ABSTRACT
Immune checkpoint blockade leads to unprecedented responses in many cancers. Although currently available agents mostly target the PD-1 and CTLA-4 pathways, agents targeting the immune checkpoint protein LAG-3 are under active clinical development, and early clinical data show that LAG-3 expression is a biomarker of response to LAG-3 blockade. To determine which cancers may benefit most from LAG-3 blockade, we performed a pan-cancer analysis of The Cancer Genome Atlas dataset to identify genomic and immunologic correlates of LAG-3 expression. High mutation burden, and expression of exogenous virus (EBV, HPV) or endogenous retrovirus (ERV3-2), were associated with overexpression of LAG-3 in multiple cancers. Although CD8+ T-cell marker (CD8A) and LAG-3 were strongly co-expressed with each other and with PD-L1 in most cancers, there were three notable exceptions: HPV+ head-neck squamous cell cancer, renal cell cancer, and glioblastoma. These results may have important implications for guiding development clinical trials of LAG-3 blockade.
KEYWORDS: Immune checkpoint blockade, LAG-3, mutation burden, virus, endogenous retrovirus
Introduction
Immune checkpoint blockade leads to impressive and durable responses in multiple cancer types, and at present, both PD-1 and CTLA-4 blockade have been FDA approved for the treatment of several cancer types.1–3 Apart from PD-1 and CTLA-4 pathways, there are several additional checkpoint genes, including LAG3 (LAG-3), the BTLA-HVEM pathway, the TIM3-GAL9 pathway, KIR3DL1 (KIR), ADORA2A (A2aR), CD276 (B7-H3) and VTCN1 (B7-H4), that can inhibit T-cell responses4 and are being investigated as therapeutic targets for immune checkpoint blockade. Clinical trials utilizing anti-LAG-3 antibodies are underway to evaluate the safety and efficacy of LAG-3 blockade alone or in combination with PD-1 blockade with or without CTLA-4 blockade.
In a small cohort (N = 61) of melanoma patients, all of whom had progressed on prior treatment with PD-L1 and/or CTLA-4 targeting antibodies, a combination of LAG-3 blockade and PD-1 blockade was recently reported to have clinically significant anti-tumor activity, with 11.5% objective response rate and 49% disease control rate.5 This finding suggests that LAG-3 is an important immune checkpoint target in melanoma, and there is a need to identify which patients may benefit most from LAG-3 inhibition, either alone or in combination with PD-1 blockade.
In this initial melanoma study, the objective response rate to combination LAG-3 blockade and PD-1 blockade was 3.5-fold higher (18% vs 5%) in patients with immunohistochemistry-based LAG-3 expression ≥1% vs <1%, although both groups had previously progressed on PD-1 blockade.5 These observations suggest that LAG-3 expression is a biomarker of response to LAG-3 blockade, like CD274 (PD-L1) expression is in case of PD-1 blockade.6 Therefore, we performed a pan-cancer analysis of The Cancer Genome Atlas (TCGA) to identify genomic and immunologic correlates of LAG-3 expression, which may potentially help predict response to LAG-3 blockade.
Materials and methods
RNA-seq data of tumors for all cancer types in TCGA were obtained from the Broad Genome Data Analysis Center (http://gdac.broadinstitute.org) and the TCGA Data Portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/), and were median adjusted and log2 transformed in the same way as previously described.7 ERBB2 focal copy number data and ESR1 mRNA expression data from the Broad Genome Data Analysis Center were used to classify breast cancer samples into clinical subtypes (ER+/HER2−, ER−/HER2−, HER2+), and these subtypes were analyzed separately. Hyper-mutation status,8 Epstein-Barr virus (EBV) status,9 Hepatitis B and C (HBV and HCV) status,10 and ERV3-2 expression data11 were obtained from recently published studies. Human papillomavirus (HPV) status was obtained from auxiliary clinical files from the TCGA Data Portal. Wilcoxon rank-sum test was used for all pairwise comparisons. Spearman Rho was used for all correlations, and P-values were calculated using either the exact permutation distributions (for small sample sizes) or large-sample approximations. All P-values are from two-sided tests, and P < .05 was used as the threshold for statistical significance. Multiple hypothesis testing correction was performed using Benjamini-Hochberg procedure. A set of 29 clear cell renal cell carcinoma (RCC) samples from Rutgers Cancer Institute of New Jersey (CINJ), obtained with consent under an IRB approved Total Cancer Care® Protocol through the Oncology Research Information Exchange Network (ORIEN) (Pro20150001762), that underwent whole exome sequencing and RNA sequencing, was used for validation. De-identified RNA sequencing data from this group were analyzed in this study.
Results
LAG-3 is co-expressed with CD8A and PD-L1 in most cancer types
Analysis of LAG-3 expression in TCGA dataset showed a wide range of expression among different cancer types (Figure 1). The highest median expression was observed in diffuse large B-cell lymphoma. Multiple solid tumors, including melanoma, cervical cancer, and head and neck squamous cell cancer had considerably high expression of LAG-3. Interestingly, outlier high expression of LAG-3 was observed in a subset of cases in several cancer types, including endometrial cancer, liver cancer, and RCC.
Figure 1.
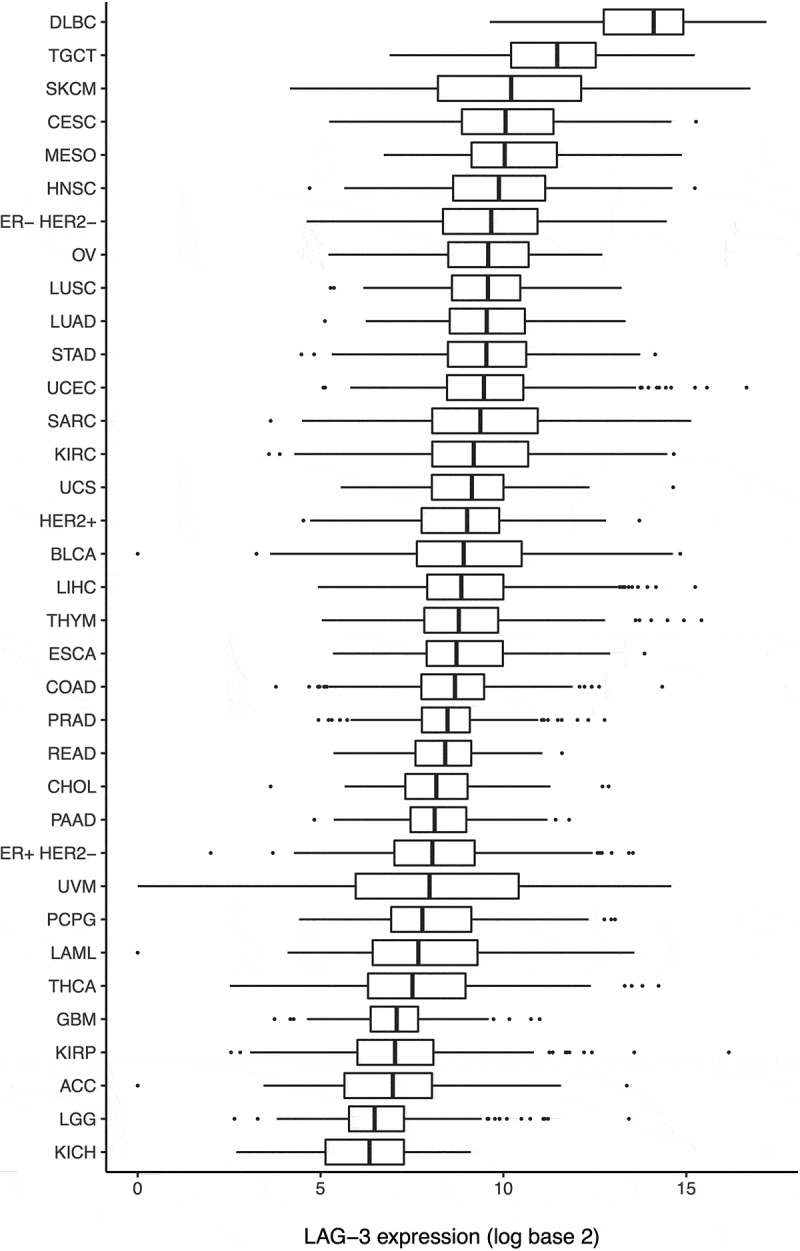
Expression of LAG-3 in various cancer types.
A pan-cancer analysis to identify genes whose expressions correlated (Spearman Rho > 0, P < .05) or anti-correlated (Spearman Rho < 0, P < .05) with LAG-3 expression showed that the number of cancer types in which a gene’s expression correlated with LAG-3 expression (minus the number of cancer types in which the gene’s expression anti-correlated with LAG-3 expression) had a bimodal distribution (Supplementary Figure 1). Pathway enrichment analysis of the genes in the second mode (i.e. genes whose expressions were correlated with LAG-3 expression in unusually high numbers of cancer types) using the ToppGene suite12 showed that these genes are involved in adaptive immune system, innate immune system, cytokine signaling, and interferon-gamma signaling (Supplementary Figure 1).
CD8+T-cells infiltrates in cancer are correlated with local immune activation and response to immune checkpoint therapy.13 To evaluate whether LAG-3 expression was associated with CD8+ T-cell infiltration, we tested for correlation between LAG-3 expression and expression of the cytotoxic T-cell marker CD8A. We found that CD8A expression was strongly correlated (Spearman Rho > 0.5) with LAG-3 expression in almost every cancer type (Figure 2(a)), suggesting that tumors with strong CD8+ T-cell infiltration tend to have high LAG3-expression. This strong correlation may be due to the expression of LAG-3 on a subset of CD8+ T cells.14-17 CD8A expression was also correlated (although less strongly, Spearman Rho < 0.5) with PD-L1 expression in most cancer types (Figure 2(b)), suggesting that the presence of cytotoxic T-cells correlates with both LAG-3 and PD-L1 expression. In fact, CD8A, PD-L1, and LAG-3 were co-expressed in most cancer types (Figure 2(c)), suggesting that the combination of LAG-3 blockade and PD-1 or PD-L1 blockade is a reasonable strategy. Similar co-expression was also observed for CD8A, CTLA-4, and LAG-3 (Supplementary Figure 2).
Figure 2.
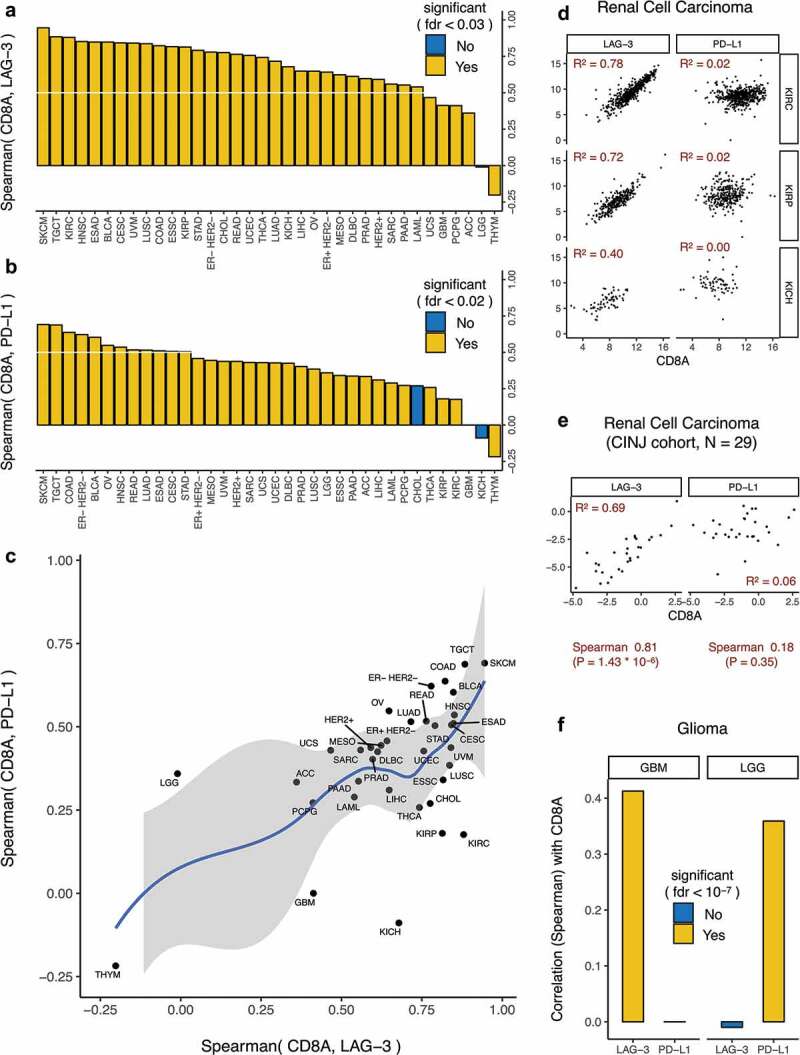
Immunologic correlates of LAG-3 expression in all cancer types.
CD8A has a stronger correlation with LAG-3 than PD-L1 in renal cancers and glioblastoma
RCC (KIRC, KIRP, KICH) and glioma (GBM, LGG) were notable exceptions to the above trend (Figure 2(c)). In RCC (Figure 2(c,d)), the correlation between CD8A and LAG-3 was much stronger than the relatively weak correlation between CD8A and PD-L1. To validate these findings, we analyzed an independent set of 29 clear cell RCC samples from CINJ that had undergone RNA sequencing. Here again there was a strong correlation between LAG-3 and CD8A expression (Figure 2(e)), while there was no correlation between PD-L1 and CD8A expression (Figure 2(e)), confirming the findings in TCGA.
In TCGA glioma data (Figure 2(c,f)), CD8A expression was correlated with LAG-3 expression (but not PD-L1 expression) in glioblastoma multiforme and PD-L1 expression (but not LAG-3 expression) in low-grade glioma, suggesting that LAG-3 blockade and PD-1 blockade may, respectively, be appropriate for tumors with CD8+ T-cell infiltration in glioblastoma multiforme and low-grade glioma. A Phase I clinical trial of LAG-3 blockade with or without PD-1 blockade in glioblastoma multiforme is currently in progress (NCT02658981), and it will be interesting to see whether LAG-3 blockade shows promising clinical activity in tumors with CD8+ T-cell infiltration.
Association between LAG-3 expression and tumor mutation burden
We recently showed8 that in eight solid cancer types, there exists a non-synonymous mutation burden threshold (iCAM), such that tumors above this threshold (iCAM+) show RNA-seq based evidence of immune activation and PD-1 or CTLA-4 pathway upregulation in TCGA dataset. Tumors classified as iCAM+ were more sensitive to PD-1 blockade and CTLA-4 blockade in publicly available datasets of melanoma, lung, and colorectal cancers compared with tumors below this threshold (iCAM−).8 To determine whether LAG-3 expression is also associated with mutation burden, we evaluated the relative expression of LAG-3 in high mutation burden (iCAM+) vs low mutation burden (iCAM−) tumors in these eight solid cancer types. In all eight cancer types, iCAM+ tumors showed significant overexpression of LAG-3 compared with iCAM− tumors (Figure 3(a)), suggesting that high mutation burden, as identified using the iCAM threshold, may also be a biomarker of response to LAG-3 blockade.
Figure 3.
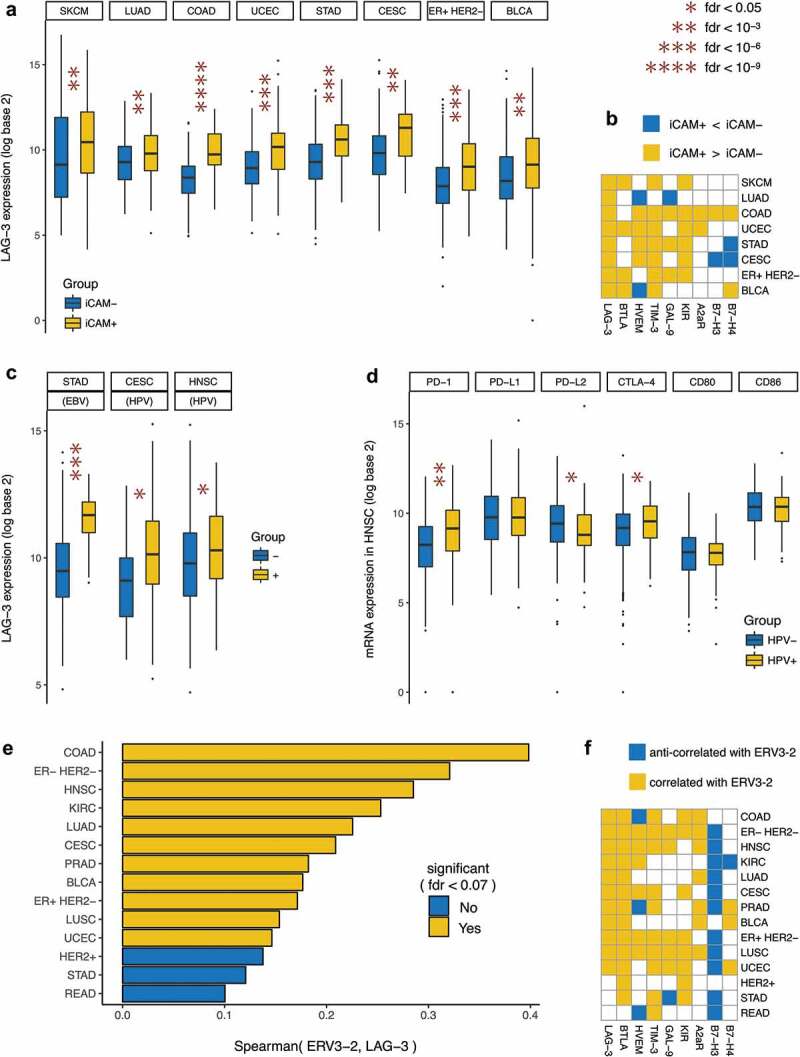
Genomic correlates of LAG-3 expression in selected cancer types.
Similar comparison for other less-studied checkpoints (Figure 3(b)) showed that unlike the PD-1 pathway, CTLA-4 pathway, and LAG-3, other checkpoints were overexpressed in iCAM+ tumors in a much more cancer type-specific manner. For example, the BTLA-HVEM pathway was upregulated in iCAM+ tumors only in endometrial cancer (UCEC), and the TIM3-GAL9 pathway was upregulated in iCAM+ tumors only in colon (COAD), gastric (STAD), and ER+ HER2− breast cancer.
LAG-3 is overexpressed in virally mediated tumors
Expression of exogenous viruses, including the presence of EBV in gastric cancer,18,19 and HPV in cervical cancer20 and head and neck squamous cell cancer20 has also been associated with evidence of immune activation. EBV expression in gastric cancer,7 NK/T-cell lymphoma,21 and Hodgkin’s disease22 has been associated with response to PD-1 blockade. To evaluate the relationship between LAG-3 expression and viral infection in tumor, we tested for association between LAG-3 expression and EBV or HPV status in these cancer types. EBV+ tumors in gastric cancer (STAD), and HPV+ tumors in cervical (CESC) and head and neck squamous cell (HNSC) cancer showed significant overexpression of LAG-3 compared with EBV− and HPV− tumors of these cancer types (Figure 3(c)). While the PD-1 and CTLA-4 pathways were also significantly upregulated in EBV+ gastric cancer7 and HPV+ cervical cancer (Supplementary Figure 3), this was not the case in HPV+ head and neck squamous cell cancer. In head and neck squamous cell cancer (Figure 3(d)), neither the ligands of PDCD1 (PD-1) nor the ligands of CTLA-4 were overexpressed in HPV+ tumors compared with HPV− tumors, which makes the overexpression of LAG-3 especially interesting. Unlike EBV+ and HPV+ tumors (Supplementary Figure 3), HBV+ and HCV+ tumors in liver cancer showed no evidence of CD8+ T-cell activation and overexpression of LAG-3 or other immune checkpoint genes (Supplementary Figure 4).
Association between LAG-3 expression and expression of potentially immunogenic ERV
The expression of normally silenced endogenous retroviral RNAs has been shown to be a potential mechanism of activation of innate immune signaling.23 We recently showed that expression of the potentially immunogenic endogenous retrovirus ERV3-2 is correlated with overall immune infiltration in 14 solid cancer types.24 To determine whether ERV expression is associated with LAG-3 expression, we tested for correlation between ERV3-2 expression and LAG-3 expression in these 14 cancer types. In 11 of the 14 cancer types, ERV3-2 expression was significantly (P < .05) correlated with LAG-3 expression (Figure 3(e)), suggesting that ERV3-2 expression may be a predictor of response to LAG-3 blockade in these cancer types. In addition to LAG-3, ERV3-2 expression was also associated with the upregulation of PD-1 and CTLA-4 pathways in most of these cancer types,24 suggesting the coordinated upregulation of multiple immune checkpoint genes in ERV-expressing tumors. In contrast, ERV3-2 expression was associated with overexpression of other checkpoint genes in a more cancer type-specific manner, such as the BTLA-HVEM pathway in only six cancer types and the TIM3-GAL9 pathway in only five cancer types (Figure 3(f)). A pan-cancer analysis to identify ERVs whose expressions correlated (Spearman Rho > 0, P < .05) or anti-correlated (Spearman Rho < 0, P < .05) with LAG-3 expression showed that ERV3-2 expression is correlated with LAG-3 expression in more cancer types than any other ERV (Supplementary Figure 5). Association between ERV3-2 expression and response to LAG-3 blockade with or without PD-1 blockade in these cancer types should be investigated in clinical trials.
Discussion
This pan-cancer analysis of LAG-3 expression shows that for most, but not all, cancers, LAG-3 expression follows the same pattern as PD-L1 expression and is similarly associated with expression of marker of cytotoxic T-cell activity.15,25 Thus, for the most part, our data suggest that LAG-3 blockade will likely benefit the same subset of cancers already shown to benefit from PD-1 pathway blockade, and suggests that clinical trials combining PD-1 and LAG-3 antibodies are rational approaches in most cancer types. The highest expression of LAG-3 is seen in B-cell lymphoma (Figure 1), and trials are currently underway for LAG-3 blockade alone and in combination with PD-1 pathway inhibitors in B-cell lymphoma (NCT02061761).
However, our analysis also showed that there are some cancers, such as RCC, where the correlation between LAG-3 and CD8A expression is much stronger than that seen for PD-L1 and CD8A. This correlation may be due to the expression of LAG-3 in a subset of CD8+ T cells.14-17 PD-L1 can be expressed both by tumor cells and immune cells, such as macrophages, in the tumor microenvironment. A lack of correlation between PD-L1 and CD8A in the setting of a high correlation between LAG-3 and CD8A in some cancers suggests that PD-L1 may not be associated with T-cell infiltrates marked by LAG-3 in these cancers. This novel observation suggests that LAG-3 blockade may have substantial clinical activity in RCC with strong CD8+ T-cell infiltration, possibly more than from PD-1 pathway blockade. This finding also suggests that LAG-3 inhibition may be effective in a subset of patients that are not responsive to PD-1/PD-L1 blockade in ccRCC. While a Phase II clinical trial of a combination of LAG-3 blockade and PD-1 blockade is already in progress in RCC (NCT02996110), our data suggest clinical trials exploring the efficacy of LAG-3 blockade monotherapy may also be warranted in RCC. Similarly, CD8A expression in glioblastoma was more strongly associated with LAG-3 expression than PD-L1 expression, suggesting that clinical trials of LAG-3 blockade should also be investigated in these aggressive cancers. Phase I clinical trial of LAG-3 blockade in glioblastoma is currently in progress (NCT02658981). As new LAG-3 inhibitors enter clinical trials, our data suggest that RCC and glioblastoma may be populations of special interest for these agents.
Since protein expression data of LAG-3 are unavailable in the TCGA dataset, the analysis presented in this paper is limited to mRNA expression data of LAG-3. However, the human protein atlas database (https://www.proteinatlas.org/ENSG00000089692-LAG3/tissue) shows that protein expression of LAG-3 is observed in appendix, lymph node, and tonsil – tissues that also have relatively high mRNA expression of LAG-3 – suggesting a concordance between mRNA expression and protein expression of LAG-3.
Anomalous results were observed in case of thymoma (THYM, Figure 2(a–c)), where CD8A expression was anti-correlated with both LAG-3 expression and PD-L1 expression. However, it should be noted that while CD8A expression is a surrogate for CD8+ T cell infiltration in most tissues, that may or may not be the case in thymoma. Unlike other tissues, thymus normally makes and stores CD8+ T cells, so the CD8A expression may represent the normally present CD8+ T cells in thymus, or real CD8+ T cell infiltration in response to cancer cells in thymoma, or some combination of both. Thus, interpretation of CD8A expression in thymic cancers is difficult, and therefore we cannot draw any conclusion in case of thymoma.
Our findings also suggest that increased mutation burden, as identified by the iCAM threshold,8 will likely be associated with response to LAG-3 blockade, similar to its known association with response to PD-1 pathway blockade. Phase II clinical trials of a combination of LAG-3 blockade and PD-1 blockade are currently in progress in non-small cell lung (NCT02750514), colon (NCT02060188), and gastric cancers (NCT02935634). It will be of great interest to see whether mutation burden is correlated with response, and whether iCAM+ tumors have high response rates in these trials. Similar trials are also warranted in skin melanoma, endometrial, cervical, bladder, and ER+ HER2− breast cancer, where we predict that high mutation burden (iCAM+) tumors may also be responsive to LAG-3 blockade.
Our results suggest that certain virally infected cancers, including EBV+ gastric cancers, and HPV+ cervical and head-and-neck cancers, also have high expression of LAG-3. A Phase I/II clinical trial of a combination of LAG-3 blockade and PD-1 blockade is currently in progress in virally mediated solid cancers (NCT02488759). Based on LAG-3 expression, our data would suggest that EBV+ tumors, in particular, may have great benefit from LAG-3 blockade, either alone or in combination with PD-1 pathway blockade. HPV+ head and neck squamous cell cancers is another group where LAG-3 blockade may be of particular benefit.
Similarly, LAG-3 expression is also correlated with abnormal expression of the potentially immunogenic endogenous retrovirus ERV3-2 in several cancer types. We previously showed that ERV3-2 expression is associated with response to PD-1 blockade in clear cell RCC,24 and these data suggest that LAG-3 blockade may also be effective in ERV3-2–expressing clear cell RCC.
Overall, analysis of TCGA data shows that LAG-3 expression is for the most part associated with known biomarkers of response to PD-1 pathway blockade, including tumor mutation burden, viral infection, and expression of immunogenic endogenous retrovirus. However, this analysis also suggests that targeting LAG-3 may be more effective than PD-1 pathway blockade in subsets of RCC, glioblastoma, and HPV+ head and neck cancers. Data from proposed and ongoing clinical trials are needed to validate these hypotheses.
Supplementary Material
Acknowledgments
The results published here are in whole or part based upon data generated by TCGA Research Network: https://www.cancer.gov/tcga. This research was supported by National Institutes of Health Grant No. P30CA072720 and an anonymous gift to precision medicine at the Rutgers Cancer Institute of New Jersey. Services, results and/or products in support of the research project were generated by the Rutgers Cancer Institute of New Jersey Biospecimen Repository and Histopathology Service Shared Resource, supported, in part, with funding from NCI-CCSG P30CA072720-5919.
Funding Statement
This work was supported by the National Cancer Institute [R01 CA243547, P30 CA072720]; Val Skinner Foundation (US); Hugs for Brady; M2GEN; U.S. Department of Defense [W81XWH1910821]; New Jersey Commission on Cancer Research (NJCCR) [DFHS18PPC022].
Disclosure of potential conflicts of interest
AP: No conflicts; JAR: No conflicts; EAS: Clinical trial research support from Astellas/Medivation; GB: No conflicts; SG: Consulted for Novartis, Roche, Foundation Medicine, Foghorn Therapeutics, and Inspirata; Owns equity in and has licensed patents to Inspirata; Spouse is an employee of Merck and has equity in Merck.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Kyi C, Postow MA.. Immune checkpoint inhibitor combinations in solid tumors: opportunities and challenges. Immunotherapy. 2016;8(7):821–7. doi: 10.2217/imt-2016-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hargadon KM, Johnson CE, Williams CJ. Immune checkpoint blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors. Int Immunopharmacol. 2018;62:29–39. doi: 10.1016/j.intimp.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Arora S, Velichinskii R, Lesh RW, Ali U, Kubiak M, Bansal P, Borghaei H, Edelman MJ, Boumber Y. 2019. Existing and emerging biomarkers for immune checkpoint immunotherapy in solid tumors. Adv Ther. doi: 10.1007/s12325-019-01051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ascierto PA, Bono P, Bhatia S, Melero I, Nyakas MS, Svane I-M, Larkin J, Gomez-Roca C, Schadendorf D, Dummer R, et al. LBA18Efficacy of BMS-986016, a monoclonal antibody that targets lymphocyte activation gene-3 (LAG-3), in combination with nivolumab in pts with melanoma who progressed during prior anti–PD-1/PD-L1 therapy (mel prior IO) in all-comer and biomarker-enriched populations. Annals Oncol. 2017;28(suppl_5). doi: 10.1093/annonc/mdx440.011. [DOI] [Google Scholar]
- 6.Khunger M, Hernandez AV, Pasupuleti V, Rakshit S, Pennell NA, Stevenson J, Mukhopadhyay S, Schalper K, Velcheti V. Programmed cell death 1 (PD-1) ligand (PD-L1) expression in solid tumors as a predictive biomarker of benefit from PD-1/PD-L1 axis inhibitors: a systematic review and meta-analysis. JCO Precision Oncol. 2017;(1):1–15. doi: 10.1200/PO.16.00030. [DOI] [PubMed] [Google Scholar]
- 7.Panda A, Mehnert JM, Hirshfield KM, Riedlinger G, Damare S, Saunders T, Kane M, Sokol L, Stein MN, Poplin E, et al. Immune activation and benefit from avelumab in EBV-positive gastric cancer. JNCI. 2017;110(3):316–320. Epub 7th ed. doi: 10.1093/jnci/djx213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda A, Betigeri A, Subramanian K, Ross JS, Pavlick DC, Ali S, Markowski P, Silk A, Kaufman HL, Lattime E, et al. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precision Oncol. 2017;(1):1–13. doi: 10.1200/PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Research Network . Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–41.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X, Zhang A, Qiu C, Wang W, Yang Y, Qiu C, Liu A, Zhu L, Yuan S, Hu H, et al. The upregulation of LAG-3 on T cells defines a subpopulation with functional exhaustion and correlates with disease progression in HIV-infected subjects. J Immunol. 2015;194(8):3873–3882. doi: 10.4049/jimmunol.1402176. [DOI] [PubMed] [Google Scholar]
- 15.Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, Netto G, Getnet D, Bruno TC, et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest. 2007;117(11):3383–3392. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solinas C, Migliori E, De Silva P, Willard-Gallo K. LAG3: the biological processes that motivate targeting this immune checkpoint molecule in human cancer. Cancers (Basel). 2019;11(8). Epub 2019/08/23. doi: 10.3390/cancers11081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyrend G, van der Gracht E, Yilmaz A, van Duikeren S, Camps M, Hollt T, Vilanova A, van Unen V, Koning F, de Miranda N, et al. PD-L1 blockade engages tumor-infiltrating lymphocytes to co-express targetable activating and inhibitory receptors. J Immunother Cancer. 2019;7(1):217. doi: 10.1186/s40425-019-0700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiaravalli AM, Feltri M, Bertolini V, Bagnoli E, Furlan D, Cerutti R, Novario R, Capella C. Intratumour T cells, their activation status and survival in gastric carcinomas characterised for microsatellite instability and Epstein-Barr virus infection. Virchows Archiv. 2006;448(3):344–353. doi: 10.1007/s00428-005-0066-4. [DOI] [PubMed] [Google Scholar]
- 19.Grogg KL, Lohse CM, Pankratz VS, Halling KC, Smyrk TC. Lymphocyte-rich gastric cancer: associations with Epstein-Barr virus, microsatellite instability, histology, and survival. Modern Patholo. 2003;16(7):641–651. doi: 10.1097/01.MP.0000076980.73826.C0. [DOI] [PubMed] [Google Scholar]
- 20.Varn FS, Schaafsma E, Wang Y, Cheng C. Genomic characterization of six virus-associated cancers identifies changes in the tumor immune microenvironment and altered genetic programs. Cancer Res. 2018;78(22):6413–6423. doi: 10.1158/0008-5472.CAN-18-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong Y-L, Chan TSY, Tan D, Kim SJ, Poon L-M, Mow B, Khong P-L, Loong F, Au-Yeung R, Iqbal J, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. 2017;129(17):2437–2442. doi: 10.1182/blood-2016-12-756841. [DOI] [PubMed] [Google Scholar]
- 22.Carbone A, Gloghini A, Carlo-Stella C. Are EBV-related and EBV-unrelated Hodgkin lymphomas different with regard to susceptibility to checkpoint blockade? Blood. 2018;132(1):17–22. doi: 10.1182/blood-2018-02-833806. [DOI] [PubMed] [Google Scholar]
- 23.Hurst TP, Magiorkinis G. Activation of the innate immune response by endogenous retroviruses. J Gen Virol. 2015;96(Pt 6):1207–1218. doi: 10.1099/jgv.0.000017. [DOI] [PubMed] [Google Scholar]
- 24.Panda A, de Cubas AA, Stein M, Riedlinger G, Kra J, Mayer T, Smith CC, Vincent BG, Serody JS, Beckermann KE, et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight. 2018;3(16):15. doi: 10.1172/jci.insight.121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichtenegger FS, Rothe M, Schnorfeil FM, Deiser K, Krupka C, Augsberger C, Schluter M, Neitz J, Subklewe M. Targeting LAG-3 and PD-1 to enhance T cell activation by antigen-presenting cells. Front Immunol. 2018;9:385. doi: 10.3389/fimmu.2018.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


