Supplemental Digital Content is available in the text
Keywords: complement, cytokine, disease activity, nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3, single nucleotide polymorphism, systemic lupus erythematosus
Abstract
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease with considerable genetic predisposition. Nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3 (NLRP3) is crucial for the innate immunity and implicated in SLE pathogenesis. Accordingly, we conducted a case-control study to find the association of NLRP3 variations with SLE susceptibility and disease activity.
Three single nucleotide polymorphisms of NLRP3 (rs3806268, rs4612666, and rs10754558) were genotyped in 400 SLE patients and 400 healthy controls; the patients were further divided into mild-to-moderate or high disease activity subgroup. Serum cytokines, complements, and autoantibodies were also detected.
We found that rs4612666 TT genotype conferred a higher risk of severe disease activity with adjusted odds ratio = 2.08, P = .02 and adjusted odds ratio = 2.34, P = .01 in the codominant and recessive model, respectively. Nevertheless, there was no association between the 3 single nucleotide polymorphisms of NLRP3 gene and SLE susceptibility. In addition, C4 decreased significantly in rs3806268 GG (P < .001) and rs4612666 TT genotype carriers (P = .03). A higher trend of interleukin-1β and interleukin-γ release were identified in rs3806268 AA and rs10754558 CC genotype carriers, respectively.
NLRP3 polymorphisms are associated with SLE disease activity and hypocomplementemia. Interleukin-1β and interleukin-γ levels in SLE patients are correlated with NLRP3 variants as well.
1. Introduction
Systemic lupus erythematosus (SLE) is a prototypic chronic autoimmune disease characterized by intermittent stages of activity and quiescence. Different individuals exhibit distinct patterns of disease activity,[1] resulting in protean clinical manifestations from slight skin lesions to life-threatening multi-organ damages. Minimizing disease activity and improving patient life quality are the basic goals for treatment at present, which relies on a better understanding of disease pathogenic mechanisms.
Immunological tolerance imbalance that results in inappropriate activation of the innate/ adaptive immune system is confirmed as a major driver of SLE onset and disease exacerbation.[2] Nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3 (NLRP3) is a recently identified pattern recognition receptor, primarily expressed in macrophages and dendritic cells, which can recognize varieties exogenous and endogenous stimuli, such as microbes, reactive oxygen species, nanoparticles, and so on. NLRP3 serving as the scaffolding protein could assemble with apoptosis-associated speck-like protein containing a caspase activation and recruitment domain and procaspase-1 into inflammasome.[3] Inflammasome is a proinflammatory component of the innate immune system, participating in regulating the pyroptosis and the secretion of interleukin (IL)-1β and IL-18. Growing pieces of evidence have linked NLRP3 to the onset and progression of multiple auto-inflammatory/immune diseases, including SLE.[4,5]
However, the relationship between NLRP3 level and SLE development or progression was reported inconsistently by different studies.[6,7] This disparity may partially attribute to the gain-of-function mutation in NLRP3 gene conferring protein and mRNA compensatory decreasing, and vice versa. Accordingly, exploring the gene polymorphisms may help in explaining the function of NLRP3 in SLE precisely.
Several studies have already found the association between NLRP3 polymorphisms and multiple sclerosis,[8] rheumatoid arthritis,[9] ankylosing spondylitis,[10] and other autoimmune diseases. Rs10754558 C allele was even found associated with increasing SLE risk in Latin American.[11] However, the influence of NLRP3 variants on SLE disease activity is still poorly understood, especially in the Chinese population. Patients with Asian ancestry tend to have higher levels of overall disease activity,[12] presenting with more severe renal disease and serological manifestations.[13,14] However, the biological basis of this association is still unknown. Sustained control of disease activity may contribute to avoid organ damage and premature mortality. Therefore, we consider investigating the association between NLRP3 variants and disease activity in the Chinese may contribute to selecting the high-risk patients for more aggressive treatment, and finally improving their prognosis.
For these reasons, we conducted this case-control study and found that rs4612666 of NLRP3 gene is associated with increasing SLE disease activity. Since autoantibody/cytokine production and complement activation are the main pathogenic bases of autoimmune diseases, we examined the relationship between NLRP3 polymorphisms and these biomarker levels at the same time.
2. Materials and methods
2.1. Subjects
Between January 2014 and August 2016, 400 SLE patients aged 14 to 75 (mean age 36.33 ± 12.99) with 400 age- and sex-matched controls were enrolled in the study. All the subjects are Chinese Han population presenting to the West China Hospital of Sichuan University. Patients fulfilled the 1997 American College of Rheumatology (1997) classification criteria for SLE[15] and drug-induced SLE was excluded. Besides, we used the 2002 updated version of SLEDAI (SLE disease activity index) to measure disease activity in SLE patients based on findings in the preceding 30 days.[16] In order to eliminate the influence of therapeutic decisions on the disease activity, the history of drug ingestion was taken down from every SLE outpatient. Only the ones who were newly diagnosed or withdrew immunosuppressive regimen (poor compliance) were recruited, their untreated period lasted at least 1 month. Patients were assigned into mild-to-moderate activity (SLEDAI ≤10) or high activity (SLEDAI >10) subgroup by the rheumatology specialists based on the recommendations from association guidelines.[17] The controls were eligible if physical examination and conventional laboratory tests revealed no abnormalities, and infection/autoimmune diseases were not suspected.
The study complied with the 1975 Declaration of Helsinki and was approved by the ethics committee at West China Hospital. All participants gave written informed consent before enrollment. Written informed consent was obtained from parents of the patients below 16 years, as well.
2.2. Genotyping
Three NLRP3 gene single nucleotide polymorphisms (SNPs) (rs3806268, rs4612666, and rs10754558) were selected; their basic information was shown in Supplementary Table 1. Genotyping was undertaken using the improved multiplex ligation detection reaction method (Genesky Biotechnologies Inc., Shanghai, China), and the process was detailed by our previous study.[18] Ten percent of samples were retested by sequencing to ensure accuracy.
2.3. Cytokines quantification and SLE-related laboratory examination
Serum IL-1β, IL-18, tumor necrosis factor (TNF)-α and interferon (IFN)-γ were measured using R&D Human Inflammation Assays in randomly selected 143 SLE patients. Complement 3 (C3), C4, immune globulin (Ig) G, IgA, and IgM were determined by nephelometry (Immage 800, Beckman Coulter, California, USA). Anti-nuclear antibody (ANA) and anti-double-stranded DNA antibody (anti-dsDNA) tests were performed via indirect immunofluorescence (Euroimmun, Lübeck, Germany). An ANA titer of 1:320 or greater is highly suggestive of autoimmune disease, so we regarded this titer as the cut-off point for ANA positive or negative. Similarly, we considered 1:10 as the cut-off point for anti-dsDNA. Several other antibodies related to SLE, including anti-Smith antibody, anti-SSA antibody (anti-SSA), anti-SSB antibody, and anti-ribosomal P protein antibody, were assayed through immunoblotting method with the kit from Euroimmun.
2.4. Statistical analysis
Statistical analysis was done by PLINK (v1.07),[19] SPSS (version 25, IBM Corp, Armonk, NY), and GraphPad Prism 8 (San Diego, CA). Haploview software package (version 4.2) was used to infer the linkage disequilibrium (LD) and haplotypes.
The Hardy–Weinberg equilibrium (HWE) was calculated for every SNP by exact test.[20] Association of NLRP3 polymorphisms and SLE susceptibility or disease activity was evaluated using logistic regression technique with adjustment for possible confounding factors if necessary. Categorical variables were compared using the χ2 test or Fisher exact test; continuous variables were analyzed by Student t-test or nonparametric test. Data were expressed as the mean ± standard deviation or median (interquartile range) as appropriate for continuous variables and number (percentage) for categorical variables. A 2-tailed P < .05 was considered significant.
3. Results
3.1. Subjects characteristics and baseline genotyping information
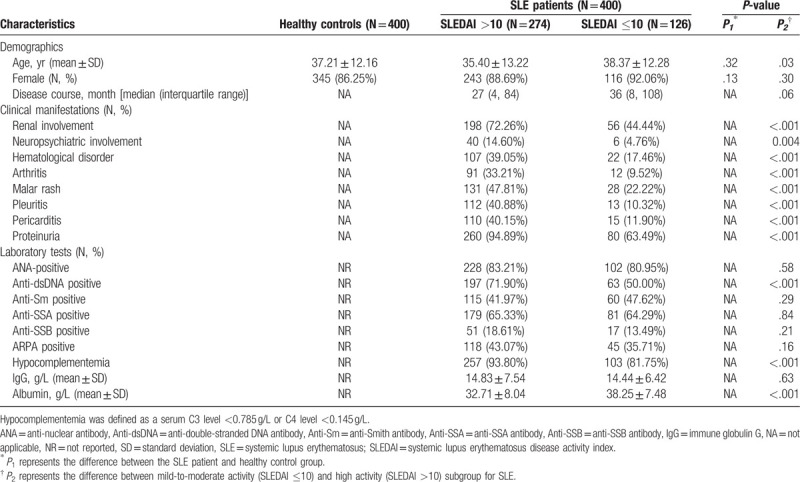
The characteristics of the participants were presented in Table 1. A total of 800 subjects were included in this study, with case and control enrolled at one to one basis. Of the SLE patients, 126 (31.50%) and 274 (68.50%) individuals were subdivided into mild-to-moderate activity or high activity groups according to their SLEDAI score. For demographic data, we only found that age showed a significant discrepancy between patients with different disease activity (P = .03). Additionally, patients with higher SLEDAI score (SLEDAI >10) showed markedly severe clinical condition from manifestations to related laboratory testing (including anti-dsDNA positive rate, the rate of C3 <0.785 g/L or C4 <0.145 g/L, and albumin level, all P < .001), excepted the positive rate of several autoantibodies (ANA, anti-Smith antibody, anti-SSA, anti-SSB antibody, anti-ribosomal P protein antibody) and the concentration of IgG.
Table 1.
Characteristics of the subjects.
For rs3806268, rs4612666, and rs10754558 in all the 800 subjects, the minor allele frequency were 0.49, 0.45, and 0.43, respectively. In the control group, rs10754558 occurred a deviation from HWE (P = .01), but the other 2 SNPs were consistent with HWE (rs3806268, P = .19; rs4612666, P = .84) (Shown in Supplementary Table 1).
3.2. NLRP3 polymorphisms are associated with SLE disease activity
As shown in Table 2, after comparing the allele or genotype distribution between SLE patients and the healthy controls, the examined SNPs showed no significant association with SLE susceptibility in the Chinese Han population. We then performed the subgroup analysis stratified by the SLEDAI score.
Table 2.
Association of NLRP3 polymorphisms with SLE susceptibility and disease activity.
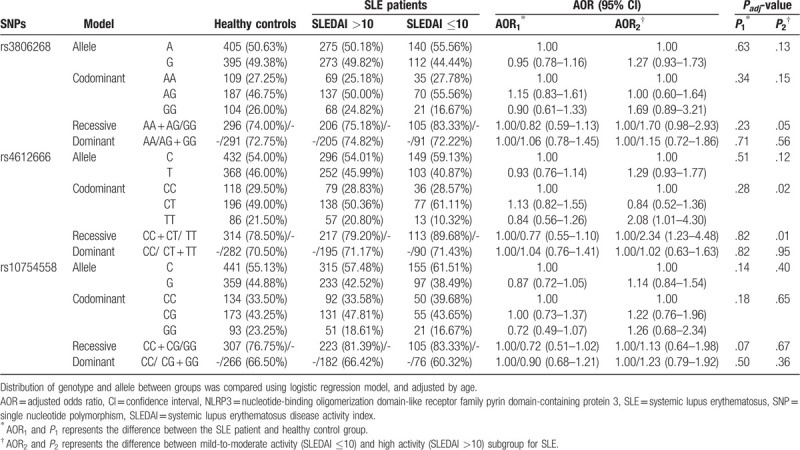
We used logistic analysis for exploring the influence of NLRP3 SNPs on SLE disease activity and adjusted for age as a possible confounding factor. Rs4612666 TT genotype conferred a higher risk of SLE progression with adjusted odds ratio (AOR) = 2.08, 95% confidence interval (CI) = 1.01–4.30, P = .02 and AOR = 2.34, 95% CI = 1.23–4.48, P = .01 in the codominant and recessive model, respectively.
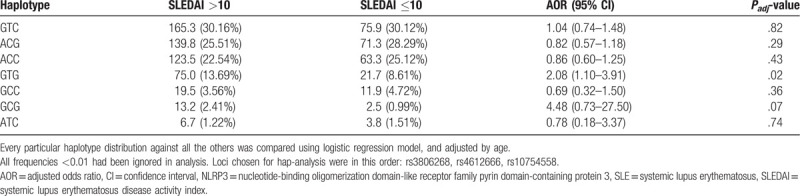
As seen in Figure 1, in the 400 SLE patients, our analysis of LD between rs3806268 and rs4612666 showed moderate (r2 = 0.70), while the other pairwise SNPs exhibited low LD (both r2 = 0.05). Seven haplotypes given in Table 3 were constructed with frequency >0.01. We compared every particular haplotype distribution against all the others, adjusting for age, finally found that haplotype GTG (in the order of rs3806268, rs4612666, and rs10754558) associated with increasing risk of SLE disease activity (the GTG frequency was 13.69% and 8.61% in the high and relatively low activity group, AOR = 2.08, 95% CI = 1.10–3.91, P = .02).
Figure 1.
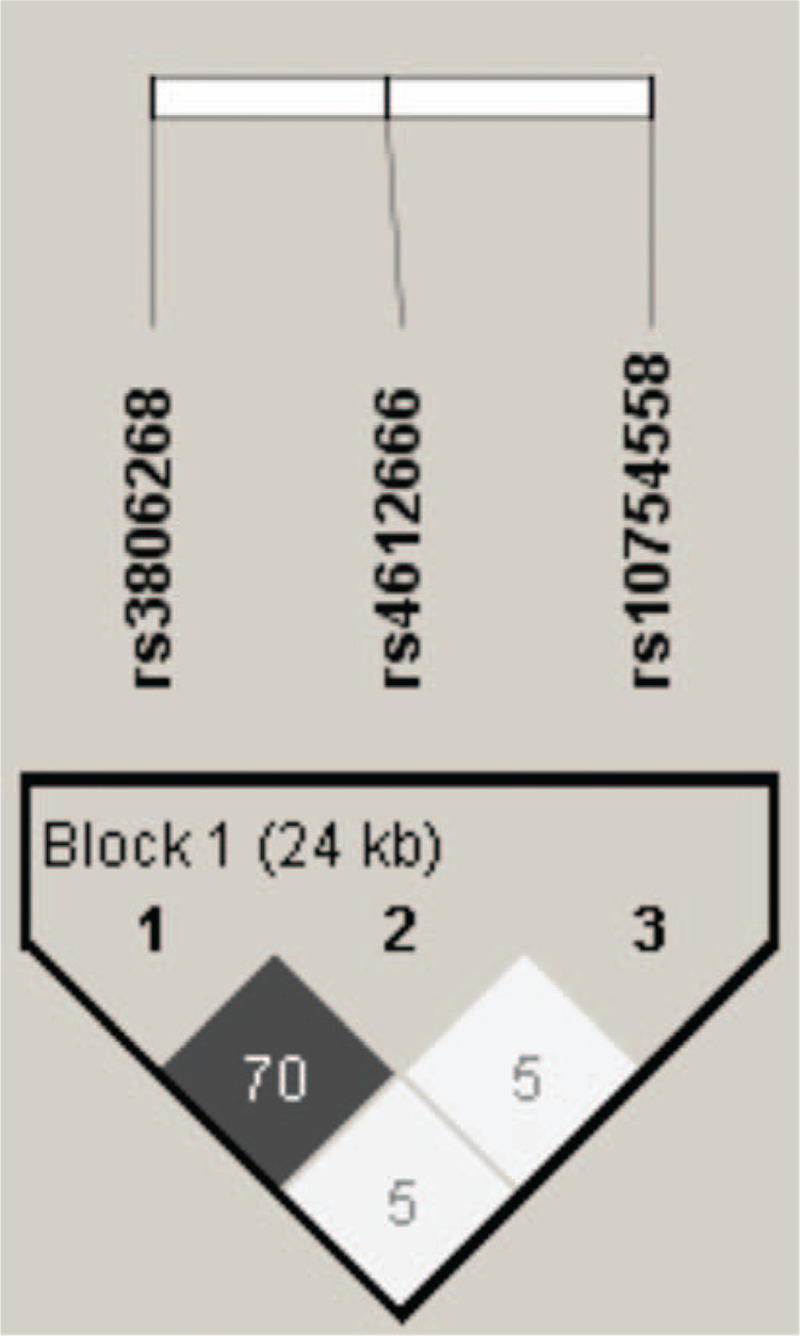
Linkage disequilibrium for SNPs of NLRP3 in 800 individuals. The linkage disequilibrium (LD) plot shows r2 values between each pair of SNPs. Rs3806268 and rs4612666 were in moderate LD (gray square).
Table 3.
Association of NLRP3 haplotypes with SLE disease activity.
3.3. Correlation of the NLRP3 polymorphisms and cytokine, complement or autoantibody expression levels
As the results above revealed that NLRP3 polymorphisms associated with SLE activity, we performed further analysis to investigate whether the production of some common SLE biomarkers differed significantly between the 3 genotypes of every single SNP.
Firstly, we compared the serum levels of IL-1β, IL-18, TNA-α, and IFN-γ in SLE patients. Only IL-1β and IFN-γ secretion exhibited statistically significant (Table 4). The expression of IL-1β was higher in the rs3806268 AA genotype carriers than the AG or GG genotype carriers (AA: 4.39 [3.15, 8.02] pg/mL vs AG: 3.15 [1.87, 5.16] pg/mL, and GG: 3.15 [1.87, 7.42] pg/mL, P = .03); IFN-γ was 8.58 (5.16, 13.69) pg/mL in the rs10754558 CC genotype carriers and that was 5.16 (1.75, 8.58) pg/mL and 5.16 (3.89, 11.56) pg/mL in the CG and GG genotype carriers, respectively, and P = .03.
Table 4.
Association of NLRP3 polymorphisms with serum cytokine levels in the SLE patients.
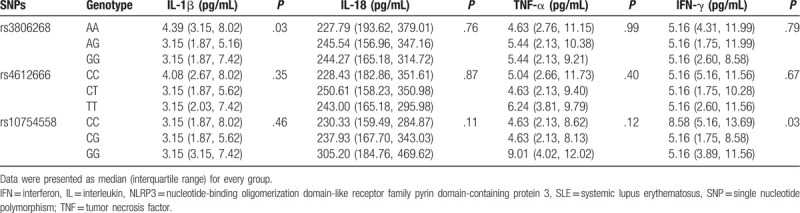
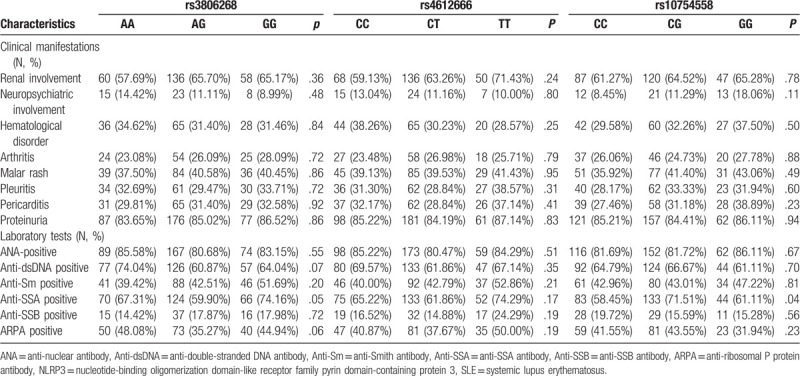
Furthermore, complements and globulins analysis found that C4 decreased significantly in rs3806268 GG and rs4612666 TT genotype (P < .001 and P = .03, respectively), details were shown in Table 5. We found no association between ANA or anti-dsDNA and NLRP3 variations. However, a higher frequency of anti-SSA positive rate was observed in rs10754558 CG genotype carriers (CG: 71.51% vs CC: 58.45% and GG: 61.11%, P = .04) (Shown in Table 6).
Table 5.
Association of NLRP3 polymorphisms with serum complement/IgG levels in the SLE patients.
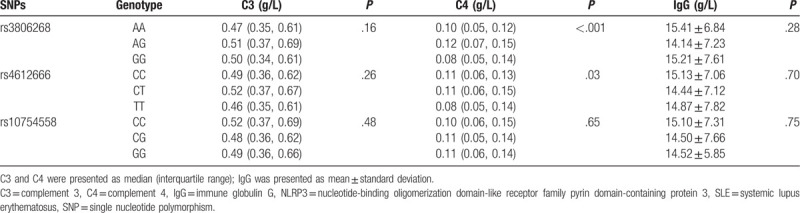
Table 6.
Association of NLRP3 polymorphisms with SLE clinical manifestations and related autoantibodies.
3.4. Association analysis of variants in NLRP3 with clinical manifestations
Table 6 illustrated the relationship of NLRP3 variants and the patterns of organ involvement in SLE. The typical clinical manifestations included arthritis, pleuritis, pericarditis, proteinuria, malar rash, and renal, neuropsychiatric or hematological disorders. There was no association between genotype distribution of every SNP and the clinical characteristics described above (all P > .05).
4. Discussion
In this research, we performed a case-control study focusing on the association of NLRP3 polymorphisms with SLE progression. We found that the rs4612666 TT genotype may aggravate the SLE disease activity. Concomitantly, haplotype GTG carrying the risk allele of rs4612666 appears to be the high-risk haplotype with a prediction of the worse cases. Furthermore, we investigated the genetic variant effect on the serum levels of several inflammation biomarkers (cytokines, autoantibodies and complements); the primary impact was found on the IL-1β, IFN-γ, and C4 levels.
Genetic factors contribute to the onset and severity of multiple autoimmune diseases, including SLE.[21] Given the treatment options are scarce at this stage, further understanding of how genetic variants affect immune function in the healthy condition and the setting of chronic inflammation is required, which may provide novel therapeutic strategies for delaying the start and controlling disease activity of SLE. NLRP3 protein includes 3 functional domains, PYD, NACHT, and LRR, composing the sensor part of the corresponding inflammasome. When activated by different stimuli, LRR firstly recognizes the signals, then NACHT mediates the self-oligomerization, and PYD interacts with apoptosis-associated speck-like protein containing a caspase activation and recruitment domain to assemble the intact inflammasome.[22] Inappropriate stimulation of NLRP3 could lead to sustained and recurrent inflammation, which is concerning diverse autoimmune pathogenesis.[3] Therefore, we focus on the association of NLRP3 polymorphisms with SLE and disease activity for the first time in the Chinese population.
Asian people are likely to have higher disease activity,[12,13] which may be determined by the genetic background. However, there is no study reported the relationship between disease severity of SLE and NLRP3 SNPs. In the current study, a more serious phenotype is highly prevalent among SLE patients with the TT genotype of rs4612666. Several previous studies demonstrated that rs4612666 TT genotype might facilitate inflammation disorders, such as large artery atherosclerosis[23] and recurrent aphthous stomatitis[24]; allele T might promote ankylosing spondylitis.[10] To some extent, these findings could support our result and suggest that rs4612666 may influence the common mechanistic pathway shared by the different inflammatory diseases.
Haplotype comprises multiple SNPs on the same chromosome, which can provide more information than single-locus analysis. Therefore, we conducted a haplotype investigation and found a positive association between GTG and SLE disease activity. GTG is a relatively common haplotype in the patients, and this particular haplotype possesses the risk allele of rs4612666, confirming the critical role of this polymorphism in risk determination.
Nevertheless, we found no association between the 3 SNPs of NLRP3 gene and SLE susceptibility. However, individuals with rs10754558 G allele encountered a decreasing risk of SLE has been described by Pontillo et al in the Brazilian.[25] The discrepancies of our researches may be driven by race and genotyping method variation. In addition, since rs10754558 biased away from HWE in the healthy control group, we consider that expanding sample size in the subsequent study may better represent the general population and reveal the actual effect of this particular SNP on SLE development in different races.
We also found that NLRP3 variants are associated with the essential biomarkers’ concentration for SLE. Complement is a vital player in innate immunity. Deficiencies in components of the classical pathway of complement, which majorly result from consumption of complement by immune complexes, carry perhaps the strongest association to SLE.[26] In the present study, we found that risk genotype (TT for rs4612666) carriers had a relative lower serum C4 concentration, suggesting that NLRP3 genetic mutations might have potential risk effects on causing hypocomplementemia, and increasing SLE disease activity significantly.
The central role of NLRP3 inflammasome is to activate caspase-1, which could convert pro-inflammatory cytokines IL-1β and IL-18 from their precursors to the mature and biologically active forms.[3] Both of the 2 ILs often correlate with the degree of autoimmune-related inflammation and have important roles in instructing adaptive immunity.[27] IL-18 is the IFN-γ inducing factor for Th1 cells, especially in the presence of IL-12; IL-1β can promote CD4+ T cells production IFN-γ in an autocrine fashion as well.[28] Wen et al have reported that serum IFN-γ is an effective biomarker of SLE disease activity.[29] In addition, TNF-α has been intensively investigated in SLE; TNF-α inhibitors, such as etanercept, certolizumab, adalimumab, and so on, are available for clinical treatment is the best demonstrating. Anti-inflammation drugs often influence TNF-α expression and NLRP3 inflammasome activation at the same time.[30,31]
All the relevant studies indicate that these cytokines’ levels may differ depending on the effect of NLRP3, and then we try to find the associations at the genetic level. A lower trend of IL-1β release was identified in the patients with GG and AG than AA genotype of rs3806268. IL-1β secretion is regulated by 2 signals: transcriptional upregulation of pro-IL-1β and activation of caspase-1; NLRP3 inflammasome is required for the latter process.[3] The increasing IL-1β production in AA genotype carriers may be directly caused by rs3806268 related NLRP3 functional alteration. Further functional studies are greatly needed to confirm this result.
Furthermore, the unexpectedly concurrent decreasing of C4 and IL-1β in rs3806268 GG genotype carriers may occur due to the complex interaction between complement system and NLRP3 inflammasome, including its downstream cytokines. The classical pathway of complement involves membrane attack complex generating. Membrane attack complex can activate the NLRP3 inflammasome and induce IL-1β and IL-18 release.[32] NLRP3 mutation might disturb this activation and the related positive feedback loop, resulting in IL-1β reducing in some particular genotypes, even if the proinflammation microenvironment is nearly identical.
Relative higher levels of IFN-γ occurred in the rs10754558 CC genotype carriers, which is consistent with the report of Pontillo et al that C allele of rs10754558 may increase the risk of SLE development. Autoantibody (Anti-dsDNA and ANA) levels were not correlated with NLRP3 SNPs in our study.
In conclusion, our study investigated the association between NLRP3 polymorphisms and the development and progression of SLE. We revealed the association of NLRP3 polymorphism (rs4612666) with increasing SLE disease activity. Serum biomarker (C4, IL-1β, and IFN-γ) expressions were associated with NLRP3 polymorphisms as well. To our knowledge, this is the first study executed in Chinese population to evaluate the relationship between NLRP3 variants and SLE disease activity. Maximizing sample size and performing function study are needed to confirm the present results in the subsequence work.
Author contributions
Junlong Zhang and Bin Yang designed the study; Junlong Zhang, Zhenzhen Su, and Qian Niu were responsible for recruitment of subjects, performed experiments, and conducted data management; Junlong Zhang, Zhenzhen Su, Zhuochun Huang, and Bin Yang performed statistical analyses and interpreted results; Zhenzhen Su and Junlong Zhang wrote the original draft; Zhenzhen Su, Junlong Zhang, and Zhuochun Huang revised the draft; all authors reviewed the manuscript.
Supplementary Material
Footnotes
Abbreviations: ANA = anti-nuclear antibody, anti-dsDNA = anti-double-stranded DNA antibody, AOR = adjusted odds ratio, CI = confidence interval, HWE = Hardy–Weinberg equilibrium, IFN = interferon, Ig = immune globulin, IL = interleukin, LD = linkage disequilibrium, NLRP3 = nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3, SLE = systemic lupus erythematosus, SLEDAI = systemic lupus erythematosus disease activity index, SNP = single nucleotide polymorphism, TNF = tumor necrosis factor.
How to cite this article: Su Z, Niu Q, Huang Z, Yang B, Zhang J. Association of nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3 polymorphisms with systemic lupus erythematosus disease activity and biomarker levels: a case-control study in Chinese population. Medicine. 2020;99:35(e21888).
This work was funded by grants from the National Natural Science Foundation of China (No. 81601830, No. 81501816, and No. 81772258) and the Science and Technology Agency of Sichuan Province (2020YFS0124, 2020YFS0126).
The authors report no conflicts of interest.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Petri M. Disease activity assessment in SLE: do we have the right instruments? Ann Rheum Dis 2007;66: Suppl 3: 61–4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tsokos GC, Lo MS, Costa Reis P, et al. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol 2016;12:716–30.. [DOI] [PubMed] [Google Scholar]
- [3].Shen HH, Yang YX, Meng X, et al. NLRP3: a promising therapeutic target for autoimmune diseases. Autoimmun Rev 2018;17:694–702.. [DOI] [PubMed] [Google Scholar]
- [4].Shao BZ, Xu ZQ, Han BZ, et al. NLRP3 inflammasome and its inhibitors: a review. Front Pharmacol 2015;6:1–9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhong Z, Sanchez-Lopez E, Karin M, et al. NLRP3 inflammasome and auto-inflammatory/immune diseases. Clin Exp Rheumatol 2016;34:12–6.. [PubMed] [Google Scholar]
- [6].Yang CA, Huang ST, Chiang BL. Sex-dependent differential activation of NLRP3 and AIM2 inflammasomes in SLE macrophages. Rheumatol (United Kingdom) 2015;54:324–31.. [DOI] [PubMed] [Google Scholar]
- [7].Ma ZZ, Sun HS, Lv JC, et al. Expression and clinical significance of the NEK7-NLRP3 inflammasome signaling pathway in patients with systemic lupus erythematosus. J Inflamm (United Kingdom) 2018;15:1–2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Imani D, Azimi A, Salehi Z, et al. Association of nod-like receptor protein-3 single nucleotide gene polymorphisms and expression with the susceptibility to relapsing–remitting multiple sclerosis. Int J Immunogenet 2018;45:329–36.. [DOI] [PubMed] [Google Scholar]
- [9].Addobbati C, da Cruz HLA, Adelino JE, et al. Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm Res 2018;67:255–64.. [DOI] [PubMed] [Google Scholar]
- [10].Zhao S, Chen H, Wu G, et al. The association of NLRP3 and TNFRSF1A polymorphisms with risk of ankylosing spondylitis and treatment efficacy of etanercept. J Clin Lab Anal 2017;31:1–3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee YH, Bae S-C. Association between functional NLRP3 polymorphisms and susceptibility to autoimmune and inflammatory diseases: a meta-analysis. Lupus 2016;25:1558–66.. [DOI] [PubMed] [Google Scholar]
- [12].Golder V, Connelly K, Staples M, et al. Association of Asian ethnicity with disease activity in SLE: an observational study from the Monash Lupus Clinic. Lupus 2013;22:1425–30.. [DOI] [PubMed] [Google Scholar]
- [13].Connelly K, Morand EF, Hoi AY. Asian ethnicity in systemic lupus erythematosus: an Australian perspective. Intern Med J 2013;43:618–24.. [DOI] [PubMed] [Google Scholar]
- [14].Sestak AL, Nath SK, Kelly JA, et al. Patients with familial and sporadic onset SLE have similar clinical profiles but vary profoundly by race. Lupus 2008;17:1004–9.. [DOI] [PubMed] [Google Scholar]
- [15].Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- [16].Touma Z, Urowitz MB, Ibañez D, et al. SLEDAI-2K 10 days versus SLEDAI-2K 30 days in a longitudinal evaluation. Lupus 2011;20:67–70.. [DOI] [PubMed] [Google Scholar]
- [17].Gordon C, Amissah-Arthur M-B, Gayed M, et al. The British Society for Rheumatology guideline for the management of systemic lupus erythematosus in adults: executive summary. Rheumatology (Oxford) 2018;57:14–8.. [DOI] [PubMed] [Google Scholar]
- [18].Zhang J, Liu X, Meng Y, et al. Autoimmune disease associated IFIH1 single nucleotide polymorphism related with IL-18 serum levels in Chinese systemic lupus erythematosus patients. Sci Rep 2018;8:1–8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 2005;76:887–93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wen L, Zhu C, Zhu Z, et al. Exome-wide association study identifies four novel loci for systemic lupus erythematosus in Han Chinese population. Ann Rheum Dis 2018;77:417. [DOI] [PubMed] [Google Scholar]
- [22].Leemans JC, Kors L, Anders HJ, et al. Pattern recognition receptors and the inflammasome in kidney disease. Nat Rev Nephrol 2014;10:398–414.. [DOI] [PubMed] [Google Scholar]
- [23].Cheng L, Yin R, Yang S, et al. Rs4612666 polymorphism of the NLRP3 gene is associated with the occurrence of large artery atherosclerotic ischemic strokes and microembolic signals. Biomed Res Int 2018;2018:6345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Slezakova S, Borilova Linhartova P, Masopustova L, et al. Association of the NOD-like receptor 3 (NLRP3) gene variability with recurrent aphthous stomatitis in the Czech population. J Oral Pathol Med 2018;47:434–9.. [DOI] [PubMed] [Google Scholar]
- [25].Pontillo A, Girardelli M, Kamada AJ, et al. Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity 2012;45:271–8.. [DOI] [PubMed] [Google Scholar]
- [26].Leffler J, Bengtsson AA, Blom AM. The complement system in systemic lupus erythematosus: an update. Ann Rheum Dis 2014;73:1601–6.. [DOI] [PubMed] [Google Scholar]
- [27].Mende R, Vincent FB, Kandane-Rathnayake R, et al. Analysis of serum interleukin (IL)-1β and IL-18 in systemic lupus erythematosus. Front Immunol 2018;9:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Arbore G, West EE, Spolski R, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science 2016;352:aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wen S, He F, Zhu X, et al. IFN-γ, CXCL16, uPAR: potential biomarkers for systemic lupus erythematosus. Clin Exp Rheumatol 2018;36:36–43.. [PubMed] [Google Scholar]
- [30].Li X, Wang M, Hong H, et al. Sophocarpine attenuates murine lupus nephritis via inhibiting NLRP3 inflammasome and NF-κB activation. Immunol Res 2018;66:521–7.. [DOI] [PubMed] [Google Scholar]
- [31].Su B, Ye H, You X, et al. Icariin alleviates murine lupus nephritis via inhibiting NF-κB activation pathway and NLRP3 inflammasome. Life Sci 2018;208:26–32.. [DOI] [PubMed] [Google Scholar]
- [32].Xie CB, Qin L, Li G, et al. Complement membrane attack complexes assemble NLRP3 inflammasomes triggering IL-1 Activation of IFN-γ-primed human endothelium. Circ Res 2019;124:1747–59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


