Abstract
The receptor for advanced glycation end products (RAGE) plays a key role in mammal physiology and in the etiology and progression of inflammatory and oxidative stress-based diseases. In adults, RAGE expression is normally high only in the lung where the protein concentrates in the basal membrane of alveolar Type I epithelial cells. In diseases, RAGE levels increase in the affected tissues and sustain chronic inflammation. RAGE exists as a membrane glycoprotein with an ectodomain, a transmembrane helix, and a short carboxyl-terminal tail, or as a soluble ectodomain that acts as a decoy receptor (sRAGE). VC1 domain is responsible for binding to the majority of RAGE ligands including advanced glycation end products (AGEs), S100 proteins, and HMGB1. To ascertain whether other ligands exist, we analyzed by MS the material pulled down by VC1 from human plasma. Twenty of 295 identified proteins were selected and associated to coagulation and complement processes and to extracellular matrix. Four of them contained a γ-carboxyl glutamic acid (Gla) domain, a calcium-binding module, and prothrombin (PT) was the most abundant. Using MicroScale thermophoresis, we quantified the interaction of PT with VC1 and sRAGE in the absence or presence of calcium that acted as a competitor. PT devoid of the Gla domain (PT des-Gla) did not bind to sRAGE, providing further evidence that the Gla domain is critical for the interaction. Finally, the presence of VC1 delayed plasma clotting in a dose-dependent manner. We propose that RAGE is involved in modulating blood coagulation presumably in conditions of lung injury.
Keywords: receptor for advanced glycation end product (RAGE), pattern recognition receptor (PRR), scavenger receptor, prothrombin, coagulation factors, mass spectrometry (MS), plasma, protein γ-carboxylation, lung injury, protein-protein interaction, lung, molecular cell biology, biophysics, complement system, Gla domain
Receptor for advanced glycation end product (RAGE) is a multi-ligand receptor of the superfamily of Igs and is related to the category of cell adhesion proteins (1). RAGE was initially identified for its capability to bind advanced glycation end products (AGEs), a highly heterogeneous class of molecules that derive from nonenzymatic reaction products of sugars with proteins (Maillard reaction) (2). Beside AGEs, RAGE binds to widely different proinflammatory ligands among those members of the S100/calgranulin protein family, high mobility group protein box-1 (HMGB1 or amphoterin), the lipopolysaccharide of bacterial cell walls, β-amyloid-peptides, and the complement C1q and C3a proteins. Further ligands identified are extracellular matrix proteins such collagen I/IV, Mac-1 (member of the β2-integrin family), and complex polymers such as heparan sulfates (reviewed in Ref. 3). In view of the large variety of ligands, RAGE was assigned to the pattern recognition receptor (PRR) family (4–7).
A wide spectrum of RAGE functions have been reported, reflecting the variety of the different ligands. RAGE is involved in the innate and adaptive immune response, in the onset and establishment of inflammation, as well as in tissue homeostasis and repair (3, 8). RAGE is encoded by AGER, a single gene unique to mammals, located in proximity of the major histocompatibility complex locus on Chr 6 where the major histocompatibility complex genes and some genes of the complement system (C4-A and C2) are located (see Ref. 9 and the human genome at the UCSC Genome Browser (10)). AGER is highly expressed in many tissues during embryo development, but in healthy adults its expression decreases in all tissues except in the lung where it is abundant but restricted to alveoli (11–14). Moreover, very low RAGE levels were detected in the heart, skeletal, and smooth muscle, thyroid gland, and adrenal cortex, in specific areas of the brain and in immune cells (dendritic, CD4+ and CD8+ T cells, and monocytes) (12, 15). Notably, RAGE expression increases during the inflammatory response due to a positive feedback loop triggered by the RAGE-dependent activation of the transcription factor NF-κB and to proinflammatory stimuli (16).
The various functions of RAGE are ascribable to several intracellular pathways that are activated depending on the specific ligand, context, and cell type (6). Activation of the signaling pathways occurs via the short cytoplasmic domain that is devoid of any enzymatic activity and oligomerization of the receptor facilitates the recruitment of adapter proteins, such as the cytoskeleton Diaphanous-1 protein (DIAPH1), TIRAP, and MyD88, essential for signaling (3, 5).
Primary signaling pathways activated by RAGE are the p21ras/mitogen-activated cascades of protein kinases (MAPKs), such as ERK1/2, JNK, and p38, NF-κB and JAK/STAT signaling pathways resulting in the activation of transcription of Type I/V collagen that triggers cell adhesion and cell spreading, taking also into account that collagen activates RAGE on the cell surface (6, 8). In addition, the binding of RAGE to Mac-1 integrin on leukocytes triggers ICAM1/VCAM1 expression and promotes leukocyte adhesion. Interestingly, heparin was reported as a ligand that inhibits RAGE signaling (17).
Besides the full-length membrane-bound receptor, a primary soluble form of RAGE is generated by alternative splicing from the AGER_v1 mRNA, named endogenous soluble (esRAGE) (18), or by ADM10-mediated protease digestion of full-length RAGE at the cell surface (19, 20). Extensive work has revealed that soluble RAGE (sRAGE) acts as a decoy for RAGE-ligands and, therefore, blocks the establishment and progression of a wide-range of pathological states in animal models such as cardiovascular diseases, diabetes, cancer, and neuronal dysfunctions (21–24).
RAGE is involved in the etiology and progression of several human diseases and in aging. In particular, RAGE has emerged as a harmful mediator of tissue damage caused by inflammation and oxidative stress, conditions that accelerate the reaction of proteins with sugars and the formation of AGEs by sustaining the activation of NF-κB causing chronic inflammatory disorders (25).
In this work, we report the results of a study aimed to identify new RAGE interactors. RAGE consists of the extracellular IgG-like domains V, C1, and C2, a single transmembrane α-helix, and a short C-terminal cytosolic domain. V and C1 form a structural integrated domain, named VC1, which is connected to the C2 domain through a hinge region. The vast majority of RAGE ligands bind to VC1. Therefore, we exploited VC1 to pulldown new RAGE ligands from human plasma. We identified proteins associated to the blood coagulation and complement processes and the extracellular matrix. Prothrombin (PT) was the major interactor. Moreover, we quantified a moderate affinity binding of PT to VC1 and a higher affinity to sRAGE. These results, along with the inhibition of blood coagulation by VC1, suggest a potential new role of sRAGE in modulating blood coagulation likely in the lung where the protein is abundantly expressed.
Results
Multiple band analysis identifies new plasma interactors of VC1
In the attempt to obtain a more comprehensive picture of the network of molecules interacting with RAGE, we used a previously described in vitro pulldown assay to identify potential new binding partners present in human plasma. Briefly, the ligand-binding domain of human RAGE (VC1) was immobilized through a C-terminal Strep-tag on streptavidin (STV)-coated magnetic beads (26). The Strep-tag has high affinity for STV allowing for stringent washing and eliminating weak or unspecific binders. The VC1 domain was expressed and secreted from the yeast Pichia pastoris (Fig. S2). VC1 was N-glycosylated and showed a remarkably high thermal stability and solubility, two important features to set up a robust assay (27). The use of VC1 in pulldown assays was validated by demonstrating its capability to capture in vitro modified AGE-human serum albumin (AGE-HSA), whereas lacking any interaction with unmodified HSA (26–28).
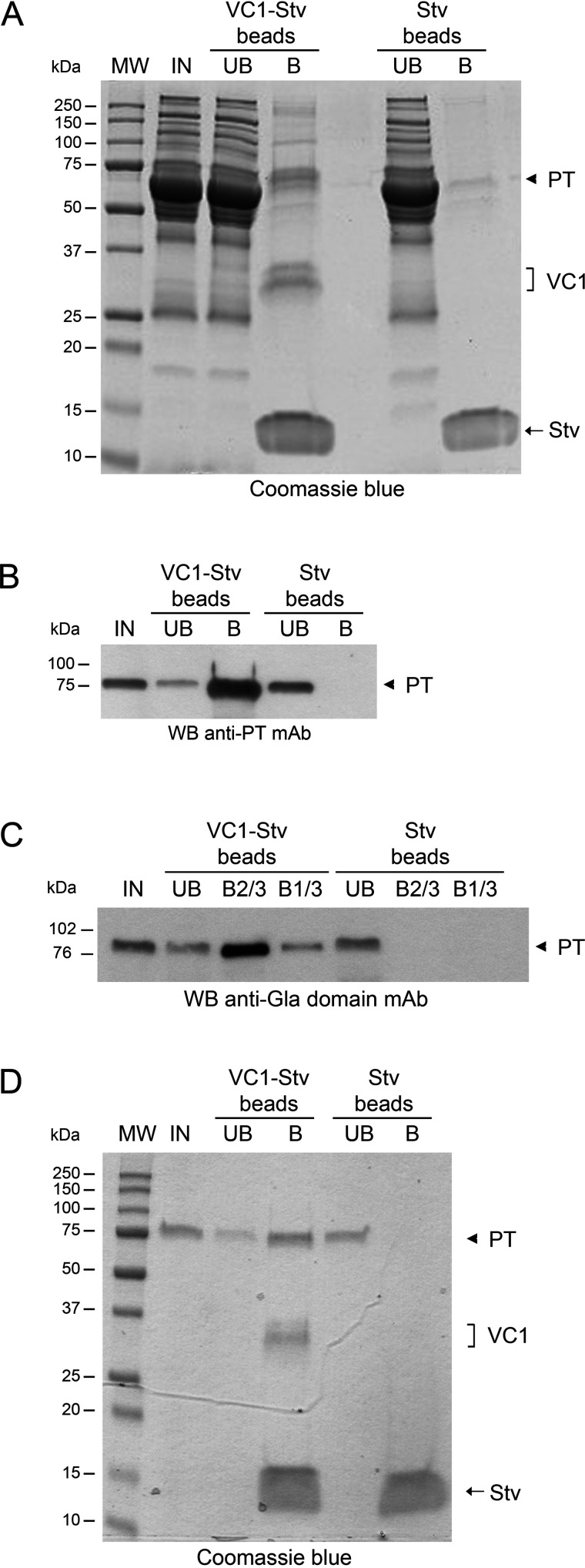
Aliquots of plasma from healthy individuals were diluted to 5 mg of protein/ml and 800 µg were used in each pulldown assay. After 2 h of incubation with plasma at 4 °C, the VC1-STV–coated beads were extensively washed and the bound material was solubilized in SDS sample buffer and analyzed by SDS-PAGE. As a control, the same procedure was applied using the STV-coated beads alone (STV-beads). A major band of about 75 kDa was reproducibly detected by SDS-PAGE and Coomassie staining in the material solubilized from VC1-STV beads, but not in the material solubilized from STV beads alone (Fig. 1A). To identify the ligands bound to VC1, we analyzed only the region of the gel above the 37-kDa marker. The abundance of STV (∼14 kDa) and VC1 (32-34 kDa) proteins, solubilized as well from the beads, did not allow the identification of ligands migrating in the range of molecular mass lower than 37 kDa (see “Experimental procedures”).
Figure 1.
PT is the most abundant RAGE-interactor detected in human plasma by a VC1 pulldown assay. A, SDS-PAGE of the fractions from pulldown assays carried out in 20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.1. IN, input; UB, unbound fraction; B, bound material obtained after boiling the beads for 5 min in SDS sample buffer. The arrowhead indicates the 75-kDa polypeptide identified as PT and present in the VC1-STV beads and absent in the control (STV beads). VC1 and STV are also indicated. Proteins were stained by Coomassie Blue. B, Western blotting (WB) analysis using anti-PT mAb of fractions from a VC1 pulldown assay. C, Western blotting analysis of a VC1 pulldown assay using anti-Gla domain mAb. B2/3 and B1/3 indicate that 2/3 and 1/3 of the total bound material were loaded. D, VC1 pulldown assay using purified PT instead of plasma. The indicated samples were analyzed by SDS-PAGE and proteins were stained by Coomassie Blue.
Gel pieces were excised and processed for peptide LC–MS fingerprinting (multiple band analysis). To increase the specificity in the identification without losing sensibility, different pulldown assays were set up as follows: (i) 5 independent pulldown assays were performed, each with plasma from a single individual; (ii) a pulldown assay from a pool of three plasma samples from other individuals; and (iii) 2 pulldown assays from the plasma of other two single subjects. The set of proteins in common to both VC1-STV and STV beads was discarded during data elaboration. The experimental design is summarized in Fig. S1.
Dataset S1 lists the human plasma proteins identified by MS (for details see “Experimental procedures”). Twenty of 295 identified proteins were selected by applying a cut-off of positive identifications in ≥3 pulldown assays over a total of 8. The subset of 20 proteins, shown in Table 1, was analyzed by the software STRING, which reveals functionally associated protein networks (29). The identified proteins converged on the complement and coagulation cascades in the KEGG classification (pathway hsa04610) with a false discovery rate of 1.05 e−18 (Fig. S3).
Table 1.
Human plasma proteins interacting with VC1 in pulldown assays
The bound material of VC1 pulldown assays from plasma samples was analyzed by LC–MS. Of 295 proteins identified and listed in supporting Dataset S1, 20 were selected using a cut-off of ≥3 positive identifications over 8 pulldown assays.
| String accession | Accession | Description | Gla domain | Molecular mass | Positive identification |
|---|---|---|---|---|---|
| kDa | |||||
| F2 | P00734 | Prothrombin | + | 70 | 8 |
| C4BPA | P04003 | C4b-binding protein α chain | 67 | 8 | |
| PROS1 | P07225 | Vitamin K-dependent protein S | + | 75.1 | 8 |
| C4A | P0C0L4 | Complement C4A | 192.7 | 8 | |
| F9 | P00740 | Coagulation factor IX | + | 51.7 | 6 |
| A2M | P01023 | α2-Macroglobulin | 163.2 | 6 | |
| VTN | P04004 | Vitronectin | 54.3 | 5 | |
| THBS1 | P07996 | Thrombospondin-1 | 129.3 | 5 | |
| ITIH2 | P19823 | Inter-α-trypsin inhibitor heavy chain H2 | 106.4 | 5 | |
| COMP | P49747 | Cartilage oligomeric matrix protein | 82.8 | 5 | |
| IGHG2 | P01859 | Immunoglobulin heavy constant γ2 | 35.9 | 4 | |
| IGHM | P01871 | Immunoglobulin heavy constant μ | 49.4 | 4 | |
| PROZ | P22891-2 | Isoform 2 of vitamin K-dependent protein Z | + | 47 | 4 |
| C3 | P01024 | Complement C3 | 187 | 3 | |
| IGHA1 | P01876 | Immunoglobulin heavy constant α1 | 37.6 | 3 | |
| FGA | P02671-1 | Fibrinogen α chain | 94.9 | 3 | |
| F5 | P12259 | Coagulation factor V | 251.5 | 3 | |
| FRK | P42685 | Tyrosine-protein kinase FRK | 58.2 | 3 | |
| ITIH3 | Q06033-1 | Inter-α-trypsin inhibitor heavy chain H3 | 99.8 | 3 | |
| TTN | Q8WZ42-12 | Isoform 12 of Titin | 3992.1 | 3 |
As indicated in Table 1, PT, the coagulation factors IX, and the anticoagulant proteins S and Z were reproducibly detected. These proteins share the Gla domain, a domain rich in glutamic acid residues that are modified by γ-carboxylation to γ-carboxylated residues (Gla). Among the other interactors, the α-chain of the complement C4b-binding protein (C4BP) and the complement C4A are notable. C4BP, an inhibitor of C4 in the complement system, is a multimeric and abundant plasma protein (25 µg/ml), containing 7 identical α-chains and a unique β-chain. Protein S (PROS1) binds C4BP through the β-chain. Approximately 60% of total PROS1 (∼15 µg/ml) circulates in complex with C4BP, whereas ∼40% (∼10 µg/ml) is in free form (30). C4A occurred in all the analyzed samples (Table 1). Prior to secretion, C4A cleavage generates three chains, the largest of which is C4b (192.7 kDa), which interacts with α-chains of C4BP. Thus, C4BP and C4A could be direct interactors or captured in complex with Protein S. Finally, C4b undergoes further processing that leads to the assembly of a complex that activates C5 and C3 convertases. Notably, C3a, a proteolytic fragment of C3 that was previously identified as a RAGE-ligand, was also identified in our study (31). In addition, α2-macroglobulin is another interesting interactor because it is highly expressed in the lung. Finally, another class of interactors is related to the extracellular matrix: vitronectin, thrombospondin-1, and the cartilage oligomeric matrix protein (also known as thrombospondin-5), and support a role in cell adhesion, a process in which RAGE is known to be involved into.
Finally, to verify whether AGE modifications contributed to the binding of the interactors to VC1, we searched for the presence of AGE/ALE modifications in the identified proteins. No AGE or ALE (advanced lipoxidation end products) modifications were detected by MS using a previously described search method (26, 32) (see “Discussion”).
Single band analysis identified the 75-kDa polypeptide as prothrombin
The major polypeptide present in pulldown assays, the 75-kDa band of Fig. 1A, was excised from Coomassie-stained gels and MS analyses identified this band as prothrombin (PT) (Table 2). Single band analysis was repeated and PT was invariably detected as the most abundant interactor of VC1.
Table 2.
Identification of the 75-kDa polypeptide as prothrombin
MS fingerprinting of the ∼75 kDa band excised from the gel of the VC1-bound material fraction separated by SDS-PAGE as shown in Fig. 1A.
| Peptide sequencea | PSMsb | XCorrc | Charged | MH+ [Da]e | ΔMf |
|---|---|---|---|---|---|
| ppm | |||||
| 101AAcLEGNcAEGLGTNY116 | 2 | 3.64 | 2 | 1699.70293 | −1.29 |
| 105EGNcAEGLGTNYR117 | 2 | 3.35 | 2 | 1440.61601 | −0.88 |
| 178RQEcSIPVcGQDQVTVAmTPR198 | 2 | 6.49 | 3 | 2448.14023 | −1.11 |
| 199SEGSSVNLSPPLEQcVPDR217 | 2 | 3.94 | 2 | 2070.97295 | −1.55 |
| 207SPPLEQcVPDR217 | 2 | 3.00 | 2 | 1297.61797 | −2.01 |
| 207SPPLEQcVPDRGQQY221 | 2 | 5.31 | 2 | 1773.82146 | −0.60 |
| 248HQDFNSAVQLVENF261 | 2 | 3.75 | 2 | 1647.77519 | −0.62 |
| 252NSAVQLVENF261 | 2 | 3.61 | 2 | 1120.56145 | −1.71 |
| 276VAGKPGDFGY285 | 2 | 3.10 | 2 | 1010.49156 | −2.63 |
| 289NYcEEAVEEETGDGLDEDSDR309 | 2 | 4.87 | 2 | 2431.93169 | 0.07 |
| 291cEEAVEEETGDGLDEDSDR309 | 2 | 5.11 | 2 | 2154.82280 | −1.15 |
| 330GSGEADcGLRPL341 | 2 | 3.73 | 2 | 1231.57231 | −1.07 |
| 364IVEGSDAEIGMSPW377 | 2 | 3.37 | 2 | 1490.67912 | −2.77 |
| 391cGASLISDR399 | 2 | 2.95 | 2 | 978.46599 | −1.40 |
| 418TENDLLVR425 | 2 | 3.17 | 2 | 959.51494 | −0.77 |
| 435ERNIEKISML444 | 1 | 3.21 | 2 | 1232.66509 | −1.39 |
| 475SDYIHPVcLPDR486 | 4 | 3.30 | 3 | 1471.69828 | −1.09 |
| 478IHPVcLPDRETAASL492 | 2 | 3.25 | 2 | 1678.85649 | −1.01 |
| 524QVVNLPIVER533 | 2 | 2.91 | 2 | 1166.68779 | −1.24 |
| 524QVVNLPIVERPVcK537 | 4 | 3.87 | 2 | 1650.93450 | −0.94 |
| 554KPDEGKR560 | 2 | 2.90 | 2 | 829.45293 | 0.29 |
| 560RGDAcEGDSGGPF572 | 2 | 4.19 | 2 | 1324.52056 | −1.34 |
| 561GDAcEGDSGGPF572 | 2 | 3.24 | 2 | 1168.41997 | −1.07 |
| 613IQKVIDQFGE622 | 2 | 3.69 | 2 | 1176.62395 | −1.72 |
aThe amino acid residues refer to the precursor sequence of prothrombin from Homo sapiens (UniProt accession number P00734, amino acids 1-622). Sequence coverage was 35.37%. Lowercase c indicates cysteine carbamidomethylation and lowercase m indicates methionine oxidation.
bPSMs: peptide spectrum matches (total number of identified peptide sequences including those redundantly identified).
cXCorr score values from Sequest algorithm.
dCharge state of the precursor ion.
eMH+ [Da] represents the singly charged mass of the precursor ions in Da.
fDifference between the theoretical mass of the peptide and the experimental mass of the precursor ion.
To confirm that the 75-kDa band indeed represented PT, we performed Western blotting analyses of the pulldown assays. A monoclonal anti-PT antibody clearly detected the 75-kDa polypeptide from VC1-STV beads, which was absent in samples prepared with STV beads alone (Fig. 1B). Thus, PT was identified as the most prominent RAGE interaction partner from plasma. Furthermore, we performed Western blotting analysis of a VC1 pulldown assay using anti-Gla mAb. As shown in Fig. 1C, the 75-kDa band was present, showing that PT is γ-carboxylated, a post-translational modification that could not be detected in the conditions used in our LC–MS analysis. Accordingly, PT is the most abundant Gla domain–containing protein captured by VC1 from plasma. This is likely related to the level of PT in the plasma that is higher than that of the other circulating Gla domain containing proteins (see also “Discussion”). Accordingly, Western blotting analysis failed to reveal other Gla domain–containing polypeptides. As expected, no band was detected in the material from the control STV beads. Hence, the 75-kDa polypeptide present in the VC1 pulldown assays is PT and is also the only Gla domain–containing protein detected in the pulldown from plasma using Western blotting analysis, taken into consideration that its level is about 1 order of magnitude higher than the level of the other detected Gla–containing proteins.
PT is a direct interactor of VC1 and binding is mediated by the Gla domain
To assess whether PT is a direct interactor of VC1, we repeated the pulldown using purified PT. As shown in the Coomassie-stained gel of Fig. 1D, VC1-STV beads captured a 75-kDa polypeptide that was absent in the material from the STV beads alone. Thus, PT is a direct interactor of VC1.
The interaction of PT with VC1 is competed by Ca2+
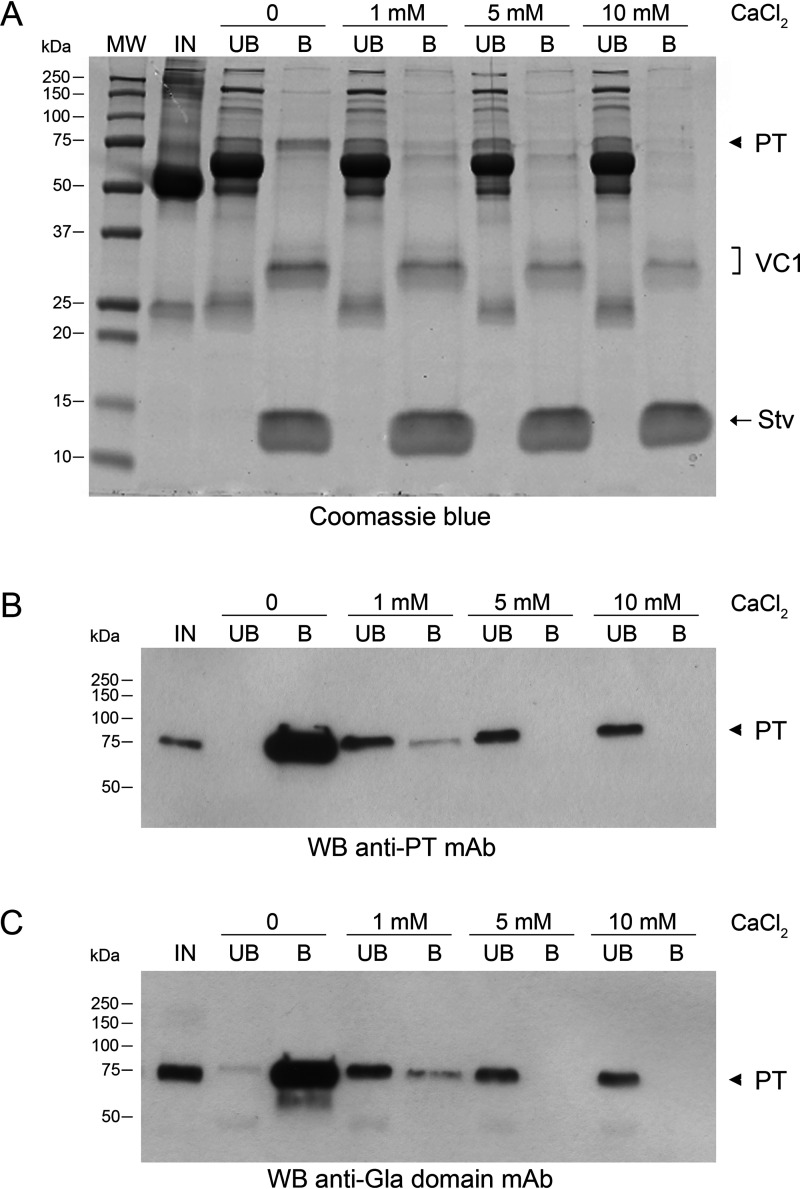
The occurrence of proteins sharing the Gla domain in the VC1 plasma interactome raised the hypothesis that the binding of VC1 to PT was mediated by this domain. The Gla domain is a calcium-binding module (Kd for calcium 0.3-0.5 mm) that in the calcium-bound form defines the binding of Gla–containing proteins to membrane phospholipids (33). In human PT, the Gla domain is an N-terminal region of 46 amino acids with 10 γ-carboxyl glutamic acid residues (Gla) that are responsible for the binding to calcium ions. Upon binding calcium ions, the negative charges of the Gla residues are neutralized and the Gla domain undergoes conformational changes that sequester calcium ions in the interior of the structure. We reasoned that if the PT-VC1 interaction was mediated by electrostatic interactions between the positively charged V domain and the highly negatively charged Gla domain, calcium ions would affect the binding. Because the human plasma was devoid of free calcium, due to the use of EDTA to inhibit coagulation, we repeated the pulldown assays after re-addition of CaCl2. As shown in Fig. 2A, in the presence of different concentrations of Ca2+, the level of PT bound to VC1 decreased in a dose-dependent manner. Both Western blotting analysis with anti-Gla and anti-PT antibodies confirmed that the 75-kDa polypeptide, whose intensity decreased, was PT (Fig. 2, B and C). To rule out that the displacement of the interacting partner by calcium was simply due to the increase of the ionic strength of the environment, we compared the effect of the addition of calcium chloride to the PT-VC1 binding reaction with those obtained by addition of NaCl in amounts that yielded the same ionic strength (i.e. 10 mm CalCl2 is equivalent to 30 mm NaCl) and no relevant effects on the VC1 pulldown were detected (data not shown).
Figure 2.
Competition in PT binding to VC1 by CaCl2. The pulldown assays were carried out in 20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.1, before and after addition to plasma of increasing concentrations of CaCl2. IN, input; UB, unbound fraction; B, bound material obtained after boiling the beads for 5 min in SDS sample buffer. A, proteins were analyzed by SDS-PAGE and stained by Coomassie Blue. B, Western blot (WB) analysis of the fractions of pulldown assays using anti-PT mAb. C, Western blot analysis of the fractions from pulldown assays using anti-Gla mAb. Arrowhead, PT. VC1, VC1 domain.
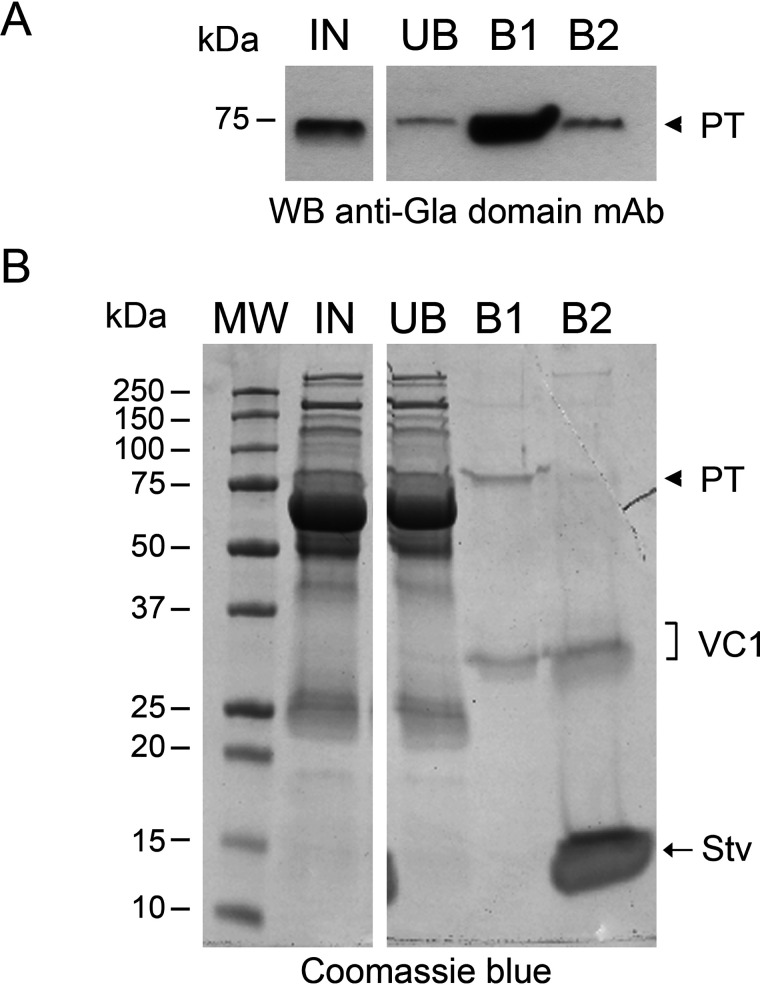
Next, we tested whether addition of calcium could displace PT already bound to VC1-STV beads. Fifty mm CaCl2 was capable to elute PT from the beads (Fig. 3A). Any residual material still bound to the resin was recovered by boiling the beads in SDS sample buffer. The majority of PT was released in the calcium-elution fraction (B1). To rule out any effect of 50 mm CaCl2 on VC1 binding to the STV beads, we stained a parallel gel with Coomassie (Fig. 3B). In the fraction eluted with calcium, where the majority of PT was released, only a tiny amount of VC1 was detected. The amount was negligible compared with that obtained by boiling the resin in SDS (Fig. 3B, lane B2 compared with B1). Thus, high concentrations of Ca2+ displace the Gla domain of PT from VC1 without significantly affecting VC1 binding to the resin.
Figure 3.
Calcium at high concentration displaces PT from the complex with VC1. A, Western blot (WB) analysis with anti-Gla domain mAb of fractions from VC1 pulldown assays carried out in 20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.1. IN, input; UB, unbound fraction; B1, bound material obtained after incubating the beads with 50 mm CaCl2; B2, residual bound material obtained by boiling beads from B1 for 5 min in SDS sample buffer. B, Coomassie-stained gel of the pulldown assays obtained in the condition described in A. The arrowhead indicates prothrombin.
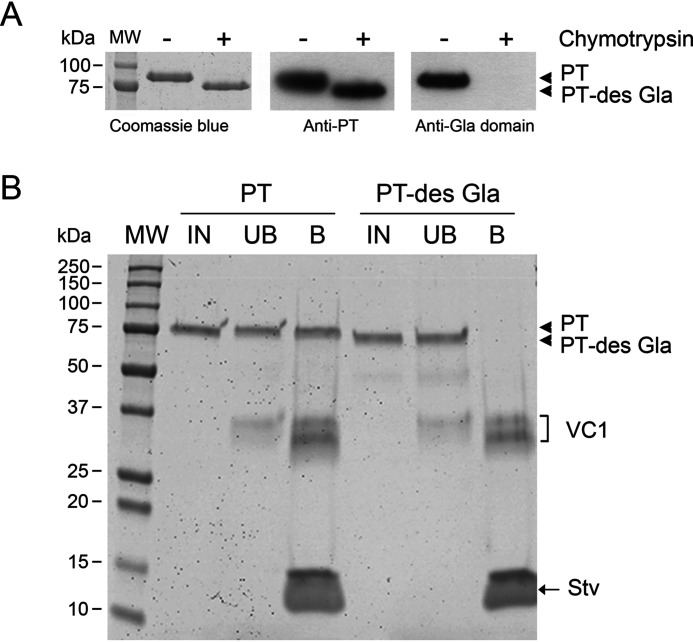
Finally, to further prove that the Gla domain was the site of interaction of PT with VC1, we repeated the pulldown assay using purified full-length PT and PT devoid of the Gla domain (PT des-Gla), obtained by treating PT with α-chymotrypsin (34) (Fig. 4). In contrast to the strong interaction of full-length PT, PT des-Gla showed no binding to VC1, corroborating the finding that the interaction of PT with RAGE occurs via the Gla domain.
Figure 4.
The Gla domain mediates the interaction of PT with VC1 in a pulldown assay. A, proteolytic treatment of purified PT removed the Gla domain and produced PT des-Gla. B, Coomassie-stained gel of VC1 pulldown assays performed in 20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.1 (no CaCl2), in the presence of PT or PT des-Gla. Fractions were labeled as: IN, input; UB, unbound fraction; B, bound material obtained after boiling the beads for 5 min in SDS sample buffer.
MST analysis of the molecular interaction of PT with VC1 or sRAGE
To further characterize the PT-RAGE interaction, we analyzed the binding of PT to VC1 by MicroScale thermophoresis (MST), a powerful technique to analyze and quantify interactions between proteins in solution (35, 36). In preliminary studies, the well-known RAGE ligands, S100B and S100A12 proteins, were used as positive control in MST and affinity values were similar to those obtained by Biacore/surface plasmon resonance assays, where one of the reactants is immobilized on a matrix (37) (data not shown).
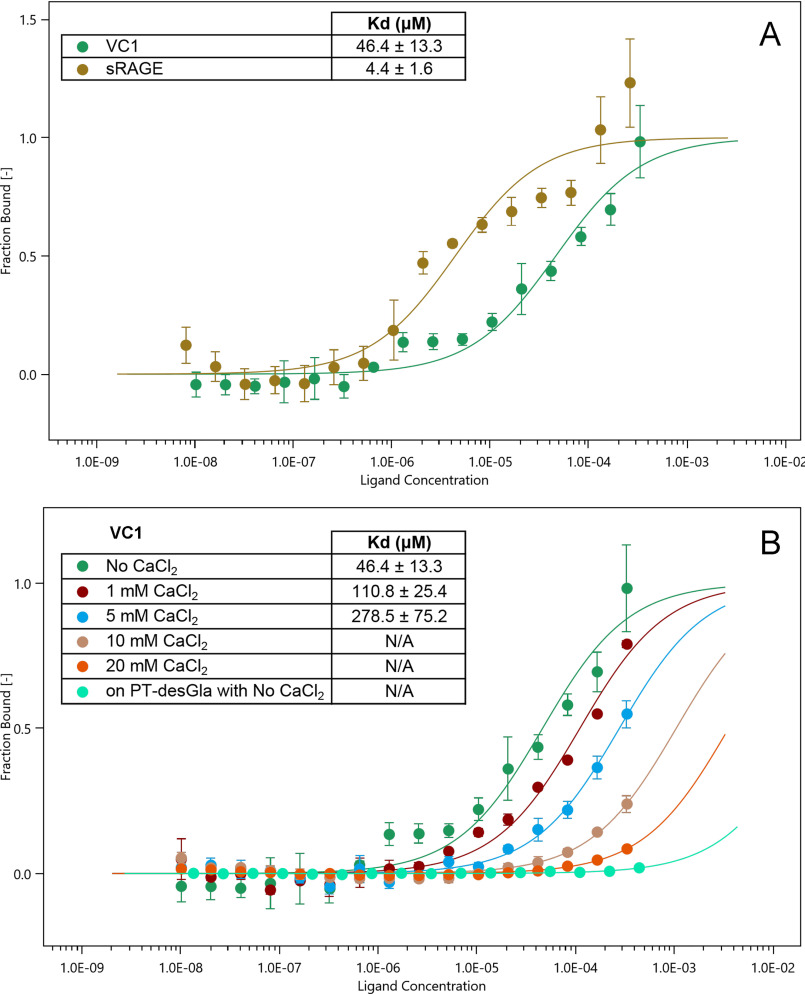
Fig. 5A shows the binding curve of fluorescently labeled PT to VC1 in the absence of calcium. MST analysis revealed the formation of a 1:1 stoichiometry complex between the two proteins with an apparent dissociation constant (Kd) of 46.4 ± 13.3 µm, indicating a moderate affinity of PT for VC1.
Figure 5.
Molecular interaction of PT with VC1 and sRAGE. A, MST measurements were carried out in 20 mm HEPES, pH 7.1, 100 mm NaCl, 0.05% Tween 20 and standard capillaries with a constant concentration of fluorescently labeled prothrombin (100 nm) mixed with varying amounts of unlabeled VC1-His-Strep (final concentrations from 10.2 nm to 333 μm) or sRAGE (from 8.1 nm to 264 μm). After 30 min incubation at 24 °C, the samples were loaded into Monolith NT.115 capillaries (NanoTemper Technologies) and measurements were performed using the Monolith NT.115 (NanoTemper Technologies) at 20% LED power and medium MST power. The values of the dissociation constant (Kd) derived from the curves for the PT-VC1 interaction (n = 5 independent measurements) and for the PT-sRAGE interaction (n = 4 independent measurements) are indicated in the inset. B, MST measurements with VC1 were performed as in A, but in the presence of increasing concentrations of CaCl2 or in the presence of fluorescently labeled PT des-Gla instead of PT (n = 3 independent measurements). N/A, not applicable. In A and B, the fraction bound is plotted against the log10 of VC1 or sRAGE molar concentrations. The error bars on individual data points represent the standard deviations among the repeats.
In addition, we analyzed the binding of PT to human recombinant sRAGE, spanning from residue 23 to 322 and corresponding to the VC1-C2 domains. sRAGE was produced as a secreted glycoprotein in P. pastoris, as described under “Experimental procedures.” Purified sRAGE migrated as a smeared polypeptide of about 45-60 kDa in agreement with its glycosylated nature (Fig. S2). The binding reaction of PT to sRAGE showed a Kd of 4.4 ± 1.6 μm, indicating that the affinity of PT for the ectodomain of RAGE is significantly higher than that for VC1 (Fig. 5A). As a next step, we tested the effect of calcium on PT binding to VC1 in MST assays. Increasing concentrations of calcium decreased the affinity of PT for VC1 and at the highest concentration, 20 mm CaCl2, the binding was almost abolished (Fig. 5B). Fig. S4 shows that also the binding of PT to sRAGE was abolished at high calcium concentration.
We also analyzed by MST the interaction of PT des-Gla with VC1 and sRAGE. PT des-Gla neither bound to VC1 (Fig. 5B) nor to sRAGE (Fig. S4) providing further evidence that the Gla domain is crucial for the interaction.
VC1 inhibits plasma clotting
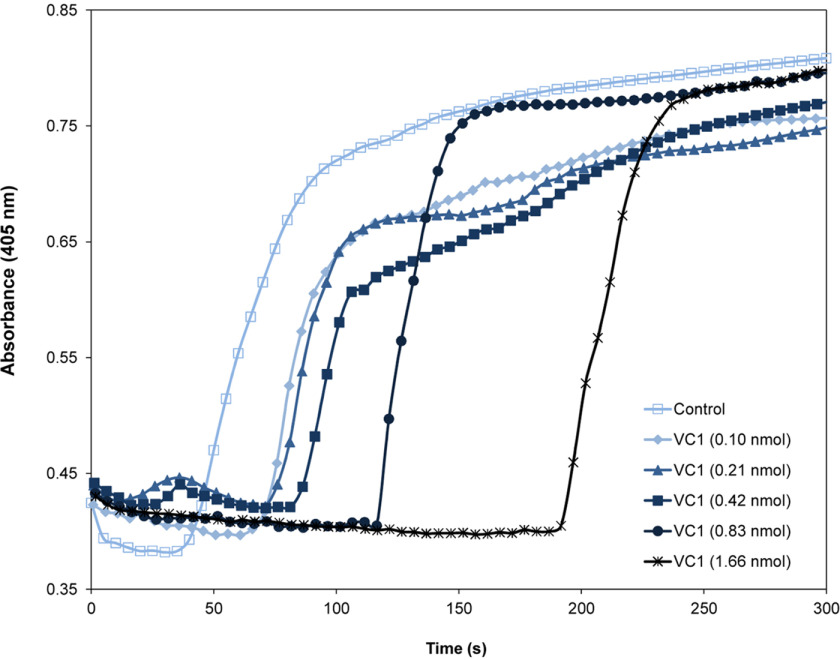
Because PT has a fundamental function in blood coagulation, we explored whether the interaction of PT with RAGE might modulate this process. To assess a potential role of VC1 in blood coagulation we measured plasma clotting by a conventional assay based on the use of activated partial thromboplastin time (aPTT) reagent to activate the intrinsic pathway. Different amounts of VC1 were added to the plasma prior the coagulation reaction was started. The clotting of the control initiated after 50 s, which is typical in these assays. Remarkably, the presence of VC1 drastically prolonged the onset of coagulation in a dose-dependent manner up to 4-fold with an onset of the clotting after ∼200 s (Fig. 6). To rule out any effect of VC1 on thrombin activity, VC1 was added to a thrombin activity assay using a substrate with a thrombin-cleavage site and activity was unaffected by VC1 (data not shown).
Figure 6.
Effects of VC1 on plasma clotting induced by Ca2+. Plasma-citrate was preincubated with various amounts of VC1 (0.1-1.66 nmol in 100 µl of plasma) and the intrinsic pathway was activated by the addition of aPTT reagent (10 µl). After addition of 20 µl of 25 mm CaCl2, plasma clotting was monitored by measuring the absorbance at 405 nm using a microtiter plate reader. Time course curves are representative of three independent experiments.
Discussion
In this work, we demonstrated that the RAGE ligand-binding domain, VC1, and sRAGE bind PT via the Gla domain. The Gla domain is highly conserved among species and is present at the N-terminal end in proteins that undergo vitamin K-dependent γ-carboxylation of glutamate residues. This post-translational modification occurs in the endoplasmic reticulum and is essential for life (38). Among the Gla-containing proteins identified in this study, four (PT, protein S, protein Z, and Factor IX) were reproducibly enriched as detected by MS analysis of VC1-interacting material from human plasma. The Gla domain–containing factor VII escaped detection in the VC1 pulldown experiments, most likely due to the tiny circulating amounts in the plasma (levels are from Ref. 39 and from Enzyme Research Laboratories Inc.). Indeed, the concentration of Factor VII is only 0.5 µg/ml compared with the levels of circulating PT (90 µg/ml), protein S (free, 10 µg/ml; total, 25 µg/ml), factor IX (5 µg/ml), and protein Z (2.2 µg/ml). Another Gla domain–containing factor, Factor X (8 µg/ml) is, nevertheless, not reported in Dataset S1, because it was present both in the material bound to VC1 and control beads, and according to the adopted criteria, was removed from the dataset of proteins specifically interacting with VC1. Regarding the interaction of VC1 with C4BP, it is remarkable to note that C4BP binds apoptotic and necrotic cells as well as DNA, to clean up after injury (30). This interaction is mediated by the Gla domain of protein S and does not affect the ability of C4BP to inhibit the complement. Notably, the known RAGE ligand C3a (31) was also identified in our analysis.
No AGE or ALE modifications were detected in the identified proteins. This is not surprising because AGEs and ALEs require a long time to form and therefore in physiological conditions accumulate in proteins with very low turnover rates such as collagen. The turnover of plasma proteins is much higher, preventing an accumulation of AGEs in amounts compatible with the affinity of a pulldown assay (39). The common RAGE ligands, HMGB1 and S100 proteins, could not be identified because members of the S100 protein family and HMGB1, are small proteins of ∼10-14 kDa (monomer) and 29 kDa, respectively, and in our study we restricted the analyses to the region of the gels containing polypeptide bands with molecular mass higher than 37 kDa. The reason of this choice was that in the elution step, STV (∼14 kDa) and VC1 (∼32-34 kDa) were released from the beads in high abundance and the identification of specific low-abundance VC1 ligands by MS, was not feasible (see “Experimental procedures”).
According to previous reports (40), proteins other than RAGE specifically interact with Gla domain–containing proteins through the Gla domain. Factor X zymogen binds to the adenovirus serotype 5 hexon protein (Ad5 hexon) via the FX Gla domain and this interaction is required for the entry of the virus into hepatocytes. Another example is the superantigen-like protein 10 (SSL10) of Staphylococcus aureus, a bacterial exoprotein that directly binds to the Gla domain of PT and factor Xa, and binding is competed by Ca2+ (41). SSL10 has two short and highly basic segments (pI ∼11 and pI ∼9.8) that are responsible for the binding of SSL10 to PT (42). The interaction likely occurs between the highly positively charged segments and the highly negatively charged Gla domain of PT. Thus, electrostatic attractions could dominate the bimolecular PT-SSL10 interaction.
The nature of interaction of VC1 with Gla domain–containing proteins seems to be similar to that reported for SSL10 and has a marked electrostatic nature. In particular, the V domain, a so called “basic trap” of RAGE, is rich in lysine and arginine residues that are positively charged at physiological pH. In addition, the structure of RAGE reveals the presence of a shallow binding region that could provide a surface of interaction with the Gla domain. The binding of PT via its Gla domain to sRAGE was confirmed by pulldown and MST experiments corroborating the interaction of PT with VC1 domain in the context of the entire ectodomain. Interestingly, sRAGE displayed a higher affinity toward PT than VC1 alone (Fig. 5A) raising the question whether C2 domain is involved in the interaction. C2 domain of RAGE displays a net negative charge and interaction with the negatively charged Gla domain of PT is rather unlikely. Nevertheless, the C2 domain might interact with other regions of PT and contribute to the stability of the complex. Furthermore, it was reported that the linker between the C1 and C2 domain as well as the C2 domain contribute to dimerization of sRAGE (43, 44), although in our MST set up and using the glycosylated sRAGE produced in P. pastoris we did not have evidence of a partial sRAGE dimerization in solution.
Ca2+ could effectively compete with RAGE-PT binding as observed for the interaction of SSL10 with PT. The competition effect of Ca2+ was clearly stronger than a simple increase of ionic strength. Gla domain has an affinity for Ca2+ ions that mask the highly negative charges of the γ-carboxylated glutamate residues and induce a transition from a disordered conformation to a structure in which the acidic residues complexed with calcium are buried inside. This contributes to confer PT the ability to bind membrane phospholipids (PS). We propose that these effects mask the Gla domain from interacting with basic proteins, such as SSL10 or the basic trap V domain of RAGE.
The physiological importance of the binding of Gla domain to calcium ions is well-established contributing to the assembly of active coagulation factors complexes: factor VIIIa-IXa that activates factor X to factor Xa and factor Va–factor Xa complex (prothrombinase complex) on the surface of membranes and leads to efficient proteolytic conversion of PT to thrombin. For this reason, interaction of the Gla domain with VC1 impairs the binding to calcium and interferes with the activation of the coagulation factors causing a delay in plasma clotting. In this respect, the Gla domain could function as a sticky segment that interacts with positively charged domains. To address the exact physiological role of the RAGE-PT interaction, further in vivo studies are required. It should be noted that the amount of circulating sRAGE in healthy subjects is too low (1.1-1.7 ng/ml) to affect blood coagulation at the ∼1.2 mm free calcium concentration typical for blood. A different scenario could be envisaged in the lung. The basal membrane of alveolar Type I (ATI) cells of the lung, rich in membrane-bound RAGE, is in direct contact with the endothelium of capillary vases and with the interstitial space, and together constitute the capillary-alveolar membrane whose integrity is fundamental for gas exchanges. In the adult, RAGE is mainly expressed in the ATI cells and it may be in contact with blood during the gas exchanges and also be present as sRAGE in the interstitial space of the capillary-alveolar membrane (13, 45). In conditions of severe lung injury as acute lung injury (ALI) and/or acute respiratory distress syndrome, losses of integrity of the alveolar-capillary membrane or changes in capillary permeability result in an extensive efflux of protein-rich edema fluid into the alveolar airspace compartment. Herein, macrophages release cytokines, tumor necrosis factor α, interleukins, and a cascade is triggered that leads to induction of coagulation in the alveolar compartment and generates a pulmonary coagulopathy (46–48). In severe ALI the plasma level of sRAGE increases and reaches a concentration of 6-12.3 ng/ml. In edema fluid of ALI/acute respiratory distress syndrome patients, sRAGE concentration is in the range of 0.9-3.2 µg/ml (49). Therefore, in these diseases, sRAGE in the alveolar space may contribute in modulating coagulation by sequestering PT from activation to thrombin. Alternatively, because thrombin plays not only a role in hemostasis but also in promoting fibrosis via thrombin receptors (50), the binding of PT to sRAGE could be beneficial in reducing/delaying thrombin signaling to fibrosis. Indeed, it is known that sRAGE exerts antifibrotic effects in a model of idiopathic pulmonary fibrosis (51). However, studies using RAGE constitutive KO homozygous mice are controversial. Lack of RAGE seems to either protect mice from lung fibrosis (50, 52) or to enhance fibrosis in some models of induced pulmonary fibrosis (51). In addition, RAGE−/− mice spontaneously develop pulmonary fibrosis with age (53). The model of disease used appears to affect the conclusions of these studies and the capability of the lung to respond to injuries. In conclusion, the new interactors identified in this work may contribute in the future to deepen the understanding of the physiological functions of RAGE and its involvement in lung pathologies.
Experimental procedures
Plasma collection
At the recruitment, blood samples from six healthy volunteers were collected between 8 and 9 a.m., following at least 6 h of fasting. To inhibit coagulation, VACUETTE® EDTA 7-ml tubes containing K3EDTA or VACUETTE® 6-ml tubes containing a buffered sodium citrate solution at the concentration of 0.109 mol/liter (3.2%) mixed at a ratio of 1 part citrate solution to 9 parts blood, were used. Plasma-EDTA or plasma-citrate samples were obtained after centrifugation at 1200 × g at room temperature for 15 min. The protein concentration of the supernatant was determined by BCA protein assay.
The studies were approved by the Ethical Committee of Ca' Granda Ospedale Maggiore Policlinico and the University of Milan. All subjects gave their informed consent to participate in the studies. The studies abide by the Declaration of Helsinki principles.
VC1 pulldown assay
The VC1 portion of human RAGE was expressed and purified with a C-terminal tandem tag (His and Strep tag) from P. pastoris culture supernatant as previously described (26). The recombinant protein was immobilized on STV-coated magnetic beads (Streptavidin Mag Sepharose™, GE Healthcare) by exploiting the affinity of the Strep tag for STV. To obtain the VC1-STV beads, 50 μg of purified VC1-His-Strep in 170 μl of buffer A (20 mm HEPES, pH 7.1, 100 mm NaCl) were added to 5 μl of packed STV-coated beads, previously equilibrated with the same buffer. 170 μl of buffer A were added to the same amount of beads to obtain the STV beads. After 1 h of incubation at 4 °C on a rotary mixer, the unbound material was carefully removed and the magnetic beads were washed twice with 500 μl of Buffer A.
For pulldown assays, plasma samples were centrifuged at 4 °C for 10 min at 16,000 rcf and then diluted to 5 mg/ml in buffer B (20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.1). The VC1-STV and the STV beads were incubated for 2 h at 4 °C with 160 μl of diluted plasma (called input). The unbound material was carefully removed and the beads were washed three times with 500 μl of buffer B. Protein bound to VC1-STV and STV were eluted boiling the beads for 5 min in 15 μl of Laemmli Sample Buffer 4× (0.25 m Tris-HCl, pH 6.8, 8% (w/v) SDS, 400 mm DTT, 40% (w/v) glycerol, and 0.04% bromphenol blue). The beads were washed with 15 μl of buffer A and the two eluates were pooled. For the pulldown assay performed in the presence of calcium, the same protocol was used, adding to buffer B different amount of CaCl2 (1, 5 or 10 mm).
sRAGE expression and purification
Human sRAGE (residues 23-322) was produced, as an untagged protein, in P. pastoris from strain KM71H transformed with pPICZαA-sRAGE (54). Cells grown in MGY (1% yeast extract, 2% peptone, 1% glycerol, 1.34% YNB, 4 × 10−5 % biotin) were diluted to 1 OD600 and induced to produce sRAGE in MEMY (1% yeast extract, 2% peptone, 0.5% methanol, 0.7% ethanol, 1.34% YNB, 4 × 10−5 % biotin) in a flask at 30 °C and 0.5% (v/v) MetOH was added daily. The culture supernatant from the 400-ml culture was collected after 72 h from induction and concentrated to ∼30 ml by ultrafiltration using a 10,000 cut-off membrane and the retentate was dialyzed overnight against 20 mm MES, pH 6. The solution was applied to a cation-exchange HITRAP SP HP column (GE Healthcare) equilibrated with the same buffer and connected to an AEKTA PRIME PLUS system (GE Healthcare). After column washing, elution was performed with a 0-1 m NaCl gradient in the same buffer. Fractions were analyzed by SDS-PAGE and fractions containing sRAGE were pooled and applied to a size exclusion chromatography Superdex 75 column (GE Healthcare) equilibrated with 20 mm MES, 300 mm NaCl, pH 6, and connected to an AEKTA PRIME PLUS system (GE Healthcare). Fractions were analyzed by SDS-PAGE and fractions containing sRAGE were pooled. Amicon centrifugal concentrators equipped with 3-kDa cutoff membranes were used to exchange the buffer with 20 mm HEPES, 100 mm NaCl, pH 7.1.
Electrophoresis and immunoblotting procedures
Input, unbound, and bound fractions obtained from pulldown experiments were analyzed by SDS-PAGE. To 3 μl of input and unbound fractions, 12 μl of MilliQ water, and 5 μl of Laemmli sample buffer 4× were added. The samples were denatured incubating for 5 min at 95 °C. SDS-PAGE was performed on Any kD™ Mini Protean® TGX™ precast gels. Protein bands were stained with Bio-Safe Coomassie Blue (Bio-Rad). Images were acquired using the calibrated densitometer GS-800 and analyzed by Quantity One software (Bio-Rad). For detection by Western blotting, proteins were transferred to nitrocellulose membranes using a Trans-Blot® Turbo™ Transfer System (Bio-Rad). Proteins were detected using a mouse mAb against thrombin (F-1) (Santa Cruz Biotechnology, dilution 1:500) or mouse mAb against human γ-carboxyglutamate (Gla) residues (Sekisui Diagnostics, catalog number 3570, dilution 1:200) in combination with the peroxidase-conjugated anti-mouse secondary antibody (Santa Cruz Biotechnology). Bound antibodies were revealed with the LiteAblot®PLUS Western blotting detection reagent (Euroclone, Italy).
Preparation of human prothrombin des-Gla
Plasma-derived prothrombin was purchased by Sekisui Diagnostics GmbH, Pfungstadt, Germany (catalog number ADG417 >95% pure by SDS-PAGE and MS analysis). Gla domain less PT (PT des-Gla) was obtained by proteolytic treatment essentially as described in Ref. 34. PT was diluted at 1 mg/ml in 50 mm HEPES, pH 8, and chymotrypsin (sequencing grade, Roche) was resuspended at 1 mg/ml in 1 mm HCl (Pierce) and then diluted to 0.025 mg/ml in the same buffer. The reaction mix containing chymotrypsin and PT at a ratio of 1/1000 and a control reaction mix were incubated for 20 min at 37 °C. Proteolytic digestion was stopped by addition of N-tosyl-l-phenylalanine chloromethylketone (400 μm) and phenylmethylsulfonyl fluoride (2 mm). Aliquots of the reaction mixtures were denatured at 100 °C for 5 min and analyzed by SDS-PAGE on 12% polyacrylamide minigels prepared using a solution of acrylamide/bisacrylamide with a ratio of 35/1. PT migrated as a single polypeptide with a Mr of 69,000 and after digestion totally converted to a slightly faster migrating band, which still reacted with anti-PT antibody but not with anti-Gla domain mAb. Thus, the whole Gla domain was removed by this treatment in agreement with previous reports (34).
In gel digestion of polypeptides
After SDS-PAGE analysis of pulldown assays, areas of the gel above the 37-kDa marker were excised using a scalpel, finely chopped, transferred to a new Eppendorf and washed with 200 µl of digestion buffer (50 mm ammonium bicarbonate). The region below the 37-kDa marker was excluded because the detection of anything other than the highly abundant streptavidin and VC1 was unfeasible, despite the high sensitivity of the high-resolution spectrometer used. The reason for this was the dynamic protein concentration range in turn ascribable to the experimental needs.
An aliquot of 200 µl of destaining solution (50% ammonium bicarbonate 50 mm, 50% acetonitrile) was added to each gel portion and heated at 37 °C for 10 min in the thermomixer (1400 rpm); the destaining solution was then discarded and this step was repeated until destaining was completed. Afterward, gel pieces were incubated with 150 µl of reducing solution (10 mm DTT in 50 mm ammonium bicarbonate) at 56 °C for 1 h and then with 150 µl of alkylating solution (55 mm iodoacetamide in 50 mm ammonium bicarbonate) at room temperature for 45 min in the dark. In-gel protein digestion was performed by two sequential digestions, one with 1 µg of sequencing grade trypsin (Roche) dissolved in digestion buffer and incubated for 16 h, followed by a second digestion with a sequencing grade chymotrypsin (1 µg) for 7 h at 25 °C in the presence of calcium chloride (10 mm). Peptide mixtures were extracted by 10 min incubations with extraction solution (acetonitrile/TFA/water; 30/3/67; v/v/v) and by an additional 10 min incubation with pure acetonitrile. The two extracts were combined and dried in a vacuum concentrator (Martin Christ). Digested peptide mixtures were then dissolved in an appropriate volume (20 µl) of 0.1% formic acid for MS analysis.
MS analyses
The flowchart of Fig. S1 outlines the entire experimental design of the MS analyses of pulldown assays. Peptide mixtures from the in-gel digestion were separated by reversed-phase (RP) nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray tandem MS (ESI-MS/MS). For each analysis 5 μl of solubilized peptides were injected onto an Acclaim PepMap™ C18 column (75 μm × 15 cm, pores 100 Å, Thermo Scientific, Waltham, MA), protected by a pre-column, namely the Acclaim PepMap™ (100 μm × 2 cm, pores 100 Å, Thermo Scientific). Samples were loaded onto the pre-column by means of the loading pump at 5 µl/min of mobile phase consisting of 99% of buffer A_LP (0.1% TFA) and 1% of buffer B_LP (0.1% HCOOH in CH3CN) for 3 min. After the loading valve switching, peptide separation was performed by the NC_pump with a 91-min linear gradient of buffer B_NP (0.1% HCOOH in CH3CN) from 1 to 40%, and a further 12 min of linear gradient from 40 to 95% (buffer B_NP). 9 min at 95% of buffer B_NP to rinse the column following the separative gradient, and the last 5 min served to re-equilibrate the column to initial conditions. The nano-chromatographic system, an UltiMate 3000 RSLCnano System (Dionex), was connected to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific Inc., Milan, Italy) equipped with a Thermo Scientific dynamic Nanospray ion source set as follows: positive ion mode, spray voltage 1.7 Kv; capillary temperature 220 °C, capillary voltage 35 V; tube lens offset 120 V. The LTQ-Orbitrap XL mass spectrometer was operated in data-dependent acquisition mode to acquire both the full MS spectra and the MS/MS spectra. Full MS spectra were acquired in “profile” mode, by the Orbitrap (FT) analyzer, in a scanning range between 300 and 1800 m/z, using a capillary temperature of 220 °C, AGC target = 5 × 105, and resolving power 60,000 (FWHM at 400 m/z). Tandem mass spectra MS/MS were acquired by the Linear Ion Trap (LTQ) in CID mode, automatically set to fragment the nine of the most intense ions in each full MS spectrum (exceeding 1 × 104 counts) under the following conditions: centroid mode, normal mode, isolation width of the precursor ion of 2.5 m/z, AGC target 1 × 104, and normalized collision energy of 35 eV. Dynamic exclusion was enabled (exclusion dynamics for 45 s for those ions observed 3 times in 30 s). Charge state screening and monoisotopic precursor selection were enabled, singly and unassigned charged ions were not fragmented. Xcalibur software (version 2.0.7, Thermo Scientific Inc., Milan, Italy) was used to control the mass spectrometer.
Protein identification
The software Proteome Discoverer (version 2.2.0.338, Thermo Scientific, USA), implemented with the algorithm SEQUEST, was used to compare the experimental full and tandem mass spectra with the theoretical ones obtained by the in silico digestion of all the proteins within the UniProt Homo sapiens proteome database (https://www.uniprot.org) (UniProt release 2017_03 - March 15, 2017, reviewed/Swiss-Prot 26,546 entries).
Trypsin and chymotrypsin were selected as the cleaving proteases, allowing a maximum of 2 missed cleavages. Peptide and fragment ion tolerances were set to 5 ppm and 10 μ, respectively. Cysteine carbamidomethylation was set as fix modification (+57.02147), whereas methionine oxidation was allowed as a variable modification. As a quality filter, only peptide with an XCorr value grater then 2.2 for doubly-charged peptides, 2.5 for triply-charged, 2.75 for quadruply charged peptide ions, and 3 for charge states quintuple or higher were considered as genuine peptide identifications. To ensure the lowest number of false positives, the mass values experimentally recorded were further processed through a combined search with the database Decoy, where the protein sequences are inverted and randomized. This operation allows the calculation of the false discovery rate for each match, so that all the proteins out of range of false discovery rate between to 0.01 (strict) and 0.05 (relaxed) were rejected.
MicroScale thermophoresis
The technique is based on thermophoresis, the directed motion of molecules in temperature gradients (36). Purified PT (Enzyme Research Laboratories) was labeled using the RED-NHS (Amine Reactive) NT-647 fluorescent dye (kit MO-L001, NanoTemper Technologies) according to the manufacturer's instructions. Unreacted dye was removed with the supplied dye removal column equilibrated with 20 mm HEPES, pH 7.1, 100 mm NaCl, 0.05% Tween 20. The labeled PT was adjusted to 200 nm with 20 mm HEPES, pH 7.1, 100 mm NaCl, 0.05% Tween 20. Unlabeled recombinant VC1-His-Strep was subjected to 16 serial dilutions in 20 mm HEPES, pH 7.1, 100 mm NaCl, 0.05% Tween 20 ranging from 666 μm to 20 nm. Unlabeled recombinant sRAGE was subjected to 16 serial dilutions in 20 mm HEPES, pH 7.1, 100 mm NaCl, 0.05% Tween 20 ranging from 528 μm to 16 nm. Labeled prothrombin and VC1 or sRAGE were mixed at a volume ratio of 1:1, incubated at 24 °C for 30 min, and then loaded into standard capillaries (Monolith NT.115 Capillaries, NanoTemper Technologies). During a MST experiment, a temperature gradient is induced by an IR laser. MST was measured using a Monolith NT.115 instrument (NanoTemper Technologies) at 24 °C. Instrument parameters were adjusted to 20% LED power and medium MST power. Data of independently pipetted measurements were analyzed (MO.Affinity Analysis software version 2.3, NanoTemper Technologies) using the signal from an MST-on time of 1 s (hot region from 4 to 5 s).
Relative fluorescence was used to quantify binding via MST. To obtain ΔF, the baseline F value (F is the fluorescence) was subtracted from all data points of the same curve. The baseline F value is equivalent to the mean F value of the unbound target, usually in capillaries 14-16, and is given by the MO.Affinity Analysis software as “unbound” value when a fit is performed. Thus, by definition, ΔF is 0 in the unbound state. The fraction bound corresponds to the value of the baseline corrected normalized fluorescence (ΔF) divided by the curve amplitude of each data point.
The fraction bound (FB) is described by the equation: Fnorm = (1-FB)Fnorm unbound + (FB)Fnorm bound, where Fnorm unbound is the normalized fluorescence of the unbound state and Fnorm bound is the normalized fluorescence of the bound state. From FB, the value Kd = [A] × [B]/[AB] of a binding reaction between partner A and partner B to form the complex AB can be derived as described in Ref. 36. In Fig. S4, where no binding is present, Fnorm was used to compare the curves and Fnorm is defined as the F1/F0 ratio where F0 corresponds to the normalized fluorescence prior to MST activation (cold region) and F1 is the normalized fluorescence after MST activation (hot region from 4 to 5 s).
Plasma clotting assay
The effect of VC1 on blood coagulation was examined by plasma clotting assay. Varying amounts of VC1 in 10 µl of 20 mm HEPES, 100 mm NaCl, pH 7.1, were added to 100 µl of citrated human plasma (Vericon 1 Normal Control Plasma, HART Biologicals) and incubated for 30 min at room temperature. After addition of 10 µl of aPTT reagent (Ultrasense EA, HART) and incubation for 2 min at 37 °C, the mixture was centrifuged at 6000 rpm at room temperature for 1 min. 110 µl of supernatant were carefully transferred to a microtiter plate. The clotting reaction was started by the addition of 20 µl of 25 mm CaCl2. The absorbance at 405 nm was monitored for 10 min every 5 s using a microplate reader (EnSight Multimode Plate Reader, PerkinElmer Life Sciences).
Data availability
MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (55) with the data set identifier PXD020355. All remaining data are contained within the article.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Delia Tarantino who kindly assisted us during the setup of MST.
This article contains supporting information.
Author contributions—G. D., G. A., and L. P. conceptualization; G. D. validation; G. D., A. A., S. D., and B. A. investigation; G. D., A. A., and G. F. methodology; L. P., G. D., G. F., A. R., and G. A. writing-review and editing; G. A. and L. P. supervision.
Funding and additional information—This work was supported by private donations (to G. A. and L. P.) and a grant from the Centro Cardiologico Monzino-IRCCS (Ricerca Corrente 2019-2021) (to A. R.). G. D. is a recipient of a postdoctoral fellowship from the University of Milan.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- RAGE
- receptor for advanced glycation end products
- PT
- prothrombin
- Gla domain
- γ-carboxyl glutamic acid domain
- AGE
- advanced glycation end product
- HMGB1
- high mobility group protein box-1
- PRR
- pattern recognition receptor
- sRAGE
- soluble RAGE
- STV
- streptavidin
- HSA
- human serum albumin
- C4BP
- C4b-binding protein
- ALE
- advanced lipoxidation end product
- MST
- MicroScale thermophoresis
- aPTT
- activated partial thromboplastin time
- ATI
- alveolar Type I
- ALI
- acute lung injury.
References
- 1. Sessa L., Gatti E., Zeni F., Antonelli A., Catucci A., Koch M., Pompilio G., Fritz G., Raucci A., and Bianchi M. E. (2014) The receptor for advanced glycation end-products (RAGE) is only present in mammals, and belongs to a family of cell adhesion molecules (CAMs). PLoS ONE 9, e86903 10.1371/journal.pone.0086903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neeper M., Schmidt A. M., Brett J., Yan S. D., Wang F., Pan Y. C., Elliston K., Stern D., and Shaw A. (1992) Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 267, 14998–15004 [PubMed] [Google Scholar]
- 3. Sorci G., Riuzzi F., Giambanco I., and Donato R. (2013) RAGE in tissue homeostasis, repair and regeneration. Biochim. Biophys. Acta 1833, 101–109 10.1016/j.bbamcr.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 4. Lin L. (2006) RAGE on the Toll road? Cell. Mol. Immunol. 3, 351–358 [PubMed] [Google Scholar]
- 5. Xie J., Reverdatto S., Frolov A., Hoffmann R., Burz D. S., and Shekhtman A. (2008) Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE). J. Biol. Chem. 283, 27255–27269 10.1074/jbc.M801622200 [DOI] [PubMed] [Google Scholar]
- 6. Kierdorf K., and Fritz G. (2013) RAGE regulation and signaling in inflammation and beyond. J. Leukoc. Biol. 94, 55–68 10.1189/jlb.1012519 [DOI] [PubMed] [Google Scholar]
- 7. Fritz G. (2011) RAGE: a single receptor fits multiple ligands. Trends Biochem. Sci. 36, 625–632 10.1016/j.tibs.2011.08.008 [DOI] [PubMed] [Google Scholar]
- 8. Oczypok E. A., Perkins T. N., and Oury T. D. (2017) All the “RAGE” in lung disease: The receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr. Respir. Rev. 23, 40–49 10.1016/j.prrv.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sugaya K., Fukagawa T., Matsumoto K., Mita K., Takahashi E., Ando A., Inoko H., and Ikemura T. (1994) Three genes in the human MHC class III region near the junction with the class II: gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics 23, 408–419 10.1006/geno.1994.1517 [DOI] [PubMed] [Google Scholar]
- 10. Haeussler M., Zweig A. S., Tyner C., Speir M. L., Rosenbloom K. R., Raney B. J., Lee C. M., Lee B. T., Hinrichs A. S., Gonzalez J. N., Gibson D., Diekhans M., Clawson H., Casper J., Barber G. P., et al. (2019) The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shirasawa M., Fujiwara N., Hirabayashi S., Ohno H., Iida J., Makita K., and Hata Y. (2004) Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells 9, 165–174 10.1111/j.1356-9597.2004.00712.x [DOI] [PubMed] [Google Scholar]
- 12. Katsuoka F., Kawakami Y., Arai T., Imuta H., Fujiwara M., Kanma H., and Yamashita K. (1997) Type II alveolar epithelial cells in lung express receptor for advanced glycation end products (RAGE) gene. Biochem. Biophys. Res. Commun. 238, 512–516 10.1006/bbrc.1997.7263 [DOI] [PubMed] [Google Scholar]
- 13. Demling N., Ehrhardt C., Kasper M., Laue M., Knels L., and Rieber E. P. (2006) Promotion of cell adherence and spreading: a novel function of RAGE, the highly selective differentiation marker of human alveolar epithelial type I cells. Cell Tissue Res. 323, 475–488 10.1007/s00441-005-0069-0 [DOI] [PubMed] [Google Scholar]
- 14. Lizotte P. P., Hanford L. E., Enghild J. J., Nozik-Grayck E., Giles B. L., and Oury T. D. (2007) Developmental expression of the receptor for advanced glycation end-products (RAGE) and its response to hyperoxia in the neonatal rat lung. BMC Develop. Biol. 7, 15 10.1186/1471-213X-7-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brett J., Schmidt A. M., Yan S. D., Zou Y. S., Weidman E., Pinsky D., Nowygrod R., Neeper M., Przysiecki C., and Shaw A. and (1993) Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am. J. Pathol. 143, 1699–1712 [PMC free article] [PubMed] [Google Scholar]
- 16. Clynes R., Moser B., Yan S. F., Ramasamy R., Herold K., and Schmidt A. M. (2007) Receptor for AGE (RAGE): weaving tangled webs within the inflammatory response. Curr. Mol. Med. 7, 743–751 10.2174/156652407783220714 [DOI] [PubMed] [Google Scholar]
- 17. Rao N. V., Argyle B., Xu X., Reynolds P. R., Walenga J. M., Prechel M., Prestwich G. D., MacArthur R. B., Walters B. B., Hoidal J. R., and Kennedy T. P. (2010) Low anticoagulant heparin targets multiple sites of inflammation, suppresses heparin-induced thrombocytopenia, and inhibits interaction of RAGE with its ligands. Am. J. Physiol. Cell Physiol 299, C97–C110 10.1152/ajpcell.00009.2010 [DOI] [PubMed] [Google Scholar]
- 18. Kalea A. Z., Reiniger N., Yang H., Arriero M., Schmidt A. M., and Hudson B. I. (2009) Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. 23, 1766–1774 10.1096/fj.08-117739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakurai S., Yamamoto Y., Tamei H., Matsuki H., Obata K., Hui L., Miura J., Osawa M., Uchigata Y., Iwamoto Y., Watanabe T., Yonekura H., and Yamamoto H. (2006) Development of an ELISA for esRAGE and its application to type 1 diabetic patients. Diabetes Res. Clin. Practice 73, 158–165 10.1016/j.diabres.2005.12.013 [DOI] [PubMed] [Google Scholar]
- 20. Raucci A., Cugusi S., Antonelli A., Barabino S. M., Monti L., Bierhaus A., Reiss K., Saftig P., and Bianchi M. E. (2008) A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10). FASEB J. 22, 3716–3727 10.1096/fj.08-109033 [DOI] [PubMed] [Google Scholar]
- 21. Taguchi A., Blood D. C., del Toro G., Canet A., Lee D. C., Qu W., Tanji N., Lu Y., Lalla E., Fu C., Hofmann M. A., Kislinger T., Ingram M., Lu A., Tanaka H., et al. (2000) Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405, 354–360 10.1038/35012626 [DOI] [PubMed] [Google Scholar]
- 22. Park L., Raman K. G., Lee K. J., Lu Y., Ferran L. J. Jr., Chow W. S., Stern D., and Schmidt A. M. (1998) Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat. Med. 4, 1025–1031 10.1038/2012 [DOI] [PubMed] [Google Scholar]
- 23. Wendt T., Harja E., Bucciarelli L., Qu W., Lu Y., Rong L. L., Jenkins D. G., Stein G., Schmidt A. M., and Yan S. F. (2006) RAGE modulates vascular inflammation and atherosclerosis in a murine model of type 2 diabetes. Atherosclerosis 185, 70–77 10.1016/j.atherosclerosis.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 24. Bucciarelli L. G., Wendt T., Qu W., Lu Y., Lalla E., Rong L. L., Goova M. T., Moser B., Kislinger T., Lee D. C., Kashyap Y., Stern D. M., and Schmidt A. M. (2002) RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E-null mice. Circulation 106, 2827–2835 10.1161/01.cir.0000039325.03698.36 [DOI] [PubMed] [Google Scholar]
- 25. Li J., and Schmidt A. M. (1997) Characterization and functional analysis of the promoter of RAGE, the receptor for advanced glycation end products. J. Biol. Chem. 272, 16498–16506 10.1074/jbc.272.26.16498 [DOI] [PubMed] [Google Scholar]
- 26. Degani G., Altomare A. A., Colzani M., Martino C., Mazzolari A., Fritz G., Vistoli G., Popolo L., and Aldini G. (2017) A capture method based on the VC1 domain reveals new binding properties of the human receptor for advanced glycation end products (RAGE). Redox Biol. 11, 275–285 10.1016/j.redox.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Degani G., Colzani M., Tettamanzi A., Sorrentino L., Aliverti A., Fritz G., Aldini G., and Popolo L. (2015) An improved expression system for the VC1 ligand binding domain of the receptor for advanced glycation end products in Pichia pastoris. Protein Expr. Purif. 114, 48–57 10.1016/j.pep.2015.06.012 [DOI] [PubMed] [Google Scholar]
- 28. Degani G., Barbiroli A., Magnelli P., Digiovanni S., Altomare A., Aldini G., and Popolo L. (2019) Insights into the effects of N-glycosylation on the characteristics of the VC1 domain of the human receptor for advanced glycation end products (RAGE) secreted by Pichia pastoris. Glycoconj. J. 36, 27–38 10.1007/s10719-018-09855-x [DOI] [PubMed] [Google Scholar]
- 29. Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., and Mering C. V. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rezende S. M., Simmonds R. E., and Lane D. A. (2004) Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood 103, 1192–1201 10.1182/blood-2003-05-1551 [DOI] [PubMed] [Google Scholar]
- 31. Ruan B. H., Li X., Winkler A. R., Cunningham K. M., Kuai J., Greco R. M., Nocka K. H., Fitz L. J., Wright J. F., Pittman D. D., Tan X. Y., Paulsen J. E., Lin L. L., and Winkler D. G. (2010) Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-α production in a receptor for advanced glycation end product-dependent manner. J. Immunol. 185, 4213–4222 10.4049/jimmunol.1000863 [DOI] [PubMed] [Google Scholar]
- 32. Mol M., Degani G., Coppa C., Baron G., Popolo L., Carini M., Aldini G., Vistoli G., and Altomare A. (2019) Advanced lipoxidation end products (ALEs) as RAGE binders: Mass spectrometric and computational studies to explain the reasons why. Redox Biol. 23, 101083 10.1016/j.redox.2018.101083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stenflo J. (1999) Contributions of Gla and EGF-like domains to the function of vitamin K-dependent coagulation factors. Crit. Rev. Eukaryotic Gene Exp. 9, 59–88 10.1615/CritRevEukaryotGeneExpr.v9.i1.50 [DOI] [PubMed] [Google Scholar]
- 34. Dode C., Rabiet M. J., Bertrand O., Labie D., and Elion J. (1980) Characterization of a proteolytically modified form of human prothorombin. Biochem. Biophys. Res. Commun. 94, 660–666 10.1016/0006-291X(80)91283-8 [DOI] [PubMed] [Google Scholar]
- 35. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., and Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Develop. Technol. 9, 342–353 10.1089/adt.2011.0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seidel S. A., Dijkman P. M., Lea W. A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J. S., Srinivasan P., Baaske P., Simeonov A., Katritch I., Melo F. A., Ladbury J. E., Schreiber G., Watts A., et al. (2013) Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 10.1016/j.ymeth.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leclerc E., Fritz G., Weibel M., Heizmann C. W., and Galichet A. (2007) S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J. Biol. Chem. 282, 31317–31331 10.1074/jbc.M703951200 [DOI] [PubMed] [Google Scholar]
- 38. Zhu A., Sun H., Raymond R. M. Jr., Furie B. C., Furie B., Bronstein M., Kaufman R. J., Westrick R., and Ginsburg D. (2007) Fatal hemorrhage in mice lacking γ-glutamyl carboxylase. Blood 109, 5270–5275 10.1182/blood-2006-12-064188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Innerhofer P., and Kienast J. (2010) Principles of perioperative coagulopathy. Best Pract. Res. Clin. Anaesthesiol. 24, 1–14 10.1016/j.bpa.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 40. Waddington S. N., McVey J. H., Bhella D., Parker A. L., Barker K., Atoda H., Pink R., Buckley S. M., Greig J. A., Denby L., Custers J., Morita T., Francischetti I. M., Monteiro R. Q., Barouch D. H., et al. (2008) Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 132, 397–409 10.1016/j.cell.2008.01.016 [DOI] [PubMed] [Google Scholar]
- 41. Itoh S., Yokoyama R., Kamoshida G., Fujiwara T., Okada H., Takii T., Tsuji T., Fujii S., Hashizume H., and Onozaki K. (2013) Staphylococcal superantigen-like protein 10 (SSL10) inhibits blood coagulation by binding to prothrombin and factor Xa via their γ-carboxyglutamic acid (Gla) domain. J. Biol. Chem. 288, 21569–21580 10.1074/jbc.M113.451419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itoh S., Takii T., Onozaki K., Tsuji T., and Hida S. (2017) Identification of the blood coagulation factor interacting sequences in staphylococcal superantigen-like protein 10. Biochem. Biophys. Res. Commun. 485, 201–208 10.1016/j.bbrc.2017.02.053 [DOI] [PubMed] [Google Scholar]
- 43. Sitkiewicz E., Tarnowski K., Poznański J., Kulma M., and Dadlez M. (2013) Oligomerization interface of RAGE receptor revealed by MS-monitored hydrogen deuterium exchange. PLoS One 8, e76353 10.1371/journal.pone.0076353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moysa A., Hammerschmid D., Szczepanowski R. H., Sobott F., and Dadlez M. (2019) Enhanced oligomerization of full-length RAGE by synergy of the interaction of its domains. Sci. Rep. 9, 20332 10.1038/s41598-019-56993-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Antonelli A., Di Maggio S., Rejman J., Sanvito F., Rossi A., Catucci A., Gorzanelli A., Bragonzi A., Bianchi M. E., and Raucci A. (2017) The shedding-derived soluble receptor for advanced glycation endproducts sustains inflammation during acute Pseudomonas aeruginosa lung infection. Biochim. Biophys. Acta 1861, 354–364 10.1016/j.bbagen.2016.11.040 [DOI] [PubMed] [Google Scholar]
- 46. Johnson E. R., and Matthay M. A. (2010) Acute lung injury: epidemiology, pathogenesis, and treatment. J. Aerosol. Med. Pulm. Drug Deliv. 23, 243–252 10.1089/jamp.2009.0775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blank R., and Napolitano L. M. (2011) Epidemiology of ARDS and ALI. Crit. Care Clin. 27, 439–458 10.1016/j.ccc.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 48. Camprubi-Rimblas M., Tantinya N., Bringue J., Guillamat-Prats R., and Artigas A. (2018) Anticoagulant therapy in acute respiratory distress syndrome. Ann. Translat. Med. 6, 36 10.21037/atm.2018.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uchida T., Shirasawa M., Ware L. B., Kojima K., Hata Y., Makita K., Mednick G., Matthay Z. A., and Matthay M. A. (2006) Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am. J. Respir. Crit. Care Med. 173, 1008–1015 10.1164/rccm.200509-1477OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Howell D. C., Goldsack N. R., Marshall R. P., McAnulty R. J., Starke R., Purdy G., Laurent G. J., and Chambers R. C. (2001) Direct thrombin inhibition reduces lung collagen, accumulation, and connective tissue growth factor mRNA levels in bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 159, 1383–1395 10.1016/S0002-9440(10)62525-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding H., Ji X., Chen R., Ma T., Tang Z., Fen Y., and Cai H. (2015) Antifibrotic properties of receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Pulm. Pharmacol. Ther. 35, 34–41 10.1016/j.pupt.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 52. He M., Kubo H., Ishizawa K., Hegab A. E., Yamamoto Y., Yamamoto H., and Yamaya M. (2007) The role of the receptor for advanced glycation end-products in lung fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1427–L1436 10.1152/ajplung.00075.2007 [DOI] [PubMed] [Google Scholar]
- 53. Englert J. M., Kliment C. R., Ramsgaard L., Milutinovic P. S., Crum L., Tobolewski J. M., and Oury T. D. (2011) Paradoxical function for the receptor for advanced glycation end products in mouse models of pulmonary fibrosis. Int. J. Clin. Exp. Pathol. 4, 241–254 [PMC free article] [PubMed] [Google Scholar]
- 54. Ostendorp T., Weibel M., Leclerc E., Kleinert P., Kroneck P. M. H., Heizmann C. W., and Fritz G. (2006) Expression and purification of the soluble isoform of human receptor for advanced glycation end products (sRAGE) from Pichia pastoris. Biochem. Biophys. Res. Commun. 347, 4–11 10.1016/j.bbrc.2006.04.077 [DOI] [PubMed] [Google Scholar]
- 55. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (55) with the data set identifier PXD020355. All remaining data are contained within the article.