Abstract
GABA type A receptors (GABAARs) mediate fast synaptic inhibition and are trafficked to functionally diverse synapses. However, the precise molecular mechanisms that regulate the synaptic targeting of these receptors are unclear. Whereas it has been previously shown that phosphorylation events in α4, β, and γ subunits of GABAARs govern their function and trafficking, phosphorylation of other subunits has not yet been demonstrated. Here, we show that the α2 subunit of GABAARs is phosphorylated at Ser-359 and enables dynamic regulation of GABAAR binding to the scaffolding proteins gephyrin and collybistin. We initially identified Ser-359 phosphorylation by MS analysis, and additional experiments revealed that it is regulated by the activities of cAMP-dependent protein kinase (PKA) and the protein phosphatase 1 (PP1) and/or PP2A. GST-based pulldowns and coimmunoprecipitation experiments demonstrate preferential binding of both gephyrin and collybistin to WT and an S359A phosphonull variant, but not to an S359D phosphomimetic variant. Furthermore, the decreased capacity of the α2 S359D variant to bind collybistin and gephyrin decreased the density of synaptic α2-containing GABAAR clusters and caused an absence of α2 enrichment in the axon initial segment. These results suggest that PKA-mediated phosphorylation and PP1/PP2A-dependent dephosphorylation of the α2 subunit play a role in the dynamic regulation of GABAAR accumulation at inhibitory synapses, thereby regulating the strength of synaptic inhibition. The MS data have been deposited to ProteomeXchange, with the data set identifier PXD019597.
Keywords: synapse, GABA receptor, GABAAR alpha 2 subunit, GABRA2, mass spectrometry (MS), protein phosphorylation, protein kinase A (PKA), protein phosphatase, gephyrin, collybistin, axon initial segment, synaptic inhibition
GABAARs mediate fast synaptic inhibition, the efficacy of which is determined by the number of inhibitory synapses (1, 2). GABAARs are ligand-gated chloride channels that form heteropentamers assembled from a combination of several subunits, including α1-6, β1–3, γ1–3, δ, ε, θ, and π (3). However, most synaptic GABAARs comprise two α1, α2, or α3 subunits, two β2 or β3 subunits, and one γ2 subunit (3, 4). Each subunit is composed of a large N-terminal domain, four transmembrane (TM) domains, and a large intracellular loop between TM3 and TM4 that is the site of multiple protein-protein interactions (5–7). Despite the high structural homology between GABAAR α-subunits, they are selectively targeted to distinct types of inhibitory synapses. Indeed, α2-containing GABAAR are enriched at synapses on the axon initial segment (AIS), whereas α1-containing receptors are more evenly distributed between the AIS and dendrites (8, 9). It has been proposed that the α-subunits play key roles in the selective subcellular targeting of GABAARs (10). However, the molecular mechanisms that regulate precise targeting of these receptors remain unknown.
Post-translational modification is one mechanism by which GABAARs are regulated. Notably, the phosphorylation of key residues on GABAARs have been previously shown to regulate protein interactions that govern trafficking and surface stability of receptors (11, 12); however, phosphorylation of GABAAR α2 subunit has not been demonstrated, and its significance is yet to be determined (13).
Protein interactions fundamental to receptor clustering at the postsynaptic domain include GABAAR α2 subunit interactions with gephyrin and collybistin. Gephyrin is the canonical inhibitory synaptic scaffold protein that is highly enriched at GABAergic postsynapses, where it colocalizes with synaptic GABAARs (14). The critical role for gephyrin in the formation of postsynaptic inhibitory synapses was highlighted through the utilization of knockout and RNAi tools, which demonstrated the loss of gephyrin and subsequent loss of synaptic GABAAR clusters (15, 16). Moreover, GABAAR mutations that reduce gephyrin binding also result in fewer synaptic clusters (17). Collybistin is a Rho guanine nucleotide exchange factor that binds both gephyrin and GABAAR α2 (18). The importance of collybistin in targeting gephyrin to the inhibitory synapse was demonstrated in collybistin-deficient mice, which exhibit a loss of gephyrin and GABAAR puncta (19). Further, mutations in GABAARs that disrupt or enhance collybistin binding resulted in corresponding decreased or increased GABAAR-containing synapses at the AIS (20, 21).
In this study, we utilized MS to identify a novel phosphorylation site at serine 359 on the GABAAR α2 subunit. This residue is situated in the large intracellular loop between TM3 and TM4 of the α2 subunit, close to the overlapping binding sites of gephyrin and collybistin. Ser-359 is phosphorylated by PKA, and dephosphorylation of this site depends on PP1/PP2A. Phosphomimetic mutation of this site decreased receptor binding to both gephyrin and collybistin, resulting in a reduced number of synaptic α2 clusters in dendrites and an absence of α2 enrichment in the AIS. These results show that the phosphoregulation of α2 by PKA and PP1/PP2A allows neurons to adjust the efficacy of gephyrin and collybistin binding, which plays a fundamental role in regulating the density of inhibitory synapses at dendrites and the enrichment of inhibitory synapses at the AIS.
Results
Identification of serine 359 on the GABAAR α2 subunit as a novel phosphorylation site
To identify novel phosphorylation sites on the GABAAR α2 subunit, we utilized previously characterized Myc/pH-sensitive GFP (pHluorin)-tagged GABAAR α2 subunit (pHα2) knock-in mice (22). Hippocampi and cortex from age- and sex-matched WT and pHα2 mice were solubilized and incubated with GFP-Trap beads to immunoprecipitate pHα2. After extensive washes, bound material was eluted and subjected to SDS-PAGE followed by Coomassie staining. An 80 kDa band corresponding to pHα2 was excised and analyzed by LC–MS/MS (Fig. 1A and Fig. S1). Phosphorylation sites were identified by the Taplin Facility using the Ascore algorithm (23). Two phosphopeptides were identified corresponding to residues Ser-359 and Ser-379 on the GABAAR α2 subunit (Fig. 1, B and C). Phosphorylated Ser-359 peptides were found in all four independent purifications, whereas Ser-379 was only found in one of four experiments. Both potential phosphorylation sites are located in the large intracellular loop of the receptor. Further, this site is conserved in the human, mouse, and rat sequence of the α2 subunit (Fig. 1D). Given the more robust phosphorylation of Ser-359, we focused on this site.
Figure 1.
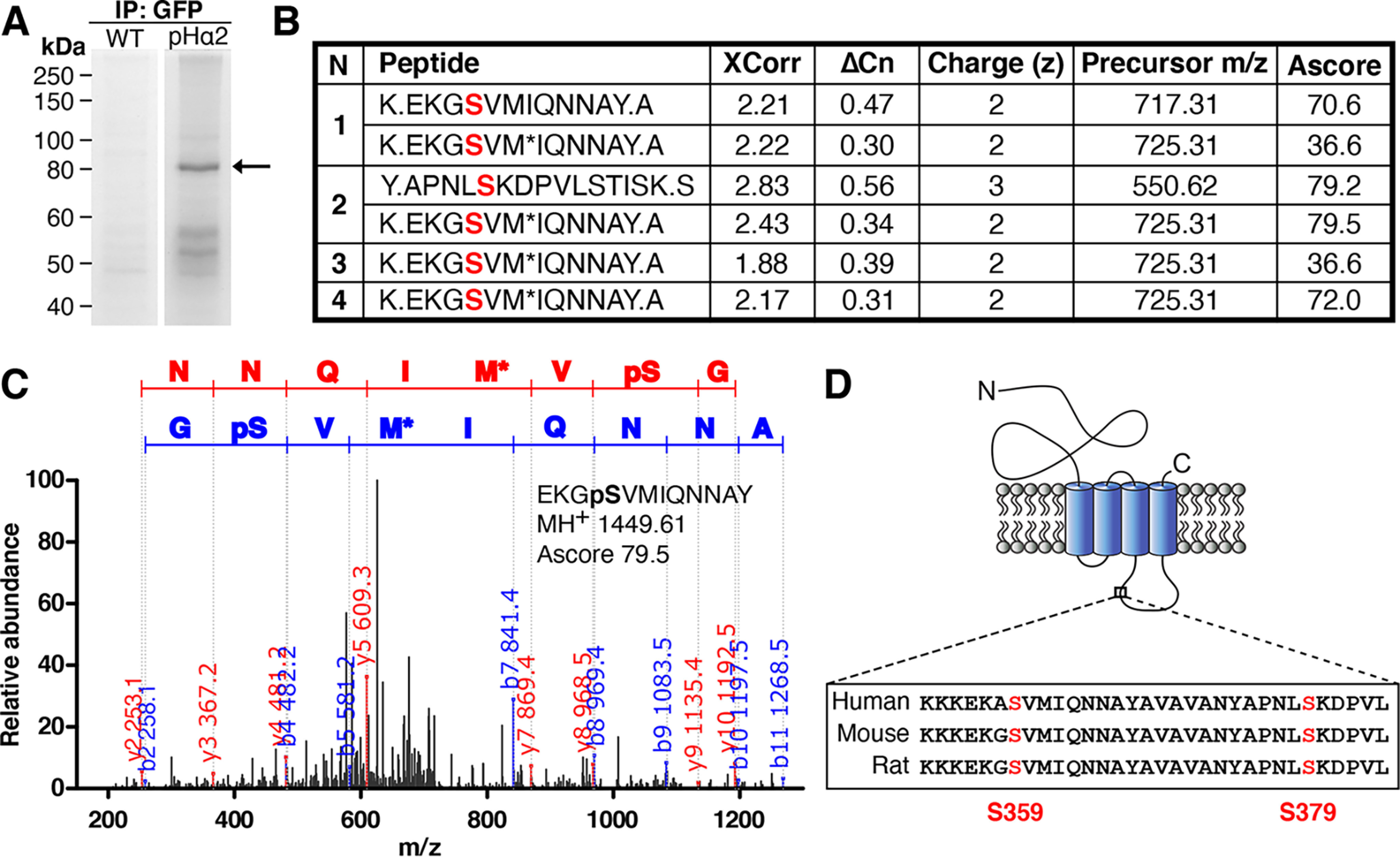
Identification of two novel phosphorylation sites on GABAAR α2 subunit at serine 359 and 379 by LC–MS/MS. A, neuronal lysates from WT and pHα2 mice were incubated with GFP-Trap and subjected to SDS-PAGE followed by Coomassie staining. The 80 kDa gel band (arrow) corresponding to pHα2 was excised and digested with both trypsin and chymotrypsin before MS analysis was performed by the Taplin MS Facility. B, GABAAR α2 phosphopeptide sequences identified by MS/MS analysis of pHα2, including the SEQUEST scores XCorr (cross-correlation) and ΔCn (delta correlation), charge state (z), precursor m/z, and Ascore values (for Ascore > 13, p < 0.05). Phosphorylated serine sites identified are colored red. M*, oxidation of methionine. A period in the peptide sequence indicates the protease cleavage sites. n = 4. C, MS/MS spectrum of the pHα2 derived for peptide EKGpSVMIQNNAY phosphorylated at Ser-359. The phosphorylated serine site is marked as pS. M*, oxidation of methionine. The observed b ion (blue) and y ion (red) peptide fragment masses are indicated on the spectrum. D, schematic depicting the GABAAR α2 subunit. The novel phosphorylated serine residues (red) are located between TM domains 3 and 4 on the large intracellular domain. Both identified serine sites are conserved in mice, rats, and humans.
Phosphorylation of purified GST-α2
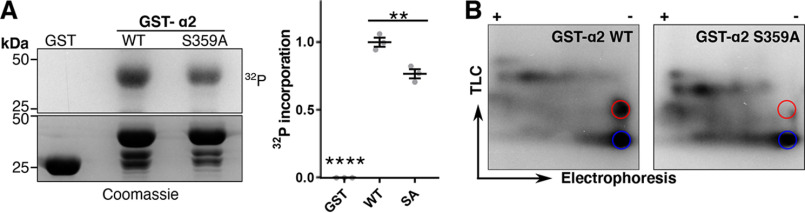
To further corroborate our MS findings, in vitro kinase assays were performed with GST alone or GST fused to the intracellular loop of the GABAAR α2 subunit. Because the kinase responsible for phosphorylating this site had yet to be identified, brain lysate was used as a source of kinase. GST, GST-α2 WT, or GST-α2 S359A phosphomutant proteins were incubated with brain lysate and [γ-32P]ATP to measure phosphory-lation levels at this site. Phosphorylation of GST alone was not detected in these experiments compared with GST-WT and GST-S359A (p < 0.0001, Fig. 2A). The mutant form of the GST-α2 subunit, where Ser-359 was converted to an alanine residue by site-specific mutagenesis, showed significantly decreased levels of phosphorylation compared with WT (WT, 1.00 ± 0.03; S359A, 0.77 ± 0.03; p = 0.0023). The decrease in phosphorylation shown for the GST-α2 S359A mutant strongly suggests that Ser-359 represents a site of phosphorylation in the α2 subunit.
Figure 2.
Kinases present in brain lysate phosphorylate GABAAR α2 at Ser-359. A, GST alone or GST fused to the intracellular loop of the α2 subunit (WT or S359A) was labeled with 10 μCi of 32P and incubated with brain lysate as a source of kinase for 7 min at 30 °C. Fusion proteins were then subjected to SDS-PAGE and visualized with autoradiography (left). Right, quantification of 32P incorporation. Data are means ± S.E. (error bars). **, p < 0.01; ****, p < 0.0001; one-way ANOVA, Tukey's post hoc test, n = 3. B, phosphopeptide map of GST-α2 WT and S359A. 32P-Labeled α2 bands from A were excised, trypsinized, blotted on TLC plates, and subjected to electrophoresis followed by ascending chromatography. Colored circles represent the two major tryptic peptides found in GST-α2 WT. The red circle highlights a tryptic peptide in GST-α2 WT that is missing in S359A. The blue circle highlights a tryptic peptide in GST-α2 WT that is still present in S359A.
Phosphorylation of this site was further examined by phosphopeptide map analysis. Two-dimensional phosphopeptide map analysis revealed two major positively charged phosphopeptides (red and blue circles, Fig. 2B), one of which corresponds to a phosphopeptide containing a phosphorylated Ser-359 (red circle). Together, these results support the MS data in the identification a novel phosphorylation site at Ser-359 in the GABAAR α2 subunit.
Serine 359 dephosphorylation is PP1/PP2A-dependent
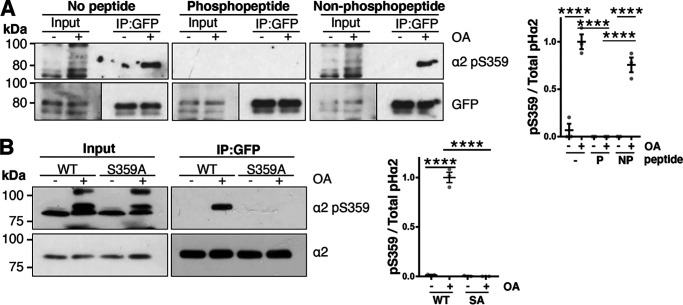
Because kinases present in brain lysate are capable of phosphorylating GABAAR α2 at Ser-359, a purified phosphorylation state–specific antibody directed toward Ser-359 was produced (PhosphoSolutions). To test the specificity of this antibody, horizontal slices from 8-week-old male pHα2 animals were treated for 30 min with 1 μm okadaic acid, a broad-spectrum inhibitor of protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A). Immunoblotting revealed that the pSer-359 antibody did not recognize the α2 subunit in crude brain lysates (Fig. 3 (A and B), input lane). Therefore, we first immunoprecipitated pHα2 from these lysates with GFP-Trap. Phosphorylation at Ser-359 was only detected following okadaic acid treatment (no peptide: DMSO, 0.069 ± 0.07; OA, 1 ± 0.08 (p < 0.0001); nonphosphopeptide: DMSO, 0.0007 ± 0.0007; OA, 0.76 ± 0.08 (p < 0.0001); Fig. 3A). Moreover, detection of the α2 Ser-359 band was prevented by incubation of the phosphoantibody with synthetic phospho-Ser-359 peptide (0.00 ± 0.00) compared with no peptide + OA and nonphosphopeptide + OA (no peptide + OA, 1 ± 0.07 (p < 0.0001); nonphosphopeptide + OA, 0.76 ± 0.08 (p < 0.0001)). However, incubation with nonphosphorylated Ser-359 peptide showed little interference with the immunodetection of the phosphorylated α2 subunit (p = 0.058). Together, these data indicate that Ser-359 dephosphory-lation depends on PP1 and/or PP2A to maintain phosphorylation at low levels under basal conditions in the mouse brain.
Figure 3.
Application of okadaic acid increases phosphorylation of GABAAR α2 Ser-359 in pHα2 mice. A, horizontal brain slices from pHα2 mice were treated with OA (1 μm) or DMSO vehicle control for 30 min. These brain lysates were then used to immunoprecipitate pHα2 and immunoblot for phosphory-lated α2 at Ser-359 (α2 pS359). Blots were incubated with α2 pS359 antibody alone (No peptide), or α2 pS359 antibody incubated with synthetic phosphorylated Ser-359 peptide (Phosphopeptide) or nonphosphorylated Ser-359 peptide (Non-phosphopeptide). Data are means ± S.E. (error bars). ****, p < 0.0001; one-way ANOVA, Tukey's post hoc test, n = 3. B, rat cortical neurons were infected at 15 DIV with lentivirus expressing pHα2 WT or the S359A mutant. At 19 DIV, neurons were treated with OA (1 μm) or DMSO vehicle control for 30 min. Cell lysates were subjected to immunoprecipitation with GFP-Trap, and immunoblots were incubated with α2 or α2 pSer-359 antibody. Data are means ± S.E. ****, p < 0.0001; one-way ANOVA, Tukey's post hoc test, n = 4.
To further validate the phosphoantibody, similar experiments were performed in neuronal cultures. Rat cortical neurons at 19 DIV expressing pHα2 WT or S359A phosphomutant were treated with okadaic acid (Fig. 3B). Similar to the experiments in slices, immunoprecipitation was required before using this antibody. Further, okadaic acid led to an increase in Ser-359 phosphorylation levels (WT−OA, 0.01 ± 0.01; WT + OA, 1 ± 0.05; p < 0.0001), which was abolished in the S359A mutant (SA + OA, 0.00 ± 0.00; p < 0.0001). Collectively, these experiments indicate that we have successfully produced a phosphospecific α2 Ser-359 antibody. Furthermore, these data corroborate that phosphorylation of this site is kept at low levels under basal conditions and confirm that its dephosphorylation is regulated by PP1 and/or PP2A.
PKA activity regulates serine 359 phosphorylation
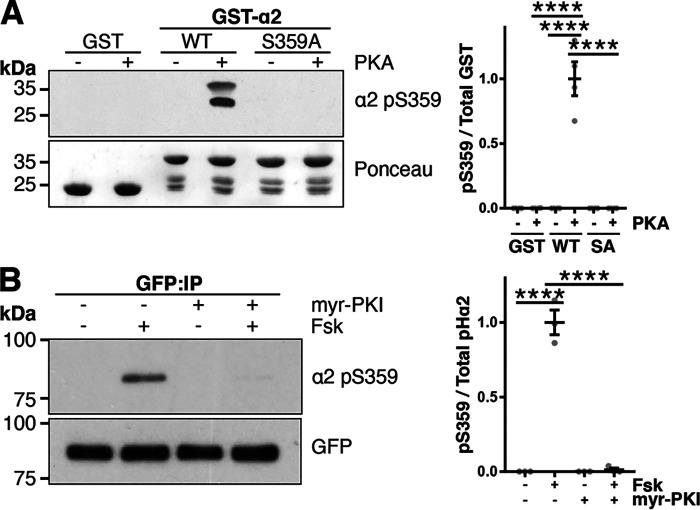
Next, we investigated which kinase phosphorylates this site. The web-based kinase prediction tool, NetPhos3.1 (24), predicted PKA as the potential kinase responsible for Ser-359 phosphorylation. To test this, we performed a kinase assay incubating the catalytic subunit of PKA with purified GST, GST-α2 WT, or GST-α2 S359A mutant. Immunoblotting showed that PKA increased Ser-359 phosphorylation in GST-α2 WT (WT − PKA, 0.00 ± 0.00; WT + PKA, 1 ± 0.13; p < 0.0001), which was not seen in GST alone (0.00 ± 0.00, p < 0.0001) or the GST-α2 S359A phosphonull mutant (0.00 ± 0.00, p < 0.0001; Fig. 4A). These results demonstrate that PKA can directly phosphorylate α2 at Ser-359. To corroborate these findings, experiments were performed in neuronal cultures. Cortical cultures infected with pHα2 WT lentivirus were incubated with 10 μm forskolin (Fsk, 10 min) to activate PKA and/or preincubated with 10 μm myristoylated PKA inhibitor (myr-PKI, 45 min). pHα2 was immunoprecipitated with GFP-Trap, and immunoblots were incubated with phospho-Ser-359 antibody to determine relative phosphorylation levels. Incubation of neurons with Fsk led to an increase in phospho-Ser-359 pHα2 levels (−Fsk, 0.01 ± 0.01; +Fsk, 1 ± 0.05; p < 0.0001), which was blocked by the PKA-specific inhibitor myr-PKI (+myr-PKI + Fsk, 0.00 ± 0.00; p < 0.0001; Fig. 4B), further suggesting that phosphorylation of Ser-359 in neurons is regulated by PKA activity.
Figure 4.
PKA phosphorylates GABAAR α2 at Ser-359. A, GST, GST-α2 WT, or GST-α2 S359A was incubated with the catalytic subunit of PKA and ATP at 30 °C for 30 min. Samples were washed and immunoblotted for pSer-359. Data are means ± S.E. (error bars). ****, p < 0.0001; one-way ANOVA, Tukey's post hoc test, n = 4. B, cortical neurons were transduced at 15 DIV with lentivirus expressing pHα2. At 19 DIV, neurons were incubated with Fsk (10 μm, 15 min) and/or pretreated with myr-PKI (10 μm, 35 min). Cell lysates were subjected to immunoprecipitation with GFP-Trap, and immunoblots were incubated with α2 pSer-359 or GFP antibody. Data are means ± S.E. ****, p < 0.0001; one-way ANOVA, Tukey's post hoc test, n = 3.
Phosphomimetic α2 S359D intracellular loop domain binds less gephyrin and collybistin in vitro
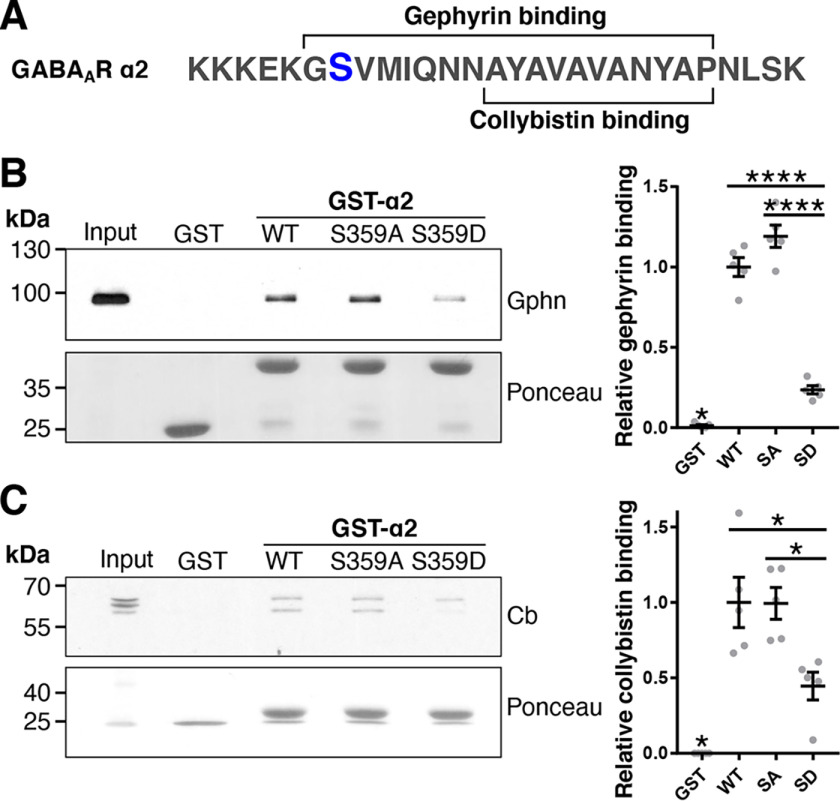
Intriguingly, the Ser-359 site is near or within the gephyrin- and collybistin-binding site in the α2 subunit (Fig. 5A) (18). We therefore asked whether Ser-359 phosphorylation affected their interaction with GABAAR α2. Pulldown assays were performed utilizing GST or GST fused to the large intracellular loop of GABAAR α2 WT, S359A phosphonull, or S359D phosphomimetic mutants to investigate binding of gephyrin and collybistin. Purified GST fusion proteins were incubated with detergent-solubilized rat cortical neuronal lysates and subjected to immunoblotting. All GST-α2 subunit loop domains bound to gephyrin or collybistin compared with GST alone (gephyrin: WT, p < 0.0001; SA, p < 0.0001; SD, p = 0.021; collybistin: WT, p < 0.0001; SA, p < 0.0001; SD, p = 0.046). However, gephyrin binding to the S359D mutant was significantly reduced (0.24 ± 0.03) compared with WT (Fig. 5B, 1.00 ± 0.06, p < 0.0001) and S359A intracellular loops (1.19 ± 0.07, p < 0.0001). Alternative splicing generates multiple collybistin isoforms that exhibit apparent molecular masses between 50 and 70 kDa, and consistent with other studies, both bands were included in our analysis (25, 26). This approach revealed that significantly more collybistin bound WT and S359A (Fig. 5C, WT, 1.00 ± 0.17 (p = 0.011); SA, 0.99 ± 0.10 (p = 0.012)) compared with S359D mutant (0.45 ± 0.19). These data indicate that phosphorylation at Ser-359 reduces gephyrin and collybistin binding to α2.
Figure 5.
Phosphomimetic mutation of α2 Ser-359 reduces binding to gephyrin (Gphn) and collybistin (Cb) in vitro. A, schematic illustrating a small section of the large intracellular loop of the GABAAR α2 subunit. The gephyrin- and collybistin-binding sites are shown, and the Ser-359 phosphorylation site is depicted in blue. B and C, detergent-solubilized cortical neuronal lysates were incubated with GST alone, GST-α2 WT, S359A phosphonull, or S359D phosphomimetic mutants. Bound proteins including gephyrin and collybistin were detected by immunoblotting. The bottom panels show Ponceau staining illustrating the amounts of GST utilized. Right, graphs show quantification of immunoblots. *, p < 0.05; ****, p < 0.0001; one-way ANOVA with Tukey's post hoc test, n = 5. Data are means ± S.E. (error bars).
Full-length α2 S359D binds less gephyrin and collybistin in neurons
We next explored the effects of the Ser-359 mutations in neurons. Cortical neurons were infected at 15 DIV with either an shRNA control lentivirus or one that knocked down endogenous GABAAR α2 (Fig. S2). Neurons were co-infected with either a GFP control virus or a virus expressing shRNA-resistant WT, S359A, or S359D forms of full-length pHα2. At 19 DIV, neuronal lysates were incubated with GFP-Trap and immunoblotted for bound proteins. Gephyrin (WT, p = 0.0003; SA, p = 0.0001; SD, p = 0.033; Fig. 6A) and collybistin (WT, p < 0.0001; SA, p < 0.0001; SD, p = 0.0006; Fig. 6B) coimmunoprecipitated significantly with pHα2 WT, pHα2 S359A, and pHα2 359D over GFP alone. Consistent with our GST-pulldown experiments, gephyrin bound WT (1.0 ± 0.04, p = 0.018) and S359A (1.13 ± 0.14, p = 0.005) significantly more than S359D (0.48 ± 0.12). Further, collybistin also bound WT (1.0 ± 0.11, p = 0.0025) and S359A (1.10 ± 0.06, p = 0.0003) more than S359D (0.56 ± 0.07). In both experiments, WT and S359A bound gephyrin (p = 0.76) and collybistin (p = 0.74) with similar efficiencies. Together these data support GST results and demonstrate that Ser-359 phosphomimetic binds gephyrin and collybistin less efficiently than WT and S359A phosphonull pHα2, suggesting that phosphorylation reduces binding of gephyrin and collybistin to α2.
Figure 6.
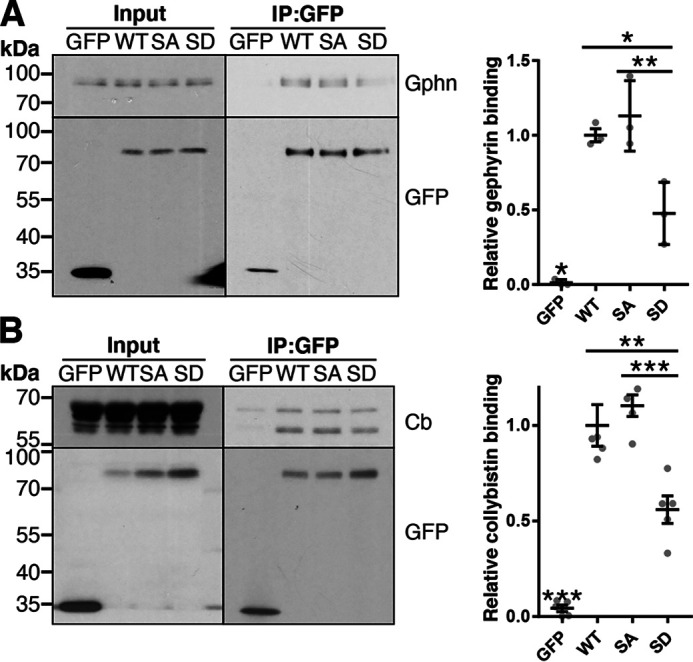
Phosphomimetic mutation of α2 Ser-359 reduces gephyrin (Gphn) and collybistin (Cb) binding in neurons. Cortical neurons were infected at 15 DIV with lentivirus expressing GFP, pHα2 WT, S359A, or S359D. These neurons were co-infected with lentivirus expressing a control shRNA (for GFP control) or shRNA targeting endogenous α2 (for WT, SA, and SD). At 19 DIV, lysates were subjected to immunoprecipitation with GFP-Trap beads, and coimmunoprecipitated proteins gephyrin (A) (n = 3) and collybistin (B) (n = 5) were detected by immunoblotting. Graphs (right) are quantification of immunoblots (left). *, p < 0.05; **, p < 0.01; ***, p < 0.001; one-way ANOVA with Tukey's post hoc test. Data are means ± S.E. (error bars).
Ser-359 phosphorylation regulates pHα2 cluster density and AIS enrichment of α2-containing GABAARs
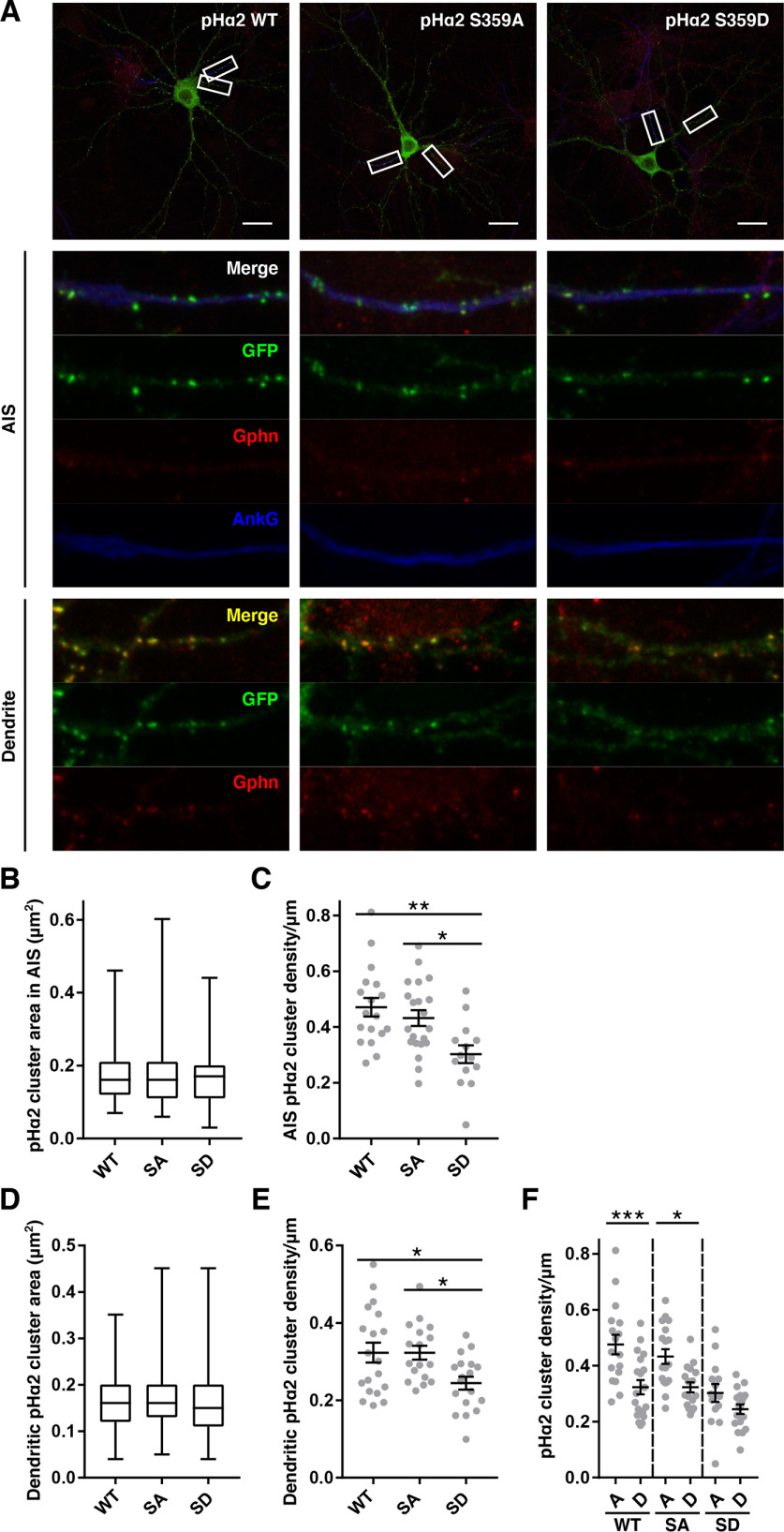
Our data show that phosphorylation at Ser-359 reduces binding to gephyrin and collybistin, both of which are crucial for GABAAR α2 synaptic clustering. To assess how synaptic clustering is affected, neurons were transfected with pHα2 WT, S359A, or S359D at 15 DIV and fixed at 19 DIV. Neurons were stained for GFP, gephyrin, and ankyrin G, a marker for the AIS. Gephyrin staining was much less prominent on the AIS compared with dendrites as reported previously (Fig. 7A) (27). There were no differences in the median area of pHα2-containing GABAAR clusters in the AIS (WT, 0.16 (IQR 0.12–2.1); SA, 0.16 (IQR 0.11–0.21); SD, 0.17 (IQR 0.11–0.20); p = 0.86; Fig. 7B) or dendrites (WT, 0.16 (IQR 0.12–0.20); SA, 0.16 (IQR 0.13–0.20); SD, 0.15 (IQR 0.11–0.20); p = 0.078; Fig. 7D) between conditions. However, the density of pHα2 puncta was significantly different between conditions in both the AIS and in dendrites. In the AIS, pHα2 cluster density/μm was significantly higher in WT (0.47 ± 0.03, p = 0.002)– and S359A (0.43 ± 0.03, p = 0.016)–expressing compared with S359D (0.30 ± 0.03)–expressing neurons (Fig. 7C). Moreover, in dendrites, pHα2 cluster density/μm was also significantly higher in WT (0.32 ± 0.03, p = 0.025)– and S359A (0.32 ± 0.02, p = 0.031)–expressing compared with S359D (0.24 ± 0.02)–expressing neurons (Fig. 7E). As expected, pHα2 cluster density/μm was enriched in the AIS compared with dendrites in pHα2 WT-expressing neurons (AIS, 0.47 ± 0.03; dendrite, 0.32 ± 0.03; p = 0.0003; Fig. 7F). This enrichment of pHα2 in the AIS was still present in S359A (AIS, 0.43 ± 0.03; dendrite, 0.32 ± 0.02; p = 0.010)–expressing but was lost in S359D (AIS, 0.30 ± 0.03; dendrite, 0.24 ± 0.02; p = 0.38)–expressing neurons. Together these data suggest that the decreased binding to gephyrin and collybistin observed in pHα2 S359D results in fewer pHα2 clusters in both the dendrites and AISs. Furthermore, the enrichment of α2-containing GABAAR clusters in the AIS is lost upon phosphorylation of Ser-359.
Figure 7.
Phosphomimetic mutations of α2 Ser-359 reduce pHα2 cluster density in neurons. A, top, hippocampal neurons transfected at 15 DIV with pHα2 WT, S359A, or S359D and fixed at 19 DIV. Neurons were stained with anti-GFP (green), anti-gephyrin (red), and anti-ankyrin G (blue) antibodies. Scale bar, 20 μm. White boxes are magnified in the images below depicting a 20-μm section of AIS and dendrite. B and D, quantification of pHα2 cluster area in the AIS and dendrites, respectively. Box-and-whisker plots, minimum, lower quartile, median, upper quartile, and maximum values; Kruskal–Wallis test. C and E, quantification of pHα2 cluster density in the AIS and dendrite, respectively. Data are means ± S.E. (error bars) *, p < 0.05; **, p < 0.01; one-way ANOVA with Tukey's post hoc test. F, quantification of pHα2 cluster density in the AIS (A) and dendrite (D) as shown in C and E. n = 3 individual cell culture preparations (AIS: 18 WT, 21 SA, and 14 SD cells; dendrite: 19 WT, 17 SA, and 18 SD cells). Data are means ± S.E. (error bars). *, p < 0.05; ***, p < 0.001; one-way ANOVA with Sidak's post hoc test.
Discussion
Identification of Ser-359 as a novel phosphorylation site on the GABAAR α2 subunit
Efficient phasic inhibition requires the accumulation of specific GABAAR subtypes at precise postsynaptic specializations. To determine the contribution of phosphorylation to this process, we examined whether the GABAAR α2 subunits were phosphorylated endogenously by utilizing a previously characterized pHα2 mouse that allows rapid receptor purification using GFP-Trap (22). MS analysis identified novel phosphorylation sites at Ser-359 and Ser-379 within the predicted major intracellular domain of the GABAAR α2 subunit. Notably, these sites flank the collybistin- and gephyrin-binding site (18). Our studies focused on Ser-359 as its phosphorylation was more reproducibly detected compared with Ser-379.
Ser-359 dephosphorylation is PP1/PP2A-dependent
To further study α2 phosphorylation, we created a phosphospecific antibody against Ser-359. Application of okadaic acid to brain slices from pHα2 mice showed that blocking PP1/PP2A led to a marked increase in endogenous α2 Ser-359 phosphorylation detected using this antibody. Additionally, in neuronal culture, this increase in phosphorylation by OA was completely abolished in the phosphomutant S359A, highlighting the specificity of this newly developed antibody and confirming Ser-359 as the first phosphorylation site identified in the α2 subunit (13). These experiments also demonstrated that α2 Ser-359 phosphorylation is continuously maintained at low levels under basal conditions by constitutive cycles of phosphory-lation and dephosphorylation. Potentially, this mechanism would allow neurons to rapidly control inhibitory synaptic receptor content and fine-tune synaptic responsiveness by dynamically altering the balance between these opposing processes.
Ser-359 phosphorylation is regulated by PKA
GST-α2 kinase assays demonstrated that Ser-359 is directly phosphorylated by PKA. These results were corroborated in neurons, where PKA activation by forskolin led to increases in phosphorylation that were abrogated by the addition of the PKA-specific inhibitor myr-PKI.
Interestingly, PKA and PP1/PP2A have been previously implicated in the regulation of GABAARs through phosphorylation of the β3 subunit, which forms receptors with α2 (3, 28), to control receptor surface expression (29–31). However, how multiple phosphorylation sites on various subunits are independently regulated, and how this influences the fate of the assembled receptors, remains to be established. Potentially, proteins that facilitate phosphorylation and dephosphorylation, such as protein kinase A–anchoring protein 79/150 (AKAP70/150), receptor-activated protein kinase C 1 (RACK1), and phospholipase C–related inactive protein type 1 (PRIP1) may assist in the precise spatio-temporal targeting of kinases and phosphatases to specific receptor subunits (31–33), although further work will be required to address this possibility directly.
GABAAR α2 S359D phosphomimetic mutant binds less efficiently to gephyrin and collybistin
Because the α2 Ser-359 phosphorylation site was near the gephyrin- and collybistin-binding site, we next addressed the role of phosphorylation at this site using in vitro binding assays as well as immunoprecipitation. As evidenced by binding assays, both gephyrin and collybistin bound preferentially to GST-α2 WT and the S359A phosphonull mutant compared with the S359D phosphomimetic. Considering that the phosphorylation of this site is kept at low levels, we might expect WT and S359A to behave in a similar fashion. Correspondingly, when pHα2 was immunoprecipitated from neuronal lysates, levels of coimmunoprecipitation of both gephyrin and collybistin were reduced in S359D phosphomimetic mutants compared with WT and S359A phosphonull mutants.
pHα2 WT and S359A phosphonull expressing neurons have a higher density of α2 clusters in dendrites and AISs
Because previous studies have shown that decreased binding to gephyrin and collybistin resulted in a decreased density of clusters of α1-containing GABAARs in dendrites and α2-containing GABAARs in the AIS, respectively (17, 20), we decided to ascertain the consequences of phosphorylation on the synaptic accumulation of GABAAR α2 by measuring the size and density of pHα2 clusters in different neuronal domains. Although there was no difference in cluster area, there were significant differences in cluster density. In both the AISs and dendrites, there was a decrease in pHα2 cluster density with S359D phosphomimetic mutant, compared with WT and S359A mutant, which we attribute to the fact that S359D has a decreased capacity to bind both collybistin and gephyrin. In dendrites, the fewer synaptic α2 puncta observed in S359D-expressing neurons is likely attributable to the decrease in gephyrin's ability to mediate the transient stabilization of these receptors at the synapse, as previously observed in α1 mutants with compromised binding to gephyrin (17). The decrease in α2 clustering at the AIS is likely due to decreased collybistin binding, because the S359D mutant displays a similar loss of AIS clusters to that reported for an α2 mutant with decreased ability to bind collybistin (20). Moreover, gephyrin-α2 subunit associations are weaker at the AIS, indicating that gephyrin has a comparatively minor role in AIS clustering of α2-containing GABAARs (34). Indeed, this seems to be the case in our study, as we also noticed a previously reported (27) lower-intensity gephyrin staining in the AIS compared with dendrites.
Collybistin immunostaining is technically challenging, but published studies suggest that this protein is not exclusively found at synapses or at the AIS; rather, it is associated with intracellular membranes throughout the cells (20). Thus, collybistin may not act as a simple scaffold protein but may regulate GABAAR intracellular trafficking to facilitate its targeting to synapses.
Enrichment of α2 in the AIS is lost in pHα2 S359D phosphomimetic-expressing neurons
Consistent with a previous report (8), we observed an enrichment of α2 clusters at the AIS compared with dendrites. Further, this enrichment is maintained in pHα2 S359A but lost in S359D-expressing neurons, suggesting that α2 S359D is found at similar levels in both dendrites and the AIS and that phosphorylation of this site limits the enrichment of α2 clusters normally observed at the AIS. Thus, this residue appears to reduce the stability of α2-GABAARs at synaptic sites across the neuron, including at the AIS. This novel regulatory mechanism sheds light on the ways in which neurons control inhibitory neurotransmission.
GABAergic neurotransmission must be tightly and precisely controlled to shape excitatory neurotransmission and generate normal network activity patterns. In the absence of such control, pathological hyperexcitability can result (20, 35–37). In the case of Ser-359, the phosphoregulation of this site would allow neurons to rapidly alter the affinity of α2-containing GABAARs for their stabilization at synapses; thus, neurons could rapidly respond to local changes in their own excitability and the excitability of the network around them, fine-tuning the density of α2-GABAAR synaptic clusters and adjusting the inhibitory drive according to their present circumstances. This novel mechanism for rapid response becomes even more integral as it pertains to inhibition at the AIS. The AIS is the site of action potential generation (38), and inhibitory neurotransmission in this area—almost exclusively mediated by synaptic α2-GABAARs—has a tremendous effect on neuronal excitability (8, 39, 40). Furthermore, the AISs of forebrain pyramidal neurons, such as the ones studied here, are solely innervated by inhibitory chandelier cell interneurons (41). Each chandelier cell is thought to innervate the AISs of hundreds of pyramidal cells, allowing for the coordination of activity across large networks and giving rise to oscillatory activity (42–44). Further, we have previously shown that disturbing the stability of α2-GABAARs at axo-axonic synapses has large consequences for inhibitory neurotransmission and network excitability (20, 21).
In summary, we have identified that phosphorylation of Ser-359 within the α2 subunit acts to reduce the binding to the inhibitory scaffold molecules collybistin and gephyrin, resulting in reduced GABAAR accumulation at synapses. This novel regulatory mechanism may allow neurons to fine-tune the activity of a subset of synapses enriched in α2 subunit-containing GABAARs.
Experimental procedures
Animals
All n numbers refer to the number of animals or the number of individual cell culture preparations. Animal protocols were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Animals (Scientific Procedures) Act 1986 and approved by both the Institutional Animal Care and Use Committee of Tufts University and the Animal Services Unit at Bristol University. Myc-pHluorin GABAAR α2 knock-in (pHα2) mice have been described previously (22).
Primary neuronal cultures
Cultures were prepared from fetuses at embryonic day 18 from pregnant Wistar rats. Pregnant dams were sacrificed by approved Schedule 1 methods. Hippocampi and cortices were extracted and washed with Hanks' balanced salt solution (Gibco) and incubated with trypsin (Gibco) for 9 or 15 min, respectively. Cells were further washed with Hanks' balanced salt solution and then dissociated with plating medium (1% penicillin/streptomycin (Sigma), 2% B27 (Gibco), 2 mm GlutaMax (Gibco), and 5% horse serum (HS; Gibco) in Neurobasal medium (Gibco)). 190,000 hippocampal cells were plated on poly-l-lysine (Sigma)–treated coverslips in plating medium. 500,000 cortical cells were plated on poly-l-lysine–treated dishes in plating medium before being placed in a humidified incubator (37 °C, 5% CO2). Two hours after plating neurons, plating medium was replaced with feeding medium (1% penicillin/streptomycin, 2% B27, and 800 μm GlutaMax in Neurobasal medium). After 7 days, 1 ml of feeding medium was supplemented to each dish.
Hippocampal slice preparation
Transverse slices were prepared from 8-week-old male WT and pHα2 mice. Brains were quickly removed from isoflurane-anesthetized mice and put in ice-cold sucrose-based cutting solution containing 87 mm NaCl, 3 mm KCl, 7 mm MgCl2⋅6H2O, 1.25 mm NaH2PO4, 0.5 mm CaCl2, 25 mm NaHCO3, 50 mm sucrose, and 25 mm glucose. 400 μm slices were cut with a vibratome (VT1000S, Leica Microsystems) and were allowed to recover for 1 h in warm (35 °C), oxygenated (95% O2, 5% CO2) artificial cerebrospinal fluid solution containing 126 mm NaCl, 2 mm MgCl2, 2 mm CaCl2, 2.5 mm KCl, 1.25 mm NaH2PO4, 26 mm NaHCO3, 1.5 mm pyruvate, 1 mm l-glutamine, and 10 mm glucose.
Antibodies for Western blots
The phospho-α2 Ser-359 antibody was produced by PhosphoSolutions (Aurora, CO, USA). The following antibodies were used for Western blotting: anti-GABAAR α2 (1:500, Phosphosolutions, #822-GA2C), anti-collybistin (1:500 or 1:2000, Synaptic Systems, #261-003), anti-gephyrin (1:1000–1:8000, C13B11, Synaptic Systems, #147111), anti-GFP (1:1000 or 1:10,000, Synaptic Systems, #132002), and anti-horseradish peroxidase–conjugated secondary antibody (1:10,000, Jackson Immunoresearch, #715035150 and #715035152).
Antibodies for imaging
Antibodies for imaging were as follows: anti-gephyrin (1:1000; Synaptic Systems, #147021), anti-Ankyrin G (1:1000; Synaptic Systems, #386003), anti-GFP (1:5000; Abcam, #ab13970), anti-chicken Alexa 488 (1:3000; Life Technologies, Inc., #A11039), anti-mouse Alexa 568 (1:2000; Life Technologies, #A11004), and anti-rabbit Cy5 (1:1000; Stratech, #711-175-152-JIR).
Expression constructs
Rat α2 was cloned from cDNA derived from rat cortical cultures and cloned into pcdna3.1(−) with primers CAC CTC GAG GCC ACC ATG AGG ACA AAA TTG AGC ACT TGC (forward) and CAC GGA TCC TCA AGG ACT AAC CCC TAA TAC AGG (reverse). For pHα2 in pcdna3.1(−), pHluorin was inserted between amino acids 4 and 5 of rat Gabra2 (22) by amplifying megaprimers with primers TGG TGC TGG CTA ACA TCC AAG AAA GTA AAG GAG AAG AAC TTT TC (forward) and GGT GAT ATT ATT TTT AGC CTC ATC TTT GTA TAG TTC ATC CAT GCC (reverse). shRNA-resistant pHα2 was generated by the mutation of the third base of three consecutive codons with primers T ATC GCT GTT TGT TAC GCC TTC GTC TTC TCT GCC TTA ATT GAA (forward). Site-directed mutagenesis was used to create phoshomimetic and phosphonull mutants: S359A, primer GAC AAG AAA AAA GAG AAA GGC GCC GTC ATG ATA CAG AAC AAC G; S359D, primer GAC AAG AAA AAA GAG AAA GGC GAC GTC ATG ATA CAG AAC AAC G. pHα2 in pcdna3.1(−) was cloned into a lentiviral vector pXLG3 WPRE px (45) under the SFFV promoter: CAC ACT AGT GCC ACC ATG AGG ACA AAA TTG AGC ACT TGC (forward) and CAC GGA TCC TCA AGG ACT AAC CCC TAA TAC AGG (reverse). Knockdown constructs were cloned into the pXLG3 vector under the H1 promoter (short hairpin shGabra2 target sequence, GCG TTT GTG TTC TCT GCC TTA; nonspecific, AAC GTA CGC GGA ATA CTT CGA). For GST fusion, the GST-GABAAR α2 construct encoding the large intracellular loop of the subunit was made from the full-length rat construct mentioned above using primers CAC GAA TTC AAT TAC TTC ACG AAA AGA GGA TGG (forward) and CAC CTC GAG TCT GTC GAT TTT GCT GAC ACT GTT (reverse).
MS analysis
pHα2 was immunoprecipitated from 8–10-week-old mice with GFP-Trap as described previously (22). Trypsin and chymotrypsin digestion, LC–MS/MS, and data analysis were performed by the Taplin Mass Spectrometry Facility (Harvard) using peak list–generating software, ReAdW.exe (version 4.3.1), search engine SEQUEST (version 28, rev 13). In all cases, enzymes were not specified for the search. Peptides were filtered based on SEQUEST scores (XCorr and ΔCn) and mass accuracy. Peptides were required to be tryptic, although there were no filters specified for chymotrypsin. Fixed modifications considered were 57.0215 Da on cysteine. Variable modifications considered were methionine oxidation (15.9949 Da) and phosphorylation on serine, threonine, and tyrosine (73.9663 Da). The mass tolerance was set to 2 Da for precursor ions and 1 Da for fragment ions. The threshold for accepting individual spectra was that the precursor ppm values were <5 ppm from expected, along with manual interpretation. Peptides produced by trypsin would be required to be tryptic peptides. The minimum XCorr values for peptides charged +1, +2, +3, and +4 were >1.5, >1.5, >1.5, and >2, respectively. Each phosphopeptide was analyzed using the Ascore algorithm to determine the confidence of a given phosphorylation site (23). Sites were confidently assigned when Ascore values were >13 (p < 0.05).
GST fusion protein production and pulldown assay
GST fusion proteins were produced as described previously (22). 19 DIV rat cortical neuronal lysates were incubated with GST fused to the large intracellular loop of GABAAR α2 as performed previously (22). GST pulldown assays for gephyrin utilized 500 μg of neuronal lysate/condition, and 2% input was loaded; for collybistin, 750 μg of neuronal lysate was utilized per condition, and 4% input was loaded.
Kinase assay and phosphopeptide mapping
GST, GST-α2 WT, and GST-α2 S359A immobilized on GSH-Sepharose beads (GE Healthcare) were phosphorylated with 10 μCi of [γ-32P]ATP (BLU502A, PerkinElmer Life Sciences) and 15 μg of brain lysate (as a source of kinase) for 7 min at 30 °C. Protein samples were separated by SDS-PAGE and visualized by autoradiography. Bands were analyzed with ImageJ (National Institutes of Health). Phosphopeptide mapping was performed as described (46). Briefly, 32P-labeled bands from kinase assays were excised, dried, and digested with trypsin (0.3 mg/ml in 50 mm NH4HCO3). Samples were spotted on a cellulose TLC plate and subjected to two-dimensional phosphopeptide mapping, first by electrophoresis and then by ascending chromatography. Dried TLC plates were exposed to X-ray film. To detect phosphorylation by PKA, GST-α2 WT and GST-α2 S359A immobilized on GSH-Sepharose beads were incubated (30 °C, 30 min) with 0.75 μg of PKA catalytic subunit (New England Biolabs) and NEBuffer for protein kinases supplemented with 200 μm ATP (New England Biolabs). Beads were washed four times (2500 × g, 2 min, 4 °C) in lysis buffer (described below) with 0.1% SDS.
Immunoprecipitation (IP)
To detect phosphorylated Ser-359 after okadaic acid (Tocris) and forskolin (Tocris)/myristoylated-PKI treatment (Tocris), mouse brain/rat neuronal cells were lysed with lysis buffer containing 20 mm Tris-HCl, pH 8, 130 mm NaCl, 1% Triton X-100, 5 mm EDTA, 10 mm NaF, 2 mm Na3VO4, 10 mm Na4P2O7 and supplemented with 0.1% SDS. Samples were spun at 16,100 × g for 20 min at 4 °C, and the supernatant (or lysate) was incubated with GFP-Trap (Chromotek) overnight, followed by four quick washes (2500 × g, 2 min, 4 °C) in 1.5 ml of lysis buffer (with 0.1% SDS). To detect bound gephyrin and collybistin, neurons were lysed and solubilized in lysis buffer, supplemented with protease inhibitors (cOmplete, Roche Applied Science). Samples were spun at 16,100 × g for 20 min, and lysate was incubated with GFP-Trap overnight, after which beads were quickly washed four times (2500 × g, 2 min, 4 °C) in 1.5 ml of lysis buffer. Bound proteins were detected by Western blotting. For coimmunoprecipitations, 2% input was loaded, and 500 μg or 2 mg of neuronal lysate was used per immunoprecipitation experiment for gephyrin and collybistin, respectively.
Western blotting and analysis
Proteins separated by SDS-PAGE (8 or 10% gel) were transferred to polyvinylidene difluoride membranes and blocked with 6% milk in PBST for 1 h. Membranes were incubated in primary antibody (1 h, 5% milk), followed by three 5-min washes, before incubation with horseradish peroxidase–conjugated secondary antibodies for 1 h. After a further five 5-min washes, blots were developed on film (CL-Xposure, Thermo Scientific). For membranes incubated with phosphorylated and nonphosphorylated peptides, 2 μg of peptide were added to primary antibodies. Collybistin exists in multiple isoforms (25, 26) and appears as a doublet in immunoblots. For the purposes of analysis, both bands were taken into account. Films were analyzed using ImageJ. Multiple comparisons were performed with one-way ANOVA followed by Tukey's post hoc test. Post hoc p values are stated in the text, and one-way ANOVA F and p values can be found in Fig. S3.
Neuronal transfection
Hippocampal neurons were Lipofectamine 2000 (Invitrogen) transfected at 15 DIV. Neuronal coverslips were rinsed in plain Neurobasal medium and transferred to fresh dishes with 1 ml of prewarmed Neurobasal medium and placed in the incubator. Two tubes containing 200 μl of Neurobasal medium were prepared. In tube 1, 1 μg of DNA and in tube 2, 1.5 μl of Lipofectamine were added. Both tubes were vortexed and incubated at room temperature for 5 min, after which tubes were mixed together and incubated at room temperature for a further 20 min to allow complex formation. The transfection mix was added to cells and placed in the incubator for 45 min, after which they were rinsed with Neurobasal medium. Coverslips were replaced into their original dishes and left in the incubator until required.
Immunocytochemistry
At 19 DIV, neurons were fixed in 2% formaldehyde, 5% sucrose solution in HBS: 20 mm HEPES, pH 7.4, 150 m NaCl, 5 mm KCl, 1.8 mm CaCl2, 0.8 mm MgCl2, 5 mm glucose for 20 min. After three quick washes in HBS, coverslips were incubated with 50 μm NH4Cl for 10 min. After a further three quick washes with HBS, coverslips were blocked and permeabilized in 0.1% Triton X-100, 1% BSA, 10% HS in HBS for 1 h. Neurons were stained with primary antibodies overnight (0.1% Triton X-100, 1% BSA, 2% HS in HBS at 4 °C) and washed three times for 10 min with HBS (supplemented with 0.1% Triton X-100) before incubating and staining with secondary antibodies for 1.5 h. Coverslips were washed three times for 10 min in HBS (supplemented with 0.1% Triton X-100) and once in HBS before being mounted on slides (Fluoromount-G, Southern Biotech).
Image acquisition and analysis
Images were captured on a confocal laser-scanning microscope (SP5-AOBS, Leica Microsystems) attached to an inverted epifluorescence microscope (DMI 6000, Lecia Microsystems) with a ×63, numerical aperture 1.4, oil immersion objective (Plan Apochromate BL, Leica Microsystems). High-resolution images (2048 × 2048, mean 2) were taken as a z-series of an average of ∼7-8 z-stacks, taken at 0.5-μm intervals. Acquisition parameters were kept the same for all scans within each experiment. Images were collecting using the SP5 system's acquisition software, and analysis was performed using ImageJ software (47, 48). Image stacks were projected using a maximum projection algorithm. For each neuron, one 20-μm section of axon initial segment and one or two 20-μm sections of secondary dendrites were analyzed. Clusters of pHα2 on the AIS were measured if they were localized with the ankyrin G marker. Clusters on the dendrite were considered synaptic if colocalized or apposed to gephyrin puncta. To examine the GABAAR α2 puncta, clusters were manually outlined, and their area was measured by ImageJ. Cluster density was defined as the number of puncta/μm of AIS or dendrite. Multiple comparisons were performed with Kruskal–Wallis or one-way ANOVA followed by Sidak's post hoc test. Post hoc p values are stated in the text, and Kruskall–Wallis and one-way ANOVA H, F, and p values can be found in Fig. S3.
Lentiviral production
Lentivirus was produced in HEK293T cells maintained in DMEM (D5796, Sigma) supplemented with 10% fetal bovine serum (F7524, Sigma) (45). Briefly, cells were washed with plain DMEM, polyethyleneimine transfected with 10 μg of lentiviral transfer plasmid pXLG3 WPRE px encoding the expression or knockdown of the protein of interest, 7.5 μg of packaging plasmid pΔ8.9, and 2.5 μg of envelope plasmid pMD2.G. After 4 h, transfection medium was replaced with complete HEK medium or neuronal feeding medium. Viral particles were harvested after ∼48 h, spun at 3000 × g for 10 min, dispensed into single-use aliquots, and stored at −80 °C until use.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (49) with the data set identifier PXD019597.
Acknowledgments
We thank the Medical Research Council and the Wolfson Foundation for establishing the Wolfson Bioimaging Facility. We thank R. Tomaino (Taplin MS Facility) for support with MS data.
This article contains supporting information.
Author contributions—Y. N. and S. J. M. conceptualization; Y. N., K. A. W., and S. J. M. resources; Y. N. and S. J. M. data curation; Y. N. formal analysis; Y. N., J. M. H., K. A. W., and S. J. M. supervision; Y. N. and S. J. M. funding acquisition; Y. N. and D. H. M. investigation; Y. N. and D. H. M. methodology; Y. N., A. J. N., K. A. W., and S. J. M. writing-original draft; Y. N., J. M. H., K. A. W., and S. J. M. project administration; Y. N., A. J. N., K. A. W., and S. J. M. writing-review and editing; J. M. H., K. A. W., and S. J. M. validation.
Funding and additional information—S. J. M. was supported by NINDS, National Institutes of Health, Grants NS087662, NS081986, NS108378, NS101888, NS103865, and NS111338 and NIMH, National Institutes of Health, Grant MH118263. Work in the laboratory of J. M. H. was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and Leverhulme Trust. S. J. M. is also supported by the Yale/NIDA Neuroproteomics Center and NIDA, National Institutes of Health, Grant DA018343. Work in the laboratory of J. M. H. was supported by the BBSRC and Leverhulme Trust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—S. J. M. serves as a consultant for AstraZeneca and SAGE Therapeutics, relationships that are regulated by Tufts University. S. J. M. holds stock in SAGE Therapeutics.
- GABAAR
- γ-aminobutyric acid type A receptor
- AIS
- axon initial segment
- DIV
- days in vitro
- Fsk
- forskolin
- GST
- glutathione S-transferase
- HBS
- HEPES-buffered saline
- HS
- horse serum
- IP
- immunoprecipitation
- IQR
- interquartile range
- myr-PKI
- myristoylated PKA inhibitor
- OA
- okadaic acid
- pHα2
- pHluorin-tagged GABAAR α2 subunit
- PKA
- cAMP-dependent protein kinase A
- PP1
- protein phosphatase 1
- PP2A
- protein phosphatase 2A
- SA
- S359A phosphonull mutant
- SD
- S359D phosphomimetic mutant
- shRNA
- short hairpin RNA
- TM
- transmembrane
- ANOVA
- analysis of variance.
References
- 1. Nusser Z., Cull-Candy S., and Farrant M. (1997) Differences in synaptic GABAA receptor number underlie variation in GABA mini amplitude. Neuron 19, 697–709 10.1016/S0896-6273(00)80382-7 [DOI] [PubMed] [Google Scholar]
- 2. Nusser Z., Hájos N., Somogyi P., and Mody I. (1998) Increased number of synaptic GABAA receptors underlies potentiation at hippocampal inhibitory synapses. Nature 395, 172–177 10.1038/25999 [DOI] [PubMed] [Google Scholar]
- 3. Olsen R. W., and Sieghart W. (2008) International Union of Pharmacology. LXX. Subtypes of γ-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol. Rev. 60, 243–260 10.1124/pr.108.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luscher B., Fuchs T., and Kilpatrick C. L. (2011) GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409 10.1016/j.neuron.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charych E. I., Liu F., Moss S. J., and Brandon N. J. (2009) GABAA receptors and their associated proteins: implications in the etiology and treatment of schizophrenia and related disorders. Neuropharmacology 57, 481–495 10.1016/j.neuropharm.2009.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacob T. C., Moss S. J., and Jurd R. (2008) GABAA receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat. Rev. Neurosci. 9, 331–343 10.1038/nrn2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z. W., and Olsen R. W. (2007) GABAA receptor associated proteins: a key factor regulating GABAA receptor function. J. Neurochem. 100, 279–294 10.1111/j.1471-4159.2006.04206.x [DOI] [PubMed] [Google Scholar]
- 8. Nusser Z., Sieghart W., Benke D., Fritschy J. M., and Somogyi P. (1996) Differential synaptic localization of two major gamma-aminobutyric acid type A receptor alpha subunits on hippocampal pyramidal cells. Proc. Natl. Acad. Sci. U. S. A. 93, 11939–11944 10.1073/pnas.93.21.11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nyíri G., Freund T. F., and Somogyi P. (2001) Input-dependent synaptic targeting of α2-subunit-containing GABAA receptors in synapses of hippocampal pyramidal cells of the rat. Eur. J. Neurosci. 13, 428–442 10.1046/j.1460-9568.2001.01407.x [DOI] [PubMed] [Google Scholar]
- 10. Fritschy J. M., Johnson D. K., Mohler H., and Rudolph U. (1998) Independent assembly and subcellular targeting of GABAA-receptor subtypes demonstrated in mouse hippocampal and olfactory neurons in vivo. Neurosci. Lett. 249, 99–102 10.1016/S0304-3940(98)00397-8 [DOI] [PubMed] [Google Scholar]
- 11. Abramian A. M., Comenencia-Ortiz E., Modgil A., Vien T. N., Nakamura Y., Moore Y. E., Maguire J. L., Terunuma M., Davies P. A., and Moss S. J. (2014) Neurosteroids promote phosphorylation and membrane insertion of extrasynaptic GABAA receptors. Proc. Natl. Acad. Sci. U. S. A. 111, 7132–7137 10.1073/pnas.1403285111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kittler J. T., Chen G., Honing S., Bogdanov Y., McAinsh K., Arancibia-Carcamo I. L., Jovanovic J. N., Pangalos M. N., Haucke V., Yan Z., and Moss S. J. (2005) Phospho-dependent binding of the clathrin AP2 adaptor complex to GABAA receptors regulates the efficacy of inhibitory synaptic transmission. Proc. Natl. Acad. Sci. U. S. A. 102, 14871–14876 10.1073/pnas.0506653102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura Y., Darnieder L. M., Deeb T. Z., and Moss S. J. (2015) Regulation of GABAARs by phosphorylation. Adv. Pharmacol. 72, 97–146 10.1016/bs.apha.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sassoè-Pognetto M., Panzanelli P., Sieghart W., and Fritschy J.-M. (2000) Colocalization of multiple GABAA receptor subtypes with gephyrin at postsynaptic sites. J. Comp. Neurol. 420, 481–498 [DOI] [PubMed] [Google Scholar]
- 15. Kneussel M., Brandstätter J. H., Laube B., Stahl S., Müller U., and Betz H. (1999) Loss of postsynaptic GABAA receptor clustering in gephyrin-deficient mice. J. Neurosci. 19, 9289–9297 10.1523/JNEUROSCI.19-21-09289.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacob T. C., Bogdanov Y. D., Magnus C., Saliba R. S., Kittler J. T., Haydon P. G., and Moss S. J. (2005) Gephyrin regulates the cell surface dynamics of synaptic GABAA receptors. J. Neurosci. 25, 10469–10478 10.1523/JNEUROSCI.2267-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mukherjee J., Kretschmannova K., Gouzer G., Maric H. M., Ramsden S., Tretter V., Harvey K., Davies P. A., Triller A., Schindelin H., and Moss S. J. (2011) The residence time of GABAARs at inhibitory synapses is determined by direct binding of the receptor α1 subunit to gephyrin. J. Neurosci. 31, 14677–14687 10.1523/JNEUROSCI.2001-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saiepour L., Fuchs C., Patrizi A., Sassoè-Pognetto M., Harvey R. J., and Harvey K. (2010) Complex role of collybistin and gephyrin in GABAA receptor clustering. J. Biol. Chem. 285, 29623–29631 10.1074/jbc.M110.121368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Papadopoulos T., Korte M., Eulenburg V., Kubota H., Retiounskaia M., Harvey R. J., Harvey K., O'Sullivan G. A., Laube B., Hülsmann S., Geiger J. R., and Betz H. (2007) Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 26, 3888–3899 10.1038/sj.emboj.7601819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hines R. M., Maric H. M., Hines D. J., Modgil A., Panzanelli P., Nakamura Y., Nathanson A. J., Cross A., Deeb T., Brandon N. J., Davies P., Fritschy J. M., Schindelin H., and Moss S. J. (2018) Developmental seizures and mortality result from reducing GABAA receptor α2-subunit interaction with collybistin. Nat. Commun. 9, 3130 10.1038/s41467-018-05481-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nathanson A. J., Zhang Y., Smalley J. L., Ollerhead T. A., Rodriguez Santos M. A., Andrews P. M., Wobst H. J., Moore Y. E., Brandon N. J., Hines R. M., Davies P. A., and Moss S. J. (2019) Identification of a core amino acid motif within the α subunit of GABAARs that promotes inhibitory synaptogenesis and resilience to seizures. Cell Rep. 28, 670–681.e8 10.1016/j.celrep.2019.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakamura Y., Morrow D. H., Modgil A., Huyghe D., Deeb T. Z., Lumb M. J., Davies P. A., and Moss S. J. (2016) Proteomic characterization of inhibitory synapses using a novel pHluorin-tagged γ-aminobutyric acid receptor, type A (GABAA), α2 subunit knock-in mouse. J. Biol. Chem. 291, 12394–12407 10.1074/jbc.M116.724443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beausoleil S. A., Villén J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- 24. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., and Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 25. Chiou T. T., Bonhomme B., Jin H., Miralles C. P., Xiao H., Fu Z., Harvey R. J., Harvey K., Vicini S., and De Blas A. L. (2011) Differential regulation of the postsynaptic clustering of γ-aminobutyric acid type A (GABAA) receptors by collybistin isoforms. J. Biol. Chem. 286, 22456–22468 10.1074/jbc.M111.236190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harvey K., Duguid I. C., Alldred M. J., Beatty S. E., Ward H., Keep N. H., Lingenfelter S. E., Pearce B. R., Lundgren J., Owen M. J., Smart T. G., Lüscher B., Rees M. I., and Harvey R. J. (2004) The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J. Neurosci. 24, 5816–5826 10.1523/JNEUROSCI.1184-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panzanelli P., Gunn B. G., Schlatter M. C., Benke D., Tyagarajan S. K., Scheiffele P., Belelli D., Lambert J. J., Rudolph U., and Fritschy J. M. (2011) Distinct mechanisms regulate GABAA receptor and gephyrin clustering at perisomatic and axo-axonic synapses on CA1 pyramidal cells. J. Physiol. 589, 4959–4980 10.1113/jphysiol.2011.216028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McKernan R. M., and Whiting P. J. (1996) Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 19, 139–143 10.1016/S0166-2236(96)80023-3 [DOI] [PubMed] [Google Scholar]
- 29. McDonald B. J., Amato A., Connolly C. N., Benke D., Moss S. J., and Smart T. G. (1998) Adjacent phosphorylation sites on GABAA receptor β subunits determine regulation by cAMP-dependent protein kinase. Nat. Neurosci. 1, 23–28 10.1038/223 [DOI] [PubMed] [Google Scholar]
- 30. McDonald B. J., and Moss S. J. (1997) Conserved phosphorylation of the intracellular domains of GABAA receptor β2 and β3 subunits by cAMP-dependent protein kinase, cGMP-dependent protein kinase protein kinase C and Ca2+/calmodulin type II-dependent protein kinase. Neuropharmacology 36, 1377–1385 10.1016/S0028-3908(97)00111-1 [DOI] [PubMed] [Google Scholar]
- 31. Terunuma M., Jang I. S., Ha S. H., Kittler J. T., Kanematsu T., Jovanovic J. N., Nakayama K. I., Akaike N., Ryu S. H., Moss S. J., and Hirata M. (2004) GABAA receptor phospho-dependent modulation is regulated by phospholipase C-related inactive protein type 1, a novel protein phosphatase 1 anchoring protein. J. Neurosci. 24, 7074–7084 10.1523/JNEUROSCI.1323-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brandon N. J., Jovanovic J. N., Colledge M., Kittler J. T., Brandon J. M., Scott J. D., and Moss S. J. (2003) A-kinase anchoring protein 79/150 facilitates the phosphorylation of GABAA receptors by cAMP-dependent protein kinase via selective interaction with receptor beta subunits. Mol. Cell Neurosci. 22, 87–97 10.1016/S1044-7431(02)00017-9 [DOI] [PubMed] [Google Scholar]
- 33. Brandon N. J., Jovanovic J. N., Smart T. G., and Moss S. J. (2002) Receptor for activated C kinase-1 facilitates protein kinase C-dependent phosphorylation and functional modulation of GABAA receptors with the activation of G-protein-coupled receptors. J. Neurosci. 22, 6353–6361 10.1523/JNEUROSCI.22-15-06353.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao Y., and Heldt S. A. (2016) Enrichment of GABAA receptor α-subunits on the axonal initial segment shows regional differences. Front. Cell Neurosci. 10, 39 10.3389/fncel.2016.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fritschy J. M. (2008) Epilepsy, E/I balance and GABAA receptor plasticity. Front. Mol. Neurosci. 1, 5 10.3389/neuro.02.005.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klausberger T., and Somogyi P. (2008) Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57 10.1126/science.1149381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roux L., and Buzsáki G. (2015) Tasks for inhibitory interneurons in intact brain circuits. Neuropharmacology 88, 10–-23 10.1016/j.neuropharm.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edwards C., and Ottoson D. (1958) The site of impulse initiation in a nerve cell of a crustacean stretch receptor. J. Physiol. 143, 138–148 10.1113/jphysiol.1958.sp006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu Y., Stornetta R. L., and Zhu J. J. (2004) Chandelier cells control excessive cortical excitation: characteristics of whisker-evoked synaptic responses of layer 2/3 nonpyramidal and pyramidal neurons. J. Neurosci. 24, 5101–5108 10.1523/JNEUROSCI.0544-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glickfeld L. L., Roberts J. D., Somogyi P., and Scanziani M. (2009) Interneurons hyperpolarize pyramidal cells along their entire somatodendritic axis. Nat. Neurosci. 12, 21–23 10.1038/nn.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Somogyi P., Nunzi M. G., Gorio A., and Smith A. D. (1983) A new type of specific interneuron in the monkey hippocampus forming synapses exclusively with the axon initial segments of pyramidal cells. Brain Res. 259, 137–142 10.1016/0006-8993(83)91076-4 [DOI] [PubMed] [Google Scholar]
- 42. Li X. G., Somogyi P., Tepper J. M., and Buzsáki G. (1992) Axonal and dendritic arborization of an intracellularly labeled chandelier cell in the CA1 region of rat hippocampus. Exp. Brain Res. 90, 519–525 10.1007/BF00230934 [DOI] [PubMed] [Google Scholar]
- 43. Cobb S. R., Buhl E. H., Halasy K., Paulsen O., and Somogyi P. (1995) Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature 378, 75–78 10.1038/378075a0 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., Zhang P., and Wyskiel D. R. (2016) Chandelier cells in functional and dysfunctional neural circuits. Front. Neural Circuits 10, 33 10.3389/fncir.2016.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rocca D. L., Wilkinson K. A., and Henley J. M. (2017) SUMOylation of FOXP1 regulates transcriptional repression via CtBP1 to drive dendritic morphogenesis. Sci. Rep. 7, 877 10.1038/s41598-017-00707-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Millar N. S., Moss S. J., and Green W. N. (1995) Assembly, post-translational processing, and subcellular localization of ion channels. in Ion Channels: A Practical Approach (Ashley R. H., ed) pp. 191–217, IRL Press, Oxford [Google Scholar]
- 47. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (49) with the data set identifier PXD019597.