Abstract
Imaging plays a key role in the management of brain tumors, including the diagnosis, prognosis, and treatment response assessment. Radiomics and deep learning approaches, along with various advanced physiologic imaging parameters, hold great potential for aiding radiological assessments in neuro-oncology. The ongoing development of new technology needs to be validated in clinical trials and incorporated into the clinical workflow. However, none of the potential neuro-oncological applications for radiomics and deep learning has yet been realized in clinical practice. In this review, we summarize the current applications of radiomics and deep learning in neuro-oncology and discuss challenges in relation to evidence-based medicine and reporting guidelines, as well as potential applications in clinical workflows and routine clinical practice.
Keywords: Radiomics, Deep learning, Neuro-oncology, Clinical workflow
INTRODUCTION
Radiomics involves the extraction of high-dimensional quantitative data reflecting imaging phenotypes. The methods currently adopted in oncologic imaging studies rely strongly on machine learning (1,2). Deep learning, a form of machine learning involving a special type of artificial neural network (3), is gaining much attention for its use in image-based pattern recognition in oncologic imaging. The goals of radiomics and deep learning studies in oncology are as follows: 1) classification of tumors, 2) providing links to genomics, 3) prediction of outcomes, and 4) response monitoring. Potential applications for deep learning and radiomics have been discussed and published in leading peer-reviewed neuro-oncologic journals, but none of these potential applications has yet been realized in clinical practice. This may be because of the rare occurrence of many brain tumors and the low availability of the source material, with magnetic resonance imaging (MRI) being used as the main diagnostic tool. More importantly, the implementation of deep learning and radiomics is technically challenging and complicated to integrate into the clinical workflow, and there is not enough consideration of evidence-based medicine in deep learning and radiomics research. Because “if it was never used, it did not exist” (4), this article reviews the potential clinical applications of radiomics and deep learning, and discusses the special considerations in neuro-oncology, with the hope of yielding more actionable change to clinical workflows.
Trends of Radiomics and Deep Learning Research in Neuro-Oncology
Deep learning involves a special type of artificial neural network that resembles the human cognitive system and is a form of machine learning. Deep learning methods, especially convolutional neural networks (CNNs), excel at pattern recognition and finding complex patterns in imaging data, and are generally more effective than previous algorithms (3). Deep learning algorithms use a data-driven approach and do not require prior definition by human experts.
Radiomics approaches involve the extraction of hundreds of quantitative features from images to assess the entire three-dimensional (3D) tumor phenotype. The resulting features may then be used as quantitative imaging biomarkers. Briefly, morphologic features (volume/shape), histogram features (first-order), texture features (second-order), and transform-based features are the most commonly used radiomics features (1,5). Typically, machine-learning techniques are subsequently applied to the extracted radiomics features in a feature selection step to reduce the dimensions of the data.
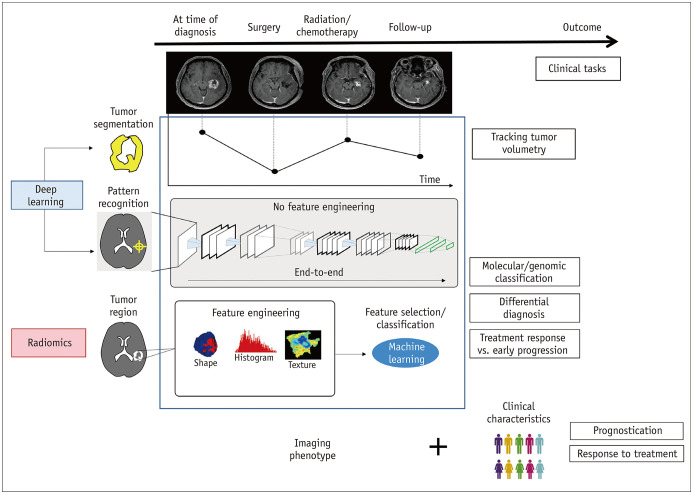
Figure 1 demonstrates imaging-based radiomics and deep learning tasks in neuro-oncology. Extensive research articles using deep learning and radiomics in neuro-oncology have been published. The clinical situations include the differential diagnosis of neoplasms, prognosis determination at initial imaging, distinguishing tumor recurrence from post-treatment contrast enhancement due to pseudoprogression or early tumor progression in glioblastoma (6) on post-therapeutic imaging after first-line treatment regimens, and predicting the treatment response to anti-angiogenic (7,8) and immunotherapies (9) at the time of recurrence. Representative applications of deep learning and radiomics in neuro-oncology are summarized in Table 1. Later in this review, we will demonstrate the utility of radiogenomics as a supplement to preexisting diagnostic techniques.
Fig. 1. Imaging-based radiomics and deep learning tasks in neuro-oncology.
Deep learning can be applied to automated tumor segmentation to track tumor volumetry and pattern recognition to conduct various end-to-end classification tasks. Radiomics approaches using engineered features and machine learning-based feature selection have also been applied to radiogenomics classification tasks, differential diagnoses, and diagnosis of early tumor progression. Imaging phenotypes identified using deep learning and radiomics could ultimately be combined with clinical characteristics to assess prognosis and treatment response of individual patients.
Table 1. Representative Applications of Radiomics and DL in Neuro-Oncology.
| Topic | Methods | References | Data (Train:Test) | Outcomes | Test Performance | External Validation | Imaging Modalities |
|---|---|---|---|---|---|---|---|
| Differential diagnosis | Radiomics | Artzi et al. (10) | 439 (351:88) | Glioblastoma, metastasis | Accuracy = 85% | No | Conventional MRI |
| AUC = 0.98 | |||||||
| Radiomics | Kang et al. (11) | 154 (112:42) | PCNSL, glioblastoma | AUC = 0.94 (external validation) | Yes | Conventional MRI, DWI | |
| Radiomics | Kniep et al. (12) | 189 | Metastasis cell type | AUC = 0.64 for non-small cell lung cancer AUC 0.82 for melanoma | No | Conventional MRI | |
| Prognostication | Radiomics | Kickingereder et al. (14) | 119 (179:40) | Glioblastoma survival | C-index = 0.696 (radiomics + clinical) | No | Conventional MRI |
| C-index = 0.637 (radiomics only) | |||||||
| Radiomics | Kickingereder et al. (16) | 181 (120:61) | Glioblastoma survival | IBS = 0.103 (molecular + clinical + radiomics) | No | Conventional MRI | |
| IBS = 0.127 (radiomics only) | |||||||
| Radiomics | Bae et al. (15) | 217 (split-sample) | Glioblastoma survival | Integrated AUC = 0.652 | No | Conventional MRI | |
| DL | Lao et al. (19) | 102 (75:37) | Glioblastoma survival | C-index = 0.739 (clinical + radiomics) | No | Conventional MRI | |
| C-index = 0.710 (radiomics only) | |||||||
| DL | Nie et al. (20) | 69 | WHO grade II and III | Accuracy = 89.9% (low- and high-risk) | No | Conventional MRI | |
| Pseudoprogression vs. progression | Radiomics | Kim et al. (6) | 118 (61:57) | Glioblastoma pseudoprogression | AUC = 0.96 (internal validation), 0.85 (external validation) | Yes | Conventional MRI, DWI, DSC |
| DL | Hu et al. (21) | 31 | Glioblastoma pseudoprogression | AUC = 0.95 | No | Conventional MRI, DWI, DSC | |
| DL | Qian et al. (24) | 35 | Glioblastoma pseudoprogression | AUC = 0.867 | No | DTI | |
| DL | Jang et al. (25) | 78 (59:19) | Glioblastoma pseudoprogression | AUC = 0.83 | Yes | Conventional MRI | |
| Treatment response assessment | Radiomics | Kickingereder et al. (7) | 172 (112:60) | Glioblastoma | Stratification between low- and high-risk | No | Conventional MRI |
| Anti-angiogenic treatment | HR = 1.85 (PFS) | ||||||
| HR = 2.60 (OS) | |||||||
| Radiomics | Grossmann et al. (8) | 293 (126:167) | Glioblastoma | Stratification between low- and high-risk | Yes | Conventional MRI | |
| Anti-angiogenic treatment | HR = 4.5 (PFS) | ||||||
| HR = 2.5 (OS) | |||||||
| Radiomics | Bhatia et al. (9) | 88 | Metastasis (melanoma) | HR = 0.68 (OS) | No | Conventional MRI | |
| Immune checkpoint inhibitor |
AUC = area under receiver operating characteristic curve, Conventional MRI = to T1-weighted, T2-weighted, fluid-attenuated inversion recovery, or contrast-enhanced T1-weighted imaging, DL = deep learning, DSC = dynamic susceptibility contrast, DTI = diffusion-tensor imaging, DWI = diffusion-weighted imaging, fMRI = functional MRI, HR = hazard ratio, IBS = integrated Brier score, OS = overall survival, PCNSL = primary central nervous system lymphoma, PFS = progression-free survival, WHO = World Health Organization
Initial Diagnosis: Differential Diagnosis Using Radiomics
Radiomics
Radiomics has been applied to cases of differential diagnoses that were challenging even for radiologists. Furthermore, studies have assessed the added value of radiomics over the performance of radiologists. The differential diagnosis between a single brain metastasis and glioblastoma for a solitary contrast-enhancing (CE) mass is challenging. However, radiomics based on conventional post-contrast T1-weighted imaging showed an accuracy of 85% and an area under the receiver operating characteristic curve (AUC) of 0.96 (10). Primary central nervous system lymphoma (PCNSL) may exhibit internal necrosis and heterogeneous contrast enhancement that mimics glioblastoma. The potential of radiomics from the apparent diffusion coefficient (ADC) has been shown to differentiate PCNSL from glioblastoma (11), showing high performance on both internal (AUC 0.984) and external (AUC 0.944) validation sets. Radiomics was shown to be feasible for classifying the subtype of brain metastases (12), with its performance ranging from an AUC of 0.64 for non-small-cell lung cancer to 0.82 for melanoma. As diagnostic decision-making for patients with unknown primary lesion sites requires extensive steps, early differential diagnoses may be helpful to narrow down the diagnostic processes. However, few studies have been conducted regarding using deep learning for differential diagnoses, which may be due to the low availability of MRI source data from patients.
Survival Prediction
Radiomics
The prognostication of glioblastoma is of particular interest, as the prognosis for patients with glioblastoma remains poor, with a median survival of 15 months (13). The poor prognosis may arise from intratumoral heterogeneity, with resistance to treatment occurring both spatially and temporally. Radiomics allows for the noninvasive quantitative characterization of tumors, and studies have shown the added value of radiomics to the well-established prognostic markers of age, the extent of surgery, Karnofsky performance score, and methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) gene. Using standard-of-care imaging, radiomic analysis improved survival prediction when combined with clinical data (C-index improving from 0.637 to 0.696) (14) or genetic data (15,16). One study emphasizes the automatization and reproducibility of a radiomics model (17).
Deep Learning
To overcome the problem of small datasets, transfer learning, and fine-tuning has been adopted (18) for survival prediction in glioma patients. A deep feature-based radiomics model using transfer learning to extract a large number of deep features from hidden layers of the CNN was applied to glioblastoma (19) and showed a C-index of 0.710 using radiomics features alone and 0.739 using radiomics and clinical predictors. Using multimodal imaging, including conventional MRI, functional MRI, and diffusion-tensor imaging (DTI), a deep learning architecture involving a 3D CNN extracted defining features from high-grade gliomas (20). This CNN showed an accuracy of 89.9% for predicting overall survival (OS) in 69 patients with high-grade gliomas, although it was without independent validation. The above findings suggest the feasibility of deep learning methods coupled with traditional radiomics features and machine learning classifiers for predicting survival. Nonetheless, studies are limited to showing the technical feasibility of deep learning applied for glioma survival prediction with small datasets, with or without an independent validation set. The proof of real-world performance on a large dataset is ultimately needed.
Follow-Up: Pseudoprogression vs. Tumor Progression
Radiomics
The differentiation of true progressive disease from pseudoprogression is challenging on conventional MRI. Before the advent of radiomics, machine learning classifiers (21) and clustering (22) methods have been adopted to solve this question using multiparametric imaging parameters, not with radiomics but with the quantitative value obtained from diffusion-weighted and perfusion-weighted imaging. Using high-dimensional radiomics features, multiparametric MRI can include physiologic information that helps improve the differentiation of pseudoprogression from tumor progression. MR perfusion radiomics using relative cerebral blood volume (CBV) and the capillary permeability measure Ktrans showed the diagnostic ability for pseudoprogression in a multiinstitution setting (23). A multi-parametric radiomics model including the ADC and CBV showed better performance on external validation (AUC, 0.90) than a single ADC or CBV parameter (AUC, 0.57–0.79), or a radiomics model based on T1-weighted images (T1WIs), or T2-weighted images (T2WIs; AUC, 0.76) (6).
Deep Learning
Deep learning applications have adopted multiparametric MRI, with both conventional and physiologic MRIs being frequently used. One study used longitudinal DTI, dictionary learning, and feature pooling to characterize two conditions (24), and showed an AUC of 0.867 for 35 patients without an independent validation set. More recently, a CNN with long short-term memory (25) was applied to differentiate tumor progression from pseudoprogression based on post-contrast T1-weighted MRI data from two institutions, with 59 patients in the training set and 19 patients in the external validation set, yielding an AUC of 0.83. The above deep learning applications were designed as proof-of-concept studies with small numbers of patients and a cross-sectional design.
Before Second-Line Treatment: Predicting Treatment Response Using Radiomics
The anti-angiogenic agent bevacizumab is the single most widely used second-line therapeutic option for recurrent glioblastoma. However, no validated imaging biomarker is available to identify patients for whom this agent is likely beneficial. Two articles demonstrated the application of radiomics approaches to anti-angiogenic treatment. Using 4842 radiomics features of T1WI, T2WI, and contrast-enhanced T1WI and analyzed with a supervised principal-component analysis, Kickingereder et al. (7) stratified patients into those with a low- or high-risk of recurrent glioblastoma (hazard ratio [HR] = 1.85 for progression-free survival [PFS] and HR = 2.60 for OS), and the low-risk group (172 patients, training:validation = 2:1). Using 65 radiomics features extracted from T1WI, T2WI, and contrast-enhanced T1WI of the multicenter BRAIN trial data, another study (8) showed successful stratification of recurrent glioblastoma patients according to OS (HR = 2.5) and PFS (HR = 4.5).
A recent study (9) showed the potential of the radiomics approach with contrast-enhanced T1WI brain metastasis treated with immune checkpoint inhibitors. In a total of 196 melanoma brain metastases from 88 patients, multiple radiomics features were found to be associated with OS, and the Laplacian of the Gaussian features best explained the OS (HR = 0.68), which was also confirmed in a validation dataset consisting of 17 patients.
Evidence-Based Medicine: Challenges for Radiomics and Deep Learning in the Clinical Workflow
The critical assessment of deep learning and radiomics studies should be based on the widely recognized principles of evidence-based medicine. Although these requirements are not always paid much attention in technical papers, they have paramount importance in clinical studies and applications. Prediction models developed from radiomics and deep learning aim to support clinical decision-making by the use of large amounts of complex data, and the validation and transparent reporting of results is important.
Validation in Deep Learning and Radiomics
For external validation, it is recommended to use adequately sized datasets that are collected either from newly recruited patients or from institutions other than those that provided the training data from which the deep learning and radiomics was developed (26).
A prospective diagnostic cohort design is a way to adequately represent the manifestation spectrum of target patients in real-world clinical settings, with a before-after study design being applied (27). In this design, patients undergo evaluation both without (before) and with (after) a diagnostic or predictive deep learning/radiomics tool, and the clinical decisions, instead of patient outcomes, are compared between the two diagnostic evaluation sessions (28,29).
One study evaluated radiomics studies (30) published up until 2018 using the search terms “radiomics” and “radiogenomics” in the PubMed, MEDLINE, and EMBASE databases. Among 77 studies with impact factors above 7.0, only 18.2% (14 studies) performed external validation. Only three studies performed prospective validation, but even this was a post-hoc analysis of the prospective registry or database. None of the studies used a diagnostic cohort design or performed a cost-effectiveness analysis.
A similar trend was found for deep learning studies in an article that evaluated the design characteristics of general artificial intelligence (AI) studies (31) using the search terms “artificial intelligence” OR “machine learning” OR “deep learning” OR “convolutional neural network” for articles published up to 2018 in the PubMed, MEDLINE, and EMBASE databases. This review found that most studies that evaluated the performance of AI algorithms for the diagnostic analysis of medical images were designed as proof-of-concept technical feasibility studies. Only 31 (6%) of the 516 studies performed external validation, and none of the 31 studies adopted all three design features of a diagnostic cohort design, inclusion of multiple institutions, and prospective data collection for external validation.
A model built with radiomics, and deep learning should not be limited to the patient population used for model construction. A high standard for the validation of radiomics and deep learning must be encouraged to transfer the techniques to routine clinical practice and prevent them from being stranded in the research domain (26).
Reporting Guidelines
Radiomics and deep learning applications in radiology may be sensitive to the characteristics of the study population, predictors, and choice of the reference standard. Comprehensive and transparent reporting is required to ensure that the performance of prediction models is generalizable. For predictive models, the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement (32) emphasizes the transparent reporting of the study settings, outcome follow-up intervals, and precise definitions of the study design elements and how outcomes were defined and measured.
In a review of 77 radiomics studies (30), the mean number of TRIPOD items reported was 18.51 ± 3.96 (mean ± standard deviation; range, 11–26). All articles clearly defined all predictors (item 7a) and described how predictors were handled (item 10a), but no article described the recalibration of the coefficients in the preexisting model in the methods (item 10e) or results (item 17) section. The radiomics studies showed less than 25% adherence to “explicitly describe the development/validation of the model or both in ‘title’ (2.6%, item 1), ‘abstract’ (7.8%, item 2), ‘or introduction’ (16.9%, item 3b).” Furthermore, the radiomics studies did not adhere to “calculate sample size” (6.5%, item 8) and “describe missing data” (11.7%, item 9).
Recently, the importance of adhering to established methodological standards has been identified for machine learning techniques. Although many aspects of the TRIPOD statement apply to radiomics/deep learning-based prediction model studies, TRIPOD focuses on regression-based prediction model approaches (33). TRIPOD has not been widely adopted by the AI community. A new initiative to develop a version of the TRIPOD statement specific to machine learning will be announced in the near future.
Comparative Accuracy: Potential Applications of Radiomics and Deep Learning in Clinical Workflow in Neuro-Oncology
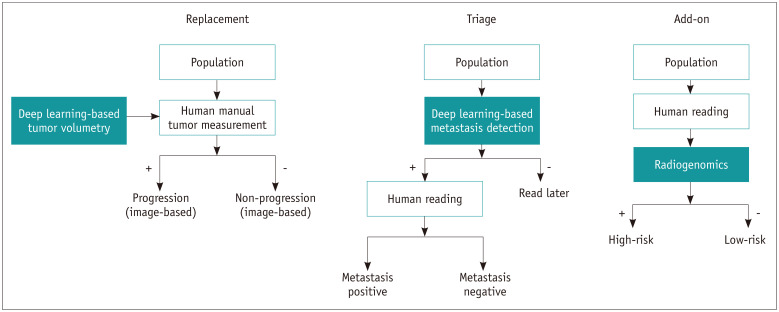
To be adopted in clinical practice, deep learning and radiomics must address unmet needs or improve existing situations. A conceptual framework for evaluation of the comparative accuracy of new diagnostic tests was developed by Bossuyt et al. (34), and it consists of the following: 1) replacement situation, 2) triage situation, and 3) add-on situation. The replacement situation is when a new test differs from existing methods in various ways, including improved accuracy, ease, comfort, and speed, and may replace existing tests. In triage, the new test is used before the existing test or testing pathway, and only patients with a particular result on the triage test continue on the testing pathway. This may be less accurate than existing tests and is not meant to replace them. Add-on test scenarios may also be positioned after the existing pathway, and the use of these tests may be limited to a subgroup of patients. The new test is more accurate but is otherwise less attractive than the existing tests. The matched clinical scenarios in neuro-oncology are tumor segmentation to replace manual segmentation, detection-based triage in brain metastasis, and radiomics to supplement existing information. Figure 2 demonstrates radiomics and deep learning in the clinical workflow and their roles in existing diagnostic pathways.
Fig. 2. Radiomics and deep learning in clinical workflow and their roles in existing diagnostic pathways.
Replacement: deep learning-based tumor volumetry has potential to replace human-derived manual tumor volumetry for defining image-based progression and non-progression. Triage: deep learning-based metastasis detection has potential to triage patients and identify those whose imaging needs to be read first and may increase radiologists' specificity and reduce tiredness. Add-on: radiogenomics applications have potential to stratify further high-risk and low-risk groups and may help guide patient management. Adapted from Bossuyt et al. BMJ 2006;332:1089-1092 (34).
Replacement: Deep Learning-Based Automatic Tumor Volumetry and Tumor Response Assessment
Deep learning-based segmentation may replace manual segmentation. This would be beneficial as computers do not tire and can provide fast and reproducible segmentation. Deep learning-based segmentation approaches for brain MRI are gaining interest in their self-learning and generalization abilities with large amounts of data (35,36,37). Most neuro-oncologic imaging studies are based on MRI, which is particularly challenging with respect to the generalizability and robustness of radiomics analysis, as MRI has non-standardized pixel values and typically shows large variations in signal intensities. The robust segmentation required for the determination of tissue volumes imposes a great challenge for quantitative imaging analysis (5). Even expert readers show high inter-reader variability in segmentation, which can affect quantitative analysis (38,39) and the extraction of high-dimensional radiomics features (1,2,40,41).
Deep learning-based segmentation of brain tumors has been a long-standing topic of interest in the field of neuro-oncology and has mainly been driven through the Brain Tumor Segmentation (BraTS) challenge (42). Specifically, the annual BraTS challenge provides an important forum for evaluating state-of-the-art methods for the volumetric segmentation of brain tumors using multiparametric MRI scans. However, despite recent developments, clinical translation of the volumetric assessment of brain tumors (e.g., for assessing treatment response) is lacking. Currently, the Response Assessment in Neuro-Oncology (RANO) criteria, which primarily rely on manual two-dimensional (2D) measurements of CE target lesions on MRI, are the standard method for assessing the treatment response of brain tumors. Underlying the use of RANO is the assumption that tumors grow spherically and that the 2D measurement of a lesion's largest diameter on MRI is a surrogate marker of tumor volume. However, this assumption is not always accurate in clinical practice, as brain tumors frequently display very complicated shapes and anisotropic growth, influenced in part by the surrounding anatomic boundaries, host tissue–tumor interface, and/or treatment-related effects (e.g., areas of necrosis and surgical cavities). Consequently, there has been a long-standing interest in using volumetric assessment of tumor burden (43,44,45), with studies indicating that volumetric measurements may be more reliable and accurate than 2D measurements (46,47). Nevertheless, although volumetric assessment may arguably be one of the most quintessential parameters for the accurate assessment of tumor burden and response (48), it lacks practicality in a clinical setting. Whereas 2D measurements of tumor diameter can be performed quickly and without dedicated software, volumetric measurements require sophisticated and time-consuming post-processing of MRI data with dedicated software (7,16,49).
Recently, the importance and meaningful clinical use of deep learning-based BraTS were demonstrated in terms of the automated and quantitative assessment of treatment response (49). Specifically, it was shown that automated deep learning-based volumetric assessment was highly accurate, with DICE coefficients of more than 0.90 for the segmentation of CE tumor and non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) signal abnormalities on independent multicenter testing (49). Using deep learning-based tumor volumes for quantitative volumetric tumor response assessment, it was shown that 1) the reliability (i.e., agreement in the quantitative volumetrically defined time to progression [comparison of the radiologist's ground truth with the automated assessment with deep learning]) was significantly higher (by a margin of 36%) than the RANO assessment, and 2) that the automated volumetrically defined time to progression was a better surrogate endpoint compared with RANO for the OS of the patients (49). These findings provide evidence that the automated and reliable assessment of tumor response in neuro-oncology with a high-throughput of cases is feasible, and clinically important for providing high-quality imaging endpoints.
Triage: Detecting Metastasis Using Deep Learning
Deep learning can be applied to the radiologists' routine workflow to reduce the effort required for existing tests and improve the overall workflow. Previous studies suggested that in a triage framework, radiologists could control for false-positive classifications through manual inspection, thereby leaving the triaging system to prioritize, review and benefit the workflow, even without accuracy improvements (50,51). One study demonstrated the utility of CNN-based analysis of computed tomography for triage of acute neurological illnesses in the emergency department (52). After applying CNN-based triage, the urgent cases appeared earlier in the queue in the prioritized list than in the routine list without the application of CNN.
In neuro-oncology, computer-assisted triage may augment the clinician's detection and shorten the reading time. The detection of brain metastases is a tedious and time-consuming task for many radiologists, particularly with the growing use of thin-section multi-sequence 3D imaging. Technical developments have shown the feasibility of using 3D template matching (53,54) and CNNs (55). One study (54) showed improvement of the radiologist's performance for detecting metastasis less than 100 mm-size from 89.83% to 100%. Imaging only-based detection and diagnosis of brain metastases without radiologists would be unacceptable under the current regulations. Furthermore, algorithms will generally be overfitted to the training data and the performance will be suboptimal. Instead, an algorithm for automated detection of brain metastases may be initially used, with radiologists then validating the result to ensure that the final diagnoses are correct. Even high-quality weak labels would lead to improved classifier performance, and consequently to superior triage results.
Add-On: Super-Diagnostics for Genetic Mutations Using Deep Learning and Radiomics
Other new tests may be positioned after the existing pathway. The use of these tests may be limited to a subgroup of patients, for example, when the new test is more accurate but otherwise less attractive than the existing test. Add-on tests can increase the sensitivity of the existing pathway, possibly at the expense of specificity. Image-based diagnosis of genetic mutations is of great interest for glioma, and new deep learning or radiomics applications may provide more accurate diagnosis than preexisting methods.
The key genomic landscapes seen in diffuse gliomas (56) are isocitrate dehydrogenase (IDH) mutation, 1p/19q codeletions, MGMT-promoter methylation status, and epidermal growth factor receptor (EGFR) amplification mutation. According to the most recent 2016 World Health Organization classification, the hallmark molecular parameters for better classification of glioma are IDH and 1p/19q co-deletion status (56). MGMT methylation status reflects the ability of tumor cells to repair DNA damage from temozolomide (57). EGFR amplification reflects the molecular pathogenesis of glioblastomas (58). Deep learning and radiomics tend to be focused on a single genomic or molecular classification, with IDH mutation status being the most popular topic, having been investigated with T2/FLAIR imaging (59), contrast-enhanced T1WI (60), diffusion- and perfusion-imaging (61), and in combination with location information (62). Recent radiomics (63,64,65) and deep learning studies (66,67,68,69) have focused more on the characterization of multiple tumoral genetic mutations, rather than single genetic mutations, to provide an integrated view.
Radiomics
Recently, copy number variations and the DNA methylation profile have been adopted in radiogenomics research, with Kickingereder et al. (70) showing that the machine learning analysis of multiparametric MRI including diffusion-, perfusion-, and susceptibility-weighted imaging could potentially predict DNA methylation status and hallmark copy number variations in 152 glioblastomas. Hu et al. (71) demonstrated radiomics-based heterogeneity using 48 image-guided biopsies from glioblastomas and showed significant image correlations with T1WI, T2WI, and contrast-enhanced T1WI for predicting platelet-derived growth factor receptor A (77.1%), EGFR (75%), cyclin-dependent kinase inhibitor 2A (87.5%), and retinoblastoma transcriptional corepressor 1 (87.5%). The feasibility of predicting the core signaling pathway of glioblastoma was shown by Park et al. (72), who showed that copy number variations, insertion/deletion, and single nucleotide variations from next-generation sequencing could be predicted using multiparametric MRI radiomics. These approaches used multimodal imaging parameters to provide information on various genomic alterations.
Deep Learning
With the application of a CNN to 256 brain MRIs from the Cancer Imaging Archives dataset (67), a study showed a prediction accuracy of 94% for IDH status, 92% for 1p/19q co-deletion status, and 83% for MGMT-promoter methylation status. Deep features obtained from DTI and dynamic susceptibility contrast imaging (208 training sets and 53 validation sets) were clustered (73), and different imaging subtypes of glioblastoma were stratified by IDH-1, MGMT, and EGFR mutations. Radiogenomic assessment beyond routine imaging diagnosis should stratify low- and high-risk patients and may further guide patient consultations and therapeutic plans.
Infrastructure to Be Integrated into the Clinical Workflow
Ultimately, the driver for clinical adoption may reside in the implementation and availability of AI applications integrated into the picture archiving and communication system (PACS) at the reading station. The integration of concise and standardized AI interpretation results directly into the Digital Imaging and Communications in Medicine standard could enable easier portability among PACSs.
The current AI frameworks are primarily accessible by computer scientists. While this is acceptable for interdisciplinary groups that have both clinical and technical experts, the adoption of such frameworks by primarily clinical sites and for clinical deployment will require substantial effort to improve the transparency of the methods, the accessibility of the frameworks, and enable prospective large-scale on-demand application in clinical routine. A prime example is the open-source Extensible Neuroimaging Archive Toolkit (XNAT; www.xnat.org), which provides the necessary software infrastructure for importing, archiving, processing, and securely distributing neuroimaging studies (74). Specifically, it also enables the integration of custom processing pipelines (e.g., deep learning-based BraTS through the XNAT Container Service Plugin) for large-scale evaluations (e.g., in clinical routine). Successful clinical translation of a deep learning-based processing pipeline and its integration within the XNAT framework was recently shown for automated quantitative (volumetric) tumor response assessment of MRI in neuro-oncology (www.neuroAI-HD.org) (49). Similar future endeavors with open-source deposition of AI-based processing pipelines (e.g., radiomic or deep learning-based prediction models, and deep learning-based BraTS tools) will be important for independent large-scale validation and clinical adoption of AI-based processing pipelines.
Existing Drawbacks and Perspectives
The most distinct drawbacks of radiomics and deep learning from the feature-based perspective are that the features are data-driven, and there is no direct biological linkage. For example, while there are many existing data-driven features for IDH mutation based on deep learning and radiomics, these features are complex and not intuitive. Contrastingly, MR spectroscopy directly measures the byproduct of IDH mutation, 2-hydroxyglutarate, while the T2/FLAIR mismatch sign gives intuitive information on IDH mutation status. This is one reason why the utilization of ‘complex’ radiomics is more directed towards providing added value to pre-existing imaging diagnostics, rather than replacing them, especially from the perspective of comparative accuracy.
Another drawback to deep learning is that it is data-dependent. Deep learning is ‘data-hungry,’ and a large amount of good quality data is required to maximize the accuracy of deep learning models. Insufficient data may mean that performance is insufficient to replace or augment pre-existing imaging diagnostics. Changing a model-learning method with meta-learner (75) or increasing the quantity of training data using synthetic data augmentation may improve model performance. From the perspective of comparative accuracy, deep learning will be more directed towards triaging and replacing mundane human tasks.
CONCLUSION
Radiomics and deep learning are active research areas in the field of neuro-oncology, and many studies have shown their potential for future clinical implementation. To be incorporated into existing clinical workflows, studies are recommended to adhere to evidence-based medicine and use data that are validated in the real-world setting. Automated tumor volumetry, the detection of brain metastases, and radiogenomics are highly likely to become end-uses for radiomics and deep learning in clinical practice. Finally, integrated infrastructure for the clinical translation of radiomics and deep learning will ultimately be needed.
Footnotes
This research was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (grant number: NRF-2020R1A2B5B01001707 and NRF-2020R1A2C4001748).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topol E. Deep medicine: how artificial intelligence can make healthcare human again. 1st ed. NewYork: Basic Books; 2019. [Google Scholar]
- 5.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Park JE, Jo Y, Shim WH, Nam SJ, Kim JH, et al. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2019;21:404–414. doi: 10.1093/neuonc/noy133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT, et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22:5765–5771. doi: 10.1158/1078-0432.CCR-16-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA, et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;19:1688–1697. doi: 10.1093/neuonc/nox092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatia A, Birger M, Veeraraghavan H, Um H, Tixier F, McKenney AS, et al. MRI radiomic features are associated with survival in melanoma brain metastases treated with immune checkpoint inhibitors. Neuro Oncol. 2019;21:1578–1586. doi: 10.1093/neuonc/noz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artzi M, Bressler I, Ben Bashat D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging. 2019;50:519–528. doi: 10.1002/jmri.26643. [DOI] [PubMed] [Google Scholar]
- 11.Kang D, Park JE, Kim YH, Kim JH, Oh JY, Kim J, et al. Diffusion radiomics as a diagnostic model for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol. 2018;20:1251–1261. doi: 10.1093/neuonc/noy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kniep HC, Madesta F, Schneider T, Hanning U, Schönfeld MH, Schön G, et al. Radiomics of brain MRI: utility in prediction of metastatic tumor type. Radiology. 2019;290:479–487. doi: 10.1148/radiol.2018180946. [DOI] [PubMed] [Google Scholar]
- 13.Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kickingereder P, Burth S, Wick A, Götz M, Eidel O, Schlemmer HP, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 15.Bae S, Choi YS, Ahn SS, Chang JH, Kang SG, Kim EH, et al. Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology. 2018;289:797–806. doi: 10.1148/radiol.2018180200. [DOI] [PubMed] [Google Scholar]
- 16.Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Götz M, Wick A, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20:848–857. doi: 10.1093/neuonc/nox188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, et al. A fullyautomatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep. 2017;7:14331. doi: 10.1038/s41598-017-14753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–1359. [Google Scholar]
- 19.Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie D, Zhang H, Adeli E, Liu L, Shen D. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. Med Image Comput Comput Assist Interv. 2016;9901:212–220. doi: 10.1007/978-3-319-46723-8_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging. 2011;33:296–305. doi: 10.1002/jmri.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JE, Kim HS, Goh MJ, Kim SJ, Kim JH. Pseudoprogression in patients with glioblastoma: assessment by using volume-weighted voxel-based multiparametric clustering of MR imaging data in an independent test set. Radiology. 2015;275:792–802. doi: 10.1148/radiol.14141414. [DOI] [PubMed] [Google Scholar]
- 23.Elshafeey N, Kotrotsou A, Hassan A, Elshafei N, Hassan I, Ahmed S, et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun. 2019;10:3170. doi: 10.1038/s41467-019-11007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian X, Tan H, Zhang J, Zhao W, Chan MD, Zhou X. Stratification of pseudoprogression and true progression of glioblastoma multiform based on longitudinal diffusion tensor imaging without segmentation. Med Phys. 2016;43:5889. doi: 10.1118/1.4963812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang BS, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Scientific Reports. 2018;8:12516. doi: 10.1038/s41598-018-31007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang A, Tam R, Cadrin-Chenevert A, Guest W, Chong J, Barfett J, et al. Canadian association of radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J. 2018;69:120–135. doi: 10.1016/j.carj.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 28.Guyatt GH, Tugwell PX, Feeny DH, Drummond MF, Haynes RB. The role of before-after studies of therapeutic impact in the evaluation of diagnostic technologies. J Chronic Dis. 1986;39:295–304. doi: 10.1016/0021-9681(86)90051-2. [DOI] [PubMed] [Google Scholar]
- 29.Knottnerus JA, Buntinx F. The evidence base of clinical diagnosis: theory and methods of diagnostic research. 2nd ed. London: BMJ Books; 2011. [Google Scholar]
- 30.Park JE, Kim D, Kim HS, Park SY, Kim JY, Cho SJ, et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2020;30:523–536. doi: 10.1007/s00330-019-06360-z. [DOI] [PubMed] [Google Scholar]
- 31.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20:405–410. doi: 10.3348/kjr.2019.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 33.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- 34.Bossuyt PM, Irwig L, Craig J, Glasziou P. Comparative accuracy: assessing new tests against existing diagnostic pathways. BMJ. 2006;332:1089–1092. doi: 10.1136/bmj.332.7549.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weston AD, Korfiatis P, Kline TL, Philbrick KA, Kostandy P, Sakinis T, et al. Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology. 2019;290:669–679. doi: 10.1148/radiol.2018181432. [DOI] [PubMed] [Google Scholar]
- 36.Lin L, Dou Q, Jin YM, Zhou GQ, Tang YQ, Chen WL, et al. Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology. 2019;291:677–686. doi: 10.1148/radiol.2019182012. [DOI] [PubMed] [Google Scholar]
- 37.Norman B, Pedoia V, Majumdar S. Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177–185. doi: 10.1148/radiol.2018172322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. Plos One. 2014 Jul 15; doi: 10.1371/journal.pone.0102107. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavic M, Bogowicz M, Wurms X, Glatz S, Finazzi T, Riesterer O, et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018;57:1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 40.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 41.Park JE, Kim HS. Radiomics as a quantitative imaging biomarker: practical considerations and the current standpoint in neuro-oncologic Studies. Nucl Med Mol Imaging. 2018;52:99–108. doi: 10.1007/s13139-017-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. Cornell University; 2018. [updated April 2019]. [Accessed March 3, 2020]. Available at: https://arxiv.org/abs/1811.02629. [Google Scholar]
- 43.Yang D. Standardized MRI assessment of high-grade glioma response: a review of the essential elements and pitfalls of the RANO criteria. Neurooncol Pract. 2016;3:59–67. doi: 10.1093/nop/npv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Bent MJ, Wefel JS, Schiff D, Taphoorn MJ, Jaeckle K, Junck L, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011;12:583–593. doi: 10.1016/S1470-2045(11)70057-2. [DOI] [PubMed] [Google Scholar]
- 45.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 46.Chow DS, Qi J, Guo X, Miloushev VZ, Iwamoto FM, Bruce JN, et al. Semiautomated volumetric measurement on postcontrast MR imaging for analysis of recurrent and residual disease in glioblastoma multiforme. AJNR Am J Neuroradiol. 2014;35:498–503. doi: 10.3174/ajnr.A3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorensen AG, Patel S, Harmath C, Bridges S, Synnott J, Sievers A, et al. Comparison of diameter and perimeter methods for tumor volume calculation. J Clin Oncol. 2001;19:551–557. doi: 10.1200/JCO.2001.19.2.551. [DOI] [PubMed] [Google Scholar]
- 48.Korn RL, Crowley JJ. Overview: progression-free survival as an endpoint in clinical trials with solid tumors. Clin Cancer Res. 2013;19:2607–2612. doi: 10.1158/1078-0432.CCR-12-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20:728–740. doi: 10.1016/S1470-2045(19)30098-1. [DOI] [PubMed] [Google Scholar]
- 50.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph. 2007;31:198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehman CD, Wellman RD, Buist DS, Kerlikowske K, Tosteson AN, Miglioretti DL. Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med. 2015;175:1828–1837. doi: 10.1001/jamainternmed.2015.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titano JJ, Badgeley M, Schefflein J, Pain M, Su A, Cai M, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med. 2018;24:1337–1341. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 53.Ambrosini RD, Wang P, O'Dell WG. Computer-aided detection of metastatic brain tumors using automated three-dimensional template matching. J Magn Reson Imaging. 2010;31:85–93. doi: 10.1002/jmri.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez-Ramírez Ú, Arana E, Moratal D. Brain metastases detection on MR by means of three-dimensional tumor-appearance template matching. J Magn Reson Imaging. 2016;44:642–652. doi: 10.1002/jmri.25207. [DOI] [PubMed] [Google Scholar]
- 55.Jun Y, Eo T, Kim T, Shin H, Hwang D, Bae SH, et al. Deeplearned 3D black-blood imaging using automatic labelling technique and 3D convolutional neural networks for detecting metastatic brain tumors. Scientific Reports. 2018;8:9450. doi: 10.1038/s41598-018-27742-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 57.Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–2197. doi: 10.1200/JCO.2007.14.8163. [DOI] [PubMed] [Google Scholar]
- 58.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J. 2013;280:5350–5370. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 59.Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27:3509–3522. doi: 10.1007/s00330-016-4653-3. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Tian Q, Wang L, Liu Y, Li B, Liang Z, et al. Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging. 2018;48:916–926. doi: 10.1002/jmri.25960. [DOI] [PubMed] [Google Scholar]
- 61.Kim M, Jung SY, Park JE, Jo Y, Park SY, Nam SJ, et al. Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur Radiol. 2020;30:2142–2151. doi: 10.1007/s00330-019-06548-3. [DOI] [PubMed] [Google Scholar]
- 62.Arita H, Kinoshita M, Kawaguchi A, Takahashi M, Narita Y, Terakawa Y, et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci Rep. 2018;8:11773. doi: 10.1038/s41598-018-30273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eichinger P, Alberts E, Delbridge C, Trebeschi S, Valentinitsch A, Bette S, et al. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep. 2017;7:13396. doi: 10.1038/s41598-017-13679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39:1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput. 2018;23:331–342. [PMC free article] [PubMed] [Google Scholar]
- 69.Liang S, Zhang R, Liang D, Song T, Ai T, Xia C, et al. Multimodal 3D DenseNet for IDH genotype prediction in gliomas. Genes (Basel) 2018;9:382. doi: 10.3390/genes9080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281:907–918. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 71.Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017;19:128–137. doi: 10.1093/neuonc/now135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park JE, Kim HS, Park SY, Nam SJ, Chun SM, Jo Y, et al. Prediction of core signaling pathway by using diffusionand perfusion-based MRI radiomics and next-generation sequencing in isocitrate dehydrogenase wild-type glioblastoma. Radiology. 2020;294:388–397. doi: 10.1148/radiol.2019190913. [DOI] [PubMed] [Google Scholar]
- 73.Rathore S, Akbari H, Rozycki M, Abdullah KG, Nasrallah MP, Binder ZA, et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci Rep. 2018;8:5087. doi: 10.1038/s41598-018-22739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 75.Ravi S, Larochelle H. Optimization as a model for few-shot learning. OpenReview.net Web site; 2016. Nov 05, [Accessed May 27, 2019]. https://openreview.net/forum?id=rJY0-Kcll¬eId=ryq49XyLg. [Google Scholar]