Abstract
MicroRNAs (miRNAs) refers to a small, short non-coding RNA of endogenous class. They have shown to have an increasingly altered expression in many types of cancer, including colorectal cancer (CRC).
In the present study, miRNA TaqManMGB and qRT-PCR was used to quantify the expression and clinical significance of 3 mature human miRNA in 82 pairs of colorectal adenocarcinoma tissues and normal adjacent tissue samples (NATS) collected from patients of the south-east part of Romania. Differences between CRC and NATS were analyzed using Wilcoxon test, while correlations between miRNAs expression levels and clinicopathological features were examined using non-parametric tests. In addition, the ability of selected miRNAs to function as biomarkers and, as potential indicators in CRC prognosis was also examined.
When the miRNA expression was compared in CRC related NATS, miR-143, and miR-145 were significantly underexpressed (4.99 ± –1.02 vs –5.66 ± –1.66, P < .001; –4.85 ± –0.59 vs –9.27 ± –1.51, P < .001, respectively), while the pattern of miR-92a was significantly overexpressed (–5.55 ± –2.83 vs –4.92 ± –2.44, P < .001). Moreover, the expression levels of selected miRNAs were identified to be correlated with gradual increases in fold change expression with the depth of tumor invasion, lymph node invasion, and maximal increases with distant metastasis. Furthermore, the receiver operating characteristic analysis demonstrated that potential diagnostic of miR-143, miR-145, and miR-92a in discriminating CRC from NATS, with the area under the curve of 0.74, 0.85, and 0.84 respectively. The Kaplan–Meier and the log-rank test showed that a high level of miR-92a and low levels of miR-143 and miR-145 predicted poor survival rate in our cohorts.
In conclusion, we can summarize that miR-145 and miR-143 are decreased, while miR-92 is increased in CRC compared to NATS, and associated with different stages of CRC pathogenesis. Thus, the expression of selected miRNAs can represent potential diagnostic and prognostic tools in patients with CRC from Romania.
Keywords: colorectal cancer, miR-92a, miR-143, miR-145, real-time quantitative polymerase chain reaction analysis, expression profiling
1. Introduction
Among all ranges of tumors, colorectal cancer (CRC) is one of the most common malignancies and a significant public health burden. Moreover, the incidence rate of CRC is 9.7%, thereby making it the third most common form of cancer worldwide and it is the fourth leading cause of cancer-related mortality.[1] As per the findings of the Global Burden of Cancer Reports in the Romania population, the relative incidence of CRC increased from 8.660 in 2012 to 11.076 in 2018, thus representing the second most common malignancy, after lung/bronchus cancers in men and breast cancer in women.[2] CRC is a multifactorial disease characterized by a sequential process associated with alteration of the molecular architecture in oncogene or tumor suppressor gene regulatory networks, mainly due to genomic mutation or epigenetic alterations and the involvement of small noncoding RNA species, named microRNAs (miRNAs) and long noncoding RNAs.[3,4]
In this regard, numerous studies and resources have been devoted to elucidating the molecular mechanisms of the CRC, however, the underlying mechanisms are not yet well understood. Therefore, improving the survival rate of patients with CRC requires a better understanding of tumor biology as well as the development of novel therapeutic and diagnostic strategies. A rapidly developing field of cancer research is the examination of miRNAs genes in CRC carcinogenesis. Molecules that are widespread and differentially expressed in CRC samples when compared to normal non-cancerous samples and may provide new insights into the mechanisms involved.
Functionally, miRNAs represent a novel class of small, non-coding RNAs (ncRNAs), found in plants, animals, and humans that use endogenous RNA interference pathways in order to modulate gene expression networks. Typically, miRNA genes are initially transcribed into the nucleus as longer primary transcripts guided by RNA polymerase II (pri-miRNAs), which are subsequently enzymatically cleaved by the Drosha into small miRNAs precursor (pre-miRNA). These pre-miRNAs are comprised of 70 nucleotides with hairpin stem-loop structures and are translocated into the cytoplasm through the assistance of Exportin-5 to undergo final maturation, within a functional miRNA to approximately 22 nucleotides catalyzed via RNase III endonuclease Dicer.[5] Predominantly, miRNAs exert their functionality in post-transcriptional modulation of gene expression through direct binding to the 3’ untranslated region (UTR) of specific messenger RNA targets, thereby leading to cleavage and degradation or suppression of translation.[5] The latest version of the miRBase database contains – 1.917 entries of mature human miRNAs (http://www.mirbase.org), which can be classified into clusters and families based on seed sequence or genomic relatedness, able to regulate the expression of one-third of human protein-coding genes.
It is now widely accepted that the altered functionality of miRNAs plays an important role in various biological and cancer-related processes, such as control of cellular homeostasis, differentiation, cell growth, and apoptosis. In colorectal tumorigenesis, the specific expression patterns of human miRNAs have been used by many researchers to understand the involvement of these regulatory molecules in the diagnosis and prognosis of this type of cancer.[6] Some of these deregulated mature miRNAs, due to high tissue specificity, altered stability, and unique expression in tumor development, might help distinguish CRC from other colon-related diseases, representing a new field of molecular diagnosis and prognosis of CRC. For instance, deregulation of miR-92a, miR-143, and miR-145 have been documented in plenty of pathophysiological events, including neoplastic diseases.[22,24] This suggests that miRNAs clusters function as oncogenic, respectively tumor suppressor genes and their inadequate expression is the result of excessive or deficient processing, and that ultimately leads to altered cellular homeostasis, essential events of tumorigenesis. In light of these data, miRNA genes have relevant biological and biomedical consequences in the detection and evolution of cancer, and their inadequate expression is an almost universal feature in human malignancies.
The present research aimed to investigate the signature of 3 mature human miRNAs involving miR-143, miR-145, and miR-92a in 82 pairs of colorectal adenocarcinoma tissues in the normal adjacent tissue samples (NATS) collected from patients in south-east Romania. The ability of the selected miRNAs to function as potential biomarkers, discriminating between CRC and NATS and, their potential as indicators in CRC prognosis was also examined. The miRNAs were selected based on previous studies on their clinical relevance in complex mechanisms of carcinogenesis.[17–27]
2. Materials and methods
2.1. Case selection
This study included 82 selected patients diagnosed with CRC at the Pathology Department of the Clinical Emergency County Hospital in Constanta, Romania. the Local Ethics Commission for the Approval of Clinical and Research Developmental Studies approved the study and all eligible patients provided written informed consent. Immediately after the surgical resection, CRC tissues and NATS (located at least 5 cm from the tumor site) were stabilized in RNAlater solution (Invitrogen by Thermo Fisher Scientific, USA) and frozen at –80 °C until further processing. A section of each sample (tumor and non-tumor) was stained with hematoxylin and eosin and evaluated by an experienced pathologist. All tumor specimens used in this study were histologically classified as colon or rectum adenocarcinoma. The histological tumor stage and differentiation grade was classified using the tumor-node-metastasis staging system of the American Joint Committee on Cancer, in accordance with standards set by World Health Organization.[7] Subsequently, clinicopathological features of patients were obtained from observation sheets and pathology reports and included age, sex, tumor location, tumor-node-metastasis stage, tumor differentiation degree, and eventual metastasis.
2.2. RNA extraction
Total RNA including miRNA molecules was isolated from the tissue samples using a miRNeasy kit (Qiagen, Germany) closely following the manufacturer's recommendations. We started with 30 mg of tissue which was thoroughly homogenized in 750 μl QIAzol Lysis Reagent for 90 seconds. Thereafter, 140 μL of chloroform was added to tissue homogenate and after 5 minutes incubation at room temperature, the sample was centrifuged for 15 minutes at 12.000 rpm at 4 °C. The upper aqueous phase containing RNA was transferred and precipitated in a new Eppendorf tube by adding 1.5 volumes of 100% ethanol. Approximately 650 μL of the precipitated sample was transferred to a RNeasy Mini column placed in an appropriate collection tube and centrifuged at 12.000 rpm for 1 minute at room temperature. After centrifugation, the filtrate was discarded and then 700 μL wash buffer RW1 was pipetted and centrifuged at 12.000 rpm for 1 minute. Next, 500 μL of wash buffer RPE was pipetted and centrifuged at 12.000 rpm for 1 minute at room temperature. To dry the membrane, the column was centrifuged at maximum speed for 1 minute. Following this, the column was placed in a new tapered collection tube, and 30 μL RNase-free water was added and centrifuged at maximum speed for 1 minute to collect an eluate.
The purity and yield of the RNA solutions were assessed by measuring the optical density at 260/280 nm using a NanoDrop One Spectrophotometer (Thermo Fisher Scientific, USA), where a ratio A260/A280 = 2 to 2.1, and A260/A230 > 2 was considered acceptable. The concentration of the samples was measured with Qubit 3.0 Fluorometer (Thermo Fisher Scientific) using the Qubit RNA HR (High-Range) Assay Kit. Furthermore, the RNA integrity number was conducted using the 2200 TapeStation Bioanalyzer (Agilent Technologies GmbH, Germany) with an RNA HS ScreenTape kit.
2.3. Reverse transcription of miRNA to complementary cDNA and qPCR reaction
Selected human miRNAs were reverse-transcribed to complementary DNA (cDNA) using the TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA). Each reaction was initiated using an RNA-specific stem-looped for reverse transcription (RT) (Table 1). The RNA concentration was set between 1 to 10 ng per 15 μL of RT reaction. Each 15 μL RT reaction consist of 7 μL master mix (0.15 μL dNTP mix, 1 μL Multiscribe RT enzyme, 1.5 μL 10 x RT Buffer, 0.19 μL RNase inhibitor and 4.16 μL Nuclease-free water), 3 μL primer, and 5 μL RNA sample. Samples were incubated in a thermocycler with the following parameters: 16 °C for 30 minutes, 42 °C for 30 minutes, 85 °C for 5 mintes, and then cooled to 4 °C.
Table 1.
The mature miRNA sequences.

The complementary DNA strand for selected targets of miRNAs was synthesized using a specific sequence TaqManMGB Assay (Applied Biosystems, San Diego, CA). For the 20 μL reaction mix, 10 μL of TaqMan 2 × Universal PCR Master Mix was added to 1.33 μL of the product from the RT reaction, 7.67 μL of RNase-free dH2O, and 1 μL of TaqMan Small RNA assay (20X). The quantitative real-time polymerase chain reaction analysis (qPCR) was performed in triplicate for each sample using the ABI 7500 Fast qPCR instrument for 40 cycles, where each cycle contained denaturation step at 95 °C for 3 seconds, and an annealing step at 60 °C for 30 seconds, followed by the extension of the primers with cleavage of the probe. Fluorescence was detected at the end of each cycle. A negative control without a template was used with all the qRT-PCR runs.
The average of cycle threshold (Ct) values obtained from triplicates of each miRNAs and endogenous control (miR-26b/RNU44) was calculated using an automatic baseline/threshold setting (7500 Fast Real-Time PCR software, version 2.3, Applied Biosystems, San Diego, CA) in concordance with the equation of Livak, Fold-change (FC) = 2−ΔΔCt, where ΔΔCt = ΔCt (CtmiR target – CtmiR26b/RUN44) tumoral tissue – ΔCt (CtmiR target – CtmiR26b/RNU44)normal tissue.[8] A fold change value <1 meant that the miRNAs were downregulated, where a value > 1 meant that the miRNAs were upregulated in the CRC relative to NATS. Thus, the results were expressed as FC in comparison with the calibrator sample, which was considered the normal value and assumed to equal 1.
2.4. Statistical Analysis
Data obtained were analyzed and graphs were constructed using SPSS version 20.0 software (SPSS, Chicago, IL) and MedCalc version 19.0.3 software (MedCalc, Ostend, Belgium). Differences between CRC and NATS were analyzed by the Wilcoxon test, while correlations between miRNAs expression levels as well as clinicopathological features were examined using the Mann–Whitney U test for 2 independent groups and Kruskal–Wallis H test for three independent groups. Survival rates for each miRNA were estimated using the Kaplan–Meier method and differences between low and high expression were calculated by using log-rank tests. The diagnostic efficacy of selected miRNAs to function as prognostic biomarkers were evaluated by using Receiver operating characteristics (ROC). Similarly, the area under the curve(AUC) was plotted to assess to evaluate the power of selected miRNAs to functions as a diagnostic tool in order to discriminate CRC from NATS. Sensitivity and specificity were then defined by the optimal cut-off point, which refers to the maximized value of the area under the ROC (Youden index). The univariate prognostic analysis revealed the parameters which affected the prognosis of CRC patients, as miRNAs expression levels and clinicopathological characteristics.
3. Results
3.1. Dysregulated expression levels of miRNAs in tumors and NATS from CRC patients
In the present study, 82 patients with colorectal adenocarcinoma along with their NATS, were investigated. This included 30 of proximal colon cases, 30 of distal colon cases, as well as 22 of rectal cases. In order to evaluate the miRNAs expression patterns in tumor tissue, the quantitative reverse transcription real-time polymerase chain reaction (qRT-PCR) was used. Additionally, it was observed that miR-143 and miR-145 were down-regulated, whereas miR-92a was up-regulated (Fig. 1).
Figure 1.
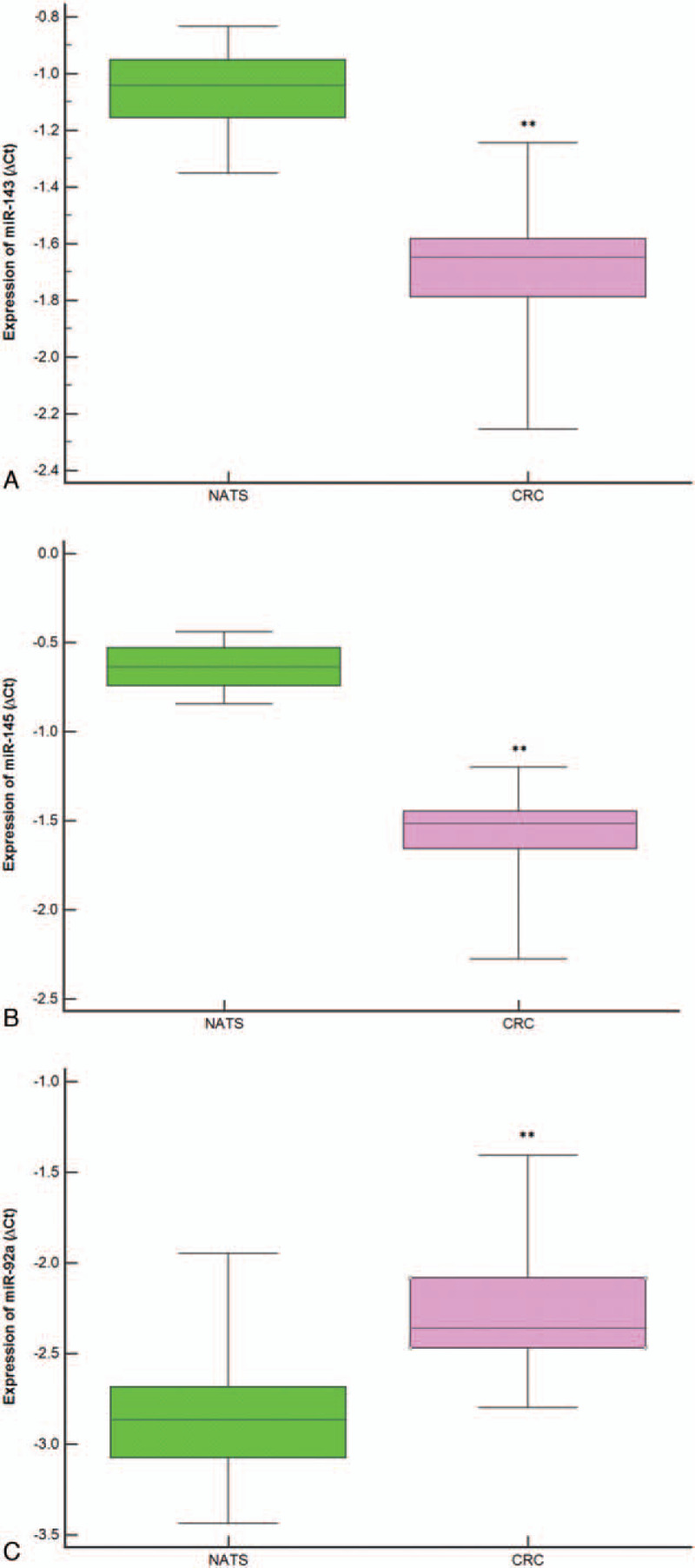
Expression levels of each microRNAs (ΔCt) in colorectal cancer tissue relative to normal adjacent tissue samples are normalized using RNU44 and miR-26b as endogenous controls. The statistically significant difference between colorectal cancer tissue and normal adjacent tissue samples samples was calculated using the Wilcoxon test (∗∗P < .001).
Consequently, the expression levels of miR-143 and miR-145 were found to decrease in 75.60% of CRC cases (–4.99 ± –1.02 vs –5.66 ± –1.66, P < .001) and 83% of cases, respectively (–4.85 ± –0.59 vs –9.27 ± –1.51, P < .001). On the other hand, the mean FC level expressions of miR-143 and miR-145 in CRC samples were downregulated around 8.21 times less and around 11.66 times less, respectively. In a similar vein, miR-92a expression level was upregulated in 78.00% of cases of CRC as compared to NATS (–5.55 ± –2.83 vs –4.92 ± –2.44, P < .001), with its fold increase being 2.32.
3.2. Selected miRNAs expression and association with clinicopathological characteristics
Associations with clinicopathological characteristics were determined in order to explore the clinical relevance of selected miRNAs. Some of these clinicopathological features were grouped, such as histological grade (G1, G2, and G3), pT stage (T1–T2 and T3–T4), pN stage (N0 and N1–N2) and pM stage (M0 and M1). As illustrated in Table 2, the higher expression of miR-92a was found to be closely associated with the depth of tumor invasion (P = .03), the involvement of regional lymph node (P < .001), and distant metastasis (P < .001).
Table 2.
Analysis of miR-143, miR-145, and miR-92a with the clinicopathologic features of CRC patients.
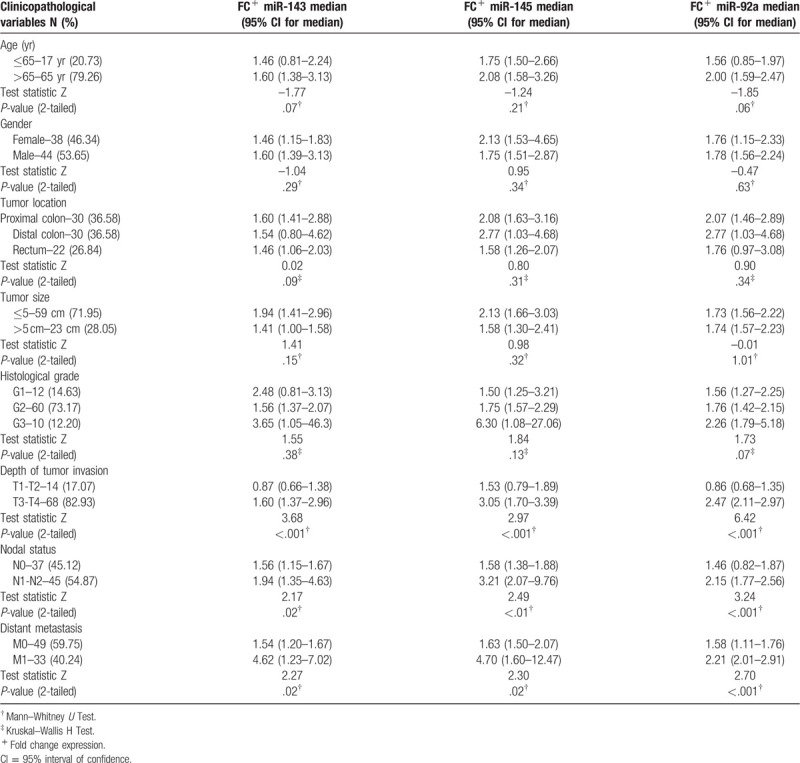
In addition, expression levels of miR-143 and miR-145 were tended to be associated with late-stage tumor invasion (P = .04, P < .001, respectively), involvement of lymph's node (P = .02, P < .001, respectively) and with distant metastasis (P < .001) as illustrated in Figure 2.
Figure 2.
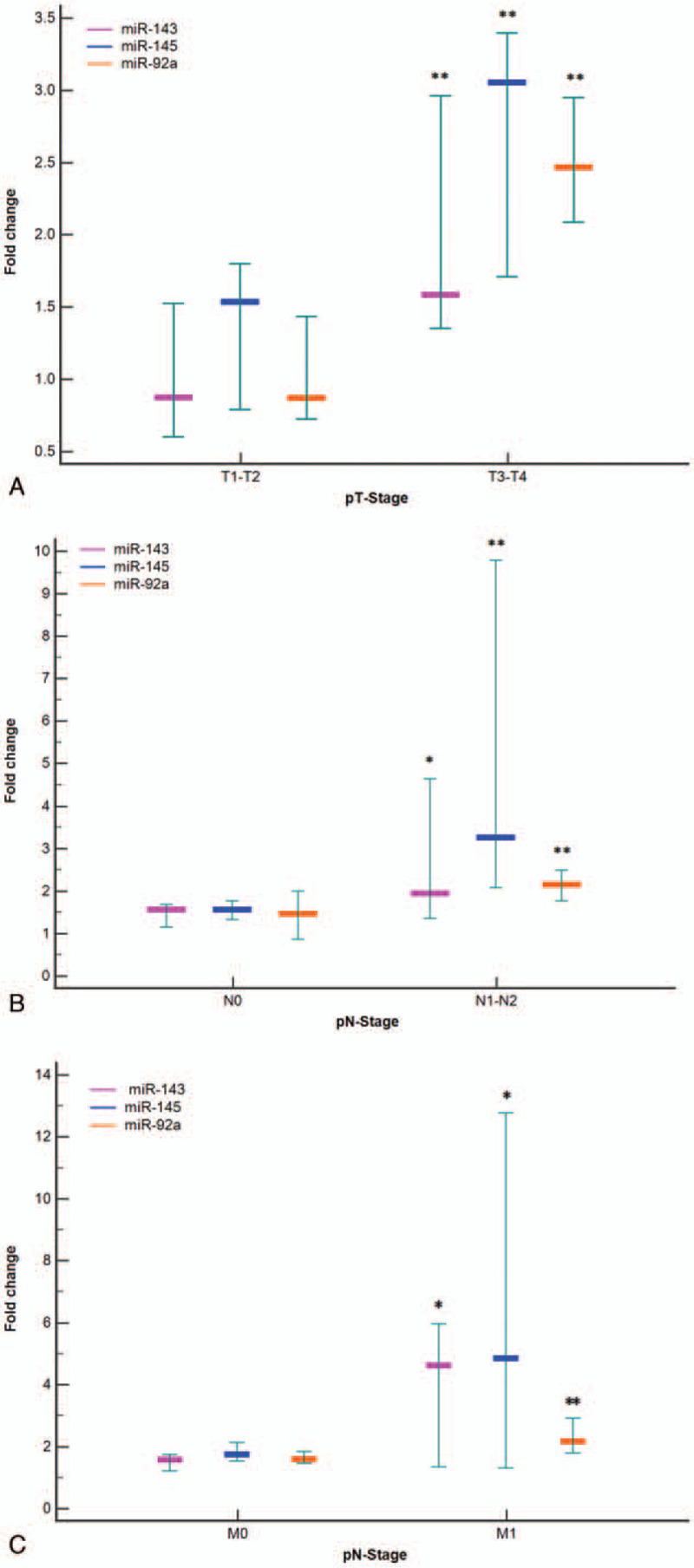
Fold change of miR-143, miR-145, and miR-92a in Romanian patients with colorectal cancer tissue according to different clinical stages. A boxplot showing fold change represented as median and 95% interval of confidence of the median, exhibited that the expressions of analyzed microRNAs increased with increasing pT stage- pT-stage (Fig. A), nodal metastasis - pN-stage (Fig. B), and distant metastasis - pM-stage (Fig. C). All experiments were conducted in triplicate (∗P < .05, ∗∗P < .001).
Moreover, no significant statistical differences were observed between selected miRNAs expression levels and gender, age, locations of tumor, tumor size, or tumor differentiation, respectively.
3.3. Correlation between deregulated expression levels of miRNAs and prognosis of CRC patients
In our study, the survival condition of the patients was followed and recorded, beginning from surgery to death or until the date of the last observation (censored data). The follow-up period ranging from 12 to 60 months and was monitored through the Oncology Department. In accordance with the relative expression levels of miR-92a, miR-143, and miR-145, patients were divided based on median values into high-expression groups and low-expression groups. The median survival time in CRC patients with high expression levels of miR-92a was found to be 36 months, which was significantly statistically lower than that median survival time in the low expression group (51 months, χ2 = 10.28; P < .01) (Fig. 3).
Figure 3.
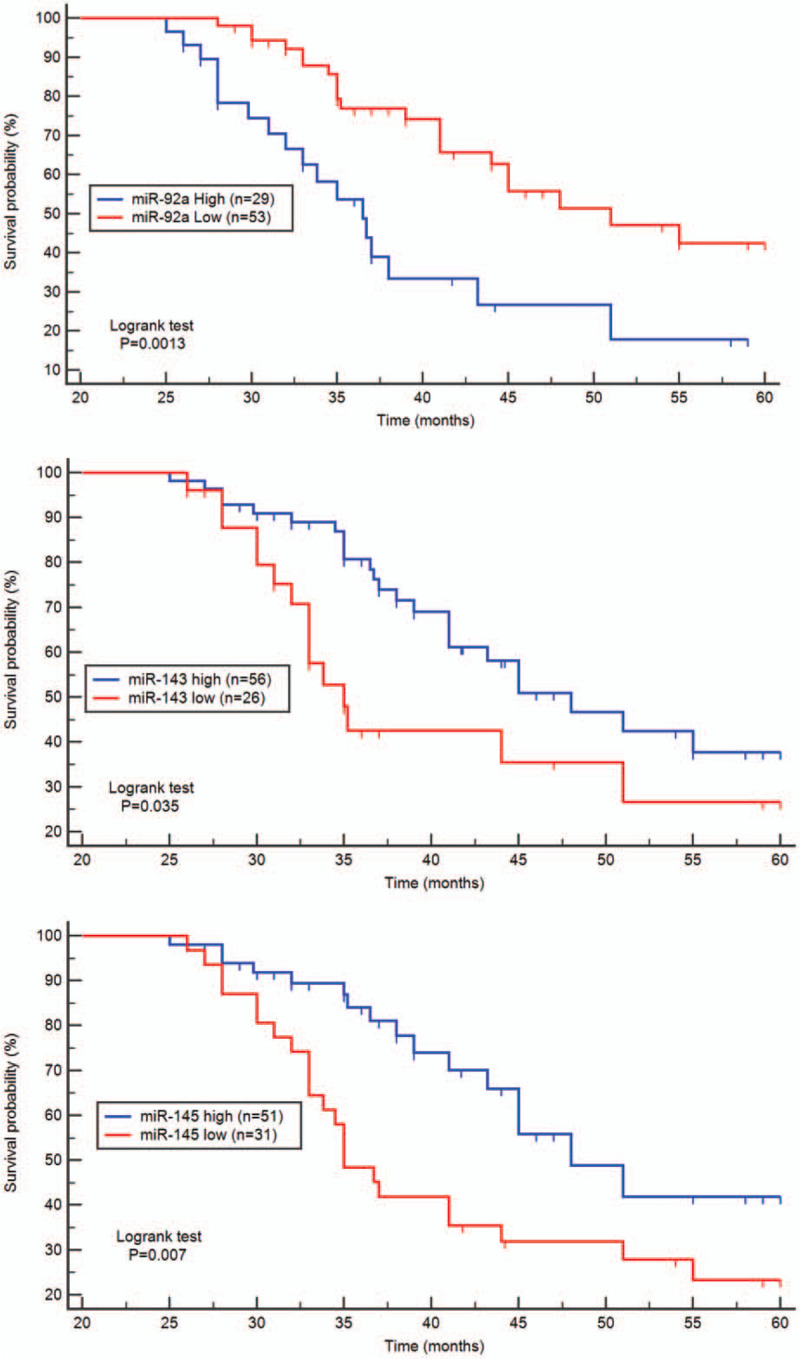
Kaplan–Meier survival curves for CRC patients. CRC patients with high miR-92a expression levels had a significant statistically poorer prognosis than those with low expression (P < .001). Moreover, the 5 yr survival probability in the low expression groups of miR-143 and miR-145 had a significant statistically poorer prognosis than in the high expression groups (P < .01, P < .001 respectively).
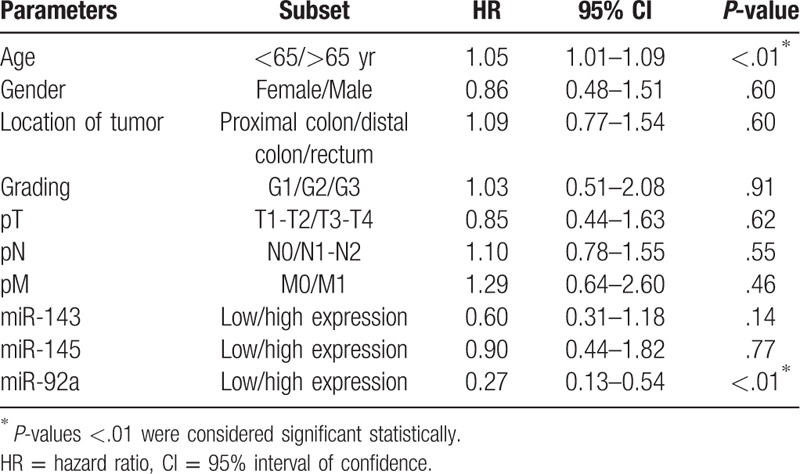
In addition, the survival probability of 5 years in the high expression group of miR-143 was 48 months significantly higher than that in the low expression group (36 months, χ2 = 4.43; P = .03). Moreover, the median survival rates of CRC patients in the low expression group of miR-145 was 35 months, significantly lower than that in the high expression group (48 months, χ2 = 7.05; P < .01). According to the univariate analysis of the overall survival rate, the level of miR-92a expression was an independent prognostic indicator for CRC (P < .01, Table 3).
Table 3.
Logistic regression of prognostic values of microRNAs associated with the clinicopathological features in colorectal cancer tissue patients.
3.4. ROC analysis
Analysis of the ROC curves and AUCs revealed that expression levels of miR-92a, miR-143, and miR-145 could help distinguish tumor tissue from normal adjacent tissues with very good specificity and sensitivity (Fig. 4). With regard to the AUC of miR-92a was 0.84 (95% interval of confidence (CI): 0.77 – 0.89, P < .001), the specificity was 80.49% and the sensitivity was 71.95% at a cut off value of 0.52. For miR-143 the specificity was 80.49% and the sensitivity was 60.98% at a cut off value of 0.41 and AUC of 0.74% (95% CI: 0.67 – 0.81, P < .001). Meanwhile the miR-145 exhibited a specificity of 85.37% and a sensitivity of 82.93% at a cut off value of 0.68 and AUC of 0.85 (95% CI: 0.78 – 0.91; P < .001).
Figure 4.
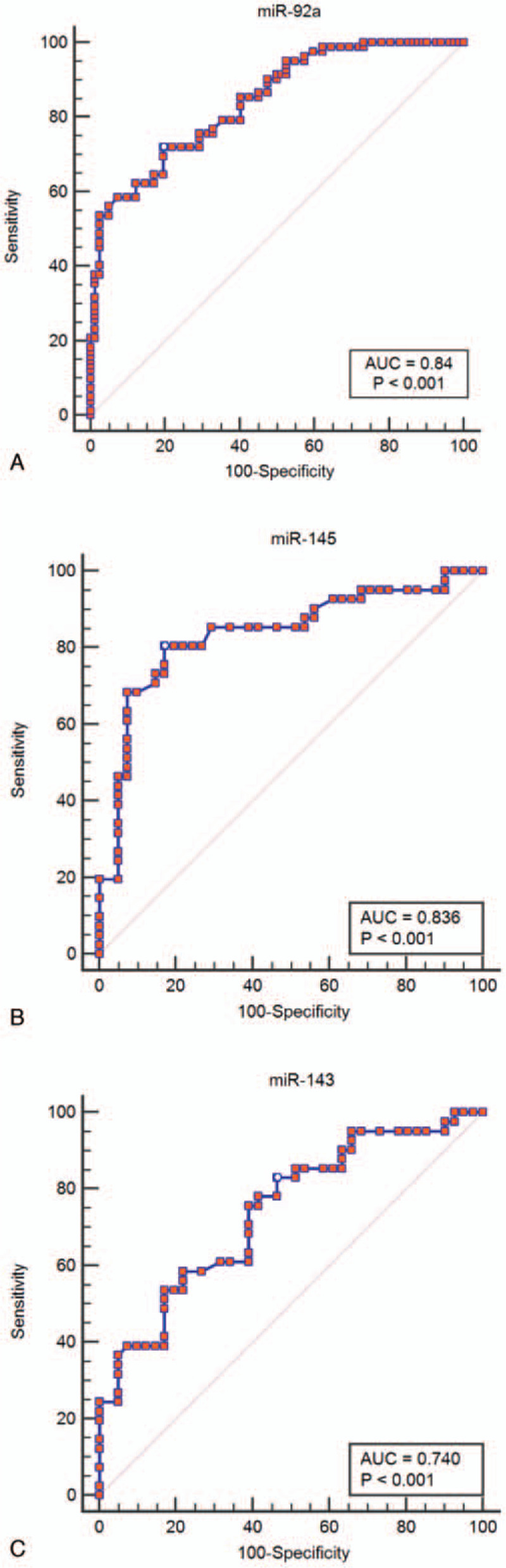
Receiver operating characteristic curves were analyzed to evaluate the miR-92a, miR-143, miR-145 expression levels as potential biomarkers in colorectal cancer tissue detection. area under the curve of upregulated miR-92a (A) expression was 0.84 (sensitivity: 71.95%, specificity: 80.49%; P < .001). For downregulated miR-145 (B), and miR-143 (C) expressions, the area under the curves were 0.85 (sensitivity: 82.93%, specificity: 85.37%; P < .001), and 0.74 (sensitivity: 60.98%, specificity: 80.49%, P < .001).
4. Discussion
CRC continues to be a major cause of mortality induced by cancer worldwide. However, since conventional strategies for CRC treatments have not yet been deemed satisfactory, and an ideal therapeutic target should be associated with the causality of disease. Over the past few decades, numerous studies and resources have identified the involvement of ncRNAs in carcinogenesis and tumor progression.[9] Among all types of ncRNAs, miRNAs have received particular attention and have been proposed as useful diagnostic and prognostic biomarkers. This is attributed to the fact that their profile of tumor tissue has been found to have a close association with a tissue of origin, due to their ability to resist degradation by endogenous ribonuclease, as well as their ease of quantitation involving several methods (e.g., qRT-PCR, microarray or sequencing technology).[10–13]
Dysregulated expressions of miRNAs with oncogenic or tumor suppressor activities have been reported in cancer development, metastasis, angiogenesis and drug resistance.[14,15] In CRC, a large variety of miRNAs are up- or down-regulated as compared to normal tissues. MiRNAs that are consistently found to be down-regulated in CRC act as tumor suppressor genes and are accordingly termed “mirsupps.” In contrast, miRNAs that are consistently found to be upregulated in CRC act as oncogenes and are referred to as “oncomirs”. Against this backdrop, studying the specific function of miRNAs in human carcinogenesis will help characterize new targets for cancer research, diagnosis, and treatment of cancer at the molecular level.
In the present study, we demonstrated that the expression profiles of miRNAs were significantly altered in the selected group of CRC patients from the south-eastern region of Romania. This was done by using the qRT-PCR method and miRNA specific hydrolysis probes TaqManMGB. Moreover, 2 miRNAs, namely, the miR-143 and miR-145, exhibited significantly lower expression, while 1 miRNA, namely the miR-92a, showed increased expression in CRC than in the NATS.
The miR-143 and miR-145 genes are situated in close proximity to each other in a ∼1.7 kb region on chromosome 5q32-33. In addition, they are co-transcribed together from a single bicistronic unit, thereby suggesting that they originate from the same primary transcript and they could be involved in similar functions.[16] Michael et al were the first authors to report the correlation between miRNAs and CRC by uncovering the down-regulation of miR-143 and miR-145 in both precancerous and cancer neoplasia tissues as compared to normal colon epithelium in CRC patients.[17]
In most studies, miR-143 and miR-145 have been presented as independent miRNAs and were not considered as concomitant re-expression genes. On the other hand, co-expression of both miRNAs has been shown to be an early event in the course of cancer development, with anti-oncogenic activities by targeting the same genes, or different genes that regulating the same pathway.[18] Functionally, miR-143/miR-145 cluster expression regulates cell proliferation and differentiation in the Lovo cells by inhibiting the KRAS gene.[19] It is notable that miR-143 and miR-145 could suppress cancer cell proliferation through target sites of the 3’-UTR of insulin-like growth factor 1 receptor and regulate their expression.[20]
In our study, an 8.21-fold decreased expression level of miR-143 in the colorectal samples was observed, as compared to the NATS. However, in the present study in consonance with other authors, we found a significant association between expression of miR-143 and different stages of CRC pathogenesis, including depth of invasion, lymph node metastasis, and distant metastasis.[21,22]
Also, miR-145 expression was decreased 11.66-folds in CRC tissue compared with NATS. Furthermore, we found that miR-145 expression was correlated with a depth of the tumor invasion, lymph node involvement, and distant metastases in CRC patients. Our observation that miR-143 and miR-145 were downregulated in CRC patients was confirmed by other reports.[23–26]
On the other hand, many studies had indicated that dysregulated miR-143, and miR-145 expressions in CRC, are often associated with the CRC progression, and metastasis, which may create a new opportunity to develop the ways of disease mechanism understand, and the potential diagnostic and/or the prognostic biomarkers. Moreover, our obtained results for the expression levels of these miRNAs from CRC samples, are suggesting their use as biomarkers for the diagnostic of the disease with a good performance indicators values (miR-143: 80.49% of specificity and 60.98% of sensitivity; miR-145: 85.37% of specificity and 82.93% of sensitivity, respectively). In addition, the 5 years survival probability time of CRC patients with low expression of miR-143 was shorter than that with the high expression of miR-143. The same difference in survival time was observed in patients with low expression of miR-143 who had a poor prognosis and it can be used as a potential indicator of CRC prognosis.
In CRC research, the role of miR-92a becomes important, as they belong to the miR-17-92 polycistronic cluster, which resides at chromosome 13q13, a region known to be frequently implicated in the proliferation of cancer cells, by suppressing carcinoma cells apoptosis whilst accelerating the progression of the tumor in CRC.[27] Abnormal expression of miR-92a has been known to play a decisive role in various diseases, including breast cancer, CRC, and leukemia.[22,28,29]
Meanwhile, the upregulation of miR-92a plays a pivotal role In CRC pathogenesis, which leads to up-regulation of β-catenin and vimentin, along with the down-regulation of E-cadherin during the regulation of epithelial-mesenchymal transition by specifically targeting phosphatase and tensin homolog, which denotes inhibitory enzyme that is known to block the pathway of PI3K/Akt.[30]
Our study showed that miR-92a was upregulated in CRC, noticing a 2.32-fold increase in colorectal tumors of patients as compared to usual adjacent tissue samples. Furthermore, heightened expression of miR-92a was found to have a significant correlation with the late stage of tumor invasion, lymph node metastasis, and development of distant metastasis in CRC patients.
According to the research carried out by Zhou et al, the upregulation of miR-92a has prognosis prediction in CRC patients; moreover, it was linked to worsening clinical variables and poor rate of survival.[31] Similar conclusions were drawn by our study, which demonstrated that the CRC patients mean survival time was 36 months with a high expression of miR-92a, which is significantly lower as compared to low expression 51 months, respectively. This implies the use of miR-92a as a potentially feasible indicator of prognosis CRC. Meanwhile, Chen et al showed that as an oncomir, diagnostic biomarker miR-92a plays a pivotal role in CRC. In addition, its regulating network may potentially facilitate the mechanism of CRC pathogenesis.[34] When it comes to distinguishing between NATS and CRS, the high specificity (80.49%) and sensitivity (71.95%) of miR-92a expression indicate the potential use of this miRNA as a potential biomarker for CRC diagnosis, which is in alignment with the views of earlier authors.[32,33]
Notably, our study has some limitations. First, because of the small number of cases investigated. The second limitation in agreement with the assumption that abnormal expression levels of selected messenger RNAs were examined only in tissue samples without matching their expression in cell cultures and sera. Therefore, further investigations are needed to confirm our findings.
5. Conclusion
In conclusion, we may summarize that patterns of selected miRNAs are differentially expressed in colorectal adenocarcinoma compared with their normal counterparts with 2 miRNAs decreased (miR-145 and miR-143) and 1 increased (miR-92). Furthermore, the expression levels of selected miRNAs were correlated with gradual increases in fold change expression with different stages of CRC pathogenesis, including depth of tumor invasion, lymph node invasion, and maximal increases with distant metastasis. In addition, ROC analysis and AUC demonstrates the ability of selected miRNAs to discriminating CRC from NATS with good performance of specificity and sensitivity. Prognosis indicators of CRC were also examined using Kaplan–Meier curves and the log-rank test which showed that 5-year overall survival of patients with a high level of miR-92a and low levels of miR-143 and miR-145 are significantly associated with poor prognosis in our cohorts.
Author contributions
All authors contributed equally to this study.
Conceptualization: Mariana Aşchie, Costel Brînzan, Anca Mitroi.
Data curation: Costel Brînzan.
Formal analysis: Costel Brînzan.
Investigation: Costel Brînzan, Anca Mitroi.
Methodology: Costel Brînzan, Anca Mitroi, Georgeta Cozaru.
Software: Costel Brînzan.
Supervision: Mariana Aşchie, Eugen Dumitru.
Validation: Georgeta Cozaru.
Writing – original draft: Costel Brînzan, Georgeta Cozaru.
Writing – review & editing: Costel Brînzan, Anca Mitroi, Eugen Dumitru.
Footnotes
Abbreviations: AUC = area under the curve, CI = 95% interval of confidence, CRC = colorectal cancer, miRNA = microRNA, ncRNAs = non-coding RNAs, NATS = normal adjacent tissue samples, qRT-PCR = real-time quantitative polymerase chain reaction analysis, ROC = receiver operating characteristic, RT = room temperature, TNM = tumor-node-metastasis, UTR = 3’ noncoding region, WHO = World Health Organization.
How to cite this article: Brînzan C, Aşchie M, Cozaru G, Dumitru E, Mitroi A. The diagnostic value of miR-92a, -143, and -145 expression levels in patients with colorectal adenocarcinoma from Romania. Medicine. 2020;99:35(e21895).
All authors contributed equally to this work and are the co-first authors.
Experiments of molecular biology was performed at the Research Center for the Morphologic and Genetic Study in Malignant Pathology, CEDMOG, from Ovidius University.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Siegel R, Desantis C, Jemal A. Colorectal cancer statistics. CA Cancer J Clin 2014;64:104–17.. [DOI] [PubMed] [Google Scholar]
- [2]. World Health Organization - cancer country profiles 2014, Romania, http://www.who.int/cancer/country-profiles/rou_en.pdf?ua=1MaL. Accessed November 25, 2019. [Google Scholar]
- [3].Brînzan C, Aşchie M, Grasa CN, et al. The mutation profiles of Kras and Braf genes in a Romanian colorectal cancer cohort. Rev Chim 2019;70:1346–50.. [Google Scholar]
- [4].Brînzan C, Aşchie M, Matei E, et al. Molecular expression profiles of selected microRNAs in colorectal adenocarcinoma in patients from south-eastern part of Romania. Medicine 2019;98:e18122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97.. [DOI] [PubMed] [Google Scholar]
- [6].Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res 2011;717:1–8.. [DOI] [PubMed] [Google Scholar]
- [7].Edge SB, Byrd DR, Compton CC, et al. AJCC cancer staging manual-seventh edition. Springer 2010;7:143–59.. [Google Scholar]
- [8].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001;25:402–8.. [DOI] [PubMed] [Google Scholar]
- [9].Seton-Rogers S. Non-coding RNAs: the cancer X factor. Nat Rev Cancer 2013;13:224–5.. [DOI] [PubMed] [Google Scholar]
- [10].Xiao B, Guo J, Miao Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta 2009;400:97–102.. [DOI] [PubMed] [Google Scholar]
- [11].Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008;299:425–36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–66.. [DOI] [PubMed] [Google Scholar]
- [13].Waldman SA, Terzic A. Translating microRNA discovery into clinical biomarkers in cancer. JAMA 2007;297:1923–5.. [DOI] [PubMed] [Google Scholar]
- [14].Esquela-Kerscher A, Slack FJ. Oncomirs- microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–69.. [DOI] [PubMed] [Google Scholar]
- [15].Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther 2008;15:341–55.. [DOI] [PubMed] [Google Scholar]
- [16].Cordes KR, Sheehy NT, White MP, et al. MiR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009;460:705–10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Michael MZ, O’ Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003;1:882–91.. [PubMed] [Google Scholar]
- [18].Sempere LF, Christensen M, Silahtaroglu A, et al. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 2007;67:11612–20.. [DOI] [PubMed] [Google Scholar]
- [19].Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009;28:1385–92.. [DOI] [PubMed] [Google Scholar]
- [20].Su J, Liang H, Yao W, et al. MiR-143 and miR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS One 2014;9:e1144–220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Secil AK, Tunca B, Tezcan G, et al. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J Surg Res 2014;191:113–22.. [DOI] [PubMed] [Google Scholar]
- [22].Nishida N, Nagahara M, Sato T, et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 2012;18:3054–70.. [DOI] [PubMed] [Google Scholar]
- [23].Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 2010;17:398–408.. [DOI] [PubMed] [Google Scholar]
- [24].Bandres E, Cubedo E, Agirre X, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 2006;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lanza G, Ferracin M, Gafa R, et al. MRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 2007;6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA 2006;103:3687–92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol 2012;36:e61–7.. [DOI] [PubMed] [Google Scholar]
- [28].Sharifi M, Salehi R, Gheisari Y, et al. Inhibition of microRNA miR-92a induces apoptosis and necrosis in human acute promyelocytic leukemia. Adv Biomed Res 2014;3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Si H, Sun X, Chen Y, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol 2013;139:223–9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lu C, Shan Z, Hong J, et al. MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. Int J Oncol 2017;51:235–44.. [DOI] [PubMed] [Google Scholar]
- [31].Zhou T, Zhang G, Liu Z, et al. Over expression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis 2013;28:19–24.. [DOI] [PubMed] [Google Scholar]
- [32].Yang X, Zeng Z, Hou Y, et al. MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta-analysis. PLoS One 2014;9:e88745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chang PY, Chen CC, Chang YS, et al. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget 2016;7:10663–75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen E, Li Q, Wang H, et al. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed Pharmacother 2018;106:1370–7.. [DOI] [PubMed] [Google Scholar]


