Abstract
CD8+ tissue-resident memory T (TRM) cells provide frontline immunity in mucosal tissues. The mechanisms regulating CD8+ TRM maintenance, heterogeneity, and protective and pathological functions are largely elusive. Here, we identify a population of CD8+ TRM cells that is maintained by major histocompatibility complex class I (MHC-I) signaling, and CD80 and CD86 costimulation after acute influenza infection. These TRM cells have both exhausted-like phenotypes and memory features and provide heterologous immunity against secondary infection. PD-L1 blockade after the resolution of primary infection promotes the rejuvenation of these exhausted-like TRM cells, restoring protective immunity at the cost of promoting postinfection inflammatory and fibrotic sequelae. Thus, PD-1 serves to limit the pathogenic capacity of exhausted-like TRM cells at the memory phase. Our data indicate that TRM cell exhaustion is the result of a tissue-specific cellular adaptation that balances fibrotic sequelae with protective immunity.
INTRODUCTION
CD8+ memory T cells offer long-term protection against pathogen reinfection. In addition to circulating central and effector memory T cells, tissue-resident memory T (TRM) cells are a recently described memory subset that mainly reside in nonlymphoid organs and offer immediate protection by coordinating local innate and adaptive immunity (1, 2). TRM cells are phenotypically and transcriptionally distinct from circulating memory T cells. CD69, which antagonizes T cell recirculation and egress from tissues (3), is a key lineage-defining marker that distinguishes TRM cells and circulating memory T cells (4). In addition to CD69, a subset of CD8+ TRM cells also express the integrin molecule CD103, which enhances the tethering of TRM cells to epithelium via E-cadherin. Tissue-derived cues, particularly transforming growth factor–β (TGF-β), are critical for TRM cell development and/or maintenance (5). Although local antigen recognition in the tissue is not absolutely required for TRM cell formation, tissue antigen reencounter by effector CD8+ T cells after their priming in the draining lymphoid organs does facilitate optimal TRM cell development (6–8). Although recent studies have suggested that TRM cells are maintained independently of T cell receptor (TCR) signaling in the skin (9), much remains to be defined regarding the mechanisms regulating the maintenance and long-term survival of TRM cells in various tissues. Conventional circulating memory CD8+ T cells are maintained in a major histocompatibility complex class I (MHC-I)–independent manner; however, whether MHC-I and/or TCR signaling contributes to TRM cell maintenance and/or function after pathogen clearance is less clear.
In contrast to circulating memory T cells, TRM cells exhibit higher levels of expression of multiple effector cytokines and cytolytic molecules including interferon-γ (IFN-γ) and tumor necrosis factor–α (TNF-α) (10). The heightened expression of these molecules confers enhanced antimicrobial activity of TRM cells (10) but could potentially cause bystander inflammation and injury in the tissue. The mechanisms balancing the protective and potentially injurious effects of TRM cells during tissue homeostasis are largely unexplored. TRM cells express T cell inhibitory molecules, including programmed cell death protein 1 (PD-1) on their surface (11). It is conceivable that the expression of these molecules may restrict the capacity of TRM cells to promote tissue pathogenesis (11, 12).
Influenza infection in mice leads to the formation of TRM cells that confer resistance to reinfection, particularly in the lungs (13, 14). After influenza virus infection in the C57BL/6 background, two major H-2Db restricted CD8+ T cell epitopes against nucleoprotein peptide 366–374 (NP366–374) or polymerase peptide 224–233 (PA224–233) are displayed (15–17). Previous studies have demonstrated that there is a marked difference in the immunodominance hierarchy between NP366–374 and PA224–233 T cells during primary and secondary CD8+ T cell responses. NP366–374 and PA224–233 epitope–specific CD8+ T cells appear in equivalent proportions during the primary CD8+ T cell responses, whereas NP366–374 T cells expand to a much greater proportion and dominate in secondary responses (16, 18–20). Antigen load, duration of antigen presentation, and differences in antigen-presenting cells (APCs) were proposed to regulate the differential responses of NP and PA epitopes (15, 21, 22). Furthermore, NP366–374 and PA224–233 TRM cells exhibited distinct molecular signatures after secondary virus challenge (23). Of note, NP antigen was detectable in the lungs weeks after infectious viral clearance (24–26). How the chronic antigen reservoir regulates the phenotype, maintenance, and function of influenza-specific TRM cells has not been explored.
Here, we report that NP366–374 TRM cells in the lung receive chronic local TCR stimulation weeks after the clearance of infectious influenza virus. Consequently, these NP366–374 TRM cells adopt both conventional memory CD8+ T cell and exhausted-like features after influenza virus infection. Unlike conventional circulating memory CD8+ T cells that are maintained in an MHC-I–independent manner, these exhausted-like TRM cells are sustained by persistent TCR–peptide/ MHC-I (pMHC-I) signaling as the depletion of H-2Db at 28 days post-infection (d.p.i.) selectively impairs the maintenance of these TRM cells. Likewise, B7-CD28 signaling blockade at the memory phase (starting at 21 d.p.i.) abrogates the persistence of exhausted-like TRM cells and consequently impairs TRM cell–mediated secondary heterologous immunity. In contrast, programmed death-ligand 1 (PD-L1) blockade at the memory phase promotes exhausted-like TRM cell expansion and rejuvenation. This augmentation of TRM cell function leads to enhanced secondary protection yet causes chronic tissue fibrotic sequelae after the resolution of the acute infection. Our data suggest that memory CD8+ T cells can adopt a tissue-specific cellular adaptation to balance fibrotic sequelae and secondary immunity.
RESULTS
Epitope-specific manifestation of an exhaustion gene signature in lung TRM cells after acute influenza infection
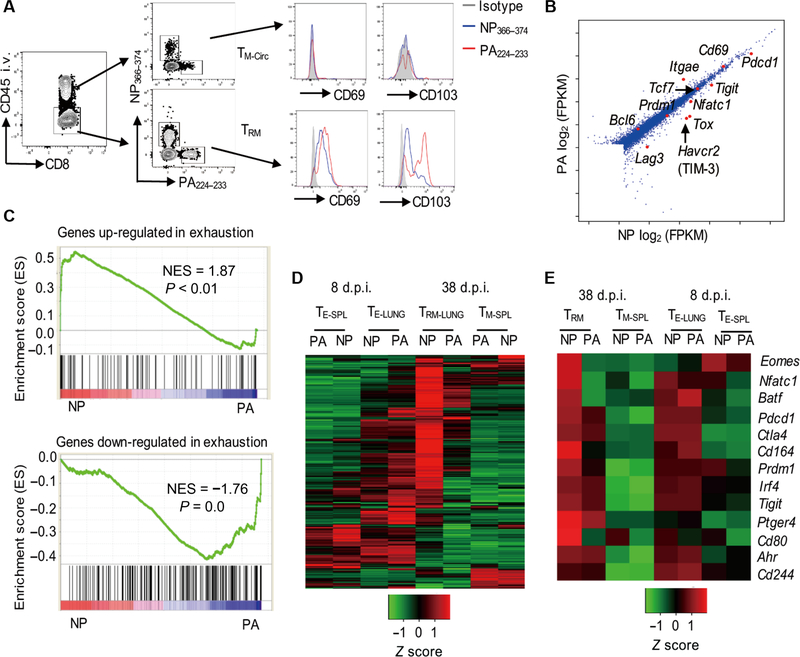
In C57BL/6 mice, influenza virus (A/PR8/34) infection is cleared from infected lungs around 10 d.p.i. (27–29). To assess the generation of TRM cells in the lung after influenza infection, we examined lung CD8+ TRM cell responses against NP366–374 and PA224–233 6 weeks after infection. We used an in vivo antibody (Ab) labeling approach, in which fluorescently coupled CD45 Ab was administered intravenously to mice 5 min before tissue harvesting to distinguish lung- circulating (intravenous Ab+) and lung-resident (intravenous Ab−) CD8+ T cells (30). Lung-resident CD8+ NP366–374 and PA224–233 TRM cells expressed comparably higher levels of CD69 than the circulating NP366–374 and PA224–233 T cells (Fig. 1A). Most of the PA224–233 TRM cells expressed CD103, but only a minor fraction of NP366–374 TRM cells was CD103+ (Fig. 1A and fig. S1, A and B). These data suggest that there may be epitope-specific heterogeneity between NP366–374 and PA224–233 TRM cells after acute influenza virus infection. To explore this idea, we performed RNA sequencing (RNA-seq) analysis of sorted lung NP366–37 and PA224–233 TRM cells 6 weeks after infection. Consistent with the surface molecule expression, NP366–374 and PA224–233 TRM cells expressed comparable CD69, but Itgae (encoding CD103) was lower in NP366–37 TRM cells (Fig. 1B). NP366–374 TRM cells showed elevated expression of a number of coinhibitory molecules—including Pdcd1, Havcr2, Tigit, and Lag3 (Fig. 1B)—and transcription factors that are typically associated with exhausted T cells generated after chronic viral infection— such as Tox, Nfatc1, and Batf (Fig. 1B) (31–33). Gene set enrichment analysis (GSEA) demonstrated that NP366–374 TRM cells were positively enriched with the genes up-regulated, and negatively enriched with the genes down- regulated, in exhausted CD8+ T cells after chronic viral infection (34), compared with PA224–233 TRM cells (Fig. 1C). To further profile the kinetics of the gene expression patterns of NP366–374 and PA224–23 CD8+ T cells, we sorted CD8+ T cells from spleens and lungs at effector (day 8) and memory (day 38) stages and performed NanoString endogenous mRNA analysis on the expression of 560 immunological genes in those effector or memory T cells without the need for amplification (Fig. 1D). We found that the immune gene expression patterns between NP366–374 and PA224–233 T cells at lung effector or splenic memory were quite similar(Fig. 1D). However, NP366–374 TRM cells and PA224–233 TRM cells had drastic differences in immune-associated gene expression patterns (Fig. 1D). Consistent with the RNA-seq data, NP366–374 TRM cells expressed higher levels of genes associated with T cell exhaustion compared with PA224–233 TRM cells (Fig. 1E). Both NP366–374 and PA224–233 lung effector cells expressed higher exhaustion-associated genes than effector T cells in spleen, a feature of effector T cell “exhaustion” or “impairment” previously described during respiratory viral infections (Fig. 1E) (35–38). Those exhausted genes were maintained or even further up-regulated in lung NP366–374 TRM cells at 38 d.p.i. (Fig. 1E). In contrast, those exhaustion-associated genes were generally down-regulated in PA224–233 TRM cells compared with day 8 effector T cells in the lungs (Fig. 1E). These observations suggest that there are distinct gene expression patterns in two epitope-specific polyclonal TRM cell populations and there exists an exhaustion-like gene pattern in a population of lung TRM cells after acute influenza infection reflective of those CD8+ T cells from chronic infections.
Fig. 1. Epitope-specific manifestation of exhaustion gene signature in lung TRM cells.
WT C57BL/6 mice were infected with influenza PR8. Spleens or lungs were harvested after intravenous (i.v.) administration of CD45 Ab at the indicated d.p.i. (A) Expression of CD69 and CD103 on lung NP366–374 or PA224–233 circulating memory (intravenous Ab+, TM-Circ) cells or TRM cells (intravenous Ab−) by flow cytometry at 40 d.p.i. (n=4). (B and C) Transcriptional profiles of NP366–374 and PA224–233 TRM cells were determined by RNA-seq at 42 d.p.i. (pooled from 16 mice). (B) Differential gene expression between NP366–374 TRM cells and PA224–233 TRM cells. FPKM, fragments per kilobase million. (C) GSEA showing positive enrichment in NP366–374 TRM cells of the genes up-regulated in exhausted CD8+ T cells (top) or showing negative enrichment in NP366–374 TRM cells of the genes down-regulated in exhausted CD8+ T cells (bottom). NES, normalized enrichment score. (D and E) Expression of immune-related genes in NP366–374 (NP) or PA224–233 (PA) lung effector (TE-LUNG) and TRM cells, and spleen effector (TE-SPL) and memory (TM-SPL) cells were determined by NanoString at 8 (effector) or 38 (memory) d.p.i. (pooled from 4 to 12 mice per group). (D) Heat map representing expression levels of 560 immune-associated genes. (E) Heat map representing expression levels of exhaustion-associated genes.
Exhausted-like NP366–374 TRM cells coexhibit exhaustion and memory features
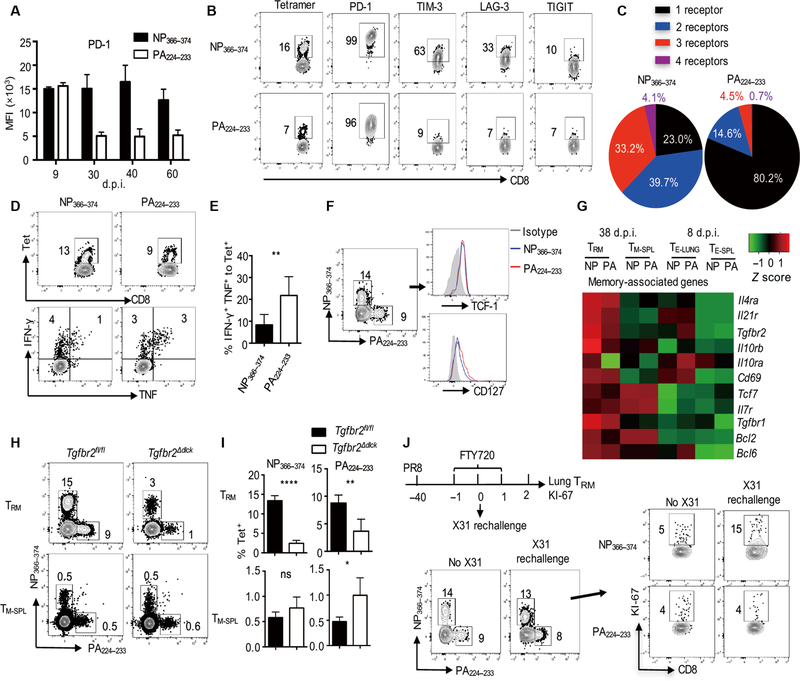
On the basis of the gene expression data, we next examined the surface expression of inhibitory molecules. We first tracked the kinetics of PD-1 expression on lung NP366–374 and PA224–233 T effector or TRM cells over time. During the effector phase, NP366–374 and PA224–233 TRM cells had similar amounts of PD-1 expression (Fig. 2A). However, lung-resident NP366–374 T cells maintained PD-1 expression that was lost on PA224–233 TRM cells at the memory stage (30 to 60 d.p.i.) (Fig. 2A). NP366–374 TRM cells also exhibited higher PD-1 expression than H2Kb-restricted T cells against the PB1 703–711 epitope (PB1703–711 TRM cells) (fig. S1C) and ovalbumin (OVA)–specific OT-I TRM cells [after infection with recombinant influenza A PR8 expressing OVA323–339 epitope (PR8-OVA)] (fig. S1D), as well as higher than their splenic or lung-circulating counterparts (fig. S1E). Thus, NP366–374 TRM cells appear to exhibit unique characteristics of high PD-1 expression at the memory stage.
Fig. 2. Exhausted-like TRM cells coexhibit exhausted and memory T cell features.
(A to G) WT C57BL/6 mice were infected with influenza PR8. Spleens and lungs were harvested after intravenous administration of CD45 Ab at the indicated d.p.i. (A) PD-1 expression levels [mean fluorescence intensity (MFI)] on intravenous Ab− lung NP366–374 or PA224–233 T cells were assessed by flow cytometry at the indicated d.p.i. (B) Expression of inhibitory receptors on lung NP366–374 or PA224–233 TRM cells was assessed by flow cytometry at 40 d.p.i. (C) Total numbers of inhibitory receptors expressed on lung NP366–374 or PA224–233 TRM cells were assessed by flow cytometry at 40 d.p.i. (D and E) IFN-γ and TNF-α production by TRM cells was assessed by flow cytometry after ex vivo stimulation with the NP366–374 or PA224–233 peptide at 40d.p.i. (D) Representative plots of tetramer (Tet), IFN-γ, and TNF-α staining in lung-resident (intravenous Ab−) CD8+ cells. (E) Frequencies of IFN-γ+ TNF-α+ cells were normalized to the frequencies of tetramer+ TRM cells in resident CD8+ cells. Percentages of IFN-γ+ TNF-α+ cells in tetramer+ TRM cells were assessed. (F) NP366–374 or PA224–233 TRM cell TCF-1 and CD127 expression was assessed by flow cytometry at 42 d.p.i. (G) Expression of CD8+ memory-associated genes in lung effector (TE-LUNG) or TRM cells, and spleen effector (TE-SPL) or memory (TM-SPL) cells was determined by NanoString at 8 or 38 d.p.i. (H and I) Tgfbr2fl/fl or Tgfbr2Δdlck mice were infected with influenza PR8. Spleens and lungs were harvested after intravenous administration of CD45 Ab at 42 d.p.i. (H) Representative flow cytometry plots. (I) Frequencies of NP366–374 or PA224–233 TRM and TM-SPL cells. (J) WT C57BL/6 mice were infected with influenza PR8, treated with FTY720 (39 to 41 d.p.i.), and then rechallenged with influenza X31 at 40 d.p.i. KI-67 expression in NP366–374 or PA224–233 TRM cells was assessed by flow cytometry before and 2 days after rechallenge. Representative of two to three experiments (n=2 to 7) except (G). Data are mean ± SD; ns, not significant. *P < 0.05, **P < 0.01, ****P < 0.0001, unpaired two-tailed t test.
We next simultaneously evaluated the expression of multiple inhibitory receptors including PD-1, T-cell immunoglobulin and mucin-domain containing-3 (TIM-3), lymphocyte-activation gene 3(LAG-3),and T cell immunoreceptor with Ig and ITIM domains (TIGIT) on TRM cells. As previously reported (4, 11), both NP366–374 and PA224–233 TRM cells were PD-1+ cells (Fig. 2B). However, NP366–374 TRM cells expressed much higher PD-1 and a large proportion of the cells simultaneously expressed two or three more coinhibitory receptors on their cell surface revealed by Boolean gating (Fig. 2, B and C, and fig. S2, A to C). Incontrast, most of the PA224–233 TRM cells only expressed PD-1 (Fig. 2C and fig. S2C). PB1703–711 TRM cells also exhibited much lower TIM-3 expression compared with NP366–374 TRM cells (fig. S2D). Thus, compared with PA224–233 or PB1703–71 TRM cells, NP366–374 TRM cells coexpressed multiple coinhibitory receptors. Similar findings were also observed in influenza X31 virus infection, although to a lesser extent than influenza PR8 infection (fig. S3, A to C). The coexpression of multiple coinhibitory receptors on NP366–374 TRM cells suggests that these cells may have features similar to exhausted CD8+ T cells observed during chronic viral infection (39). Another hallmark of exhausted CD8+ T cells is diminished production of effector cytokines, particularly TNF-α, in response to antigenic stimulation (39, 40). We therefore examined lung TRM cell cytokine production after ex vivo peptide stimulation. NP366–37 TRM cells produced less IFN-γ and TNF-α compared with PA224–23 TRM cells, particularly when normalized to antigen-specific tetramer+ cells (Fig. 2, D and E, and fig. S3D), suggesting that NP366–374 TRM cells are less sensitive to TCR stimulation. These data indicate that NP366–374 TRM cells exhibit features of exhausted CD8+ T cells.
However, NP366–374 TRM cells expressed memory CD8+ T cell markers T cell factor 1 (TCF-1) and CD127 (Fig. 2F) (41), similar to the levels found in PA224–233 TRM cells. Furthermore, we observed comparable levels of memory-associated genes between NP366–374 and PA224–23 TRM cells (Fig. 2G). TGF-β signaling has been shown to be important in the development of TRM cells in various tissues (5, 42). To address the role of TGF-β signaling in epitope-specific TRM cell development, we infected wild-type (WT) (Tgfbr2fl/fl) or dLck-cre Tgfbr2fl/fl (Tgfbr2Δdlck) mice with influenza virus. Tgfbr2 deficiency did not impair CD8+ T cell priming in the secondary lymphoid organ at the effector phase (9 d.p.i.) (fig. S4A) but resulted in decreased frequency and numbers of both NP366–374 and PA224–233 TRM cells at 6 weeks after infection (Fig. 2, H and I, and fig. S4B). Impaired expression of CD103 on NP366–374 or PA224–233 TRM cells was also observed in the absence of TGF-β signaling (fig. S4C). These data suggest that similar to “conventional” PA224–233 TRM cells, NP366–374 TRM cells also require TGF-β signaling for their formation. A key feature of memory T cells is their ability to expand upon secondary antigenic challenge. Recent reports have revealed that TRM cells are able to proliferate in situ after reinfection (43, 44). We therefore examined whether NP366–374 TRM cells could respond to and proliferate in the lung upon heterologous influenza rechallenge. We blocked lymphocyte circulation 6 weeks after infection by the injection of FTY720 at 1 day before reinfection (fig. S4D). We then infected the mice with influenza A/X31 (H3N2), which differs in the surface proteins but shares internal proteins with influenza A/PR8, and examined TRM cell activation and proliferation 2 days after rechallenge (Fig. 2J). Influenza X31 reinfection stimulated CD69 up-regulation on lung NP366–374 TRM cells, suggesting that these cells received and responded to antigenic signals invivo after secondary influenza X31 challenge (fig. S4E). There was a marked increase in TRM cell proliferation, specifically the NP366–374 TRM cell population (Fig. 2J and fig. S4, F and G). These data cor-related well with previous findings that NP366–374 memory T cells dominate over PA224–233 memory T cells in the secondary expansion and provide heterologous immunity (15, 18, 19). Collectively, our results demonstrate that NP366–374 TRM cells exhibit both exhausted and memory T cell features. We termed this population of memory cells “exhausted-like TRM cells.”
Exhausted-like TRM cells receive persistent in situ TCR stimulation
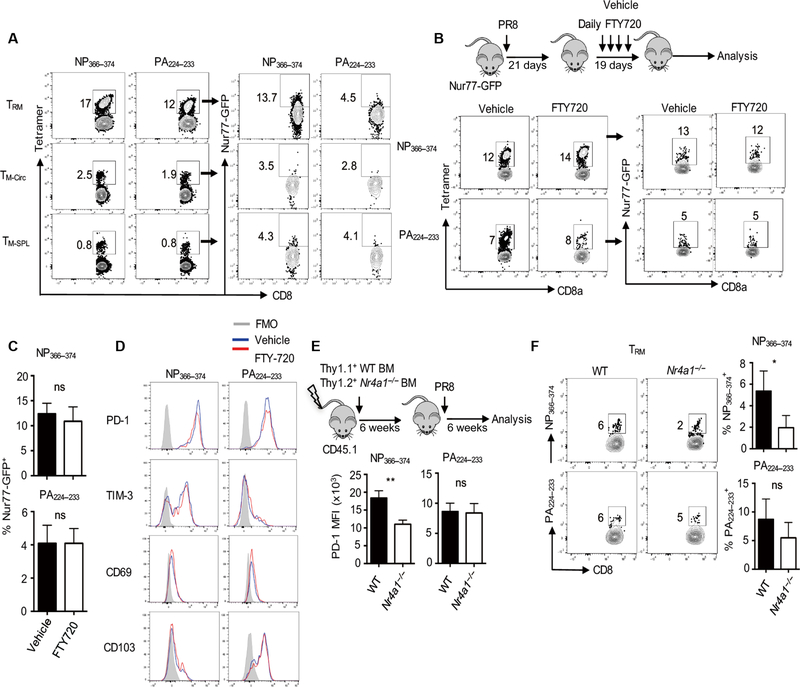
We next sought to determine the underlying mechanisms regulating the formation and maintenance of exhausted-like TRM cells. Persistent TCR signaling is involved in the development of exhausted CD8+ T cells during chronic viral infections (39). We first explored whether NP366–374 TRM cells still received TCR signaling at the memory stage. Using Nur77-GFP transgenic mice that report on the activation of TCR signaling, we examined Nur77-GFP expression in TRM cells, and circulating and splenic memory T cell populations after infection with influenza PR8. A greater proportion of exhausted-like NP366–37 TRM cells expressed Nur77-GFP compared with PA224–233 TRM cells at 6 weeks after infection (Fig. 3A). NP366–374 TRM cells had higher Nur77-GFP expression than NP366–374 memory cells in the spleen or in the lung vasculature (Fig. 3A), suggesting that NP366–374 TRM cells may receive long-term TCR stimulation in situ. This is consistent with the fact that NP protein is more abundant than PA protein during influenza infection, which may lead to NP antigen persistence in the lung long after viral clearance (15, 25).
Fig. 3. TCR signaling is required for exhausted-like TRM cell formation and maintenance.
(A) Nur77-GFP mice were infected with influenza PR8. Spleens and lungs were harvested after intravenous administration of CD45 Ab. Green fluorescent protein (GFP) expression in TRM cells, lung-circulating memory (TM-Circ, intravenous Ab+), and TM-SPL cells was assessed by flow cytometry at 40 d.p.i. (B to D) Nur77-GFP mice were infected with influenza PR8 and received vehicle or FTY720 daily starting at 21 d.p.i. Mice were euthanized after intravenous administration of CD45 Ab at 40 d.p.i. (B) Schematic of experimental design (top) and representative flow cytometry plots of Nur77-GFP expression in NP366–374 or PA224–233 TRM cells. (C) Quantification of percentages of Nur77-GFP+ cells in NP366–374 or PA224–233 TRM cells after vehicle or FTY720 treatment. (D) PD-1, TIM-3, CD69, or CD103 expression on NP366–374 or PA224–233 TRM cells after vehicle or FTY720 treatment was assessed by flow cytometry. (E and F) Thy1.1+ C57BL/6 WT and Thy1.2+ Nr4a1−/− (Nur77) mixed bone marrow (BM) chimeric mice were infected with influenza PR8. Spleens and lungs were harvested after intravenous administration of CD45 Ab at 40 d.p.i. (E) PD-1 expression on NP366–374 or PA224–233 TRM cells was determined by flow cytometry. (F) Representative plots (left) and percentages (right) of NP366–374 or PA224–233 TRM cells in Thy1.1+ WT or Thy1.2+ Nr4a1−/−-resident CD8+ T cells. Representative of two to three experiments (n = 3 to 5). Data are mean ± SD; ns, not significant. *P < 0.05, **P < 0.01, unpaired two-tailed t test.
Residual antigen presentation in the draining mediastinal lymph nodes (mLN) by migratory dendritic cells can influence the phenotype and localization of memory T cells (24, 26). It is thus possible that NP366–374 CD8+ T cells may receive sustained and stronger antigen stimulation than PA224–233 memory T cells in the mLN and then migrate into the lung to develop into exhausted-like TRM cells. We injected FTY720 to WT mice daily starting at 3 weeks after infection to block T cell migration (45) and then checked TRM cell phenotype at 40 d.p.i. (Fig. 3B). Long-term FTY720 treatment suppressed T cell migration (fig. S5A). FTY720 treatment did not alter TRM cell Nur77-GFP expression or PD-1, TIM-3, CD69, or CD103 expression (Fig. 3, C and D), suggesting that T cell migration at the memory stage is probably not required for the development of exhausted-like TRM cells. These data also indicated that local TCR signaling may be responsible for the development of exhausted-like TRM cells. To further explore this idea, we wondered whether local inoculation of PA224–233 peptide could lead to the induction of the exhausted-like phenotype in PA224–233 TRM cells. PA224–233 TRM cells acquired high levels of PD-1 expression after intranasal PA224–23 but not NP366–374 peptide administration at 35 d.p.i. (fig. S5, B and C). PA224–233 peptide stimulation also suppressed CD103 but not CD69 expression (fig. S5, D to F). These data suggest that lung in situ antigen presentation and resulting TCR signaling likely induce the development of epitope-specific exhausted-like TRM cells. Nur77-GFP+ cells had higher PD-1 expression (fig. S6A), suggesting that Nur77 (encoded by Nr4a1 gene) itself may be involved in the generation of exhausted-like TRM cells. To explore this idea, we created WT and Nr4a1−/− 1:1 mixed bone marrow chimeric mice and infected these mice with influenza PR8 (Fig. 3E). We found that Nur77 deficiency decreased PD-1 expression and frequencies of exhausted-like NP366–37 TRM cells, but not those of PA224–233 TRM cells or splenic memory T cells (Fig. 3, E and F, and fig. S6, B to D), suggesting that TCR-induced Nur77 expression is of specific relevance for the development of exhausted-like TRM cells.
Persistent MHC-I-dependent signaling drives the formation and maintenance of exhausted-like TRM cells
We next sought to determine whether the exhaustion of NP366–37 TRM cells is dependent on NP antigen dose. We used an NP mutant influenza PR8 virus, in which the asparagine at the fifth position of the NP366–374 epitope was replaced with glutamine (N370Q). The N370Q mutation prevents the loading of NP366–374 peptide to MHC-I (46). This point mutation did not markedly affect influenza viral fitness and lung pathogenesis (fig. S7A) (46). We mixed a higher dose of this “mutant” NP virus [~200 plaque-forming units (pfu) per mouse] with lower doses of the “WT” NP virus (~40 or ~10 pfu). In this case, the initial WT NP epitope/antigen amount is limited compared with a high-dose WT virus (~200 pfu) infection, but the viral-induced inflammation and disease progression are equivalent. We found that host morbidity after WT influenza PR8 virus and the mixed virus (NP mutant PR8/WT PR8) was similar (fig. S7A). However, PD-1 or TIM-3 expression was decreased on NP366–374 TRM cells from mixed virus–infected lungs, suggesting that antigen dose is a determinant of TRM cell exhaustion phenotype (fig. S7B). Influenza virus infection leads to the chronic deposition of antigen in the lung for about 2 to 3 months after infection (24, 26). Consistent with the timing of complete antigen clearance in the lung, we found that Nur77-GFP and inhibitory receptor expression on exhausted-like TRM cells were diminished at 120 d.p.i. (fig. S8).
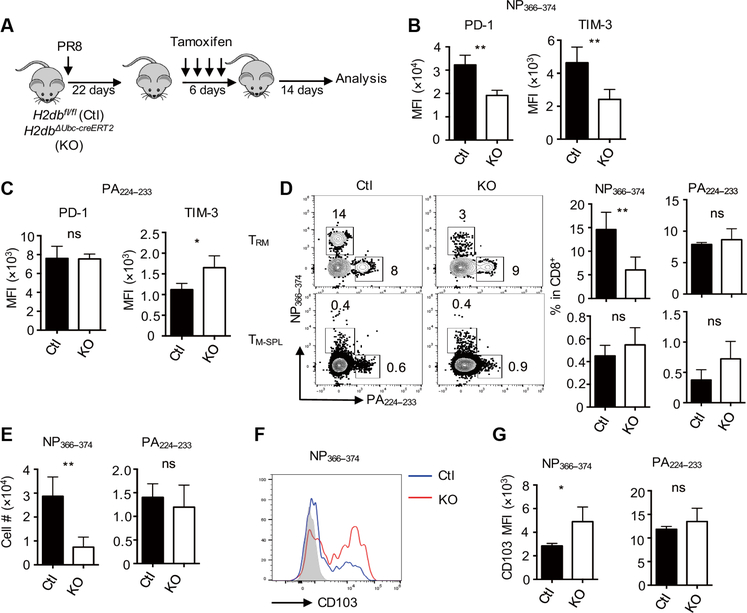
The above data suggested that TRM cell exhaustion is likely caused by persistent antigen presentation in the respiratory tract. Thus, we sought to determine whether the development of exhausted-like NP366–374 TRM cells requires persistent MHC-I–dependent signaling. We bred MHC-I–deficient mice with the transgenic mice harboring H2db floxed allele (H2dbfl/fl). We then crossed the mice with Ubc-cre ERT2 transgenic mice (H2dbΔUbc-cre ERT2, KO), allowing tamoxifen-inducible ubiquitous depletion of H-2Db molecules (fig. S9A). Tamoxifen treatment before infection inhibited the development of influenza-specific CD8+ T cell responses in H2dbΔUbc-cre ERT2 mice (fig. S9, B to D), confirming the validity of the mouse model. We then infected control or H2dbΔUbc-cre ERT2 mice with influenzaand depleted H-2Db around 4 weeks after infection (Fig. 4A). We examined TRM cell responses 2 weeks after H-2Db ablation. Tamoxifen injection caused H-2Db depletion in H2dbDUbc-cre ERT2 mice (fig. S9E). H-2Db ablation led to diminished expression of PD-1 and other coinhibitory receptors (including TIM-3 and TIGIT), specifically on exhausted-like NP366–374 TRM cells, but not on PA224–233 TRM cells (Fig. 4, B and C, and fig. S9F). H-2Db depletion decreased the frequencies and numbers of NP366–373 TRM cells but not those of PA224–233 TRM cells (Fig. 4, D and E). H-2Db ablation did not cause the loss of splenic NP366–374 or PA224–233 memory T cells (Fig. 4 D), whichis consistent with the notion that the maintenance of splenic memory T cells is MHC-I independent (47). H-2Db depletion resulted in increased CD103 expression but did not alter CD69 expression on exhausted-like TRM cells (Fig. 4, F and G, and fig. S9G).
Fig. 4. Persistent TCR–pMHC-I signaling drives the formation and maintenance of exhausted-like TRM cells.
H2dbfl/fl (Ctl) and H2dbΔUbc-creERT2 (KO) mice were infected with influenza PR8 and then treated with tamoxifen starting at 22 d.p.i. Spleens and lungs were harvested after intravenous administration of CD45 Ab at 42 d.p.i. (A) Schematic of the experimental design. (B) Average PD-1 or TIM-3 expression levels (MFI) on NP366–374 TRM cells evaluated by flow cytometry. (C) Average PD-1 or TIM-3 expression levels (MFI) on PA224–233 TRM cells evaluated by flow cytometry. (D) Representative plots and average frequencies of TRM cells (top) or TM-SPL cells (bottom) in lung-resident or splenic CD8+ T cells, respectively. (E) Cell numbers of NP366–374 or PA224–233 TRM cells. (F) Representative plot of CD103 expression on NP366–374 TRM cells. (G) Average CD103 expression levels (MFI) on NP366–374 or PA224–233 TRM cells were evaluated by flow cytometry. Representative of three experiments (n=4 mice per group). Data are mean ± SD; ns, not significant. *P < 0.05, **P < 0.01, unpaired two-tailed t test.
These data suggest that TCR–pMHC-I–dependent signaling at the memory phase is required for the development and maintenance of polyclonal exhausted-like TRM cells but not their splenic counterparts with TCRs against the same influenza epitope or local resident T cells with a TCR repertoire against the PA224–233 epitope. Consistent with this notion, NP366–374 TRM cells gradually declined over time, whereas PA224–233 TRM cells or splenic NP366–374 and PA224–233 memory cells were more stably maintained (fig. S10, A to C). To determine the consequence of exhausted-like NP366–374 TRM cell loss over time, we rechallenged the influenza PR8–infected mice with influenza X31 at 40 or 120 days after primary influenza PR8 infection in the presence of FTY720. Influenza X31 infection resulted in greater weight loss in those mice that were previously infected with influenza PR8 at 120 d.p.i. compared with those mice that were infected with influenza PR8 at 40 days prior, indicating that the loss of exhausted-like TRM cells may lead to impaired TRM cell–mediated protection against heterologous virus rechallenge (fig. S10D).
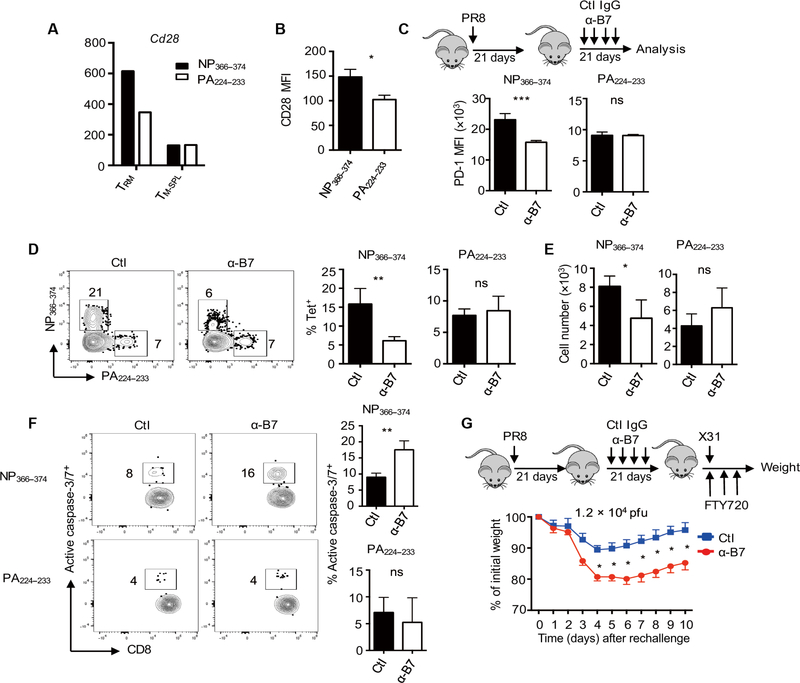
CD28 signaling is required for the maintenance of exhausted-like TRM cells
CD28 costimulation is required for naïve T cell expansion and memory T cell programming (48, 49), but its function in the maintenance of circulating or resident memory CD8+ T cells is not clear. Given that persistent pMHC-I–TCR signaling is required for the exhausted-like TRM cell maintenance, we sought to determine whether CD28 costimulation is required for the maintenance of those cells. In our NanoString gene expression data, CD28 was up-regulated in exhausted-like NP366–374 TRM cells compared with PA224–233 TRM cells (Fig. 5A). The gene expression data were confirmed by flow cytometry analysis of surface CD28 protein on NP366–374 and PA224–233 TRM cells (Fig. 5B). To determine the function of CD28 signaling in the development and/or maintenance of exhausted-like TRM cells after the resolution of primary infection, we blocked CD28-B7 interaction through the administration of anti-B7.1 (CD80) plus anti-B7.2 (CD86) (α-B7) starting at 21 d.p.i. (Fig. 5C). We then determined TRM phenotype and responses at 6 weeks after infection. Blockade of B7 signaling decreased Nur77-GFP expression in NP366–374 TRM cells (fig. S11, A and B). B7 Ab treatment also decreased PD-1 expression on NP366–374 TRM cells but not PA224–233 TRM cells (Fig. 5C). Furthermore, B7 Ab treatment markedly decreased both the frequency and cell numbers of exhausted-like NP366–374 TRM cells but not those of PA224–233 TRM cells (Fig. 5, D and E). Blockade of CTLA-4, the other receptor for B7.1 and B7.2, did not impair NP366–374 TRM cell maintenance (fig. S11D), suggesting that the blockade of CD28 signaling resulting from a-B7 treatment is responsible for the maintenance of exhausted-like NP366–374 TRM cells. Mechanistically, B7 blockade decreased NP366–374 TRM cell survival and proliferation as reflected by enhanced active caspase-3/7 activity and decreased KI-67 staining, respectively, after a-B7 treatment (Fig. 5F and fig. S11E). These data suggest that persistent CD28 signaling at the memory stage is required for the maintenance of exhausted-like TRM cells.
Fig. 5. B7 blockade diminishes exhausted-like TRM cell maintenance.
(A) WT mice were infected with influenza PR8. CD28 gene expression in NP366–374 or PA224–233 TRM cells or spleen memory T cells was determined by NanoString at 38 d.p.i. (B) WT mice were infected with influenza PR8. CD28 expression levels (MFI) on TRM cells were assessed by flow cytometry at 42 d.p.i. (C to F) WT mice were infected with influenza PR8 and B7 costimulation was blocked through the administration of α-B7.1 plus α-B7.2 at 21 d.p.i. Spleens or lungs were harvested after intravenous administration of CD45 Ab at 42 d.p.i. (C) Schematic of experimental design and PD-1 expression levels (MFI) on TRM cells. (D) Frequencies of NP366–374 or PA224–233 TRM cells in total lung-resident (intravenous Ab−) CD8+ T cells. (E) Cell numbers of NP366–374 or PA224–233 TRM cells. (F) Representative plots (left) and frequencies (right) of active caspase-3/7+ cells in NP366–374 or PA224–233 TRM cells. (G) WT mice were infected with influenza PR8 with or without B7 blockade at 21 d.p.i. and then rechallenged with X31 (1.2 × 104 pfu) at 42 d.p.i. in the presence of FTY720. Percentages of original weight after rechallenge were assessed daily. Representative of two to three experiments except (A) (n = 3 to 4 mice per group). Data are mean ± SD; ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed t test.
To explore whether impaired maintenance of exhausted-like TRM cells could decrease host resistance to heterologous immunity, we infected WT mice with influenza PR8 and then treated the mice with α-B7 as above. We injected the mice with FTY720 and subsequently rechallenged them with influenza X31 virus (1.2 × 104 pfu) at 6 weeks after infection (Fig. 5G). Mice receiving B7 blockade lost significantly more weight compared with control mice after influenza X31 infection (Fig. 5G). These data indicate that B7-CD28 costimulation is essential for the maintenance of exhausted-like TRM cells, which provide protective immunity against heterologous reinfection.
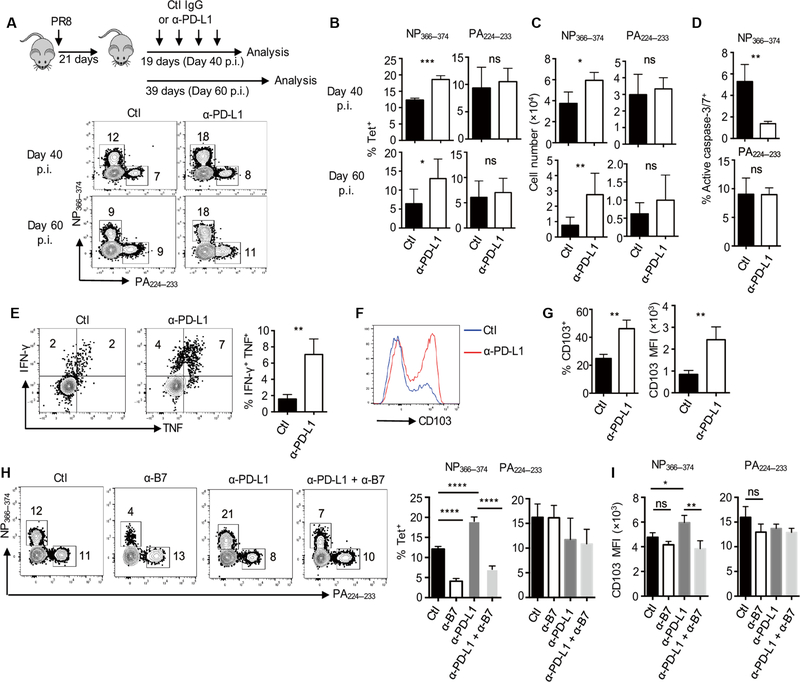
PD-L1 blockade promotes rejuvenation of exhausted-like TRM cells
Blockade of PD-1 and PD-L1 interaction promoted exhausted cell expansion and rejuvenation during chronic viral infection (40). We wondered whether the inhibition of PD-L1–PD-1 interaction could rejuvenate exhausted-like TRM cells after the resolution of acute influenza virus infection. We infected the WT mice with influenza PR8 and treated the mice with a-PD-L1 from 21 to 37 d.p.i. (Fig. 6A). We then evaluated TRM cell responses at 40 or 60 d.p.i. PD-L1 blockade increased both the frequency and cell numbers of exhausted-like NP366–37 TRM cells but not PA224–233 TRM cells or splenic NP366–374 and PA224–233 memory T cells at 40 or 60 d.p.i. (Fig. 6, A to C, and fig. S12, A to C). Consistent with the enhanced maintenance of NP366–374 TRM cells, α-PD-L1 blockade enhanced their survival (Fig. 6D and fig. S12, D and E). In addition to treating the mice with α-PD-L1 starting at 21 d.p.i., we performed additional experiments by injecting α-PD-L1 to the infected mice starting at 42 d.p.i. and then assessed TRM cell responses at 70 d.p.i. The blockade of PD-L1–PD-1 interaction in this setting also increased the percentages and cell numbers of NP366–374 TRM cells but not PA224–233 TRM cells in the lung (fig. S12, F to H), suggesting that PD-L1 signaling inhibits the magnitude of NP366–374 TRM cell responses at the memory stage.
Fig. 6. PD-L1 blockade rejuvenates exhausted-like TRM cells.
(A to G) WT mice were infected with influenza PR8 and received control IgG (Ctl) or a-PD-L1 from 21 to 37 d.p.i. Spleens and lungs were harvested after intravenous administration of CD45 Ab at the indicated d.p.i. (A) Experimental design and representative plots of NP366–37 or PA224–233 TRM cells at 40 or 60 d.p.i. (B) Frequencies of NP366–374 or PA224–233 TRM cells in total resident (intravenous Ab−) CD8+ T cells at 40 or 60 d.p.i. (C) Cell numbers of NP366–374 or PA224–233 TRM cells at 40 or 60 d.p.i. (D) Frequencies of active caspase-3/7+ cells in NP366–374 TRM cells at 35 d.p.i. (E) IFN-γ and TNF-α production by TRM cells was determined after ex vivo NP366–374 peptide stimulation at 40 d.p.i. Left panel, representative plots. Right panel, average frequencies of IFN-γ and TNF-α double-positive cells with or without PD-L1 blockade. (F and G) CD103 expression on NP366–374 lung TRM cells was determined at 40 d.p.i. Representative plots (F) and frequenciesof CD103+ cells (G, left) or CD103 expression levels (MFI) (G, right) in NP366–374 TRM cells. (H and I) WT mice were infected with influenza PR8 and received control Ab, α-PD-L1, and/or α-B7 as indicated starting at 21 d.p.i. Lungs were harvested after intravenous administration of CD45 Ab at 40 d.p.i. (H) Representative plots and percentages of TRM cells in lung-resident (intravenous Ab−) CD8+ T cells. (I) CD103 expression levels (MFI) on TRM cells were determined by flow cytometry. Representative of two to four experiments (n = 3 to 6). Mean ± SD; ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 unpaired two-tailed t test or one-way analysis of variance (ANOVA) with Tukey multiple comparison test.
We next examined whether PD-L1 blockade affected TRM cell cytokine production. We found that PD-L1 blockade increased IFN-γ and TNF-α production by NP366–374 TRM cells (Fig. 6E and fig. S13, A to E) but had no effects on the IFN-γ and TNF-α production of PA224–233 TRM cells (fig. S13, F to K). This was true even when we normalized the percentages of IFN-γ and/or TNF-α production to antigen-specific tetramer+ cell numbers (Fig. 6E). Thus, PD-L1 blockade promoted exhausted-like TRM cell rejuvenation. In addition, PD-L1 blockade led to increased percentages of CD103+ cells and greater per-cell CD103 expression levels in exhausted-like TRM cells (Fig. 6, F and G) but did not affect CD103 expression on PA224–233 TRM cells (fig. S14, A and B). PD-L1 blockade did not alter CD69 expression on either NP366–374 or PA224–233 TRM cells (fig. S14C).
Because CD28 is required for the long-term maintenance of exhausted-like TRM cells and CD28 signaling was recently shown as a major target of PD-1 blockade (50, 51), we investigated whether the rejuvenation of exhausted-like TRM cells by PD-L1 blockade at the memory phase was dependent on CD28 signaling. We infected WT mice with influenza PR8 and then administered control Ab, α-PD-L1, α-B7, or α-PD-L1 and α-B7 starting at 21 d.p.i. Co-blockade of CD28 signaling abrogated the effects of PD-L1 blockade on exhausted-like NP366–374 TRM cell maintenance (Fig. 6H and fig. S15). Similarly, B7 blockade decreased CD103 levels on exhausted-like TRM cells after a-PD-L1 blockade (Fig. 6I). These data suggest that CD28 signaling is important for the effects of PD-L1 blockade on the maintenance and rejuvenation of exhausted-like TRM cells.
Exhausted-like TRM cells balance protective immunity and fibrotic sequelae
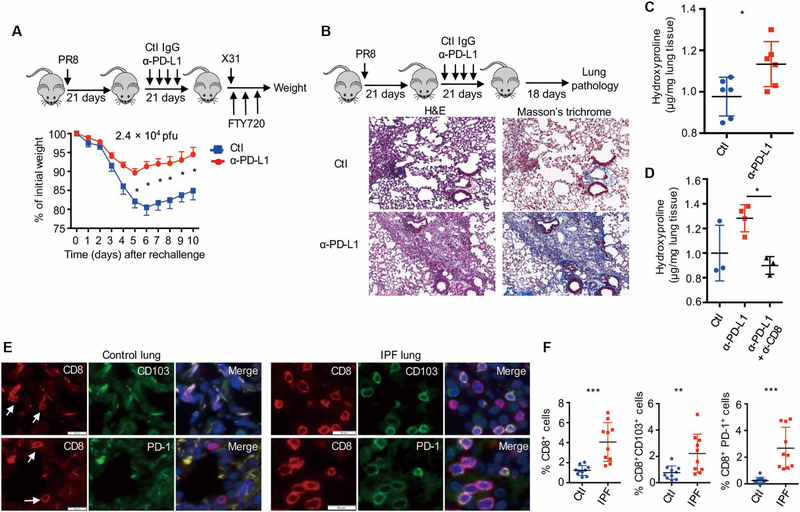
Because exhausted-like NP366–374 TRM cells were important in TRM cell–mediated heterologous immunity and PD-L1 blockade at the memory phase increased cell numbers and function of these cells, we examined whether PD-L1 blockade increased TRM cell–mediated immunity to influenza reinfection. We infected WT mice with influenza PR8 and blocked PD-L1 starting at 21 d.p.i. We then treated the mice with FTY720 and subsequently challenged the mice with a high dose of influenza X31 (2.4 × 104 pfu) at 6 weeks after infection. Mice with PD-L1 blockade lost significantly less weight than mice receiving immunoglobulin G control Ab (Fig. 7A). Similar results were obtained with mice receiving primary influenza X31 infection and PD-L1 blockade followed by secondary influenza PR8 infection (fig. S16A). These data suggest that the rejuvenation of exhausted-like TRM cells enhances TRM cell–mediated heterologous protection against influenza reinfection.
Fig. 7. TRM cell exhaustion balances protective immunity and fibrotic sequelae.
(A) WT mice were infected with influenza PR8 and received control IgG (Ctl) or α-PD-L1 from 21 to 37 d.p.i. Mice were rechallenged with influenza X31 (2.4 × 104 pfu) in the presence of FTY720 at 42 d.p.i. Percentages of original weight were determined daily after rechallenge. (B to D) WT mice were infected with influenza PR8 and received control IgG (Ctl) or α-PD-L1 from 21 to 37 d.p.i. Lung pathology and hydroxyproline levels were determined at 60 d.p.i. (B) Hematoxylin and eosin (H&E) and Masson’s trichrome C staining of lung sections. (C) Hydroxyproline levels (micrograms per milligram of lung tissue) of the lungs. (D) Hydroxyproline levels of the lungs from mice received control Ab, α-PD-L1, or α-PD-L1 plus α-CD8 (CD8 depletion). (E and F) CD8, PD-1, and CD103 staining was performed on lung sections from control (n = 10) or patients with IPF (n = 10). (E) Representative of CD8, PD-1, and CD103 staining. Blue, DAPI (4',6-diamidino-2-phenylindole). (F) Frequencies of CD8+ cells, CD8+ CD103+, or CD8+ PD-1+ cells in DAPI+ cells of control or IPF lungs. (A to D) Representative of two to five experiments (n = 3 to 6). Mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, unpaired two-tailed t test.
TRM cells in the lung undergo enhanced cell death and wane over time after influenza infection (52, 53). This transient nature of influenza-induced lung TRM cells was proposed as a protective mechanism intended to prevent pathology, but this has not been proven (54). Because PD-L1 blockade promoted rejuvenation and maintenance in a population of TRM cells, we explored whether this could promote pathologic responses in the lung. PD-L1 blockade from 21 to 42 d.p.i. increased tissue damage and inflammation at 60 d.p.i. (Fig. 7B and fig. S16B). PD-L1 blockade resulted in enhanced lung collagen deposition as revealed by Masson’s trichrome staining and hydroxyproline assay (surrogate measure of lung collagen content) (Fig. 7, B and C). The enhanced fibrotic sequelae after PD-L1 blockade were observed at 90 d.p.i. (fig. S16, C and D), indicating that PD-L1 blockade may cause tissue injury and persistent fibrotic sequelae after the resolution of acute influenza infection. The increased injurious and fibrotic responses after PD-L1 blockade were CD8+ T cell dependent as CD8+ T cell depletion abrogated the effects of PD-L1 blockade (Fig. 7D and fig. S17). These data suggest that TRM cell expansion and/or rejuvenation potentially contributed to pulmonary pathology and fibrosis.
To further explore this idea in human fibrotic lung disease, we stained the lungs from control or patients with idiopathic pulmonary fibrosis (IPF) with CD8, PD-1, and CD103. IPF lungs had increased tissue CD8+ T cells and CD8+ T cells coexpressing PD-1 or CD103 (Fig. 7, E and F), suggesting that patients with IPF have an enhanced TRM cell presence. Consistent with the notion, ITGAE (CD103) and PDCD1 (PD-1) genes were significantly up-regulated in the lungs of patients with interstitial lung diseases (ILDs, mostly IPF) in a large publicly available dataset (fig. S18). These data indicate that CD8+ TRM cells may be involved in the development and/or progress of human lung fibrosis. In summary, our data suggested that although exhausted-like properties render NP366–374 TRM cells less protective against reinfection, this TRM cell exhaustion prevents the host from developing overt injury resulting in fibrotic sequelae after acute influenza virus infection.
DISCUSSION
Memory CD8+ T cells are heterogeneous and display subsets of diversity with respect to their trafficking, metabolism, epigenetic regulation, and longevity (55). Here, we have described two types of TRM cell populations that arise from acute influenza virus infection. The conventional PA224–233 TRM cells are relatively long-lived and maintained in a TCR–pMHC-I–independent manner. An exhausted-like NP366–374 TRM cell population that exhibit both exhausted and memory features also develop from influenza virus infection. They are phenotypically exhausted but retain responsiveness in situ to secondary infections and also afford protection against heterotypic viruses with conserved CD8+ T cell epitopes. Compared with conventional TRM cells (for example, PA224–233 TRM cells), the maintenance of these exhausted-like TRM cells is dependent on persistent TCR–pMHC-I and CD28-B7 costimulatory signals. These TRM cells have pathogenic and fibrogenic potential if their activities are unchecked after the release of PD-1–imposed suppression. These data indicate that polyclonal CD8+ memory T cells of different specificities may exhibit distinct transcriptional, phenotypic, and functional differences in the same tissue although they derive from the same infection. This type of epitope-specific variation and regulation of memory cells was observed previously in lymphoid organs (15).
The development of those exhausted-like features in NP366–374 TRM cells is likely due to the active adaptation of those cells to the local antigen containing lung environment even weeks after the clearance of the infectious virus. It is well documented that acute influenza virus infection leads to persistent influenza antigen deposition in the lung and continuous antigen presentation is observed in the draining lymph nodes executed via migratory lung dendritic cells (24, 26). Studies have suggested that the continuous antigen presentation may affect the quantity and quality of CD8+ memory T cells in secondary lymphoid organs (26). It is possible that exhausted-like NP366–374 TRM cells migrate continuously from the draining LN where they receive antigenic signaling. However, evidence presented here supports the idea that local antigen presentation in the lung, rather than in the draining lymph nodes, may be responsible for the development of exhausted-like TRM cells. First, compared with the resident NP366–374 TRM cells, NP366–374 cells in the lung vasculature were Nur77-GFP negative and express lower levels of PD-1, suggesting that exhausted-like NP366–374 TRM cells receive antigen presentation and CD28 signaling after the establishment of residence in the lung. Second, long-term blockade of T cell circulation via FTY720 treatment does not affect Nur77, PD-1, or other coinhibitory receptor expression on NP366–374 TRM cells, suggesting that T cell circulation at the memory stage is not required for the development of exhausted-like TRM cells. The fact that exhausted-like phenotypes are only observed in NP366–374 but not in PA224–233 TRM cells is probably due to the antigen dose and/or the duration of antigen presentation. Influenza virus contains a higher number of NP molecules (560 molecules per virion) than PA molecules (8 molecules per virion) (15). At later times after infection, when virally infected cells are cleared and the majority of antigen is in the form of cellular debris and neutralized virions, NP proteins would be present in greater amounts than PA (15). Therefore, the probability of processing and presentation of NP by lung APCs at the memory stage would be much higher than PA. Nevertheless, future studies are warranted to distinguish the roles of lung local antigen presentation versus draining LN peptide presentation in driving the development of exhausted-like TRM cells, to identify the exact cell types maintaining the residual antigen, and to determine the precise APC populations required for the maintenance of exhausted-like TRM cells.
The pathogenic cascade of lung fibrosis is thought to be initiated by repetitive microinjuries to the alveolar epithelium (56). Because of the enhanced expression of effector cytokines and cytotoxic molecules, TRM cells could certainly be capable of initiating and/or amplifying epithelial injury if triggered by antigenic and/or rejuvenating signals such as PD-1 blockade. Whether TRM cell activation is involved in the development and/or exacerbations of human pulmonary fibrosis requires future studies. Influenza and other viral infections have been associated with both the development and exacerbation of established pulmonary fibrosis (57–60). It is tempting to speculate that the activation and/or rejuvenation of preexisting viral-specific exhausted-like TRM cells may contribute to the development of the lung injury and pathology. These ideas are supported by a recent report that PD-1–mediated inactivation of human CD8+ T cells alleviates lung fibrosis development in a humanized mouse model of bleomycin-induced pulmonary fibrosis (61). The degree of the pulmonary fibrosis observed in the influenza-infected mice after PD-L1 blockade is much milder than the acute lung fibrosis that develops after bleomycin inoculation. Whether these fibrotic sequelae could result in significant changes in lung function requires further studies.
Some limitations of this report are worth noting. In our study, we have only assessed TRM cell phenotypes for a limited number of CD8+ T cell epitopes after influenza infection, and it is possible that there is an even broader spectrum of TRM phenotypes. Furthermore, only respiratory mucosal tissue was examined in this study. Whether exhaustion-like TRM cells are present in the lungs or other mucosal tissues after infections with distinct pathogens warrants further studies. Nevertheless, TRM cell exhaustion observed in our study is likely an active adaptation of the cells delicately maintaining tissue immune memory while simultaneously preventing the development of excessive fibrotic sequelae. Potential activation of lung TRM cells may result in amplified inflammatory, injurious, and fibrotic signals contributing to the development and/or exacerbation of preexisting fibrotic respiratory diseases. It is thus important to identify strategies to specifically promote the protective function while, if possible, restraining the pathological function of TRM cells during vaccination and/or immunotherapies.
MATERIALS AND METHODS
Mouse and infection
WT C57BL/6, dLck-cre, Tgfbr2fl/fl, CD90.1 (Thy1.1), CD45.1, Ubc-creERT2, OT-I, and Nr4a1−/− mice were originally purchased from the Jackson Laboratory and bred in-house. Tgfbr2Δdlck mice were generated by breeding dLck-cre to Tgfbr2fl/fl mice. Control mice for Tgfbr2Δdlck mice are littermates without dLck-cre transgene (Tgfbr2fl/fl). To generate H2dbfl/fl transgenic mice, LoxP sites were inserted into a Db transgene. The transgene was introduced to C57BL/6 mice by the Mayo Clinic Transgenic Mouse Core (Rochester, MN). These animals were then backcrossed onto an MHC-I–deficient background until they lacked endogenous MHC-I. To generate H2dbΔUbc-creERT2 mice, MHC-I–deficient H2dbfl/fl transgenic mice were bred to Ubc-creERT2 transgenic mice to generate Ubc-creERT2/H2dbfl/fl double transgenic mice on an MHC-I background (H2dbΔUbc-creERT2) (fig. S4A). Nur77-GFP reporter mice were a gift from G. Rajagopalan (Mayo Clinic) and bred in-house. All animal experiments were performed in animal housing facilities at the Indiana University School of Medicine (IUSM, Indianapolis, IN) or the Mayo Clinic (Rochester, MN). Sex-matched and age-matched 9- to 12-week-old mice of both sexes were used in the experiments. All animal experiments were approved by the IUSM or the Mayo Clinic Institutional Animal Care and Use Committees. Influenza A/PR8/34 virus harboring point mutation that abrogates the binding of MHC-I to the NP366–374 peptide (PR8-NP mutant) was generated as described before (46). For influenza virus infection, influenza WT PR8 virus (~200 pfu per mouse unless stated in the text in the primary infection and ~1 × 104 pfu per mouse in the secondary infection), PR8-NP mutant virus (~200 pfu per mouse), PR8 expressing ovalbumin peptide (PR8-OVA) (~1200 pfu per mouse), or X31 (~800 pfu per mouse in the primary infection and ~1.2 × 104 or ~2.4 × 104 pfu per mouse in the secondary infection as indicated in the text) was diluted in fetal bovine serum (FBS)–free Dulbecco’s modified Eagle’s medium (Corning) on ice and inoculated in anesthetized mice through intranasal route as described before (28).
Human lung tissue sections
Archived human surgical lung biopsy specimens from individuals diagnosed with usual interstitial pneumonia (UIP) on biopsy (clinically IPF; n = 10) were obtained from the Mayo Clinic tissue bank. The diagnosis of UIP/IPF was based on standard criteria (62). Control lung tissue was obtained from surgical biopsy specimens acquired from patients undergoing lung biopsy for benign indications: primarily benign lung nodules. The control lung tissue samples used were from the lung tissue adjacent to the resected lung nodule. The control subjects (n = 10) did not have any evidence of interstitial lung diseases. The use of human lung tissue in this study was approved by the Mayo Clinic Institutional Review Board committee (protocol number 18–004030).
Immunofluorescence staining
Staining for two groups of combination of either CD8/CD103 or CD8/PD-1 was performed on formalin-fixed paraffin-embedded (FFPE) lung tissue slides. FFPE slides were deparaffinized in CitriSolv for 30 min and then immersed in alcohol series from 100, 95, 85, and 75% to distill H2O for 5 min each for tissue hydration. For antigen retrieval, hydrated slides were steamed for 20 min in 1 mM EDTA. The slides were then blocked with 10% normal goat serum phosphate-buffered saline (PBS) for 30 min at room temperature (RT) and then were incubated with either rabbit anti-CD103 (Abcam) or rabbit anti–PD-1 (Cell Signaling) overnight at 4°C.8After rinsing in 0.1% PBST (PBS with Tween 20) solution, the slides were incubated with Alexa Fluor 488–conjugated goat anti-rabbit secondary Ab (Life Technologies). After rinsing with 0.1% PBST, the slides were then incubated with Alexa Fluor 647–conjugated mouse anti- CD8 (BioLegend) for 60 min at RT. After stringent washing in 0.1% PBST, slides were aired before mounting with 4’,6-diamidino-2- phenylindole for nuclei counterstain. Tissue staining for the Ab mixture was reviewed and representative images were captured in Olympus cellSens Dimension system. Fifteen representative image fields were captured for each patient for quantification purposes.
RNA-seq and data analysis
Total RNA was isolated from sorted pooled NP366–374 or PA224–233 TRM population from 16 mice (Qiagen). High-quality total RNA was used to generate the RNA-seq library. cDNA synthesis, end-repair, A-base addition, and ligation of the Illumina indexed adapters were performed according to the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA). The concentration and size distribution of the completed libraries were determined using an Agilent Bioanalyzer DNA 1000 chip (Santa Clara, CA) and Qubit fluorometry (Invitrogen, Carlsbad, CA). Paired-end libraries were sequenced on an Illumina HiSeq 4000 following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit. Base calling was performed using Illumina’s Real-Time Analysis (RTA) software (version 2.5.2). Paired-end RNA-seq reads were aligned to the mouse reference genome (GRCm38/mm10) using RNA-seq spliced read mapper Tophat2 (v2.1.1) (63). Pre- and post-alignment quality controls, gene level raw read count, and normalized read count (i.e., FPKM) were performed using RSeQC package (v2.3.6) with the NCBI mouse RefSeq gene model (64). We further calculated the logFC (fold change) by dividing FPKMNP/FPKMPA with additional restrictions on FPKM values as min (FPKMNP, FPKMPA) > 0 and max (FPKMNP, FPKMPA) > 5, and genes were sorted by logFC for GSEA analysis (http://www.broadinstitute.org/gsea/). Gene lists of up-regulated and down-regulated genes in exhausted CD8+ T cells during chronic viral infection (compared with memory CD8+ T cells in acute viral infection) are adapted from GSE9650 (34). RNA-seq data were deposited in Gene Expression Omnibus (GEO) database (GEO number: GSE115786).
Hydroxyproline assay
Lung tissue was hydrolyzed in 1 ml of 6 M HCl at 95°C overnight. The hydrolysate was cooled down to RT and centrifuged for 10 min at 13,000g. The black particles on the surface of the hydrolysate were removed by vacuum sucking. Two and a half microliters of each sample was added to an indicated well of assay plate. Hydroxyproline standard solution was purchased from Sigma-Aldrich. Standard solution (1 mg/ml) was diluted 10 times, and 0, 2, 4, 6, 8, and 10 ml of diluted standard solution were added into different wells in the assay plate for generation of standard curve. Then, the assay plate was placed in a 60°C oven to dry samples. One hundred microliters of freshly made chloramine-T solution (2.0 ml of n-propanol, 0.282 g of chloramine-T, and 2.0 ml of H2O in 20 ml of citrate acetate buffer) was added into each well, and the mixture was incubated at RT for 20 min. Then, 100 ml of fresh Ehrlich solution (4.5 g of 4-dimethylaminobenzaldehyde in 18.6 ml of n-propanol and 7.8 ml of perchloric acid) was added into each well. After that, the assay plate was placed at a 60°C oven for 60 min before reading at 560-nm wavelength in a Thermax plate reader.
Intravascular CD8+ T cell labeling
Mice were injected intravenously with 1.5 mg of anti-CD45 diluted in 200 μl of sterile PBS as previously described (30). Mice were euthanized, and tissues were collected 5 min after injection of the intravenous Ab. Tissues were dissociated in 37°C for 30 min with gentleMACS (Miltenyi Biotec). Lung-circulating CD8+ are defined by intravenous Ab+, and lung-resident CD8+ are defined by intravenous Ab−.
OTI cell transfer
One million splenocytes from Thy1.1+ OTI mice were transferred into WT (Thy1.2+) congenic mice. Then, the mice were infected with PR8-OVA virus 24 hours later. At 6 weeks after infection, mice were injected with intravenous CD8• Ab, and lungs were collected for TRM analysis. TRM phenotype of transferred OTI cells and endogenous NP366–374 or PA224–233 populations were based on Thy1.1 and Thy1.2 staining separation.
Flow cytometry analysis
Fluorescence-activated cell sorting (FACS) Abs were primarily purchased from BioLegend, BD Biosciences, or eBioscience. H-2Db Ab was purchased from Accurate Chem. The clone numbers of those Abs are as follows: CD8α(53–6.7), CD8β(YTS156.7.7), CD45(30-F11), CD45.1(A20), CD90.1(OX-7), CD90.2(53–2.1), PD-1(29F.1A12), TIM-3 (RMT3–23), Lag-3(C9B7W), TIGIT(1G9), CD103(2E7), CD69(H1.2F3), CD49a(TS2/7), CD127(A7R34), TCF-1(7F11A10), IFN-γ(XMG1.2), TNF(MP6-XT22), KI-67(SolA15), and H-2Db(B22–249.R1). The dilution of surface staining Abs was 1:200, and dilution of intracellular staining Abs was 1:100. H-2Db-NP366–374 and H-2Db-PA224–2 tetramers were from the National Institutes of Health tetramer facility. After Ab staining, cells were acquired through an 11-color Attune NxT system (Life Technologies). Data were then analyzed by FlowJo software (Tree Star).
Intracelluar staining
Cell suspensions were stained with the indicated surface marker, and staining was performed at 4°C for 30 min. Cells were washed twice with FACS buffer (PBS, 2 mM EDTA, 2% FBS, and 0.09% sodium azide) before fixation and permeabilization with either Perm Fix and Perm Wash (BD Bioscience, for cytokine staining) or the Foxp3 transcription factor staining buffer set (eBioscience, for KI-67 and TCF-1 staining) for 1 hour at RT in the dark. Cells were washed twice with perm wash (BD Bioscience or eBioscience); stained with Abs against TCF-1, KI-67, IFN-γ, and TNF for at least 30 min at RT; and washed twice with perm wash before flow cytometry acquisition (29).
Apoptotic cell detection
CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit (Life Technologies) was used to detect active caspase activity inside the cells. Lung cells were incubated with CellEvent Caspase-3/7 green detection reagent for 25 min at 37°C as described in the manual. Annexin V Apoptosis Detection Kit (BioLegend) was used to detect phosphatidylserine on apoptotic cell surface. Lung cells were stained with annexin V and 7-aminoactinomycin D (7-AAD) for 15 min at RT according to the manual.
Tamoxifen treatment
To induce gene recombination in H2dbΔUbc-creERT2 mice, tamoxifen (Sigma-Aldrich) was dissolved in warm sunflower oil (Sigma-Aldrich) and administered via daily intraperitoneal injection for six consecutive times. Each application was 2 mg per mouse at a concentration of 20 mg/ml.
FTY720 treatment
For the influenza rechallenge experiment, mice were treated daily with FTY720 (1 mg/kg) by intraperitoneal injection starting at 1 day before rechallenge. For the chronic blockade of memory T cell migration, mice were treated daily with FTY720 (1 mg/kg) starting at 21 d.p.i. until 40 d.p.i., when mice were euthanized for TRM analysis.
Ab depletion and blockade in vivo
Anti-CD8, anti–PD-L1, anti-B7.1, anti-B7.2, and control Abs were purchased from Bio X Cell. For PD-L1 or CTLA-4 blockade experiments, WT B6 mice were infected with influenza and received intraperitoneal injection of control or blocking Abs at a dose of 500 μg per mouse for the first time at 21 d.p.i. Mice then received intraperitoneal injection of Abs every 4 days (250 mg per mouse) thereafter. For B7 blockade experiments, WT B6 mice were infected with influenza and received anti-B7.1 (200 μg per mouse) and anti-B7.2 (200 μg per mouse) treatment every 3 days starting at 21 d.p.i. or as stated in the text. For CD8 T cell depletion, mice received intraperitoneal injection of 400 μg per mouse once a week starting at 21 d.p.i.
Bone marrow chimera
To generate WT and Nr4a1−/− mixed bone marrow chimera, we injected CD45.1 mice with Busulfan (Sigma) at 100 mg/kg for four consecutive days. Mice were then reconstituted with Thy1.1+ WT bone marrow cells mixed with Thy1.2+ Nr4a1−/− bone marrow cells (1:1 ratio). Mice were rested for 6 weeks before infection with influenza. At 6 weeks after infection, mice were euthanized for the analysis of TRM. WT CD8+ T cells are identified as CD8+CD45.1−CD45.2+Thy1.1+ Thy1.2−, and Nr4a1−/− CD8+ T cells are identified as CD8+CD45.1− CD45.2+Thy1.1−Thy1.2+.
Peptide restimulation in vitro
Lung tissues were dissociated with gentleMACS (Miltenyi). Cell suspensions were restimulated with NP366–374 or PA224–233 peptide (100 ng/ml) (AnaSpec) for 5 hours in the presence of GolgiStop (BD Biosciences) (65). After restimulation, cells were first stained with surface markers and then were fixed and permeabilized using Perm Fix/Wash kits as described in the protocol.
Peptide inoculation in vivo
WT mice were infected with influenza PR8. At 35 d.p.i., mice were intranasally inoculated with PBS, PA224–233 peptide (10 μg per mouse), or NP366–374 peptide (10 μg per mouse) (AnaSpec) dissolved in PBS. At 37 d.p.i., mice were injected with intravenous CD8β Ab, and lungs were collected for TRM phenotype analysis.
Lung histopathology
After euthanasia, mice were perfused with PBS (10 ml) via the right ventricle. Paraformaldehyde (10%) (PF) was then gently instilled into the lung and left inflated for 1 min before excising and moving the lobe to 10% PF for 48 hours followed by transfer to ethanol (70%). Samples were shipped to the Mayo Clinic Histology Core Lab (Scottsdale, AZ) where they were embedded in paraffin, and 5-mm sections were cut for hematoxylin and eosin and Masson’s trichrome stain.Slideswere then digitallyscannedbytheMayo ClinicPathology Research Core (Rochester, MN) at 400× resolution with the Aperio system (Leica).
NanoString analysis
Total RNA from sorted T cell populations (n = 4 to 12 mice per group) was extracted with mini RNA Kit (Qiagen). Equal amounts of total RNA from different cells were used for the assay. Hybridization reaction was established by following the instruction of the manufacturer. Aliquots of Reporter CodeSet and Capture ProbeSet were thawed at RT. Then, a master mix was created by adding 70 μl of hybridization buffer to the tube containing the reporter codeset. Eight microliters of this master mix was added to each of the tubes for different samples; 5 μl (50 ng) of the total RNA sample was added into each tube. Then, 2 μl of the well-mixed Capture probeset was added to each tube and placed in the preheated 65°C thermal cycler. All the sample mixes were incubated for 16 hours at 65°C for completion of hybridization. The samples were then loaded into the sample hole in the cartridge and loaded into the NanoString nCounter SPRINT Profiler machine (NanoString). When the corresponding Reporter Library File (RLF) running is finished, the raw data were downloaded and analyzed with NanoString Software nSolver 3.0 (NanoString). mRNA counts were processed to account for hybridization efficiency, background noise, and sample content, and were normalized using the geometric mean of housekeeping genes. Fold changes were calculated comparing the experimental group to their appropriate controls. Heat map was generated by MeV software.
Statistical analysis
The means of two groups were compared with nonpaired two-tailed Student’s t test. To compare the means of more than two groups, one-way ANOVA with Tukey multiple comparison test was performed. All statistical analyses were performed using Prism 6 software (GraphPad Software). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Fig. S1. Epitope-specific expression of CD103 and PD-1 on TRM cells.
Fig. S2. Epitope-specific expression of inhibitory receptors on TRM cells.
Fig. S3. Exhausted-like TRM cells are present in the X31 influenza model.
Fig. S4. Exhausted-like TRM cells respond effectively to recall in situ.
Fig. S5. Cognate peptide inoculation directly promotes PD-1 expression on PA224–233 TRM cells.
Fig. S6. Nur77 (Nr4a1) is required for the induction and/or maintenance of exhausted-like TRM cells.
Fig. S7. Limiting NP antigen dose decreases the exhaustion phenotype.
Fig. S8. TCR signaling and inhibitor receptor expression on exhausted-like TRM cells are lost 4 months after infection.
Fig. S9. Persistent MHC-I engagement maintains exhausted-like TRM phenotypes and cells.
Fig. S10. NP366–374 TRM cells, PD-1 expression, and heterologous protection are lost over time.
Fig. S11. Continuous CD28 signaling is required for the maintenance of NP366–374 TRM cells.
Fig. S12. PD-L1 blockade promotes exhausted-like TRM cell survival and CD103 expression.
Fig. S13. PD-L1 blockade promotes cytokine production of exhausted-like TRM cells to antigenic restimulation.
Fig. S14. PD-L1 blockade promotes CD103 expression.
Fig. S15. B7 signaling is required for the effects of PD-L1 blockade.
Fig. S16. PD-L1 blockade promotes heterologous immunity and causes tissue pathology.
Fig. S17. CD8 T cells are responsible for the development of tissue pathology after α-PD-L1 blockade.
Fig. S18. ILD lungs exhibit elevated PD-1, CD103, and TIM-3 expression.
Table S1. Raw data.
Acknowledgments
We thank T. Braciale and H. Dong for critical reading of the manuscript and G. Rajagopalan for Nur77-GFP mice. We thank the NIH Tetramer Core Facility, Mayo Clinic Genomic Core, Pathology Core, and Flow Cytometry Core for reagents and technical assistance.
Funding: This work was supported by grants from NIH RO1 AI112844, RO1 AG047156, and RO1 HL126647 to J.S.; T32AG049672 to N.P.G.; R01 HL62150 and NHLBI contract 268201600004I-0–26800001-1 to A.H.L.; R01 AI057459 and R01 AI129241 to M.H.K.; R56 NS094150 and R01 NS103212 to A.J.J.; RO1 AI125701 and R21 AI139721 to N.Z.; R01HL122559 to J.E.K.; Huvis Foundation grant to R.V.; Mayo Clinic Kogod Aging Center Pilot grant and Mayo Clinic Center for Biomedical Discovery discretionary fund to J.S.; and CRI (Cancer Research Institute) Clinic and Laboratory Integration Program to N. Z.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data are deposited on GEO database (GEO number: GSE115786).
REFERENCES AND NOTES
- 1.Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song J-Y, Jacobs H, Haanen JB, Schumacher TN, Skin-resident memory CD8+T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D, Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skon CN, Lee J-Y, Anderson KG, Masopust D, Hogquist KA, Jameson SC, Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho S-H, Lerner H, Friedman AL, Shen Y, Farber DL, Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang N, Bevan MJ, Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity 39, 687–696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muschaweckh A, Buchholz VR, Fellenzer A, Hessel C, König P-A, Tao S, Tao R, Heikenwälder M, Busch DH, Korn T, Kastenmüller W, Drexler I, Gasteiger G, Antigen-dependent competition shapes the local repertoire of tissue-resident memory CD8+ T cells. J. Exp. Med. 213, 3075–3086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan TN, Mooster JL, Kilgore AM, Osborn JF, Nolz JC, Local antigen in nonlymphoid tissue promotes resident memory CD8+ T cell formation during viral infection. J. Exp. Med. 213, 951–966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies B, Prier JE, Jones CM, Gebhardt T, Carbone FR, Mackay LK, Cutting edge: Tissue-resident memory T cells generated by multiple immunizations or localized deposition provide enhanced immunity. J. Immunol. 198, 2233–2237 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Lauron EJ, Yang L, Harvey IB, Sojka DK, Williams GD, Paley MA, Bern MD, Park E, Victorino F, Boon ACM, Yokoyama WM, Viral MHCI inhibition evades tissue-resident memory T cell formation and responses. J. Exp. Med. 216, 117–132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behr FM, Chuwonpad A, Stark R, van Gisbergen KPJM, Armed and ready: Transcriptional regulation of tissue-resident memory CD8 T cells. Front. Immunol 9, 1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, Brasser G, Jongejan A, Jonkers RE, Nota B, Basak O, Clevers HC, Moerland PD, Amsen D, van Lier RAW, Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Wijeyesinghe S, Masopust D, Resident memory T cells are a Notch above the rest. Nat. Immunol. 17, 1337–1338 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Takamura S, Persistence in temporary lung niches: A survival strategy of lung-resident memory CD8+ T cells. Viral Immunol. 30, 438–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cauley LS, Environmental cues orchestrate regional immune surveillance and protection by pulmonary CTLs. J. Leukoc. Biol. 100, 905–912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballesteros-Tato A, León B, Lee BO, Lund FE, Randall TD, Epitope-specific regulation of memory programming by differential duration of antigen presentation to influenza-specific CD8+ T cells. Immunity 41, 127–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC, Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 12, 48–54 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan S, Thomas PG, Balancing immune protection and immune pathology by CD8+ T-cell responses to influenza infection. Front. Immunol. 7, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belz GT, Xie W, Doherty PC, Diversity of epitope and cytokine profiles for primary and secondary influenza A virus-specific CD8+ T cell responses. J. Immunol. 166, 4627–4633 (2001). [DOI] [PubMed] [Google Scholar]
- 19.van Gisbergen KP, Klarenbeek PL, Kragten NAM, Unger P-PA, Nieuwenhuis MBB, Wensveen FM, ten Brinke A, Tak PP, Eldering E, Nolte MA, van Lier RAW, The costimulatory molecule CD27 maintains clonally diverse CD8+ T cell responses of low antigen affinity to protect against viral variants. Immunity 35, 97–108 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Braciale TJ, Role of T cell immunity in recovery from influenza virus infection. Curr. Opin. Virol. 3, 425–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowe SR, Turner SJ, Miller SC, Roberts AD, Rappolo RA, Doherty PC, Ely KH, Woodland DL, Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J. Exp. Med. 198, 399–410 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC, A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc. Natl. Acad. Sci. U.S.A. 103, 994–999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizawa A, Bi K, Keskin DB, Zhang G, Reinhold B, Reinherz EL, TCR-pMHC encounter differentially regulates transcriptomes of tissue-resident CD8 T cells. Eur. J. Immunol. 48, 128–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zammit DJ, Turner DL, Klonowski KD, Lefrançois L, Cauley LS, Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity 24, 439–449 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi Y, Onodera T, Yamada Y, Daio R, Tsuiji M, Inoue T, Kobayashi K, Kurosaki T, Ato M, Takahashi Y, Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 212, 1709–1723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TS, Hufford MM, Sun J, Fu Y-X, Braciale TJ, Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J. Exp. Med. 207, 1161–1172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hufford MM, Kim TS, Sun J, Braciale TJ, Antiviral CD8+ T cell effector activities in situ are regulated by target cell type. J. Exp. Med. 208, 167–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Madan R, Karp CL, Braciale TJ, Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15, 277–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J, Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D, Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page N, Klimek B, De Roo M, Steinbach K, Soldati H, Lemeille S, Wagner I, Kreutzfeldt M, Di Liberto G, Vincenti I, Lingner T, Salinas G, Brück W, Simons M, Murr R, Kaye J, Zehn D, Pinschewer DD, Merkler D, Expression of the DNA-binding factor TOX promotes the encephalitogenic potential of microbe-induced autoreactive CD8+ T cells. Immunity 48, 937–950.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN, Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16, 1147–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez GJ, Pereira RM, Äijö T, Kim EY, Marangoni F, Pipkin ME, Togher S, Heissmeyer V, Zhang YC, Crotty S, Lamperti ED, Ansel KM, Mempel TR, Lähdesmäki H, Hogan PG, Rao A, The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42, 265–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R, Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Erickson JJ, Gilchuk P, Hastings AK, Tollefson SJ, Johnson M, Downing MB, Boyd KL, Johnson JE, Kim AS, Joyce S, Williams JV, Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J. Clin. Invest. 122, 2967–2982 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erickson JJ, Rogers MC, Tollefson SJ, Boyd KL, Williams JV, Multiple inhibitory pathways contribute to lung CD8+ T cell impairment and protect against immunopathology during acute viral respiratory infection. J. Immunol. 197, 233–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutigliano JA, Sharma S, Morris MY, Oguin III TH, McClaren JL, Doherty PC, Thomas PG, Highly pathological influenza A virus infection is associated with augmented expression of PD-1 by functionally compromised virus-specific CD8+ T cells. J. Virol. 88, 1636–1651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao S, Jiang L, Moser EK, Jewett LB, Wright J, Du J, Zhou B, Davis SD, Krupp NL, Braciale TJ, Sun J, Control of pathogenic effector T-cell activities in situ by PD-L1 expression on respiratory inflammatory dendritic cells during respiratory syncytial virus infection. Mucosal Immunol 8, 746–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wherry EJ, Kurachi M, Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R, Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Kaech SM, Cui W, Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA, TGF-β-dependent CD103 expression by CD8+ T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J. Exp. Med. 201, 1647–1657 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, Fife BT, Masopust D, Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, Carbone FR, Tscharke DC, Heath WR, Mueller SN, Mackay LK, Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19, 183–191 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Turner DL, Bickham KL, Thome JJ, Kim CY, D'Ovidio F, Wherry EJ, Farber DL, Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7, 501–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webby RJ, Andreansky S, Stambas J, Rehg JE, Webster RG, Doherty PC, Turner SJ, Protection and compensation in the influenza virus-specific CD8+ T cell response. Proc. Natl. Acad. Sci. U.S.A. 100, 7235–7240 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R, Persistence of memory CD8 T cells in MHC class I-deficient mice. Science 286, 1377–1381 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Borowski AB, Boesteanu AC, Mueller YM, Carafides C, Topham DJ, Altman JD, Jennings SR, Katsikis PD, Memory CD8+ T cells require CD28 costimulation. J. Immunol. 179, 6494–6503 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA, CD28 costimulation: From mechanism to therapy. Immunity 44, 973–988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD, T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science 355, 1428–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL, Sharpe AH, Freeman GJ, Blazar BR, Turka LA, Owonikoko TK, Pillai RN, Ramalingam SS, Araki K, Ahmed R, Rescue of exhausted CD8 T cells by PD-1–targeted therapies is CD28-dependent. Science 355, 1423–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reagin KL, Klonowski KD, Incomplete memories: The natural suppression of tissue-resident memory CD8 T cells in the lung. Front. Immunol. 9, 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slutter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, Harty JT, Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2, eaag2031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Braeckel-Budimir N, Harty JT, Influenza-induced lung TRM: Not all memories last forever. Immunol. Cell Biol. 95, 651–655 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Jameson SC, Masopust D, Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desai O, Winkler J, Minasyan M, Herzog EL, The role of immune and inflammatory cells in idiopathic pulmonary fibrosis. Front. Med 5, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mineo G, Ciccarese F, Modolon C, Landini MP, Valentino M, Zompatori M, Post-ARDS pulmonary fibrosis in patients with H1N1 pneumonia: Role of follow-up CT. Radiol. Med 117, 185–200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umeda Y, Morikawa M, Anzai M, Sumida Y, Kadowaki M, Ameshima S, Ishizaki T, Acute exacerbation of idiopathic pulmonary fibrosis after pandemic influenza A (H1N1) vaccination. Intern. Med. 49, 2333–2336 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Naik PK, Moore BB, Viral infection and aging as cofactors for the development of pulmonary fibrosis. Expert Rev. Respir. Med. 4, 759–771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma BB, Singh V, Flu and pulmonary fibrosis. Lung India 30, 95–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni K, Liu M, Zheng J, Wen L, Chen Q, Xiang Z, Lam K-T, Liu Y, Chan GC-F, Lau Y-L, Tu W, PD-1/PD-L1 pathway mediates the alleviation of pulmonary fibrosis by human mesenchymal stem cells in humanized mice. Am. J. Respir. Cell Mol. Biol. 58, 684–695 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier J-F, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King TE Jr., Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ, ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis, An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183, 788–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, Wang S, Li W, RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ, CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat. Immunol. 12, 327–334 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Epitope-specific expression of CD103 and PD-1 on TRM cells.
Fig. S2. Epitope-specific expression of inhibitory receptors on TRM cells.
Fig. S3. Exhausted-like TRM cells are present in the X31 influenza model.
Fig. S4. Exhausted-like TRM cells respond effectively to recall in situ.
Fig. S5. Cognate peptide inoculation directly promotes PD-1 expression on PA224–233 TRM cells.
Fig. S6. Nur77 (Nr4a1) is required for the induction and/or maintenance of exhausted-like TRM cells.
Fig. S7. Limiting NP antigen dose decreases the exhaustion phenotype.
Fig. S8. TCR signaling and inhibitor receptor expression on exhausted-like TRM cells are lost 4 months after infection.
Fig. S9. Persistent MHC-I engagement maintains exhausted-like TRM phenotypes and cells.
Fig. S10. NP366–374 TRM cells, PD-1 expression, and heterologous protection are lost over time.
Fig. S11. Continuous CD28 signaling is required for the maintenance of NP366–374 TRM cells.
Fig. S12. PD-L1 blockade promotes exhausted-like TRM cell survival and CD103 expression.
Fig. S13. PD-L1 blockade promotes cytokine production of exhausted-like TRM cells to antigenic restimulation.
Fig. S14. PD-L1 blockade promotes CD103 expression.
Fig. S15. B7 signaling is required for the effects of PD-L1 blockade.
Fig. S16. PD-L1 blockade promotes heterologous immunity and causes tissue pathology.
Fig. S17. CD8 T cells are responsible for the development of tissue pathology after α-PD-L1 blockade.
Fig. S18. ILD lungs exhibit elevated PD-1, CD103, and TIM-3 expression.
Table S1. Raw data.