Some cell lines exhibit transcriptomic, epigenomic, and mutational signatures of a different cancer type than originally annotated.
Abstract
Cell lines are commonly used as cancer models. The tissue of origin provides context for understanding biological mechanisms and predicting therapy response. We therefore systematically examined whether cancer cell lines exhibit features matching the presumed cancer type of origin. Gene expression and DNA methylation classifiers trained on ~9000 tumors identified 35 (of 614 examined) cell lines that better matched a different tissue or cell type than the one originally assigned. Mutational patterns further supported most reassignments. For instance, cell lines identified as originating from the skin often exhibited a UV mutational signature. We cataloged 366 “golden set” cell lines in which transcriptomic and epigenomic profiles strongly resemble the cancer type of origin, further proposing their assignments to subtypes. Accounting for the uncertain tissue of origin in cell line panels can change the interpretation of drug screening and genetic screening data, revealing previously unknown genomic determinants of sensitivity or resistance.
INTRODUCTION
Cell lines are an important research tool, often used in place of primary cells and intact organisms to study biological processes. Cell lines are used for various applications such as testing drug metabolism and cytotoxicity, study of gene function, generation of artificial tissues, and synthesis of biological compounds (1). In cancer research, cell lines derived from tumors are commonly used as models, because they are presumed to carry the genomic and epigenomic alterations that arise in tumors (2). To understand the response of tumors to therapy, many studies have linked genetic and/or epigenetic alterations with drug response across cell line panels, generating datasets such as the Genomics of Drug Sensitivity in Cancer (GDSC) (3) and the Cancer Cell Line Encyclopedia (CCLE) (4). These efforts have advanced our understanding of tumor biology by generating a massive resource of genomic, transcriptomic, epigenomic, and drug response data for hundreds of cell lines (2).
As a model for cancer, cell lines are cost-effective, convenient, and amenable to high-throughput screening (1, 2). However, a major question associated with the use of cell lines is whether they are representative of the cancer they are meant to model, which may be complicated by issues of misidentification (1, 2, 5).
Misidentified cell lines may lead to inconsistent conclusions across studies using the affected cell lines. For instance, the cell lines referred to as HEp-2 and INT 407 in the literature are commonly cross-contaminated with HeLa (cervical cancer) cells, rather than being laryngeal cancer and normal intestinal epithelium cells, respectively (6, 7). Because of the potential for contamination, demonstrating cell line identity via genetic markers is now a routine quality-control step. Current resources based on large-scale cancer cell panels are therefore largely unaffected by this issue (4).
However, even if the genetic identity of the cell line is correct, its properties may not match the cancer type it is meant to model. In particular, the tissue of origin might be incorrect. One way in which this error could arise is that tumors thought to originate in a certain tissue might be metastatic lesions originating from a distal site (8). Cell lines derived from these tumors would have a different tissue/cell type identity than that assigned at isolation, constituting a case of mislabeling. It is conceivable that, also in the case of primary tumors, ambiguous histological or anatomical features may cause the tumor type or subtype to be misdiagnosed and therefore also for a cell line derived from that tumor. Furthermore, the process of establishing the culture might select for a rare cell type that is not representative of the tumor isolate on the whole, meaning that the derived cell line would again effectively be mislabeled with a different cell type (9). In addition to the initial changes upon adaptation to culture, cell lines evolve over time due to selection and due to genetic drift, potentially diverging from the characteristics of the originating tissue (1).
Tissue and/or cell type is a key determinant of response of cultured cells to a variety of experimental conditions, including drug exposure and genetic perturbation (10, 11). Therefore, having accurate information on the tissue and cell type identity of a tumoral cell line is important for interpreting the experimental results obtained using these cell lines.
Recent work has examined cell line panels of certain cancer types, showing discrepancies between the features of cell lines and corresponding tumor types or subtypes. For example, a gene expression analysis of lung tumors and lung cell lines (9) suggested that some lung adenocarcinoma cell lines did not resemble adenocarcinoma tumors but instead clustered with other lung tumor subtypes (small cell and squamous cell tumors). A study of high-grade serous ovarian cancer cell lines that used gene expression, driver gene mutations, and copy number alteration (CNA) data reported that two frequently used cell lines showed poor genetic similarity to molecular profiles of this ovarian cancer subtype (12). A study of a panel of renal cancer cell lines compared their CNA to kidney tumors, finding that some cell lines used as models of the clear cell carcinoma more closely resemble papillary renal cell cancer (13). These examples highlight the need to systematically identify the cell lines whose genotype and/or molecular phenotypes do not resemble the characteristics of the matched human tumor type. A major challenge in the use of human tumor molecular data to classify cell lines are the widespread global changes in gene regulation between cell lines and tumors that arise in cell culture conditions.
We performed a systematic analysis that aligned mRNA expression and DNA methylation data between ~600 cancer cell lines and ~9000 tumors from 22 different cancer types, adjusting for global differences in transcriptomes and epigenomes. Classifiers trained on human tumor mRNA and DNA methylation profiles were used to identify those cell lines whose genomic and epigenomic profiles are highly consistent with human tumors of their declared cancer type of origin. Conversely, we used the same classifiers to identify those cell lines that might be mislabeled with an incorrect cancer type or that might have diverged from their original tissue and/or cell type identity. Our data suggest that tens of cell lines might be epigenetically and/or genetically not consistent with their stated tissue or cell type of origin, which is an important consideration for experiments that use these cell lines. We demonstrate this by reanalyzing associations between drug sensitivity and genetic variation in a large panel of cell lines. After explicitly accounting for putative cases of cell lines with mislabeled tissue identity, many previously unknown associations of genes with drug sensitivity or resistance were revealed.
RESULTS
Identification of tissue/cell type of origin for cell lines by a joint analysis with tumors
The tissue that originated a tumor is well known to be a major determinant of drug responses, including drugs targeted to specific genetic mutations, both in vitro (10, 11) and in vivo (14, 15). Tissue of origin is an important factor in shaping the networks of genetic interactions in cancer (16), and it also determines the phenotypes resulting from genetic perturbation (11). Therefore, ascertaining the tissue/cell type identity of cell lines is relevant for interpreting results of various experiments. For this reason, here, we have systematically examined the global features of the transcriptome and epigenome to identify the tissue of origin of tumoral cell lines.
During the process of adaptation to cell culture, the cells undergo changes in gene regulation that affect many genes (17, 18). The global alterations in gene expression and DNA methylation mean that it is not straightforward to directly compare cell line transcriptomes and epigenomes with data obtained from tumors. To adjust for these cell culture–associated shifts, we introduce a computational methodology—HyperTracker—which can unify transcriptome, epigenome, and mutational data across tumors and cell lines and provide robust predictions of tissue, cell type, and subtype identity.
In particular, we collected gene expression [RNA sequencing (RNA-Seq)] and DNA methylation data (microarrays) for 9681 and 9039 human tumors, respectively (TCGA), and additionally for 614 cell lines (CL) of various solid cancer types and 69 CL of blood tumors. For gene expression data (GE), we examined transcript-per-million (TPM) normalized counts for the 12,419 genes, where RNA-Seq data could be linked between cell lines and tumors. For DNA methylation data (MET), we examined beta values for 10,141 probes from methylation arrays after selecting a single probe per gene promoter with the highest variance across the dataset. To align human tumor and cell line data, we quantile-normalized the data and applied ComBat, a batch effect correction method (19), which is highly performant compared to other related methods (20). In brief, ComBat estimates parameters for location and scale adjustment of each batch (TCGA and CL in our case) for each gene. Then, it removes the variability that is particular to the CL but not present in TCGA, while retaining the intra-dataset variability of the tumors, which should presumably be evident in both the tumor and in the cell line datasets.
A principal components analysis (PCA) in the data (before and after adjustment) suggests that there were strong global differences between TCGA and CL, which are largely removed by our approach (Fig. 1A and fig. S1A). To quantify this, we calculated the effect size (Cohen’s d statistic) for each gene/probe between TCGA and CL before and after adjustment. It can be observed that these differences are reduced to the minimum after adjustment (fig. S1C). In addition, we trained a classification model that predicts the CL versus TCGA origin of the data points based on GE and MET (Fig. 1B and fig. S1B). The model is able to distinguish CL versus TCGA perfectly when using the preadjustment datasets [area under the receiver operating characteristic (ROC) curve (AUC) = 1/area under the precision-recall (PR) curve (AUPRC) = 1], while the post-adjustment datasets do not perform better than random (~0.5 for AUC and ~0 for AUPRC), suggesting that the cell type–specific signal has been largely removed. Last, we tested the optimal number of features (genes/probes) using tumor classifiers and calculating the accuracy in the cell line data (table S1A); we selected 5000 features with the highest SD for subsequent analyses.
Fig. 1. Methodology for data alignment and cancer type classification.
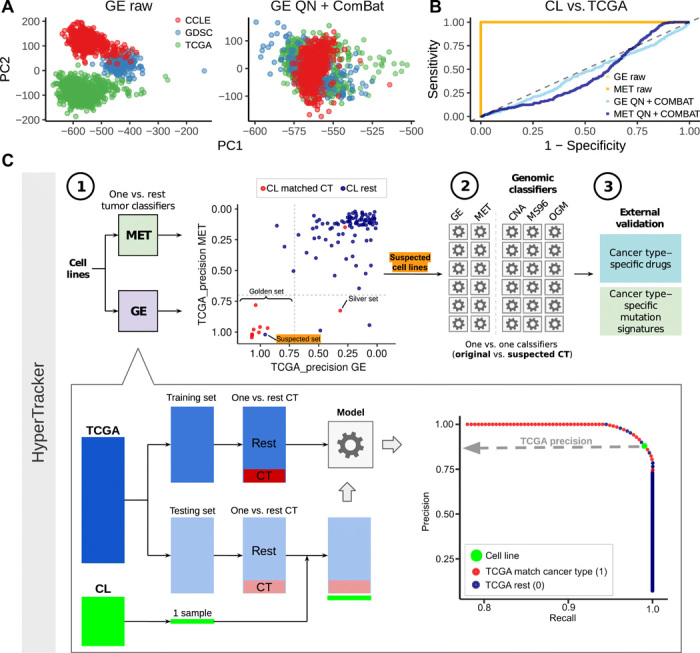
(A) Principal component (PC) 1 and PC2 of a PCA, in the gene expression (GE) data, before adjustment for batch effects (raw data) and after adjustment [quantile normalization (QN) + ComBat] [see fig. S1 for PCA of DNA methylation data (MET)]. Colors represent the dataset sources (GDSC and CCLE are two sources for the cell line data, and TCGA is the source for the tumor data). (B) ROC curves for classifying tumors versus cell lines in the data before adjustment (orange) and after adjustment (blue) for GE and MET. (C) Schematic overview of the HyperTracker methodology. First, we systematically identified possible mislabeled cell lines using GE and MET data, independently. Second, we used various types of mutation-based data to corroborate the predictions. Third, we further validated the cell lines (CL) suspected to originate from skin using independent data, such as drug sensitivity. CT, cancer type.
Once the data were aligned, we set out to determine which cell lines have tissue identity not matching the declared tissue of origin (henceforth, “suspect set”) and, conversely, which cell lines have largely retained their tissue identity (henceforth, “golden set”), by comparing against a large set of tumors from 17 tissues in the TCGA (Fig. 1C). Using TCGA data, we derived one-versus-rest classification models (using ridge regression), separately for the GE and the MET data. These two data types were used because they yielded higher accuracies for the one-versus-rest setting than the four different mutation-based classifier types we tested (fig. S1G); the mutation-based classifiers were nonetheless useful for subsequent validation analyses (see below). Some pairs of cancer types were considered jointly based on their overall similarity, for example, stomach adenocarcinoma (TCGA code: STAD) and esophageal adenocarcinoma (subset of samples from TCGA code: ESAD); see Materials and Methods for a full list. Our study examines solid cancer types and blood cancers in separate analyses.
Differential tumor purity across the TCGA does not have a notable bearing on our tissue classification: Accuracies of models from higher-purity tumor samples were similar to models from other tumor samples (fig. S1D). Moreover, introducing GE data of healthy tissues [from Genotype-Tissue Expression (GTEx)] into a joint analysis with the TCGA tumor data did not further improve the GE classifier (fig. S1F). GE data from healthy tissues, considered by themselves, were less informative for assigning cancer cell lines to tissues of origin than GE data from tumors (fig. S1E), consistent with a tumoral origin of the cell lines.
Next, we obtained predictions of cancer type identity for each cell line. For every cancer type, we split TCGA data randomly into training and testing sets, and we used the calculated PR curve of the TCGA testing dataset to obtain the precision score for every cell line (details in Materials and Methods; all TCGA-derived precision score values are listed in table S1, B and C). The higher the precision, the more likely the cell line is to belong to that particular cancer type. As expected, most of the cancer type labels of the cell lines match the declared tissue of origin of that cell line—they tend to cluster at high precision values for the cognate cancer type (red dots in Fig. 2A and fig. S2). However, among these many correctly classified cell lines (red dots), there are some with similarly high precision scores, but which were originally annotated as belonging to another cancer type (Fig. 2A, blue dots with labels shown). A clustering analysis of the GE and MET values for the genes with the highest weights in the classification models (Fig. 2B and fig. S3) showed that the samples generally cluster by cancer type, but not by CL versus TCGA label. Moreover, we observed that the suspect cell lines (i.e., cell lines with highly confident precision scores to a different cancer type) tend to cluster with the newly assigned cancer type, rather than with the original one (Fig. 2B).
Fig. 2. Detection of cell lines suspected to be mislabeled with a different cancer type.
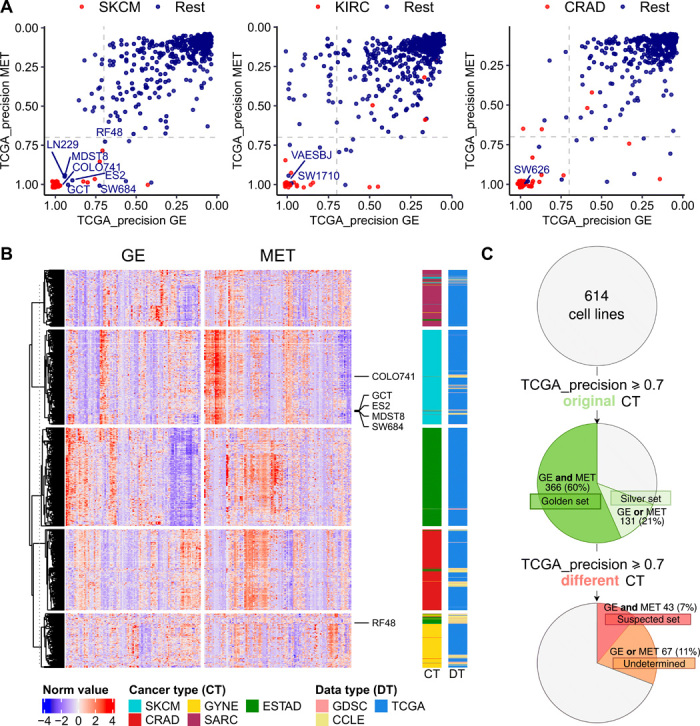
(A) TCGA-based precision scores for 614 cell lines were calculated in the MET and GE cancer type classifiers (one-versus-rest) and for 69 blood cancer cell lines. The higher the precision, the higher the confidence that the sample belongs to that particular cancer type (here, showing cases of SKCM, KIRC, and CRAD from left to right; see fig. S2 for the other cancer types). The cell lines that were originally annotated as the cancer type that is being tested are shown in red, and the rest in blue. (B) Heat map showing the 25 genes (GE) and CpG probes (MET) with the highest absolute values of ridge regression coefficients for each of the cancer types in the plot in one-versus-rest classifiers. The suspected skin cell lines are labeled. The cancer types shown are the suspected cancer type [melanoma (SKCM) in this case] and, additionally, the originally declared cancer types of the suspected cell lines [here, esophagus and stomach cancer (ESTAD), sarcoma (SARC), colorectal cancer (CRAD), and ovarian and uterus cancer (GYNE)]. See fig. S3 for the heat maps for the rest of the suspected cell lines. (C) Overview of the results from the systematic mislabeling testing of all cell lines. Cell lines with a TCGA_precision ≥ 0.7 to its original cancer type in (i) both in GE and in MET are assigned to the golden set group and (ii) either in GE or in MET are assigned to the silver set. If, however, the TCGA-based precision ≥ 0.7 to a different cancer type in GE and in MET, the cell line is assigned to the suspect set.
In further analyses, we designated as the golden set those cell lines that have precision ≥0.7 (see fig. S4 and Materials and Methods for threshold selection) for both GE and, independently, MET in their originally declared cancer type (n = 366 of 614 examined cell lines, 60%). For these cell lines, two independent types of evidence—transcriptomes and epigenomes—support that they match their expected cancer type well, suggesting that these cell lines would be preferred experimental models. Further, we designated as the “silver set” those cell lines that have precision ≥0.7 for one classifier (either GE or MET but not both) (n = 131 of 614, 21%). From the remaining 117 cell lines, we selected as suspect set those CL that exhibit a precision of ≥0.7 for both GE and for MET, but for a different cancer type than declared for that cell line (n = 43 of 614, 7% of analyzed cell lines) (Fig. 2C and fig. S4C). This set of cell lines may consist either of mislabeled cell lines, where the cancer type of origin is different than initially thought, or of heavily diverged cell lines, where the genomic and/or epigenomic alterations accumulating during culture have overridden the original cancer type identity. Notably, cell line cross-contamination issues (21) cannot commonly underlie the trends we observe, because the repositories that provided GE and MET data have used genetic markers to ascertain the identity of the cell lines (4). The fact that two classifiers based on independent data types—one transcriptomic and one epigenomic—reached the same predictions adds confidence that these are bona fide cases of mistaken tissue/cell type identity. In case of blood cancer classifiers, which were highly accurate in distinguishing myeloid and lymphoid lineages, there were no cell lines suspected to be mislabeled between the lineages (fig. S2B).
Validation of individual examples of suspected mislabeled cell lines using genomic classifiers
We detected 43 cell lines that bear both transcriptomic and epigenomic features of a different cancer type than the one they were originally annotated with. We next turned to support individual examples of reassigned tissue identity by analyzing independent data. In particular, we used genomic sequence–based classifiers, which are able to predict the tissue of origin based on somatic mutation patterns (22, 23), in particular, the trinucleotide mutation spectra and the presence of oncogenic mutations and CNA profiles (22, 23). In this validation setting, we applied these genomic classifiers to a problem of one-versus-one classification, where we contrasted the originally assigned cancer type versus the newly proposed cancer type for each reassignment. We found that these one-versus-one classifiers based on genomic data had satisfactory accuracy with whole-exome sequencing (WES) datasets that we used [fig. S5; notably, our recent work (22) suggests that whole-genome sequences, when available, would provide more powerful classifiers that draw on regional mutation density (RMD) patterns].
For the 43 examples of suspect cell line tissue identity, we first derived one-versus-one classification models separately for GE and MET and prioritized reassignments that were consistently observed across multiple runs of the classification algorithm. We randomized the labels to obtain a background model of expected values of the consistency score for the reclassification (Fig. 3B and fig. S6A). From the 43 suspect cell lines, 35 are consistently reassigned to the other tissue (score > 10) (Fig. 3A and fig. S6B). Next, we calculated the same score for the genomic classifiers (based on mutations and CNA, as described above) on these 35 suspect cell lines (Fig. 3A).
Fig. 3. Additional evidence supporting tissue identity of the suspected mislabeled cell lines.
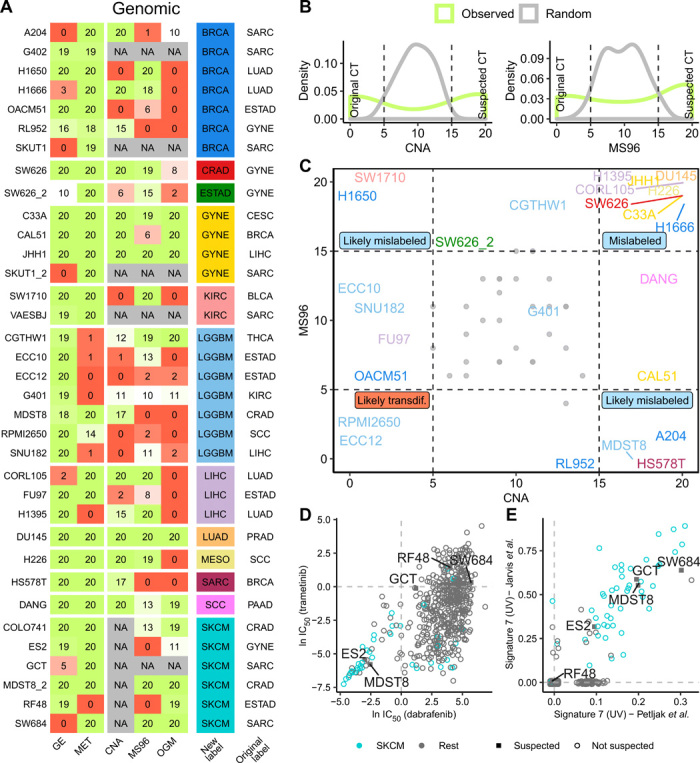
(A) Prediction consistency score (0 to 20) for each suspected cell line for 20 runs of one-versus-one classifiers that predicted suspected versus original cancer type in GE, MET, CNA, trinucleotide mutation spectrum (MS96), and oncogenic mutations (OGM). A value of 20 means that the cell line is predicted as suspected consistently in the 20 runs of the classification algorithm, and a value of 0 means that it is predicted as original cancer type. (B) Histograms of the consistency scores for CNA and MS96 classifiers for the models based on actual data and a baseline expectation on randomized data. (C) Prediction scores for MS96 and CNA for the suspected cell lines. Colors represent the suspected cancer type [see column “new” in (A)]. Gray dots represent the random values. (D) Drug sensitivity (IC50) for mutant BRAF-targeting drugs dabrafenib and trametinib for 614 cell lines. Cell lines originally labeled as skin cancer are shown in turquoise, and skin-suspected cell lines are marked with a square and their name. (E) Burden of UV-associated mutation signature 7 (estimated from two different sources) in 614 cell lines. Cell lines originally labeled as skin cancer are shown in turquoise, and skin-suspected cell lines are marked with a square and the name label.
Of these, approximately two-thirds (n = 22 cell lines) received high support for the new tissue label by one or more genomic classifiers [Fig. 3A; consistency score ≥ 15, corresponding to randomization-based false discovery rates (FDRs) of 0, 0, and 18% for the CNA, OGM, and MS96 classifiers, respectively; Fig. 3B]. These data suggest that 22 cell lines are candidates for assignment to another cancer type, based on converging evidence from the levels of the genome, epigenome, and transcriptome, which provides higher confidence. Reassuringly, this list contains two cell lines that have been previously shown to be misclassified: SW626, which was initially annotated as ovarian cancer but later discovered to be derived from colon cancer (24), and COLO741, which was originally thought to be a colon adenocarcinoma cell line but later shown to originate from a melanoma (25). The fact that these two known examples were detected and reassigned to the correct cancer type provides evidence that our HyperTracker method is overall reliable.
The two plausible reasons why a cell line thought to originate from one cell type would be reassigned to a different cell type are (i) that, at the time of isolation, the cell line was not of the type that it was thought to be (mislabeling) and (ii) that, during prolonged cell culture, the cell line diverged greatly and now resembles another cell type (transdifferentiation). Our data allow us to examine how prevalent each case is: Mislabeling is expected to be reflected equally in both the epigenome and the genome, while transdifferentiation is expected to be reflected more strongly in the (presumably more malleable) epigenome, and less so in the genome, which retains the mutations from the original tumor. We suggest that mislabeling at isolation is a much more common scenario (Fig. 3C, many reassigned cell lines are in the upper right corner). However, it is possible that there exist individual examples of cell lines that have effectively transdifferentiated during culture, because their genomic features are consistent with the original tissue identity, while the epigenomic features are consistent with another tissue (Fig. 3C, lower left corner; e.g., the RPMI2650 and OACM51 cell lines are possible candidates).
Validation of cell lines suspected to originate from the skin
From the previous analysis, we identified six cell lines that are reassigned from various cancer types to skin cancer. We note that, of skin cancers, the TCGA study contains only melanoma but not the nonmelanoma skin cancers, so we were not able to distinguish between cell type identities of different skin cancers.
To further support that these cell lines are skin cells, we performed an independent analysis based on mutational signatures. Large-scale analyses of trinucleotide mutation spectra across human tumors have revealed at least 30 different types of mutational signatures (26). Of these, signature 7 (C>T changes in CC and TC contexts) was associated with exposure to ultraviolet (UV) light and is highly abundant in sun-exposed melanoma tumors (27). The same signatures were recently estimated in cancer cell lines (28, 29), which enabled us to use the existence of UV-linked signature 7 to examine whether these cell lines originated from the skin. On the basis of mutational burden of signature 7, the known melanoma cell lines are clearly separated from the rest (Fig. 3E), meaning the approach can distinguish skin-derived cells. Among the melanoma cell lines with high mutational burden of signature 7, we found four of five of the suspected cell lines (Fig. 3E), in particular, GCT, SW684, ES2, and MDST8 are very likely skin cell lines, and not sarcoma, sarcoma, ovarian cancer, or colorectal cancer, respectively, as originally thought. For the sixth suspected cell line COLO741, the mutational signature data are not available; however, COLO741 has previously been reported to express skin-specific genes (25).
The RF48 cell line (originally considered stomach, here putatively reassigned to skin) does not exhibit the UV signature or the DNA methylation patterns of skin; therefore, a highly confident call cannot be made. Nonetheless, a pattern of cancer driver mutations in RF48 suggests that it is not a stomach cell line (Fig. 3A). Previous work based on gene expression proposed a lymphoid origin for RF48 (30).
Next, we sought to substantiate these findings using drug sensitivity data. In particular, two drugs (dabrafenib and trametinib) that target mutant BRAF are approved for treating melanoma in the clinic. These drugs are known to have poor efficacy in other cancer types bearing BRAF mutations, such as in colon cancers (31). Therefore, sensitivity to these drugs adds confidence that we are looking at a melanoma cell line (note that the converse does not necessarily hold here: resistance does not imply that the cell line is not a melanoma). Therefore, we compared the median inhibitory concentration (IC50) of these two drugs for all cell lines (Fig. 3D). As expected, many melanoma cell lines cluster at low values of IC50 for the two drugs, meaning that these cells are sensitive to the drug. This includes two of five of our suspected cell lines (ES2 and MDST8), providing further supporting evidence that these are of skin, likely melanoma, origin.
In conclusion, out of six cell lines suspected to originate from skin, four were confirmed by the UV mutational signatures and two were additionally confirmed by the drug sensitivity to BRAF/MEK (mitogen-activated protein kinase kinase) inhibitors. This notable example demonstrates how the transcriptome- and epigenome-based tissue/cell type classifiers are able to link cultured human cell lines with their correct cancer type of origin.
In addition to these examples of skin cell lines, we were able to support several other cancer type reassignments using drug sensitivity (results are summarized in table S1D) (32). The DANG cell line is consistent with squamous cell carcinoma of the lung or of the head and neck (SCC), rather than with its original assignment of pancreatic adenocarcinoma (notably, this reassignment is also observed with multiple genomic classifiers; Fig. 3A). Similarly, SW1710 may be a kidney, rather than a bladder, cell line, based on the original reassignment via transcriptome and epigenome, supported by the mutation patterns (Fig. 3A) and additionally supported in the global analysis of drug responses (table S1D). We note that such analyses of drug screening data can be applied to distinguish only certain pairs of tissues and not all reassignments can be reliably validated in this test (see AUC scores in table S1D).
Identification of subtypes for cell lines using multi-omics analyses
Tumors are heterogeneous, and major differences exist between tumor samples of the same cancer type. To manage this variability, cancer types are subdivided on the basis of their molecular characteristics, including global patterns in gene expression and DNA methylation (33–35). However, with the exception of a few tumor types, prominently breast cancer, molecular subtypes are still being established or refined, often with the goal of better predicting disease progression in response to particular treatments. Because drug screens and genetic screens performed on cell line panels have the goal of serving as models for actual tumors, it is useful to be able to transfer the subtype assignments from tumors to cell lines, thereby establishing which cell lines are the most appropriate model for each cancer subtype.
Previously, molecular subtypes from tumors have been inferred in cell lines using different strategies, often based on gene expression, for example, in breast (36), colorectal (37), and renal cancer (13). In a recent pan-cancer study, subtypes have been assigned to a set of 600 cell lines (38).
Our approach to assign subtypes to cell lines is to apply the same strategies that yielded accurate cancer type classifiers: first, the integration of transcriptomic and epigenomic data to boost confidence in the predictions, and second, careful adjustment of the datasets to make them comparable between TCGA tumors and cell lines (fig. S1). An important consideration here is the lack of known labels needed to assess accuracy; thus, assignments should be treated as tentative. However, for breast cancer cell lines, the subtype labels are available and can be used as a benchmark (36).
We examined subtypes proposed for 15 cancer types in the TCGA and generated subtype classifiers (see Materials and Methods) for each cancer type. The combination of both data types (GE and MET) achieved a higher cross-validation accuracy in the TCGA (median AUPRC across cancer types: 0.81) than GE (0.76) or MET (0.72) individually. Therefore, we used the combined datasets to generate subtype classifiers and propose assignments of the cell lines to cancer subtypes. Most were uniquely assigned to a single subtype (fig. S7A); we used only those in further analysis. As a benchmark, we calculated the accuracy for the breast cancer cell lines with subtypes available (fig. S7B): The median AUPRC across subtypes for CL is 0.83. This suggests acceptable performance in obtaining tentative subtype assignments for cell lines in all 15 cancer types, which we provide in table S1E. This resource is complementary to a recent set of subtype predictions for nine cancer types based on transcriptomes (38).
Next, we examined whether the relative prevalence of subtypes is similar between tumors and cell line panels of the same cancer type. Cell line panels of some cancer types have good representation of subtypes, for instance, lung squamous cell cancer, head and neck squamous cell cancer, lung adenocarcinoma, and gastric/esophageal cancers (fig. S7C). However, the converse is the case for liver, skin, and thyroid cancer cell lines, where a single subtype predominates in cell line panels, unlike in tumors (statistics are listed in table S1F). In addition, we observe suboptimal representation (half of the tumor subtypes are not represented) in the kidney, bladder, and brain cancer cell line panels, when considering the 463 cell lines we analyzed. This suggests that, in some cancer types more than others, the commonly used cell line panels do not represent the diversity of molecular subtypes in tumors well. One possible reason for this is the relative ease of culturing certain subtypes compared to others (2).
Accounting for mislabeled cell lines reveals associations in drug screening data
We detected 35 cell lines that may have a tissue or cell type identity different than the one originally assigned to them. Because the cell type is an important determinant of drug response in cancer cell lines and in tumors (10), we hypothesized that the inclusion of this new tissue information when searching for genetic determinants of drug sensitivity may change the results. In a comprehensive study, Iorio et al. (10) searched for associations between drug response and cancer functional events (CFEs): recurrent mutations, CNA, and hypermethylation events present in human tumors. Here, we used GDSCTools (39) to reproduce the results of that study, however, after filtering the cell lines to those that better represent the cancer type in question. In particular, we repeated the same analysis using for each tissue (i) all the cell lines, (ii) only the cell lines in the golden set (G), and (iii) as a less stringent filtering criterion, only the cell lines in the golden set and silver set (G&S). In addition, as controls, we included a random subset of cell lines that matches (iv) the number of cell lines in golden set (r_G) and (v) the number in “golden and silver set” combined (r_G&S).
For most of the cancer types, we observed that one of the filtered subsets recovered a higher number of significant [at FDR ≤ 25%, as applied in the original study (10)] associations of CFE with drug sensitivity or resistance than were recovered using all cell lines (fig. S8A and table S1G). For instance, for glioblastoma, using the golden set cell lines, we found 23 associations, which were not recovered from the entire cell line panel or from the random subset controls (Fig. 4B). For example, this recovers the positive association of CDKN2A loss with camptothecin sensitivity (Fig. 4C), which was previously reported in an independent analysis of the NCI-60 cell line panel screening data (40). Similarly, for pancreatic adenocarcinoma, benefits were observed by focusing on cell lines that resemble the corresponding cancer type better: Using only the golden set plus silver set cell lines, 10 significant, previously unknown associations were found (Fig. 4B). For instance, we detected that SMAD4-mutant cell lines are more resistant to piperlongumine, a natural product that exerts antitumor properties via multiple pathways (41). Mutations in the tumor suppressor gene EP300 were associated with higher sensitivity to three drugs in pancreatic cancer cell lines (Fig. 4D). The observation that more associations were found despite using a somewhat lower number of cell lines (thus less statistical power) emphasizes the importance of focusing on the cell lines that more closely model the tissue and/or cell type of origin of the cognate tumor.
Fig. 4. Drug sensitivity association testing using high-confidence sets of cell lines.
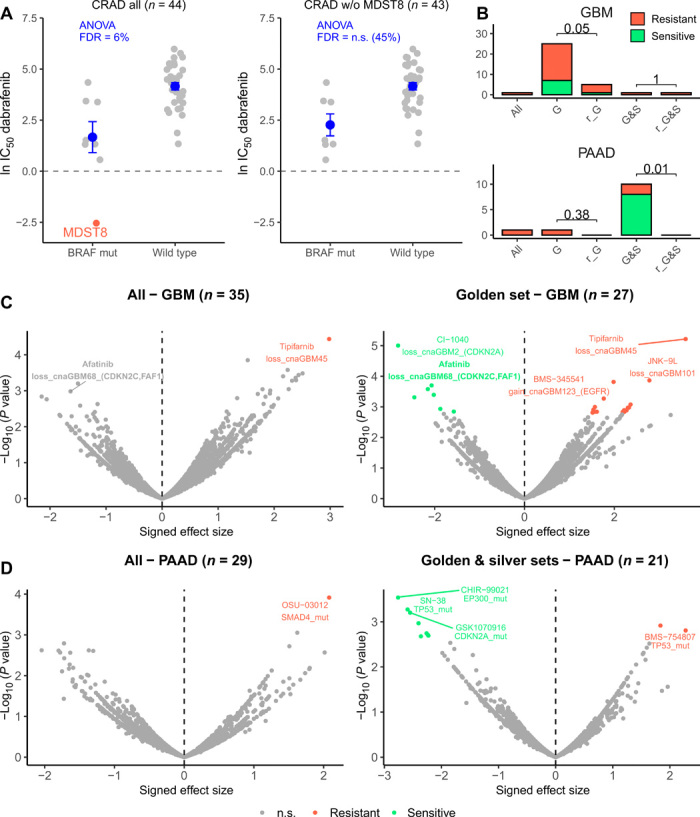
(A) Drug sensitivity (IC50) to dabrafenib in all colorectal (CRAD) cell lines (left) and all CRAD cell lines except MDST8 (right), which is suspected of being skin cancer. Cell lines with a BRAF mutation and without (wild type) are compared. ANOVA FDR for this association (dabrafenib sensitivity with BRAF mutation status) is shown in blue for both datasets. Horizontal line is shown at 0, because score < 0 implies sensitivity to the drug. Dots and error bars represent the mean and SEM. (B) Number of significant associations between “CFEs” (includes mutations and CNAs in cancer genes) and drug associations detected (at FDR 25%) in the ANOVA test for all cell lines (“all”), cell lines in the golden set (“G”), cell lines in the golden plus silver sets (“G&S”), random subset of cell lines that match the number in the golden set (“r_G”), and random subset of cell lines that match the number in the golden plus silver sets (“r_G&S”). For the random subsets, the number of significant associations is calculated from 10 random selections and median shown. P values for a sign test (one-tailed) between the number of associations in the G/G&S and in r_G/r_G&S are shown. See fig. S7 for the remaining cancer types. (C) Differential sensitivity of drugs was analyzed by ANOVA for all brain cancer cell lines (left) and the brain cancer cell lines in the golden set only (right). Each point is an association between the sensitivity of a drug and a genetic feature (CFE). (D) Differential sensitivity of drugs was analyzed by ANOVA for all pancreatic (PAAD) cell lines (left) and PAAD cell lines in the golden and silver set only (right). Each point is an association between the sensitivity of a drug and a genetic feature (CFE). n.s., not significant.
Colorectal cancer provides an illustrative example of the importance of removing nonrepresentative cell lines from drug screening efforts. In the original study (10), 50 colorectal adenocarcinoma (CRAD) cell lines were tested. Of those, we strongly suspected that MDST8 derives from skin. To test the influence of this individual mislabeled cell line, we have performed association testing with all colorectal cell lines and again after excluding MDST8. For the association between dabrafenib response and BRAF mutation status, we observed that CRAD cell lines in general (i.e., irrespective of BRAF mutation) are not sensitive to dabrafenib, except for MDST8, which is strongly sensitive to dabrafenib (Fig. 4A). The FDR of the analysis of variance (ANOVA) analysis when using all (allegedly) colorectal cell lines is significant at 6%, while when removing MDST8 the FDR for BRAF-dabrafenib association in colon becomes nonsignificant (45%). Therefore, in this case, the presence of a single mislabeled cell line is sufficient to cause the appearance of an association between a drug and a feature, which is likely not relevant for this cancer type. This is evidenced in the clinical responses of patients: BRAF-mutant melanoma patients respond well to dabrafenib, while colorectal tumors with the same BRAF V600 mutation are not sensitive to BRAF or MEK inhibitor monotherapy (31).
Accounting for mislabeled cell lines in genetic screening data analyses
Motivated by the associations revealed by reanalyzing the drug screening data, we asked whether the same extends to genetic screening data in cancer cell lines, because results in genetic screens also depend on cell lineage (11). To further investigate, we analyzed CRISPR screening data from Project Score and Project Achilles (see Materials and Methods), from which 347 cell lines overlap our tested cell lines. Then, we applied the same association testing method, which was, however, underpowered, because the number of available overlapping cell lines was smaller. Nonetheless, in colorectal and ovarian cancer (which have the largest number of cell lines in this dataset), we observed that by focusing only on the golden set and/or silver set, the number of recovered associations increased (as a control, there were no increases in randomly chosen cell line subsets of the same size; fig. S8B and table S1H).
To illustrate the importance of removing suspect cell lines in gene dependency screenings, we provide two examples of associations that were originally not significant because of the presence of a mislabeled cell line. For ovarian cancer, the presence of SW626 [mislabeled cell line confirmed by the literature (24)] prevents finding the association between MED8 dependency and a copy number gain in the region containing ASXL1 [“cnaOV72” in (10)] as significant (fig. S9A). Similarly, for colorectal cancer, the presence of MDST8 (mislabeled cell line confirmed by the UV mutational signature) prevents detection of the association between TUBB4B dependency and a copy number gain in the region containing STK4 (“cnaCOREAD32”) (fig. S9B). Next, a significant association between WRN dependency and MLL2 (also known as KMT2D) gene mutation is recovered only with the filtered cell lines in ovarian cancer (fig. S9C). This WRN-MLL2 association has been recently reported using a different set of cell lines (from Project Score) (42) that partially overlap our set.
Last, our reanalyses of drug screening and genetic screening data revealed an interesting association independently supported in both drug and genetic data. The drug afatinib inhibits the epidermal growth factor receptor (EGFR) protein and is clinically indicated for EGFR-mutated lung cancer; however, in EGFR-altered glioblastoma, afatinib is generally not considered to elicit a response (43). Consistently, afatinib sensitivity was associated with EGFR alterations in lung cancer previously (10), as well as in our reanalysis (FDR lung G&S sets = 0.6%), but not in the brain cell line panel (all cell lines, FDR ≥ 25%). However, using the focused (golden set) of brain cancer cell lines revealed a significant association (ANOVA FDR = 15%; fig. S9D) between afatinib sensitivity and a different genetic alteration: copy number loss in a region at 1p32.3 containing the CDKN2C and FAF1 genes [“cnaGBM68” in (10)]. The same loss at 1p32.3 is associated with sensitivity to genetic knockout of EGFR in brain cell line panels in two independent large-scale genetic screens (Project Scores and Project Achilles; fig. S9, F and G) and to pharmacological inhibition in another drug screen (PRISM; fig. S9E). The meta-analysis of the two drug screens and two genetic screens suggests high strength of combined evidence (P = 0.00094, Fisher’s method of combining P values) linking the CNA loss at 1p32.3 (chr1: 51169045-51472178) with sensitivity to pharmacological or genetic EGFR inhibition in brain cells, suggesting a candidate marker for follow-up work.
In summary, the presence of cell lines with dubious or incorrect labels of tissue identity may strongly affect association studies of drug or CRISPR screening data in two different ways. First, the presence of mislabeled cell lines can cause the appearance of spurious associations that do not reflect the biology of the cancer type of interest. Second, the presence of mislabeled or divergent cell lines can prevent the recovery of true associations.
DISCUSSION
Cell lines are commonly used as models for tumors; however, it is an open question how to best apply the available cell line panels to learn about cancer biology. The availability of genomic data from large tumor cohorts and from cell line panels has spurred efforts to find which cell lines are closer to tumors by their transcriptomic (9, 37, 38) and/or genomic features (12, 13), presumably making better models, and which are more distant from examples of actual tumors, thus making less good models.
Our work addresses a different question: We attempt to detect the cancer type (i.e., tissue and/or cell type) that originated the cell line to ascertain whether this matches the declared origin of the cell line. A mismatch may conceivably stem from the sampling step, for instance, a metastasis might have a different tissue of origin than thought at the time of surgical collection. The work-up after collecting the tumor sample may have inaccurately assigned the cancer type, based on unclear histological or anatomical features. Another possibility is that the mismatch might stem from the step of adaptation to cell culture, where a minority cell type that is not representative of the tumor prevails over other tumoral cells. We consider these to be cases of cell line mislabeling during isolation. In addition, we would also detect cases where the cell line might have acquired some properties of a different tissue/cell type during extended periods in culture; however, our analyses (Fig. 3C) suggest that this is a less common occurrence, although individual examples cannot be ruled out.
This phenomenon of tissue/cell type mislabeling is distinct from well-known and widespread cell line misidentification issues (21), where one cell line (often HeLa) was mistakenly used in place of another cell line originating from a different individual, commonly due to cross-contamination. The cell line panels that provided data used in our analyses (GDSC and CCLE) have authenticated their cell lines (4, 42); thus, misidentification/cross-contamination cannot underlie our observations of mislabeling of the cancer type of origin. [We note that there were rare cases of misidentified cells reported in these panels (42); however, these do not overlap our results.]
Methodologically improving over previous work, we introduce the HyperTracker framework that performs global analyses, which independently examine transcriptomic, epigenomic, and mutational features. In addition, we carefully adjust for the known bulk differences between cell lines and tumors, which might have resulted, e.g., from impurities in tumors or from altered expression of cell cycle–related genes in cell lines (38). Parallel analyses of different omics data provide increased confidence in our inferences, which suggested, remarkably, that 5.7% (35 of total 614 considered cell lines) exhibit significant transcriptomic and epigenomic features of a different tissue/cell type than the declared cell type of origin. For 3.6% (22 cell lines), these reassignments to a different cancer type were additionally supported in at least one type of genomic evidence. This increased confidence that these were examples of cell lines with mislabeled (or, less likely, diverged) tissue/cell type identity. Notable examples are cell lines GCT, SW684, ES2, and MDST8 that we predict to originate from the skin, based on the presence of the UV mutational signature, in addition to strong evidence in transcriptome/epigenome data. These cases are reminiscent of the recent reports of UV mutational signatures found in some cases of presumed lung cancers, suggesting that they may instead be metastases originating from sun-exposed skin (44).
In interpreting our data, an important consideration is that the cancer sample types in TCGA may not necessarily reflect the full diversity of rarer subtypes within a cancer type, which may cause some ambiguous predictions. For instance, the ECC10 and ECC12 cell lines are assigned to STAD cancer type (stomach adenocarcinoma) when matched with TCGA tumors. These cell lines originate from gastric small cell neuroendocrine carcinomas. This might explain why, in our analysis, gene expression patterns point toward brain tissues, while mutational features suggest stomach cancer. In such cases of disagreement between different types of features, a future use of a more exhaustive set of reference tumor data, which includes rarer cancer types, may resolve the ambiguity and improve confidence in predictions.
The genomic classifiers we used here were based on whole-exome sequences and were overall less powerful than the transcriptome/DNA methylation classifiers in our data (figs. S1G and S5). Recent work by us and others (22, 23) suggest that analyzing whole-genome sequences of these cell lines would permit use of additional, highly predictive features based on RMD of chromosomal domains. This may provide further genomic evidence for the identity of the cell of origin for the 35 suspected cell lines. Furthermore, targeted experimental follow-up work on these cell lines will provide further evidence to support or refute our predictions, which are based on global, multi-omics analyses.
Knowing the correct tissue-of-origin label for a cell line is important, because this has a strong bearing on the response of the cell line to drug treatment and to genetic perturbation. We demonstrate the implications of this general principle to analyses of drug and genetic screening data: By accounting for suspect cell lines, the power to discover determinants of sensitivity to pharmacological and to genetic perturbation may increase substantially for some cancer types, such as brain, lung, and pancreatic cancers. Therefore, when designing future screening efforts, it is important not only to increase the number of cell lines to gain more power but also to focus on the cell lines that best reflect the tissue and/or cell type of interest.
MATERIALS AND METHODS
Omics data collection and preparation
DNA methylation data
We downloaded DNA methylation data as beta values (platform: Illumina HumanMethylation450) from the GDC Data Portal (45) for TCGA samples and from GDSC (3) for CL samples. We filtered out all probes outside promoter regions and probes with not available (NA) values in more than 100 samples. For the probes in promoter regions, we selected only one probe per gene that had the highest SD across samples. We transformed the beta values to m-values (log2 ratio of the intensities of methylated probe versus unmethylated probe). In total, this yielded 10,141 features for 942 CL samples and 8453 TCGA samples.
Gene expression data
We downloaded gene expression data as TPM from GDC Data Portal (45) for TCGA samples, from GDSC (3) and the CCLE (4) for CL samples, and from GTEx Portal for healthy data. We filtered out genes with NA values in more than 100 samples and selected the overlapping genes between the four sources. We removed low expressed genes (TPM < 1 in 90% of the samples) and applied a square-root transformation to the TPM data. In total, we have 12,419 features for 942 CL samples and 9149 TCGA samples.
For both DNA methylation (MET) and gene expression (GE), we created datasets of different sizes: 1000, 2000, 3000, 5000, and 8000 features by selecting the genes/probes with the highest SD across TCGA samples. In addition, we downloaded tumor purity data from Aran et al. (46) and used consensus measurement of purity estimations (CPE) for creating the groups of high- versus low-purity samples. High-purity samples were defined as CPE > 0.75, and for the lower-purity group, we took a subset of samples (same number of samples that are in high-purity subset) with the lowest purity.
CNA data
We downloaded CNA data (computed by gene) from GDC Data Portal (45) for TCGA samples and from DepMap (47) for CL samples. In total, we have 20,491 features for 942 CL samples and 9188 TCGA samples. To reduce the dataset, we selected CNA events in 299 known cancer driver genes (48).
Mutation data
For human tumors, we downloaded mutation data as WES MC3 dataset (49) from the GDC Data Portal for TCGA samples. For cell lines, aligned short reads (bam files) were obtained from the European Genome-phenome Archive (ID number: EGAD00001001039). Variant calling was performed using Strelka (version 2.8.4) with default parameters. Variant annotation was performed using ANNOVAR (version 2017-07-16). In samples where Strelka was unable to run, a realignment was performed using Picard tools (version 2.18.7) to convert the bams to FASTQ, and following that, the alignment was performed by executing bwa sampe (version 0.7.16a) with default parameters. The resulting bam files were sorted and indexed using Picard tools. To account for germline variants, we removed all mutations that were present in the gnomAD v2.1.1 database (50) at an allele frequency of ≥0.001 in any of the populations. Last, using the filtered somatic mutations, we calculated three sets of mutational features: RMD, mutation spectra (MS96), and oncogenic mutations (OGM) as described by Salvadores et al. (22). RMD features did not exhibit high accuracy when applied to exome-sequencing data and so were not considered further in this analysis.
For the cell line samples, we matched their cancer types to the TCGA cancer types using the cell line metadata from GDSC (3) and manually annotated those that did not have a TCGA cancer type label using Cellosaurus (51). Next, we selected the cell lines from solid tumors that had a matching cancer type in TCGA, yielding a total of 614 cell lines from 22 cancer types. Blood cancers (LAML and DLBC) are tested separately, because the cell lines there commonly grow in suspension, making them less likely to be confounded with solid tumors. For further analysis, we merged the cancer types that were overall similar: HNSC with LUSC and ESCC (jointly known as SCC), GBM with LGG (LGGBM), STAD with ESAD (ESTAD), and OV with UCEC (GYNE).
The identification of the cell line samples was performed by the laboratories generating the cell line databases, using short tandem repeat analysis (4, 42). They reported a few commonly misidentified cell lines: Ca9-22, RIKEN, MKN28, KP-1N, OVMIU, and SK-MG-1 (42). These cell lines do not overlap with our suspected samples, and additionally, the misidentification does not affect tissue or cancer type of origin.
Data alignment between tumors and cell lines
For the alignment of TCGA and CL data, we first applied quantile normalization (R package preprocessCore 1.46.0) and next applied ComBat (R package sva 3.32.1), a batch effect correction method. We used ComBat as if our dataset was the TCGA and CL data combined, and the batch effects labels were whether a sample belongs to TCGA or CL (for MET) or a sample belongs to TCGA, GDSC, or CLLE (for GE). We applied this method for GE, MET, CNA, MS96, and RMD. For validation, we calculated a PCA, subsampling TCGA data to match the number of CL samples (stratified by cancer types). In addition, we calculated Elastic Net classifiers to predict (in the processed dataset) TCGA versus CL and calculated the AUC and AUPRC to check whether the process of alignment is being successful or not. In addition to the chosen adjustment method, we tested other approaches based on canonical correlation analysis, partial least squares, and PCA, which did not exceed accuracy of ComBat and therefore were not examined further.
Cancer type classifiers
For the TCGA dataset, we generated ridge regression model for predicting the cancer type in a one-versus-rest manner (using cv.glmnet function with alpha = 0 and family = binomial, R package glmnet 2.0.18). To calculate the accuracy, we trained classifiers in the TCGA dataset and tested in the CL dataset. In particular, we calculated the AUC and the AUPRC for each cancer type versus the rest (all the rest of cancer types combined).
TCGA_precision score
For each cell line, we calculated a TCGA_precision score of belonging to a particular cancer type. For this, we divided the TCGA data into two datasets (training and testing) of the same size, keeping the cancer type proportions. For each cancer type, we trained classifiers in the TCGA training dataset, and we introduced the cell lines one by one with the testing data and calculated the PR curve (TCGA testing + 1CL). We set the cell line precision score for that specific cancer type as the precision at the threshold where the cell line is situated in the PR curve. Overall, for every cell line, we obtained 17 precision scores, 1 for each possible cancer type. We repeated this procedure five times and calculated the median precision for every case to get more robust values. In addition, when training for one cancer type (label = 1) versus the rest of cancer types combined (label = 0), we made some exceptions and removed those cancer types that are similar, and therefore, the classifier is not good at separating them (e.g., when we calculated precision for ESTAD, we removed from the rest CRAD and PAAD, all hidden cases in table S1I). This is conservative with respect to reassigning cell lines to another cancer type; however, some resolution is traded off, because the more closely related cancer types are, by design, not distinguished. We have further attempted to reclassify cell lines within these hidden tissues and the combined ones. However, when using one-versus-one classifiers, the accuracy is not good enough for distinguishing the two cancer types in the cell lines. We selected TCGA-based precision score ≥ 0.7 as a threshold to separate the different sets (golden, silver, and suspected sets), because this cutoff value approximately maximizes the F1 score for cell line classification (fig. S4A). At the TCGA-based precision threshold ≥0.7, the FDRs estimated on the original cell line labels for gene expression and DNA methylation classifiers would be 28 and 22%, respectively (fig. S4A); because the original labels are not always correct, these FDR estimates are conservative. Also, by visual inspection of the distribution densities of putatively correct predictions (originally labeled as the matching cancer type) and putatively incorrect predictions (originally labeled as another cancer type) for cell line tissue labels, one can appreciate how a TCGA_precision ≥70% threshold separates well the two groups (fig. S4B).
Once we have a list of suspected cell lines, we have an “original” cancer type and a “suspected” cancer type. Therefore, we generated one-versus-one classifiers (original versus suspected) using TCGA dataset (balancing the classes), and for each suspected cell line, we checked whether it is predicted as original or suspected. We repeated this prediction 20 times and counted the number of times a cell line is predicted as suspected. Therefore, we defined a consistency score (range between 0 and 20) for every cell line, where 0 means never predicted as suspected and 20 always predicted as suspected. As a control, we repeated the same procedure with randomized cancer type labels, providing estimates of false discovery for different consistency score thresholds (Fig. 3B and fig. S6A). We calculated this prediction score for GE, MET, CNA, OGM, and MS96 datasets. For calculating the FDR at a score ≥ 15, we applied the following formula: FDR = FP/(FP + TP), where FP is the number of cell lines with score ≥ 15 in the randomized data and TP is the number of cell lines with score ≥ 15 in the actual data.
Independent validation
We downloaded drug sensitivity for the CL from the GDSC database (3). From the provided drugs, we selected trametinib and dabrafenib, U.S. Food and Drug Administration–approved drugs for melanoma treatment. We compared IC50 values for these two drugs for all cancer types.
We downloaded mutational signatures from cell lines available from Jarvis et al. (29) and Petljak et al. (28), and we compared the exposures of all cell lines for signature 7 (UV light). In Petljak et al. dataset, signature 7 is divided into signature 7a, b, c, and d. Therefore, we used the sum of exposures across all four subtypes of signature 7.
We downloaded another set of drug screening data (PRISM 19Q3) (32) for the CL dataset. For the suspected cell lines, we generated one-versus-one classifiers (using cv.glmnet function with alpha = 0 and family = binomial, R package glmnet 2.0.18) for predicting original versus suspected cancer type, based on the drug sensitivity data. We performed 20 runs of each case and counted how many times it is predicted as suspected (consistency score, which can vary from 0 to 20). In addition, we calculated the AUC for each classifier.
Subtype classifiers
We downloaded cancer subtypes for the TCGA samples from the R package TCGAbiolinks 2.12.6, which comprises many available molecular subtype classifications that have been described by TCGA-related publications [mRNA, DNA methylation, protein, miRNA, CNA, integrative (iCluster), and others]. From those, we selected the “subtype_selected,” which is the classification that was chosen as the representative one in that cancer type (usually mRNA or integrative; see documentation of PanCancerAtlas_subtypes function for the complete list). We combined the GE and MET datasets to predict subtypes. For these data, we generated ridge regression model for predicting the subtypes in a one-versus-rest manner (using cv.glmnet function with alpha = 0 and family = binomial, R package glmnet 2.0.18) within each cancer type. We trained models in TCGA, and we predicted subtypes for the cell lines. In addition, we used cell line’s subtypes for breast cancer from a previous paper (36) to calculate the confusion matrix and the AUPRC.
We performed a chi-square test (R package stats 3.6.0) and calculated the Cramer’s V statistic (R package lsr 0.5) for checking whether the proportion of subtypes between TCGA and CL is maintained for each cancer type.
Drug and CRISPR screening data
We downloaded drug sensitivity and CFE data from Iorio et al. (10). CFEs are a collection of recurrent mutations, CNA, and hypermethylation events present in human tumors (10). We used GDSCTools (39) to search for associations between the drugs and the CFEs in every cancer as they did. In particular, we performed this analysis using for each tissue (i) all the cell lines, (ii) only the cell lines in the golden set (G), and (iii) only the cell lines in the golden and silver set (G&S). In addition, as controls, we included a random subset of all cell lines matching (iv) the number of cell lines in the golden set (r_G) and (v) the number in golden and silver set combined (r_G&S). We counted the number of significant hits (at FDR ≤ 25%) for each of the cancer types. For the controls, we repeated the subsampling 10 times and took the median of significant hits. We compared the number of hits for all the cell lines (same as in Iorio et al. study) with the number of hits for the different subsets of cell lines according to our grouping. In addition, we performed a sign test (R package BSDA 1.2.0) comparing the significant hits in the G/G&S subsets versus the significant hits over 10 runs in the random G/random G&S and calculated the P value for all cancer types (alternative = “less”).
Similarly, we downloaded gene dependency data from Project Score (42) and Project Achilles (52) processed with the Project Score pipeline and combined them. From a total of 696 unique cell lines, 357 overlap with the 614 cell lines tested with our method. For those 357 tested cell lines, we repeated the same procedure as described above for the drug sensitivity data.
Supplementary Material
Acknowledgments
The results published here are, in whole or part, based on data generated by the TCGA Research Network, the Genomics of Drug Sensitivity in Cancer (GDSC) Project, the Cancer Cell Line Encyclopedia (CCLE) Project, the Cancer Dependency Map (DepMap) Project (in particular, Project Score), and the Genotype-Tissue Expression (GTEx) Project. In addition, we thank all the members from the Genome Data Science group for insightful discussions. Funding: F.S. is funded by the ERC StG 757700 HYPER-INSIGHT and by MINECO grant BFU2017-89833-P. M.S. is funded by the FPU2017 fellowship of the Spanish government. We acknowledge funding from the Severo Ochoa Center of Excellence award to the IRB Barcelona. Author contributions: M.S.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing, revision, and editing; F.F.-T.: data curation and methodology; F.S.: conceptualization, funding acquisition, methodology, project administration, supervision, writing, revision, and editing. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data can be downloaded from the publicly available databases as described in Materials and Methods. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/27/eaba1862/DC1
REFERENCES AND NOTES
- 1.Kaur G., Dufour J. M., Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2, 1–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodspeed A., Heiser L. M., Gray J. W., Costello J. C., Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol. Cancer Res. 14, 3–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W., Soares J., Greninger P., Edelman E. J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J. A., Thompson I. R., Ramaswamy S., Futreal P. A., Haber D. A., Stratton M. R., Benes C., McDermott U., Garnett M. J., Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geraghty R. J., Capes-Davis A., Davis J. M., Downward J., Freshney R. I., Knezevic I., Lovell-Badge R., Masters J. R. W., Meredith J., Stacey G. N., Thraves P., Vias M.; Cancer Research UK , Guidelines for the use of cell lines in biomedical research. Br. J. Cancer 111, 1021–1046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan L., Glänzel W., Korch C., Capes-Davis A., Widespread use of misidentified cell line KB (HeLa): Incorrect attribution and its impact revealed through mining the scientific literature. Cancer Res. 77, 2784–2788 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Neimark J., Line of attack. Science 347, 938–940 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Campbell B. B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R., Davidson S., Edwards M., Elvin J. A., Hodel K. P., Zahurancik W. J., Suo Z., Lipman T., Wimmer K., Kratz C. P., Bowers D. C., Laetsch T. W., Dunn G. P., Johanns T. M., Grimmer M. R., Smirnov I. V., Larouche V., Samuel D., Bronsema A., Osborn M., Stearns D., Raman P., Cole K. A., Storm P. B., Yalon M., Opocher E., Mason G., Thomas G. A., Sabel M., George B., Ziegler D. S., Lindhorst S., Issai V. M., Constantini S., Toledano H., Elhasid R., Farah R., Dvir R., Dirks P., Huang A., Galati M. A., Chung J., Ramaswamy V., Irwin M. S., Aronson M., Durno C., Taylor M. D., Rechavi G., Maris J. M., Bouffet E., Hawkins C., Costello J. F., Meyn M. S., Pursell Z. F., Malkin D., Tabori U., Shlien A., Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virtanen C., Ishikawa Y., Honjoh D., Kimura M., Shimane M., Miyoshi T., Nomura H., Jones M. H., Integrated classification of lung tumors and cell lines by expression profiling. Proc. Natl. Acad. Sci. U.S.A. 99, 12357–12362 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio F., Knijnenburg T. A., Vis D. J., Bignell G. R., Menden M. P., Schubert M., Aben N., Gonçalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X., Egan R. K., Liu Q., Mironenko T., Mitropoulos X., Richardson L., Wang J., Zhang T., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N. S., Haber D. A., Stratton M. R., Benes C. H., Wessels L. F. A., Saez-Rodriguez J., McDermott U., Garnett M. J., A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E., Dede M., Lenoir W. F., Wang G., Srinivasan S., Colic M., Hart T., A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci. Alliance 2, e201800278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domcke S., Sinha R., Levine D. A., Sander C., Schultz N., Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 4, 2126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha R., Winer A. G., Chevinsky M., Jakubowski C., Chen Y.-B., Dong Y., Tickoo S. K., Reuter V. E., Russo P., Coleman J. A., Sander C., Hsieh J. J., Hakimi A. A., Analysis of renal cancer cell lines from two major resources enables genomics-guided cell line selection. Nat. Commun. 8, 15165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson P., Bandlamudi C., Cheng M. L., Srinivasan P., Chavan S. S., Friedman N. D., Rosen E. Y., Richards A. L., Bouvier N., Selcuklu S. D., Bielski C. M., Abida W., Mandelker D., Birsoy O., Zhang L., Zehir A., Donoghue M. T. A., Baselga J., Offit K., Scher H. I., O’Reilly E. M., Stadler Z. K., Schultz N., Socci N. D., Viale A., Ladanyi M., Robson M. E., Hyman D. M., Berger M. F., Solit D. B., Taylor B. S., Tumour lineage shapes BRCA-mediated phenotypes. Nature 571, 576–579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopetz S., Desai J. R., Chan E. T., Hecht J. R., O’dwyer P. J., Maru D. M., Morris V. K., Janku F., Dasari A., Chung W., Issa J. P. J., Gibbs P., James B. P., Powis G., Nolop K. B., Bhattacharya S., Saltz L. L., Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J. Clin. Oncol. 2015, 4032–4038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S., Lehner B., Epigenetic epistatic interactions constrain the evolution of gene expression. Mol. Syst. Biol. 9, 645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukk M., Kapushesky M., Nikkilä J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A., A global map of human gene expression. Nat. Biotechnol. 28, 322–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varley K. E., Gertz J., Bowling K. M., Parker S. L., Reddy T. E., Pauli-Behn F., Cross M. K., Williams B. A., Stamatoyannopoulos J. A., Crawford G. E., Absher D. M., Wold B. J., Myers R. M., Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson W. E., Li C., Rabinovic A., Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Grennan K., Badner J., Zhang D., Gershon E., Jin L., Liu C., Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLOS ONE 6, e17238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horbach S. P. J. M., Halffman W., The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLOS ONE 12, e0186281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvadores M., Mas-Ponte D., Supek F., Passenger mutations accurately classify human tumors. PLoS Comput. Biol. 15, e1006953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao W., Atwal G., Polak P., Karlic R., Cuppen E., Danyi A., de Ridder J., van Herpen C., Lolkema M. P., Steeghs N., Getz G., Morris Q. D., Stein L. D.; PCAWG Tumor Subtypes and Clinical Translation Working Group , A deep learning system can accurately classify primary and metastatic cancers based on patterns of passenger mutations. Nat. Commun. 11, 214494 (2019). [Google Scholar]
- 24.Furlong M. T., Hough C. D., Sherman-Baust C. A., Pizer E. S., Morin P. J., Evidence for the colonic origin of ovarian cancer cell line SW626. J. Natl. Cancer Inst. 91, 1327–1328 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A., The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat. Commun. 6, 7002 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Alexandrov L. B., Jones P. H., Wedge D. C., Sale J. E., Campbell P. J., Nik-Zainal S., Stratton M. R., Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayward N. K., Wilmott J. S., Waddell N., Johansson P. A., Field M. A., Nones K., Patch A.-M., Kakavand H., Alexandrov L. B., Burke H., Jakrot V., Kazakoff S., Holmes O., Leonard C., Sabarinathan R., Mularoni L., Wood S., Xu Q., Waddell N., Tembe V., Pupo G. M., De Paoli-Iseppi R., Vilain R. E., Shang P., Lau L. M. S., Dagg R. A., Schramm S.-J., Pritchard A., Dutton-Regester K., Newell F., Fitzgerald A., Shang C. A., Grimmond S. M., Pickett H. A., Yang J. Y., Stretch J. R., Behren A., Kefford R. F., Hersey P., Long G. V., Cebon J., Shackleton M., Spillane A. J., Saw R. P. M., López-Bigas N., Pearson J. V., Thompson J. F., Scolyer R. A., Mann G. J., Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Petljak M., Alexandrov L. B., Brammeld J. S., Price S., Wedge D. C., Grossmann S., Dawson K. J., Ju Y. S., Iorio F., Tubio J. M. C., Koh C. C., Georgakopoulos-Soares I., Rodríguez-Martín B., Otlu B., O’Meara S., Butler A. P., Menzies A., Bhosle S. G., Raine K., Jones D. R., Teague J. W., Beal K., Latimer C., O’Neill L., Zamora J., Anderson E., Patel N., Maddison M., Ng B. L., Graham J., Garnett M. J., McDermott U., Nik-Zainal S., Campbell P. J., Stratton M. R., Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarvis M. C., Ebrahimi D., Temiz N. A., Harris R. S., Mutation signatures including APOBEC in cancer cell lines. JNCI Cancer Spectr. 2, pky002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji J., Chen X., Leung S. Y., Chi J.-T. A., Chu K. M., Yuen S. T., Li R., Chan A. S. Y., Li J., Dunphy N., So S., Comprehensive analysis of the gene expression profiles in human gastric cancer cell lines. Oncogene 21, 6549–6556 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Corcoran R. B., Atreya C. E., Falchook G. S., Kwak E. L., Ryan D. P., Bendell J. C., Hamid O., Messersmith W. A., Daud A., Kurzrock R., Pierobon M., Sun P., Cunningham E., Little S., Orford K., Motwani M., Bai Y., Patel K., Venook A. P., Kopetz S., Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600–mutant colorectal cancer. J. Clin. Oncol. 33, 4023–4031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. M. Corsello, R. T. Nagari, R. D. Spangler, J. Rossen, M. Kocak, J. G. Bryan, R. Humeidi, D. Peck, X. Wu, A. A. Tang, V. M. Wang, S. A. Bender, E. Lemire, R. Narayan, P. Montgomery, U. Ben-David, Y. Chen, M. G. Rees, N. J. Lyons, J. M. McFarland, B. T. Wong, L. Wang, N. Dumont, P. J. O’Hearn, E. Stefan, J. G. Doench, H. Greulich, M. Meyerson, F. Vazquez, A. Subramanian, J. A. Roth, J. A. Bittker, J. S. Boehm, C. C. Mader, A. Tsherniak, T. R. Golub, Non-oncology drugs are a source of previously unappreciated anti-cancer activity. bioRxiv 730119 [Preprint]. 9 August 2019. 10.1101/730119. [DOI]
- 33.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceccarelli M., Barthel F. P., Malta T. M., Sabedot T. S., Salama S. R., Murray B. A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S. M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., Rao A. A., Grifford M., Cherniack A. D., Zhang H., Poisson L., Carlotti C. G., da C. Tirapelli D. P., Rao A., Mikkelsen T., Lau C. C., Yung W. K. A., Rabadan R., Huse J., Brat D. J., Lehman N. L., Barnholtz-Sloan J. S., Zheng S., Hess K., Rao G., Meyerson M., Beroukhim R., Cooper L., Akbani R., Wrensch M., Haussler D., Aldape K. D., Laird P. W., Gutmann D. H., Anjum S., Arachchi H., Auman J. T., Balasundaram M., Balu S., Barnett G., Baylin S., Bell S., Benz C., Bir N., Black K. L., Bodenheimer T., Boice L., Bootwalla M. S., Bowen J., Bristow C. A., Butterfield Y. S. N., Chen Q.-R., Chin L., Cho J., Chuah E., Chudamani S., Coetzee S. G., Cohen M. L., Colman H., Couce M., D’Angelo F., Davidsen T., Davis A., Demchok J. A., Devine K., Ding L., Duell R., Elder J. B., Eschbacher J. M., Fehrenbach A., Ferguson M., Frazer S., Fuller G., Fulop J., Gabriel S. B., Garofano L., Gastier-Foster J. M., Gehlenborg N., Gerken M., Getz G., Giannini C., Gibson W. J., Hadjipanayis A., Hayes D. N., Heiman D. I., Hermes B., Hilty J., Hoadley K. A., Hoyle A. P., Huang M., Jefferys S. R., Jones C. D., Jones S. J. M., Ju Z., Kastl A., Kendler A., Kim J., Kucherlapati R., Lai P. H., Lawrence M. S., Lee S., Leraas K. M., Lichtenberg T. M., Lin P., Liu Y., Liu J., Ljubimova J. Y., Lu Y., Ma Y., Maglinte D. T., Mahadeshwar H. S., Marra M. A., McGraw M., McPherson C., Meng S., Mieczkowski P. A., Miller C. R., Mills G. B., Moore R. A., Mose L. E., Mungall A. J., Naresh R., Naska T., Neder L., Noble M. S., Noss A., O’Neill B. P., Ostrom Q. T., Palmer C., Pantazi A., Parfenov M., Park P. J., Parker J. S., Perou C. M., Pierson C. R., Pihl T., Protopopov A., Radenbaugh A., Ramirez N. C., Rathmell W. K., Ren X., Roach J., Robertson A. G., Saksena G., Schein J. E., Schumacher S. E., Seidman J., Senecal K., Seth S., Shen H., Shi Y., Shih J., Shimmel K., Sicotte H., Sifri S., Silva T., Simons J. V., Singh R., Skelly T., Sloan A. E., Sofia H. J., Soloway M. G., Song X., Sougnez C., Souza C., Staugaitis S. M., Sun H., Sun C., Tan D., Tang J., Tang Y., Thorne L., Trevisan F. A., Triche T., Van Den Berg D. J., Veluvolu U., Voet D., Wan Y., Wang Z., Warnick R., Weinstein J. N., Weisenberger D. J., Wilkerson M. D., Williams F., Wise L., Wolinsky Y., Wu J., Xu A. W., Yang L., Yang L., Zack T. I., Zenklusen J. C., Zhang J., Zhang W., Zhang J., Zmuda E., Noushmehr H., Iavarone A., Verhaak R. G. W., Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Sethi N. S., Hinoue T., Schneider B. G., Cherniack A. D., Sanchez-Vega F., Seoane J. A., Farshidfar F., Bowlby R., Islam M., Kim J., Chatila W., Akbani R., Kanchi R. S., Rabkin C. S., Willis J. E., Wang K. K., McCall S. J., Mishra L., Ojesina A. I., Bullman S., Pedamallu C. S., Lazar A. J., Sakai R., Caesar-Johnson S. J., Demchok J. A., Felau I., Kasapi M., Ferguson M. L., Hutter C. M., Sofia H. J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J. C., Zhang J., Chudamani S., Liu J., Lolla L., Naresh R., Pihl T., Sun Q., Wan Y., Wu Y., Cho J., DeFreitas T., Frazer S., Gehlenborg N., Getz G., Heiman D. I., Kim J., Lawrence M. S., Lin P., Meier S., Noble M. S., Saksena G., Voet D., Zhang H., Bernard B., Chambwe N., Dhankani V., Knijnenburg T., Kramer R., Leinonen K., Liu Y., Miller M., Reynolds S., Shmulevich I., Thorsson V., Zhang W., Akbani R., Broom B. M., Hegde A. M., Ju Z., Kanchi R. S., Korkut A., Li J., Liang H., Ling S., Liu W., Lu Y., Mills G. B., Ng K.-S., Rao A., Ryan M., Wang J., Weinstein J. N., Zhang J., Abeshouse A., Armenia J., Chakravarty D., Chatila W. K., Bruijn I., Gao J., Gross B. E., Heins Z. J., Kundra R., La K., Ladanyi M., Luna A., Nissan M. G., Ochoa A., Phillips S. M., Reznik E., Sanchez-Vega F., Sander C., Schultz N., Sheridan R., Sumer S. O., Sun Y., Taylor B. S., Wang J., Zhang H., Anur P., Peto M., Spellman P., Benz C., Stuart J. M., Wong C. K., Yau C., Hayes D. N., Parker J. S., Wilkerson M. D., Ally A., Balasundaram M., Bowlby R., Brooks D., Carlsen R., Chuah E., Dhalla N., Holt R., Jones S. J. M., Kasaian K., Lee D., Ma Y., Marra M. A., Mayo M., Moore R. A., Mungall A. J., Mungall K., Robertson A. G., Sadeghi S., Schein J. E., Sipahimalani P., Tam A., Thiessen N., Tse K., Wong T., Berger A. C., Beroukhim R., Cherniack A. D., Cibulskis C., Gabriel S. B., Gao G. F., Ha G., Meyerson M., Schumacher S. E., Shih J., Kucherlapati M. H., Kucherlapati R. S., Baylin S., Cope L., Danilova L., Bootwalla M. S., Lai P. H., Maglinte D. T., Berg D. J. V. D., Weisenberger D. J., Auman J. T., Balu S., Bodenheimer T., Fan C., Hoadley K. A., Hoyle A. P., Jefferys S. R., Jones C. D., Meng S., Mieczkowski P. A., Mose L. E., Perou A. H., Perou C. M., Roach J., Shi Y., Simons J. V., Skelly T., Soloway M. G., Tan D., Veluvolu U., Fan H., Hinoue T., Laird P. W., Shen H., Zhou W., Bellair M., Chang K., Covington K., Creighton C. J., Dinh H., Doddapaneni H., Donehower L. A., Drummond J., Gibbs R. A., Glenn R., Hale W., Han Y., Hu J., Korchina V., Lee S., Lewis L., Li W., Liu X., Morgan M., Morton D., Muzny D., Santibanez J., Sheth M., Shinbrot E., Wang L., Wang M., Wheeler D. A., Xi L., Zhao F., Hess J., Appelbaum E. L., Bailey M., Cordes M. G., Ding L., Fronick C. C., Fulton L. A., Fulton R. S., Kandoth C., Mardis E. R., McLellan M. D., Miller C. A., Schmidt H. K., Wilson R. K., Crain D., Curley E., Gardner J., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton C., Shelton T., Sherman M., Thompson E., Yena P., Bowen J., Gastier-Foster J. M., Gerken M., Leraas K. M., Lichtenberg T. M., Ramirez N. C., Wise L., Zmuda E., Corcoran N., Costello T., Hovens C., Carvalho A. L., de Carvalho A. C., Fregnani J. H., Longatto-Filho A., Reis R. M., Scapulatempo-Neto C., Silveira H. C. S., Vidal D. O., Burnette A., Eschbacher J., Hermes B., Noss A., Singh R., Anderson M. L., Castro P. D., Ittmann M., Huntsman D., Kohl B., Le X., Thorp R., Andry C., Duffy E. R., Lyadov V., Paklina O., Setdikova G., Shabunin A., Tavobilov M., McPherson C., Warnick R., Berkowitz R., Cramer D., Feltmate C., Horowitz N., Kibel A., Muto M., Raut C. P., Malykh A., Barnholtz-Sloan J. S., Barrett W., Devine K., Fulop J., Ostrom Q. T., Shimmel K., Wolinsky Y., Sloan A. E., Rose A. D., Giuliante F., Goodman M., Karlan B. Y., Hagedorn C. H., Eckman J., Harr J., Myers J., Tucker K., Zach L. A., Deyarmin B., Hu H., Kvecher L., Larson C., Mural R. J., Somiari S., Vicha A., Zelinka T., Bennett J., Iacocca M., Rabeno B., Swanson P., Latour M., Lacombe L., Têtu B., Bergeron A., McGraw M., Staugaitis S. M., Chabot J., Hibshoosh H., Sepulveda A., Su T., Wang T., Potapova O., Voronina O., Desjardins L., Mariani O., Roman-Roman S., Sastre X., Stern M.-H., Cheng F., Signoretti S., Berchuck A., Bigner D., Lipp E., Marks J., McCall S., McLendon R., Secord A., Sharp A., Behera M., Brat D. J., Chen A., Delman K., Force S., Khuri F., Magliocca K., Maithel S., Olson J. J., Owonikoko T., Pickens A., Ramalingam S., Shin D. M., Sica G., Meir E. G. V., Zhang H., Eijckenboom W., Gillis A., Korpershoek E., Looijenga L., Oosterhuis W., Stoop H., van Kessel K. E., Zwarthoff E. C., Calatozzolo C., Cuppini L., Cuzzubbo S., DiMeco F., Finocchiaro G., Mattei L., Perin A., Pollo B., Chen C., Houck J., Lohavanichbutr P., Hartmann A., Stoehr C., Stoehr R., Taubert H., Wach S., Wullich B., Kycler W., Murawa D., Wiznerowicz M., Chung K., Edenfield W. J., Martin J., Baudin E., Bubley G., Bueno R., Rienzo A. D., Richards W. G., Kalkanis S., Mikkelsen T., Noushmehr H., Scarpace L., Girard N., Aymerich M., Campo E., Giné E., Guillermo A. L., Bang N. V., Hanh P. T., Phu B. D., Tang Y., Colman H., Evason K., Dottino P. R., Martignetti J. A., Gabra H., Juhl H., Akeredolu T., Stepa S., Hoon D., Ahn K., Kang K. J., Beuschlein F., Breggia A., Birrer M., Bell D., Borad M., Bryce A. H., Castle E., Chandan V., Cheville J., Copland J. A., Farnell M., Flotte T., Giama N., Ho T., Kendrick M., Kocher J.-P., Kopp K., Moser C., Nagorney D., O’Brien D., O’Neill B. P., Patel T., Petersen G., Que F., Rivera M., Roberts L., Smallridge R., Smyrk T., Stanton M., Thompson R. H., Torbenson M., Yang J. D., Zhang L., Brimo F., Ajani J. A., Gonzalez A. M. A., Behrens C., Bondaruk J., Broaddus R., Czerniak B., Esmaeli B., Fujimoto J., Gershenwald J., Guo C., Lazar A. J., Logothetis C., Meric-Bernstam F., Moran C., Ramondetta L., Rice D., Sood A., Tamboli P., Thompson T., Troncoso P., Tsao A., Wistuba I., Carter C., Haydu L., Hersey P., Jakrot V., Kakavand H., Kefford R., Lee K., Long G., Mann G., Quinn M., Saw R., Scolyer R., Shannon K., Spillane A., Stretch J., Synott M., Thompson J., Wilmott J., Al-Ahmadie H., Chan T. A., Ghossein R., Gopalan A., Levine D. A., Reuter V., Singer S., Singh B., Tien N. V., Broudy T., Mirsaidi C., Nair P., Drwiega P., Miller J., Smith J., Zaren H., Park J.-W., Hung N. P., Kebebew E., Linehan W. M., Metwalli A. R., Pacak K., Pinto P. A., Schiffman M., Schmidt L. S., Vocke C. D., Wentzensen N., Worrell R., Yang H., Moncrieff M., Goparaju C., Melamed J., Pass H., Botnariuc N., Caraman I., Cernat M., Chemencedji I., Clipca A., Doruc S., Gorincioi G., Mura S., Pirtac M., Stancul I., Tcaciuc D., Albert M., Alexopoulou I., Arnaout A., Bartlett J., Engel J., Gilbert S., Parfitt J., Sekhon H., Thomas G., Rassl D. M., Rintoul R. C., Bifulco C., Tamakawa R., Urba W., Hayward N., Timmers H., Antenucci A., Facciolo F., Grazi G., Marino M., Merola R., de Krijger R., Gimenez-Roqueplo A.-P., Piché A., Chevalier S., McKercher G., Birsoy K., Barnett G., Brewer C., Farver C., Naska T., Pennell N. A., Raymond D., Schilero C., Smolenski K., Williams F., Morrison C., Borgia J. A., Liptay M. J., Pool M., Seder C. W., Junker K., Omberg L., Dinkin M., Manikhas G., Alvaro D., Bragazzi M. C., Cardinale V., Carpino G., Gaudio E., Chesla D., Cottingham S., Dubina M., Moiseenko F., Dhanasekaran R., Becker K.-F., Janssen K.-P., Slotta-Huspenina J., Abdel-Rahman M. H., Aziz D., Bell S., Cebulla C. M., Davis A., Duell R., Elder J. B., Hilty J., Kumar B., Lang J., Lehman N. L., Mandt R., Nguyen P., Pilarski R., Rai K., Schoenfield L., Senecal K., Wakely P., Hansen P., Lechan R., Powers J., Tischler A., Grizzle W. E., Sexton K. C., Kastl A., Henderson J., Porten S., Waldmann J., Fassnacht M., Asa S. L., Schadendorf D., Couce M., Graefen M., Huland H., Sauter G., Schlomm T., Simon R., Tennstedt P., Olabode O., Nelson M., Bathe O., Carroll P. R., Chan J. M., Disaia P., Glenn P., Kelley R. K., Landen C. N., Phillips J., Prados M., Simko J., Smith-McCune K., VandenBerg S., Roggin K., Fehrenbach A., Kendler A., Sifri S., Steele R., Jimeno A., Carey F., Forgie I., Mannelli M., Carney M., Hernandez B., Campos B., Herold-Mende C., Jungk C., Unterberg A., von Deimling A., Bossler A., Galbraith J., Jacobus L., Knudson M., Knutson T., Ma D., Milhem M., Sigmund R., Godwin A. K., Madan R., Rosenthal H. G., Adebamowo C., Adebamowo S. N., Boussioutas A., Beer D., Giordano T., Mes-Masson A.-M., Saad F., Bocklage T., Landrum L., Mannel R., Moore K., Moxley K., Postier R., Walker J., Zuna R., Feldman M., Valdivieso F., Dhir R., Luketich J., Pinero E. M. M., Quintero-Aguilo M., Carlotti C. G., Santos J. S. D., Kemp R., Sankarankuty A., Tirapelli D., Catto J., Agnew K., Swisher E., Creaney J., Robinson B., Shelley C. S., Godwin E. M., Kendall S., Shipman C., Bradford C., Carey T., Haddad A., Moyer J., Peterson L., Prince M., Rozek L., Wolf G., Bowman R., Fong K. M., Yang I., Korst R., Rathmell W. K., Fantacone-Campbell J. L., Hooke J. A., Kovatich A. J., Shriver C. D., DiPersio J., Drake B., Govindan R., Heath S., Ley T., Tine B. V., Westervelt P., Rubin M. A., Lee J. I., Aredes N. D., Mariamidze A., Thorsson V., Bass A. J., Laird P. W., Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell 33, 721–735.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J.-P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A., Wang N. J., Kuo W.-L., Stilwell J. L., Pinkel D., Albertson D. G., Waldman F. M., McCormick F., Dickson R. B., Johnson M. D., Lippman M., Ethier S., Gazdar A., Gray J. W., A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronen J., Hayat S., Akalin A., Evaluation of colorectal cancer subtypes and cell lines using deep learning. Life Sci. Alliance 2, e201900517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D. M., van ‘t Veer L. J., Butte A. J., Goldstein T., Sirota M., Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 10, 3574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cokelaer T., Chen E., Iorio F., Menden M. P., Lightfoot H., Saez-Rodriguez J., Garnett M. J., GDSCTools for mining pharmacogenomic interactions in cancer. Bioinformatics 34, 1226–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikediobi O. N., Reimers M., Durinck S., Blower P. E., Futreal A. P., Stratton M. R., Weinstein J. N., In vitro differential sensitivity of melanomas to phenothiazines is based on the presence of codon 600 BRAF mutation. Mol. Cancer Ther. 7, 1337–1346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi Y., Kasukabe T., Kumakura S., Piperlongumine rapidly induces the death of human pancreatic cancer cells mainly through the induction of ferroptosis. Int. J. Oncol. 52, 1011–1022 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Behan F. M., Iorio F., Picco G., Gonçalves E., Beaver C. M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D. A., McRae R., Pooley R., Wilkinson P., van der Meer D., Dow D., Buser-Doepner C., Bertotti A., Trusolino L., Stronach E. A., Saez-Rodriguez J., Yusa K., Garnett M. J., Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Westphal M., Maire C. L., Lamszus K., EGFR as a target for glioblastoma treatment: An unfulfilled promise. CNS Drugs 31, 723–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell J. D., Alexandrov A., Kim J., Wala J., Berger A. H., Pedamallu C. S., Shukla S. A., Guo G., Brooks A. N., Murray B. A., Imielinski M., Hu X., Ling S., Akbani R., Rosenberg M., Cibulskis C., Ramachandran A., Collisson E. A., Kwiatkowski D. J., Lawrence M. S., Weinstein J. N., Verhaak R. G. W., Wu C. J., Hammerman P. S., Cherniack A. D., Getz G., Network C. G. A. R., Artyomov M. N., Schreiber R., Govindan R., Meyerson M., Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grossman R. L., Heath A. P., Ferretti V., Varmus H. E., Lowy D. R., Kibbe W. A., Staudt L. M., Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aran D., Sirota M., Butte A. J., Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Cell Line Encyclopedia Consortium; Genomics of Drug Sensitivity in Cancer Concortium , Pharmacogenomic agreement between two cancer cell line data sets. Nature 528, 84–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Ng P. K.-S., Jeong K. J., Cao S., Wang Z., Gao J., Gao Q., Wang F., Liu E. M., Mularoni L., Rubio-Perez C., Nagarajan N., Cortés-Ciriano I., Zhou D. C., Liang W.-W., Hess J. M., Yellapantula V. D., Tamborero D., Gonzalez-Perez A., Suphavilai C., Ko J. Y., Khurana E., Park P. J., Van Allen E. M., Liang H.; MC3 Working Group; Cancer Genome Atlas Research Network, Lawrence M. S., Godzik A., Lopez-Bigas N., Stuart J., Wheeler D., Getz G., Chen K., Lazar A. J., Mills G. B., Karchin R., Ding L., Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellrott K., Bailey M. H., Saksena G., Covington K. R., Kandoth C., Stewart C., Hess J., Ma S., Chiotti K. E., McLellan M., Sofia H. J., Hutter C., Getz G., Wheeler D., Ding L., Caesar-Johnson S. J., Demchok J. A., Felau I., Kasapi M., Ferguson M. L., Hutter C. M., Sofia H. J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J. C., Zhang J., Chudamani S., Liu J., Lolla L., Naresh R., Pihl T., Sun Q., Wan Y., Wu Y., Cho J., DeFreitas T., Frazer S., Gehlenborg N., Getz G., Heiman D. I., Kim J., Lawrence M. S., Lin P., Meier S., Noble M. S., Saksena G., Voet D., Zhang H., Bernard B., Chambwe N., Dhankani V., Knijnenburg T., Kramer R., Leinonen K., Liu Y., Miller M., Reynolds S., Shmulevich I., Thorsson V., Zhang W., Akbani R., Broom B. M., Hegde A. M., Ju Z., Kanchi R. S., Korkut A., Li J., Liang H., Ling S., Liu W., Lu Y., Mills G. B., Ng K.-S., Rao A., Ryan M., Wang J., Weinstein J. N., Zhang J., Abeshouse A., Armenia J., Chakravarty D., Chatila W. K., de Bruijn I., Gao J., Gross B. E., Heins Z. J., Kundra R., La K., Ladanyi M., Luna A., Nissan M. G., Ochoa A., Phillips S. M., Reznik E., Sanchez-Vega F., Sander C., Schultz N., Sheridan R., Sumer S. O., Sun Y., Taylor B. S., Wang J., Zhang H., Anur P., Peto M., Spellman P., Benz C., Stuart J. M., Wong C. K., Yau C., Hayes D. N., Parker M. D. W., Ally A., Balasundaram M., Bowlby R., Brooks D., Carlsen R., Chuah E., Dhalla N., Holt R., Jones S. J. M., Kasaian K., Lee D., Ma Y., Marra M. A., Mayo M., Moore R. A., Mungall A. J., Mungall K., Robertson A. G., Sadeghi S., Schein J. E., Sipahimalani P., Tam A., Thiessen N., Tse K., Wong T., Berger A. C., Beroukhim R., Cherniack A. D., Cibulskis C., Gabriel S. B., Gao G. F., Ha G., Meyerson M., Schumacher S. E., Shih J., Kucherlapati M. H., Kucherlapati R. S., Baylin S., Cope L., Danilova L., Bootwalla M. S., Lai P. H., Maglinte D. T., Van Den Berg D. J., Weisenberger D. J., Auman J. T., Balu S., Bodenheimer T., Fan C., Hoadley K. A., Hoyle A. P., Jefferys S. R., Jones C. D., Meng S., Mieczkowski P. A., Mose L. E., Perou A. H., Perou C. M., Roach J., Shi Y., Simons J. V., Skelly T., Soloway M. G., Tan D., Veluvolu U., Fan H., Hinoue T., Laird P. W., Shen H., Zhou W., Bellair M., Chang K., Covington K., Creighton C. J., Dinh H., Doddapaneni H., Donehower L. A., Drummond J., Gibbs R. A., Glenn R., Hale W., Han Y., Hu J., Korchina V., Lee S., Lewis L., Li W., Liu X., Morgan M., Morton D., Muzny D., Santibanez J., Sheth M., Shinbrot E., Wang L., Wang M., Wheeler D. A., Xi L., Zhao F., Hess J., Appelbaum E. L., Bailey M., Cordes M. G., Ding L., Fronick C. C., Fulton L. A., Fulton R. S., Kandoth C., Mardis E. R., McLellan M. D., Miller C. A., Schmidt H. K., Wilson R. K., Crain D., Curley E., Gardner J., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton C., Shelton T., Sherman M., Thompson E., Yena P., Bowen J., Gastier-Foster J. M., Gerken M., Leraas K. M., Lichtenberg T. M., Ramirez N. C., Wise L., Zmuda E., Corcoran N., Costello T., Hovens C., Carvalho A. L., de Carvalho A. C., Fregnani J. H., Longatto-Filho A., Reis R. M., Scapulatempo-Neto C., Silveira H. C. S., Vidal D. O., Burnette A., Eschbacher J., Hermes B., Noss A., Singh R., Anderson M. L., Castro P. D., Ittmann M., Huntsman D., Kohl B., Le X., Thorp R., Andry C., Duffy E. R., Lyadov V., Paklina O., Setdikova G., Shabunin A., Tavobilov M., McPherson C., Warnick R., Berkowitz R., Cramer D., Feltmate C., Horowitz N., Kibel A., Muto M., Raut C. P., Malykh A., Barnholtz-Sloan J. S., Barrett W., Devine K., Fulop J., Ostrom Q. T., Shimmel K., Wolinsky Y., Sloan A. E., De Rose A., Giuliante F., Goodman M., Karlan B. Y., Hagedorn C. H., Eckman J., Harr J., Myers J., Tucker K., Zach L. A., Deyarmin B., Hu H., Kvecher L., Larson C., Mural R. J., Somiari S., Vicha A., Zelinka T., Bennett J., Iacocca M., Rabeno B., Swanson P., Latour M., Lacombe L., Têtu B., Bergeron A., McGraw M., Staugaitis S. M., Chabot J., Hibshoosh H., Sepulveda A., Su T., Wang T., Potapova O., Voronina O., Desjardins L., Mariani O., Roman-Roman S., Sastre X., Stern M.-H., Cheng F., Signoretti S., Berchuck A., Bigner D., Lipp E., Marks J., McCall S., McLendon R., Secord A., Sharp A., Behera M., Brat D. J., Chen A., Delman K., Force S., Khuri F., Magliocca K., Maithel S., Olson J. J., Owonikoko T., Pickens A., Ramalingam S., Shin D. M., Sica G., Van Meir E. G., Zhang H., Eijckenboom W., Gillis A., Korpershoek E., Looijenga L., Oosterhuis W., Stoop H., van Kessel K. E., Zwarthoff E. C., Calatozzolo C., Cuppini L., Cuzzubbo S., DiMeco F., Finocchiaro G., Mattei L., Perin A., Pollo B., Chen C., Houck J., Lohavanichbutr P., Hartmann A., Stoehr C., Stoehr R., Taubert H., Wach S., Wullich B., Kycler W., Murawa D., Wiznerowicz M., Chung K., Edenfield W. J., Martin J., Baudin E., Bubley G., Bueno R., De Rienzo A., Richards W. G., Kalkanis S., Mikkelsen T., Noushmehr H., Scarpace L., Girard N., Aymerich M., Campo E., Giné E., Guillermo A. L., Van Bang N., Hanh P. T., Phu B. D., Tang Y., Colman H., Evason K., Dottino P. R., Martignetti J. A., Gabra H., Juhl H., Akeredolu T., Stepa S., Hoon D., Ahn K., Kang K. J., Beuschlein F., Breggia A., Birrer M., Bell D., Borad M., Bryce A. H., Castle E., Chandan V., Cheville J., Copland J. A., Farnell M., Flotte T., Giama N., Ho T., Kendrick M., Kocher J.-P., Kopp K., Moser C., Nagorney D., O’Brien D., O’Neill B. P., Patel T., Petersen G., Que F., Rivera M., Roberts L., Smallridge R., Smyrk T., Stanton M., Thompson R. H., Torbenson M., Yang J. D., Zhang L., Brimo F., Ajani J. A., Gonzalez A. M. A., Behrens C., Bondaruk J., Broaddus R., Czerniak B., Esmaeli B., Fujimoto J., Gershenwald J., Guo C., Lazar A. J., Logothetis C., Meric-Bernstam F., Moran C., Ramondetta L., Rice D., Sood A., Tamboli P., Thompson T., Troncoso P., Tsao A., Wistuba I., Carter C., Haydu L., Hersey P., Jakrot V., Kakavand H., Kefford R., Lee K., Long G., Mann G., Quinn M., Saw R., Scolyer R., Shannon K., Spillane A., Stretch J., Synott M., Thompson J., Wilmott J., Al-Ahmadie H., Chan T. A., Ghossein R., Gopalan A., Levine D. A., Reuter V., Singer S., Singh B., Tien N. V., Broudy T., Mirsaidi C., Nair P., Drwiega P., Miller J., Smith J., Zaren H., Park J.-W., Hung N. P., Kebebew E., Linehan W. M., Metwalli A. R., Pacak K., Pinto P. A., Schiffman M., Schmidt L. S., Vocke C. D., Wentzensen N., Worrell R., Yang H., Moncrieff M., Goparaju C., Melamed J., Pass H., Botnariuc N., Caraman I., Cernat M., Chemencedji I., Clipca A., Doruc S., Gorincioi G., Mura S., Pirtac M., Stancul I., Tcaciuc D., Albert M., Alexopoulou I., Arnaout A., Bartlett J., Engel J., Gilbert S., Parfitt J., Sekhon H., Thomas G., Rassl D. M., Rintoul R. C., Bifulco C., Tamakawa R., Urba W., Hayward N., Timmers H., Antenucci A., Facciolo F., Grazi G., Marino M., Merola R., de Krijger R., Gimenez-Roqueplo A.-P., Piché A., Chevalier S., McKercher G., Birsoy K., Barnett G., Brewer C., Farver C., Naska T., Pennell N. A., Raymond D., Schilero C., Smolenski K., Williams F., Morrison C., Borgia J. A., Liptay M. J., Pool M., Seder C. W., Junker K., Omberg L., Dinkin M., Manikhas G., Alvaro D., Bragazzi M. C., Cardinale V., Carpino G., Gaudio E., Chesla D., Cottingham S., Dubina M., Moiseenko F., Dhanasekaran R., Becker K.-F., Janssen K.-P., Slotta-Huspenina J., Abdel-Rahman M. H., Aziz D., Bell S., Cebulla C. M., Davis A., Duell R., Elder J. B., Hilty J., Kumar B., Lang J., Lehman N. L., Mandt R., Nguyen P., Pilarski R., Rai K., Schoenfield L., Senecal K., Wakely P., Hansen P., Lechan R., Powers J., Tischler A., Grizzle W. E., Sexton K. C., Kastl A., Henderson J., Porten S., Waldmann J., Fassnacht M., Asa S. L., Schadendorf D., Couce M., Graefen M., Huland H., Sauter G., Schlomm T., Simon R., Tennstedt P., Olabode O., Nelson M., Bathe O., Carroll P. R., Chan J. M., Disaia P., Glenn P., Kelley R. K., Landen C. N., Phillips J., Prados M., Simko J., Smith-McCune K., VandenBerg S., Roggin K., Fehrenbach A., Kendler A., Sifri S., Steele R., Jimeno A., Carey F., Forgie I., Mannelli M., Carney M., Hernandez B., Campos B., Herold-Mende C., Jungk C., Unterberg A., von Deimling A., Bossler A., Galbraith J., Jacobus L., Knudson M., Knutson T., Ma D., Milhem M., Sigmund R., Godwin A. K., Madan R., Rosenthal H. G., Adebamowo C., Adebamowo S. N., Boussioutas A., Beer D., Giordano T., Mes-Masson A.-M., Saad F., Bocklage T., Landrum L., Mannel R., Moore K., Moxley K., Postier R., Walker J., Zuna R., Feldman M., Valdivieso F., Dhir R., Luketich J., Pinero E. M. M., Quintero-Aguilo M., Carlotti C. G., Santos J. S. D., Kemp R., Sankarankuty A., Tirapelli D., Catto J., Agnew K., Swisher E., Creaney J., Robinson B., Shelley C. S., Godwin E. M., Kendall S., Shipman C., Bradford C., Carey T., Haddad A., Moyer J., Peterson L., Prince M., Rozek L., Wolf G., Bowman R., Fong K. M., Yang I., Korst R., Rathmell W. K., Fantacone-Campbell J. L., Hooke J. A., Kovatich A. J., Shriver C. D., DiPersio J., Drake B., Govindan R., Heath S., Ley T., Van Tine B., Westervelt P., Rubin M. A., Lee J. I., Aredes N. D., Mariamidze A., Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Systems 6, 271–281.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski, The Genome Aggregation Database Consortium, B. M. Neale, M. J. Daly, D. G. MacArthur, Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 531210 [Preprint]. 13 August 2019. 10.1101/531210. [DOI]
- 51.Bairoch A., The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 29, 25–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers R. M., Bryan J. G., McFarland J. M., Weir B. A., Sizemore A. E., Xu H., Dharia N. V., Montgomery P. G., Cowley G. S., Pantel S., Goodale A., Lee Y., Ali L. D., Jiang G., Lubonja R., Harrington W. F., Strickland M., Wu T., Hawes D. C., Zhivich V. A., Wyatt M. R., Kalani Z., Chang J. J., Okamoto M., Stegmaier K., Golub T. R., Boehm J. S., Vazquez F., Root D. E., Hahn W. C., Tsherniak A., Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/27/eaba1862/DC1


