Abstract
Background and Aims
Genome-wide association studies [GWASs] of European populations have identified numerous susceptibility loci for Crohn’s disease [CD]. Susceptibility genes differ by ethnicity, however, so GWASs specific for Asian populations are required. This study aimed to clarify the Japanese-specific genetic background for CD by a GWAS using the Japonica array [JPA] and subsequent imputation with the 1KJPN reference panel.
Methods
Two independent Japanese case/control sets (Tohoku region [379 CD patients, 1621 controls] and Kyushu region [334 CD patients, 462 controls]) were included. GWASs were performed separately for each population, followed by a meta-analysis. Two additional replication sets [254 + 516 CD patients and 287 + 565 controls] were analysed for top hit single nucleotide polymorphisms [SNPs] from novel genomic regions.
Results
Genotype data of 4 335 144 SNPs from 713 Japanese CD patients and 2083 controls were analysed. SNPs located in TNFSF15 (rs78898421, Pmeta = 2.59 × 10−26, odds ratio [OR] = 2.10), HLA-DQB1 [rs184950714, pmeta = 3.56 × 10−19, OR = 2.05], ZNF365, and 4p14 loci were significantly associated with CD in Japanese individuals. Replication analyses were performed for four novel candidate loci [p <1 × 10−6], and rs488200 located upstream of RAP1A was significantly associated with CD [pcombined = 4.36 × 10−8, OR = 1.31]. Transcriptome analysis of CD4+ effector memory T cells from lamina propria mononuclear cells of CD patients revealed a significant association of rs488200 with RAP1A expression.
Conclusions
RAP1A is a novel susceptibility locus for CD in the Japanese population.
Keywords: RAP1A, Crohn’s disease, susceptibility gene
1. Introduction
Inflammatory bowel disease [IBD] is characterised by chronic gastrointestinal tract inflammation. There are two common forms of IBD: Crohn’s disease [CD] and ulcerative colitis, distinguished by the distribution of inflammation and other symptoms. There is a strong genetic component to IBD susceptibility. Indeed, genome-wide association studies [GWASs], meta-analyses, and trans-ethnic analyses have identified more than 200 candidate susceptibility loci for IBD.1–3
Recent advances in screening for IBD susceptibility genes include ImmunoChip, a platform of approximately 200 000 single nucleotide polymorphisms [SNPs] designed for fine-mapping known loci associated with immune-mediated diseases.4 However, ImmunoChip was designed from the pilot results of the 1000 Genomes Project and is therefore based on populations of European ancestry. Thus, it was not designed for use in individuals of non-European ancestry, such as Japanese individuals. Furthermore, numerous studies have confirmed ethnic differences in CD susceptibility ‘genes’. For example, NOD2 variants associated with CD in Caucasians5,6 are not polymorphic in Japanese populations.7,8 In addition, ATG16L1 T300A is polymorphic in Japanese populations but is not associated with CD susceptibility in Japanese individuals.9,10 On the other hand, TNFSF15 has been established as an East Asian dominant CD susceptibility gene.11,12 Therefore, although there have been significant advances in our knowledge of the genetic architecture of IBD, these are largely based on studies of European populations, so studies designed specifically to address the genetic background for IBD in East Asian populations are required.
Tohoku University and Tohoku Medical Megabank Organisation created a 1KJPN panel of genomic variations obtained through whole-genome resequencing of 1070 Japanese individuals.13 We subsequently developed the Japonica array [JPA], which is optimised for genotyping and imputing individuals of Japanese ancestry.14 The array contains 659 253 SNPs, including tag SNPs for imputation as well as SNPs related to phenotypes from previously reported GWASs and pharmacogenomics studies. The tag SNPs were selected on the basis of the 1KJPN panel data, allowing imputation of ungenotyped SNPs with high accuracy.
The aim of this study was to clarify the Japanese-specific genetic background of CD. We used our high-quality reference panel data and a powerful genotyping array to complete the first CD GWAS specifically designed for the Japanese population. Additionally, genotype–phenotype analyses were performed to clarify the relationships between novel susceptibility loci and clinical subphenotypes of CD.
2. Materials and Methods
2.1. Subjects
In total, we analysed 1483 CD patients and 2935 healthy controls from two independent case/control sample sets for GWASs and two replication sets. The two GWAS sample sets were from Tohoku in northeast Japan and Kyushu in southwest Japan. The Tohoku cohort [GWAS_Tohoku] included 410 CD patients recruited at Tohoku University Hospital from May 1998 to May 2015 and 1720 controls who participated in the cohort study for the Tohoku Medical Megabank Project.15 The Kyushu cohort [GWAS_Kyushu] included 349 CD patients recruited at Kyushu University and 16 affiliated hospitals and 468 controls working as medical staff at the National Hospital Organisation [NHO] Nagasaki Medical Centre.
The first replication sample set [Replication 1] included 254 patients with CD and 287 healthy controls from the Kyushu and Tohoku regions. The second independent replication sample set [Replication 2] included 516 patients with CD who participated in the MENDEL study,16 and 565 controls from the Tokyo [n = 418] and Kyushu [n = 147] regions. These four cohorts were non-overlapping. All CD patients were of Japanese ethnicity and diagnosed on the basis of clinical symptoms and endoscopic, radiographic, and histological findings according to conventional criteria proposed by the Japanese Ministry of Health, Labour and Welfare.17 Patients with CD were stratified according to age at diagnosis, disease location, and progression, following the Montreal classification.18 Patient characteristics of the GWAS samples are summarised in Supplementary Table S1, available as Supplementary data at ECCO-JCC online.
CD4+ effector memory T [Tem] cells among lamina propria mononuclear cells [LPMCs] were isolated from the surgical specimens of seven Japanese CD patients undergoing bowel resection at Tohoku University Hospital, from July 2015 to September 2016. Patient characteristics and medications are shown in Supplementary Table S2, available as Supplementary data at ECCO-JCC online.
The Ethics Committees of Tohoku University School of Medicine and Kyushu University Graduate School of Medical Sciences approved this study. The study was performed in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments. All participants provided informed consent.
2.2. Genotyping
Genomic DNA was extracted from peripheral blood leukocytes by standard phenol–chloroform precipitation using the NA1000 Automated Nucleic Acid Extraction Machine [Kurabo, Osaka, Japan] or PAXgene DNA Kit [BD Biosciences, Franklin Lakes, NJ]. Genome-wide SNP genotypes were determined using the Japonica Array [Toshiba, Tokyo, Japan]. Genotype calling was conducted using Affymetrix Power Tools [version 1.18.2; Thermo Fisher Scientific Inc., Waltham, MA]. Quality control [QC] criteria were dish QC ≥0.82 and sample call rate ≥97%, as recommended by Affymetrix. SNPs were categorised by cluster separation using the SNPolisher package [version 1.5.2; Thermo Fisher Scientific Inc.]. Subsequent analyses were based on 614 597 autosomal SNPs categorised as ‘Recommended’. Identity by descent probabilities [PI_HAT] were estimated using plink v1.90a software,19 and cryptic relatives were detected by the maximum unrelated set identification [IMUS] method implemented in PRIMUS v1.8.020 using a minimum PI_HAT value of 0.1. For QC, SNPs with call rates <97% and samples with genotyping rates <97% or samples of cryptic relatives were excluded from GWASs [Figure 1].
Figure 1.
Analysis flow of this study. Tohoku and Kyushu sample sets were analysed separately. Genotyping was performed using the Japonica array [JPA]. Untyped genotypes were imputed with the 1KJPN panel as a reference. Disease associations were analysed in each sample set using logistic regression, and a meta-analysis was performed including both genome-wide association studies [GWASs]. Finally, two independent replication analyses were performed with novel candidates.
Four novel candidate variants with p <1 × 10−6 were analysed in the first replication sample set [Replication 1] using TaqMan SNP Genotyping Assays [Applied Biosystems, Foster City, CA] according to the manufacturer’s protocol, on a Light Cycler 96 System [Roche, Basel, Switzerland]. As an additional replication dataset [Replication 2], genotype frequencies of these four candidates were obtained from whole-genome sequenced data of healthy controls [Supplementary Method, available as Supplementary data at ECCO-JCC online] and from imputed genotype data of CD cases analysed in the MENDEL study.16
2.3. Imputation
Untyped genotypes were imputed in GWAS samples using IMPUTE2 [version 2.3.2]21 and 1070 healthy individuals from Japan (1KJPN panel, which includes >20 million single nucleotide variants [SNVs]) as a reference dataset.13 SNPs with Hardy–Weinberg equilibrium [HWE] p-value < 1 × 10−4, call rate <0.99, or minor allele frequency <0.005 were filtered out as SNP QC for prephasing. A total of 564 190 SNPs on autosomal chromosomes were phased using SHAPEIT [v2.r644]22 as follows: --burn 10, --prune 10, and --main 25. Phased genotypes were imputed with IMPUTE2 using the 1KJPN panel as follows: -Ne 2000, -k hap 1000, -burnin 15, and -iter 50. This yielded data for 24 344 327 variants. SNVs with low imputation quality [posterior genotype probability of <0.8], low call rate of <0.97, minor allele frequency <0.05, and HWE-p <1 × 10−6 were excluded. Further analysis was performed using genotype data of >4 million SNVs from 713 Japanese CD cases and 2083 controls [Figure 1].
2.4. Principal component analysis
Differences between the Tohoku and Kyushu sample sets and population outliers were detected by principal component analysis [PCA] of linkage disequilibrium [LD]-independent SNPs from 2796 GWAS samples using PLINK1.90 software [www.cog-genomics.org/plink/1.9/].19 LD pruning was conducted using PLINK1.90 as follows: -indep-pairwise 50 5 0.1. The samples appeared almost homogeneous in the plot showing the top two principal components [PC1 and PC2] of each sample [Supplementary Figure S1A, available as Supplementary data at ECCO-JCC online]. However, closer observation revealed detectable differences between the Tohoku and Kyushu samples, which are identified in the PC1 histogram [Supplementary Figure S1B, available as Supplementary data at ECCO-JCC online].
2.5. Cell isolation and culture
Lamina propria mononuclear cells [LPMCs] from the diseased parts of resected small intestine or colon were isolated by sequential treatment with dithiothreitol [DTT], ethylenediaminetetraacetic acid [EDTA], and collagenase as previously described,23 followed by selection using Ficoll-Hypaque [GE Healthcare, Tokyo, Japan]. CD4+ cells were separated from LPMCs by negative selection using Easy Sep Magnet [STEMCELL Technology, Grenoble, France] with Easy Sep Human CD4+ T cell Enrichment [STEMCELL Technology]. Tem cells were then isolated from the total CD4+ T cell population using a FACSAria II cell sorter [BD Biosciences] after staining with anti-CD3-FITC, CD4-PE, CD45RO-APC, CD197 [CCR7]-BV421, and 7ADD-Cell Viability Solution [BD Biosciences]. Sorting efficiency was always >98%. Total RNA was extracted from isolated Tem cells using the AllPrep DNA/RNA mini kit [Qiagen, Dusseldorf, Germany] according to the manufacturer’s instructions.
2.6. Library preparation and sequencing
Total RNA quality and quantity were assessed by rRNA band integrity analysis using the Agilent RNA 6000 Chip [Agilent Technologies, Palo Alto, CA]. cDNA was synthesised using the SMART-Seq v4 Ultra Low Input RNA Kit [Clontech Laboratories, Inc., Mountainview, CA]. We used 500 pg total RNA as the template to ensure full-length cDNA synthesis containing the 5′-end and direct polymerase chain reaction [PCR] adapter addition to both ends of the first-strand cDNA. Amplified cDNA was validated using the Agilent 2100 BioAnalyzer High Sensitivity DNA Chip [Agilent Technologies]. Amplification yielded approximately 2–10 ng cDNA with fragments lengths of 400–10 000 bp. Full-length cDNA was processed using the Low Input Library Prep Kit [Clontech Laboratories]. Before generating the final library, the amplified cDNA samples were sheared using a Covaris S2 sonicator [Covaris Inc., Woburn, MA]. Fragments of 200–500 bp were ligated to adapters from Illumina [San Diego, CA] and amplified by PCR. Libraries were quantified using the Agilent 2100 Bioanalyzer [Agilent Technologies] and KAPA Library Quantification Kit [Kapa Biosystems, Wilmington, MA]. The resulting purified libraries were applied to a flow cell [Illumina] for cluster generation and sequenced using 100 bp paired-end reads on an Illumina Hiseq2500 sequencer, according to the manufacturer’s protocol.
2.7. Transcriptome analysis
Adapter and low-quality sequences were removed using cutadapt [v1.2.1],24 and poly-A/T sequences were eliminated using PRINSEQ [v0.19.2].25 For gene expression analysis, the trimmed reads were aligned to the reference human genome [GRCh37/hg19] using TopHat [v2.0.13].26 Mapped reads were assembled using Cufflinks [v2.2.1],27 and the transcripts across all samples were merged using the Cuffmerge utility of the Cufflinks package. Fragments per kilobase per million map reads [FPKM] were calculated using Cuffquant.
2.8. Statistical analysis
In light of PCA showing characteristic differences between Tohoku and Kyushu sample sets, we conducted two separate GWASs, followed by a meta-analysis [fixed-effect model] of all results using 4 335 144 variants on autosomal chromosomes. GWASs were evaluated by logistic regression with sex as a covariate using PLINK v 1.90 software.19 SNPs with pmeta <5 × 10−8 were considered of genome-wide significance and SNPs with pmeta <1 × 10−6 were considered to be nominally significant. SNPs located within 250 kbp were considered to be in one region. In addition, all SNPs in the human leukocyte antigen region were considered to be at one location. After GWASs, the representative top hit SNPs from novel candidate regions were selected for replication analysis. We compared the allele frequencies, dominant model, and recessive model between CD and control groups using the chi square test. Finally, a meta-analysis of the four results [GWAS_Tohoku, GWAS_Kyushu and two replications] was performed, and SNPs with pcombined <5 × 10−8 were considered to be of genome-wide significance. Heterogeneity among the studies was evaluated using Cochrane’s Q statistic. Deviations from HWE were evaluated using the chi square test.
Correlations between genotype and expression levels of Tem cell genes located around associated novel SNPs [±200 kbp] were analysed using linear regression. Manhattan plots were generated using the R package qqman,28 and regional association plots were generated using the LocusZoom application.29 All statistical analyses except genome-wide linear regression were performed using R software [version 3.2.5] [http://www.r-project.org/].
2.9. Pathway analysis
SNPs with pmeta <5 × 10−4 were used for interaction network construction and gene enrichment analysis. These SNPs were first annotated to corresponding genes, and the genes were further annotated using multiple biologically functional databases, including the human protein reference database [www.hprd.org], Reactome, NCI/Nature pathway interaction database, and others. The final networks were constructed from interactions identified in any of the databases. Pathway and gene set enrichment analyses were performed using STRING [http://string-db.org/]30 and cytoscape [http://www.cytoscape.org].31
3 Results
3.1. Discovery GWAS meta-analysis identified four novel candidate loci
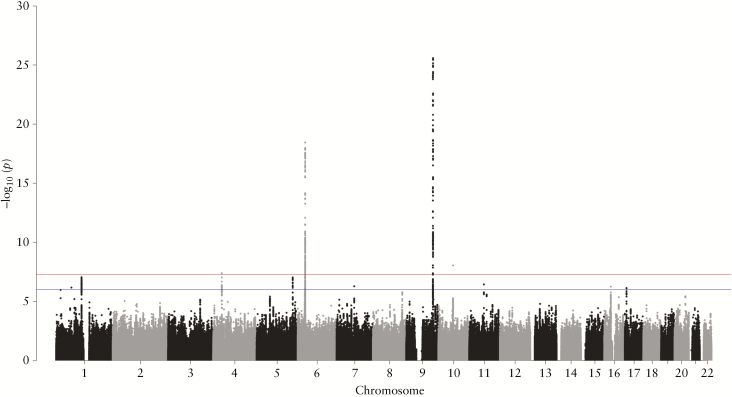
The Manhattan plot and Q-Q plot of the meta-analysis for the two GWASs [GWAS_Tohoku and GWAS_Kyushu] are shown in Figure 2 and in Supplementary Figure S2, available as Supplementary data at ECCO-JCC online. The genomic inflation factors [lambda GC] of GWAS_Tohoku, GWAS_Kyushu, and GWAS_meta analysis were 1.0898, 1.0591, and 1.0802, respectively. A total of 585 variants with statistical significance or nominal significance [pmeta <1 × 10−6] could be grouped into 11 loci [Table 1]. Seven of the 11 loci were located within or close to regions previously associated with CD. Of these, TNFSF1511,12 [rs78898421, pmeta = 2.59 × 10−26, OR = 0.48], HLA-DQB132,33 [rs184950714, pmeta = 3.56 × 10−19, OR = 2.05], ZNF3652 [rs224136, pmeta = 9.04 × 10−9, OR = 0.63] and 4p143,34 [rs73243351, pmeta = 4.04 × 10−8, OR = 1.53] were also significantly associated with CD in Japanese individuals in our cohorts [pmeta <5 × 10−8], and the other three known loci, IL12B,2,35,36 IL27,2,10 and IL23R G149R,37–39 showed nominal significance [Supplementary Figure S3, available as Supplementary data at ECCO-JCC online]. The remaining four loci identified [CD_5, 7, 8 and 11] are all novel candidates [Supplementary Figure S4, available as Supplementary data at ECCO-JCC online].
Figure 2.
Manhattan plot of meta-analysis results for two CD GWASs in Japanese individuals. Single nucleotide polymorphisms are plotted according to chromosomal location, with the -log10[P] from the meta-analysis of two GWASs. The red line indicates the threshold for genome-wide significance [pmeta = 5 × 10−8]. The blue line indicates the threshold for nominal significance [pmeta = 1 × 10−6]. CD, Crohn’s disease; GWAS, genome-wide association study.
Table 1.
Meta-analysis of two independent GWASs for Japanese CD.
| Locus ID | Chr | Rangea [Mbp] | No. of SNPsb | Novel [ref] or known | Genes | Top SNP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP | Genotyped or imputed | GWAS_Tohoku | GWAS_Kyushu | GWAS_Meta | ||||||||||||
| p-value | OR [95%CI] | p-value | OR [95%CI] | MAF | p meta | OR | Qc | |||||||||
| Ctrl | CD | |||||||||||||||
| CD_1 | 9 | 117.4–117.7 | 170 | 11 | TNFSF15, TNFSF8 | rs78898421 | Imputed | 2.67E-17 | 2.08 [1.75–2.44] | 1.47E-10 | 2.13 [1.69–2.70] | 47.0% | 63.1% | 2.59E-26 | 2.10 | 0.8328 |
| CD_2 | 6 | 31.1–32.9 | 318 | 32,33 | HLA-DQB1 | rs184950714 | Imputed | 9.40E-12 | 1.92 [1.59–2.32] | 3.13E-09 | 2.36 [1.78–3.14] | 15.6% | 27.7% | 3.56E-19 | 2.05 | 0.2378 |
| CD_3 | 10 | 64.5 | 1 | 2,42 | ZNF365 | rs224136 | Genotyped | 2.14E-08 | 0.57 [0.47–0.70] | 3.80E-02 | 0.75 [0.58–0.98] | 29.7% | 21.9% | 9.04E-09 | 0.63 | 0.1046 |
| CD_4 | 4 | 38.3–38.4 | 15 | 3,34 | rs73243351 | Imputed | 8.37E-04 | 1.37 [1.14–1.65] | 1.20E-06 | 1.96 [1.49–2.57] | 20.3% | 27.7% | 4.04E-08 | 1.53 | 0.0320 | |
| CD_5 | 1 | 112.1–112.3 | 64 | Novel | RAP1A, FAM212B | rs488200 | Imputed | 8.74E-06 | 1.44 [1.22–1.68] | 2.99E-03 | 1.43 [1.13–1.80] | 44.5% | 50.9% | 8.85E-08 | 1.43 | 0.9653 |
| CD_6 | 5 | 158.8 | 11 | 2,35 | IL12B | rs56167332 | Genotyped | 8.76E-09 | 1.58 [1.35–1.85] | 3.14E-01 | 1.13 [0.89–1.43] | 38.1% | 46.0% | 9.24E-08 | 1.43 | 0.0184 |
| CD_7 | 11 | 64.9 | 1 | Novel | SYVN1, CAPN1 | rs11227126 | Imputed | 1.17E-04 | 1.76 [1.32–2.34] | 6.01E-04 | 2.23 [1.41–3.52] | 5.6% | 9.4% | 3.65E-07 | 1.88 | 0.3899 |
| CD_8 | 7 | 76.9 | 1 | Novel | GSAP, FGL2 | rs4727354 | Imputed | 1.16E-07 | 2.32 [1.70–3.17] | 3.78E-01 | 1.27 [0.75–2.17] | 4.1% | 7.1% | 5.17E-07 | 1.99 | 0.0558 |
| CD_9 | 16 | 28.5 | 1 | 2,10 | IL27 | rs56354901 | Genotyped | 3.14E-04 | 1.49 [1.20–1.85] | 2.61E-04 | 1.87 [1.34–2.62] | 12.2% | 16.6% | 5.61E-07 | 1.59 | 0.2599 |
| CD_10 | 1 | 67.6 | 1 | 2,37-39 | IL23R | rs76418789 | Genotyped | 1.68E-06 | 0.33 [0.21–0.52] | 3.09E-02 | 0.58 [0.36–0.95] | 7.7% | 3.6% | 6.59E-07 | 0.43 | 0.0914 |
| CD_11 | 17 | 7.1 | 2 | Novel | DLG4, ACADVL | rs2521985 | Imputed | 2.45E-05 | 0.70 [0.59–0.83] | 9.18E-03 | 0.72 [0.56–0.92] | 39.3% | 32.0% | 7.23E-07 | 0.70 | 0.8481 |
Chr, chromosome; MAF, minor allele frequency; SNP, single nucleotide polymorphism; CD, Crohn’s disease; GWAS, genome-wide association study; CI, confidence interval.
aPositions are based on the Genome Reference Consortium human build 37 [GRCh37].
bNumber of SNPs with pmeta <1 × 10−6.
c p-values for Cochrane’s Q statistic.
3.2. Novel association of rs488200 with CD
Among novel loci, CD_5 exhibited the strongest association with CD. It was tagged by rs488200 [pmeta = 8.85 × 10−8, OR = 1.43] and located downstream of RAP1A and FAM212B [Figure 3]. Two replication analyses of the top hit SNPs from the four novel loci [Table 2] revealed a significant association of rs488200 in CD_5 with CD [p = 2.11 × 10−2]. Further, combined analysis [Tohoku GWAS, Kyushu GWAS, Replication 1, Replication 2, and combined] yielded a statistically significant genome-wide association with CD in Japanese individuals [pcombined = 4.36 × 10−8, OR = 1.31].
Figure 3.
Locus zoom plots of pmeta around the CD_5 locus. The top associated SNP [rs488200] is shown as a purple circle and the remaining SNPs as circles or squares by whether the SNPs were imputed or genotyped, with colours indicating the level of linkage disequilibrium [R2] with rs488200. CD, Crohn’s disease; SNP, single nucleotide polymorphism.
Table 2.
Replication and combined analysis of four novel candidate SNPs.
| Locus ID | Chr | Top SNP | Samples | Genotype counts & associations | A1 allele frequencies & associations | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP | A1/A2 | Control | CD | HWE-p | p-value | Control | CD | p-value | OR | Qa | |||||||||
| A1/A1 | A1/A2 | A2/A2 | A1/A1 | A1/A2 | A2/A2 | Ctrl | CD | Dominant | Recessive | ||||||||||
| CD_5 | 1 | rs488200 | T/A | GWAS_Tohoku | 324 | 820 | 480 | 108 | 182 | 85 | 0.438 | 0.619 | 7.79E-03 | 1.92E-04 | 45.2% | 53.1% | 8.74E-06 | 1.44 [1.22–1.68] | |
| GWAS_Kyushu | 75 | 239 | 140 | 77 | 165 | 89 | 0.111 | 0.975 | 2.30E-01 | 1.87E-02 | 42.8% | 48.2% | 2.99E-03 | 1.43 [1.13–1.80] | |||||
| Replication 1 | 57 | 115 | 88 | 59 | 130 | 59 | 0.098 | 0.446 | 1.28E-02 | 6.16E-01 | 44.0% | 50.0% | 5.70E-02 | 1.27 [0.99–1.63] | |||||
| Replication 2 | 128 | 279 | 158 | 128 | 263 | 122 | 0.820 | 0.563 | 1.18E-01 | 3.76E-01 | 47.3% | 50.6% | 1.33E-01 | 1.14 [0.96–1.35] | |||||
| Replication_Combined | 185 | 394 | 246 | 187 | 393 | 181 | 0.255 | 0.363 | 6.73E-03 | 3.11E-01 | 46.3% | 50.4% | 2.11E-02 | 1.18 | 0.4692 | ||||
| All Combined | 584 | 1453 | 866 | 372 | 740 | 355 | 0.581 | 0.811 | 9.53E-05 | 7.42E-05 | 45.1% | 50.6% | 4.36E-08 | 1.31 | 0.2143 | ||||
| CD_7 | 11 | rs11227126 | T/A | GWAS_Tohoku | 3 | 188 | 1447 | 2 | 71 | 304 | 0.224 | 0.320 | 7.72E-05 | 2.43E-01 | 5.9% | 9.9% | 1.17E-04 | 1.76 [1.32–2.34] | |
| GWAS_Kyushu | 2 | 39 | 414 | 1 | 58 | 272 | 0.305 | 0.252 | 3.18E-04 | 7.59E-01 | 4.7% | 9.1% | 6.01E-04 | 2.23 [1.41–3.52] | |||||
| Replication 1 | 2 | 36 | 242 | 3 | 38 | 213 | 0.607 | 0.385 | 4.04E-01 | 5.80E-01 | 7.1% | 8.7% | 3.57E-01 | 1.23 [0.79–1.93] | |||||
| Replication 2 | 1 | 94 | 470 | 2 | 75 | 437 | 0.096 | 0.520 | 4.11E-01 | 5.20E-01 | 8.5% | 7.7% | 4.91E-01 | 0.90 [0.66–1.22] | |||||
| Replication_Combined | 3 | 130 | 712 | 5 | 113 | 650 | 0.250 | 0.971 | 8.37E-01 | 4.07E-01 | 8.0% | 8.0% | 9.66E-01 | 0.97 | 0.2506 | ||||
| All Combined | 8 | 357 | 2573 | 8 | 242 | 1226 | 0.221 | 0.287 | 1.46E-04 | 2.64E-01 | 6.3% | 8.7% | 2.60E-04 | 1.39 | 0.0022 | ||||
| CD_8 | 7 | rs4727354 | T/G | GWAS_Tohoku | 4 | 123 | 1509 | 2 | 62 | 313 | 0.376 | 0.567 | 8.01E-08 | 3.70E-01 | 4.0% | 8.8% | 1.16E-07 | 2.32 [1.70–3.17] | |
| GWAS_Kyushu | 0 | 42 | 410 | 0 | 36 | 296 | 0.300 | 0.296 | 4.74E-01 | NA | 4.6% | 5.4% | 3.78E-01 | 1.27 [0.75–2.17] | |||||
| Replication 1 | 0 | 22 | 256 | 0 | 33 | 220 | 0.492 | 0.267 | 5.49E-02 | NA | 4.0% | 6.5% | 5.96E-02 | 1.69 [0.97–2.95] | |||||
| Replication 2 | 0 | 59 | 506 | 1 | 51 | 459 | 0.190 | 0.736 | 8.86E-01 | NA | 5.2% | 5.2% | 9.71E-01 | 0.99 [0.68–1.45] | |||||
| Replication_Combined | 0 | 81 | 762 | 1 | 84 | 679 | 0.143 | 0.333 | 3.30E-01 | NA | 4.8% | 5.6% | 2.94E-01 | 1.19 | 0.1196 | ||||
| All Combined | 4 | 246 | 2681 | 3 | 182 | 1288 | 0.508 | 0.188 | 1.41E-05 | NA | 4.3% | 6.4% | 7.45E-06 | 1.59 | 0.0065 | ||||
| CD_11 | 17 | rs2521985 | T/C | GWAS_Tohoku | 269 | 757 | 602 | 48 | 141 | 186 | 0.235 | 0.012 | 7.33E-06 | 7.58E-02 | 39.8% | 31.6% | 2.45E-05 | 0.70 [0.59–0.83] | |
| GWAS_Kyushu | 55 | 237 | 162 | 35 | 149 | 146 | 0.025 | 0.740 | 1.56E-02 | 5.13E-01 | 38.2% | 33.2% | 9.18E-03 | 0.72 [0.56–0.92] | |||||
| Replication 1 | 42 | 133 | 112 | 33 | 109 | 109 | 0.805 | 0.488 | 3.01E-01 | 6.20E-01 | 37.8% | 34.9% | 3.17E-01 | 0.88 [0.69–1.13] | |||||
| Replication 2 | 55 | 186 | 177 | 67 | 216 | 214 | 0.578 | 0.290 | 8.95E-01 | 9.89E-01 | 35.4% | 35.2% | 9.28E-01 | 0.99 [0.83–1.19] | |||||
| Replication_Combined | 97 | 319 | 289 | 100 | 325 | 323 | 0.550 | 0.206 | 4.81E-01 | 7.82E-01 | 36.4% | 35.1% | 5.21E-01 | 0.95 | 0.4462 | ||||
| All Combined | 421 | 1313 | 1053 | 183 | 615 | 655 | 0.663 | 0.055 | 2.52E-05 | 1.28E-01 | 38.7% | 33.8% | 5.36E-05 | 0.81 | 0.0256 | ||||
HWE, Hardy–Weinberg equilibrium; OR, odds ratio; N/A, not available; SNP, single nucleotide polymorphism; CD, Crohn’s disease; GWAS, genome-wide association study.
a p-values for Cochrane’s Q statistic.
3.3. Expression level of RAP1A in Tem cells was associated with the rs488200 genotype
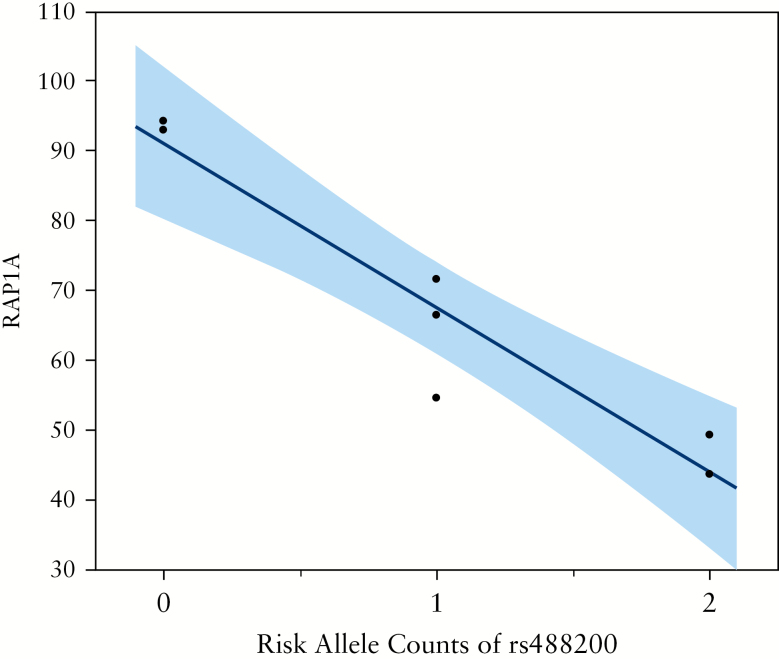
We selected 10 transcripts from genes located within ±200 kbp of rs488200, to investigate the effects of rs488200 variants on the expression levels of neighbouring genes. However, we were unable to detect the expression of six of these genes in Tem cells [mean FPKM <1.0], so only four genes were analysed. RAP1A was significantly associated with rs488200 genotype [Supplementary Table S3, available as Supplementary data at ECCO-JCC online], and the CD risk allele [T allele] was associated with decreased expression of RAP1A in Tem cells [p = 9.27 × 10−4, adjusted R2 = 0.888] [Figure 4].
Figure 4.
Scatter plot of rs488200 risk allele counts and the expression level of RAP1A in CD4+ effector memory T [Tem] cells. The blue line indicates the regression line, and the blue area indicates the 95% confidence limits. A strong association can be observed between the rs488200 genotype and RAP1A expression.
3.4. The cell adhesion molecule pathway was significantly associated with CD in Japanese individuals
Pathway and gene set enrichment analyses using the two GWAS meta-analysis results showed an association with KEGG pathways, including ‘cell adhesion molecules’ [CAMs], ‘antigen processing and presentation’, and ‘intestinal immune network for IgA production’ [FDR = 8.26 × 10−6, 4.27 × 10−5 and 9.85 × 10−5, respectively] [Supplementary Table S4, available as Supplementary data at ECCO-JCC online]. An interaction network analysis showed direct connections of RAP1A with several kinases or kinase pathway modulators, including PTPN1 [protein tyrosine phosphatase, non-receptor type 1], CSK [C-terminal Src kinase], RASA2 [Ras p21 protein activator 2], and KSR1 [kinase suppressor of Ras 1] [Figure 5].
Figure 5.
Interaction network derived from the pathway analysis according to results of the genome-wide association study [GWAS] meta-analysis.
3.5. All novel candidate SNPs showed Japanese-specific associations with CD
To confirm whether these novel candidates are Japanese-specific, we assessed the results of a recent meta-analysis on European CD imputed GWASs by the International IBD Genetics Consortium3 [Table 3]. None of the novel candidates, including rs488200, were associated with CD in individuals of European ancestry, suggesting that these are Japanese-specific candidate susceptibility genes [p = 0.295, OR = 1.03]. Of the previously identified Japanese CD-associated SNPs, the effect of almost every one was in the same direction as in European populations but did not reach statistical significance. For instance, rs78898421 from the CD_1 locus located near the TNFSF15 gene is very rare in the European population [risk allele frequencies: 0.74% in controls and 1.01% in CD patients].
Table 3.
Comparison of the associations between Japanese and European CD for top hit candidate loci in Japanese CD patients.
| LocusID | Genes | Top SNP | A1/A2 | A1 Allele Frequencies | p-values | Odds ratios | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Japanese | European | Japanese | European | Japanese | European | ||||||
| Ctrl | CD | Ctrl | CD | ||||||||
| Previously reported loci | |||||||||||
| CD_1 | TNFSF15, TNFSF8 | rs78898421 | T/G | 47.0% | 63.1% | 0.74% | 1.01% | 2.59E-26 | 8.49E-02 | 2.10 | 1.26 |
| CD_2 | HLA-DQB1 | rs184950714 | A/G | 15.6% | 27.7% | 4.5% | 4.7% | 3.56E-19 | 7.80E-01 | 2.05 | 1.02 |
| CD_3 | ZNF365 | rs224136 | T/C | 29.7% | 21.9% | 15.8% | 13.5% | 9.04E-09 | 1.76E-11 | 0.63 | 0.80 |
| CD_4 | rs73243351 | A/G | 20.3% | 27.7% | 9.2% | 10.3% | 4.04E-08 | 5.26E-04 | 1.53 | 1.14 | |
| CD_6 | IL12B | rs56167332 | A/C | 38.1% | 46.0% | 33.5% | 36.4% | 9.24E-08 | 2.28E-11 | 1.43 | 1.19 |
| CD_9 | IL27 | rs56354901 | C/T | 12.2% | 16.6% | 31.5% | 34.1% | 5.61E-07 | 6.20E-07 | 1.59 | 1.14 |
| CD_10 | IL23R | rs76418789 | A/G | 7.7% | 3.6% | 0.64% | 0.60% | 6.59E-07 | 4.87E-01 | 0.43 | 0.86 |
| Novel loci | |||||||||||
| CD_5 | RAP1A, FAM212B | rs488200 | T/A | 44.5% | 50.9% | 36.6% | 37.0% | 8.85E-08 | 2.95E-01 | 1.43 | 1.03 |
| CD_7 | SYVN1, CAPN1 | rs11227126 | T/A | 5.6% | 9.4% | 22.7% | 22.8% | 3.65E-07 | 3.08E-01 | 1.88 | 1.03 |
| CD_8 | GSAP, FGL2 | rs4727354 | T/G | 4.1% | 7.1% | 15.1% | 14.9% | 5.17E-07 | 9.62E-01 | 1.99 | 1.00 |
| CD_11 | DLG4, ACADVL | rs2521985 | T/C | 39.3% | 32.0% | 34.8% | 34.4% | 7.23E-07 | 7.65E-01 | 0.70 | 0.99 |
SNP, single nucleotide polymorphism; CD, Crohn’s disease.
4. Discussion
In this study, a GWAS of Japanese CD patients using a population-optimised genotyping array and imputation identified four significant and seven candidate loci, including four novel loci. When the four novel candidates were examined by additional replication analysis, only rs488200 from the CD_5 locus was significantly associated with Japanese CD. We also examined the cis-transcriptional effect of rs488200, and found that the expression level of RAP1A in Tem cells was significantly associated with rs488200 genotype.
The first GWAS of CD in Japan by Yamazaki et al. reported associations of TNFSF15 variants with Japanese and Caucasian CD patients.11 Subsequently, several GWASs and meta-analyses were performed in Caucasian CD patients.1–3,40 However, the genetic background of CD in Japanese individuals remains unclear for a number of reasons. Sample size is a significant issue in Japan, owing to lower CD prevalence rates than in populations of European ancestry. Although a few GWASs of CD in East Asia have been published,11,34,41 those from Japan and Korea genotyped only a limited number of cases using a genome-wide chip. The genotyping platform is also an important issue when analysing susceptibility genes in non-Caucasian populations.42 The recent evolution of genotyping technology and extension of reference genome data for imputation from the 1000 Genomes Project43,44 enabled us to perform a GWAS in East Asians using a regular GWAS array. However, most of the genotyping arrays were not designed for imputation in Asian populations, and the reference panel for East Asia in the 1000 Genomes Project included an extremely limited number of patients of Japanese ancestry. Population-matched reference data are critical for imputation accuracy, especially for SNPs with lower allele frequencies. The JPA is a population-optimised array specific for the Japanese population.14 In addition, this is the best approach to obtain whole-genome imputation data using the results in combination with the 1KJPN panel, which contains the largest number of haplotypes of Japanese ancestry.13 The present study is thus an important first step in clarifying the Japanese-specific genetic background of CD because we analysed a new sample set from Tohoku region with a new SNP array, and performed imputation with a robust reference panel.
In addition to novel loci, we confirmed the associations of several known loci with CD in Japanese individuals. All seven known loci have been associated with CD or IBD in Caucasians. Among them, TNFSF15 is known as the East Asian dominant susceptibility gene,11 and 4p143,34 and IL23R G149R37–39 were also found first in an East Asian or trans-ethnic analysis. The associations of ZNF365, IL12B, and IL27 were replicated from previous studies of Japanese and/or Korean CD patients.10,35,36,42 Therefore, the associations of six loci and HLA are shared with CD in Caucasians and East Asians, although the effect sizes may be different.
One of the novel susceptibility loci was tagged by rs488200. Associated SNPs at this locus were synonymous, so we hypothesised that the SNPs conferred susceptibility to CD by altering the expression levels of neighbourhood genes. This locus includes the RAP1A gene, which is associated with lymphocyte homing, a process considered very important in the pathogenesis of CD.45 Therefore, we examined the effect of rs488200 on the expression levels of neighbouring genes, especially RAP1A, in Tem cells from LPMCs of CD patients. Despite the relatively small number of samples, there was a strong association of rs488200 with RAP1A expression, consistent with available eQTL from PBMCs46 or transformed fibroblasts.47 We also checked the Genotype-Tissue Expression [GTEx] Project database, but there were no data on rs488200. However, some variants around rs488200 with pmeta <1 × 10−6 showed significant associations with RAP1A expression in adipose tissue, tibial nerve, transformed fibroblasts, and whole blood [Supplementary Table S5, available as Supplementary data at ECCO-JCC online]. The intronic SNP rs500184 with Pmeta = 1.23 × 10−7 was located at the predicted promoter region including the transcriptional start site of the RAP1A gene [Supplementary Figure S5, available as Supplementary data at ECCO-JCC online].
Rap1 is a small GTPase with two highly homologous isoforms [Rap1A and Rap1B with 95% identity] encoded by separate genes [RAP1A and RAP1B].48 Rap1A is reported to be the more critical isoform for endothelial barrier function. The ‘rolling’ interaction is an important mechanism for homing of gut-specific lymphocytes. Tethering of the lymphocyte CAM selectin to its ligand holds the lymphocyte lightly on the endothelium wall, facilitating trans-endothelial migration into the tissue. Ishihara et al. reported that Rap1-GDP suppresses L-selectin-dependent rolling by limiting tether formation, and that Rap1 regulates T cell homing to the gut.45 In addition, they showed that Rap1-deficient mice developed spontaneous colitis. We found that an increase in the CD risk allele count of rs488200 was associated with decreased expression of RAP1A in Tem cells, consistent with findings in mice. This genetic association of CD with RAP1A is also consistent with the notion that altered lymphocyte homing to the gut contributes to the pathogenesis of CD. The efficacy of anti-integrin therapies, including natalizumab, vedolizumab, and etrolizumab, provides further evidence to support this hypothesis and suggests an opportunity to personalise IBD management, if drugs that work through these mechanisms are more effective in individuals harbouring susceptibility alleles in these same pathways.49–51
Pathway analysis using our GWAS results indicated the importance of CAMs in the association with CD, consistent with the well-described association between CD and the integrin pathway. In addition, the association of antigen processing and presentation pathway, also known as a major pathway in the pathogenesis of CD, was confirmed. Network analysis suggested a direct interaction of RAP1A with KSR1, a kinase suppressor of Ras 1 identified as an IBD locus in a recent European ancestry fine-mapping study.52 However, the functional relationship between these two genes is unclear.
We also examined the associations of these candidate SNPs with disease phenotypes, but found no significant associations. As the sample size of our study was too small to perform detailed subgroup analyses, further studies with larger sample sizes are required to identify specific genotype−disease phenotype associations.
No novel candidate identified in this study, including rs488200, has been associated with CD in individuals of European ancestry. Additionally, some of the top hit SNPs, even in previously reported loci, were rare variants and/or were not associated with CD in Europeans. These findings suggest that RAP1A could be a Japanese-specific susceptibility gene and that even shared loci may contain distinct CD-associated alleles. Population-specific susceptibility genes [including NOD2 in Caucasians] are important not only for screening of risk within populations but also because they may help identify novel therapeutic options for CD.
There are some limitations in this study. We used imputed genotype data in the GWAS, and the accuracy of the imputation using 1KJPN, using the samples in this study, was not verified due to scarcity of sample. Additionally, we adopted the meta-analysis approach taking into consideration the population substructures between Tohoku and Kyushu samples; however, there are other approaches for analysis [i.e. a linear mixed model] using which different results might be obtained.
In conclusion, we performed a GWAS of Japanese CD using a population-optimised genotyping array and imputation, and for the first time report the novel susceptibility gene RAP1A. The risk allele [T] is significantly associated with lower expression of RAP1A in Tem cells from LPMCs of Japanese CD patients. Additional studies and functional analyses are needed to determine the effect size on CD risk, the pathogenic mechanisms, and whether the presence of the risk allele is an important influence on therapeutic response.
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP15H04805, by AMED under Grant Number JP18kk0305002, and in part by the Tohoku Medical Megabank Project [Special Account for reconstruction from the Great East Japan Earthquake].
Conflict of Interest
MN reports grants from Obtain, the research grant from Toshiba Corporation during the study period, outside the submitted work; AH reports grants from AbbVie GK, outside the submitted work; TMreports grants from Mitsubishi Tanabe Pharma Corporation, EA pharma Co., Nippon Kayaku Co., Janssen Pharmaceutical K.K., AbbVie GK, and Kyorin Pharmaceutical Co., outside the submitted work. The other authors have no conflicts of interest to declare.
Supplementary Material
Acknowledgments
SNP genotyping was supported in part by the Centre of Innovation Program from the Japan Science and Technology Agency, JST. Computational resources were provided in part by the ToMMo supercomputer system. We would like to thank the member of MENDEL study group for sharing the genetic data.
Conference presentation: part of this work was presented as an oral presentation at Digestive Disease Week, Chicago, IL, May 2017.
Author Contributions
Y. Kakuta, Y. Kawai, MN, and Y. Kinouchi designed the study. T. Naito, Y. Kawai, AH, JU, YF, DO, HC, JY, and Y. Kakuta acquired data. AH, JU, YF, T. Nakano, YI, RI, DO, HN, SM, KY, NY, HC, YS, MO, RM, MK, HS, Y. Kanazawa, TK, KE, and ME recruited patients. Y. Kakuta, Y. Kawai, ZL, DL, DMcG, and MN analysed data. Y. Kakuta, Y. Kinouchi, AM, TS, and DMcGn drafted the manuscript. MN, MEi, KT, and TM provided samples.
References
- 1. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC]. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011;13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 6. Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 7. Inoue N, Tamura K, Kinouchi Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology 2002;123:86–91. [DOI] [PubMed] [Google Scholar]
- 8. Yamazaki K, Takazoe M, Tanaka T, Kazumori T, Nakamura Y. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn’s disease. J Hum Genet 2002;47:469–72. [DOI] [PubMed] [Google Scholar]
- 9. Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet 2007;52:575–83. [DOI] [PubMed] [Google Scholar]
- 10. Hirano A, Yamazaki K, Umeno J, et al. Association study of 71 European Crohn’s disease susceptibility loci in a Japanese population. Inflamm Bowel Dis 2013;19:526–33. [DOI] [PubMed] [Google Scholar]
- 11. Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet 2005;14:3499–506. [DOI] [PubMed] [Google Scholar]
- 12. Kakuta Y, Kinouchi Y, Negoro K, Takahashi S, Shimosegawa T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut 2006;55:1527–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagasaki M, Yasuda J, Katsuoka F, et al. ; ToMMo Japanese Reference Panel Project. Rare variant discovery by deep whole-genome sequencing of 1070 Japanese individuals. Nat Commun 2015;6:8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawai Y, Mimori T, Kojima K, et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet 2015;60:581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuriyama S, Yaegashi N, Nagami F, et al. The Tohoku medical megabank project: design and mission. J Epidemiol 2016;26:493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kakuta Y, Kawai Y, Okamoto D, et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: a multicenter study. J Gastroenterol 2018, Jun 19. doi: 10.1007/s00535-018-1486-7. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsui T, Hirai F, Hisabe T. Proposed diagnostic criteria for Crohn’s disease. Annual reports of the research group of intractable inflammatory bowel disease granted by the Ministry of Health, Labour, and Welfare of Japan. 2011; 52–4.
- 18. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staples J, Nickerson DA, Below JE. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet Epidemiol 2013;37:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2011;9:179–81. [DOI] [PubMed] [Google Scholar]
- 23. Fiocchi C, Youngman KR. Isolation of human intestinal mucosal mononuclear cells. Curr Protoc Immunol 2001;Chapter 7:Unit 7.30. [DOI] [PubMed] [Google Scholar]
- 24. Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011;17:, 10–2.
- 25. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turner SD. Qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv 005165 2014. http://biorxiv.org/content/early/2014/05/14/005165.abstract [Google Scholar]
- 29. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakajima A, Matsuhashi N, Kodama T, Yazaki Y, Takazoe M, Kimura A. HLA-linked susceptibility and resistance genes in Crohn’s disease. Gastroenterology 1995;109:1462–7. [DOI] [PubMed] [Google Scholar]
- 33. Yoshitake S, Kimura A, Okada M, Yao T, Sasazuki T. HLA class II alleles in Japanese patients with inflammatory bowel disease. Tissue Antigens 1999;53:350–8. [DOI] [PubMed] [Google Scholar]
- 34. Yang SK, Hong M, Zhao W, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 2014;63:80–7. [DOI] [PubMed] [Google Scholar]
- 35. Yamazaki K, Takahashi A, Takazoe M, et al. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn’s disease in Japanese patients. Gut 2009;58:228–32. [DOI] [PubMed] [Google Scholar]
- 36. Kakuta Y, Kimura T, Negoro K, et al. Increased expression of IL12b mRNA transcribed from the risk haplotype for Crohn’s disease is a risk factor for disease relapse in Japanese patients. J Gastroenterol 2017;52(12):1230–9. [DOI] [PubMed] [Google Scholar]
- 37. Kim SW, Kim ES, Moon CM, et al. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut 2011;60:1527–36. [DOI] [PubMed] [Google Scholar]
- 38. Onodera K, Arimura Y, Isshiki H, et al. Low-frequency IL23R coding variant associated with Crohn’s disease susceptibility in Japanese subjects identified by personal genomics analysis. PLoS One 2015;10:e0137801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong SN, Park C, Park SJ, et al. ; IBD Study Group of the Korean Association for the Study of Intestinal Diseases [KASID]. Deep resequencing of 131 Crohn’s disease associated genes in pooled DNA confirmed three reported variants and identified eight novel variants. Gut 2016;65:788–96. [DOI] [PubMed] [Google Scholar]
- 40. Barrett JC, Hansoul S, Nicolae DL, et al. ; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamazaki K, Umeno J, Takahashi A, et al. A genome-wide association study identifies 2 susceptibility loci for Crohn’s disease in a Japanese population. Gastroenterology 2013;144:781–8. [DOI] [PubMed] [Google Scholar]
- 42. Yang SK, Hong M, Choi H, et al. Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm Bowel Dis 2015;21:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abecasis GR, Altshuler D, Auton A, et al. ; 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ishihara S, Nishikimi A, Umemoto E, Miyasaka M, Saegusa M, Katagiri K. Dual functions of Rap1 are crucial for T-cell homeostasis and prevention of spontaneous colitis. Nat Commun 2015;6:8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Westra HJ, Arends D, Esko T, et al. Cell specific eQTL analysis without sorting cells. PLoS Genet 2015;11:e1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aguet F, Brown AA, Castel S, et al. Local genetic effects on gene expression across 44 human tissues. bioRxiv 074450 2016. [Google Scholar]
- 48. Wittchen ES, Aghajanian A, Burridge K. Isoform-specific differences between Rap1A and Rap1B GTPases in the formation of endothelial cell junctions. Small GTPases 2011;2:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sandborn WJ, Feagan BG, Rutgeerts P, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med 2013;369:711–21. [DOI] [PubMed] [Google Scholar]
- 50. Ghosh S, Goldin E, Gordon FH, et al. ; Natalizumab Pan-European Study Group. Natalizumab for active Crohn’s disease. N Engl J Med 2003;348:24–32. [DOI] [PubMed] [Google Scholar]
- 51. Luthra P, Peyrin-Biroulet L, Ford AC. Systematic review and meta-analysis: opportunistic infections and malignancies during treatment with anti-integrin antibodies in inflammatory bowel disease. Aliment Pharmacol Ther 2015;41:1227–36. [DOI] [PubMed] [Google Scholar]
- 52. Huang H, Fang M, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.