and clinically suspected COVID-19, it was noted that both patients showing positive RT-PCR results in tears had critical cases of COVID-19.15
Sample collection has been described using conjunctival swabs and Schirmer paper strips.3 , 13, 14, 15 , 17, 18, 19 To the best of our knowledge, in the published literature, either the precorneal tear film of a single eye was tested, or in the case of bilateral tear collection, the samples were transferred in separate VTMs from each eye. The diagnostic sensitivity of the precorneal tear film could be marred by the insufficient sample volume.27
To overcome the lack of uniformity in the study populations in the available studies, we exclusively included adults with moderate and severe COVID-19 who showed positive results on naso-oropharyngeal RT-PCR analysis. The quantity of ocular samples was increased by inoculation of tears from both the eyes in a single VTM. The viral load is known to fall during the second and third week of symptoms23; therefore, the ocular sampling was performed within 48 hours of confirmatory naso-oropharyngeal swab. Conjunctival swab has been considered the gold standard for tear collection and evaluation of viral RNA. To determine the best method for collection of tears, we used the methods described previously, namely, conjunctival swab (group 2) and Schirmer paper strips (group 3), along with a combination of conjunctival swab with Schirmer test strips (group 1).
In our study, 18 of 75 patients (24%) showed positive RT-PCR results in tears by at least one of the methods. The closest to this outcome is a report from Iran, in which tear samples with positive results were found in 3 of 30 (10%) patients with severe, laboratory-confirmed COVID-19. The additional 13 patients in their study showed negative nasopharyngeal swab results.18 In a retrospective analysis of 38 patients from Hubei, only 28 patients showed positive RT-PCR results from a nasopharyngeal swab, 4 of whom had moderate, 2 of whom had severe, and 6 of whom had critical COVID-19. Tears showed positive results for viral RNA in 2 patients with critical COVID-19 (7%).15 In 121 patients from a study in Wuhan, only 3 (2.4%) showed positive results in tears; 52.1% of patients had mild to moderate disease and 47.9% of patients had severe or critical laboratory-confirmed COVID-19.14 Only a single patient showed positive conjunctival swab results of 45 (2.23%) in an Indian study. However, these patients included 14 with asymptomatic disease, including the patient with positive RT-PCR results in tears.19
Seah et al3 carried out consecutive sampling in 17 patients but did not detect viral RNA in any of the tear samples. However, 52 of 64 samples in their study were collected in the second and third week of onset of symptoms. In a cross-sectional study by Zhang et al,28 of 102 COVID-19 patients, only 1 showed tear positivity. Their average time of sample collection was 18.15 days, ranging from 6 to 46 days. In our series, the tear samples showing positive results had been collected from 4th to the 9th day, with the median being 5th day from onset of symptoms.
Ocular manifestations of COVID-19 have been observed along with other clinical features. Patients also have demonstrated solely conjunctivitis and keratoconjunctivitis.29 , 30 Zhou et al14 reported ocular symptoms in 8 of 121 patients (6.6%), in the form of itching (n = 5; 62.5%), redness (n = 3; 37.5%), tearing (n = 3; 37.5%), discharge (n = 2; 25%), and foreign body sensation (n = 2; 25%). Of the 3 patients with positive RT-PCR results in tears, only 1 showed ocular symptoms; the remaining 2 were asymptomatic. In another study, 12 of 38 patients showed ocular involvement in the form of conjunctival hyperemia, chemosis, epiphora, and increased secretions, of whom 2 showed positive results in tears.15 Karimi et al,18 in their series of 43 patients, observed that of the 3 patients with positive tear sample results, 1 had conjunctivitis, 1 had foreign body sensation, and the third was asymptomatic. Citing a low viral load in noninflamed tissues, ocular involvement has been suggested as a prerequisite for viral shedding in tears.13 The absence of ocular signs and symptoms in any of patients with positive tear results in our series and also in other studies implies that viral shedding in tears is not always related to ocular inflammation, as proposed previously.14 , 18 , 19 Collection of tears and analysis of viral RNA from both eyes probably led to increased yield, although no ocular abnormalities were found in any of our cases.
Most of the researchers have used conjunctival swabs for tear collection,13, 14, 15 , 17, 18, 19 except Seah et al,3 who used Schirmer paper strips. In our study, an equal number of samples showed positive results in groups 1 and 2. The number of samples with positive results in group 3 was less than that in groups 1 and 2, although this finding was statistically insignificant. This implied that use of conjunctival swabs alone is justified for tear collection.
The viral load in tears can be assessed indirectly by the Ct value of the E gene. It was observed that if the Ct value of the E gene was less, the viral RNA could be detected by all the 3 methods. Probably in patients with severe viremia early in the course of the disease, high viral shedding in tears occurs, reflected by low Ct value of E gene and detection by all the 3 methods.
Also, in patients in whom only a single sample from any one group showed positive results, the Ct value was more than 32, and except for a single patient (patient 18), all such patients were in groups 1 or 2. This indicated that the viral load in the sample collected by the Schirmer test strip (group 3) was less and also that the ability to detect a sample with a lesser viral load is more with the conjunctival swab alone (group 2) or in combination with Schirmer test strips (group 1). To our knowledge, none of the previous studies have given the Ct value of positive tear samples until now.
No statistical correlation of viral shedding in tears with patient age or systemic comorbidity was found in the present study. The efficiency of real-time RT-PCR analysis depends on adequate amounts of viral RNA in the collected sample. A high prevalence of 24% in our study could be attributed to various factors that led to a higher viral load in the analyzed specimen. A consistent study population of patients with moderate to severe laboratory-confirmed COVID-19, collection of tear samples in the early course of disease, and inoculation of samples from both the eyes in a single VTM could have contributed to the increase in detection rates.
The limitations of this study were the noninclusion of patients with mild and asymptomatic COVID-19, the small sample size, and the 1-time sampling. We could not correlate the Ct values of nasopharyngeal and tear samples because of the nonavailability of Ct values from nasopharyngeal swabs of patients, because confirmatory nasopharyngeal real-time RT-PCR analysis had been carried out in many patients in another laboratory 1 day before admission to our institution.
To conclude, conjunctival swab is a satisfactory method of tear collection for assessment of the presence of SARS-CoV-2 by real-time RT-PCR analysis. The respiratory tract is not the only transmission route, and considerable viral shedding occurs in the precorneal tear film in patients with moderate to severe COVID-19, thus implying that besides N95 respirators, use of goggles and face shields by healthcare workers should be mandatory when interacting with patients with moderate to severe COVID-19 to reduce the transmission of SARS-CoV-2.
Manuscript no. D-20-01746.
Footnotes
Supplemental material available atwww.aaojournal.org.
Disclosure(s): All authors have completed and submitted the ICMJE disclosures form. The author(s) have no proprietary or commercial interest in any materials discussed in this article.
HUMAN SUBJECTS: Human subjects were included in this study. After obtaining the institutional ethics committee clearance at Maulana Azad Medical College, the trial was registered (CTRI/2020/05/025291). In accordance with the Declaration of Helsinki, written informed consent was obtained from all the patients.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Arora, Goel
Analysis and interpretation: Arora, Goel, Saxena, Manchanda
Data collection: Arora, Goel, Kumar, Chhabra, Saxena, Manchanda, Pumma
Obtained funding: N/A; Study was performed as part of the authors' regular employment duties. No additional funding was provided.
Overall responsibility: Arora, Goel
Supplementary Data
References
- 1.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 2.Liang L., Wu P. There may be virus in conjunctival secretion of patients with COVID-19. Acta Ophthalmol. 2020;98(3):223. doi: 10.1111/aos.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seah I.Y.J., Anderson D.E., Kang A.E.Z., et al. Assessing viral shedding and infectivity of tears in coronavirus disease 2019 (COVID-19) patients. Ophthalmology. 2020;127(7):977–979. doi: 10.1016/j.ophtha.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadhu S., Agrawal R., Pyare R., et al. COVID-19: limiting the risks for eye care professionals. Ocul Immunol Inflamm. 2020;28(5):714–720. doi: 10.1080/09273948.2020.1755442. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L., Xu Z., Castiglione G.M., et al. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf. 2020;18(4):537–544. doi: 10.1016/j.jtos.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma D., Chen C.B., Jhanji V., et al. Expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in human primary conjunctival and pterygium cell lines and in mouse cornea. Eye (Lond) 2020;34(7):1212–1219. doi: 10.1038/s41433-020-0939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seah I., Agrawal R. Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul Immunol Inflamm. 2020;28(3):391–395. doi: 10.1080/09273948.2020.1738501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia J., Tong J., Liu M., et al. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J Med Virol. 2020;92(6):589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Duan C., Zeng Y., et al. Ocular findings and proportion with conjunctival SARS-COV-2 in COVID-19 patients. Ophthalmology. 2020;127(7):982–983. doi: 10.1016/j.ophtha.2020.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P., Duan F., Luo C., et al. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020;138(5):575–578. doi: 10.1001/jamaophthalmol.2020.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun C., Wang Y., Liu G., Liu Z. Role of the eye in transmitting human coronavirus: what we know and what we do not know. Front Public Health. 2020;8:155. doi: 10.3389/fpubh.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Zeng Y., Tong Y., Chen C. Ophthalmologic evidence against the interpersonal transmission of 2019 novel coronavirus through conjunctiva. medRxi. 2020 Available at: www.medrxiv.org; Accessed 10.06.20. [Google Scholar]
- 18.Karimi S., Arabi A., Shahraki T., Safi S. Detection of severe acute respiratory syndrome coronavirus-2 in the tears of patients with coronavirus disease 2019. Eye. 2020;34(7):1220–1223. doi: 10.1038/s41433-020-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar K., Prakash A.A., Gangasagara S.B., et al. Presence of viral RNA of SARS-CoV-2 in conjunctival swab specimens of COVID-19 patients. Indian J Ophthalmol. 2020;68:1015–1017. doi: 10.4103/ijo.IJO_1287_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Directorate General of Health Services (EMR Division), Ministry of Health and Family Welfare, Government of India. Revised guidelines on clinical management of COVID-19. https://www.mohfw.gov.in/pdf/RevisedNationalClinicalManagementGuidelineforCOVID1931032020.pdf March 31, 2020. Accessed 10.06.20.
- 21.ICMR-National Institute of Virology (ICMR-NIV), Pune Standard operating procedure for detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by rRT-PCR: first-line screening assay. https://www.icmr.gov.in/pdf/covid/labs/1_SOP_for_First_Line_Screening_Assay_for_2019_nCoV.pdf Accessed 10.06.20.
- 22.ICMR-National Institute of Virology (ICMR-NIV), Pune Standard operating procedure for detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by rRT-PCR: confirmation assay. https://www.icmr.gov.in/pdf/covid/labs/2_SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf Accessed 10.06.20.
- 23.Zou L., Ruan F., Huang M., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napoli P.E., Nioi M., d’Aloja E., Fossarello M. The ocular surface and the coronavirus disease 2019: does a dual ‘ocular route’ Exist? J Clin Med. 2020;9(5):1269. doi: 10.3390/jcm9051269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C.W., Liu X.F., Jia Z.F. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395(10224):e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colavita F., Lapa D., Carletti F., et al. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann Intern Med. 2020;173(3):242–243. doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seittzman G.D., Doan T. No time for tears. Ophthalmology. 2020;127(7):980–981. doi: 10.1016/j.ophtha.2020.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Chen X., Chen L., et al. The evidence of SARS-CoV-2 infection on ocular surface. Ocul Surf. 2020;18:360–362. doi: 10.1016/j.jtos.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheema M., Aghazadeh H., Nazarali S., et al. Keratoconjunctivitis as the initial medical presentation of the novel coronavirus disease 2019 (COVID-19) Can J Ophthalmol. 2020;55(4):e125–e129. doi: 10.1016/j.jcjo.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daruich A., Martin D., Bremond-Gignac D. Ocular manifestation as first sign of coronavirus disease 2019 (COVID-19): interest of telemedicine during the pandemic context. J Fr Ophthalmol. 2020;43(5):389–391. doi: 10.1016/j.jfo.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.