SUMMARY
Clear cell renal cell carcinoma (ccRCC) is the most common kidney cancer and has very few mutations that are shared between different patients. To better understand the intratumoral genetics underlying mutations of ccRCC, we carried out single-cell exome sequencing on a ccRCC tumor and its adjacent kidney tissue. Our data indicate that this tumor was unlikely to have resulted from mutations in VHL and PBRM1. Quantitative population genetic analysis indicates that the tumor did not contain any significant clonal subpopulations and also showed that mutations that had different allele frequencies within the population also had different mutation spectrums. Analyses of these data allowed us to delineate a detailed intratumoral genetic landscape at a single-cell level. Our pilot study demonstrates that ccRCC may be more genetically complex than previously thought and provides information that can lead to new ways to investigate individual tumors, with the aim of developing more effective cellular targeted therapies.
INTRODUCTION
Renal cell carcinoma accounts for about 209,000 new cancer cases and 102,000 deaths worldwide per year (Rini et al., 2009), of which ~80% are clear cell renal cell carcinoma (ccRCC) (Ng et al., 2008). Previous studies have shown that ccRCC is a genetically distinct adult carcinoma with a relatively low mutation rate (Greenman et al., 2007). Sequencing analysis of such tumors has revealed that there are few common mutations shared between different ccRCC patients, including VHL and PBRM1 (Dalgliesh et al., 2010; Varela et al., 2011). However, the intratumoral heterogeneity of ccRCC remains unknown, and quantification of the heterogeneity remains a difficult task especially in those tumors without mutations in VHL and PBRM1.
A major difficulty in determining causative mutations in many cancers relates to the fact that mutational analyses are carried out on DNA from tumor tissues obtained through surgery, and such samples may thus include adjacent noncancerous cells as well as a mixture of cancer cells that may be at different mutational stages, given that there is an accumulation of mutations during cancer progression. Thus, even with current sequencing technology, exploring cancer biology using DNA from mixed cancer tissue DNA (Baudot et al., 2009) can be extremely difficult.
One way to circumvent the problem of mixed cell types in a tumor sample would be to carry out sequencing on single cells isolated from a tumor. Recently, Navin et al. developed a method called single-nucleus sequencing, whereby they isolated single-cell nuclei from breast cancer tissue and performed low-coverage sequencing to investigate DNA copy number variation between the isolated cells (Navin et al., 2011). Their results indicated that single-cell sequencing is a promising way to infer specific intratumoral genetic changes; however, it was not possible to use this method to assess specific single nucleotide changes.
To quantitatively investigate the intratumoral heterogeneity of ccRCC, we selected a 59-year-old Chinese male with ccRCC to carry out exome sequencing of 25 single cells from the tumor and adjacent noncancer tissue. Exome sequencing of the tumor and the matched normal tissue showed that this tumor was unlikely to be related to mutations in VHL and PBRM1, indicating that recurrent mutations identified in a patient population may not be relevant with regard to a single patient or tumor. This emphasizes the importance of assessing and diagnosing cancers and patients at an individual level to determine the most efficacious treatment. Our single-cell exome sequencing data allowed us to carry out a population genetic analysis on the tumor. We did not observe any significant clonal subpopulations within this tumor. We also found that most of the somatic mutations occurred only in a small fraction of the cells and that mutations with different allele frequencies showed very different mutation spectrums. This may be the result of selection during tumor progression. We also compared the results from the single-cell sequencing with a screen for mutations in a cohort of 98 ccRCC patients (Guo et al., 2012), enabling the identification of potential key genes, including AHNAK and SRGAP3, in the establishment of this tumor. Our data provide the first intratumoral genetic landscape at a single-cell level and reveal genetic characteristics of this tumor that indicate that ccRCC may be more genetically complex than previously thought.
RESULTS
Exome Sequencing Indicated that Tumor Development Was Unrelated to the Presence of VHL and PBRM1 Mutations
We obtained a sample of the tumor present in the left kidney, classified as stage IV according to the 2002 AJCC TNM classification of renal cell carcinoma (Figure S1 available online), and a sample of adjacent kidney tissue. We first carried out whole-exome sequencing of the cancer and normal adjacent kidney tissue of this patient. More than 95% of the target regions in cancer tissue sequencing were covered sufficiently for confident variant calling (defined as ≥ 20×) (Table 1).
Table 1.
Exome Sequencing Coverage of Cancer and Normal Control Tissue
| Sample ID | Human All Exons Coverage (%) | VHL Exons Coverage (%) | PBRM1 Exons Coverage (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ≥1× | ≥10× | ≥20× | ≥1× | ≥10× | ≥20× | ≥1× | ≥10× | ≥20× | |
| RC-T | 98.94 | 96.79 | 95.01 | 100 | 89.80 | 80.88 | 100 | 100 | 100 |
| RN-T | 97.05 | 80.72 | 65.70 | 100 | 66.67 | 64.25 | 100 | 99.65 | 89.76 |
“RC-T” and “RN-T” are cancer tissue and normal control tissue, respectively.
Given the commonly identified renal cancer mutations in VHL and PBRM1, we first checked the sequence quality of the whole exons of VHL and PBRM1. The capture data covered 100% of the exons of these genes, with > 80% of the gene exon regions covered in the cancer tissue sequencing, which provided sufficient coverage for confident variant calling (defined as ≥ 20×) (Table 1) and allowed us to confidently detect somatic mutations in the coding regions of VHL and PBRM1. We found no mutation in the coding region of VHL and three somatic mutations in PBRM1 of extremely low frequency (average mutant allele frequency of < 4%). We validated these findings using PCR and Sanger sequencing (Table S1).
Both VHL and PBRM1 are located on chromosome 3p, which has previously been reported to be a variation hot spot (Beroukhim et al., 2009; Pei et al., 2010). Given this, we further excluded the involvement of these two genes in the ccRCC of this patient by looking for loss of heterozygosity (LOH) across the whole exome. We found no signals of LOH on chromosome 3 or in other reported LOH or deletion hot spots such as chromosomes 1, 6, and 14 (Chen et al., 2009; Pei et al., 2010) (Figure S2).
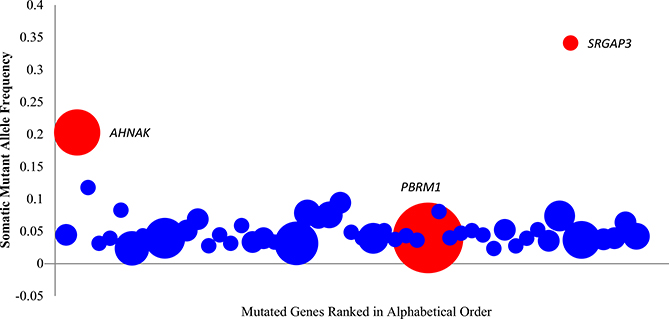
We then examined the sequence reads from this patient that contained regions with known variants observed in a ccRCC cohort of 98 patients (Guo et al., 2012). Here, we found that PBRM1, which has been reported to be recurrent in more than 20% of patients, was present at an extremely low mutant allele frequency (based on the mutant read number ratio in tissue sequencing) in this patient. Conversely, there were mutant alleles with a high frequency in several other genes (e.g., AHNAK and SRGAP3) that have been shown to be infrequently mutated in ccRCC at the population level (Guo et al., 2012) (Figure 1).
Figure 1. Somatic Mutant Allele Frequency from the Individual Patient that Contained Known Variants in the Large ccRCC Cohort of 98 Patients in Guo et al. (2012).
Mutant sites selected in cancer tissue here are covered by more than ten normal reads without any mutations in the normal tissue. The vertical axis showed the average somatic mutant allele frequency in a gene. The horizontal axis showed that mutant sites contained genes, which also present in the 98 patient cohort ranked by alphabet. The area of the circle represents the observed mutation frequency in the 98 patient cohort of each gene. AHNAK, SRGAP3, and PBRM1 were highlighted in red. For further description of the genetic analysis of cancer and normal tissue sequencing data, see also Table 1, Figure S1, and Figure S2.
These analyses showed that the cancer in this patient was unlikely to be related to the presence of VHL and PBRM1 mutations. We therefore went on to perform further genetic analyses on this VHL/PBRM1-negative ccRCC patient to investigate the other genetic mechanisms that may underlie this type of ccRCC.
Single-Cell Exome Sequencing and Population Somatic Mutation Calling and Validation
To obtain the most detailed cellular genetic information on this tumor, we carried out single-cell exome sequencing (Hou et al., 2012, in this issue of Cell). In total, we sequenced 20 single-cell exomes from the tumor and 5 single-cell exomes from the adjacent normal tissue. On average, more than 80% of the target regions were covered sufficiently for confident population variant calling (defined as ≥ 5×) (Li et al., 2010; Yi et al., 2010) (Table S2). We then performed single-cell analysis using a modified bioinformatics pipeline, as shown in Figure S3 (Hou et al., 2012).
After determining the false positive and false negative rates as previously described (Hou et al., same issue) (Figures S4 and S5), we identified 260 somatic mutation sites in the coding region between the cancer and normal population (average ~78.9 mutations per single cancer cell) and only 12 somatic mutations within the normal control population (average ~20.4 mutations per single normal cell), indicating that our somatic mutation calling in the cancer cells was not due to amplification errors (kappa square test, p = 1.4 × 105). Of the 260 somatic mutant alleles, 93.64% were covered by at least 10 reads, which indicated that the called heterozygous somatic mutations are of sufficient confidence (Hou et al., 2012). Comparison of the somatic mutation frequency of the 260 somatic mutations from the single-cell sequence data with that of the somatic mutation frequency in our whole-cancer tissue sample showed a high correlation (r2 ≈ 0.8) (Figure S6). We further validated our somatic mutation calling accuracy using PCR sequencing by randomly selecting 35 somatic mutation sites from three different cells. We were able to amplify a total of 85 sites, and of these, 82 of the somatic mutations (96.47%) were confirmed by PCR-based capillary sequencing (Tables S3A–S3C). This further indicates the high accuracy of our mutation calling and that it was suitable for use in further analyses.
Population Analysis Indicates that Three of the Individual Tumor Cells Were Normal Cells
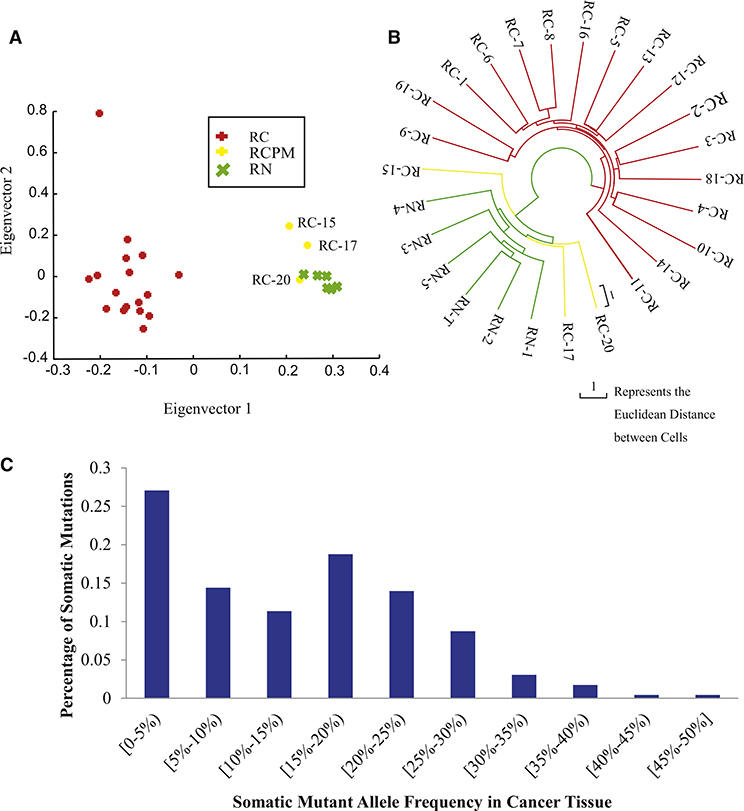
Because surgically removed tumors can contain both normal and cancer cells, we next determined whether the single cells that we isolated from the tumor sample were indeed tumor cells. We performed principle component analysis (PCA) mutation profiling on the 260 somatic mutation sites and found that cells RC15, RC17, and RC20 clustered tightly with the adjacent noncancer tissue (Figure 2A), indicating that these are normal cells rather than cancer cells and that these were not included in further analyses of tumor heterogeneity.
Figure 2. Somatic Mutation and Single-Cell Population Analysis in This ccRCC Patient.
(A) Principle component analysis (PCA) of cancer cells (RC, red), normal control cells (RN, green), and normal cells picked as cancer cells (RCPM, yellow) based on principle component analysis (PCA).
(B) Neighbor joining phylogenetic tree constructed using sites of somatic mutation data by Euclidean distance; cancer cells (RC, red), normal control cells (RN, green), and normal cells picked as cancer cells (RCPM, yellow) are presented here. RN-T here represents the normal tissue DNA as control. After filtering the three normal cells picked as cancer cells, we identified 229 somatic mutations (see Table S3 for details).
(C) Mutant allele frequency spectrum somatic mutations in 17 cancer cells. Based on Fisher’s exact test, we separated the mutations into common mutations and rare mutations (see Table S3 for details).
For single-cell data evaluation and details of somatic mutation calling, see also Figures S3, S4, S5, and S6 and Tables S2, S3, S4.
The ccRCC Tumor Has No Apparent Cell Subpopulations
Using the above PCA analysis to compare mutation patterns between the individual cells, we observed that the normal cells clustered tightly, whereas the cancer cells were more diverse and showed no obvious cell subpopulations. To assess the timescale between the appearance of somatic mutations in the individual cells relative to one other, we carried out a phylogenetic analysis. We built a tree based on the modified neighbor joining (NJ) method (Saitou and Nei, 1987) using the 260 identified somatic mutation sites, with the normal cells serving as the common node at the root of the tree. The cancer cells were significantly separated from each other after emerging from their common node. These data were consistent with those of the PCA profiling, as it again indicates that there were no subpopulations of cancer cells within the cancer tissue (Figure 2B). The branch that separated the cancer cells and the normal cells was very short in comparison to the branches separating the cancer cells, suggesting that the time for generating genetic changes that result in normal cells progressing to cancer cells is very short. However, the diversity that we observe between the individual cancer cells is large, which most likely reflects the accumulation of passenger mutations that were closely linked to changes specifically related to cancer progression.
The phylogenetic tree analysis also confirmed the PCA data with RC15, RC17, and RC20 cells, which clustered tightly with adjacent noncancerous tissue, indicating that they are more likely to be normal cells rather than cancer cells; we therefore removed these from the cancer cell data set prior to further analyses. Removal of these cells resulted in a final number of 229 somatic mutation sites between the cancer cell population and the normal controls (Table S4).
Quantification Analysis of Intratumoral Heterogeneity
To investigate the intratumoral mutation landscape, we first evaluated the intratumoral somatic mutation frequency. To reduce the influence of missing data of individual single cells, we defined somatic mutation frequency of the 229 sites using the mutant reads ratio in cancer tissue sequencing data. The frequencies showed two distinct peaks, with one in a frequency range of 0%–5% (low frequency), indicating that the cancer tissue contained many rare mutations that were only present in a few of the cancer cells. The other peak was at a frequency range of 15%–20%, indicating that there were no dominant clones in the cancer tissue and that there was significant intratumoral heterogeneity of this cancer tissue (Figure 2C and Table S4). According to this allele frequency spectrum, we defined a commonly shared (tissue common) mutation site as one with more than 20% mutant allele frequency in tumor tissue (p = 0.000009, kappa square test). The rest of mutation sites that were not commonly shared (cell specific) were designated as rare mutation sites. Strikingly, more than 70% of the mutations were cell-specific sites, and less than 30% were tissue common sites (Table S4). This result is consistent with general observations from sequencing of cancer tissues that one can identify more mutations at greater sequencing depth. This therefore indicated that, if sequencing is carried out at a depth lower than 50-fold, detection of rare mutations would be difficult.
Mutations of Different Frequencies Have Different Mutation Mechanisms
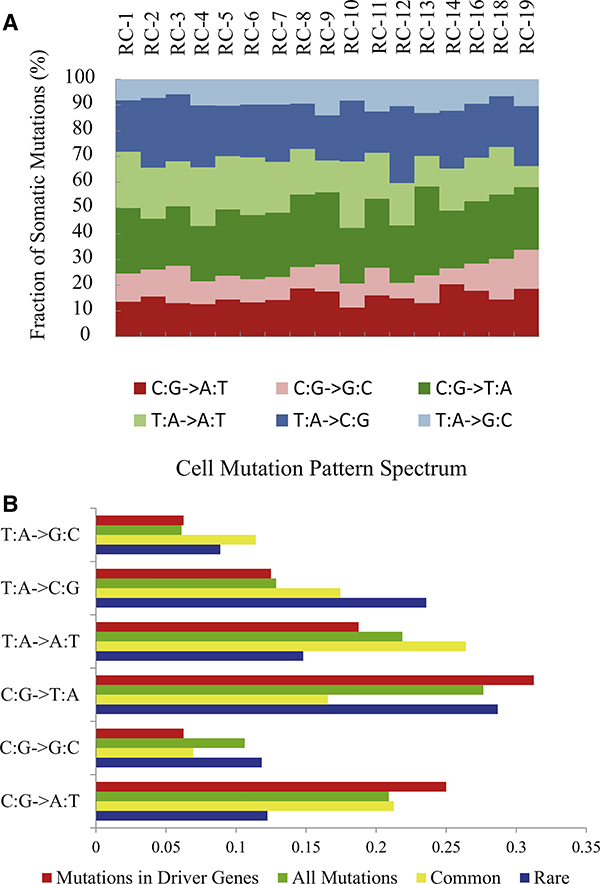
Our data also allowed us to investigate the specific types of mutation mechanisms that occurred within the ccRCC tumor. Analysis of all of our somatic mutations showed a preference for C·G → T·A, which is similar to the previously reported mutation mechanism pattern seen in ccRCC (Dalgliesh et al., 2010) and other cancers (Greenman et al., 2007) (Figure 3A). Strikingly, however, when we assessed rare mutations and the common mutations separately, we found that the type of mutation mechanism was different. Here, we found that the patterns of the rare mutation sites were primarily as those seen above, whereas the common mutation sites showed an increasing percentage of transversion mutations (Figure 3B). This confirms the mutation spectrum seen in a renal cancer patient population exome analysis (Guo et al., 2012), in which we also found an increasing percentage of transversion mutations (Figure 3B). We suggest that the presence of such a mutation pattern could potentially be used as a metric of ccRCC cancer progression.
Figure 3. Somatic Mutation Pattern Spectrums.
(A) Somatic mutation pattern spectrum of individual ccRCC cells.
(B) Somatic mutation pattern spectrum of rare mutations (blue) and common mutations (yellow) compared with spectrum of driver mutations (red) and all nonsynonymous mutations (green) in the 98 patient cohort.
See also Table S4.
Intratumoral Genetic Landscape at the Single-Cell Level
We identified 120 somatic mutations in the coding regions and assessed their potential functional impact by looking at differences between nonsynonymous (NS) and synonymous (S) mutations in all of the cells. The NS/S ratio was 4.0, which was relatively higher than that reported in previous reports of ccRCC and other tumor types (Ding et al., 2010; Lee et al., 2010; Ley et al., 2008; Pleasance et al., 2010a, 2010b; Varela et al., 2011). This may be due to single-cell sequencing having a higher sensitivity for identifying somatic mutations.
Given that we carried out single-cell exome sequencing, we were not only able to observe the genes with mutations, but could also assess the frequency at which a mutant allele was present among the different cells. This can provide greater information on the relative impact of each change on tumorigenesis, beyond that possible with whole-tumor genome sequencing.
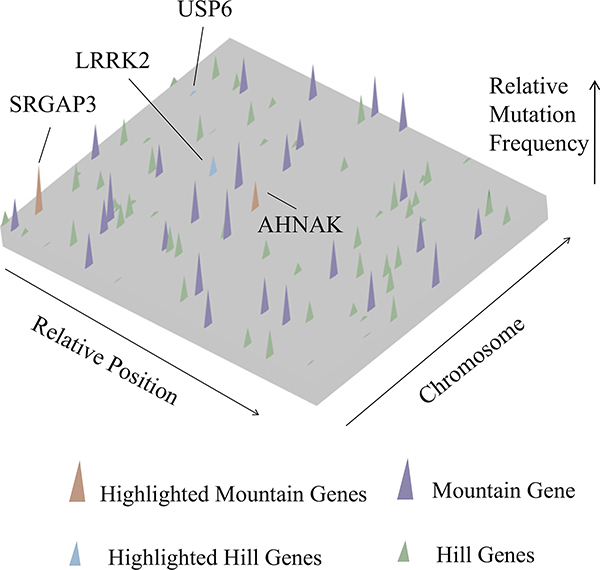
To investigate the frequency of nonsynonymous somatic mutations, we carried out an analysis that graphically shows the intratumoral mutational landscape of the single ccRCC patient, representing the mutational heterogeneity within a single cancer tissue. Our landscape display shows a small number of mutant genes that are present in a large fraction of individual cells (which we term “mountains”) and a significantly greater number of genes mutant in only one or a few cells (“hills”). We defined a gene as a “mountain” if the gene contains at least one nonsynonymous common mutation site and defined the genes of the rest as “hill.” We detected 28 mountain genes (Table S5, in purple or red) and 66 hill genes (Table S5, in blue or green). Our analysis showed that the “mountains” and “hills” were evenly distributed in the patient genome and had no significant bias for any chromosome (Figure 4).
Figure 4. Intratumoral Gene Mutation Landscape of an Individual ccRCC Patient.
Nonsynonymous somatic mutations are plotted in two-dimensional space, which represents chromosomal positions of mutant genes. Chromosomal positions and mutant genes were ranked as indicated by arrows beside the plane. Each mutant gene is scaled by a relative position (0–1) on its chromosome; thus, its position on different chromosomes was normalized by its total length. Higher peaks (purple) —peak heights assigned a value of mutant reads ratio—indicate the 28 identified mountain genes. The shorter peaks (green), with peak heights assigned a value of mutant reads ratio, show the 66 identified hill genes. Genes recurrently mutated in the large patient cohort are marked in red (mountain) and blue (hill). See also Table S5 for details.
Mutated Genes and Their Potential Roles in This ccRCC Tumor
Five (two mountain and three hill genes [SRGAP3, NIPBL, UBE4A, USP6, and SH3GL1]) of the 94 identified genes were present in either the Cancer Gene Census (http://www.sanger.ac.uk/genetics/CGP/Census/) data set or have been reported as ccRCC mutations in previous work (Dalgliesh et al., 2010; Varela et al., 2011). Additionally, amino acid analysis and SIFT (Kumar et al., 2009) predictions indicate that four of these five genes (SRGAP3, NIPBL, UBE4A, and SH3GL1) contain a truncating or likely functionally damaging mutation.
Of note, four of the 94 identified genes (two mountain genes [AHNAK and SRGAP3] and two hill genes [LRRK2 and USP6]) were also present in a cohort of 99 ccRCC patients (this included the 98 ccRCC patients [Guo et al., 2012] and the individual patient reported here; see Table 2 and Figure 5A for details). Genes that are recurrently mutated (mutated in more than 2% patients) in this cohort are particularly attractive as potential driving factors for cancer initiation and development in this patient.
Table 2.
Key Genes Identified in This Patient
| Gene Name | Mutations | Patient Prevalence (%)a | P Valueb (Passenger Probability) | Mutant Allele Frequency in Cancer Tissue | Mutant Cell Number | Mountain/Hilla |
|---|---|---|---|---|---|---|
| AHNAK | g.chr11:62042132G > A; p.P5445 > S | 5% | 9.29 × 10−9 | 20% | 12 | M |
| LRRK2 | g.chr12:38985956A > G; p.I1294 > V | 4% | 4.28 × 10−4 | 8% | 8 | H |
| SRGAP3 | g.chr3:9041948T > A; p.R535a | 2% | 2.92 × 10−1 | 34% | 16 | M |
| USP6 | g.chr17:4976948C > G; p.T72 > R | 2% | 3.26 × 10−1 | 1.99% | 3 | H |
Patient prevalence means the mutant genes recurred in the 99 ccRCC patients (including this patient); M/H represents mountain or hill gene.
Significance of the observed mutation rate over the expected mutation rate in Guo et al. (2012).
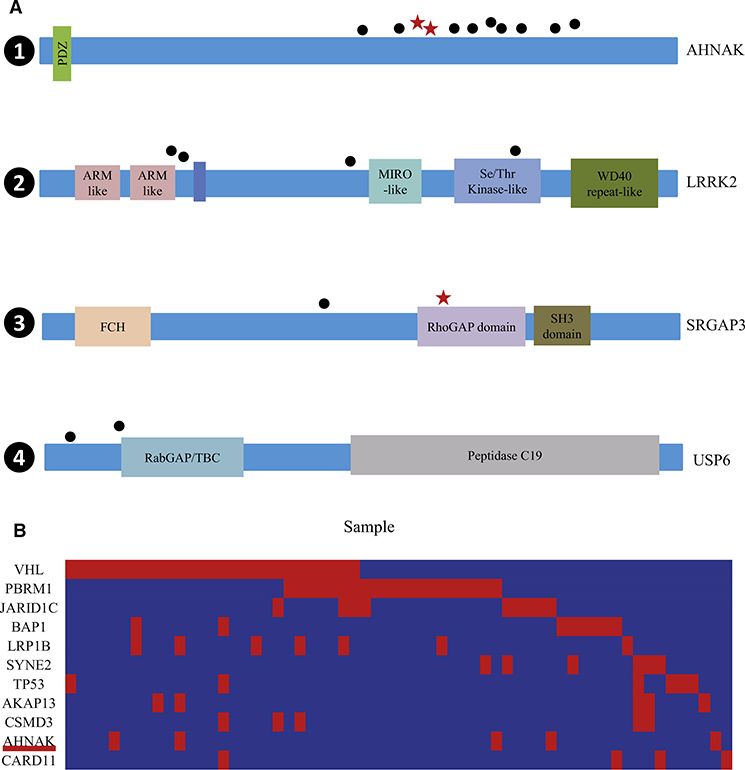
Figure 5. Analysis of Genes of This Patient that Were Recurrently Mutated in the ccRCC Patient Cohort.
Analysis includes 98 patients from Guo et al. (2012) and the patient of this single-cell exome sequencing study.
(A) Somatic mutations in AHNAK, LRRK2, SRGAP3, and USP6. The types and relative positions of confirmed somatic mutations are shown in the transcripts of AHNAK (1), LRRK2 (2), SRGAP3 (3), and USP6 (4) using the following symbols: red stars, nonsense mutations; bullets, missense mutations. Domains and motifs in each encoded protein product are indicated. ARM-like, Armadillo-like helical; MIRO-like, mitochondrial Rho-like; Se/Thr Kinase-like domain, serine/threonine protein kinase-like domain; WD40 repeat-like, WD40/YVTN repeat-like-containing domain; FCH, Fps/Fes/Fer/CIP4 homology; RhoGAP domain, Rho GTPase-activating protein domain; SH3 domain, Src homology 3 domain; RabGAP/TBC, Rab-GAP/TBC domain; Peptidase C19, ubiquitin carboxyl-terminal hydrolase 2.
See also Table 2 for details.
(B) Concurrent and mutually exclusive mutations observed in the genes known to be frequently mutated in this cohort. For each gene (row) indicated, tumors (columns) with or without mutations are labeled in red or blue, respectively. We selected only genes harboring nonsilent mutations in at least five subjects for this analysis.
One of the mountain genes of particular interest was AHNAK, which recurred in 5 of 99 (~5%) of the ccRCC patients (Figure 5A). AHNAK is expressed as a 17.5 kilobase mRNA in several cellular lineages but is typically repressed in cell lines that are derived from human neuroblastomas and from several other types of tumors (Shtivelman et al., 1992). The Ahnak protein activates protein kinase C (PKC) through dissociation of the PKC-protein phosphatase 2A complex (Lee et al., 2008). The Ahnak protein also has been reported to be involved in a downstream signaling pathway that regulates the expression of genes that transform renal fibroblasts into more active myofibroblasts as characterized by enhanced proliferation and contractility (Zhang et al., 2009). In addition, evidence has indicated that Ahnak serves as a lysine acetylation target that is involved in chromatin remodeling (Choudhary et al., 2009; Deribe et al., 2009). We note that AHNAK also showed interaction (score: 0.027; see Experimental Procedures for details) with the HIF1A gene, which plays a critical role in transcriptional gene activation involved in ccRCC angiogenesis (Maxwell, 2005; Poon et al., 2009), by the protein-protein prediction method.
In this regard, we further investigated the potential role of AHNAK in this ccRCC patient by looking for correlations among mutations in the known frequently altered genes (genes harboring at least five nonsilent mutations) in the large ccRCC patient cohort (Guo et al., 2012) with these biological findings in mind. Of note, we observed positive correlations between AHNAK and genes that have been shown to be involved in lysine acetylation/histone deacetylases or chromatin remodeling (p < 0.01; including VHL [Geng et al., 2011; Qian et al., 2006], PBRM1 [Bourachot et al., 2003; Hargreaves and Crabtree, 2011], and JARID1C [Jensen et al., 2005]) (Figure 5B). This indicated that mutations in AHNAK may alter the lysine acetylation/histone deacetylases or chromatin remodeling-related pathways and may further lead to abnormal EGFR pathways that finally make the kidney cells proliferate malignantly.
To gain insight on the potential biological functional characteristics of the potential cancer genes that we identified in this tumor— genes that had not previously been identified as mutated in ccRCC or other cancers—we carried out a gene ontology (GO) (Ashburner et al., 2000) analysis. We found that these genes were enriched in the categories of cell-cycle regulation, cell or genome structure maintenance, and vascular development, which are pathways commonly altered during tumorigenesis for cancer initiation and development of metastasis (Table S5).
In addition to the genes noted above, there were other hill genes that had been previously reported to have a functional correlation with cancer development (Table S5). Interestingly, we also found mutations in some drug response-related and prognostic-related genes (Table S5). For example, PABPC1 has been identified as a prognostic indicator in some cancers (Takashima et al., 2006), and RPL8 expression levels have been correlated with response to chemotherapy (Salas et al., 2009).
DISCUSSION
We present a novel genetic characterization of a VHL/PBRM1-negative ccRCC tumor by single-cell exome sequencing. Population analysis of identified somatic mutations allowed us to distinguish cancer from normal cells. Both our principle component analysis (PCA) and phylogenetic analysis demonstrated that no subpopulations could be observed in this tumor. Quantification analysis of tumor heterogeneity enabled the identification of common and rare mutations and their unique characteristics. To the best of our knowledge, the cell mutation frequency and the corresponding “mountain” and “hill” genes provide the first detailed intratumoral genetic landscape at a single-cell level.
Of interest, the mountain gene AHNAK, which is involved in chromatin-remodeling processes, was predicted to interact with HIF1A in this VHL-negative patient. In addition, the hill gene USP6 is a ubiquitin-mediated proteolysis pathway (UMPP) gene that is able to initiate tumorigenesis by inducing the production of matrix metalloproteinases following NF-κB activation (Ye et al., 2010). These recurrent chromatin remodeling-related and UMPP-related genes identified here confirm that genes in these two important biological processes that are frequently mutated in a large patient cohort (Dalgliesh et al., 2010; Guo et al., 2012; Varela et al., 2011) may potentially drive cancer progression, as they are seen in an individual patient lacking mutations in the known drivers of kidney cancer.
The mutations in hill genes (like USP6 [Figure 5A] and LRRK2) that are present in only a small number of cells appear to play roles in cellular modification, including ubiquitination processes and GTPase activation, suggesting that the hill genes may initiate a variety of processes that promote progression once the cells have undergone mutations that initiate cancer formation.
Among the mutated genes in this ccRCC patient, TUBB is potentially interesting, as it plays a role in structure maintenance (Hall et al., 1983); the truncating somatic mutation that we identified in six cancer cells could result in abnormal microtubule development and could conceivably contribute to the instability of cancer cells. The CCKBR gene, mutated in 12 cells, may also be of interest for future study, as it encodes a G protein-coupled receptor for gastrin and cholecystokinin (CCK) (Pisegna et al., 1992). These regulatory peptides of the brain and gastrointestinal tract had a miss-spliced transcript variant, including an intron observed in cells from colorectal and pancreatic tumors (Caplin et al., 2000; Yu et al., 2006). Thus, alterations in CCKBR function could have an impact on cell proliferation processes. A final gene worth noting is the SULT1A1 gene, which mediates metabolic activation of carcinogenic N-hydroxyarylamines to DNA-binding products and serves as a modulating factor in cancer risk (Liang et al., 2004; Tang et al., 2003; Zheng et al., 2003).
In this article, we present a pilot study and analysis of how biological insights may be derived from single-cell exome sequencing of individual solid tumors. Single-cell exome sequencing can provide detailed information on individual tumor development and on its specific cell lineage origin. Though gene discovery is an essential part of understanding cancer biology, an important area for cancer research is understanding the development of drug resistance and tumor relapse, which may best be investigated on an individual basis (Audenet et al., 2011). Single-cell sequencing analysis of individual tumors may provide a good way forward for such studies, especially for genetically complex tumors. Both clonal evolution and cancer stem cell models suggest that drug resistance and tumor relapse are the result of intratumoral heterogeneity and subpopulation diversity of cancer cells (Marusyk and Polyak, 2010; Visvader, 2011). In leukemia, data have shown that intratumoral clonal diversity and architecture are important for diagnosis, relapse, and outcome. This emphasizes the importance of targeted therapy based on knowledge of the genetic and functional makeup of intratumoral clones (Dalgliesh et al., 2010; Varela et al., 2011). Our single-cell analysis of the mutant genes and their frequency in the ccRCC tumor of this patient underscore the potential capability of the single-cell exome sequencing for comprehensive analyses of drug resistance and tumor relapse at an individual level. Future work using similar single-cell sequence analyses on numerous individual tumors may also aid in unraveling and revealing differences and commonalities in tumor development in a cross-section of patients, which could add another level to cancer diagnosis and more effective targeted therapy.
EXPERIMENTAL PROCEDURES
Case Report
The patient in our study was a 59-year-old Chinese male with clear cell renal cell carcinoma on left kidney classified as stage IV (T4N0M0) according to the 2002 AJCC TNM classification of renal cell carcinoma. A signed written consent was obtained before recruitment for the study, according to the regulations of the institutional ethics review boards. Fresh samples were obtained from this patient in Peking University Shenzhen Hospital on March 17, 2010.
Collection of Single Cells and Cell Lysis
Single cells from this fresh carcinoma were isolated immediately under inverted microscope (Nikon Instruments Co., Ltd.) by a mouth-controlled pipetting system. Then, each cell was transferred into a precooled PCR tube containing cell lysis solution on ice. After that, sample in each tube was incubated in a thermo cycler for 10 min at 65°C. A physiological saline blank was done parallel as a negative control.
Multiple Displacement Amplification and Storage
Whole-genome amplification (WGA) was achieved on these samples using REPLI-g Mini Kit according to the manufacturer’s manual (QIAGEN GmbH). A reaction of a total volume of 50 μl was performed at 30°C for 16 hr and then terminated at 65°C for 10 min. Amplified DNA products were stored at −20°C.
Concentration Measurement and Amplification Coverage Estimation
The concentration of MDA products was measured using the Qubit Quantization Platform (Invitrogen Life Science). Ten housekeeping genes located on different chromosomes were selected for PCR check of coverage of amplified products. The MDA products amplified successfully by at least eight housekeeping genes were selected for further procedures.
Library Preparation and Sequencing
Exome capture was performed following the procedure of Agilent SureSelect Platform. The libraries were prepared following the protocol of Illumina library preparation procedures. The sequencing processes were performed on Illumina Hiseq 2000 platform.
Public Data Used
The human (Homo sapiens) reference genome sequence (Hg18) and its annotation files were downloaded from UCSC Genome Bioinformatics (http://genome.ucsc.edu/). The target region files of exome capture were downloaded from Agilent website (http://www.genomics.agilent.com).
Reads Mapping
Linker and adaptor sequences were masked before mapping. Short read pairs were mapped to the NCBI Build 36 Human Reference Genome using SOAPaligner version 2.20 (http://soap.genomics.org.cn/soapaligner.html) with a maximum of three mismatches, nongap mapping model, and seed length 32. The insert size distribution of each library was checked by Eland contained in the Solexa Pipeline, and the parameter of insert size range was set according to the Eland survey results. Reads that could only be mapped to a unique exome capture target region were selected for consensus sequence identification.
Consensus Sequence Calling
In each cell, consensus sequence was called using SOAPSNP version 1.03 using Hg18 as reference (http://soap.genomics.org.cn/soapsnp.html). To confirm the best parameters of calling SNPs, the relationship between false positive rate (FPR) and sequencing quality and depth was evaluated. First, sites of quality of 99 and with depth between 25× and 40× were selected in normal control mix DNA consensus sequence as control. We then looked at the distribution of the discord rate between these sites and those in normal control cells. Thus, the FPR represents the inconsistent rate of these sites between normal single cells and the normal mixed tissue. As shown in Figures S4 and S5, at sequencing depths greater than 6×, the FPR is sharply reduced, and at consensus sequence quality scores greater than 20, the FPR is also sharply reduced. The consensus sequence of each cell was then filtered by the following criteria: (1) quality value should be greater than 20; (2) sequencing depth should be greater than 6; and (3) p value of the rank-sum test should be greater than 0.05 (Li et al., 2009). Allele type, which was not satisfied with these criteria, was marked as missing (“–“). Finally, all of these consensus sequences were grouped into population genotype (Lam et al., 2010; Patterson et al., 2006; Xia et al., 2009; Yi et al., 2010).
Sites of Somatic Mutation Calling and Filtering
To train the best parameter for calling somatic mutations, we further evaluated the false positive (FP) and false negative (FN) rates. For the FP evaluation, we calculated the average FP rate based on the SNPs that we called. The average FP rate is 2.67 × 10−5, which is consistent with previous reports (Ling et al., 2009; Pugh et al., 2008; Spits et al., 2006). For the FN evaluation (especially for the allele dropout, ADO), we compared the heterozygous rate between each normal single cell and the normal mixed control. The average FN here was 16.43%. Using a Binomial distribution model (considering FP as input parameter), we determined that the presence of three or more cells having a specific mutation in the cancer cell population provided sufficient confidence to call a somatic mutation site as present in the cancer cell population. To further avoid false positives, we also removed somatic mutation sites in situations in which the corresponding information in the normal mixed control was at a sequencing depth of less than 10× or if the second best base was covered by mutant reads. In total, we identified 260 sites of somatic mutation in our whole data set.
Principle Component Analysis of Cells
Principle component analysis (PCA) was performed as in the previous population genetic study (Lam et al., 2010; Xia et al., 2009) based on the sites of somatic mutation that we called in 20 cancer cells. The eigenvector decomposition of the transformed genotype data was performed using the R function “eigen,” and the significance of the eigenvectors was determined with a Tracey-Widom test implemented in the program “twstats” provided with the EIGENSOFT software (Patterson et al., 2006).
Construction of Phylogenetic Tree of Individual Tumor Cells
All sites of somatic mutation were used to calculate the genetic distances between each single-cell sample. We constructed the tree of cell by the neighbor joining (NJ) method (Saitou and Nei, 1987) based on Euclidean distance of the difference in each cell. The weight was 0 when the genotype of this cell was the same as the normal control; the weight was 0.5 when the genotype of this site was missing; and the weight was 1 when the genotype is different with the normal control. Then, the NJ tree was constructed by software PHYLIP (http://evolution.genetics.washington.edu/phylip.html).
Concurrence and Mutual Exclusion Analysis
We performed the concurrence and mutual exclusion analysis on the significantly mutated genes that showed nonsilent mutations in at least five tumor cells by permutation test, as previously described (Sathirapongsasuti et al., 2011), with minor modifications. We separated the significantly mutated genes into different biological groups and calculated the possibility of concurrence and mutual exclusion possibility between AHNAK and different biological groups according to the method described in the reference.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a National Basic Research Program of China (973 program numbers 2011CB809202 and 2011CB809203), the Chinese 863 program (numbers 2009AA022707 and 2012AA02A201), the Shenzhen Municipal Government of China (grant ZYC201005250020A), the Key Laboratory Project Supported by Shenzhen City (grants CX B200903 110066A and CXB201108250096A), and Shenzhen Key Laboratory of Gene Bank for National Life Science. This project was also supported by grants from the Innovative Research Team Project of Guangdong, the Guangdong Enterprise Key Laboratory of Human Disease Genomics, and the Promotion Program for Shenzhen Key Laboratory, Shenzhen, China (CXB200903090055A and CXB201005250016A). We also acknowledge the Ole Rømer grant from the Danish Natural Science Research Council, the Danish National Research Foundation, the National Natural Science Foundation of China, and funds from the Shenzhen Municipal Government and the Local Government of Yantian District of Shenzhen. We also acknowledge support from the Intramural Research Program of the National Cancer Institute, National Institutes of Health, USA.
ACCESSION NUMBERS
All sequencing data from this study are deposited in NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRA050201.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and five tables and can be found with this article online at doi:10.1016/j.cell.2012.02.025.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenet F, Yates DR, Cancel-Tassin G, Cussenot O, and Rouprêt M (2011). Genetic pathways involved in carcinogenesis of clear cell renal cell carcinoma: genomics towards personalized medicine. BJU Int Published online October 28, 2011. 10.1111/j.1464-410X.2011.10661.x. [DOI] [PubMed] [Google Scholar]
- Baudot A, Real FX, Izarzugaza JM, and Valencia A (2009). From cancer genomes to cancer models: bridging the gaps. EMBO Rep. 10, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. (2009). Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 69, 4674–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, and Muchardt C (2003). Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J. 22, 6505–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplin M, Savage K, Khan K, Brett B, Rode J, Varro A, and Dhillon A (2000). Expression and processing of gastrin in pancreatic adenocarcinoma. Br. J. Surg. 87, 1035–1040. [DOI] [PubMed] [Google Scholar]
- Chen M, Ye Y, Yang H, Tamboli P, Matin S, Tannir NM, Wood CG, Gu J, and Wu X (2009). Genome-wide profiling of chromosomal alterations in renal cell carcinoma using high-density single nucleotide polymorphism arrays. Int. J. Cancer 125, 2342–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, and Mann M (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. (2010). Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MH, Kalaidzidis Y, Milutinovic N, Kratchmarova I, Buerkle L, Fetchko MJ, et al. (2009). Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci. Signal 2, ra84. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, and Qian DZ (2011). HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J. Biol. Chem. 286, 38095–38102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. (2007). Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, Jia W, Li Z, He M, Sun L, et al. (2012). Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 44, 17–19. [DOI] [PubMed] [Google Scholar]
- Hall JL, Dudley L, Dobner PR, Lewis SA, and Cowan NJ (1983). Identification of two human beta-tubulin isotypes. Mol. Cell. Biol. 3, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, and Crabtree GR (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. (2012). Single-cell exome sequencing reveals monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, this issue, 873–885. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. (2005). Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, and Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Lee IH, Lim HJ, Yoon S, Seong JK, Bae DS, Rhee SG, and Bae YS (2008). Ahnak protein activates protein kinase C (PKC) through dissociation of the PKC-protein phosphatase 2A complex. J. Biol. Chem. 283, 6312–6320. [DOI] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. (2010). The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465, 473–477. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. (2008). DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, and Wang J (2009). SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, Albrechtsen A, Andersen G, Cao H, Korneliussen T, et al. (2010). Resequencing of 200 human exomes identifies an excess of low-frequency nonsynonymous coding variants. Nat. Genet. 42, 969–972. [DOI] [PubMed] [Google Scholar]
- Liang G, Miao X, Zhou Y, Tan W, and Lin D (2004). A functional polymorphism in the SULT1A1 gene (G638A) is associated with risk of lung cancer in relation to tobacco smoking. Carcinogenesis 25, 773–778. [DOI] [PubMed] [Google Scholar]
- Ling J, Zhuang G, Tazon-Vega B, Zhang C, Cao B, Rosenwaks Z, and Xu K (2009). Evaluation of genome coverage and fidelity of multiple displacement amplification from single cells by SNP array. Mol. Hum. Reprod. 15, 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, and Polyak K (2010). Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH (2005). The HIF pathway in cancer. Semin. Cell Dev. Biol. 16, 523–530. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CS, Wood CG, Silverman PM, Tannir NM, Tamboli P, and Sandler CM (2008). Renal cell carcinoma: diagnosis, staging, and surveillance. AJR Am. J. Roentgenol. 191, 1220–1232. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, and Reich D (2006). Population structure and eigenanalysis. PLoS Genet. 2, e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Feder MM, Al-Saleem T, Liu Z, Liu A, Hudes GR, Uzzo RG, and Testa JR (2010). Combined classical cytogenetics and microarray-based genomic copy number analysis reveal frequent 3;5 rearrangements in clear cell renal cell carcinoma. Genes Chromosomes Cancer 49, 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisegna JR, de Weerth A, Huppi K, and Wank SA (1992). Molecular cloning of the human brain and gastric cholecystokinin receptor: structure, functional expression and chromosomal localization. Biochem. Biophys. Res. Commun. 189, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, et al. (2010a). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. (2010b). A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E, Harris AL, and Ashcroft M (2009). Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert Rev. Mol. Med. 11, e26. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Delaney AD, Farnoud N, Flibotte S, Griffith M, Li HI, Qian H, Farinha P, Gascoyne RD, and Marra MA (2008). Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res. 36, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, and Pili R (2006). Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 66, 8814–8821. [DOI] [PubMed] [Google Scholar]
- Rini BI, Campbell SC, and Escudier B (2009). Renal cell carcinoma. Lancet 373, 1119–1132. [DOI] [PubMed] [Google Scholar]
- Saitou N, and Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Salas S, Jézéquel P, Campion L, Deville JL, Chibon F, Bartoli C, Gentet JC, Charbonnel C, Gouraud W, Voutsinos-Porche B, et al. (2009). Molecular characterization of the response to chemotherapy in conventional osteosarcomas: predictive value of HSD17B10 and IFITM2. Int. J. Cancer 125, 851–860. [DOI] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, Quackenbush J, and Nelson SF (2011). Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27, 2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Cohen FE, and Bishop JM (1992). A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc. Natl. Acad. Sci. USA 89, 5472–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, and Sermon K (2006). Whole-genome multiple displacement amplification from single cells. Nat. Protoc. 1, 1965–1970. [DOI] [PubMed] [Google Scholar]
- Takashima N, Ishiguro H, Kuwabara Y, Kimura M, Haruki N, Ando T, Kurehara H, Sugito N, Mori R, and Fujii Y (2006). Expression and prognostic roles of PABPC1 in esophageal cancer: correlation with tumor progression and postoperative survival. Oncol. Rep. 15, 667–671. [PubMed] [Google Scholar]
- Tang D, Rundle A, Mooney L, Cho S, Schnabel F, Estabrook A, Kelly A, Levine R, Hibshoosh H, and Perera F (2003). Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res. Treat. 78, 217–222. [DOI] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. (2011). Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE (2011). Cells of origin in cancer. Nature 469, 314–322. [DOI] [PubMed] [Google Scholar]
- Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, et al. (2009). Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Pringle LM, Lau AW, Riquelme DN, Wang H, Jiang T, Lev D, Welman A, Blobel GA, Oliveira AM, and Chou MM (2010). TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-kappaB. Oncogene 29, 3619–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Tong SL, Ding YM, Ding J, Fang XM, Zhang XF, Liu ZJ, Zhou YH, Liu QS, Luo HS, and Yu JP (2006). Enhanced expression of cholecystokinin-2 receptor promotes the progression of colon cancer through activation of focal adhesion kinase. Int. J. Cancer 119, 2724–2732. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kernan KA, Thomas A, Collins S, Song Y, Li L, Zhu W, Leboeuf RC, and Eddy AA (2009). A novel signaling pathway: fibroblast nicotinic receptor alpha1 binds urokinase and promotes renal fibrosis. J. Biol. Chem. 284, 29050–29064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Wang Y, Schabath MB, Grossman HB, and Wu X (2003). Sulfotransferase 1A1 (SULT1A1) polymorphism and bladder cancer risk: a case-control study. Cancer Lett. 202, 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.