Abstract
The structural maintenance of chromosomes hinge domain containing protein 1 (SMCHD1) is a large multidomain protein involved in epigenetic gene silencing. Variations in the SMCHD1 gene are associated with two debilitating human disorders, facioscapulohumeral muscular dystrophy (FSHD) and Bosma arhinia microphthalmia syndrome (BAMS). Failure of SMCHD1 to silence the D4Z4 macro-repeat array causes FSHD, yet the consequences on gene silencing of SMCHD1 variations associated with BAMS are currently unknown. Despite the interest due to these roles, our understanding of the SMCHD1 protein is in its infancy. Most knowledge of SMCHD1 function is based on its similarity to the structural maintenance of chromosomes (SMC) proteins, such as cohesin and condensin. SMC proteins and SMCHD1 share similar domain organisation and affect chromatin conformation. However, there are important differences between the domain architectures of SMC proteins and SMCHD1, which distinguish SMCHD1 as a non-canonical member of the family. In the last year, the crystal structures of the two key domains crucial to SMCHD1 function, the ATPase and hinge domains, have emerged. These structures reveal new insights into how SMCHD1 may bind and regulate chromatin structure, and address how amino acid variations in SMCHD1 may contribute to BAMS and FSHD. Here, we contrast SMCHD1 with canonical SMC proteins, and relate the ATPase and hinge domain structures to their roles in SMCHD1-mediated epigenetic silencing and disease.
Keywords: BAMS, chromatin, epigenetics, FSHD, gene silencing, SMCHD1
Introduction
The structural maintenance of chromosomes hinge domain containing protein 1 (SMCHD1) is an epigenetic regulator that controls gene expression at selective sites across the genome [1]. While its initial discovery revealed that Smchd11 is critical in the process of X-chromosome inactivation and thereby essential in female embryo viability [2], numerous studies have now recognised its role in regulating the expression of various autosomal gene clusters such as Pcdh and HoxB, in addition to monoallelically expressed targets such as selected genes within the Snrpn cluster [3–7]. The exact underlying mechanism remains unknown, but experimental evidence suggests that Smchd1 is involved in the maintenance of long range chromatin looping such that it limits promoter–enhancer interactions and therefore creates a transcriptionally repressive environment [4,8,9].
Importantly, heterozygous SMCHD1 variants are associated with autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) and the rare craniofacial disorder Bosma arhinia microphthalmia syndrome (BAMS) (Table 1) [10–12]. However, the mechanisms by which pathogenic SMCHD1 variants lead to different clinical disorders are not fully understood. The reported pathogenic variants associated with the two conditions do not typically overlap and the resulting phenotypic outcomes are entirely distinctive, suggesting different molecular mechanisms are at play. SMCHD1 is a very large and flexible dimer of ∼250 kDa protomers, which presents a major challenge in understanding SMCHD1's mechanism of action via structural approaches. Only one study to date has revealed low-resolution images of full-length Smchd1 via negative stain electron microscopy [13]. In the last year, X-ray crystallography has provided the first high-resolution structures of its two known functional regions: the N-terminal ATPase and C-terminal hinge domains [14,15].
Table 1. SMCHD1 single nucleotide polymorphisms (SNPs) resulting in missense mutations described in patients with facioscapulohumeral muscular dystrophy type 2 (FSHD2) and Bosma arhinia microphthalmia syndrome (BAMS).
| Mutation | SMCHD1 domain | Associated disease | Pubmed ID |
|---|---|---|---|
| Arg34Pro | UBL | FSHD2 | 31243061 |
| Asn104Ser | UBL | FSHD2 | 31243061 |
| Leu107Pro | UBL-ATPase linker | FSHD2, BAMS | 31243061, 29980640, 28067909 |
| Ala110Thr | UBL-ATPase linker | FSHD2 | 31243061, 25370034 |
| Met129Arg | ATPase | BAMS | 31243061 |
| Met129Lys | ATPase | BAMS | 31243061, 28067909 |
| Ala134Ser | ATPase | BAMS | 31243061, 28067911 |
| Ser135Asn | ATPase | BAMS | 31243061, 28067909, 28067911 |
| Ser135Cys | ATPase | BAMS | 31243061, 28067909, 28067911, 30698748 |
| Ser135Ile | ATPase | BAMS | 31243061, 28067909, 28067911 |
| Glu136Asp | ATPase | BAMS | 31243061, 28067909 |
| Glu136Gly | ATPase | BAMS | 31243061, 28067911, 30698748 |
| Gly137Glu | ATPase | FSHD2, BAMS | 31243061, 28067909, 25256356 |
| Asn139His | ATPase | BAMS | 31243061, 28067909 |
| Leu141Phe | ATPase | BAMS | 31243061, 28067909, 28067911 |
| Asp150His | ATPase | FSHD2 | 31243061 |
| Phe171Val | ATPase | BAMS | 31243061, 28067909 |
| Gly188Arg | ATPase | FSHD2 | 31243061 |
| Met189Val | ATPase | FSHD2 | 31243061 |
| Gln193Pro | ATPase | FSHD2 | 30698748 |
| Leu194Phe | ATPase | FSHD2 | 31243061, 25256356 |
| Lys204Glu | ATPase | FSHD2 | 31243061, 29980640 |
| Ala242Gly | ATPase | BAMS | 31243061, 28067909 |
| Ala242Thr | ATPase | FSHD2 | 31243061, 29980640 |
| His263Asp | ATPase | FSHD2 | 31243061, 25256356 |
| Glu264Lys | ATPase | FSHD2 | 31243061 |
| Tyr283Cys | ATPase | FSHD2 | 31243061, 27061275 |
| Trp324Ser | ATPase | BAMS | 31243061, 28067911 |
| Arg344Gln | ATPase | FSHD2 | 31243061, 29980640 |
| Gln345Arg | ATPase | BAMS | 31243061, 28067909 |
| His348Arg | ATPase | BAMS | 31243061, 28067909, 28067911 |
| Tyr353Cys | ATPase | FSHD2 | 31243061, 23143600 |
| Gln400Leu | Transducer | BAMS | 31243061, 28067909 |
| Asp420Val | Transducer | BAMS | 31243061, 28067909, 28067911, 30698748 |
| Gly425Arg | Transducer | FSHD2 | 31243061, 25256356 |
| Arg428Cys | Transducer | FSHD2 | 31243061 |
| Glu473Gln | Transducer | BAMS | 31243061, 28067909 |
| Gly478Glu | Transducer | FSHD2 | 31243061, 25370034 |
| Arg479Pro | Transducer | FSHD2 | 31243061, 23143600 |
| Arg479Leu | Transducer | FSHD2 | 31243061, 28744936 |
| Arg479Gln | Transducer | FSHD2 | 31243061 |
| Cys492Arg | Transducer | FSHD2 | 31243061, 23143600 |
| Lys518Glu | Transducer | BAMS | 31243061, 28067911 |
| Phe519Ser | Transducer | FSHD2 | 31243061, 29980640 |
| Thr523Lys | Transducer | BAMS | 31243061, 28067909 |
| Asn524Ser | Transducer | BAMS | 31243061, 28067909 |
| Thr527Met | Transducer | FSHD2 | 31243061, 24075187 |
| Gln551Arg | Transducer | FSHD2 | 31243061 |
| Arg552Gln | Transducer | BAMS | 31243061, 28067909, 28067911 |
| Trp596Gly | linker (strand prediction) | FSHD2 | 31243061 |
| Val615Asp | linker (strand prediction) | FSHD2 | 31243061, 25370034 |
| Pro622Leu | linker (helical prediction) | FSHD2 | 31243061 |
| Val641Leu | linker (strand prediction) | FSHD2 | 31243061 |
| Pro690Ser | linker (loop prediction) | FSHD2 | 31243061, 23143600 |
| Leu748Pro | linker (strand prediction) | FSHD2 | 31243061, 25256356 |
| Tyr774Cys | linker (loop prediction) | FSHD2 | 31243061 |
| Asp849Asn | linker (strand prediction) | FSHD2 | 31243061, 23143600 |
| Leu923Pro | linker (strand prediction) | FSHD2 | 31243061 |
| Leu978His | linker (loop prediction) | FSHD2 | 31243061 |
| Tyr981Asp | linker (strand prediction) | FSHD2 | 31243061, 27153398 |
| Gly1063Arg | linker (disorder prediction) | FSHD2 | 31243061 |
| Leu1108Pro | linker (disorder prediction) | FSHD2 | 31243061 |
| Val1114Ile | linker (disorder prediction) | FSHD2 | 31243061 |
| Val1271Leu | linker (strand prediction) | FSHD2 | 31243061 |
| Ile1300Lys | linker (disorder prediction) | FSHD2 | 31243061 |
| Gln1463Pro | linker (strand prediction) | FSHD2 | 31243061, 25370034 |
| Met1468Ile | linker (strand prediction) | FSHD2 | 31243061, 25256356 |
| Pro1485Leu | linker (disorder prediction) | FSHD2 | 31243061, 25370034 |
| Phe1554Ser | linker (helical prediction) | FSHD2 | 31243061, 23143600 |
| Asp1750Gly | SMC hinge | FSHD2 | 31243061 |
| Asp1750Val | SMC hinge | FSHD2 | 31243061 |
| Tyr1846Cys | SMC hinge | FSHD2 | 31243061 |
| Arg1866Gly | SMC hinge | FSHD2 | 31243061, 27153398 |
| Arg1866Gln | SMC hinge | FSHD2 | 31243061 |
SMCHD1 is a non-canonical SMC protein
The structural maintenance of chromosomes (SMC) family of proteins are key organisers of chromatin architecture in all living organisms [16]. SMCHD1 is considered a non-canonical member of the SMC family owing to differences in its type of ATPase domain and in its overall linear domain architecture [14,17–21] (Figure 1A,B). SMC protein complexes function via highly conserved mechanisms owing to their essential roles, such as mediating chromosome conformation throughout the cell cycle. In mammals, each of the various functional complexes comprise of a heterodimeric pair of SMC subunits, such as SMC1 and SMC3, or SMC2 and SMC4, which are the core components of cohesin and condensin, respectively [17–19,21]. It should be noted that whilst prokaryotes have homodimeric SMC proteins, in this review we focus on mammalian proteins as there are no prokaryotic equivalents of SMCHD1. Each SMC monomer has three regions: the ABC-type ATPase domain that is split between the N- and C- termini, the DNA-interacting SMC hinge domain and an extended coiled-coil linker region that bridges the two domains (Figure 1B) [21–23]. The canonical SMC primary sequence starts with the Walker A motif from the ABC-type ATPase, followed by a N-terminal α-helix that links to the SMC hinge domain. At the C-terminus of the SMC hinge there is another α-helix, which forms an antiparallel coiled-coil with the N-terminal α-helix. This coiled-coil folds the protein back on itself allowing the C-terminal Walker B motif to dimerise with the N-terminal Walker A motif to form a functional ABC-type ATPase domain.
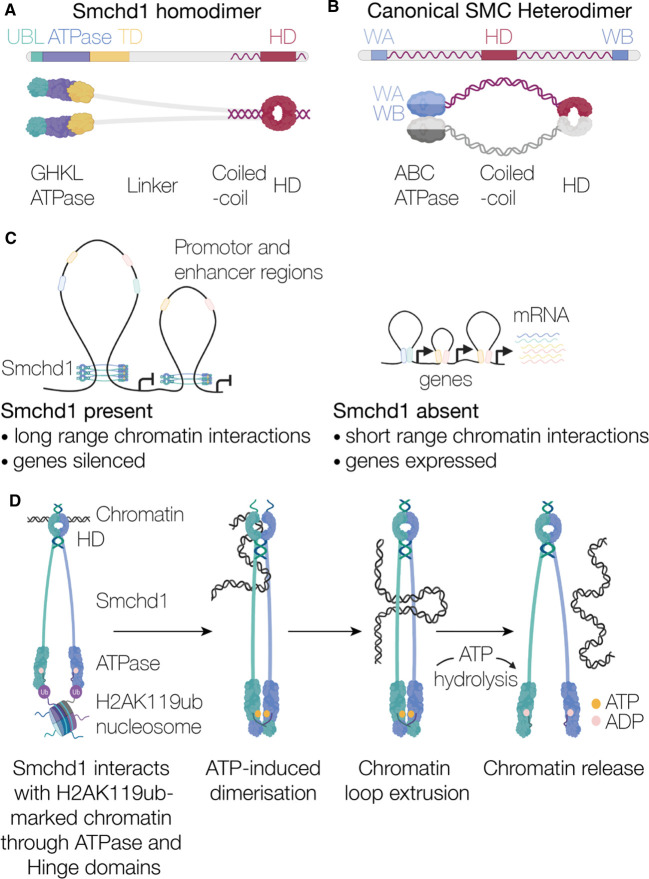
Figure 1. Overall structure and function of Smchd1.
Schematic comparing the similarities and differences between (A) Smchd1 and (B) a canonical SMC protein. (C) Smchd1 causes an increase in long range chromatin interactions silencing gene expression. (D) Hypothetical model of Smchd1 engaging with chromatin to facilitate long range DNA interactions controlled by ATP hydrolysis.
SMCHD1 also has a SMC hinge domain, however, in contrast with canonical SMC proteins, it is located at the protein's C-terminus (Figure 1A) [1,2,24]. A characteristic of canonical SMC hinge domains is the GX6GX3GG motif that lies at the hinge domain dimer interface. In SMCHD1, this sequence is one amino acid shorter, GX6GX2GG, which leads to altered structure in this region [15]. Furthermore, the SMCHD1 ATPase has a GHKL-type architecture and is located uniquely at the N-terminus instead of an ABC-type ATPase split between the N- and C- termini (Figure 1A,B) [13,14,20]. Between the ATPase and SMC hinge there is a long linker region spanning approximately 1200 amino acids. In canonical SMC proteins, this region is predominantly α-helical, but for SMCHD1 it is predicted to consist of β-strands with only the last 200 amino acids, proximal to the SMC hinge, predicted to be α-helical (Figure 1A,B). Finally, the C-terminal segment in SMCHD1 is predicted to contain a relatively short α-helix spanning approximately 100 amino acids, compared with approximately 300 amino acids for the canonical SMC proteins [25,26]. In mammals, the canonical SMC proteins heterodimerise through interactions primarily at the hinge domain interface with some interactions at the ATPase region [17–19,21]. In contrast, SMCHD1 forms a homodimer at the C-terminus through its SMC hinge domain (Figure 1A) [24]. SMCHD1 can also homodimerise through its N-terminal ATPase region (Figures 1A and 2A). This dimerisation occurs via exchange of a ubiquitin-like (UBL) domain from one protomer to the other; this UBL domain is not present in the canonical SMC proteins (Figure 1A and 2A) [14]. In addition, evidence from small-angle X-ray scattering (SAXS) and electron microscopy indicates that SMCHD1 α-helices form N- to N- and C- to C- coiled-coil pairings [13,24], providing an additional region for SMCHD1 homodimerisation.
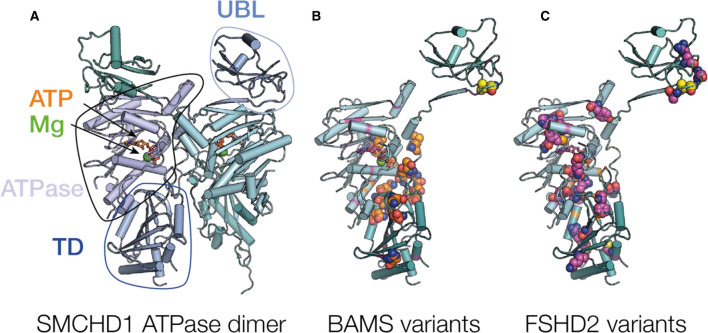
Figure 2. The SMCHD1 ATPase domain.
(A) Representation of the SMCHD1 GHKL ATPase region (PDB ID 6MW7) [14]. The image shows strand-swapped ubiquitin-like (UBL) domain (circled) bound to the GHKL ATPase from the opposing monomer. The ATPase, ATP and magnesium (Mg) binding sites, and transducer domains (TD) from the same protomer are also circles and labelled. (B,C) The locations of missense variants within the ATPase region that are associated with (B) BAMS (orange) and (C) FSHD2 (magenta) displayed on one monomer from the dimer. Missense variations associated with both diseases are shown in yellow.
The SMC heterodimers form a closed ring structure that is proposed to topologically entrap or encircle DNA [27,28] (Figure 1C,D). Despite being well-studied, their exact molecular mechanism is not established. The ‘loop extrusion’ model is the most widely accepted mechanism that describes the functions of cohesin and condensin. This model proposes they push chromatin through their central coiled-coil ring structure to form chromatin loops in an ATP-dependent manner [19,29,30]. This energy-dependent process drives loop elongation, where one or both DNA interaction sites translocate away from each other, leading to the formation of chromatin loops (Figure 1C,D). The association of cohesin with chromatin is additionally dependent on the transcription factor CCCTC-binding factor (CTCF), which acts as a barrier to the loop extrusion process by stabilising cohesin at CTCF-binding sites and creating the base of the established DNA loop [31,32]. This mechanism facilitates promoter–enhancer interactions between distal regions of the genome to help establish functional domains that are required for transcriptional regulation. The loop extrusion model has recently been demonstrated in vitro for both yeast condensin [33,34] and human cohesin [35,36], where the process was visualised via single-molecule live imaging techniques using surface-tethered DNA. While yeast condensin was observed to undergo the predicted topological entrapment of DNA where both strands are embraced by its ring-like structure, human cohesin appeared to instead interact with DNA pseudo-topologically or non-topologically; therefore, embracing either one strand of DNA only or not encircling DNA entirely [35,36]. Precisely how SMC complexes use their ATPase activity to facilitate chromosome structure rearrangements remains uncertain. However, common to all proposed mechanisms, SMC proteins use ATP hydrolysis to change conformation and entrap DNA, and this ultimately drives dynamic loop formation [16,33,37,38].
There are clear similarities between SMCHD1 and canonical SMC proteins. However, differences in gene architecture and domain organisation between SMCHD1 and canonical SMC proteins suggest that functional differences may also be present. Experimental evidence suggests that SMCHD1 retains the ability to alter chromatin structure as part of its mechanism of action, similar to canonical SMC proteins [4,8,9]. But, while canonical SMC proteins are known to assemble into functional protein complexes by forming heterodimers, SMCHD1 is only known to form homodimers. This feature along with the presence of a GHKL-type ATPase domain in SMCHD1 as opposed to ABC-type ATPases (Figure 1A,B) may indicate that it is more similar to the MORC family of proteins, which are also part of the GHKL superfamily [39]. The MORC family use ATP-binding to control dimerisation events and entrap DNA [40], forming chromatin loops by compaction, as opposed to the active loop extrusion proposed for canonical SMC proteins [33,35,36].
ATPase domain
SMCHD1 has a GHKL-type ATPase domain, a functionally diverse protein superfamily that is named for the archetypal members, Gyrase B, Hsp90 histadine kinase and MutL [20,41]. GHKL ATPases typically consist of a Bergerat fold which describes an α/β sandwich that consists of a four-stranded β-sheet and three α-helices, in addition to a unique feature — a long flexible loop known as the ATP-lid. This loop is highly variable across members of the GHKL superfamily. Different sequences and conformations among the ATP-lid distinguish different GHKL proteins, yet they all hold a conserved role in ATP-binding which suggests their functional importance. The recently solved crystal structure of a SMCHD1 ATPase construct (residues 25–580) revealed the Bergerat fold as the catalytic domain (residues 110–395) (Figure 2A) [14]. Pedersen et al. also identified a novel UBL domain at the N-terminus (residues 25–110) that undergoes a domain-swapping event between two SMCHD1 monomers via an N-terminal β-strand (residues 110–120) (Figure 2A). This is the first study to describe the dimerisation of SMCHD1's ATPase domain, which is not an unexpected finding considering many GHKL ATPases sustain the ability to homodimerise [41]. Additionally, the obligate homodimerisation of SMCHD1's hinge domain reflects potential contacts at the opposing end of the protein to where the N-terminal ATPase domain resides. This study revealed that SMCHD1 dimerisation requires not only the UBL domain, but also the presence of ATP, in addition to the transducer domain that abuts the C-terminus of the ATPase domain (Figure 2A). Surprisingly, this structure did not reveal any interface contacts between SMCHD1 monomers at the transducer domain; dimerisation appears to instead create a cavity between the transducer domains of two monomers. An explanation for the inability of SMCHD1 to dimerise upon absence of the transducer domains was not postulated. The distal C-terminal region commonly serves as a site of homodimerisation across members of the GHKL family [42–44]. It would, therefore, be interesting to further investigate whether extending the SMCHD1 ATPase construct at the C-terminus may reveal contacts between the transducer domains.
Despite providing valuable insights into the molecular structure of SMCHD1, it is important to note this structure used a catalytically inactive point mutant of the ATPase domain [3]. The E147A mutation introduced was previously shown to completely abolish the ATPase activity of Smchd1 [20], as the glutamic acid is a conserved residue across members of the GHKL superfamily that is indispensable for the ATP hydrolysis step. Because this crystal structure was solved in the presence of ATP, it is presumed the E147A mutant retains the ability to bind, but not hydrolyse, ATP. Interestingly, dimerisation triggered by ATP-binding is a common feature among GHKL ATPases [40,45–47]. This is closely followed by the closing of the ATP-lid over the active site to allow ATP hydrolysis and subsequent dissociation of the dimer. If the E147A mutant is trapped in the ATP-bound state, this phenomenon likely justifies a preferential dimerisation over wild-type SMCHD1. This idea is further supported by native PAGE analyses where a proportion of the E147A variant of SMCHD1 migrates as a higher molecular mass species under native conditions, which was interpreted as a dimer. Surprisingly, the wild-type or any other SMCHD1 variant tested remained largely monomeric. However, these results contrast chemical cross-linking experiments performed for the corresponding SMCHD1 variants, which suggest that all except FSHD2-related mutants exhibit some capacity to dimerise [14]. It, therefore, remains of outstanding interest whether the wild-type counterpart adopts a similar dimeric conformation to the E147A variant, prompting more in-depth biophysical experiments to establish dimerisation parameters for both wild-type and variant forms of SMCHD1.
All SMCHD1 variations found in BAMS patients that have been identified to date map to the N-terminal region, in addition to numerous FSHD2-associated variations in the same region, highlighting the overall importance of the N-terminal region in SMCHD1's function (Figure 2B,C and Table 1). One of the most intriguing aspects of unveiling an atomic structure of SMCHD1's ATPase domain was the ability to spatially map pathogenic variants to help provide a better understanding in how these may alter SMCHD1's function as an epigenetic regulator. Largely, it seems disease variants do not cluster in specific regions of the ATPase domain, although there seems to be a hotspot for BAMS-associated mutations located in a loop region of SMCHD1 that is situated at the dimer interface, suggesting that BAMS-associated variants possibly alter SMCHD1 dimerisation (Figure 2B,C). Prior to the release of the first crystal structure of SMCHD1's ATPase domain, a separate study mapped the location of FSHD2 and BAMS variants in SMCHD1 based on the crystal structure of the GHKL ATPase TRAP1 [48]. Their findings suggested that FSHD2 variants are almost exclusively located around the ATP-binding site, whereas the majority of BAMS variants localise to a loop region within the dimer interface, consistent with the published structure of SMCHD1. Pedersen et al. [14] briefly explored the dimerisation properties of SMCHD1 variants via native PAGE and cross-linking experiments, and concluded that dimerisation was preserved in BAMS-associated mutants but greatly reduced in FSHD2-associated mutants. This raises the possibility that SMCHD1 variations in FSHD2 patients may impact ATP-binding and dimerisation in the N-terminal region, which would be expected to diminish SMCHD1 function. Conversely, it remains to be explored whether variations identified in BAMS patients might influence other functions of SMCHD1, such as chromatin interactions or recruitment of potential protein interactors. While ATPase domain catalytic activity appears important for normal SMCHD1 function, it is compromised in some FSHD2 patients [49]. Additional FSHD2-associated variations have been identified throughout the SMCHD1 gene and are not limited to the ATPase region. Each of these variations prevents SMCHD1 silencing the D4Z4 macro-repeat array in FSHD2 patients. Therefore, defective ATPase dimerisation is one explanation for SMCHD1's loss of function in FSHD2, but there are likely multiple contributing factors that require examination in the context of the full-length protein.
Hinge domain
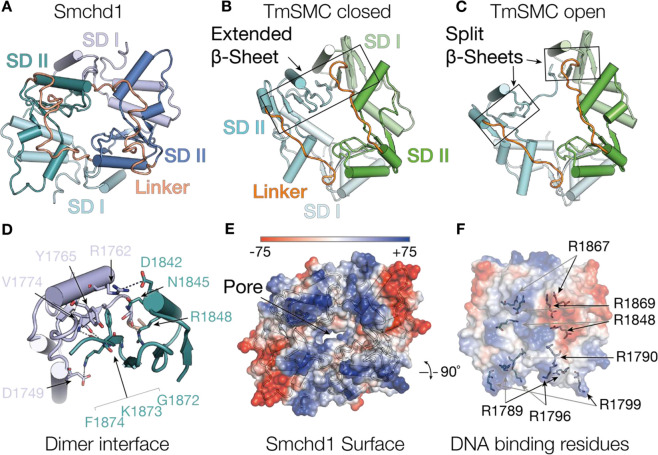
The hinge domain of SMCHD1 is located at the C-terminus of the protein. The hinge domain forms the principal interface for the assembly of SMCHD1 dimers and can interact with chromatin. We recently solved the structure of the mouse Smchd1 hinge domain, providing the first example of a homodimeric mammalian SMC hinge domain (Figure 3A) [15]. The Smchd1 hinge domain homodimer comprised two protomers related by rotational symmetry, with each monomer containing two pseudo-symmetrical subdomains bridged by an inter-subdomain linker region (Figure 3A). This structure shared most of the secondary structure features present in the canonical SMC hinge domains, however, there were distinct differences. The closest published structural homologue was from the extremophile bacteria Thermotoga maritima (Figure 3B,C) and the closest mammalian structure was from human SMC4, varying by a root mean squared deviation of 3.4 and 3.8 Å, respectively [28,50]. This large deviation in architecture is due to a change in the angle between the two subdomains. Hinge domains have two dimerisation interfaces between subdomain I and subdomain II of opposing protomers. The dimerisation interface in Smchd1 contains unique hydrogen bonding, hydrophobic and salt bridge interactions that facilitate its homodimerisation and preclude its heterodimerisation with the other mammalian SMC proteins (Figure 3D). An interesting feature of SMC hinge domains is an extended β-sheet that bridges across the dimerisation interface, typically comprising three strands from subdomain I on one protomer and five strands from subdomain II on the opposing protomer (Figure 3A–C) [28,51–54]. Published structures of the canonical SMC hinge domains adopt either open or closed conformations, (Figure 3A–C). In a closed conformation both dimerisation interfaces are intact showing two extended β-sheets (Figure 3B) [28,50]. In an open conformation, one dimerisation interface is lost resulting in one of the extended β-sheets splitting in half to form two discrete β-sheets (Figure 3C) [28,53,54]. This splitting of the extended β-sheet occurs next to the GX6GX3GG motif in subdomain II, highlighting the importance of this motif. In our structure, the Smchd1 hinge domain structure showed a closed conformation with both extended β-sheets intact (Figure 3A) [15]. It remains of outstanding interest whether the SMCHD1 hinge domain may similarly transition to open conformers, and how this might relate to its functions in epigenetic regulation.
Figure 3. The Smchd1 Hinge domain.
(A) Structure of the mouse Smchd1 hinge domain showing cartoon representation of the two protomers (green and blue) in the homodimer indicating subdomain I (SD I, pale colours) and subdomain II (SD II, dark colours) and subdomain linker region (orange). (B,C) Structures of the Thermotoga maritima SMC hinge domain showing both protomers (cyan and green) in the dimers, subdomain I (SD I, pale colours) and subdomain II (SD II, dark colours) and subdomain linker region (orange) displayed. (B) shows a closed hinge domain conformation with an extended β-sheet and (C) open conformation with a split β-sheet at one dimerisation interface. Structures are displayed in the same orientation as Smchd1, aligning on subdomain II of the blue and green protomers from smchd1 and T. maritima SMC hinge domains, respectively. (D) Cartoon representation of the Smchd1 HD dimerisation interface between SD I (pale) of one protomer to SDII from the opposing protomer (colouring as in panel (A)). Key residues involved in the interaction are indicated in stick representation with hydrogen bonding and electrostatic interactions indicated with dashed lines. (E) Surface electrostatic representation of the Smchd1 HD indicating the surface exposed positive region in the interdomain linker region. (F) Surface representation of the Smchd1 HD showing positively charged arginine residues involved in binding DNA. These include surface exposed residues from the interdomain linker region (R1789, R1796, R1799) and residues buried in the dimer pore (R1790, R1848, R1869), and R1867, which is substituted to glycine in an FSHD patient kindred. Black and grey arrows indicate residues from opposing protomers in the Smchd1 dimer.
Surface electrostatics of the Smchd1 hinge domain structure identified positive surfaces in the inter-subdomain linker region and in the central pore formed between the dimers (Figure 3E,F). In canonical SMC proteins positively charged residues can play a role in DNA binding [51,55]. This is also the case for SMCHD1, however, only positively charged residues buried in the central pore of the Smchd1 hinge domain were demonstrated to produce an altered phenotype in cellular experiments (Figure 3F) [15]. This includes the FSHD2 substitution arginine 18662 to glycine, which is part of the SMCHD1 GX6GX2GG motif, two amino acids after the first glycine (Figure 3F and Table 1). The other mutation that displayed an altered phenotype was arginine 1848 to alanine. Exactly how the SMCHD1 hinge domain interacts with chromatin and binds to DNA has not been established, but these data support a role for positively charged regions in the inter-subdomain linker and proximal to the central pore; how this may occur is discussed later.
Flanking the SMCHD1 hinge domain are two short coiled-coil regions. In the canonical SMC proteins, these coiled-coils are arranged head-to-tail and are intra-molecular. However, for Smchd1 this is unlikely to be the case and instead a head-to-head intermolecular arrangement is proposed [13,15,24]. Part of the N-terminal α-helix was included in the crystallised Smchd1 hinge domain sequence, but no coiled-coils were observed in the electron density [15]. The positioning of the hinge domain N- and C- termini did not support either coiled-coil arrangement: head-to-tail intra-molecular or head-to-head intermolecular. Negative stain electron microscopy images suggest a head-to-head arrangement of homodimeric full-length Smchd1. The observed particles are long (>40 nm) and thin with globular domains observable at each apex, which are likely to be the ATPase and hinge domain segregated at the N- and C-termini [13]. This suggests a head-to-head arrangement of hinge domain protomers within the Smchd1 homodimer, with intermolecular coiled-coils. This evidence is corroborated by SAXS, which showed an asymmetric SAXS envelope supporting this unusual SMC architecture for the non-canonical Smchd1 [24]. However, whether the N- and C-terminal α-helices can interact is not clear and therefore a complete understanding of how SMCHD1 forms a head-to-head arrangement and the role of these α-helices remains unclear.
Hypothetical models and open questions
UBL domains are generally found in E3 ubiquitin ligases or hold functions within ubiquitylation systems. For example, Parkin is an E3 ligase where its N-terminal UBL domain has been established as auto-inhibitory of its E3 ligase activity [56]. In the chromatin-modifying complex polycomb repressive complex 1 (PRC1), its core component, Bmi1 (PCGF4), which is required for H2AK119 ubiquitylation, also has a predicted ubiquitin-like fold [57]. This is suggested to undergo homo-oligomerisation, contributing to Bmi1 function in ubiquitylation [58]. Other studies show that Bmi1 directly interacts with Ring1B, the E3 ligase of the PRC1 complex, and together they bind the histone surface of nucleosome core particle to correctly orient the E2 ligase UbcH5c for H2A ubiquitylation [59]. Linking these findings to SMCHD1, recent work has reported that Smchd1's localisation to the inactive X chromosome is dependent on the ubiquitylated Histone 2A Lys119 (H2AK119ub) epigenetic mark. Removal of PRC1-mediated H2AK119ub leads to global loss of Smchd1 protein stability and genome-wide changes in gene silencing [60,61]. While a direct interaction between SMCHD1 and H2AK119ub has not been detected experimentally, it is likely that an adaptor protein that is yet to be identified may bridge this interaction. UBL domains are known to mediate a broad range of protein interactions in other systems. In the case of SMCHD1, we hypothesise that the UBL domain plays a role in its localisation to H2AK119ub-marked chromatin via specific protein interactions that are yet to be determined.
The use of human induced pluripotent stem cells has allowed the investigation of several SMCHD1 disease variants from BAMS or FSHD patients. These cells have provided a platform to test the ability of SMCHD1 to interact with the FSHD-related macro-repeat array, D4Z4 [62]. This repeat array houses the gene for the transcription factor DUX4, the driver of FSHD, whose expression is normally silenced by SMCHD1 [11]. One FSHD-associated variant, Q193P located in the SMCHD1 ATPase domain, exhibited enhanced binding to DUX4 [62]. Interestingly, the same mutation was previously shown to abrogate Smchd1's ATPase activity in vitro [49]. This phenomenon was not observed in BAMS-associated variants or controls [49,62]. Additionally, there are many FSHD2-associated variations that result in non-sense mediated decay of SMCHD1 mRNA transcripts [11]. Consequently, these variants show less SMCHD1 associated with the D4Z4 macro-repeat array and also fail to silence the array. Collectively, these data suggest that ATP hydrolysis is required for SMCHD1 to release itself from chromatin (Figure 1D), and an inability to do so results in a failure of SMCHD1-dependent epigenetic silencing at target genes. A similar mechanism in which cohesin releases from DNA relies on its ATPase activity has been previously proposed [37]. This model is based on the observation that mutations that impaired the ATPase activity of one of the cohesin component proteins, Smc1, precluded DNA release. Taken together, this idea raises the possibility that SMCHD1 may require a ‘sweet-spot’ of ATPase activity for optimal function, and too little or too much may be equally detrimental. This phenomenon is highlighted in FSHD- and BAMS-associated mutants where opposing effects on ATPase activity were observed [49].
Although it is established that the SMCHD1 hinge domain is critical for chromatin interactions, it is currently unclear exactly how the positive amino acids proximal to the central pore can play such an important role in these interactions. This region is too distal from the surface to interact directly, inferring that the hinge domain undergoes a large transition from the closed conformation observed in the Smchd1 hinge domain crystal structure to interact with chromatin. Adopting an open hinge domain conformation could expose this region and allow direct interactions with DNA and histone proteins. A similar model has recently been proposed for the human cohesin complex, where the hinge domain heterodimer was captured in an open conformation while bound to single-stranded DNA [54]. It is also unclear whether the α-helices that flank the hinge domain play a role in SMCHD1 chromatin interactions. Whilst experimental evidence reveals a head-to-head and tail-to-tail arrangement for these regions, they could potentially form head-to-tail interactions in response to a stimulus, such as DNA binding.
Almost half the SMCHD1 polypeptide is structurally and functionally uncharacterised, with the linker region between the ATPase and the start of the coiled-coil, N-terminal to the hinge domain, completely unstudied. Sequence analysis predicts a combination of β-sheet and disorder in this region, which is a stark contrast with the canonical SMC proteins which are entirely α-helical in the corresponding region. This linker region appears to be a unique feature of SMCHD1 and may hold clues to how it can convert chromatin interactions and ATP hydrolysis into gene silencing events.
Based on the evidence we have presented here, we propose that SMCHD1 is initially recruited to H2AK119ub-marked chromatin sites [60], and forms interactions with DNA via its hinge domain (Figure 1D). Interaction with DNA may trigger a conformational change in SMCHD1's hinge domain, potentially through the opening of the dimer interface, to expose buried residues (e.g. arginine 18663). This conformational change could open the central linker region between the ATPase and hinge domain to allow DNA to reel through the SMCHD1 dimer. ATP-binding is then proposed to induce ATPase dimerisation, leading to domain-swapping of the UBL domain [14]. ATP hydrolysis can then allow the ATPases to disengage from chromatin and transition back to a monomeric form, resulting in a V-like structure of SMCHD1. This would enable SMCHD1 to then directly interact with promoter and enhancer regions of genes it regulates such as the Pcdh cluster [3]. Whether SMCHD1's presence at these sites promotes chromatin interactions either via a loop extrusion or a chromatin compaction mechanism, as demonstrated for SMC proteins and MORC1, respectively, to silencing gene expression [33,35,36,40] remains of ongoing interest (Figure 1C).
In conclusion, SMCHD1 is an important epigenetic modifier that is involved in silencing genes and is required for normal development. Missense variations in the SMCHD1 gene alter its ability to silence genes and can result in the human diseases BAMS and FSHD, debilitating developmental diseases with no known treatments. Although much progress has been made into understanding SMCHD1 structure and function, many hypotheses remain to be investigated to better understand how this enigmatic protein can result in disease and, importantly, whether it can be exploited as a potential therapeutic target.
Perspectives
Highlight importance of the field: SMCHD1 is an important epigenetic modifier that is involved in silencing genes for normal cellular function. Missense substitutions in the SMCHD1 gene alter its ability to silence genes and can result in the debilitating human developmental diseases, BAMS and FSHD, with no known cures.
A summary of current thinking: SMCHD1 silences genes by initially localising to H2AK119ub-marked chromatin and further interacting with DNA through its SMC hinge domain. This promotes long range chromatin interactions and excludes transcriptional regulators from these regions silencing gene expression in these regions.
A comment on future directions: The exact mechanism by which SMCHD1 exerts its silencing function is yet to be established and the precise functions of large regions of the protein are currently unknown. Future work will establish precisely how the individual domains of SMCHD1 work in concert to regulate gene expression and whether these functions can be modulated as therapies for FSHD and BAMS.
Abbreviations
- BAMS
Bosma arhinia microphthalmia syndrome
- CTCF
CCCTC-binding factor
- FSHD
facioscapulohumeral muscular dystrophy
- FSHD
facioscapulohumeral muscular dystrophy
- PRC1
polycomb repressive complex 1
- SAXS
small-angle X-ray scattering
- SMC
structural maintenance of chromosomes
- UBL
ubiquitin like
Notes
Lower case for Smchd1 indicates mouse isoform in place of upper case (SMCHD1) for the human isoform.
Human numbering, 1867 in the mouse Smchd1 hinge domain sequence and crystal structure.
Human numbering, 1867 in the mouse sequence.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of the Walter and Eliza Hall Institute in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Funding
The authors were supported by an Australian Research Training Program scholarship (A.D.G), the Bellberry-Viertel Senior Medical Research Fellowship (M.E.B), Australian National Health and Medical Research Council fellowships (P.E.C., 1079700; J.M.M, 1105754, 1172929) and grant (to P.E.C., M.E.B. and J.M.M; 1098290). Additional support was provided by the Victorian State Government Operational Infrastructure Support, Australian National Health and Medical Research Council IRIISS grant (9000433).
References
- 1.Jansz N., Chen K., Murphy J.M. and Blewitt M.E. (2017) The epigenetic regulator SMCHD1 in development and disease. Trends Genet. 33, 233–243 10.1016/j.tig.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 2.Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M. et al. (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40, 663–669 10.1038/ng.142 [DOI] [PubMed] [Google Scholar]
- 3.Chen K., Hu J., Moore D.L., Liu R., Kessans S.A., Breslin K. et al. (2015) Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. U.S.A. 112, E3535–E3544 10.1073/pnas.1504232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansz N., Keniry A., Trussart M., Bildsoe H., Beck T., Tonks I.D. et al. (2018) Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 25, 766–777 10.1038/s41594-018-0111-z [DOI] [PubMed] [Google Scholar]
- 5.Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M. et al. (2013) Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 33, 3150–3165 10.1128/MCB.00145-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mould A.W., Pang Z., Pakusch M., Tonks I.D., Stark M., Carrie D. et al. (2013) Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin 6, 19 10.1186/1756-8935-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason A.G., Slieker R.C., Balog J., Lemmers R., Wong C.J., Yao Z. et al. (2017) SMCHD1 regulates a limited set of gene clusters on autosomal chromosomes. Skelet. Muscle 7, 12 10.1186/s13395-017-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gdula M.R., Nesterova T.B., Pintacuda G., Godwin J., Zhan Y., Ozadam H. et al. (2019) The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 10, 30 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C.Y., Jegu T., Chu H.P., Oh H.J. and Lee J.T. (2018) SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174, 406–21.e25 10.1016/j.cell.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon C.T., Xue S., Yigit G., Filali H., Chen K., Rosin N. et al. (2017) De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet. 49, 249–255 10.1038/ng.3765 [DOI] [PubMed] [Google Scholar]
- 11.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 44, 1370–1374 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I. et al. (2017) SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 49, 238–248 10.1038/ng.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brideau N.J., Coker H., Gendrel A.V., Siebert C.A., Bezstarosti K., Demmers J. et al. (2015) Independent mechanisms target SMCHD1 to trimethylated histone H3 lysine 9-modified chromatin and the inactive X chromosome. Mol. Cell. Biol. 35, 4053–4068 10.1128/MCB.00432-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen L.C., Inoue K., Kim S., Perera L. and Shaw N.D. (2019) A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1. Commun. Biol. 2, 255 10.1038/s42003-019-0499-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K., Birkinshaw R.W., Gurzau A.D., Wanigasuriya I., Wang R., Iminitoff M. et al. (2020) Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid–binding residues. Sci. Signal. 13, eaaz5599 10.1126/scisignal.aaz5599 [DOI] [PubMed] [Google Scholar]
- 16.Hassler M., Shaltiel I.A. and Haering C.H. (2018) Towards a unified model of SMC complex function. Curr. Biol. 28, R1266–R1R81 10.1016/j.cub.2018.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeppsson K., Kanno T., Shirahige K. and Sjogren C. (2014) The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15, 601–614 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- 18.Jessberger R. (2002) The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3, 767–778 10.1038/nrm930 [DOI] [PubMed] [Google Scholar]
- 19.Uhlmann F. (2016) SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 10.1038/nrm.2016.30 [DOI] [PubMed] [Google Scholar]
- 20.Chen K., Dobson R.C., Lucet I.S., Young S.N., Pearce F.G., Blewitt M.E. et al. (2016) The epigenetic regulator Smchd1 contains a functional GHKL-type ATPase domain. Biochem. J. 473, 1733–1744 10.1042/BCJ20160189 [DOI] [PubMed] [Google Scholar]
- 21.Hirano T. (2006) At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7, 311–322 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- 22.Gligoris T. and Lowe J. (2016) Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 26, 680–693 10.1016/j.tcb.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasmyth K. and Haering C.H. (2005) The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- 24.Chen K., Czabotar P.E., Blewitt M.E. and Murphy J.M. (2016) The hinge domain of the epigenetic repressor Smchd1 adopts an unconventional homodimeric configuration. Biochem. J. 473, 733–742 10.1042/BJ20151049 [DOI] [PubMed] [Google Scholar]
- 25.Beasley M., Xu H., Warren W. and McKay M. (2002) Conserved disruptions in the predicted coiled-coil domains of eukaryotic SMC complexes: implications for structure and function. Genome Res. 12, 1201–1209 10.1101/gr107302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strunnikov A.V., Larionov V.L. and Koshland D. (1993) SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123, 1635–1648 10.1083/jcb.123.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson D.E., Losada A., Erickson H.P. and Hirano T. (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156, 419–424 10.1083/jcb.200111002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haering C.H., Lowe J., Hochwagen A. and Nasmyth K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 10.1016/S1097-2765(02)00515-4 [DOI] [PubMed] [Google Scholar]
- 29.Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K. and Nasmyth K. (2003) ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 13, 1941–1953 10.1016/j.cub.2003.10.036 [DOI] [PubMed] [Google Scholar]
- 30.Nasmyth K. (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 10.1146/annurev.genet.35.102401.091334 [DOI] [PubMed] [Google Scholar]
- 31.Busslinger G.A., Stocsits R.R., van der Lelij P., Axelsson E., Tedeschi A., Galjart N. et al. (2017) Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson I.F., Goetz D., Zaczek M.P., Molodtsov M.I., Huis In ‘t Veld, P.J, Weissmann, F. et al. (2016) Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 35, 2671–2685 10.15252/embj.201695402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganji M., Shaltiel I.A., Bisht S., Kim E., Kalichava A., Haering C.H. et al. (2018) Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbatsh A.M.O., Kim E., Eeftens J.M., Raaijmakers J.A., van der Weide R.H., Garcia-Nieto A. et al. (2019) Distinct roles for condensin's two ATPase sites in chromosome condensation. Mol. Cell 76, 724–37.e5 10.1016/j.molcel.2019.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G. and Peters J.M. (2019) DNA loop extrusion by human cohesin. Science 366, 1338–1345 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- 36.Kim Y., Shi Z., Zhang H., Finkelstein I.J. and Yu H. (2019) Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruber S. (2017) Shaping chromosomes by DNA capture and release: gating the SMC rings. Curr. Opin. Cell Biol. 46, 87–93 10.1016/j.ceb.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 38.Terakawa T., Bisht S., Eeftens J.M., Dekker C., Haering C.H. and Greene E.C. (2017) The condensin complex is a mechanochemical motor that translocates along DNA. Science 358, 672–676 10.1126/science.aan6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D.Q., Nair S.S. and Kumar R. (2013) The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 8, 685–693 10.4161/epi.24976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H., Yen L., Wongpalee S.P., Kirshner J.A., Mehta N., Xue Y. et al. (2019) The gene-silencing protein MORC-1 topologically entraps DNA and forms multimeric assemblies to cause DNA compaction. Mol. Cell 75, 700–10.e6 10.1016/j.molcel.2019.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta R. and Inouye M. (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25, 24–28 10.1016/S0968-0004(99)01503-0 [DOI] [PubMed] [Google Scholar]
- 42.Richter K., Muschler P., Hainzl O. and Buchner J. (2001) Coordinated ATP hydrolysis by the Hsp90 dimer. J. Biol. Chem. 276, 33689–33696 10.1074/jbc.M103832200 [DOI] [PubMed] [Google Scholar]
- 43.Wendorff T.J., Schmidt B.H., Heslop P., Austin C.A. and Berger J.M. (2012) The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J. Mol. Biol. 424, 109–124 10.1016/j.jmb.2012.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guarne A., Ramon-Maiques S., Wolff E.M., Ghirlando R., Hu X., Miller J.H. et al. (2004) Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 23, 4134–4145 10.1038/sj.emboj.7600412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer L.M., Abhiman S. and Aravind L. (2008) Mutl homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 3, 8 10.1186/1745-6150-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbett K.D. and Berger J.M. (2005) Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure 13, 873–882 10.1016/j.str.2005.03.013 [DOI] [PubMed] [Google Scholar]
- 47.Douse C.H., Bloor S., Liu Y., Shamin M., Tchasovnikarova I.A., Timms R.T. et al. (2018) Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms. Nat. Commun. 9, 651 10.1038/s41467-018-03045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemmers R., van der Stoep N., Vliet P.J.V., Moore S.A., San Leon Granado D., Johnson K. et al. (2019) SMCHD1 mutation spectrum for facioscapulohumeral muscular dystrophy type 2 (FSHD2) and Bosma arhinia microphthalmia syndrome (BAMS) reveals disease-specific localisation of variants in the ATPase domain. J. Med. Genet. 56, 693–700 10.1136/jmedgenet-2019-106168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurzau A.D., Chen K., Xue S., Dai W., Lucet I.S., Ly T.T.N. et al. (2018) FSHD2- and BAMS-associated mutations confer opposing effects on SMCHD1 function. J. Biol. Chem. 293, 9841–9853 10.1074/jbc.RA118.003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soh Y.M., Burmann F., Shin H.C., Oda T., Jin K.S., Toseland C.P. et al. (2015) Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol. Cell 57, 290–303 10.1016/j.molcel.2014.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alt A., Dang H.Q., Wells O.S., Polo L.M., Smith M.A., McGregor G.A. et al. (2017) Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat. Commun. 8, 14011 10.1038/ncomms14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchiyama S., Kawahara K., Hosokawa Y., Fukakusa S., Oki H., Nakamura S. et al. (2015) Structural basis for dimer formation of human condensin structural maintenance of chromosome proteins and its implications for single-stranded DNA recognition. J. Biol. Chem. 290, 29461–29477 10.1074/jbc.M115.670794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamada K., Su'etsugu M., Takada H., Miyata M. and Hirano T. (2017) Overall shapes of the SMC-ScpAB complex are determined by balance between constraint and relaxation of its structural parts. Structure 25, 603–16.e4 10.1016/j.str.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 54.Shi Z., Gao H., Bai X.C. and Yu H. (2020) Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368, 1454–1459 10.1126/science.abb0981 [DOI] [PubMed] [Google Scholar]
- 55.Griese J.J., Witte G. and Hopfner K.P. (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 38, 3454–3465 10.1093/nar/gkq038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trempe J.F., Sauve V., Grenier K., Seirafi M., Tang M.Y., Menade M. et al. (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 10.1126/science.1237908 [DOI] [PubMed] [Google Scholar]
- 57.Gray F., Cho H.J., Shukla S., He S., Harris A., Boytsov B. et al. (2016) BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat. Commun. 7, 13343 10.1038/ncomms13343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Isono K., Endo T.A., Ku M., Yamada D., Suzuki R., Sharif J. et al. (2013) SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 26, 565–577 10.1016/j.devcel.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 59.McGinty R.K., Henrici R.C. and Tan S. (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 10.1038/nature13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jansz N., Nesterova T., Keniry A., Iminitoff M., Hickey P.F., Pintacuda G. et al. (2018) Smchd1 targeting to the inactive X is dependent on the xist-HnrnpK-PRC1 pathway. Cell Rep. 25, 1912–1923.e9 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- 61.Wang C.Y., Colognori D., Sunwoo H., Wang D. and Lee J.T. (2019) PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10, 2950 10.1038/s41467-019-10755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dion C., Roche S., Laberthonniere C., Broucqsault N., Mariot V., Xue S. et al. (2019) SMCHD1 is involved in de novo methylation of the DUX4-encoding D4Z4 macrosatellite. Nucleic Acids Res. 47, 2822–2839 10.1093/nar/gkz005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelley L.A., Mezulis S., Yates C.M., Wass M.N. and Sternberg M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sacconi S., Lemmers R.J., Balog J., van der Vliet P.J., Lahaut P., van Nieuwenhuizen M.P. et al. (2013) The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet. 93, 744–751 10.1016/j.ajhg.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mul K., Lemmers R., Kriek M., van der Vliet P.J., van den Boogaard M.L., Badrising U.A. et al. (2018) FSHD type 2 and Bosma arhinia microphthalmia syndrome: Two faces of the same mutation. Neurology 91, e562–ee70 10.1212/WNL.0000000000005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larsen M., Rost S., El Hajj N., Ferbert A., Deschauer M., Walter M.C. et al. (2015) Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur. J. Hum. Genet. 23, 808–816 10.1038/ejhg.2014.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemmers R.J., Goeman J.J., van der Vliet P.J., van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M. et al. (2015) Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 24, 659–669 10.1093/hmg/ddu486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen K., Puppo F., Roche S., Gaillard M.C., Chaix C., Lagarde A. et al. (2017) Molecular combing reveals complex 4q35 rearrangements in Facioscapulohumeral dystrophy. Hum. Mutat. 38, 1432–1441 10.1002/humu.23304 [DOI] [PubMed] [Google Scholar]
- 69.van den Boogaard M.L., Lemmers R., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S. et al. (2016) Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet. 98, 1020–1029 10.1016/j.ajhg.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]