Abstract
Chloride channel calcium-activated (CLCA) genes encode regulators for chloride transport across the cell membrane. As for cancer development, some CLCA genes are considered putative tumor suppressor genes. The aim of this study was to explore whether CLCA4 gene would have mutations in its nucleotide repeats in colorectal cancer (CRC). In a public database, we found that CLCA4 gene had mononucleotide repeats in the coding sequences that might be mutational targets in the cancers with microsatellite instability. For this, the current study studied 146 CRCs for mutation and expression analyses by single-strand conformation polymorphism analysis, DNA sequencing, and immunohistochemistry. Overall, we found CLCA4 frameshift mutations in 12/101 (11.8%) CRCs with high-microsatellite instability (MSI-H), but none in microsatellite stable CRCs (0/45) (P<0.01). In addition, we analyzed intratumoral heterogeneity of the CLCA4 frameshift mutations and found that 1 CRC harbored regional intratumoral heterogeneity of the CLCA4 frameshift mutation. Loss of CLCA4 protein expression was identified in 50% of CRCs. Also, cancers with MSI-H harboring CLCA4 frameshift mutations showed lower CLCA4 immunostaining than those with the wild-type. Our data indicate that the CLCA4 gene harbors alterations both in somatic mutation and expression, suggesting their roles in tumorigenesis of CRC with MSI-H.
Key Words: CLCA4, calcium-activated chloride channel, frameshift mutation, colon cancer, microsatellite instability, heterogeneity, expression
Chloride channel calcium-activated (CLCA) family proteins regulate the transport of chloride across the plasma membrane.1–3 There are 3 subtypes of human CLCAs (CLCA1, CLCA2, and CLCA4), all of which map to chromosome 1p31-p22 and share high degrees of homology in sequence and functions (calcium-activated chloride conductance), but differ significantly in their tissue distributions,1–3 CLCA2 expression is consistently lost in breast cancers and reintroduction of the CLCA2 abrogates cellular invasiveness and tumorigenicity.2,4 CLCA4 is widely expressed in gastrointestinal tissues (stomach, small intestine, and colon).1,5 CLCA4 expression is downregulated in colorectal cancer (CRC), breast cancer and head/neck cancer.5,6 Functionally, CLCA4 is stress-inducible and its overexpression inhibits colony formation of breast cancer cells.7 Also, loss of CLCA4 increases epithelial-mesenchymal transition in cancer cells.7 These data, together, suggest that CLCA inactivation is involved in cancer pathogenesis by inhibiting their tumor suppressor gene (TSG) activities.
CLCA4 mutation has been observed in many cancers at low prevalence (for example, 0.44% of CRC) (http://intogen.org/).8,9 Most of the CLCA4 mutations (77%) in the database are missense mutations and there is only 1 truncating mutation reported. Approximately 10% to 30% of CRCs exhibit defects in mismatch repair that can result in microsatellite instability (MSI).10 In a public genome database (http://genome.cse.ucsc.edu/), we found that human CLCA4 had mononucleotide repeats in the coding sequences that could be targeted for a frameshift mutation in cancers with MSI. Frameshift mutation of genes containing mononucleotide repeats is a feature of CRC with MSI.10 Tumor development begins through clonal expansion of a single cell. The resulting cell population usually becomes heterogeneous after branching sub-clonal expansions, which leads to intratumor heterogeneity (ITH).11–13 In this study, we analyzed mononucleotide repeats in the coding sequences in CLCA4 gene for the detection of somatic frameshift mutations and the mutational ITH in CRCs.
MATERIALS AND METHODS
Tissue Samples and Microdissection
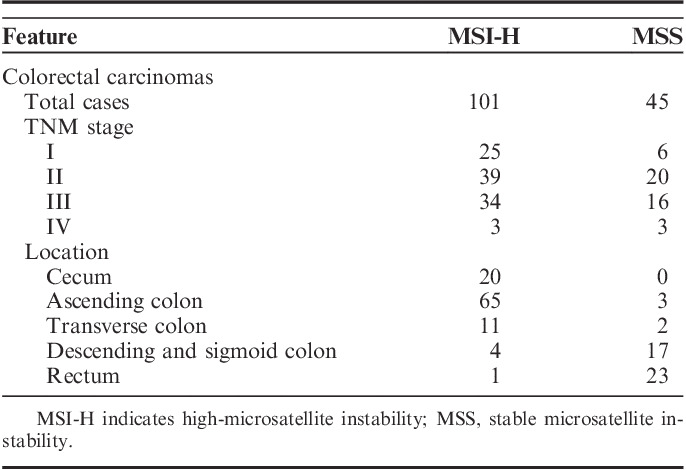
For the mutation analysis, methacarn-fixed tissues of 146 sporadic CRCs were used in this study. All of the patients were Koreans. The CRCs consisted of 101 CRCs with high microsatellite instability (MSI-H) and 45 CRCs with microsatellite stable (MSS). Normal tissues of the CRCs were prepared from normal mucosa of the matched patients. MSI status of each CRC was evaluated by the Promega system, which used 5 mononucleotide repeats (BAT25, BAT26, NR-21, NR-24, and MONO-27) using fluorescent-labeled polymerase chain reaction (PCR) and capillary electrophoresis (ABI Genetic Analyzer analysis). The tumoral MSI status was characterized as MSI-H, if ≥2 of these markers show instability, MSI-L, if one of the markers shows instability and MSS if none of the markers shows instability.14 For 54 CRCs of the total 129 CRCs, we collected 4 to 7 different tumor areas and 1 normal mucosal area from each frozen CRC specimen. Of the 54 CRCs, 16 CRCs with MSI-H were analyzed for the detection of regional ITH of the CLCA4 gene repeats. The tumor areas were 0.027 to 1 cm3 and at least 1.0 cm apart from each other. To confirm that these multi-regional biopsies were all areas of carcinoma (as opposed to areas of normal or dysplasia), they were frozen, stained with hematoxylin and eosin and examined under a light microscope. The tumor cell purities of these tissues were at least 70%.
Pathologic features of the cancers are summarized in Table 1. The histologic features of CRC with MSI-H, including mucinous histology, tumor-infiltrating lymphocytes, medullary pattern, and Crohn like inflammation, were evaluated in all blocks of all cases by a pathologist. Malignant cells and normal cells were selectively procured from hematoxylin and eosin-stained slides using a 30G1/2 hypodermic needle by microdissection as described previously.15,16 The DNA extraction was performed by a modified single-step DNA extraction method by proteinase K treatment.15,16 Briefly, the cells obtained in 20 µL of DNA extraction buffer were incubated at 52°C for 1 or 2 days. The mixture was boiled for 10 minutes to inactivate the proteinase K, and 1 µL of this solution was used as DNA template for PCR amplification. Approval of this study was obtained from the Catholic University of Korea, College of Medicine’s institutional review board for this study.
TABLE 1.
Summary of Pathologic Features of the Gastric and Colorectal Cancers
Single-Strand Conformation Polymorphism (SSCP) Analysis
We analyzed mononucleotide repeats of CLCA4 in their coding sequences (1 T8 repeat in exon 2 and 1 A8 repeat in exon 4). Genomic DNA from the microdissected cells was isolated and was amplified by PCR with specific primer pairs for exon 2 (forward and reverse, respectively: 5′-TCTAACTTCACCATCCTCACA-3′ and 5′-TTTGGCCTTTTGTAC TGAGG-3′) and exon 4 (5′-CTGTTTGTCCATGAGTGGGC-3′ and 5′-ATGGATAAAAGCCTGAAGTCAGTT-3′). Radioisotope ([32P]dCTP) was incorporated into the PCR products for detection by autoradiogram. After SSCP, mobility shifts on the SSCP gels (FMC Mutation Detection Enhancement system; Intermountain Scientific, Kaysville, UT) were determined by visual inspection. Direct DNA sequencing reactions in both forward and reverse sequences were performed in the cancers with the mobility shifts in the SSCP using a capillary automatic sequencer (3730 DNA Analyzer; Applied Biosystem, Carlsbad, CA). When mutations in the CLCA4 gene were suspected by SSCP, Sanger sequencing analysis of an independently isolated DNA from another tissue section of the same patients was performed to exclude potential artifacts originated from PCR.
Immunohistochemistry
Immunohistochemistry for CLCA4 was performed in tissue sections of the 101 CRCs with MSI-H and the 45 CRCs with MSS. We adopted the ImmPRESS System (Vector Laboratories, Burlingame, CA) using commercial antibody against a region of human CLCA4 (amino acids 306-476) (dilution 1/12; Thermo Fisher Scientific, Rockford, IL). After deparaffinization, heat-induced epitope retrieval was conducted by immersing the slides in Coplin jars filled with 10 mmol/L citrate buffer (pH 6.0) and boiling the buffer for 30 minutes in a pressure cooker (Nordic Ware, Minneapolis, MN) inside a microwave oven at 700 W; the jars were then cooled for 20 minutes. Immunohistochemistry intensity was graded as follows: 0, negative; 1+ when the cells showed weak staining in the cytoplasm; 2+, moderate; and 3+, intense. The extent was graded as follows: 0, 0% to 5%; 1, 6% to 15%; 2, 16% to 49%; 3, >50%. These 2 grades were then multiplied to generate the immunohistochemistry score (IS); IS 0 to 2 as negative and 3 or 4 or 6 or 9 as positive. The immunostaining was judged to be specific by the absence of consistent immunostaining of cells with the replacement of primary antibody with blocking reagent and reduction of immunostaining intensity as a dilution of the antibody was increased. The immunohistochemistry procedures in detail were described previously.17
RESULTS
Somatic Mutations of CLCA4
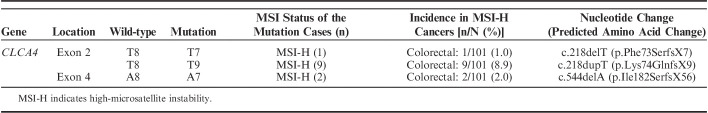
Genomic DNAs of 146 CRCs (101 with MSI-H and 45 with MSS) were studied to detect frameshift mutations within the T8 and A8 nucleotide repeats of CLCA4. In SSCP, we observed aberrant bands in 12 of the 146 CRCs (Table 2, Fig. 1). Normal tissues of matched patients exhibited no aberrant migration in SSCP, indicating that the aberrant bands had somatically risen (Fig. 1). Sanger sequencing confirmed somatic mutation of the CLCA4 gene (Fig. 1), which consisted of a deletion and a duplication mutation in the T8 and another deletion mutation in the A8 that would lead to termination of CLCA4 protein translation (Table 2). The CLCA4 mutation was found in MSI-H CRCs (12/101, 11.8%), but there was not any in MSS CRCs (0/45) (Table 2) (Fisher exact test, P=0.01). All of the CLCA4 mutations were heterozygous frameshift mutations within the repeats (Fig. 1). In the cancers with MSI-H, there was no correlation between the features of the tumors (stages, histologic grade, subtypes, mucinous histology, medullary pattern, and tumor-infiltrating lymphocytes) and presence of the mutations (P>0.05). We did not analyze the differences in patient survival between MSI-H with and without CLCA4 mutation, because survival data of many of the patients were inappropriate for the analysis.
TABLE 2.
Summary of CLCA4 Mutations in Colorectal Cancers
FIGURE 1.
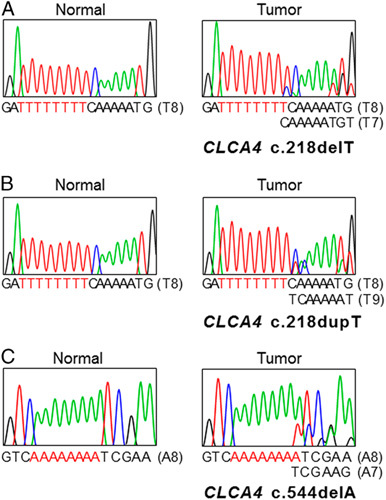
Representative single-strand conformation polymorphism and DNA sequencings of repeats in CLCA4 in colon cancers. DNA sequencing analyses (A–C) of CLCA4 from the tumor (lane T) and normal tissues (lane N). Direct DNA sequencing analyses from T8 repeat in exon 2 (A), T8 repeat in exon 2 (B) and A8 repeat in exon 4 (C) of CLCA4 show heterozygous deletion (A, C) or duplication (B) of a base in tumor tissues as compared with normal tissues.
From 96 regional fragments of 16 CRCs (4 to 7 fragments per case) with MSI-H were collected and analyzed with respect to their regional status of CLCA4 frameshift mutations in the repeats. One of the 16 CRCs (6.3%) harbored ITH of the mutations. The CRC case (#3) showed the CLCA4 deletion mutation (T8) in 1 (#3-5) of 6 regions (Table 3, Fig. 2).
TABLE 3.
Summary of CLCA4 Expression and Frameshift Mutation in Colorectal Cancers
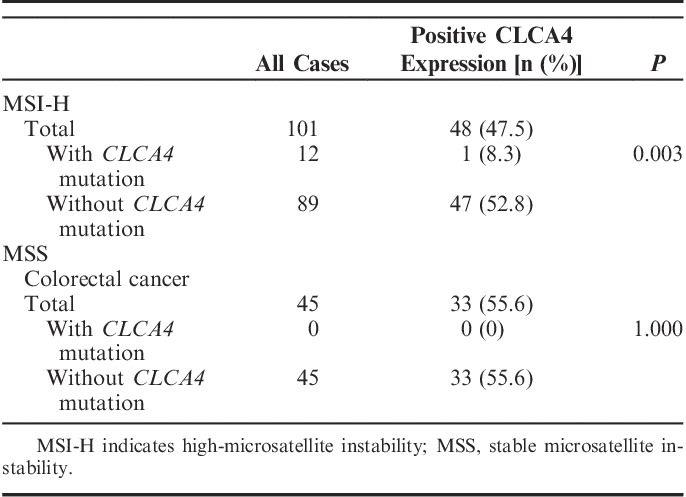
FIGURE 2.
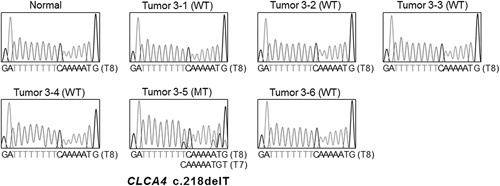
Intratumoral heterogeneity of a CLCA4 frameshift mutation in colon cancer. Direct DNA sequencings show CLCA4 c.218delT mutation (MT) in a region (3-5) and wild-type (WT) CLCA4 in the other 5 regions (3-1, 3-2, 3-3, 3-4, and 3-6).
Immunohistochemistry
The CLCA4 protein expression was analyzed in the CRCs by immunohistochemistry (Fig. 3, Table 3). The CLCA4 immunostaining was observed mainly in the cytoplasm (Fig. 3). In normal colonic mucosal epithelial cells, CLCA4 immunostaining was observed in the cytoplasm (moderate to intense intensities: IS 6 or 9, Fig. 3A) as well. The CLCA4 immunostaining (weak to moderate intensities: IS 2 to 6) was positive in 73 (50.0%) of 146 CRCs (Table 3, Fig. 3). CLCA4 expression in MSI-H CRCs (48/101) was not significantly different from that in MSS CRCs (25/45) (Fisher exact test, P<0.05). Eleven of 12 CRCs with the CLCA4 frameshift mutation were negative for CLCA4 immunostaining (Fig. 2). CRCs with MSI-H harboring CLCA4 frameshift mutations showed lower CLCA4 immunostaining than those with the wild-type (Table 3) (Fisher exact test, P=0.003). CRCs with the CLCA4 frameshift mutations (1/12) showed lower CLCA4 immunostaining than those without the mutations (72/134) (P=0.002).
FIGURE 3.
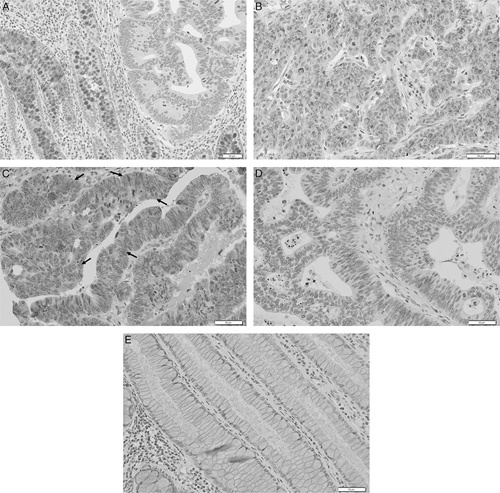
Visualization of CLCA4 expression in colorectal cancer tissues by immunohistochemistry. A, Mucosal epithelial cells of normal colonic glans (left) show positive CLCA4 immunostaining, whereas cancer cells in the neoplastic glands show negative immunostaining (right). B and C, Colon cancers shows positive CLCA4 immunostaining in the cancer cells (arrows for positive cells in C). D, Another colon cancer shows negative CLCA4 immunostaining in the cancer cells. E, In a lower concentration of anti-CLCA4 antibody (1/400), the normal mucosal cells show negative CLCA4 immunostaining.
DISCUSSION
Although TSG functions of CLCA4 have been identified, genetic alterations of CLCA4 gene are known to be rare in many cancers (http://intogen.org/). However, it is still possible that genetic alterations of CLCA4 gene could be limited to a specific subtype of human cancers such as those with MSI-H. The aim of this study was 2-fold. First, we attempted to find frameshift mutations in CLCA4 gene, which could inactivate the TSG functions of CLCA4. We found frameshift mutations of CLCA4 in about 12% of MSI-H CRCs. We identified a significant difference of CLCA4 mutation incidence between the CRCs with MSI-H and MSS, indicating that the mutations were MSI-H-specific. In addition, our study found that CLCA4 frameshift mutation harbored ITH in CRCs. Second, we analyzed CLCA4 protein expression in CRC tissues and found a significantly lower expression in CRCs with the CLCA4 frameshift mutations than those without the mutations. Together, our data may indicate that CLCA4 gene is altered in CRCs by both somatic frameshift mutation/ITH in the mononucleotide repeats and expression loss of CLCA4.
CLCA4 mutation is found in many cancers at low incidences, but it became a significantly mutated gene by the Pan-Cancer analysis.8 In the present study, we found that 11.8% of MSI-H CRCs carried the CLCA4 frameshift mutation in the repeats, but not in the MSS cases. None of these frameshift mutations have been detected in other cancers (http://intogen.org). Together with the Pan-Cancer analysis data, our data may indicate that CLCA4 frameshift mutation rarely occurs in human cancers including MSS CRCs, but may commonly occur in MSI-H CRCs. CLCA4 protein (full-length: 919 amino acids) is comprised of a signal peptide (residues 1-21), a metalloprotease domain (45-199), a von Willebrand factor type A domain (305-459), a cleavage site (697-698) and a transmembrane domain (895-915).18 In the present study, we observed 3 types of CLCA4 frameshift mutations (Table 2) that would delete amino acids after the 73rd or 74th or 182nd residue and remove most of the functional domains in the CLCA4. The frameshift mutations would truncate and disable the CLCA4 protein and hence be considered a loss-of-function mutation. In CRC, Yang et al5 found that RNA expression level of CLCA4 was significantly reduced in 41% of CRCs compared with the normal colonic mucosa. Functionally, CLCA4 inhibits cell proliferation and epithelial-mesenchymal transition.7 Our study is in agreement with these studies.5,7 that showed findings consistent with the TSG functions of CLCA4. Together with these, our data suggest that these mechanisms might play a role in tumorigenesis of MSI-H CRC by inhibiting TSG functions of CLCA4.
CLCA4 immunostaining was largely negative (11/12, 91.7%) in CRCs harboring the CLCA4 frameshift mutations (Table 3). As anti-CLCA4 antibody used in this study was developed by immunizing a peptide of amino acids 306-476 in CLCA4, which would be removed by the mutations (p.Phe73SerfsX7, p.Lys74GlnfsX9, and p.Ile182SerfsX56). Hence, the mutant CLCA4 might not be detected by the antibody. Both DNA sequencing and SSCP revealed that the CLCA4 mutation in all cases was heterozygous (Fig. 1), indicating the presence of the second allele. Negative CLCA4 immunostaining in the cases carrying the frameshift mutation might be caused by the mutation in 1 allele and by other mechanisms in the second allele such as epigenetic silencing.
In the present study, we found ITH of CLCA4 frameshift mutation (6.3%), which is in agreement with earlier reports showing genetic ITH for noncoding microsatellite markers as well as repeat sequences within coding genes.19 However, it is reasonable to conceive that other regions of this gene, as well as other genes, could harbor mutational ITH in the CRCs. ITH in the cancers may have implications for biomarker strategies. For example, low-level mutations with a metastasis potential may define clinical outcomes as the clones with ITH easily achieve clonal dominance during the progression and affect treatment efficacy.20 However, probably due to the small number of the ITH case, we did not find any significant difference and further analysis with a large cohort is required for this in future studies.
Footnotes
Supported by grants from Korea Research Foundation (2012R1A5A2047939 and 2017R1A2B2002314).
The authors declare no conflict of interest.
REFERENCES
- 1.Agnel M, Vermat T, Culouscou JM. Identification of three novel members of the calcium-dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett. 1999;455:295–301. [DOI] [PubMed] [Google Scholar]
- 2.Pauli BU, Abdel-Ghany M, Cheng HC, et al. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol. 2000;27:901–905. [DOI] [PubMed] [Google Scholar]
- 3.Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005;85:1061–1092. [DOI] [PubMed] [Google Scholar]
- 4.Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999;59:5488–5491. [PubMed] [Google Scholar]
- 5.Yang B, Cao L, Liu B, et al. The transition from proliferation to differentiation in colorectal cancer is regulated by the calcium activated chloride channel A1. PLoS One. 2013;8:e60861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye H, Yu T, Temam S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Walia V, Elble RC. Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS One. 2013;8:e83943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. [DOI] [PubMed] [Google Scholar]
- 11.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. [DOI] [PubMed] [Google Scholar]
- 12.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. [DOI] [PubMed] [Google Scholar]
- 13.Kim TM, Jung SH, Baek IP, et al. Regional biases in mutation screening due to intratumoural heterogeneity of prostate cancer. J Pathol. 2014;233:425–435. [DOI] [PubMed] [Google Scholar]
- 14.Murphy K, Zhang S, Geiger T, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi EJ, Kim MS, Song SY, et al. Intratumoral heterogeneity of frameshift mutations in MECOM gene is frequent in colorectal cancers with high microsatellite instability. Pathol Oncol Res. 2017;23:145–149. [DOI] [PubMed] [Google Scholar]
- 16.Jo YS, Choi MR, Song SY, et al. Frameshift mutations of HSPA4 and MED13 in gastric and colorectal cancers. Pathol Oncol Res. 2016;22:769–772. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Choi YJ, Je EM, et al. Frameshift mutation of WISP3 gene and its regional heterogeneity in gastric and colorectal cancers. Hum Pathol. 2016;50:146–152. [DOI] [PubMed] [Google Scholar]
- 18.Pawłowski K, Lepistö M, Meinander N, et al. Novel conserved hydrolase domain in the CLCA family of alleged calcium-activated chloride channels. Proteins. 2006;63:424–439. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Gafà R, Tibiletti MG, et al. Genetic progression in microsatellite instability high (MSI-H) colon cancers correlates with clinico-pathological parameters: a study of the TGRbetaRII, BAX, hMSH3, hMSH6, IGFIIR and BLM genes. Int J Cancer. 2000;89:230–235. [PubMed] [Google Scholar]
- 20.Almendro V, Cheng YK, Randles A, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]