Abstract
Mucin-type O-glycosylation is an essential post-translational modification required for protein secretion, extracellular matrix formation, and organ growth. O-Glycosylation is initiated by a large family of enzymes (GALNTs in mammals and PGANTs in Drosophila) that catalyze the addition of GalNAc onto the hydroxyl groups of serines or threonines in protein substrates. These enzymes contain two functional domains: a catalytic domain and a C-terminal ricin-like lectin domain comprised of three potential GalNAc recognition repeats termed α, β, and γ. The catalytic domain is responsible for binding donor and acceptor substrates and catalyzing transfer of GalNAc, whereas the lectin domain recognizes more distant extant GalNAc on previously glycosylated substrates. We previously demonstrated a novel role for the α repeat of lectin domain in influencing charged peptide preferences. Here, we further interrogate how the differentially spliced α repeat of the PGANT9A and PGANT9B O-glycosyltransferases confers distinct preferences for a variety of endogenous substrates. Through biochemical analyses and in silico modeling using preferred substrates, we find that a combination of charged residues within the α repeat and charged residues in the flexible gating loop of the catalytic domain distinctively influence the peptide substrate preferences of each splice variant. Moreover, PGANT9A and PGANT9B also display unique glycopeptide preferences. These data illustrate how changes within the noncatalytic lectin domain can alter the recognition of both peptide and glycopeptide substrates. Overall, our results elucidate a novel mechanism for modulating substrate preferences of O-glycosyltransferases via alternative splicing within specific subregions of functional domains.
Keywords: O-glycosylation, PGANT, Galnt, mucin, secretion, Drosophila, salivary gland, lectin domain, splicing, lectin, alternative splicing
Proteins undergo post-translational modifications that regulate their structure, stability, function, and interaction with other molecules. Mucin-type O-glycosylation is an evolutionarily conserved post-translational modification that is implicated in human health and disease (reviewed in Refs. 1 and 2). Indeed, the disease hyperphosphatemic familial tumoral calcinosis, which is typified by hyperphosphatemia and the development of subdermal calcified tumors, is caused by mutations in the GALNT3 gene, whose product is responsible for the initiation of O-glycosylation (3–4). Additionally, changes in O-glycosylation are also associated with other conditions, including blood lipid levels, bone density, kidney function, and the development and progression of certain types of cancer (5–13). More recently, congenital loss of GALNT2 function has been found to underlie a complex neurological syndrome (14). Therefore, a comprehensive understanding of the factors that regulate the synthesis and function of O-glycans is crucial to understanding how their alteration contributes to disease.
Biogenesis of mucin-type O-glycosylation is initiated in the Golgi through the transfer of a GalNAc monosaccharide to serine or threonine residues of target proteins. A large family of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases (i.e. Galnts or GALNTs) initiates this first sugar addition (∼20 Galnts in mammals and 10 PGANTs in Drosophila) (1, 2). Members of this family show unique spatial and temporal expression patterns during embryonic development and different expression levels in various organs and tissues, suggesting that each member has unique in vivo substrates and functional roles (1, 2, 15, 16). Structurally, members of this family are similar, with each having a conserved catalytic domain responsible for binding donor (UDP-GalNAc) and acceptor (proteins or peptides) substrates to coordinate GalNAc transfer, as well as a separate ricin-like lectin domain that potentially recognizes an extant GalNAc on previously glycosylated substrates (17, 18). The catalytic domain contains a catalytic flexible gating loop that becomes ordered and adopts a closed conformation upon acceptor and donor substrate binding (19–23). The lectin domain is unique to the GALNT enzyme family and consists of three repeats (α, β, and γ) that can potentially bind to previously glycosylated substrates. Such binding positions the acceptor site of a glycopeptide substrate into the catalytic domain to facilitate the glycosylation of additional, more distant sites (17–19, 21–25). Crystal structures of GALNT2, GALNT3, GALNT4, and GALNT12 bound to glycopeptide substrates show the glycopeptide–Thr–GalNAc bound to the α repeat of the lectin domain in an identical manner to help position the acceptor threonine into the active site of the catalytic domain at a distance (5–17 residues) from the lectin-bound glycopeptide–Thr–GalNAc (19–22).
Studies employing libraries of peptides have revealed unique amino acid preferences vicinal to the site of glycosylation for the catalytic domains of several family members (26, 27). Likewise, libraries of glycopeptide substrates have revealed isoform-specific preferences for the position of extant GalNAc residues (relative to sites of glycosylation) within the lectin domain (17, 25). Specifically, certain family members prefer that the extant remote GalNAc is C-terminal to the putative site of GalNAc addition (GALNT1, -T2, and -T14), whereas others prefer the extant remote GalNAc to be N-terminal to the site of GalNAc addition (GALNT3, -T4, -T6, and -T12) (17, 25). This selectivity is in part influenced by changes in the ∼10-amino acid flexible linker between the catalytic and lectin domains (24). Taken together, the distinctive structural features of each family member are thought to confer specific peptide and glycopeptide preferences to ensure the correct modification of unique sites, resulting in the dense but deliberate glycosylation of mucins and mucin-like proteins that consist of regions rich in serine and threonine. The high density of O-glycans found in mucins is thought to confer many biological properties, such as resistance to proteolysis, rigid rod-like structures, and the ability to form highly protective, hydrated membranes along many epithelial surfaces of the body (28–30).
Studies in model organisms have demonstrated that O-glycosylation is essential for viability (16, 31, 32) and has roles in extracellular matrix formation and secretion, cell adhesion, cell proliferation, microbiome composition, and organ growth and development (30, 33–37). Previous work in the fruit fly Drosophila melanogaster identified a member of this enzyme family (CG30463; pgant9) that influenced viability (16). Recent studies have found that pgant9 is differentially spliced within the Drosophila salivary gland to generate two isoforms (PGANT9A and PGANT9B) that differ only within the α repeat of the lectin region (38). Moreover, the expression of each isoform has differential effects on the morphology of secretory granules within the salivary gland, because of differential glycosylation of an in vivo mucin (38). Here, we further characterize the substrate specificities of PGANT9A and PGANT9B in vitro. Interestingly, we find that PGANT9A and PGANT9B have differential preferences for charged peptide substrates that are dictated by charged residues within the α repeat along with charged residues within the gating loop of the catalytic domain. Additionally, we find that each splice variant displays unique glycopeptide specificities, indicating that this lectin-domain splicing event further influences recognition of glycopeptide substrates. Our results provide the first demonstration of a developmentally regulated splicing event within the lectin domain of a GALNT/PGANT family member that can modulate both peptide and glycopeptide substrate preferences. These studies provide a new regulatory mechanism by which the specificity of O-glycosylation can be modulated in vivo and coordinated with the expression of endogenous substrates.
Results
Alternative splicing of the lectin domain α repeat alters mucin peptide preferences
pgant9 is differentially spliced in a tissue-specific manner that changes only the α repeat of the lectin region (Fig. 1A) to yield two splice variants known as pgant9A and pgant9B (38). This splicing event results in an α repeat that is highly positively charged (PGANT9A) or highly negatively charged (PGANT9B) (Fig. 1B) (38). Previous work has shown that each splice variant exhibited a preference for an oppositely charged peptide substrate (38). In that study, the negatively charged PGANT9B preferred to glycosylate the positively charged Sgs3 mucin, and the specific loss of PGANT9B resulted in defects in secretory granule morphology, likely because of alterations in the glycosylation of Sgs3. PGANT9A showed little activity on the positively charged region of Sgs3 but robustly glycosylated this region when positively charged amino acids were changed to negatively charged ones (38). This led to the suggestion that this splicing event may exist to ensure proper glycosylation of endogenous substrates containing charged regions. To further explore whether PGANT9A and PGANT9B have distinct preferences for other endogenous substrates, we examined their activity toward a variety of mucins expressed in Drosophila (Fig. 1, C and D, and Fig. S1). We created peptide substrates based on the repetitive regions unique to each mucin and performed enzymatic assays using purified PGANT9A and PGANT9B expressed in Pichia pastoris (38). As shown in Fig. 1C, both PGANT9A and PGANT9B glycosylated a number of mucin-based peptides, including Muc18B, Muc11A, Sgs1, and Muc55B, in addition to the previously characterized Sgs3. Based on these results, we next tested whether PGANT9A and PGANT9B preferences for substrates containing charged residues could be predictably altered by varying peptide charge. We therefore changed each charged residue to an oppositely charged residue and performed in vitro enzyme assays (Fig. 2, A–D, and Fig. S2). As shown previously, PGANT9B robustly glycosylates the endogenous Sgs3 substrate (which is positively charged) but has reduced activity on the negatively charged version (Sgs3-E) (Fig. 2A) (38). In contrast, PGANT9A prefers the negatively charged version (Sgs3-E) and does not act substantially on the positively charged Sgs3 (Fig. 2A) (38). Similarly, Sgs1 (which is preferred by PGANT9B), can become a more robust substrate for PGANT9A by changing positively charged residues to negatively charged ones (Sgs1-E) (Fig. 2B and Fig. S2). Interestingly, however, this change had little effect on the activity of PGANT9B (Fig. 2B and Fig. S2). Likewise, the preference of PGANT9A for Muc18B or Muc11A was dramatically reduced when negatively charged residues were changed to positively charged ones (Muc18B-K or Muc11A-K) (Fig. 2, C and D). However, the activity of PGANT9B was not as dramatically altered with these charged residue changes (Fig. 2, C and D, and Fig. S2). Taken together, our results provide evidence that PGANT9A has a strong preference for negatively charged mucin-based substrates and that this preference can be predictably altered by altering substrate charge. In contrast, the preference of PGANT9B for positively charged substrates was not as striking.
Figure 1.
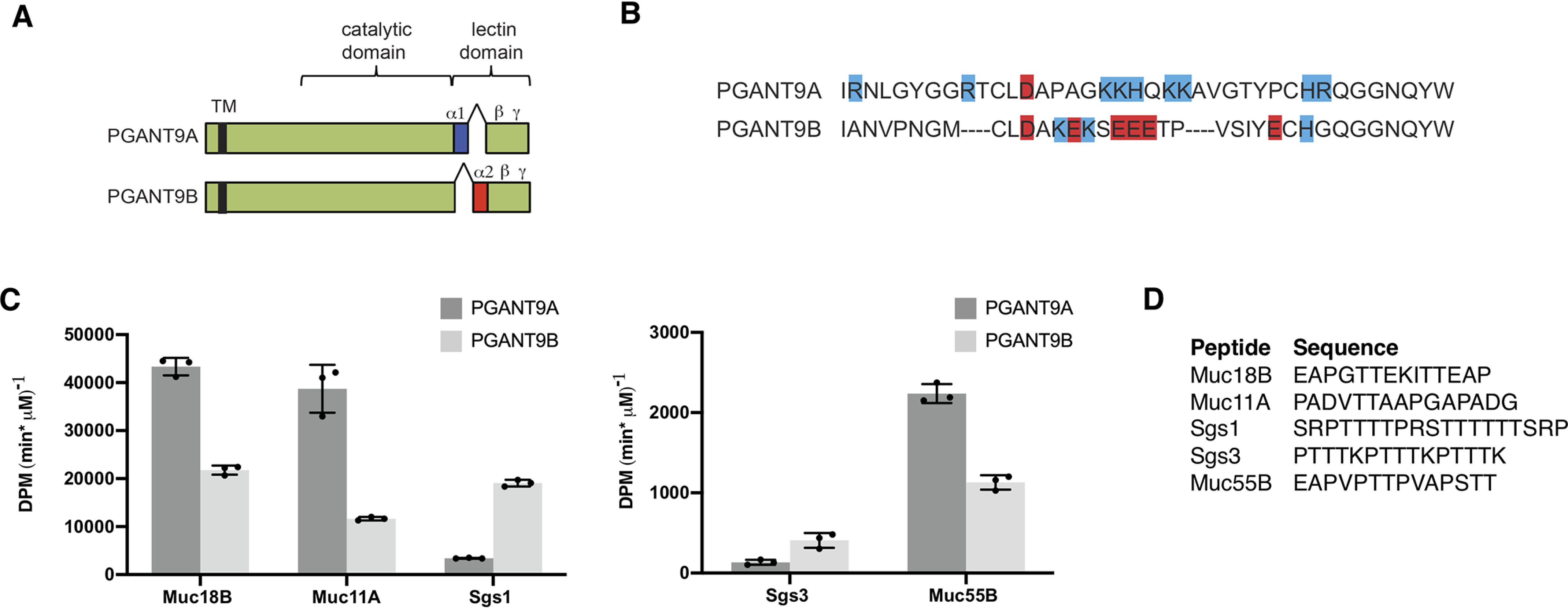
PGANT9A and PGANT9B glycosylate diverse mucin-based substrates. A, PGANT9A and PGANT9B are identical with the exception of the differentially spliced α repeat of the lectin domain. B, amino acid sequences of the differentially spliced α repeats are shown. Positively charged residues are highlighted blue, and negatively charged residues are highlighted red. C, peptides based on endogenous mucin substrates were used in in vitro assays to determine enzymatic activity of PGANT9A and PGANT9B. Both PGANT9A and PGANT9B were able to glycosylate a number of mucin-based peptides. D, peptides used in each reaction are shown. Each data point represents an individual assay. Error bars show S.D. Each set of assays was repeated three times.
Figure 2.

Peptide preference can be altered by modifying peptide charge. Peptides and their oppositely charged versions were used for in vitro enzymatic assays. PGANT9A and PGANT9B were tested with Sgs3 and Sgs3-E (A), Sgs1 and Sgs1-E (B), Muc18B and Muc18B-K (C), and Muc11A and Muc11A-K (D). The peptides used in each reaction are shown to the right. Each data point represents an individual assay. Error bars show S.D. Each set of assays was repeated three times. Student's t test was used to calculate p values. *, p < 0.05; **, p < 0.01.
To further understand why PGANT9A is apparently more sensitive to substrate charge than PGANT9B, we next examined which charged residues have the most influence on the activities of PGANT9A or PGANT9B. Using a peptide based on the previously characterized Sgs3 mucin sequence (which contains regularly spaced charged residues every 5 amino acids), we altered each charged residue individually and then tested PGANT9A or PGANT9B activity (Fig. 3, A–C, and Fig. S3). PGANT9A activity predictably increased with the addition of each negatively charged residue at each position in the peptide substrate (Fig. 3A). Unexpectedly, PGANT9B activity increased dramatically with the addition of a negative residue at amino acid position 5 in the peptide (Fig. 3B). These results suggest that a negative charge at this particular position is somehow increasing the ability of PGANT9B to glycosylate this substrate, even though the α repeat of PGANT9B is negatively charged.
Figure 3.

Position of negatively charged residue dramatically affects PGANT9B activity. A and B, peptides based on Sgs3 were used to test the effect of a negatively charged residue on the activity of PGANT9A (A) and PGANT9B (B) in vitro. Each data point represents an individual assay. C, peptides used in each reaction are shown. Error bars show S.D. Each set of assays was repeated three times. Statistical comparisons were performed between Sgs3 and each peptide variant using the Student's t test to calculate p values. *, p < 0.05; **, p < 0.01.
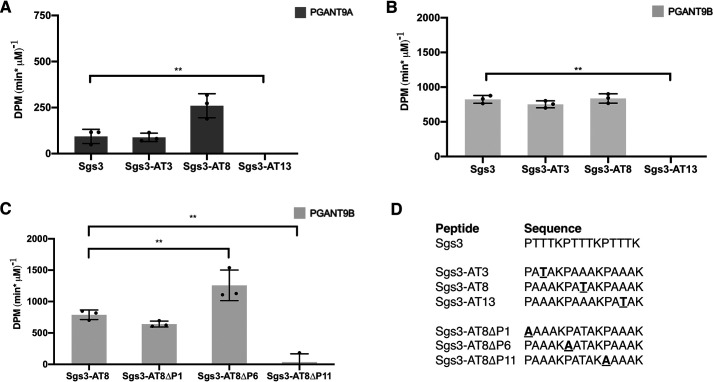
To understand how this might be happening, we designed peptides based on the Sgs3 sequence with only one acceptor site to see where PGANT9A or PGANT9B prefer to glycosylate (Fig. 4 and Fig. S4). Based on these single-site acceptors, PGANT9A or PGANT9B will glycosylate a threonine in the center of the peptide (threonine 8) or near the N terminus (threonine 3). Neither splice variant acted on a threonine at the C terminus (threonine 13) (Fig. 4, A and B). To further explore substrate preference, we next examined which prolines influenced enzymatic activity on single-site acceptors. Previous studies have shown that most GALNTs have a conserved proline-binding pocket within the active site that interacts with prolines at the +1 and +3 position of the acceptor threonine to coordinate substrate binding (25, 26). Indeed, the previously solved structures of PGANT9A and PGANT9B also predict that such a binding pocket exists within their catalytic domains (38). To test this, we mutated each proline in the Sgs3-AT8 single-site acceptor and performed assays with PGANT9A or PGANT9B. Although PGANT9A showed no activity against any of these substrates (likely because of its very low activity against the Sgs3-AT8 single-site acceptor), PGANT9B still had robust activity against substrates in which the N-terminal (proline 1) or central proline (proline 6) were changed to alanine (Fig. 4, C and D, and Fig. S4). However, PGANT9B activity was completely abrogated when the C-terminal proline (proline 11) was mutated (Fig. 4, C and D). These results suggest that this proline 11 likely sits in the proline pocket of PGANT9B to position threonine 8 in the catalytic center for glycosylation.
Figure 4.
PGANT9A and PGANT9B preferred sites of addition. A and B, single-site acceptors based on the Sgs3 sequence were used to determine the preferred sites of addition by PGANT9A (A) and PGANT9B (B). C, peptides based on the preferred single-site acceptor, Sgs3-AT8, were designed to determine whether proline is important for activity. There was no activity detected for PGANT9A with these peptides. D, peptides used in each reaction are shown. Error bars show S.D. Each set of assays was repeated three times. Statistical comparisons were performed between the first peptide shown and each other variant using the Student's t test to calculate p values. **, p < 0.01.
In silico modeling reveals contributions of the α repeat and gating loop in influencing charged peptide preferences
Based on this in vitro data, we modeled the Sgs3 peptide and the single charge variant (Sgs3-1E5) into the crystal structure of PGANT9B using threonine 8 as the acceptor and placing the +3 proline (proline 11) in the conserved proline binding pocket (Fig. 5, A and B). We also modeled the negatively charged variant (Sgs3-E, which PGANT9A prefers) into PGANT9A using the same parameters (Fig. 5C). Interestingly, these models predict that a positively charged region (AA 437–450) in the catalytic flexible gating loop of PGANT9A and PGANT9B (Fig. 5, D and E) will lie in close proximity to the N terminus of the peptide substrate (Fig. 5, A–C). Within that region are four consecutive residues that contain a net positive charge in most human and fly isoforms, corresponding to RKRS in PGANT9A and PGANT9B (AA 440–443) (Fig. 5, D and E). However, PGANT9A and PGANT9B also contain additional nonconserved positively charged residues (Lys446 and Arg448) within this gating loop (starred residues in Fig. 5E). The net positive charge in the gating loop may explain why a negative charge in the N terminus of Sgs3-1E5 would enhance activity for PGANT9B (Fig. 5, A versus B), independent of the negative charges within its lectin domain α repeat, which are predicted to interact with the C terminus of the peptide. Likewise, modeling Sgs3-E into the crystal structure of PGANT9A shows the negatively charged peptide interacting with both the positively charged gating loop and the positively charged α repeat (Fig. 5C). Taken together, these results support a more complex model where the substrate preferences of PGANT9A and PGANT9B are further influenced by a positively charged region within the catalytic gating loop. The combination of the positive charge in the gating loop along with the positive charge of the α repeat are likely responsible for the strong preference for negatively charged substrates seen for PGANT9A. Likewise, the combination of positive charge in the gating loop combined with negative charge in the α repeat of PGANT9B may explain why it is less sensitive to substrate charge.
Figure 5.

In silico modeling of substrates in PGANT9A and PGANT9B. A and B, PGANT9B bound to the positively charged Sgs3 peptide (A) and bound to the Sgs3-1E5 peptide (B) containing a negative charge (E5) at the N terminus. The catalytic domain of PGANT9B is shown in green, and the lectin domain is pink. Both the catalytic loop (His437–Gly450) and the α repeat are shown as electrostatic potential surfaces, where blue indicates electropositive potential, and red indicates electronegative potential. The peptides are shown as electrostatic potential spheres, and the Lys (K5 in A) or Glu (E5 in B) at position 5 is shown as a stick. C, model of PGANT9A bound to the negatively charged Sgs3-E peptide, where the catalytic domain is shown in wheat, and the lectin domain is blue. D, a sequence analysis of the catalytic flexible loop of human and Drosophila GalNAc-Ts showing that a cluster of positive charges RKRH are conserved in the loop, corresponding to RKRS at AA 440–443 of PGANT9A and PGANT9B shown in E. E, the preference of a negative charge at the N terminus of the peptide is dictated by the positive charges in the catalytic flexible loop, highlighted as blue letters. Starred residues indicate additional positive charges in the catalytic loop of PGANT9A and PGANT9B that are not highly conserved in Drosophila and human isoforms, as shown in D.
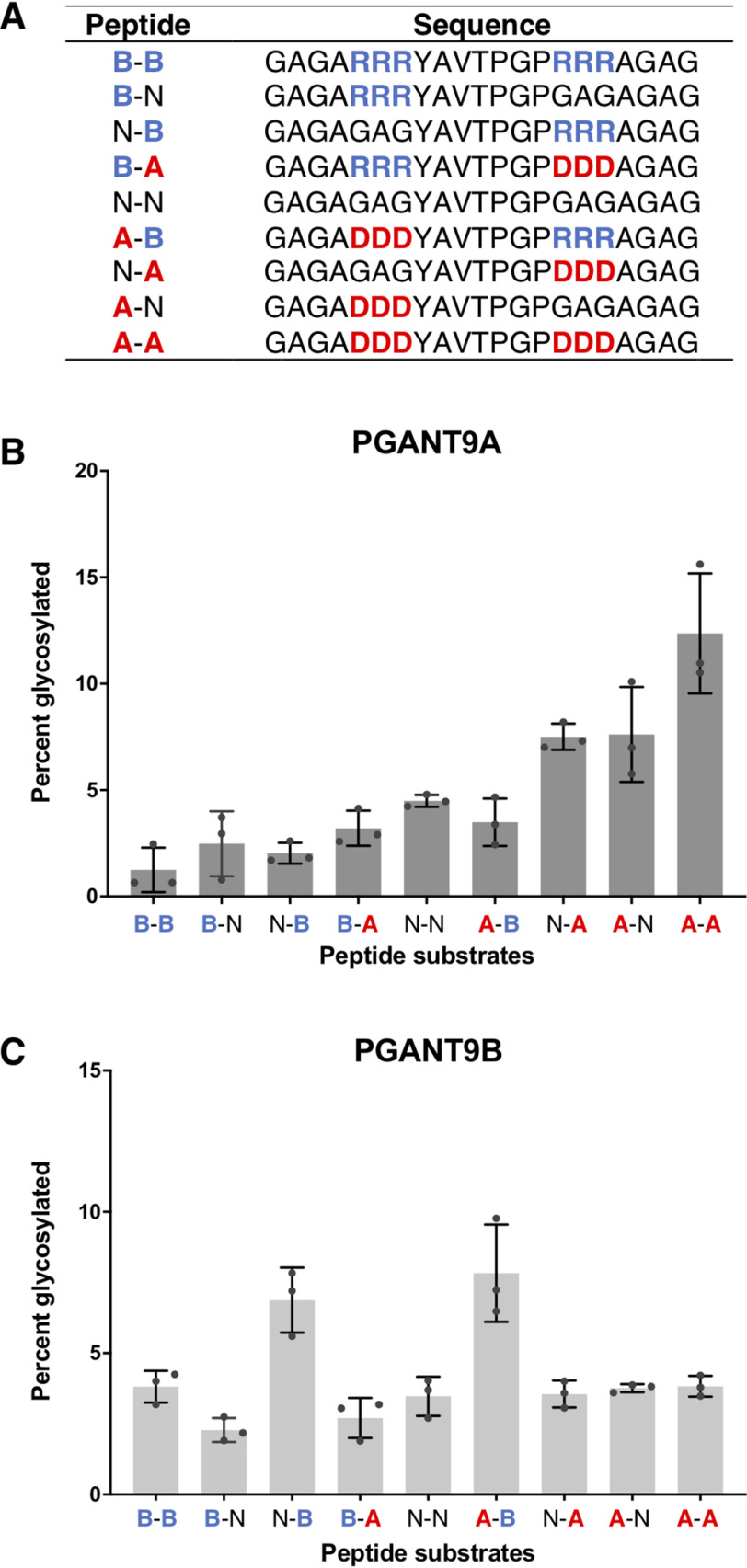
To further test this model, we created a series of peptide substrates that varied in charge at positions N-terminal and C-terminal to the site of glycosylation (Fig. 6A) to interrogate PGANT9A and PGANT9B charge preferences. The N-terminal and C-terminal regions contained either neutral (N), acidic (A), or basic (B) patches flanking the acceptor threonine (Fig. 6A). This array of nine peptide variants was incubated with either PGANT9A or PGANT9B, and glycosylation was quantified (Fig. 6, B and C). Interestingly, PGANT9A strongly preferred peptides that contained acidic regions N- and/or C-terminal to the site of GalNAc addition, with no basic regions present (Fig. 6B and Fig. S5A). PGANT9B preferred peptides with a C-terminal basic charge that also had an acidic or neutral region in the N terminus (Fig. 6C and Fig. S5B). These results are in agreement with our in silico modeling shown in Fig. 5 and in vitro data on endogenous substrates and provide further support for a model where charge within the gating loop of the catalytic domain combines with the charge of the α repeat of the lectin domain to drive peptide substrate preferences.
Figure 6.
PGANT9A and PGANT9B display unique preferences for charged residues N- and C-terminal to the site of glycosylation. A, sequences of peptides containing neutral, acidic, or basic residues N- and C-terminal to the potential site of glycosylation are shown. B and C, percentage of glycosylation by PGANT9A (B) and PGANTB (C) against the differently charged peptides is shown. Error bars show S.D. All assays were repeated three times. Statistical analyses are shown in Fig. S5.
Alternative lectin domain α repeats confer unique glycopeptide substrate preferences
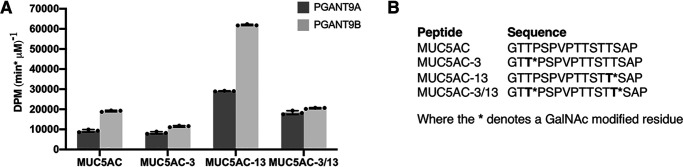
We next examined whether the differential splicing within the α repeat of the lectin domain of PGANT9A and PGANT9B altered their preferences for glycopeptide substrates. Previous studies have shown that the lectin domain is able to recognize extant GalNAc on glycopeptide substrates to influence further GalNAc addition by the catalytic domain (17, 18, 25). To test whether splicing within the α repeat of the lectin domain also influences glycopeptide preferences, we performed enzyme assays on glycopeptide substrates (Fig. 7 and Fig. S6). We noticed consistent and reproducible differences in the activities of PGANT9A and PGANT9B on certain glycopeptide substrates relative to the unglycosylated MUC5AC peptide (Fig. 7, A and B), suggesting that this splicing event may further influence glycopeptide affinity/preferences. We therefore characterized the glycopeptide preferences of each splice variant using a series of universal randomized glycopeptide substrate libraries designed to probe the role of the lectin domain in recognizing prior remote GalNAc glycosylation (25, 26). These random glycopeptide substrates contain a single GalNAc residue placed near the N or C terminus and a distant region of randomized residues, 5–17 residues away, which also contain Thr as an acceptor (Fig. 8A) (17). As shown in Fig. 8 (B and C), both PGANT9A and PGANT9B show significantly enhanced activity against both the N- and C-terminal glycopeptides (GP(T*22)R or GP(T*10)L) compared with the nonglycosylated peptides (GP(A22)R or GP(A10)L). This indicates that both N- and C-terminal extant GalNAc residues are recognized by both PGANT9A and PGANT9B lectin domains to enhance the ability of the catalytic domain to transfer GalNAc to more distant sites. Interestingly, PGANT9A and PGANT9B displayed slightly different preferences for the orientation of the glycosylated residue relative to the site where the catalytic domain would transfer additional GalNAc (Fig. 8, B–E). PGANT9B slightly prefers glycopeptide substrates where the extant GalNAc is C-terminal to the sites of glycosylation (GP(T*22)R), whereas PGANT9A slightly prefers glycopeptide substrates in which the extant GalNAc is N-terminal to the sites of GalNAc addition (GP(T*10)L) (Fig. 8, B–D). This difference is expressed as a ratio in Fig. 8D and as a schematic drawing in Fig. 8E. These data suggest that the splicing event within the α repeat of the lectin domain of PGANT9A and PGANT9B also generates unique glycopeptide substrate preferences. Taken together, our study provides the first evidence for developmentally regulated alternative splicing within the lectin domain of a member of the polypeptide GalNAc transferase family that influences both peptide and glycopeptide substrate specificity.
Figure 7.
PGANT9A and PGANT9B display unique activities on glycopeptide substrates. A, PGANT9A and PGANT9B were tested against peptide and glycopeptide substrates in in vitro enzymatic assays. B, sequences of the peptides and glycopeptides used are shown. Each data point represents an individual assay. Error bars show S.D. Each set of assays was repeated three times.
Figure 8.

PGANT9A and PGANT9B display different orientation preferences for glycopeptides. A, for lectin domain binding, peptides were designed with a single C- or N-terminal GalNAc-O-Thr residue (T*), flanked by five randomized residues (magenta) containing no acceptor, and followed by 12 randomized residues (green) including the putative sites of GalNAc addition. Control peptides have Ala residue instead of GalNac-O-Thr. B, representative time-course plots for showing the net [3H]GalNAc utilization. C, Sephadex G10 chromatograms with overnight incubations demonstrating high [3H]GalNAc transfer to (glyco)peptide substrates (fractions 27–33) with minimal hydrolysis (i.e. free GalNAc, fractions 37–43). D, ratios of transfer GP(T*22)R versus GP(T*10)L (normalized to DPM/OD). For PGANT9A, n = 12 and for PGANT9B (n = 14). E, schematic drawing of orientation preferences for PGANT9A and PGANT9B.
Discussion
Here we provide the first demonstration that a developmentally regulated splicing event controls multiple aspects of substrate preferences within members of the polypeptide GalNAc transferase family. Specifically, we show that splicing within the noncatalytic portion of the enzyme influences both peptide and glycopeptide preferences. O-Glycosylation is an essential protein modification that is catalyzed by a large family of enzymes that are each thought to have some overlapping as well as unique substrate specificities (1, 2). The identification of splicing as another mechanism to further refine the substrate preferences of an individual isoform adds a new layer of complexity to the factors that govern this essential post-translational modification.
In this study, we provide new mechanistic details of how splicing in the noncatalytic lectin domain can alter peptide preferences. Our previous work suggested a model in which the positively charged α repeat of PGANT9A, which lies in close proximity to the active site, allowed access of negatively charged substrates, whereas the negatively charged α repeat of PGANT9B allowed access of positively charged substrates (38). This model was supported by the preferences of each splice variant for one mucin-based peptide, Sgs3 (38). However, our studies herein interrogating the preferences for other endogenous substrates revealed a more complicated story. Although PGANT9A still displayed a striking preference for negatively charged peptide substrates, PGANT9B was less sensitive to substrate charge. In silico modeling of the preferred substrates for each splice variant highlighted a role for charged residues within the catalytic region of each enzyme in dictating substrate preference. In particular, a cluster of positively charged residues in the flexible catalytic gating loop of both enzymes is predicted to lie in close proximity to the N terminus of substrates docked within the active site and thus influence substrate preferences. These models predicted that the positive charges of the gating loop combined with the positive charges of the α repeat of the lectin domain of PGANT9A would result in a strong preference for negatively charged substrates as well as a strong aversion to positively charged substrates. Likewise, the positive charges of the gating loop combined with the negative charges of the α repeat of PGANT9B would result in an enzyme that could interact with substrates that contained both positively and negatively charged regions. Indeed, these models were supported by our in vitro data using peptides derived from endogenous mucin substrates. Furthermore, synthetic peptide substrates with varied charge N- and C-terminal to the site of GalNAc addition confirmed these predictions; PGANT9A strongly preferred negatively charged amino acids in both N- and C-terminal regions, whereas PGANT9B preferred negative charges in the N terminus and positive charges in the C terminus. Taken together, our biochemical and in silico modeling data support a model in which the charge within the gating loop of the catalytic domain combines with the charge of the α repeat of the lectin domain to drive peptide substrate preferences for the PGANT9 splice variants. These data highlight heretofore unknown mechanistic details governing substrate specificity that involves charge contributions from both the catalytic and lectin domains.
We also found that this splicing event influences glycopeptide preferences. Screening of a random glycopeptide library revealed differences between the preferred directionality of addition by each splice variant. Taken together, these results indicate that PGANT9A and PGANT9B have distinct glycopeptide preferences that vary among different glycopeptide substrates. How the differentially spliced α repeats influence the recognition of extant GalNAc residues in different glycopeptide substrates remains to be investigated. Toward this goal, we are currently attempting to co-crystalize each splice variant with glycopeptide substrates to determine how extant GalNAcs are recognized by the lectin domain.
Our studies highlight splicing of enzyme subregions as another mechanism whereby the substrate preferences of the PGANT/GALNT family members may be fine-tuned based on the needs of the cells/tissues where they are expressed. This mechanism of altering substrate preferences could be crucial to ensuring proper glycosylation of diverse endogenous substrates. For example, many mucins have charged residues within their serine- and threonine-rich repeats, whereas the repeat regions of other mucins are uncharged. Splicing that modulates whether a PGANT/GALNT will interact with a charged or uncharged mucin could be highly beneficial, especially in tissues where the mucin profile changes developmentally. As our current study demonstrates, charged residues in the catalytic domain, as well as the lectin domain, have the potential to influence substrate preferences. Because lectin and catalytic subdomains lie in exonic regions of other members of this family (1), it suggests that splicing events that modify these domains may be used more widely by other PGANTs/GALNTs. In fact, we and others have detected putative splice variants for other PGANT and GALNT family members (39, 40). For example, splicing of the human GALNT13 alters both the length and charged residues within the γ repeat of the lectin domain (39, 40). This splicing event results in changes in the affinities for certain glycopeptide substrates but does not appear to alter directionality of GalNAc addition to glycopeptides (39, 40). Whether splicing alters specific peptide preferences remains to be examined in more detail. Other mammalian splice variants have been predicted in silico (e.g. Galnt7, Galnt14, Galnt15, Galnt17, Galnt18); however, whether splicing occurs in vivo and how it might alter peptide and/or glycopeptide preferences remains to be determined.
In summary, we have found a new regulatory paradigm whereby transferase activity/substrate preferences can be modified in vivo via splicing of specific subdomains within the lectin region. Additionally, we have further characterized how substrate preferences are influenced by a combination of charged residues in both the lectin domain and the flexible catalytic gating loop. Moreover, we have discovered that alteration of the lectin domain influences both peptide and glycopeptide preferences. This discovery illustrates a new mechanism for modulating substrate preferences in vivo to accommodate the glycosylation demands of specific cells/tissues and highlights multi-faceted roles for the lectin domain in substrate recognition.
Experimental procedures
Enzyme assays
Expression of recombinant PGANT9A and PGANT9B was performed either using P. pastoris or COS7 cells as described previously (21, 38). Peptide substrates were synthesized by Peptide 2.0. Assays for PGANT9A and PGANT9B activity were performed as described previously (41) using PGANT9A and PGANT9B purified from Pichia. The reactions were run for 30 min at 37 °C in 25-µl final volumes consisting of the following: 25 mm acceptor substrate, 7.3 μm UDP-[14C]GalNAc (54.7 mCi/mmol; 0.02 mCi/ml), 2 mm cold UDP-GalNAc, 500 mm MnCl2, 40 mm cacodylate (pH 6.5), 40 mm 2-mercaptoethanol, and 0.1% Triton X-100. The reactions were then quenched with 30 mm EDTA. Glycosylated products were separated from unincorporated UDP-[14C]GalNAc by anion-exchange chromatography using AG1-X8 resin columns (Bio-Rad catalog no. 1401454), and product incorporation was determined by liquid scintillation counting (Beckman Coulter LS6500). Reactions without acceptor peptide were also used to generate background values that were subtracted from each experimental value. Assays for each peptide substrate were run in triplicate and repeated three times. Experimental values for each substrate were then averaged, and the standard deviations were calculated. Enzyme activity (initial rate) is expressed as DPM min−1 µm−1.
In silico modeling
Sgs3 peptides were initially manually docked into the active sites of PGANT9A or PGANTB in Coot (42) using Thr5 as the acceptor, based on experimental data. The peptides from co-crystal structures of GalNAc-T4 (PDB code 6H0B), GalNAc-T2 (PDB code 2FFU), GalNAc-T12 (PDB code 6PXU), and GalNAc-T3 (PDB code 6S24) were used to guide the positioning of the acceptor threonine5 in the active site, the +3 proline in the conserved pocket, and conserved backbone residues in the peptide. The docked peptide–enzyme complex models were then submitted to the FlexPepDock Server for model refinement (43, 44). Electrostatic potential maps and models in Fig. 6 (A–C) were generated in MacPyMOL (PyMOL Molecular Graphics System, version 2.0; Schrodinger), and Fig. 6E was generated using Seq2Logo 2.0 (45).
PGANT9A and PGANT9B activity against substrates with flanking charged residues
A series of nine differently charged peptide substrates designed to address the roles of flanking charged residues clusters were custom synthesized by RS Synthesis (Louisville, KY, USA). The invariant sequences used in these model substrates contain the +1 and +3 preference for proline seen across most O-glycoslytransferases that contain a proline pocket in the catalytic domain (including PGANT9A and PGANT9B). The additional central invariant sequences are based on prior work defining the human GALNT3 (a related ortholog of PGANT9A and PGANT9B) optimal sequence YAVTPGP (20), which was flanked by all possible combinations of the three residues clusters; RRR, DDD, and GAG (representing positive (basic, B), negative (acidic, A), and neutral control (N) clusters respectively). These were then further flanked by GAGA- and -AGAG sequences at the N and C termini, respectively, resulting in a total of nine different peptide substrates.
Identical transferase reactions were performed at the same time with PGANT9A and PGANT9B against nine different charged substrates. Final incubations contained the following: 100 mm sodium cacodylate buffer, pH 6.8, 0.1% Triton X-100, 1 mm 2-mercaptoethanol, 10 mm MnCl2, 0.2 mm UDP-[3H]GalNAc (American Radio Chemicals, St. Louis, MO, USA), 350 μm peptide substrate and media from COS7 cells expressing PGANT9A or PGANT9B. The reactions were initiated by placing 13 µl of a premade buffer mix containing acceptor, co-factor, detergent, and enzyme (90 µl of medium in a total of 155 µl) into Eppendorf tubes containing 7 µl of each peptide substrate (1 mm). The reactions were capped and incubated for 4 h in a shaking microincubator at 35 °C and quenched by the addition of two volumes of 50 mm EDTA (40 µl) on ice. The reaction time was optimized for less than 15% product formation and less than 50% donor (UDP-[3H]GalNAc) utilization. Three independent experiments were performed with each set of peptide substrates for both PGANT9A and PGANT9B.
The quenched reactions were diluted to 300 µl with 1% TFA and passed through hydrophobic macro Targa C-18 spin-columns (Nest Group Inc., Southborough, MA, USA) equilibrated with 100% acetonitrile (2 volumes of 300 µl) followed by 0.1% TFA/water (2 volumes of 300 µl). After loading the sample, the column was washed with 0.1% TFA/water (2 volumes of 200 µl) to remove free UDP-[3H]GalNAc and free [3H]GalNAc. Bound (glyco)peptide products were eluted first with 50% acetonitrile and then with 100% acetonitrile washes (each 2 volumes of 300 µl). [3H] scintillation counting (in DPM) of aliquots of the diluted incubation mix (50 µl), the combined column flow through and wash (200 µl), and the eluted (glyco)peptide products (600 µl) were performed on a Beckman LS-6500 scintillation counter. Samples were counted several times and averaged to improve the signal to noise. The percentage of substrate glycosylation was obtained from the [3H] DPM counts of the (glyco)peptide eluate relative to the initial [3H] DPM counts after correcting for the concentrations of UDP-[3H]GalNAc and peptide in the reaction mix.
Characterization of lectin domain specificity from random glycopeptide substrates
The lectin-domain probing random glycopeptide substrates GP(T*22)R and GP(T*10)L (and their nonglycosylated controls GP(A22)R and GP(A10)L) were utilized to compare the remote GalNAc-O-Thr glycopeptide specificity of PGANT9A and PGANT9B as previously described (17, 25). (Note that these glycopeptides were previously named GPIV, GPV, GPIV-C, and GPV-C, respectively, and synthesized by Sussex Research, Ottawa, Canada) (17). The reactions were carried out using 68 mm sodium cacodylate, pH 6.5, 1.8 mm 2-mercaptoethanol, 10 mm MnCl2, 50 μm UDP-[3H]GalNAc (∼6 × 108 DPM/µmol), 5 mg/ml (∼1.5 mm) of substrates GP(T*22)R, GP(A22)R, GP(T*10)L, and GP(A10)L and 100–180 μl of PGANT9A or PGANT9B (as media from COS7-expressing cells) to a total final volume of 250–300 μl. The reactions were incubated at 37 °C, and 50–80-μl aliquots were removed for analysis after incubating for 2 h, 4 h, and overnight for PGANT9A and PGANT9B. In all cases, reactions with both transferases were performed concurrently under identical reaction conditions. Glycopeptide workup and isolation was performed as described previously (25). The lyophilized pooled glycopeptide fractions were taken up in 1 ml of H2O and transferase activity quantified by [3H]GalNAc DPM content (Beckman LS-6500 scintillation counter) normalized to the 220- and 280-nm OD values. With this approach, any losses in peptide substrate during sample processing were corrected for by normalizing to the peptide OD values. The C-terminal/N-terminal random glycopeptide utilization ratios (GP(T*22)R/GP(T*10)L) were obtained from these normalized values.
Data availability
All data are contained within the article and in the supporting information.
Supplementary Material
Acknowledgments
We thank our colleagues for many helpful discussions.
This article contains supporting information.
Author contributions—C. M., S. J., L. R., E. J. P. D., and N. L. S. data curation; C. M., S. J., L. R., and E. J. P. D. validation; C. M., S. J., Z. A. S., L. R., E. J. P. D., and N. L. S. investigation; C. M., S. J., L. R., E. J. P. D., and N. L. S. visualization; C. M., S. J., and K. G. T. H. methodology; C. M., T. A. G., L. A. T., N. L. S., and K. G. T. H. writing-review and editing; Z. A. S., T. A. G., L. A. T., N. L. S., and K. G. T. H. conceptualization; Z. A. S., T. A. G., L. A. T., N. L. S., and K. G. T. H. supervision; T. A. G., L. A. T., N. L. S., and K. G. T. H. formal analysis; T. A. G., L. A. T., N. L. S., and K. G. T. H. funding acquisition; K. G. T. H. resources; K. G. T. H. writing-original draft.
Funding and additional information—This work was supported by Grants Z01-DE-000713 (to K. G. T. H.), 1-ZIA-DE000739-05 (to L. A. T.), R01-GM-113534 (to T. A. G.), and Z01-DE000754-01 (to N. L. S.) from the Intramural Research Program of the NIDCR, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- AA
- amino acid(s)
- PDB
- Protein Data Bank
- DPM
- disintegrations per minute.
References
- 1. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 10.1093/glycob/cwr182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tran D. T., and Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 10.1074/jbc.R112.418558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 10.1038/ng1358 [DOI] [PubMed] [Google Scholar]
- 4. Ichikawa S., Sorenson A. H., Austin A. M., Mackenzie D. S., Fritz T. A., Moh A., Hui S. L., and Econs M. J. (2009) Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology 150, 2543–2550 10.1210/en.2008-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guda K., Moinova H., He J., Jamison O., Ravi L., Natale L., Lutterbaugh J., Lawrence E., Lewis S., Willson J. K., Lowe J. B., Wiesner G. L., Parmigiani G., Barnholtz-Sloan J., Dawson D. W., et al. (2009) Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc. Natl. Acad. Sci. U.S.A. 106, 12921–12925 10.1073/pnas.0901454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duncan E. L., Danoy P., Kemp J. P., Leo P. J., McCloskey E., Nicholson G. C., Eastell R., Prince R. L., Eisman J. A., Jones G., Sambrook P. N., Reid I. R., Dennison E. M., Wark J., Richards J. B., et al. (2011) Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 7, e1001372 10.1371/journal.pgen.1001372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorski M., Tin A., Garnaas M., McMahon G. M., Chu A. Y., Tayo B. O., Pattaro C., Teumer A., Chasman D. I., Chalmers J., Hamet P., Tremblay J., Woodward M., Aspelund T., Eiriksdottir G., et al. (2015) Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 87, 1017–1029 10.1038/ki.2014.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., Voight B. F., Bonnycastle L. L., Jackson A. U., Crawford G., Surti A., et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41, 56–65 10.1038/ng.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khetarpal S. A., Schjoldager K. T., Christoffersen C., Raghavan A., Edmondson A. C., Reutter H. M., Ahmed B., Ouazzani R., Peloso G. M., Vitali C., Zhao W., Somasundara A. V., Millar J. S., Park Y., Fernando G., et al. (2016) Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24, 234–245 10.1016/j.cmet.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim Y. J., and Varki A. (1997) Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj. J. 14, 569–576 [DOI] [PubMed] [Google Scholar]
- 11. Ono M., and Hakomori S. (2004) Glycosylation defining cancer cell motility and invasiveness. Glycoconj. J. 20, 71–78 10.1023/B:GLYC.0000018019.22070.7d [DOI] [PubMed] [Google Scholar]
- 12. Tian E., Wang S., Zhang L., Zhang Y., Malicdan M. C., Mao Y., Christoffersen C., Tabak L. A., Schjoldager K. T., and Ten Hagen K. G. (2019) Galnt11 regulates kidney function by glycosylating the endocytosis receptor megalin to modulate ligand binding. Proc. Natl. Acad. Sci. U.S.A. 116, 25196–25202 10.1073/pnas.1909573116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 10.1038/ng.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zilmer M., Edmondson A. C., Khetarpal S. A., Alesi V., Zaki M. S., Rostasy K., Madsen C. G., Lepri F. R., Sinibaldi L., Cusmai R., Novelli A., Issa M. Y., Fenger C. D., Abou Jamra R., Reutter H., et al. (2020) Novel congenital disorder of O-linked glycosylation caused by GALNT2 loss of function. Brain 143, 1114–1126 10.1093/brain/awaa063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tian E., and Ten Hagen K. G. (2006) Expression of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology 16, 83–95 10.1093/glycob/cwj051 [DOI] [PubMed] [Google Scholar]
- 16. Tran D. T., Zhang L., Zhang Y., Tian E., Earl L. A., and Ten Hagen K. G. (2012) Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J. Biol. Chem. 287, 5243–5252 10.1074/jbc.M111.306159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerken T. A., Revoredo L., Thome J. J., Tabak L. A., Vester-Christensen M. B., Clausen H., Gahlay G. K., Jarvis D. L., Johnson R. W., Moniz H. A., and Moremen K. (2013) The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 288, 19900–19914 10.1074/jbc.M113.477877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raman J., Fritz T. A., Gerken T. A., Jamison O., Live D., Liu M., and Tabak L. A. (2008) The catalytic and lectin domains of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J. Biol. Chem. 283, 22942–22951 10.1074/jbc.M803387200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Las Rivas M., Paul Daniel E. J., Coelho H., Lira-Navarrete E., Raich L., Compañón I., Diniz A., Lagartera L., Jiménez-Barbero J., Clausen H., Rovira C., Marcelo F., Corzana F., Gerken T. A., and Hurtado-Guerrero R. (2018) Structural and mechanistic insights into the catalytic-domain–mediated short-range glycosylation preferences of GalNAc-T4. ACS Cent. Sci. 4, 1274–1290 10.1021/acscentsci.8b00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Las Rivas M., Paul Daniel E. J., Narimatsu Y., Compañón I., Kato K., Hermosilla P., Thureau A., Ceballos-Laita L., Coelho H., Bernadó P., Marcelo F., Hansen L., Maeda R., Lostao A., Corzana F., et al. (2020) Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3. Nat. Chem. Biol. 16, 351–360 10.1038/s41589-019-0444-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandez A. J., Daniel E. J. P., Mahajan S. P., Gray J. J., Gerken T. A., Tabak L. A., and Samara N. L. (2019) The structure of the colorectal cancer-associated enzyme GalNAc-T12 reveals how nonconserved residues dictate its function. Proc. Natl. Acad. Sci. U.S.A. 116, 20404–20410 10.1073/pnas.1902211116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fritz T. A., Raman J., and Tabak L. A. (2006) Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J. Biol. Chem. 281, 8613–8619 10.1074/jbc.M513590200 [DOI] [PubMed] [Google Scholar]
- 23. Lira-Navarrete E., de Las Rivas M., Compañón I., Pallarés M. C., Kong Y., Iglesias-Fernández J., Bernardes G. J. L., Peregrina J. M., Rovira C., Bernadó P., Bruscolini P., Clausen H., Lostao A., Corzana F., and Hurtado-Guerrero R. (2015) Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat. Commun. 6, 6937 10.1038/ncomms7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Las Rivas M., Lira-Navarrete E., Daniel E. J. P., Compañón I., Coelho H., Diniz A., Jiménez-Barbero J., Peregrina J. M., Clausen H., Corzana F., Marcelo F., Jiménez-Osés G., Gerken T. A., and Hurtado-Guerrero R. (2017) The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences. Nat. Commun. 8, 1959 10.1038/s41467-017-02006-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Revoredo L., Wang S., Bennett E. P., Clausen H., Moremen K. W., Jarvis D. L., Ten Hagen K. G., Tabak L. A., and Gerken T. A. (2016) Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 26, 360–376 10.1093/glycob/cwv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerken T. A., Jamison O., Perrine C. L., Collette J. C., Moinova H., Ravi L., Markowitz S. D., Shen W., Patel H., and Tabak L. A. (2011) Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 10.1074/jbc.M111.218701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gerken T. A., Raman J., Fritz T. A., and Jamison O. (2006) Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J. Biol. Chem. 281, 32403–32416 10.1074/jbc.M605149200 [DOI] [PubMed] [Google Scholar]
- 28. Johansson M. E., Sjövall H., and Hansson G. C. (2013) The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10, 352–361 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tabak L. A. (1995) In defense of the oral cavity: structure, biosynthesis, and function of salivary mucins. Annu. Rev. Physiol. 57, 547–564 10.1146/annurev.ph.57.030195.002555 [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Turner B., Ribbeck K., and Ten Hagen K. G. (2017) Loss of the mucosal barrier alters the progenitor cell niche via Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling. J. Biol. Chem. 292, 21231–21242 10.1074/jbc.M117.809848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ten Hagen K. G., and Tran D. T. (2002) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J. Biol. Chem. 277, 22616–22622 10.1074/jbc.M201807200 [DOI] [PubMed] [Google Scholar]
- 32. Tian E., and Ten Hagen K. G. (2007) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J. Biol. Chem. 282, 606–614 10.1074/jbc.M606268200 [DOI] [PubMed] [Google Scholar]
- 33. Peluso G., Tian E., Abusleme L., Munemasa T., Mukaibo T., and Ten Hagen K. G. (2020) Loss of the disease-associated glycosyltransferase Galnt3 alters Muc10 glycosylation and the composition of the oral microbiome. J. Biol. Chem. 295, 1411–1425 10.1074/jbc.RA119.009807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian E., Hoffman M. P., and Ten Hagen K. G. (2012) O-Glycosylation modulates integrin and FGF signalling by influencing the secretion of basement membrane components. Nat. Commun. 3, 869 10.1038/ncomms1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L., Syed Z. A., van Dijk Härd I., Lim J. M., Wells L., and Ten Hagen K. G. (2014) O-Glycosylation regulates polarized secretion by modulating Tango1 stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7296–7301 10.1073/pnas.1322264111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L., Tran D. T., and Ten Hagen K. G. (2010) An O-glycosyltransferase promotes cell adhesion during development by influencing secretion of an extracellular matrix integrin ligand. J. Biol. Chem. 285, 19491–19501 10.1074/jbc.M109.098145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L., Zhang Y., and Hagen K. G. (2008) A mucin-type O-glycosyltransferase modulates cell adhesion during Drosophila development. J. Biol. Chem. 283, 34076–34086 10.1074/jbc.M804267200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ji S., Samara N. L., Revoredo L., Zhang L., Tran D. T., Muirhead K., Tabak L. A., and Ten Hagen K. G. (2018) A molecular switch orchestrates enzyme specificity and secretory granule morphology. Nat. Commun. 9, 3508 10.1038/s41467-018-05978-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Festari M. F., Trajtenberg F., Berois N., Pantano S., Revoredo L., Kong Y., Solari-Saquieres P., Narimatsu Y., Freire T., Bay S., Robello C., Bénard J., Gerken T. A., Clausen H., and Osinaga E. (2017) Revisiting the human polypeptide GalNAc-T1 and T13 paralogs. Glycobiology 27, 140–153 10.1093/glycob/cww111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raman J., Guan Y., Perrine C. L., Gerken T. A., and Tabak L. A. (2012) UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases: completion of the family tree. Glycobiology 22, 768–777 10.1093/glycob/cwr183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zara J., Hagen F. K., Ten Hagen K. G., Van Wuyckhuyse B. C., and Tabak L. A. (1996) Cloning and expression of mouse UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3. Biochem. Biophys. Res. Commun. 228, 38–44 10.1006/bbrc.1996.1613 [DOI] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. London N., Raveh B., Cohen E., Fathi G., and Schueler-Furman O. (2011) Rosetta FlexPepDock web server: high resolution modeling of peptide–protein interactions. Nucleic Acids Res 39, W249–W253 10.1093/nar/gkr431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raveh B., London N., and Schueler-Furman O. (2010) Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–2040 10.1002/prot.22716 [DOI] [PubMed] [Google Scholar]
- 45. Thomsen M. C., and Nielsen M. (2012) Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res 40, W281–W287 10.1093/nar/gks469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and in the supporting information.