Abstract
The efficacy of superoxide dismutase-1 (SOD1) folding impacts neuronal loss in motor system neurodegenerative diseases. Mutations can prevent SOD1 post-translational processing leading to misfolding and cytoplasmic aggregation in familial amyotrophic lateral sclerosis (ALS). Evidence of immature, wild-type SOD1 misfolding has also been observed in sporadic ALS, non-SOD1 familial ALS and Parkinson's disease. The copper chaperone for SOD1 (hCCS) is a dedicated and specific chaperone that assists SOD1 folding and maturation to produce the active enzyme. Misfolded or misfolding prone SOD1 also interacts with heat shock proteins and macrophage migration inhibitory factor to aid folding, refolding or degradation. Recognition of specific SOD1 structures by the molecular chaperone network and timely dissociation of SOD1-chaperone complexes are, therefore, important steps in SOD1 processing. Harnessing these interactions for therapeutic benefit is actively pursued as is the modulation of SOD1 behaviour with pharmacological and peptide chaperones. This review highlights the structural and mechanistic aspects of a selection of SOD1-chaperone interactions together with their impact on disease models.
Keywords: amyotrophic lateral sclerosis, molecular chaperones, neurodegeneration, Parkinson's disease, pharmacological chaperones, protein misfolding
Introduction
The central dogma of molecular biology dictates that information flows from DNA through RNA to proteins. That information, comprising the primary sequence, is sufficient to direct protein folding which can occur in microseconds for small proteins. Folding of large, multidomain proteins or those that require post-translational modifications is slower and may require assistance. If the information fed into the folding reaction is corrupt, as is the case when coding sequence mutations are introduced, this can prevent the acquisition of a stable state.
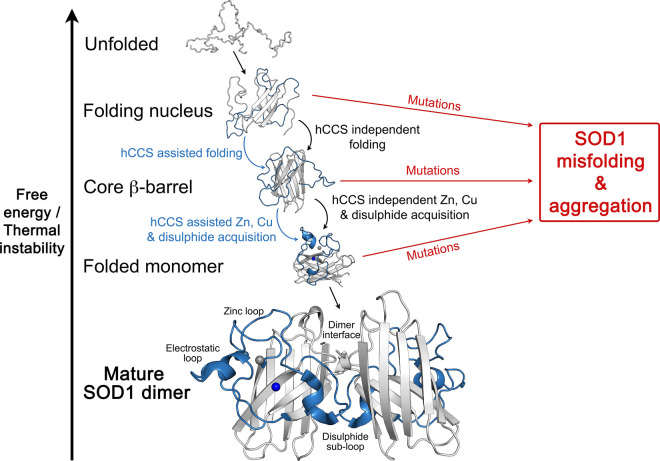
Superoxide dismutase-1 (SOD1) is a relatively small protein that is a part of the human defence against oxidative stress. Following translation, spontaneous SOD1 folding nucleates around hydrophobic residues that ultimately form the protein core [1]. A monomeric Greek-key β-barrel structure is produced as a result [2]. Zinc and electrostatic loops are predominantly disordered with sub-fractions of mature state structures populated [3]. Three post-translational modifications (PTMs) must take place for SOD1 to attain activity and stability. Zinc binding and formation of an intra-subunit disulphide bond restrain movement of the long zinc loop. This creates the SOD1 homodimer interface, together with N- and C-termini, and four inter-subunit hydrogen bonds form. [4]. The addition of copper to the SOD1 active site contributes to thermal stability and realises enzymatic activity. Without these PTMs, and resulting homodimer formation, SOD1 has reduced thermal stability, populates unfolded or high energy conformations [5] and is prone to self-association (Figure 1) [6].
Figure 1. The SOD1 folding and post-translational modification pathway.
Successive folding and PTM events increase SOD1 stability and can be accomplished through the action of the copper chaperone for SOD1 (hCCS) or independently. However, each step can be inhibited by the presence of ALS mutations. This leads to the persistence of unfolded or misfolded states that may be degraded or form intracellular aggregates. Copper and zinc ions are represented as blue and silver spheres respectively.
Mutations of the SOD1 coding gene are known to cause the motor neuron disease amyotrophic lateral sclerosis (ALS) [7]. Roughly two-thirds of SOD1 amino acids have been shown to harbour ALS-related mutations and, in general, they are found in conserved metal binding, interface and β-barrel residues. Each mutation diminishes the likelihood SOD1 will progress along its maturation pathway to populate stable and folded states (Figure 1) but the molecular and phenotypic manifestations of individual mutations can vary, reviewed [8].
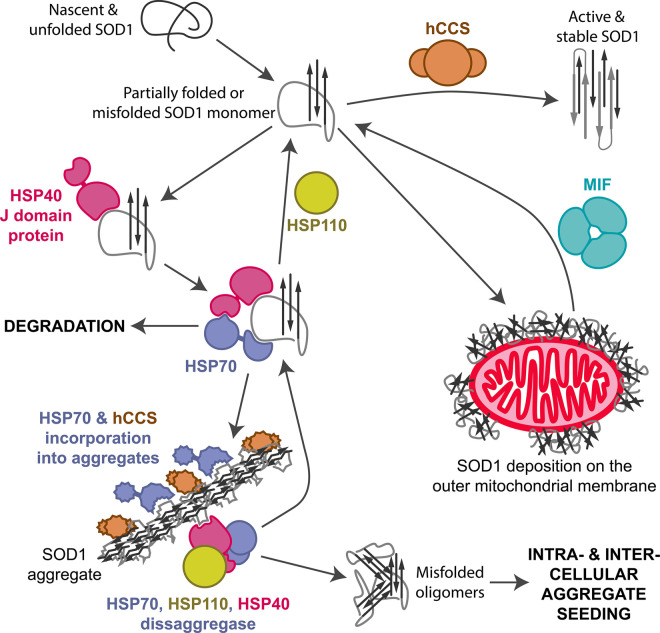
Misfolded SOD1 is found in the neurons and glia of ALS spinal cord, brainstem and hippocampus [9,10]. Parkinson's disease substantia nigra and locus coeruleus neurons also display SOD1 accumulation [11]. Misfolded SOD1 can range from monomers to the large inclusions characteristic of SOD1-ALS cases [9,12]. In addition to SOD1, inclusions also contain heat shock proteins (HSP) [13–16], ubiquitin [11,13,17], proteasomes [13] and the copper chaperone for SOD1 (hCCS) [11,13,18]. This is indicative of an effort by the cell to fold, refold or degrade a protein that has become problematic (Figure 2). Understanding how SOD1 is chaperoned to activity and why proteostatic mechanisms are ineffective in some disease states will guide us in designing interventions that assist these processes. In this review, I focus on molecular recognition and stabilisation of SOD1 by molecular chaperones and the outcomes achieved by modulation of these interactions, then describe several pharmacological and peptide chaperones that may form the basis of future therapeutics.
Figure 2. Molecular chaperone interactions with SOD1.
The copper chaperone for SOD1 (hCCS) is highly specific for SOD1 and has a role in every stage of SOD1 maturation. Macrophage migration inhibitory factor (MIF) can reduce misfolded SOD1 accumulation on the surface of mitochondria. The heat shock protein (HSP) system is involved in both SOD1 folding, degradation and possibly breakdown of aggregates. This last step may provide seeds that propagate SOD1 misfolding and aggregation within the cell or in surrounding cells. HSP interactions with SOD1 are not specific but involve high-affinity binding to hydrophobic residues including SOD1 β-strand 7.
The copper chaperone for SOD1
SOD1 is found predominantly in the cytosol where both copper binding and retention of disulphide bonds are disfavoured. Some SOD1 analogues are activated exclusively by a copper chaperone [19] while others are activated through an independent route [20]. Parkinson's disease-related protein DJ-1 has been reported to form complexes with human SOD1 and act as a copper chaperone [21,22]. However, validation of either functionality is lacking [23]. The only chaperone protein known to directly interact with and activate human SOD1 is hCCS and it does so for 80% of SOD1 molecules [24,25]. Copper can, however, be supplied to SOD1 independent of hCCS by histone H3–H4 or a route involving glutathione [26,27]. hCCS comprises copper-binding domain I, SOD1-like domain II and C-terminal domain III. The latter has a role in disulphide transfer [28]. hCCS domain II binds to SOD1 through an ATP-independent mechanism bringing functional hCCS domains I and III into the proximity of nascent SOD1. Full-length and domain II truncated hCCS form heterodimeric complexes with intra-subunit disulphide bond reduced wild-type SOD1 and every ALS mutant directly tested to date [29–32]. Dissociation constants of 42, 68, 35 and 33 nM have been measured for hCCS binding to metal-free Ala4Val, His80Arg, Gly85Arg and Gly93Ala mutant SOD1, respectively [31]. Zinc metalated SOD1 forms a heterodimer with hCCS that has higher affinity than the heterodimer formed by metal-free SOD1 [29,32]. However, experiments on Saccharomyces cerevisiae orthologues indicated heterodimer affinity progressively weakened as the SOD1 substrate was post-translationally modified [33]. This contradiction may result from the study of different orthologues or use of different techniques, but further experiments are needed to reconcile the results. What is clear, however, is that zinc binding by metal-free SOD1 does not precluded complex formation with hCCS, on the contrary, SOD1 zinc affinity is increased while the heterodimer is intact [32,34,35]. Furthermore, co-expression of hCCS domain II aids wild-type and ALS mutant Ala4Val, Ile113Thr and Gly93Ala SOD1 folding within the cytoplasm of human cells. This indicates hCCS has a molecular chaperone activity independent of SOD1 PTMs [32].
On complexation with hCCS domain II, SOD1 proton-amide chemical shifts are observed for residues in the zinc loop, electrostatic loop and almost all β-strands [32]. Thus, the stabilising effect of complexation with hCCS permeates further than the heterodimer interface. However, the largest structural discrepancy between mature SOD1 and that found bound to hCCS is the adoption of an induced fit disulphide sub-loop conformation at the heterodimer interface [30]. Substitution of hCCS Arg232 and Arg104 for SOD1 Ile151 and Asn19, respectively, create non-covalent interactions across the heterodimer interface with the SOD1 disulphide sub-loop. A further repulsive interaction between hCCS Ala231 and SOD1 Gly51 prevents the sub-loop occupying the conformation found in the mature enzyme and also assists complex dissociation once PTM events are complete [30]. These interactions are the source of hCCS specificity for disulphide reduced SOD1 and prime the substrate for PTMs. They also reduce the conformational freedom of a long, mobile loop. SOD1 thermal instability resulting from the ALS-associated Ile113Thr mutation is, therefore, diminished when complexed with hCCS [29]. Due to this stabilising interaction, overexpression of hCCS in transgenic mice and cell models reduces the formation of non-natively associated mutant SOD1 species [36–38]. These observations are consistent with hCCS protecting SOD1 from the aggregation. However, overexpression of hCCS causes extreme mitochondrial vacuolisation and very short life-span in Gly93Ala and Gly37Arg human SOD1 transgenic mice. This effect was relieved by increasing the availability of copper [39] but was completely absent from Gly86Arg mouse SOD1 (human Gly85Arg) or Leu126X truncated human SOD1 overexpressing mice [36,40]. Gly86Arg and Leu126X exist as disulphide reduced monomers [40]. Thus, toxicity differences could be explained by reduced hCCS affinity for Gly85Arg and Leu126X and poor retention within mitochondria as a result.
Macrophage migration inhibitory factor
Macrophage migration inhibitory factor (MIF) is a pro-inflammatory cytokine expressed cytoplasmically and secreted in response to numerous immunogenic stimuli. MIF is a 12.5 kDa peptide that exists in a predominantly trimeric state arranged around a central pore. Small populations of dimeric and monomeric MIF have been observed but are unstable and aggregation prone. MIF is constitutively expressed by many tissue types including motor system cells but is conspicuously absent from neuronal cell bodies despite high-level MIF mRNA transcription [41,42]. In the absence of MIF, misfolded mutant SOD1 deposits are found on the cytoplasmic surface of mitochondria while, conversely, overexpression of MIF reduces mutant SOD1 association with both endoplasmic reticulum and mitochondrial membranes thereby maintaining mitochondrial function [42–44]. This translates into increased survival of mutant SOD1 expressing motor neurons when MIF production is increased. Overexpression of MIF in mutant SOD1 transgenic mice also reduced the formation of non-detergent buffer insoluble SOD1 species [43]. These observations are indicative that MIF harbours a SOD1 chaperone role. The proof of concept that MIF-coding DNA can be delivered to affected neuronal cells has been accomplished. Adeno-associated virus (AAV) delivery to spinal neurons reduced SOD1 misfolding, delayed disease onset, slowed disease progression and extended life expectancy in Gly93Ala and Gly37Arg mutant SOD1 transgenic mice [45]. It is important to note, however, that intraspinal injection of AAV transporting MIF encoding DNA occurred one day after birth, long before ALS-like symptoms would be evident.
MIF has a thiol oxidoreductase activity which is dependent on the conformation of the MIF trimer as dictated by the oxidation state of Cys80 which, in turn, regulates Cys56 and Cys59 competence for disulphide exchange [46]. Oxidoreductase activity seems a probable source of the SOD1 chaperone activity. Co-expression of an oxidoreductase knockout Cys59Ser MIF with Gly93Ala and Gly85Arg SOD1 resulted in only partial protection against SOD1 misfolding when expression level was high [42,47]. Furthermore, direct comparison indicated the Cys59Ser oxidoreductase knockout mutant did not supress SOD1 misfolding and cell death to the same extent as wild-type MIF. However, Cys59Ser MIF marginally prevented cell death when compared with cells where no MIF was overexpressed [47]. These results are indicative that, like hCCS, MIF-catalysed SOD1 disulphide formation is not wholly responsible for the SOD1 chaperone role. Preventing MIF trimer dissociation through an engineered inter-subunit disulphide between Cys80 and a mutated Asn110Cys site [48] does, however, consistently reduce mutant SOD1 misfolding and related cell death [44,47]. Trimeric MIF appears, therefore, to be the chaperone active species.
There is currently no indication of the interaction surfaces through which MIF prevents SOD1 misfolding. However, the necessity for trimeric MIF may reside in the increased surface area through which SOD1 can interact. A physical interaction with substrate is a prerequisite of molecular chaperone activity and a 367 nM affinity for as isolated i.e. complete zinc and 87% copper metalated Gly93Ala SOD1 [49] has been recorded for wild-type MIF [42]. In contrast, wild-type MIF has a 1.3 μM affinity for metal-free wild-type SOD1 but 548 nM when the trimer-locked Asn110Cys variant is assayed [47]. Thus, MIF appears to have a lower affinity for the metal-free SOD1 species that are likely to misfold, aggregate and cause toxicity than the metalated and more stable species.
Heat shock proteins
Mutant SOD1 is known to interact with many of the constitutive and induced groups of chaperone proteins that comprise the proteostasis network (Figure 2). Indeed, HSP70 and HSP27 are so deeply woven into the mutant SOD1 life cycle that they, along with ubiquitin, are components of the protein aggregates that are common to Parkinson's disease, SOD1-related ALS and mutant SOD1 transgenic animal models [11,13–16]. Mutation of HSPs is also implicated in the pathogenesis of ALS [50], Parkinson's disease [51] and other motor system diseases. The centrality of protein quality control malfunction in neurodegeneration is, therefore, clear, as is targeting it for potential therapeutic benefit in ALS [52].
HSP70
Hydrophobic residues are sequestered within the core of proteins when folded and exclusion of water contributes to protein stability. Exposure of these hydrophobic regions can prelude non-native self-association, as is the case for SOD1 [53]. The HSP70 group of molecular chaperones are purposed to prevent those aggregation events and are ubiquitously conserved. Together with folding they have critically important roles in protein refolding, prevention of aggregation and degradation, reviewed [54]. As such they must interact with a broad range of substrates. To do so, they bind short stretches of hydrophobic amino acids through a conformational sampling procedure [55,56]. HSP70 proteins and a host of co-chaperones function by reducing exposure of aggregation-prone regions while allowing secondary structure to form prior to long-range contacts [57].
Mutant SOD1 is favoured as an HSP70 substrate over wild-type SOD1 in vivo [58]. HSP70 group chaperones HSPA1 and HSPA8 have been shown to bind peptides representing the SOD1 zinc loop and β-strand 7. HSPA8, which is the most highly expressed HSP70 [59], has 7.1 μM affinity for a β-strand 7 peptide that is buried within the SOD1 core behind metal binding and disulphide loops in the mature state [60]. An absence of post-translation modifications would, therefore, be necessary for this binding site to be exposed. While β-strand 7 is not a structured component of SOD1 aggregates, reviewed [8], it is part of the folding nucleus that orchestrates SOD1 β-barrel formation [1]. Thus, the timely termination of this interaction is exceptionally important. Sequestering β-strand 7 while β-strands 1-3 scan conformations on a spectrum between stable and aggregation-prone may drag HSP70 proteins into growing mutant SOD1 aggregates [15]. Overexpression of several HSP70 chaperones along with Ala4Val SOD1 in cell models has been shown to increase the abundance of mutant SOD1 aggregates [61]. This phenomenon was ascribed to the nucleotide exchange factor (NEF) favoured to bind individual HSP70 proteins. Conversely, overexpression of HSP70 member HSPA1A consistently decreased aggregation through an interaction with HSPH2 NEF, an HSP110 group member [61]. Binding of the correct HSP70 chaperone to mutant SOD1 is, therefore, an important indicator of the fate, and therefore the toxicity, of a mutant SOD1 molecule.
Promoting misfolded or aggregated mutant SOD1 clearance by increasing the abundance of individual HSP70 chaperones has yielded mixed results perhaps because of the variability described above. Expression of murine inducible HSP70 in primary motor neurons along with Gly93Ala SOD1 reduced the percentage of cells with aggregates and increased cell viability [62]. Conversely, overexpression of HSP70 in SOD1 mutant transgenic mice had little or no effect on disease course [15,63]. A second approach has been a global up-regulation of HSP70 chaperones via the heat shock factor-1 (HSF1) transcription factor. Overexpression of SIRT1 was able to activate HSF1, up-regulate HSP70 and reduce the abundance of non-native SOD1 dimers. While marginal effects were observed on disease readouts, late-stage disease was significantly extended in low Gly93Ala SOD1 expressing mice [64]. Treatment of Gly93Ala SOD1 transgenic mice with the HSF1-binding small molecule arimoclomol also yielded increased maximum life expectancy from 20 to 23 weeks regardless of whether treatment was initiated at presymptompatic, early or late post-symptomatic time points. This paralleled a reduction in the number of spinal cord ubiquitin-positive SOD1 aggregates [65,66]. The success of arimoclomol studies has led to several clinical trials to test its therapeutic use in ALS; a large phase 3 trial will conclude in 2021 but a smaller trial has already shown arimoclomol to have favourable toxicity profile and promising efficacy [67].
HSP110
HSP110 co-chaperones are a sub-group of NEFs that promote the release of ADP from HSP70 proteins. This results in switching to a low-affinity state and substrate dissociation. Unlike other NEFs, HSP110 co-chaperones are highly similar to HSP70 proteins and form heterodimers with their target HSP70 through their respective nucleotide-binding domains. All three HSP110 proteins have been shown to copurify with mutant SOD1 [58,61]. However, it is not currently known if HSP110 co-chaperones have a direct, functional interaction with unfolded protein or whether copurification is a result of associations through HSP70. Overexpression of the HSPA4L human HSP110 did increase survival time in Gly85Arg and Gly93Ala SOD1 transgenic mice. Interestingly, high copy number (>270) Gly85Arg SOD1 mice were impervious to rescue by HSPA4L, whereas low copy number (<270) showed two and 6-month median and maximum life extension, respectively [63].
HSP40 (DNAJ or J domain proteins)
HSP40 proteins are a large group of modular proteins that are categorised by the presence of a J domain which mediates interactions with HSP70 proteins. Outside the J domain, HSP40s are heterogenous and can comprise many different domain structures and functionalities but their primary function is to present substrates to HSP70 and stimulate ATP hydrolysis [68] (Figure 2). DNAJB1 interacts with HSP70 when co-expressed with Ala4Val SOD1 and suppresses aggregation. Conversely, expressing the DNAJB1 His32Gln mutant, known to reduce stimulation of HSP70 ATPase, together with Ala4Val SOD1 led to a massive increase in SOD1 aggregation [61]. Furthermore, DNAJB2, DNAJB7 and the autophagy-inducer DNAJC13 are also suppressors of mutant SOD1 accumulation into aggregates [61,69,70]. DNAJB2 has been shown to have a direct interaction with misfolded Gly93Ala SOD1 in transgenic mouse spinal cord homogenates and target it for proteasomal degradation [70]. While, conversely, a DNAJB2 distal hereditary motor neuropathy-associated mutation increases mutant SOD1 aggregation [71].
HSP disaggregases
S. cerevisiae HSP104 is a protein disaggregase that functions in cooperation with HSP70 and HSP40 proteins [72]. Overexpression of S. cerevisiae HSP104 in mouse N2a cells can free mutant human SOD1 from large, condensed protein aggregates returning it to a wild-type-like dispersion. However, the SOD1 released is not natively structured but small oligomers at least trimeric in size [73]. While metazoans, including humans, do not produce directly related HSP104 othologous proteins, HSP70, HSP110 and HSP40 group chaperones can function together as a disaggregation machine [74]. This system is able to resolublise luciferase aggregates in nematode muscle but facilitates the transfer of Parkinson's disease-related α-synuclein species between adjacent tissues where it is competent to increase aggregation and reduce motility [75]. Like α-synuclein, SOD1 aggregation is assisted by fragmentation, both in cell-free and transgenic mouse models, and spreads through extracellular spaces [76–78]. There is growing evidence that relatively low molecular mass soluble species are the instigator of cell death and disease whereas large, insoluble aggregates are inert and possibly protective [79,80]. It may be, therefore, that disaggregation systems are an inadvertent source of hazardous, misfolded SOD1 oligomers (Figure 2).
Pharmacological chaperones
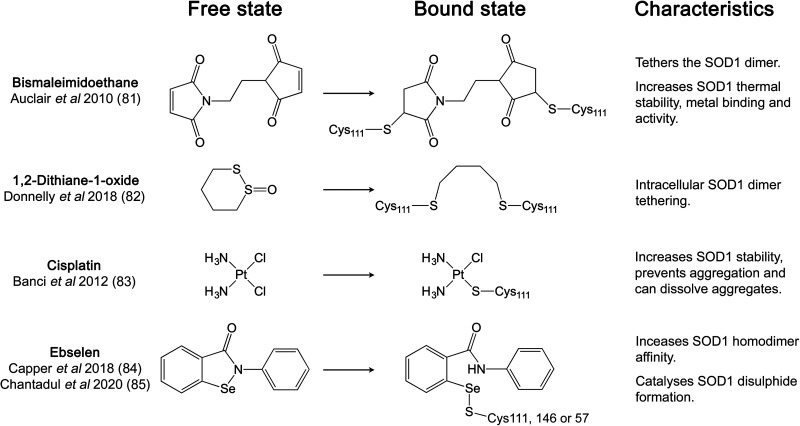
Covalent binding of small molecules at Cys111 has been a frequently utilised approach for SOD1 pharmacological chaperone discovery due to its accessibility, reactivity and potential for linking or stabilising the SOD1 homodimer (Figure 3). The first example of this approach was the use of maleimide linkers to span the 9 Å gap across the SOD1 dimer interface between Cys111 sulphydryls thereby covalently tethering monomers within a SOD1 homodimer. These modifications yielded exceptional mutant SOD1 thermal stability increases, along with the restoration of Gly85Arg SOD1 metal binding and activity [81]. Similarly, cyclic disulphides have been shown to tether opposing Cys111 residues, prevent dead-end single thiol modifications and are cell penetrable. Mono-S-oxo derivatives including 1,2-dithiane-1-oxide were able to bind and tether 95% of SOD1 monomers as dimers within cells over 30 min with an EC50 of 5 μM [82]. Taking another approach, cisplatin increases metal-free SOD1 thermal stability and is able to prevent or reverse SOD1 aggregation within neuron-like cells without subunit tethering [83].
Figure 3. Pharmacological chaperones targeting SOD1 Cys111.
Structures of each chaperone before and after covalent binding to SOD1 sulphydryls.
Ebselen
Ebselen is a small molecule organoselenium with antioxidant and neuroprotective properties. It is thiol reactive and forms a selenylsulphide bond at SOD1 Cys111 through the nucleophilic attack of the free sulphydryl. Two ebselen molecules are able to bind opposing Cys111 residues in the SOD1 dimer interface groove [84]. The hydrophobic SOD1 homodimer interface is expanded through aromatic π–π stacking of adjacent ligands. Mutant SOD1 homodimer affinity is increased as a result, particularly for Ala4Val SOD1 which is known to have a propensity for monomerisation [84]. Similar binding pose and homodimer affinity were obtained with ebsulphur [84]. Derivatisation of both compounds to enhance Cys111 reactivity and further extend the subunit interface has also been demonstrated.
In each of the above crystallographic studies, no ligand densities were observed close to Cys6 within the SOD1 β-barrel. However, ebselen binding to wild-type or Ala4Val SOD1 yielded a reduction in thermal stability. Analysis of melt curves indicated an unstable sub-population that was transformed into a 5.6 an 8.8°C stabilisation on binding to Cys6Ser and Ala4Val/Cys6Ser SOD1, respectively [85]. Derivatives that maintain a compact, low mass structure resulted in higher thermal stability than extended forms [85]. When Cys6 is present, however, a population of SOD1 molecules appear receptive to ligand binding within the β-barrel. This highlights the promiscuity of ebselen; an assessment of stable ebselen binding within cells indicated over 400 protein targets [86]. Furthermore, the selenylsulphide bond is prone to cleavage by cytoplasmic reductants unless it is protected. As a consequence stable cytoplasmic binding to SOD1 Cys111 has not been observed [84,86]. However, ebselen is a thiol oxidase and was able to catalyse formation of the SOD1 intra-subunit disulphide bond within cells in the absence of overexpressed hCCS [84]. This likely happens by the formation of a selenylsulphide bonding intermediate on either Cys57 or Cys146 which is then resolved through selenyl-thiol exchange to create the SOD1 disulphide. This process prevented the accumulation of unfolded Gly93Ala and Ala4Val SOD1 mutants through an oxidative folding mechanism [84]. As the formation of the SOD1 disulphide has a direct effect on homodimer affinity, ebselen is able to increase homodimerisation within cells when applied exogenously at 20 μM [87]. SOD1 has proven difficult as a target for occupancy-driven drug development due to the lack of cavity binding sites. This event-driven approach focusing on SOD1 post-translation modifications offers a new strategy for structural stabilisation. However, target engagement and specificity may still prove difficult given the abundance of free thiols within cells.
Chaperone peptides
Several peptide-based chaperone strategies have been applied to aid SOD1 folding and inhibit its aggregation including the repurposing of stability patches on the metal-free SOD1 molecule to block self-association [88]. Another interesting approach based on click chemistry screened penta-peptides for binding to a modified SOD1 electrostatic loop peptide and yielded several binding interactions that were then refined for SOD1 selectivity [89]. A peptide (HGGF4-fluorophenylalanineQ) bound mature and metal-free SOD1 at the electrostatic loop with 8.0 and 0.94 μM affinity respectively, indicating a preference for the conformationally dynamic, misfolding prone species. Presence of the peptide accelerated wild-type and Gly93Ala SOD1 folding rate 2.5- and 1.5-fold, respectively, and binding reduced the hydrodynamic radius of metal-free wild-type SOD1 consistent with structural stabilisation [89].
Perspectives
SOD1 misfolding is observed in ALS and Parkinson's disease. Targeted engagement of SOD1 through the action of native or synthetic molecular and pharmacological chaperones has the potential to improve disease outcomes for many individuals.
Disease-related mutations and lack of post-translational mutations result in SOD1 misfolding and accumulation of protein aggregates within neuronal cells. Cellular chaperone systems have several points of contact with SOD1. These interactions are not always effective and can result in chaperone incorporation into SOD1 aggregates. Despite this, intervention strategies based on the modulation of native molecular chaperones have elicited beneficial effects on disease characteristics. The small molecule arimoclomol has shown that a pharmacological route is also practicable.
Combinations of HSP system mechanics along with hCCS or MIF binding mechanisms will create new molecular chaperones for SOD1. A more detailed understanding of the structural nature of SOD1 misfolding and which routes through the chaperone network are effective for particular misfolding states is a necessary prerequisite. Promising work on pharmacological and peptide chaperones for SOD1 now has to address specificity and target engagement. We can also expect that one strategy will not be effective for all SOD1 mutants or states, so personalised or combination approaches will be necessary.
Abbreviations
- AAV
Adeno-associated virus
- ALS
amyotrophic lateral sclerosis
- HSF1
heat shock factor-1
- HSP
heat shock proteins
- MIF
macrophage migration inhibitory factor
- NEF
nucleotide exchange factor
- PTMs
post-translational modifications
- SOD1
superoxide dismutase-1
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
This work was funded by the Motor Neurone Disease Association (Wright/Oct18/969-799).
Open Access
Open access for this article was enabled by the participation of University of Liverpool in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC
References
- 1.Nordlund A. and Oliveberg M. (2006) Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc. Natl. Acad. Sci. U.S.A. 103, 10218–10223 10.1073/pnas.0601696103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y. and O'Halloran T.V. (2004) The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 279, 47998–48003 10.1074/jbc.M406021200 [DOI] [PubMed] [Google Scholar]
- 3.Sekhar A., Rumfeldt J.A., Broom H.R., Doyle C.M., Bouvignies G., Meiering E.M. et al. (2015) Thermal fluctuations of immature SOD1 lead to separate folding and misfolding pathways. eLife 4, e07296 10.7554/eLife.07296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hörnberg A., Logan D.T., Marklund S.L. and Oliveberg M. (2007) The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J. Mol. Biol. 365, 333–342 10.1016/j.jmb.2006.09.048 [DOI] [PubMed] [Google Scholar]
- 5.Culik R.M., Sekhar A., Nagesh J., Deol H., Rumfeldt J.A.O., Meiering E.M. et al. (2018) Effects of maturation on the conformational free-energy landscape of SOD1. Proc. Natl. Acad. Sci. U.S.A. 115, E2546–E2555 10.1073/pnas.1721022115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa Y., Kaneko K., Yamanaka K., O'Halloran T.V. and Nukina N. (2008) Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 283, 24167–24176 10.1074/jbc.M802083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A. et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 8.Wright G.S.A., Antonyuk S.V. and Hasnain S.S. (2019) The biophysics of superoxide dismutase-1 and amyotrophic lateral sclerosis. Q. Rev. Biophys. 52, e12 10.1017/S003358351900012X [DOI] [PubMed] [Google Scholar]
- 9.Rakhit R., Robertson J., Vande Velde C., Horne P., Ruth D.M., Griffin J. et al. (2007) An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat. Med. 13, 754–759 10.1038/nm1559 [DOI] [PubMed] [Google Scholar]
- 10.Stamenković S., Dučić T., Stamenković V., Kranz A. and Andjus P.R. (2017) Imaging of glial cell morphology, SOD1 distribution and elemental composition in the brainstem and hippocampus of the ALS hSOD1G93A rat. Neuroscience 357, 37–55 10.1016/j.neuroscience.2017.05.041 [DOI] [PubMed] [Google Scholar]
- 11.Trist B.G., Davies K.M., Cottam V., Genoud S., Ortega R., Roudeau S. et al. (2017) Amyotrophic lateral sclerosis-like superoxide dismutase 1 proteinopathy is associated with neuronal loss in Parkinson's disease brain. Acta Neuropathol. 134, 113–127 10.1007/s00401-017-1726-6 [DOI] [PubMed] [Google Scholar]
- 12.Shibata N., Hirano A., Kobayashi M., Asayama K., Umahara T. and Ikemoto A. (1993) Immunohistochemical demonstration of Cu/Zn superoxide dismutase in the spinal cord of patients with familial amyotrophic lateral sclerosis. Acta Histochem. Cytochem. 26, 619–622 10.1267/ahc.26.619 [DOI] [Google Scholar]
- 13.Watanabe M., Dykes-Hoberg M., Culotta V.C., Price D.L., Wong P.C. and Rothstein J.D. (2001) Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 8, 933–941 10.1006/nbdi.2001.0443 [DOI] [PubMed] [Google Scholar]
- 14.Weisberg S.J., Lyakhovetsky R., Werdiger A., Gitler A.D., Soen Y. and Kaganovich D. (2012) Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc. Natl. Acad. Sci. U.S.A. 109, 15811–15816 10.1073/pnas.1205829109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Shinobu L.A., Ward C.M., Young D. and Cleveland D.W. (2005) Elevation of the Hsp70 chaperone does not effect toxicity in mouse models of familial amyotrophic lateral sclerosis. J. Neurochem. 93, 875–882 10.1111/j.1471-4159.2005.03054.x [DOI] [PubMed] [Google Scholar]
- 16.Mateju D., Franzmann T.M., Patel A., Kopach A., Boczek E.E., Maharana S. et al. (2017) An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrawell N.E., Lambert-Smith I., Mitchell K., McKenna J., McAlary L., Ciryam P. et al. (2018) SOD1A4V aggregation alters ubiquitin homeostasis in a cell model of ALS. J. Cell Sci. 131, jcs209122 10.1242/jcs.209122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zetterström P., Graffmo K.S., Andersen P.M., Brännström T. and Marklund S.L. (2011) Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic amyotrophic lateral sclerosis (ALS) model mice. J. Biol. Chem. 286, 20130–20136 10.1074/jbc.M111.218842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culotta V.C., Klomp L.W., Strain J., Casareno R.L., Krems B. and Gitlin J.D. (1997) The copper chaperone for superoxide dismutase. J. Biol. Chem. 272, 23469–23472 10.1074/jbc.272.38.23469 [DOI] [PubMed] [Google Scholar]
- 20.Jensen L.T. and Culotta V.C. (2005) Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J. Biol. Chem. 280, 41373–41379 10.1074/jbc.M509142200 [DOI] [PubMed] [Google Scholar]
- 21.Yamashita S., Mori A., Kimura E., Mita S., Maeda Y., Hirano T. et al. (2010) DJ-1 forms complexes with mutant SOD1 and ameliorates its toxicity. J. Neurochem. 113, 860–870 10.1111/j.1471-4159.2010.06658.x [DOI] [PubMed] [Google Scholar]
- 22.Girotto S., Cendron L., Bisaglia M., Tessari I., Mammi S., Zanotti G. et al. (2014) DJ-1 is a copper chaperone acting on SOD1 activation. J. Biol. Chem. 289, 10887–10899 10.1074/jbc.M113.535112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbieri L., Luchinat E. and Banci L. (2018) Intracellular metal binding and redox behavior of human DJ-1. J. Biol. Inorg. Chem. 23, 61–69 10.1007/s00775-017-1509-5 [DOI] [PubMed] [Google Scholar]
- 24.Subramaniam J.R., Lyons W.E., Liu J., Bartnikas T.B., Rothstein J., Price D.L. et al. (2002) Mutant SOD1 causes motor neuron disease independent of copper chaperone-mediated copper loading. Nat. Neurosci. 5, 301–307 10.1038/nn823 [DOI] [PubMed] [Google Scholar]
- 25.Wong P.C., Waggoner D., Subramaniam J.R., Tessarollo L., Bartnikas T.B., Culotta V.C. et al. (2000) Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 97, 2886–2891 10.1073/pnas.040461197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll M.C., Girouard J.B., Ulloa J.L., Subramaniam J.R., Wong P.C., Valentine J.S. et al. (2004) Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc. Natl. Acad. Sci. U.S.A. 101, 5964–5969 10.1073/pnas.0308298101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attar N., Campos O.A., Vogelauer M., Cheng C., Xue Y., Schmollinger S. et al. (2020) The histone H3-H4 tetramer is a copper reductase enzyme. Science 369, 59–64 10.1126/science.aba8740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banci L., Bertini I., Cantini F., Kozyreva T., Massagni C., Palumaa P. et al. (2012) Human superoxide dismutase 1 (hSOD1) maturation through interaction with human copper chaperone for SOD1 (hCCS). Proc. Natl. Acad. Sci. U.S.A. 109, 13555–13560 10.1073/pnas.1207493109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright G.S.A., Antonyuk S.V. and Hasnain S.S. (2016) A faulty interaction between SOD1 and hCCS in neurodegenerative disease. Sci. Rep. 6, 27691 10.1038/srep27691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sala F.A., Wright G.S.A., Antonyuk S.V., Garratt R.C. and Hasnain S.S. (2019) Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS. PLOS Biol. 17, e3000141 10.1371/journal.pbio.3000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd S.D., Ullrich M.S., Calvo J.S., Behnia F., Meloni G. and Winkler D.D. (2020) Mutations in superoxide dismutase 1 (Sod1) linked to familial amyotrophic lateral sclerosis can disrupt high-affinity zinc-binding promoted by the copper chaperone for Sod1 (Ccs). Molecules 25, 1086 10.3390/molecules25051086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luchinat E., Barbieri L. and Banci L. (2017) A molecular chaperone activity of CCS restores the maturation of SOD1 fALS mutants. Sci. Rep. 7, 17433 10.1038/s41598-017-17815-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyd S.D., Liu L., Bulla L. and Winkler D.D. (2018) Quantifying the interaction between copper-zinc superoxide dismutase (Sod1) and its copper chaperone (Ccs1). J. Proteomics Bioinform. 11, 473 10.4172/jpb.1000473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd S.D., Calvo J.S., Liu L., Ullrich M.S., Skopp A., Meloni G. et al. (2019) The yeast copper chaperone for copper-zinc superoxide dismutase (CCS1) is a multifunctional chaperone promoting all levels of SOD1 maturation. J. Biol. Chem. 294, 1956–1966 10.1074/jbc.RA118.005283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luchinat E., Barbieri L., Rubino J.T., Kozyreva T., Cantini F. and Banci L. (2014) In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 5, 1–10 10.1038/ncomms6502 [DOI] [PubMed] [Google Scholar]
- 36.Son M., Puttaparthi K., Kawamata H., Rajendran B., Boyer P.J., Manfredi G. et al. (2007) Overexpression of CCS in G93A-SOD1 mice leads to accelerated neurological deficits with severe mitochondrial pathology. Proc. Natl. Acad. Sci. U.S.A. 104, 6072–6077 10.1073/pnas.0610923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proescher J.B., Son M., Elliott J.L. and Culotta V.C. (2008) Biological effects of CCS in the absence of SOD1 enzyme activation: implications for disease in a mouse model for ALS. Hum. Mol. Genet. 17, 1728–1737 10.1093/hmg/ddn063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H.K., Chung Y.W., Chock P.B. and Yim M.B. (2011) Effect of CCS on the accumulation of FALS SOD1 mutant-containing aggregates and on mitochondrial translocation of SOD1 mutants: implication of a free radical hypothesis. Arch. Biochem. Biophys. 509, 177–185 10.1016/j.abb.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams J.R., Trias E., Beilby P.R., Lopez N.I., Labut E.M., Samuel Bradford C. et al. (2016) Copper delivery to the CNS by CuATSM effectively treats motor neuron disease in SOD(G93A) mice co-expressing the copper-chaperone-for-SOD. Neurobiol. Dis. 89, 1–9 10.1016/j.nbd.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Son M., Fu Q., Puttaparthi K., Matthews C.M. and Elliott J.L. (2009) Redox susceptibility of SOD1 mutants is associated with the differential response to CCS over-expression in vivo. Neurobiol. Dis. 34, 155–162 10.1016/j.nbd.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacher M., Meinhardt A., Lan H.Y., Dhabhar F.S., Mu W., Metz C.N. et al. (1998) MIF expression in the rat brain: implications for neuronal function. Mol. Med. 4, 217–230 10.1007/BF03401919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Israelson A., Ditsworth D., Sun S., Song S., Liang J., Hruska-Plochan M. et al. (2015) Macrophage migration inhibitory factor as a chaperone inhibiting accumulation of misfolded SOD1. Neuron 86, 218–232 10.1016/j.neuron.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leyton-Jaimes M.F., Benaim C., Abu-Hamad S., Kahn J., Guetta A., Bucala R. et al. (2016) Endogenous macrophage migration inhibitory factor reduces the accumulation and toxicity of misfolded SOD1 in a mouse model of ALS. Proc. Natl. Acad. Sci. U.S.A. 113, 10198–10203 10.1073/pnas.1604600113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Zhao D., Xian M., Wang Z., Bi E., Su P. et al. (2020) MIF as a biomarker and therapeutic target for overcoming resistance to proteasome inhibitors in human myeloma. Blood in press 10.1182/blood.2020005795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leyton-Jaimes M.F., Kahn J. and Israelson A. (2019) AAV2/9-mediated overexpression of MIF inhibits SOD1 misfolding, delays disease onset, and extends survival in mouse models of ALS. Proc. Natl. Acad. Sci. U.S.A. 116, 14755–14760 10.1073/pnas.1904665116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schinagl A., Kerschbaumer R.J., Sabarth N., Douillard P., Scholz P., Voelkel D. et al. (2018) Role of the cysteine 81 residue of macrophage migration inhibitory factor as a molecular redox switch. Biochemistry 57, 1523–1532 10.1021/acs.biochem.7b01156 [DOI] [PubMed] [Google Scholar]
- 47.Shvil N., Banerjee V., Zoltsman G., Shani T., Kahn J., Abu-Hamad S. et al. (2018) MIF inhibits the formation and toxicity of misfolded SOD1 amyloid aggregates: implications for familial ALS. Cell Death Dis. 9, 107 10.1038/s41419-017-0130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouertatani-Sakouhi H., El-Turk F., Fauvet B., Cho M.-K., Karpinar D.P., Roy D.L. et al. (2010) Identification and characterization of novel classes of macrophage migration inhibitory factor (MIF) inhibitors with distinct mechanisms of action. J. Biol. Chem. 285, 26581–26598 10.1074/jbc.M110.113951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayward L.J., Rodriguez J.A., Kim J.W., Tiwari A., Goto J.J., Cabelli D.E. et al. (2002) Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 277, 15923–15931 10.1074/jbc.M112087200 [DOI] [PubMed] [Google Scholar]
- 50.Farhan S.M.K., Howrigan D.P., Abbott L.E., Klim J.R., Topp S.D., Byrnes A.E. et al. (2019) Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat. Neurosci. 22, 1966–1974 10.1038/s41593-019-0530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roosen D.A., Blauwendraat C., Cookson M.R. and Lewis P.A. (2019) DNAJC proteins and pathways to parkinsonism. FEBS J. 286, 3080–3094 10.1111/febs.14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliott E., Bailey O., Waldron F.M., Hardingham G.E., Chandran S. and Gregory J.M. (2020) Therapeutic targeting of proteostasis in amyotrophic lateral sclerosis-a systematic review and meta-analysis of preclinical research. Front. Neurosci. 14, 511 10.3389/fnins.2020.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Münch C. and Bertolotti A. (2010) Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J. Mol. Biol. 399, 512–525 10.1016/j.jmb.2010.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenzweig R., Nillegoda N.B., Mayer M.P. and Bukau B. (2019) The Hsp70 chaperone network. Nat. Rev. Mol. Cell. Biol. 20, 665–680 10.1038/s41580-019-0133-3 [DOI] [PubMed] [Google Scholar]
- 55.Sekhar A., Velyvis A., Zoltsman G., Rosenzweig R., Bouvignies G. and Kay L.E. (2018) Conserved conformational selection mechanism of Hsp70 chaperone-substrate interactions. eLife 7, e32764 10.7554/eLife.32764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rüdiger S., Germeroth L., Schneider-Mergener J. and Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 10.1093/emboj/16.7.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekhar A., Rosenzweig R., Bouvignies G. and Kay L.E. (2016) Hsp70 biases the folding pathways of client proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E2794–E2801 10.1073/pnas.1601846113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Farr G.W., Zeiss C.J., Rodriguez-Gil D.J., Wilson J.H., Furtak K. et al. (2009) Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc. Natl. Acad. Sci. U.S.A. 106, 1392–1397 10.1073/pnas.0813045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brocchieri L., Conway de Macario E. and and Macario A.J. (2008) Hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 8, 19 10.1186/1471-2148-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Claes F., Rudyak S., Laird A.S., Louros N., Beerten J., Debulpaep M. et al. (2019) Exposure of a cryptic Hsp70 binding site determines the cytotoxicity of the ALS-associated SOD1-mutant A4V. Protein Eng. Des. Sel. 32, 443–457 10.1093/protein/gzaa008 [DOI] [PubMed] [Google Scholar]
- 61.Serlidaki D., van Waarde M.A.W.H., Rohland L., Wentink A.S., Dekker S.L., Kamphuis M.J. et al. (2020) Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J. Biol. Chem. 295, 7301–7316 10.1074/jbc.RA119.012449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruening W., Roy J., Giasson B., Figlewicz D.A., Mushynski W.E. and Durham H.D. (1999) Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J. Neurochem. 72, 693–699 10.1046/j.1471-4159.1999.0720693.x [DOI] [PubMed] [Google Scholar]
- 63.Nagy M., Fenton W.A., Li D., Furtak K. and Horwich A.L. (2016) Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc. Natl. Acad. Sci. U.S.A. 113, 5424–5428 10.1073/pnas.1604885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe S., Ageta-Ishihara N., Nagatsu S., Takao K., Komine O., Endo F. et al. (2014) SIRT1 overexpression ameliorates a mouse model of SOD1-linked amyotrophic lateral sclerosis via HSF1/HSP70i chaperone system. Mol. Brain 7, 62 10.1186/s13041-014-0062-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalmar B., Novoselov S., Gray A., Cheetham M.E., Margulis B. and Greensmith L. (2008) Late stage treatment with arimoclomol delays disease progression and prevents protein aggregation in the SOD1 mouse model of ALS. J. Neurochem. 107, 339–350 10.1111/j.1471-4159.2008.05595.x [DOI] [PubMed] [Google Scholar]
- 66.Kieran D., Kalmar B., Dick J.R.T., Riddoch-Contreras J., Burnstock G. and Greensmith L. (2004) Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat. Med. 10, 402–405 10.1038/nm1021 [DOI] [PubMed] [Google Scholar]
- 67.Benatar M., Wuu J., Andersen P.M., Atassi N., David W., Cudkowicz M. et al. (2018) Randomized, double-blind, placebo-controlled trial of arimoclomol in rapidly progressive SOD1 ALS. Neurology 90, e565–e574 10.1212/WNL.0000000000004960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kityk R., Kopp J. and Mayer M.P. (2018) Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 Chaperones. Mol. Cell 69, 227–237.e4 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 69.Besemer A.S., Maus J., Ax M.D.A., Stein A., Vo S., Freese C. et al. (2020) Receptor-mediated endocytosis 8 (RME-8)/DNAJC13 is a novel positive modulator of autophagy and stabilizes cellular protein homeostasis. Cell. Mol. Life Sci. 10.1007/s00018-020-03521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Novoselov S.S., Mustill W.J., Gray A.L., Dick J.R., Kanuga N., Kalmar B. et al. (2013) Molecular chaperone mediated late-stage neuroprotection in the SOD1G93A mouse model of amyotrophic lateral sclerosis. PLoS One 8, e73944 10.1371/journal.pone.0073944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blumen S.C., Astord S., Robin V., Vignaud L., Toumi N., Cieslik A. et al. (2012) A rare recessive distal hereditary motor neuropathy with HSJ1 chaperone mutation. Ann. Neurol. 71, 509–519 10.1002/ana.22684 [DOI] [PubMed] [Google Scholar]
- 72.Glover J.R. and Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- 73.Kim Y., Park J.-H., Jang J.-Y., Rhim H. and Kang S. (2013) Characterization and Hsp104-induced artificial clearance of familial ALS-related SOD1 aggregates. Biochem. Biophys. Res. Commun. 434, 521–526 10.1016/j.bbrc.2013.03.107 [DOI] [PubMed] [Google Scholar]
- 74.Nillegoda N.B., Kirstein J., Szlachcic A., Berynskyy M., Stank A., Stengel F. et al. (2015) Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tittelmeier J., Sandhof C.A., Ries H.M., Druffel-Augustin S., Mogk A., Bukau B. et al. (2020) The HSP110/HSP70 disaggregation system generates spreading-competent toxic α-synuclein species. EMBO J. 39, e103954 10.15252/embj.2019103954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang L., Zetterström P., Brännström T., Marklund S.L., Danielsson J. and Oliveberg M. (2015) SOD1 aggregation in ALS mice shows simplistic test tube behavior. Proc. Natl. Acad. Sci. U.S.A. 112, 9878–9883 10.1073/pnas.1503328112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ayers J.I., Fromholt S.E., O'Neal V.M., Diamond J.H. and Borchelt D.R. (2016) Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 131, 103–114 10.1007/s00401-015-1514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grad L.I., Yerbury J.J., Turner B.J., Guest W.C., Pokrishevsky E., O'Neill M.A. et al. (2014) Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 3620–3625 10.1073/pnas.1312245111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu C., Beck M.V., Griffith J.D., Deshmukh M. and Dokholyan N.V. (2018) Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 115, 4661–4665 10.1073/pnas.1800187115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gill C., Phelan J.P., Hatzipetros T., Kidd J.D., Tassinari V.R., Levine B. et al. (2019) SOD1-positive aggregate accumulation in the CNS predicts slower disease progression and increased longevity in a mutant SOD1 mouse model of ALS. Sci. Rep. 9, 6724 10.1038/s41598-019-43164-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Auclair J.R., Boggio K.J., Petsko G.A., Ringe D. and Agar J.N. (2010) Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc Natl. Acad. Sci. U.S.A. 107, 21394–21399 10.1073/pnas.1015463107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Donnelly D.P., Dowgiallo M.G., Salisbury J.P., Aluri K.C., Iyengar S., Chaudhari M. et al. (2018) Cyclic thiosulfinates and cyclic disulfides selectively cross-link thiols while avoiding modification of lone thiols. J. Am. Chem. Soc. 140, 7377–7380 10.1021/jacs.8b01136 [DOI] [PubMed] [Google Scholar]
- 83.Banci L., Bertini I., Blaževitš O., Calderone V., Cantini F., Mao J. et al. (2012) Interaction of cisplatin with human superoxide dismutase. J. Am. Chem. Soc. 134, 7009–7014 10.1021/ja211591n [DOI] [PubMed] [Google Scholar]
- 84.Capper M.J., Wright G.S.A., Barbieri L., Luchinat E., Mercatelli E., McAlary L. et al. (2018) The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation. Nat. Commun. 9, 1693 10.1038/s41467-018-04114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chantadul V., Wright G.S.A., Amporndanai K., Shahid M., Antonyuk S.V., Washbourn G. et al. (2020) Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy. Commun. Biol. 3, 1–10 10.1038/s42003-020-0826-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Z., Jiang Z., Chen N., Shi Q., Tong L., Kong F. et al. (2018) Target discovery of ebselen with a biotinylated probe. Chem. Commun. 54, 9506–9509 10.1039/C8CC04258F [DOI] [PubMed] [Google Scholar]
- 87.Oh-hashi K. and Hirata Y. (2020) Elucidation of the molecular characteristics of wild-type and ALS-LINKED mutant SOD1 using the nanoLuc complementation reporter system. Appl. Biochem. Biotechnol. 190, 674–685 10.1007/s12010-019-03114-x [DOI] [PubMed] [Google Scholar]
- 88.Banerjee V., Shani T., Katzman B., Vyazmensky M., Papo N., Israelson A. et al. (2016) Superoxide dismutase 1 (SOD1)-derived peptide inhibits amyloid aggregation of familial amyotrophic lateral sclerosis SOD1 mutants. ACS Chem. Neurosci. 7, 1595–1606 10.1021/acschemneuro.6b00227 [DOI] [PubMed] [Google Scholar]
- 89.Bunck D.N., Atsavapranee B., Museth A.K., VanderVelde D. and Heath J.R. (2018) Modulating the folding landscape of superoxide dismutase 1 with targeted molecular binders. Angew. Chem. 57, 6212–6215 10.1002/anie.201802269 [DOI] [PMC free article] [PubMed] [Google Scholar]