Collaboration broadens the “root economics space” ranging from “do-it-yourself” to “outsourcing” to mycorrhizal partners.
Abstract
Plant economics run on carbon and nutrients instead of money. Leaf strategies aboveground span an economic spectrum from “live fast and die young” to “slow and steady,” but the economy defined by root strategies belowground remains unclear. Here, we take a holistic view of the belowground economy and show that root-mycorrhizal collaboration can short circuit a one-dimensional economic spectrum, providing an entire space of economic possibilities. Root trait data from 1810 species across the globe confirm a classical fast-slow “conservation” gradient but show that most variation is explained by an orthogonal “collaboration” gradient, ranging from “do-it-yourself” resource uptake to “outsourcing” of resource uptake to mycorrhizal fungi. This broadened “root economics space” provides a solid foundation for predictive understanding of belowground responses to changing environmental conditions.
INTRODUCTION
The diversity of plant traits across the globe shapes ecosystem functioning (1). Seeking general patterns, ecologists have used economic theory to explain trait variation in leaves as the aboveground plant organs for resource acquisition by photosynthesis (1–3). Aboveground plant strategies thereby fall along a “leaf economics spectrum” (2) from cheaply constructed but short-lived leaves optimized for “fast” resource acquisition to more expensive but persistent leaves with a “slower” rate of return over longer time scale.
Fine roots acquire resources from the soil and are often considered the belowground equivalent of leaves (4). Therefore, fine-root trait variation has been hypothesized to follow a similar one-dimensional spectrum (1, 5). At one side of this spectrum, plants with a “fast” belowground resource acquisition strategy are expected to construct long, narrow-diameter roots with minimal biomass investment but high metabolic rates (1, 4, 6). At the opposite side of the spectrum, plants with a “slow” strategy are expected to achieve longer life span and prolonged return on investment by constructing thicker-diameter, denser roots (4, 7).
However, mixed empirical results caused ecologists to question whether variation in root traits can be adequately explained by a one-dimensional “fast-slow” economics spectrum (1, 5, 8–10). Instead, when taken together, earlier results reveal that root trait variation might be driven by multiple evolutionary pressures (8, 11–14). Here, we aim to settle this debate by presenting a new conceptual framework of root economics that better captures the complexity of belowground resource acquisition strategies. First, we integrated existing knowledge to build a conceptual understanding of the covariation among four key root traits (Table 1 and Fig. 1). Second, we tested our conceptual model against root traits of 1810 plant species from a global database. Third, we investigated generality of the concept across all biomes of the world, different plant growth forms, and symbiotic partnerships. All analyses were phylogenetically informed using fine-root trait data from the Global Root Trait (GRooT) database (15).
Table 1. Rationale of the conceptual framework of root trait correlations depicted in Fig. 1.
Expected correlations are based on mathematical and ecological rationale and empirical support from the literature. De facto correlations (see also fig. S1) are phylogenetically informed correlation coefficients of species subsets with the respective trait coverage. D, root diameter; SRL, specific root length; RTD, root tissue density; N, root nitrogen content; CF, cortex fraction.
| Trait pair | Expected correlation | Rationale | Empirical support | De facto correlation | P | n species |
| SRL - D | Negative | A thicker root is shorter per unit mass. |
(9, 14, 18–20) | −0.70 | <0.0001 | 1402 |
| RTD - N | Negative | RTD increases with cell wall stabilization, which is poor in nitrogen. |
(9, 14, 19) | −0.26 | <0.0001 | 851 |
| CF - D | Positive | CF increases with increasing D at a higher rate than stele fraction. |
(10, 19, 22) | 0.22 | <0.0001 | 317 |
| SRL - RTD | Negative | A root with a higher tissue density is shorter per unit mass. |
(14) | −0.23 | <0.0001 | 1284 |
| RTD - CF | Negative | Cortex tissue is less dense than stele tissue. |
(19) | −0.20 | 0.0002 | 304 |
| RTD - D | Negative | D scales positively with CF. Cortex tissue is less dense than stele tissue. |
(14, 19) | −0.20 | <0.0001 | 1318 |
Fig. 1. Conceptual framework of the root economics space.
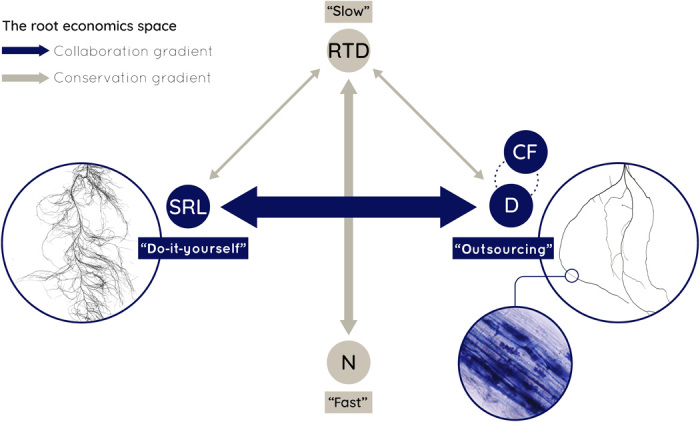
On the basis of this concept, we hypothesize (i) a collaboration gradient ranging from do-it-yourself soil exploration by high specific root length (SRL) to outsourcing by investing carbon into the mycorrhizal partner and hence extraradical hypheae, which requires a large cortex fraction (CF) and root diameter (D) and (ii) a conservation gradient ranging from roots with high root tissue density (RTD) that show a slow resource return on investment but are long-lived and well-protected, to fast roots with a high nitrogen content (N) and metabolic rate for fast resource return on investment but a short life span. Arrows indicate negative correlations between the single traits (see Table 1).
The currency of root economics is the carbon input required to construct fine roots that explore the soil for resource acquisition. Specific root length (SRL)—the root length per unit mass—therefore reflects the rate of return per unit of investment and is a function of both root diameter (D) and root tissue density (RTD)—the root mass per unit of root volume—following
Although this equation (6) is a simplification when sampling heterogeneous fine-root populations (16), it implies that SRL increases with decreasing D and/or RTD. Besides efficient soil exploration, plants have to maintain a high metabolic rate to assure fast resource acquisition leading to high nitrogen content (N) in the fine roots (1, 17). While strong negative relationships between SRL and D (9, 14, 18–20) and between RTD and N (9, 14, 19) have been observed, the relationships between SRL and RTD (19, 21, 22) and between D and N (10) have been less clear. In fact, observations across a wide range of species suggest that plants can construct roots with many combinations of SRL and RTD (9, 14), indicating complex trait interactions inconsistent with a one-dimensional root economics spectrum (8–11, 14).
A growing body of literature (8, 11–14) indicates that this root trait complexity may result from the range of belowground resource uptake strategies. In contrast to aboveground photosynthesis, which is solely conducted by plant organs, belowground many species have the ability to outsource resource acquisition. This gradient of plant collaboration strategies ranges from “do-it-yourself” acquisition by cheap roots for efficient soil exploration to “outsourcing” acquisition via the investment of carbon in a mycorrhizal fungal partner for the return of limiting resources. However, these outsourcing strategies have consequences for root traits. This is particularly true for arbuscular mycorrhizal fungi (AMF) because plants must increase their root cortical area, and hence their D, to provide the intraradical habitat for their fungal partner (19, 23, 24). This is generalizable for plant symbiosis with AMF, the most widespread type of mycorrhizal fungi (24) and also well documented for ectomycorrhizal (EM) fungi (25). Using this body of literature as our foundation, we developed an overarching concept of root economics based on the understanding that plants can optimize resource uptake by investing carbon either in thin roots that efficiently explore the soil themselves (14) or in a mycorrhizal fungal partner, which often requires a thick root for efficient symbiosis (Fig. 1).
This conceptualized collaboration gradient from do-it-yourself to outsourcing challenges the traditional spectrum of root economics that assumes D to increase with RTD for tissue conservation. Both scaling laws and empirical data (22) show that as D increases, root cortex area increases at a faster rate than stele area such that D scales positively with the cortex fraction (CF) (19) [although patterns can vary among growth forms (10)]. The parenchymatous cortical tissue has a lower carbon content and dry weight than the stele tissue, which transports nutrients and water through lignified cells (26, 27). Thus, CF and RTD will be negatively correlated (Table 1). Furthermore, since D and CF are closely positively correlated and increase in unison with mycorrhizal symbiosis, D should be negatively correlated with RTD. These relationships contradict the assumption of a one-dimensional root economics spectrum, where plants with a slow strategy are expected to construct roots that are both thick and dense, and advocate for a multidimensional space of root trait variation.
RESULTS AND DISCUSSION
By testing pairwise correlations of all traits, we confirmed the bivariate relationships underlying our new concept of a belowground economics trait space with two main dimensions (Table 1). The strongest negative correlation was found between SRL and D (R = −0.70), representing the “collaboration” gradient from do-it-yourself to outsourcing. We also found a negative correlation between RTD and root N (R = −0.26) as observed in previous studies (9, 14, 19), which corresponds to a “conservation” gradient, representing the traditional trade-off between fast and slow return on investment (Fig. 1).
On a subset of 748 species with complete information on the four main root traits (SRL, D, RTD, and root N), we could confirm these two distinct and largely independent gradients in a phylogenetically informed principal component analysis (PCA) where the first two axes encompass a plane with a cumulative explanatory power of 77% of all root trait variation. Henceforth, we refer to these gradients as the main dimensions of the “root economics space” (Fig. 2A). The first PCA axis (44% of total trait variation) represents a gradient from SRL to D, confirming our conceptualized collaboration gradient and suggesting that it actually represents the main source of root trait variation. The second PCA axis (33% of total trait variation) represents the conservation gradient from root N to RTD (table S1).
Fig. 2. The root economics space.
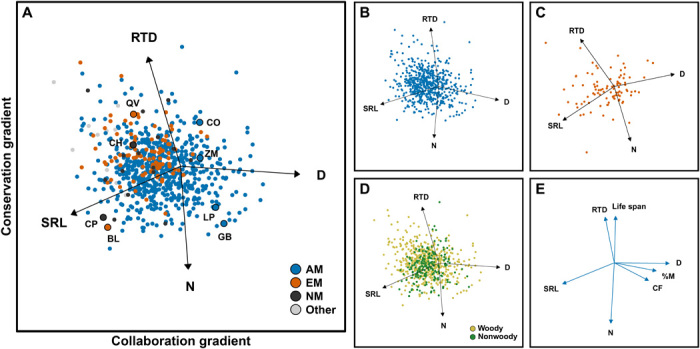
Phylogenetically informed principal component analyses (PCAs) of core traits of (A) 748 global species, (B) 621 arbuscular mycorrhizal (AM) species (blue), and (C) 94 ectomycorrhizal (EM) species (red). NM, nonmycorrhizal. The collaboration gradient (44%) ranges from do-it-yourself roots with high SRL to outsourcing roots with “thick diameters” (D). The conservation gradient (33%) ranges from fast (N) to slow (RTD). For each corner of the root economics space, in (A) we highlight two representative plant species: QV, Quercus virginiana Mill.; CH, Carex humilis Leyss.; CO, Cornus officinalis Siebold & Zucc.; ZM, Zea mays L.; LP, Lathyrus pratensis L.; GB, Ginkgo biloba L.; BL, Betula lenta L.; CP, Cardamine pratensis L. (D) Woody (ocher) and nonwoody (green) species show no distinct pattern within the root economics space (see fig. S4 and table S4). (E) PCA based on bivariate trait relationships. The percentage mycorrhizal colonization (%M) and the CF are positively correlated with D along the collaboration gradient, while root life span is negatively correlated with N along the conservation gradient. See table S1 for PCA scores.
Species associated with AMF were the largest group in the database and were distributed over the entire trait space (Fig. 2A) but differed significantly from both nonmycorrhizal (NM) and EM species (table S4). NM plants aggregated on the do-it-yourself side of the collaboration gradient and on the slow side of the conservation gradient. EM plants showed less variation along the collaboration gradient than AM plants with a tendency toward do-it-yourself and slow as well. A high RTD, indicative of a slow strategy, might partly originate from the fact that EM species are often woody (28) or it might reflect a general slow nutrient cycling in ecosystems dominated by EM species (29, 30). The tendency for EM plants toward do-it-yourself roots with high SRL likely results not only from the nature of the EM symbiosis that is less dependent on cortex area but also from its more recent evolution, as evolutionarily younger species tend to have thinner roots (14, 23, 27, 31). Even so, PCAs that solely represent the root traits of either AM or EM plant species (Fig. 2, B and C, and table S1) show the same dimensions of variation as in the global dataset, highlighting the existence of the same trade-offs within each mycorrhizal type.
Plant species associated with N2-fixing bacteria differed from other species (table S4) by being located on the fast side of the conservation gradient as their roots are rich in N (fig. S2A). Nevertheless, we could still confirm the collaboration gradient as the first PCA axis within this species set (fig. S2, B and C, and table S1). The importance of the collaboration gradient within N2-fixing species might derive from the large P demands of plants associated with N2-fixing bacteria, which leads to either high mycorrhizal dependency or alternative do-it-yourself strategies like cluster roots (24). Investigating different plant growth forms, we found that woody plants span a wider range of variation than nonwoody plants within the global trait space (Fig. 2 and table S4). Still, the two gradients of the root economics space exist within both woody and nonwoody plants (fig S4), indicating that there is wide variation and very similar trade-offs operating irrespective of growth form. Last, the two dimensions of the root economics space are present irrespective of biome (Fig. 3 and table S1), which did not differ from each other in their location within the global trait space (table S4).
Fig. 3. The root economics space is present in different biomes.
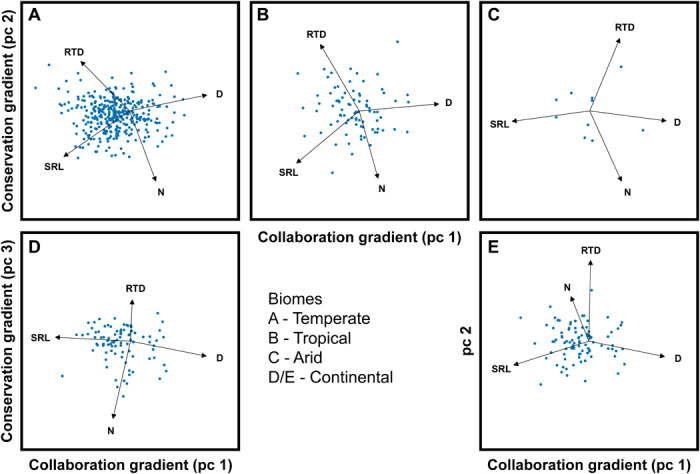
Root traits and trait relations are known to vary across biomes (14). We found no respective between group variation within the root economics space (table S4). Still, to test whether the concept is broadly generalizable, we present separate PCAs for biomes spanning arid to tropical. We found that the root economics space was apparent in all of the biomes represented by our species (panels A, B, C, and D). In continental systems, the conservation gradient was represented by principal component 3 (D) instead of principal component 2 (E). See table S1 for principal component analyses. pc, principal component.
To confirm our ecological interpretation of the proposed gradients, we added traits to the PCA that act as proxies for ecological functions (Fig. 2E and table S2). We used percent root length colonized by AMF (%M) as a proxy for the strength of the mycorrhizal symbiosis (32) and CF as a general proxy for the ability of a species to host mycorrhizal fungi (19, 33, 34). We found both %M and CF to be associated with the outsourcing side of the collaboration gradient. To test whether the proposed conservation gradient aligns with the classical fast-slow economics spectrum, we used root life span as a proxy for short- or long-term investment of plant carbon (1, 35–37). We found that longer life span was indeed associated with the slow side of the conservation gradient, which is consistent with reports of negative relationships between root life span and N (1, 35, 37).
The decrease in D over evolutionary time (14, 31) suggests a reduced dependence of plants on mycorrhizal fungi. We found that the collaboration gradient was indeed phylogenetically conserved, showing an evolutionary transition from outsourcing to do-it-yourself (Fig. 4 and tables S3 and S5). In contrast, the fast-slow trade-off of the conservation gradient was less pronounced across all plant families in our database (Fig. 4) and also less phylogenetically conserved (table S3). Terrestrial plants coevolved with AMF, causing the mycorrhizal symbiosis to be evolutionarily stable (38, 39). This might explain the finding that the consequences for root morphology and anatomy—being associated with the collaboration gradient—are phylogenetically conserved at high levels. Different explanations have been proposed as to why D gradually decreased with evolutionary time, including a decline in atmospheric CO2 (13), leading to higher water demands and a reduction in the dependence on mycorrhizal fungi (14). Remarkably, this trend very rarely resulted in a complete loss of the mycorrhizal symbiosis but instead led to varying degrees of outsourcing. Following this line of reasoning, evolutionary history might be the reason why the collaboration gradient is the main source of variation in root traits. As the ability to outsource is a major difference between above- and belowground economics, the importance of the collaboration gradient might be the key to explaining decoupling of root and leaf traits, leading to inconsistencies within the plant economics spectrum found in the past (9, 11, 27, 40).
Fig. 4. The collaboration gradient is phylogenetically conserved.
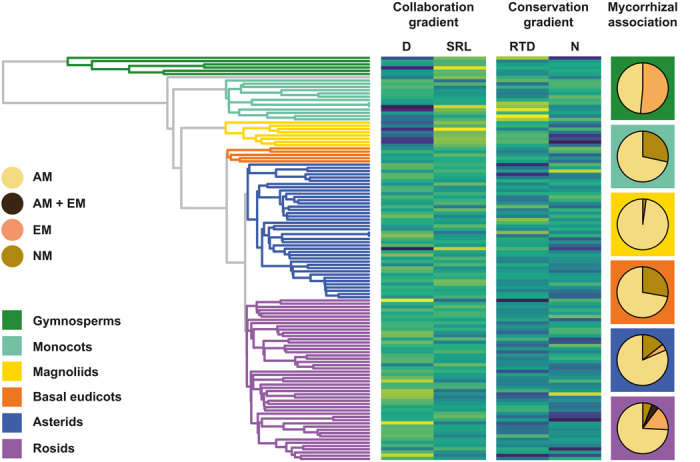
On the left, we display the phylogenetic tree of 1810 species aggregated at a family level with the standardized family mean trait values of the four core traits (center) ranging from low (yellow) to medium (green) to high (blue). The collaboration gradient shows a strong phylogenetic pattern (λ = 0.8, P < 0.001) with a transition from families with thick D to those with high SRL. The phylogenetic signal in the conservation gradient is less pronounced (λ = 0.5, P < 0.001), although still significant (see also table S3). For detailed information about specific clades, see table S5, and for family distribution across clades, see table S6. Pie charts (right) depict the fraction of different mycorrhizal association types within the broader plant phylogenetic clades (indicated by corresponding background colors).
CONCLUSION
Together, we provide a conceptual framework explaining the mechanistic basis behind root trait covariation and show its ubiquity across biomes, growth forms, and symbiotic partnerships. The root economics space synthesizes recent evidence to illustrate why root trait variation cannot be adequately explained by a one-dimensional spectrum (8, 9, 11, 14, 19, 41). Plant outsourcing of belowground resource acquisition through collaboration with mycorrhizal fungal partners is not just an “extra” dimension in the root economics space but rather is the main dimension of root trait variation, which is fundamentally different from the aboveground economy. This collaboration gradient from do-it-yourself to outsourcing represents an investment in soil exploration by either the root itself or its mycorrhizal fungal partners. It is independent from the conservation gradient, which represents the well-known concept of fast versus slow return on investment. Thus, both gradients depict different facets of root economics and, rather than a single one-dimensional spectrum, encompass a whole root economics space of plant strategies for belowground resource acquisition.
MATERIALS AND METHODS
Database
All analyses presented here are based on the GRooT database (15). The GRooT database combines root trait observations from the Fine-Root Ecology Database (FRED) (42) and TRY (43) with additional datasets providing data measured on individual plants for which taxonomical information is available. It includes data on both coarse and fine roots. For the objective of this study, we selected fine roots only, as coarse roots are usually not absorptive and therefore less relevant in the context of root economics (42, 44). We treated roots as fine roots if they met at least one of the following criteria: (i) they were of root orders 1 to 3, (ii) they were classified as “fine roots” by the initial authors, or (iii) their diameter was smaller than 2 mm. Data measured on dead roots were excluded from the analyses. Furthermore, we excluded ferns (Polypodiopsida) because of their very special root morphology that is hardly comparable with vascular plants (45, 46). We only selected data where species-level information was available. During the past decade, a set of root traits was found to be highly informative of root economics: SRL, D, RTD, and root N (8–11, 14, 20, 27). Hence, we focused our main analyses on those four traits. In addition, we analyzed the percentage of root length colonized by mycorrhizal fungi (%M) – while > 99 % of these data refer to arbuscular mycorrhizal colonization - and the root area occupied by the cortex, i.e., CF as proxies for the strength of mycorrhizal symbiosis, as well as mean root life span. We checked the values of these traits for outliers and excluded values of RTD exceeding 1.0 in further analyses.
Categorical data from GRooT such as main biome type (tropical, temperate, continental, arid, or polar) following the Köppen-Geiger classification, plant woodiness (woody, nonwoody, or facultatively woody), mycorrhizal association (NM, arbuscular mycorrhiza, ectomycorrhiza, or other (e.g., ericoid mycorrhiza), and nitrogen-fixing ability (fixers or nonfixers) were used in the downstream analysis and testing of our conceptual framework. GRooT includes mycorrhizal association data from FungalRoot (47), which did not cover our entire species set. To achieve full data cover, we filled the gaps and did minor annotations based on the following general rules:
(i)Mycorrhizal association is constant within species, hence excluding a facultative mycorrhizal type. In cases where a lack of mycorrhizal colonization has been reported only under specific environmental conditions less suitable for the mycorrhizal symbiosis, we assigned the species as mycorrhizal, while in cases with intraradical hyphae but no evidence for a symbiotic interface, we assigned species to be NM.
(ii)Almost all plants have one type of mycorrhizal association as the dominant one. Therefore, dual mycorrhizal association was only assigned if species show no clear dominance toward one type.
(iii)The mycorrhizal association type is usually constant within a monophyletic genus and often within a family (24, 47, 48). Therefore, we filled remaining gaps with the respective mycorrhizal association type of sister species.
Data processing
All data processing and analyses were done using R 3.6.1 (49). In this study, we analyzed how different root traits are related to each other at the level of plant species; hence, a first step was to calculate species mean values. As root traits were measured in different studies varying in design (e.g., on in situ grown plants versus plants growing in pots), and because most traits varied several orders of magnitude, several steps of data processing were required before calculating species mean trait values. First, to obtain normal distributions, we log-transformed each trait, except for %M and CF, which were scaled to the range of 0 to 1 and arcsine square root transformed. We then Z transformed each trait to a mean of 0 and an SD of 1 to assure variance homogeneity. Furthermore, we corrected for main study design (measurements on plants in situ, in pots, or hydroponics) and the publication in which the trait measurements were first reported (as a proxy for other study specific factors, e.g., plant age, soil conditions, or sample handling). This was done by building a linear mixed model for each trait, where the trait was treated as the response variable, study design as a fixed factor, and publication as random factor. We used residuals of these models in further analyses.
Within some species, the categorical traits woodiness and biome had different data entries (e.g., because the species occurs both in temperate and continental biomes). In those cases, we categorized the species in the biome in which it had most observations, and we categorized its woodiness by its most commonly observed entry in further analyses.
In total, we analyzed information of 1547, 1662, 1361, and 1158 species for D, SRL, RTD, and N, respectively. Scientific names in GRooT are standardized among datasets and brought up to date by querying species names using the Taxonomic Name Resolution Service v4.0 (http://tnrs.iplantcollaborative.org/) (50). We constructed a phylogenetic tree including all species using the backbone phylogeny from Zanne et al. (51) and adding additional missing species with the function “add.tips” from the package “phangorn” (52). We calculated Pagel’s λ using the package “picante” and evaluated the strength of the phylogenetic signal for each trait; a large (close to the upper bound of 1.0) Pagel’s λ value indicates higher phylogenetic conservatism (53), whereas a low (close to 0.0) value indicates a lack of phylogenetic conservatism.
Analyses
As all traits exerted strong phylogenetic signal (table S3), we used phylogenetically informed methods for all analyses. We first assessed bivariate relationships between the four core traits (D, SRL, RTD, and N) and CF to build our conceptual framework (Table 1 and Fig. 1), and we also tested for relationships of these traits with %M and root life span (fig. S1). Sample sizes varied for these bivariate correlations, depending on the number of species with complete information for both involved traits, and ranged from 19 (for the correlation between %M and root life span) to 1402 (for the correlation between D and SRL) (fig. S1). In total, we used 1810 species for these bivariate correlations. We fit phylogenetic generalized least square models using the “pgls” function in the R package “caper” (54, 55) to each pair of traits to conduct phylogenetically corrected regression analyses. Phylogenetically corrected correlation coefficients (r values) were then calculated by taking the square root of the adjusted model r2 and by multiplying this with −1 if the regression coefficient was negative. In cases where the adjusted model r2 was negative, we assigned an r coefficient of 0.
We used phylogenetically informed PCA to identify main dimensions of variation among root economic traits. A phylogenetic PCA was performed for the four core traits D, SRL, RTD, and N using the “phyl.pca” function of the “phytools” package (56). There were 748 species that had complete data for these four core traits. The eigenanalysis uses the correlation structure of the phylogeny to inform its estimates of eigenvalues and eigenvectors (56, 57). To assess whether the PCA results and hence the dimensions of the root economics space were sensitive to biome type, mycorrhizal association, woodiness, or nitrogen-fixing ability, we repeated the above analysis for subsets of different biomes [tropics, temperate, continental, and arid—the polar biome was represented by too few species (n = 5) to perform a reliable analysis], mycorrhizal association type (arbuscular mycorrhiza versus ectomycorrhiza), woodiness (woody versus nonwoody), and nitrogen-fixing ability (present or absent).
We assessed whether roots from species with different mycorrhizal associations (arbuscular mycorrhiza, ectomycorrhiza, arbuscular mycorrhiza, and ectomycorrhiza, i.e., intraspecific variation in mycorrhizal association type, ericoid mycorrhiza, and nonmycorrhiza associated), species from different biomes (temperate, tropical, arid, and continental), woody or nonwoody species, and species that either did or did not associate with bacteria able to fix nitrogen, differed significantly from each other in the global multidimensional PCA space, i.e., in their first two PCA axes (which jointly explained 77% of all trait variation). This was done using a permutational multiple analysis of variance, in which the first two PCA axes were treated as the response variables and mycorrhizal association type, biome, woodiness, or ability to fix nitrogen as the fixed factor. We used Euclidean pairwise distances in PCA space among species and calculated 999 permutations using the “pairwise.adonis” function in the “pairwiseAdonis” package (58). To test for the significance of differences between different categories of mycorrhizal associations and biomes, we used false discovery rates (59) to reduce the likelihood of type I errors due to multiple testing.
Furthermore, we investigated multivariate trait space for seven traits, i.e., the four core traits from the above-described PCAs (D, SRL, RTD, and N) supplemented by three additional traits: CF, %M, and root life span. Observation-based PCA requires each replicate (species) to have complete data for all traits, but there were few species with a complete set of all seven traits. Therefore, we performed an alternative dimensionality reduction analysis based on pairwise correlations between traits. For this analysis, we computed phylogenetically informed pairwise correlations between each of the 21 trait combinations, where each trait combination included a slightly different set of species for which traits were available (fig. S1). We then performed a standard eigenanalysis on this positive definite matrix of phylogenetic correlation coefficients (60).
Visual examination of the distribution of traits across the phylogeny was obtained using the function “phylo.heatmap” in the package “phytools” (56). We further examined the phylogenetic trends observed across broader phylogenetic clades of seed plants using a randomization test to quantitatively compare individual clade trait values to the rest of the phylogeny. The test determines whether the mean trait value observed in a clade deviates significantly from the population mean under the null hypothesis that the trait has a random phylogenetic distribution. To do so, we created an algorithm in R that selected clades sequentially at each node. Because of the large number of species, we selected particular nodes that enveloped important phylogenetic clades with at least 30 species (tree tips) included. For each clade, we calculated the observed mean and kurtosis values as measures of central tendency and dispersion values within clades, respectively. Then, we generated a series of 999 random values shuffling trait values among the tips of the original tree. Significance was calculated after estimating if the observed clade mean or kurtosis were outside the 95% confidence intervals of the clade estimations using the randomized datasets. In the case of the kurtosis, values higher than the randomized mean were interpreted as evidence of underdispersion in the clade (leptokurtic distribution), whereas lower values were considered sign of overdispersion (platykurtic distribution).
Supplementary Material
Acknowledgments
We would like to thank I. Mansour for text editing. Funding: This paper is a joint effort of the working group sROOT supported by sDiv, the Synthesis Centre of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118). The sROOT workshops and L.M. were also supported by NWO-Vidi grant 864.14.006. J.B. was supported by DFG grants RI-1815/20-1 and RI 1815/22-1. C.M.I., M.L.M., and FRED were supported by the U.S. Department of Energy’s Office of Science, Biological and Environmental Research Program. Author contributions: J.B., A.W., and L.M. conceived the idea for the project. All authors were involved in collecting datasets, developing the conceptual framework, and interpreting the results. J.B., F.v.d.P., D.C.L., N.G.-R., O.J.V.-B., and L.M.Y. performed the statistical analyses. T.W.K. annotated the mycorrhizal associations. J.B., A.W., C.M.I., and L.M. wrote the first draft of the manuscript. All authors commented on and agreed with the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data analyzed in the study originate from the GRooT database (15), which is publicly available on GitHub (https://github.com/GRooT-Database/GRooT-Data). The R script including all analyses and figure preparations is available from the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/27/eaba3756/DC1
REFERENCES AND NOTES
- 1.Reich P. B., The world-wide `fast-slow’ plant economics spectrum: A traits manifesto. J. Ecol. 102, 275–301 (2014). [Google Scholar]
- 2.Wright I. J., Reich P. B., Westoby M., Ackerly D. D., Baruch Z., Bongers F., Cavender-Bares J., Chapin T., Cornelissen J. H. C., Diemer M., Flexas J., Garnier E., Groom P. K., Gulias J., Hikosaka K., Lamont B. B., Lee T., Lee W., Lusk C., Midgley J. J., Navas M.-L., Niinemets U., Oleksyn J., Osada N., Poorter H., Poot P., Prior L., Pyankov V. I., Roumet C., Thomas S. C., Tjoelker M. G., Veneklaas E. J., Villar R., The worldwide leaf economics spectrum. Nature 428, 821–827 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Díaz S., Kattge J., Cornelissen J. H. C., Wright I. J., Lavorel S., Dray S., Reu B., Kleyer M., Wirth C., Prentice I. C., Garnier E., Bönisch G., Westoby M., Poorter H., Reich P. B., Moles A. T., Dickie J., Gillison A. N., Zanne A. E., Chave J., Wright S. J., Sheremet’ev S. N., Jactel H., Baraloto C., Cerabolini B., Pierce S., Shipley B., Kirkup D., Casanoves F., Joswig J. S., Günther A., Falczuk V., Rüger N., Mahecha M. D., Gorné L. D., The global spectrum of plant form and function. Nature 529, 167–171 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Eissenstat D. M., Costs and benefits of constructing roots of small diameter. J. Plant Nutr. 15, 763–782 (1992). [Google Scholar]
- 5.Freschet G. T., Cornelissen J. H. C., van Logtestijn R. S. P., Aerts R., Evidence of the ‘plant economics spectrum’ in a subarctic flora. J. Ecol. 98, 362–373 (2010). [Google Scholar]
- 6.Ostonen I., Püttsepp Ü., Biel C., Alberton O., Bakker M. R., Lõhmus K., Majdi H., Metcalfe D., Olsthoorn A. F. M., Pronk A., Vanguelova E., Weih M., Brunner I., Specific root length as an indicator of environmental change. Plant Biosyst. 141, 426–442 (2007). [Google Scholar]
- 7.Ryser P., Eek L., Consequences of phenotypic plasticity vs. interspecific differences in leaf and root traits for acquisition of aboveground and belowground resources. Am. J. Bot. 87, 402–411 (2000). [PubMed] [Google Scholar]
- 8.Weemstra M., Mommer L., Visser E. J. W., van Ruijven J., Kuyper T. W., Mohren G. M. J., Sterck F. J., Towards a multidimensional root trait framework: A tree root review. New Phytol. 211, 1159–1169 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kramer-Walter K. R., Bellingham P. J., Millar T. R., Smissen R. D., Richardson S. J., Laughlin D. C., Root traits are multidimensional: Specific root length is independent from root tissue density and the plant economic spectrum. J. Ecol. 104, 1299–1310 (2016). [Google Scholar]
- 10.Kong D., Wang J., Wu H., Valverde-Barrantes O. J., Wang R., Zeng H., Kardol P., Zhang H., Feng Y., Nonlinearity of root trait relationships and the root economics spectrum. Nat. Commun. 10, 2203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann J., Ryo M., Prati D., Hempel S., Rillig M. C., Roots traits are more than analogues of leaf traits : The case for diaspore mass. New Phytol. 216, 1130–1139 (2017). [DOI] [PubMed] [Google Scholar]
- 12.McCormack M. L., Iversen C. M., Physical and functional constraints on viable belowground acquisition strategies. Front. Plant Sci. 10, 1215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comas L. H., Mueller K. E., Taylor L. L., Midford P. E., Callahan H. S., Beerling D. J., Evolutionary patterns and biogeochemical significance of angiosperm root traits. Int. J. Plant Sci. 173, 584–595 (2012). [Google Scholar]
- 14.Ma Z., Guo D., Xu X., Lu M., Bardgett R. D., Eissenstat D. M., McCormack M. L., Hedin L. O., Evolutionary history resolves global organization of root functional traits. Nature 555, 94–97 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Guerrero-Ramirez N., Mommer L., Freschet G. T., Iversen C. M., McCormack M. L., Kattge J., Poorter H., van der Plas F., Bergmann J., Kuyper T. W., York L. M., Bruelheide H., Laughlin D. C., Meier I. C., Roumet C., Semchenko M., Sweeney C. J., van Ruijven J., Valverde-Barrantes O. J., Aubin I., Catford J. A., Manning P., Martin A., Milla R., Minden V., Pausas J. G., Smith S. W., Soudzilovskaia N. A., Ammer C., Butterfield B., Craine J., Cornelissen J. H. C., de Vries F. T., Isaac M. E., Kramer K., König C., Lamb E. G., Onipchenko V. G., Peñuelas J., Reich P. B., Rillig M. C., Sack L., Shipley B., Tedersoo L., Valladares F., van Bodegom P., Weigelt P., Wright J. P., Weigelt A., Global Root Traits (GRooT) Database. bioRxiv 10.1101/2020.05.17.095851, (2020). [Google Scholar]
- 16.Rose L., Pitfalls in root trait calculations: How ignoring diameter heterogeneity can lead to overestimation of functional traits. Front. Plant Sci. 8, 898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom A. J., Chapin F. S. III, Mooney H. A., Resource limitation in plants–An economic analogy. Annu. Rev. Ecol. Syst. 16, 363–392 (1985). [Google Scholar]
- 18.Chen W., Zeng H., Eissenstat D. M., Guo D., Variation of first-order root traits across climatic gradients and evolutionary trends in geological time. Glob. Ecol. Biogeogr. 22, 846–856 (2013). [Google Scholar]
- 19.Kong D., Ma C., Zhang Q., Li L., Chen X., Zeng H., Guo D., Leading dimensions in absorptive root trait variation across 96 subtropical forest species. New Phytol. 203, 863–872 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Roumet C., Birouste M., Picon-Cochard C., Ghestem M., Osman N., Vrignon-Brenas S., Cao K., Stokes A., Root structure - function relationships in 74 species: Evidence of a root economics spectrum related to carbon economy. New Phytol. 210, 815–826 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Valverde-Barrantes O. J., Blackwood C. B., Root traits are multidimensional: Specific root length is independent from root tissue density and the plant economic spectrum: Commentary on Kramer-Walter et al. (2016). J. Ecol. 104, 1311–1313 (2016). [Google Scholar]
- 22.Valverde-Barrantes O. J., Horning A. L., Smemo K. A., Blackwood C. B., Phylogenetically structured traits in root systems influence arbuscular mycorrhizal colonization in woody angiosperms. Plant and Soil 404, 1–12 (2016). [Google Scholar]
- 23.Brundrett M. C., Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Brundrett M. C., Tedersoo L., Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 220, 1108–1115 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Horan D. P., Chilvers G. A., Lapeyrie F. F., Time sequence of the infection process eucalypt ectomycorrhizas. New Phytol. 109, 451–458 (1988). [Google Scholar]
- 26.Hummel I., Vile D., Violle C., Devaux J., Ricci B., Blanchard A., Garnier É., Roumet C., Relating root structure and anatomy to whole-plant functioning in 14 herbaceous Mediterranean species. New Phytol. 173, 313–321 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Valverde-Barrantes O. J., Freschet G. T., Roumet C., Blackwood C. B., A worldview of root traits: The influence of ancestry, growth form, climate and mycorrhizal association on the functional trait variation of fine-root tissues in seed plants. New Phytol. 215, 1562–1573 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Cairney J. W. G., Evolution of mycorrhiza systems. Naturwissenschaften 87, 467–475 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Lin G., McCormack M. L., Ma C., Guo D., Similar below-ground carbon cycling dynamics but contrasting modes of nitrogen cycling between arbuscular mycorrhizal and ectomycorrhizal forests. New Phytol. 213, 1440–1451 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Phillips R. P., Brzostek E., Midgley M. G., The mycorrhizal-associated nutrient economy: A new framework for predicting carbon-nutrient couplings in temperate forests. New Phytol. 199, 41–51 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Comas L. H., Callahan H. S., Midford P. E., Patterns in root traits of woody species hosting arbuscular and ectomycorrhizas: Implications for the evolution of belowground strategies. Ecol. Evol. 4, 2979–2990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treseder K. K., The extent of mycorrhizal colonization of roots and its influence on plant growth and phosphorus content. Plant and Soil 371, 1–13 (2013). [Google Scholar]
- 33.Laliberté E., Below-ground frontiers in trait-based plant ecology. New Phytol. 213, 1597–1603 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Wang R., Wang Q., Zhao N., Xu Z., Zhu X., Jiao C., Yu G., He N., Different phylogenetic and environmental controls of first-order root morphological and nutrient traits: Evidence of multidimensional root traits. Funct. Ecol. 32, 29–39 (2017). [Google Scholar]
- 35.Tjoelker M. G., Craine J. M., Wedin D., Reich P. B., Tilman D., Linking leaf and root trait syndromes among 39 grassland and savannah species. New Phytol. 167, 493–508 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Mommer L., Weemstra M., The role of roots in the resource economics spectrum. New Phytol. 195, 725–727 (2012). [DOI] [PubMed] [Google Scholar]
- 37.McCormack M. L., Adams T. S., Smithwick E. A. H., Eissenstat D. M., Predicting fine root lifespan from plant functional traits in temperate trees. New Phytol. 195, 823–831 (2012). [DOI] [PubMed] [Google Scholar]
- 38.van der Heijden M. G. A., Martin F. M., Selosse M.-A., Sanders I. R., Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytol. 205, 1406–1423 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Maherali H., Oberle B., Stevens P. F., Cornwell W. K., McGlinn D. J., Mutualism persistence and abandonment during the evolution of the mycorrhizal symbiosis. Am. Nat. 188, E113–E125 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Weemstra M., Sterck F. J., Visser E. J. W., Kuyper T. W., Goudzwaard L., Mommer L., Fine-root trait plasticity of beech (Fagus sylvatica) and spruce (Picea abies) forests on two contrasting soils. Plant and Soil 415, 175–188 (2016). [Google Scholar]
- 41.Fort F., Volaire F., Guilioni L., Barkaoui K., Navas M.-L., Roumet C., Root traits are related to plant water-use among rangeland Mediterranean species. Funct. Ecol. 31, 1700–1709 (2017). [Google Scholar]
- 42.Iversen C. M., McCormack M. L., Powell A. S., Blackwood C. B., Freschet G. T., Kattge J., Roumet C., Stover D. B., Soudzilovskaia N. A., Valverde-Barrantes O. J., van Bodegom P. M., Violle C., A global fine-root ecology database to address below-ground challenges in plant ecology. New Phytol. 215, 15–26 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Kattge J., Bönisch G., Díaz S., Lavorel S., Prentice I. C., Leadley P., Tautenhahn S., Werner G. D. A., Aakala T., Abedi M., Acosta A. T. R., Adamidis G. C., Adamson K., Aiba M., Albert C. H., Alcántara J. M., Carolina Alcázar C., Aleixo I., Ali H., Amiaud B., Ammer C., Amoroso M. M., Anand M., Anderson C., Anten N., Antos J., Apgaua D. M. G., Ashman T.-L., Asmara D. H., Asner G. P., Aspinwall M., Atkin O., Aubin I., Baastrup-Spohr L., Bahalkeh K., Bahn M., Baker T., Baker W. J., Bakker J. P., Baldocchi D., Baltzer J., Banerjee A., Baranger A., Barlow J., Barneche D. R., Baruch Z., Bastianelli D., Battles J., Bauerle W., Bauters M., Bazzato E., Beckmann M., Beeckman H., Beierkuhnlein C., Bekker R., Belfry G., Belluau M., Beloiu M., Benavides R., Benomar L., Berdugo-Lattke M. L., Berenguer E., Bergamin R., Bergmann J., Carlucci M. B., Berner L., Bernhardt-Römermann M., Bigler C., Bjorkman A. D., Blackman C., Blanco C., Blonder B., Blumenthal D., Bocanegra-González K. T., Boeckx P., Bohlman S., Böhning-Gaese K., Boisvert-Marsh L., Bond W., Bond-Lamberty B., Boom A., Boonman C. C. F., Bordin K., Boughton E. H., Boukili V., Bowman D. M. J. S., Bravo S., Brendel M. R., Broadley M. R., Brown K. A., Bruelheide H., Brumnich F., Bruun H. H., Bruy D., Buchanan S. W., Bucher S. F., Buchmann N., Buitenwerf R., Bunker D. E., Bürger J., Burrascano S., Burslem D. F. R. P., Butterfield B. J., Byun C., Marques M., Scalon M. C., Caccianiga M., Cadotte M., Cailleret M., Camac J., Camarero J. J., Campany C., Campetella G., Campos J. A., Cano-Arboleda L., Canullo R., Carbognani M., Carvalho F., Casanoves F., Castagneyrol B., Catford J. A., Cavender-Bares J., Cerabolini B. E. L., Cervellini M., Chacón-Madrigal E., Chapin K., Chapin F. S., Chelli S., Chen S.-C., Chen A., Cherubini P., Chianucci F., Choat B., Chung K.-S., Chytrý M., Ciccarelli D., Coll L., Collins C. G., Conti L., Coomes D., Cornelissen J. H. C., Cornwell W. K., Corona P., Coyea M., Craine J., Craven D., Cromsigt J. P. G. M., Csecserits A., Cufar K., Cuntz M., da Silva A. C., Dahlin K. M., Dainese M., Dalke I., Fratte M. D., Dang-Le A. T., Danihelka J., Dannoura M., Dawson S., de Beer A. J., De Frutos A., De Long J. R., Dechant B., Delagrange S., Delpierre N., Derroire G., Dias A. S., Diaz-Toribio M. H., Dimitrakopoulos P. G., Dobrowolski M., Doktor D., Dřevojan P., Dong N., Dransfield J., Dressler S., Duarte L., Ducouret E., Dullinger S., Durka W., Duursma R., Dymova O., E-Vojtkó A., Eckstein R. L., Ejtehadi H., Elser J., Emilio T., Engemann K., Erfanian M. B., Erfmeier A., Esquivel-Muelbert A., Esser G., Estiarte M., Domingues T. F., Fagan W. F., Fagúndez J., Falster D. S., Fan Y., Fang J., Farris E., Fazlioglu F., Feng Y., Fernandez-Mendez F., Ferrara C., Ferreira J., Fidelis A., Finegan B., Firn J., Flowers T. J., Flynn D. F. B., Fontana V., Forey E., Forgiarini C., François L., Frangipani M., Frank D., Frenette-Dussault C., Freschet G. T., Fry E. L., Fyllas N. M., Mazzochini G. G., Gachet S., Gallagher R., Ganade G., Ganga F., García-Palacios P., Gargaglione V., Garnier E., Garrido J. L., de Gasper A. L., Gea-Izquierdo G., Gibson D., Gillison A. N., Giroldo A., Glasenhardt M.-C., Gleason S., Gliesch M., Goldberg E., Göldel B., Gonzalez-Akre E., Gonzalez-Andujar J. L., González-Melo A., González-Robles A., Graae B. J., Granda E., Graves S., Green W. A., Gregor T., Gross N., Guerin G. R., Günther A., Gutiérrez A. G., Haddock L., Haines A., Hall J., Hambuckers A., Han W., Harrison S. P., Hattingh W., Hawes J. E., He T., He P., Heberling J. M., Helm A., Hempel S., Hentschel J., Hérault B., Hereş A.-M., Herz K., Heuertz M., Hickler T., Hietz P., Higuchi P., Hipp A. L., Hirons A., Hock M., Hogan J. A., Holl K., Honnay O., Hornstein D., Hou E., Hough-Snee N., Hovstad K. A., Ichie T., Igić B., Illa E., Isaac M., Ishihara M., Ivanov L., Ivanova L., Iversen C. M., Izquierdo J., Jackson R. B., Jackson B., Jactel H., Jagodzinski A. M., Jandt U., Jansen S., Jenkins T., Jentsch A., Jespersen J. R. P., Jiang G.-F., Johansen J. L., Johnson D., Jokela E. J., Joly C. A., Jordan G. J., Joseph G. S., Junaedi D., Junker R. R., Justes E., Kabzems R., Kane J., Kaplan Z., Kattenborn T., Kavelenova L., Kearsley E., Kempel A., Kenzo T., Kerkhoff A., Khalil M. I., Kinlock N. L., Kissling W. D., Kitajima K., Kitzberger T., Kjøller R., Klein T., Kleyer M., Klimešová J., Klipel J., Kloeppel B., Klotz S., Knops J. M. H., Kohyama T., Koike F., Kollmann J., Komac B., Komatsu K., König C., Kraft N. J. B., Kramer K., Kreft H., Kühn I., Kumarathunge D., Kuppler J., Kurokawa H., Kurosawa Y., Kuyah S., Laclau J.-P., Lafleur B., Lallai E., Lamb E., Lamprecht A., Larkin D. J., Laughlin D., Le Bagousse-Pinguet Y., le Maire G., le Roux P. C., le Roux E., Lee T., Lens F., Lewis S. L., Lhotsky B., Li Y., Li X., Lichstein J. W., Liebergesell M., Lim J. Y., Lin Y.-S., Linares J. C., Liu C., Liu D., Liu U., Livingstone S., Llusià J., Lohbeck M., López-García Á., Lopez-Gonzalez G., Lososová Z., Louault F., Lukács B. A., Lukeš P., Luo Y., Lussu M., Ma S., Pereira C. M. R., Mack M., Maire V., Mäkelä A., Mäkinen H., Malhado A. C. M., Mallik A., Manning P., Manzoni S., Marchetti Z., Marchino L., Marcilio-Silva V., Marcon E., Marignani M., Markesteijn L., Martin A., Martínez-Garza C., Martínez-Vilalta J., Mašková T., Mason K., Mason N., Massad T. J., Masse J., Mayrose I., Carthy J. M., McCormack M. L., Culloh K. M., Mc Fadden I. R., Mc Gill B. J., Mc Partland M. Y., Medeiros J. S., Medlyn B., Meerts P., Mehrabi Z., Meir P., Melo F. P. L., Mencuccini M., Meredieu C., Messier J., Mészáros I., Metsaranta J., Michaletz S. T., Michelaki C., Migalina S., Milla R., Miller J. E. D., Minden V., Ming R., Mokany K., Moles A. T., Molnár V. A., Molofsky J., Molz M., Montgomery R. A., Monty A., Moravcová L., Moreno-Martínez A., Moretti M., Mori A. S., Mori S., Morris D., Morrison J., Mucina L., Mueller S., Muir C. D., Müller S. C., Munoz F., Myers-Smith I. H., Myster R. W., Nagano M., Naidu S., Narayanan A., Natesan B., Negoita L., Nelson A. S., Neuschulz E. L., Ni J., Niedrist G., Nieto J., Niinemets Ü., Nolan R., Nottebrock H., Nouvellon Y., Novakovskiy A.; Nuttrient Network, Nystuen K. O., O’Grady A., O’Hara K., O’Reilly-Nugent A., Oakley S., Oberhuber W., Ohtsuka T., Oliveira R., Öllerer K., Olson M. E., Onipchenko V., Onoda Y., Onstein R. E., Ordonez J. C., Osada N., Ostonen I., Ottaviani G., Otto S., Overbeck G. E., Ozinga W. A., Pahl A. T., Paine C. E. T., Pakeman R. J., Papageorgiou A. C., Parfionova E., Pärtel M., Patacca M., Paula S., Paule J., Pauli H., Pausas J. G., Peco B., Penuelas J., Perea A., Peri P. L., Petisco-Souza A. C., Petraglia A., Petritan A. M., Phillips O. L., Pierce S., Pillar V. D., Pisek J., Pomogaybin A., Poorter H., Portsmuth A., Poschlod P., Potvin C., Pounds D., Powell A. S., Power S. A., Prinzing A., Puglielli G., Pyšek P., Raevel V., Rammig A., Ransijn J., Ray C. A., Reich P. B., Reichstein M., Reid D. E. B., Réjou-Méchain M., de Dios V. R., Ribeiro S., Richardson S., Riibak K., Rillig M. C., Riviera F., Robert E. M. R., Roberts S., Robroek B., Roddy A., Rodrigues A. V., Rogers A., Rollinson E., Rolo V., Römermann C., Ronzhina D., Roscher C., Rosell J. A., Rosenfield M. F., Rossi C., Roy D. B., Royer-Tardif S., Rüger N., Ruiz-Peinado R., Rumpf S. B., Rusch G. M., Ryo M., Sack L., Saldaña A., Salgado-Negret B., Salguero-Gomez R., Santa-Regina I., Santacruz-García A. C., Santos J., Sardans J., Schamp B., Scherer-Lorenzen M., Schleuning M., Schmid B., Schmidt M., Schmitt S., Schneider J. V., Schowanek S. D., Schrader J., Schrodt F., Schuldt B., Schurr F., Garvizu G. S., Semchenko M., Seymour C., Sfair J. C., Sharpe J. M., Sheppard C. S., Sheremetiev S., Shiodera S., Shipley B., Shovon T. A., Siebenkäs A., Sierra C., Silva V., Silva M., Sitzia T., Sjöman H., Slot M., Smith N. G., Sodhi D., Soltis P., Soltis D., Somers B., Sonnier G., Sørensen M. V., Sosinski E. E. Jr., Soudzilovskaia N. A., Souza A. F., Spasojevic M., Sperandii M. G., Stan A. B., Stegen J., Steinbauer K., Stephan J. G., Sterck F., Stojanovic D. B., Strydom T., Suarez M. L., Svenning J.-C., Svitková I., Svitok M., Svoboda M., Swaine E., Swenson N., Tabarelli M., Takagi K., Tappeiner U., Tarifa R., Tauugourdeau S., Tavsanoglu C., te Beest M., Tedersoo L., Thiffault N., Thom D., Thomas E., Thompson K., Thornton P. E., Thuiller W., Tichý L., Tissue D., Tjoelker M. G., Tng D. Y. P., Tobias J., Török P., Tarin T., Torres-Ruiz J. M., Tóthmérész B., Treurnicht M., Trivellone V., Trolliet F., Trotsiuk V., Tsakalos J. L., Tsiripidis I., Tysklind N., Umehara T., Usoltsev V., Vadeboncoeur M., Vaezi J., Valladares F., Vamosi J., van Bodegom P. M., van Breugel M., van Cleemput E., van de Weg M., van der Merwe S., van der Plas F., van der Sande M. T., van Kleunen M., van Meerbeek K., Vanderwel M., Vanselow K. A., Vårhammar A., Varone L., Valderrama M. Y. V., Vassilev K., Vellend M., Veneklaas E. J., Verbeeck H., Verheyen K., Vibrans A., Vieira I., Villacís J., Violle C., Vivek P., Wagner K., Waldram M., Waldron A., Walker A. P., Waller M., Walther G., Wang H., Wang F., Wang W., Watkins H., Watkins J., Weber U., Weedon J. T., Wei L., Weigelt P., Weiher E., Wells A. W., Wellstein C., Wenk E., Westoby M., Westwood A., White P. J., Whitten M., Williams M., Winkler D. E., Winter K., Womack C., Wright I. J., Wright S. J., Wright J., Pinho B. X., Ximenes F., Yamada T., Yamaji K., Yanai R., Yankov N., Yguel B., Zanini K. J., Zanne A. E., Zelený D., Zhao Y.-P., Zheng J., Zheng J., Ziemińska K., Zirbel C. R., Zizka G., Zo-Bi I. C., Zotz G., Wirth C., TRY plant trait database – Enhanced coverage and open access. Glob. Chang. Biol. 26, 119–188 (2019). [DOI] [PubMed] [Google Scholar]
- 44.McCormack M. L., Dickie I. A., Eissenstat D. M., Fahey T. J., Fernandez C. W., Guo D., Helmisaari H.-S., Hobbie E. A., Iversen C. M., Jackson R. B., Leppälammi-Kujansuu J., Norby R. J., Phillips R. P., Pregitzer K. S., Pritchard S. G., Rewald B., Zadworny M., Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol. 207, 505–518 (2015). [DOI] [PubMed] [Google Scholar]
- 45.W. Troll, Vergleichende Morphologie der Pflanzen (Verlag der Gebrüder Borntraeger, 1943).
- 46.P. Raven, R. F. Evert, S. E. Eichhorn, Biology of plants (W.H. Freeman and Company Publisher, ed. 8, 2013). [Google Scholar]
- 47.Soudzilovskaia N. A., Vaessen S., Barcelo M., He J., Rahimlou S., Abarenkov K., Brundrett M. C., Gomes S. I. F., Merckx V., Tedersoo L., FungalRoot: Global online database of plant mycorrhizal associations. bioRxiv 10.1101/717488, (2019). [DOI] [PubMed] [Google Scholar]
- 48.Brundrett M., Tedersoo L., Misdiagnosis of mycorrhizas and inappropriate recycling of data can lead to false conclusions. New Phytol. 221, 18–24 (2019). [DOI] [PubMed] [Google Scholar]
- 49.R Core Team, R: A language and environment for statistical computing (2019).
- 50.Boyle B., Hopkins N., Lu Z., Raygoza Garay J. A., Mozzherin D., Rees T., Matasci N., Narro M. L., Piel W. H., Mckay S. J., Lowry S., Freeland C., Peet R. K., Enquist B. J., The taxonomic name resolution service: An online tool for automated standardization of plant names. BMC Bioinformatics 14, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanne A. E., Tank D. C., Cornwell W. K., Eastman J. M., Smith S. A., FitzJohn R. G., McGlinn D. J., O’Meara B. C., Moles A. T., Reich P. B., Royer D. L., Soltis D. E., Stevens P. F., Westoby M., Wright I. J., Aarssen L., Bertin R. I., Calaminus A., Govaerts R., Hemmings F., Leishman M. R., Oleksyn J., Soltis P. S., Swenson N. G., Warman L., Beaulieu J. M., Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Schliep K. P., Phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagel M., Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
- 54.D. Orme, R. Freckleton, D. Thomas, T. Petzoldt, S. Fritz, N. Isaac, W. Pearse, Caper: Comparative Analyses of Phylogenetics and Evolution in R (2018).
- 55.Freckleton R. P., Harvey P. H., Pagel M., Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 160, 712–726 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 57.Revell L. J., Size-correction and principal components for interspecific comparative studies. Evolution 63, 3258–3268 (2009). [DOI] [PubMed] [Google Scholar]
- 58.P. Martinez Arbizu, pairwiseAdonis: Pairwise Multilevel Comparison using Adonis (R Packag. version 0.3, 2019).
- 59.Benjamini Y., Hochberg Y., Controlling the false discovery rate : A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57, 289–300 (1995). [Google Scholar]
- 60.D. C. Lay, Linear algebra and its applications (Pearson, 2006).
- 61.Freschet G. T., Valverde-Barrantes O. J., Tucker C. M., Craine J. M., Mccormack M. L., Violle C., Fort F., Blackwood C. B., Urban-Mead K. R., Iversen C. M., Bonis A., Comas L. H., Cornelissen J. H. C., Dong M., Guo D., Hobbie S. E., Holdaway R. J., Kembel S. W., Makita N., Onipchenko V. G., Picon-Cochard C., Reich P. B., de la Riva E. G., Smith S. W., Soudzilovskaia N. A., Tjoelker M. G., Wardle D. A., Roumet C., Climate, soil and plant functional types as drivers of global fine-root trait variation. J. Ecol. 105, 1182–1196 (2017). [Google Scholar]
- 62.van Velzen R., Holmer R., Bu F., Rutten L., van Zeijl A., Liu W., Santuari L., Cao Q., Sharma T., Shen D., Roswanjaya Y., Wardhani T. A. K., Kalhor M. S., Jansen J., van den Hoogen J., Güngör B., Hartog M., Hontelez J., Verver J., Yang W.-C., Schijlen E., Repin R., Schilthuizen M., Schranz M. E., Heidstra R., Miyata K., Fedorova E., Kohlen W., Bisseling T., Smit S., Geurts R., Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. U.S.A. 115, E4700–E4709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/27/eaba3756/DC1


