Abstract
The WRKY proteins are a large family of transcription factors that play important roles in stress responses and plant development. However, the roles of most WRKYs in strawberry are not well known. In this study, FvWRKY71 was isolated from the woodland strawberry ‘Ruegen’. FvWRKY71 was highly expressed in the shoot apex and red fruit. Subcellular localization analysis showed that FvWRKY71 was located in the nucleus. Transactivation analysis showed that FvWRKY71 presented transcriptional activation activity in yeast. Overexpression of FvWRKY71 in Arabidopsis and woodland strawberry revealed early flowering in the transgenic plants compared with the wild-type control. Gene expression analysis indicated that the transcript levels of the flowering time and development integrator genes AP1, LFY, FT, AGL42, FUL, FPF1, SEP1, SEP2, and SEP3 were increased in FvWRKY71-overexpressing Arabidopsis and strawberry plants compared with the wild-type controls, which may result in accelerated flowering in transgenic plants. Furthermore, FvWRKY71 was proven to directly bind to the W-boxes (TTGACT/C) of the FvFUL, FvSEP1, FvAGL42, FvLFY, and FvFPF1 promoters in vitro and in vivo. Taken together, our results reveal a transcriptional regulatory cascade of FvWRKY71 involved in promoting flowering in woodland strawberry.
Subject terms: Flowering, Plant sciences
Introduction
The WRKY proteins are a family of plant transcription factors (TFs). Members of the family harbor one or two conserved WRKY domains, which contain a conserved WRKYGQK heptapeptide sequence at the N-terminus and a C2H2 or C2HC zinc-finger protein motif at the C-terminus1. Based on the number of WRKY domains and the structure of the C-terminal zinc-finger motif, the WRKY family is divided into three categories: groups I, II, and III. Group II proteins are further classified as IIa, IIb, IIc, IId, or IIe proteins based on the primary amino acid sequence2. It has been more than 20 years since the identification of the first WRKY protein1. Since that time, diverse biological functions of WRKY proteins in different species have been studied3.
WRKY proteins play crucial roles in plant signaling, the regulation of plant development and the responses to various biological and abiotic stresses4. In Arabidopsis, most WRKY genes have been shown to be involved in defense against pathogens2. For example, AtWRKY33 can modulate host defenses against Alternaria brassicicola and Botrytis cinerea infection5,6. AtWRKY46 forms a complex with AtWRKY70 and AtWRKY53 to positively regulate basal resistance to Pseudomonas syringae7. AtWRKY8, AtWRKY25, AtWRKY33, and AtWRKY63 are involved in the response to abiotic stress8–11. AtWRKY6, AtWRKY44, AtWRKY57, and AtWRKY71 are involved in plant developmental processes2,12–14. In rice, at least five OsWRKY genes have been demonstrated to participate in the defense response against pathogens15,16. In addition, many OsWRKY genes are regulators of the response to abiotic stresses, such as heat, drought, and salt16–18. Most WRKY genes are involved in the stress response, but some participate in the determination of flowering time. In Arabidopsis, WRKY75 positively regulates flowering via the GA-mediated signaling pathway19. AtWRKY12 is a positive factor in the regulation of flowering, whereas AtWRKY13 is a negative factor in the modulation of flowering under short-day conditions. Further analysis indicated that both WRKY12 and WRKY13 can directly bind to the promoter of FUL and produce different effects on their downstream target genes20. The heterologous overexpression of WRKY12 in Miscanthus lutarioriparius in Arabidopsis promotes flowering. Consistent with the early flowering phenotype, the expression levels of the APETALA 1 (AP1), FLOWERING LOCUS T (FT), LEAFY (LFY), and FRUITFULL (FUL) genes are significantly increased in transgenic Arabidopsis plants21. AtWRKY71 accelerates flowering by regulating the flowering genes FT and LFY22. The overexpression of CpWRKY71 in Arabidopsis also causes an early flowering, precocious leaf senescence phenotype23. The potential pathways of the WRKY TFs involved in flowering are not fully understood.
The molecular mechanism of flowering has been extensively studied in the annual model plant Arabidopsis, whereas less is known about the molecular control of flowering in perennial species, such as strawberry. Strawberry plants can be divided into two main groups based on their flowering habits: seasonal flowering (SF) and perpetual flowering (PF) strawberry. SF strawberry flowers under short days and is induced to flower in autumn24. SF patterns exist in diploid woodland strawberry (Fragaria vesca) and cultivated strawberry (Fragaria × ananassa), which also exhibit genomes with a high degree of colinearity25–27. In F. vesca, SF is caused by a single repressor gene: SEASONAL FLOWERING LOCUS (SFL)28. TERMINAL FLOWER1 (TFL1), a candidate gene for SFL, is activated to inhibit strawberry flowering in summer and is suppressed to induce flower initiation in autumn29. The knockdown of FvSOC1 produces a continuous flowering phenotype, which is similar to mutant of the floral repressor FvTFL130. In SF strawberry, FvFT1 and FaFT1 are homologous genes of FT in Arabidopsis. PF strawberry produces new inflorescences continuously throughout the growing season from spring until late autumn. In the diploid Fragaria vesca, the PF trait is caused by a recessive mutation in the SFL gene31. A major locus controlling PF in cultivated strawberry is the perpetual flowering and runnering (PFRU) locus, which modulates the balance between sexual and asexual plant reproduction27,31. FaPFRU shows opposite effects on flowering and runnering, exerting a positive effect on flowering and a negative effect on runnering, indicating that the two traits are genetically linked and share common physiological control31.
To date, the only available information on the WRKY family in woodland strawberry has come from the genome-wide analysis of the expression of WRKY genes in different developmental stages or under biotic and abiotic stresses32,33 and the potential roles of the WRKY genes in woodland strawberry are largely unknown. In the annual model plant Arabidopsis, WRKY71 can promote flowering. However, whether WRKY71 presents a similar function or regulatory mechanism in flowering in perennial plants, such as strawberries, is still largely unknown. In addition, the expression of FvWRKY46, another name of FvWRKY71, is upregulated during fruit development and ripening based on RNA-seq and RT-qPCR results32, which indicates that FvWRKY71 may play a role in regulating fruit development and ripening, in addition to promoting flowering. To investigate the role of WRKY71 in F. vesca, the FvWRKY71 gene was overexpressed in Arabidopsis and woodland strawberry, and we found that FvWRKY71 conferred early flowering. We also found that FvWRKY71 accelerated flowering through the direct activation of different flowering-related genes between strawberry and Arabidopsis. These data will enrich the regulatory network of WRKY71 in different plants and provide some information on the promotion of flowering in Rosaceae plants.
Results
Structural analysis of FvWRKY71
To investigate the function of the FvWRKY71 protein, we first identified and cloned FvWRKY71 from the woodland strawberry ‘Ruegen’ by RT-PCR using the specific primers shown in Supplemental Table S1. The coding sequence of FvWRKY71 was 1107 bp in length, which encoded a protein of 368 amino acids. The estimated molecular weight and isoelectric point were 41.62 and 6.46 kDa, respectively.
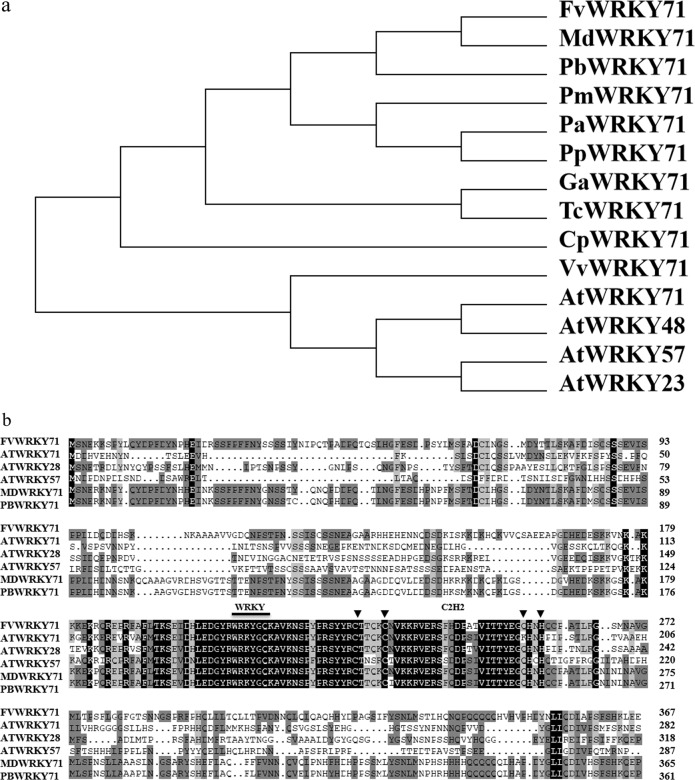
To examine the evolutionary relationship between FvWRKY71 and its orthologs from multiple species, we performed multiple-sequence alignment using their full-length amino acid sequences. A phylogenetic tree was constructed through neighbor-joining analysis (Fig. 1a). The phylogenetic tree results indicated that FvWRKY71 exhibited the closest genetic relationship to the WRKY71 proteins from apple and pear, which also belong to the Rosaceae family (Fig. 1a). FvWRKY71 was also homologous to Arabidopsis WRKY71, WRKY48, WRKY57, and WRKY23 according to BLAST analysis in the TAIR database.
Fig. 1. Phylogenetic and structural analyses of FvWRKY71.
a Phylogenetic tree based on the alignment of amino acid sequences among WRKY proteins. b Multiple alignment of WRKY proteins from higher plants
To better understand the conserved domain of FvWRKY71, we used DNAMAN software to carry out amino acid sequence analysis. FvWRKY71 was shown to contain a conserved WRKYGQ domain and a conserved C2H2-type zinc-finger motif (Fig. 1b). Therefore, FvWRKY71 identified in this study contains one WRKY domain and belongs to group II34 (Fig. 1a).
Expression patterns of FvWRKY71 in strawberry
Different developmental stages and different organs were selected to detect the expression patterns of the FvWRKY71 gene by RT-qPCR (Fig. 2a). The expression levels in the flowers, shoot apex, and red fruits were approximately 2-, 6- and 17-fold higher than that in the roots, respectively. Lower expression levels were found in the roots, petioles, leaves, and green fruits. (Fig. 2a).
Fig. 2. Expression pattern and subcellular localization of FvWRKY71.
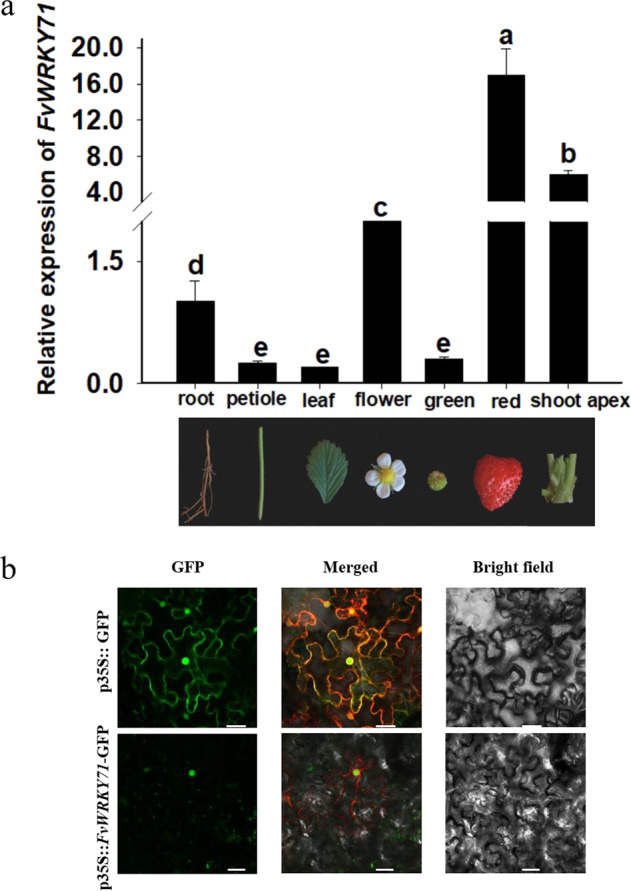
a Expression pattern of FvWRKY71 in different organs. RT-qPCR analysis was used to test the relative expression level of FvWRKY71 in various organs. Values are the mean ± SD from three independent experiments with three biological replicates. b Subcellular localization of the FvWRKY71 protein. p35S::GFP was transformed into tobacco (top). p35S::FvWRKY71-GFP was transformed into tobacco (bottom), and the fusion protein was located in the nucleus. Bar = 30 μm
Subcellular localization of FvWRKY71
To examine the subcellular localization of the FvWRKY71 protein, we transiently expressed the recombinant FvWRKY71-GFP protein in Nicotiana benthamiana leaves. The fusion protein signal was examined by confocal microscopy. As shown in Fig. 2b, the FvWRKY71-GFP fusion protein only existed in the nucleus, while the green signal of the GFP control was distributed in the nucleus and the cell membrane. The results indicate that FvWRKY71 localizes to the nucleus (Fig. 2b).
Transcriptional activation activity of FvWRKY71 in yeast
A yeast system was used to test whether FvWRKY71 exhibited transcriptional activation activity. The complete coding region of FvWRKY71 was fused to the GAL4 DNA-binding domain in the pGBT9 vector, and the construct was then transformed into Y2H Gold cells. The empty pGBT9 vector served as a control. As shown in Fig. 3a, yeast cells transformed with the pBD-FvWRKY71 fusion vector grew well on both SD/-Trp and SD/-Trp/-Leu/-Ade media. They also exhibited blue colonies on alpha-galactosidase plates (Fig. 3a), which indicated that they presented alpha-galactosidase activity. In contrast, the control yeast strain was only viable on SD/-Trp medium. These data showed that FvWRKY71 exhibited transcriptional activation activity in yeast. To further elucidate which segments presented transcriptional activation activity, we transformed the N-terminus and C-terminus of FvWRKY71 in the pGBT9 vector into yeast cells. The results showed that the N-terminus showed transcriptional activation activity, while the C-terminus did not (Fig. 3b).
Fig. 3. Transactivation activation activation analysis of FvWRKY71.
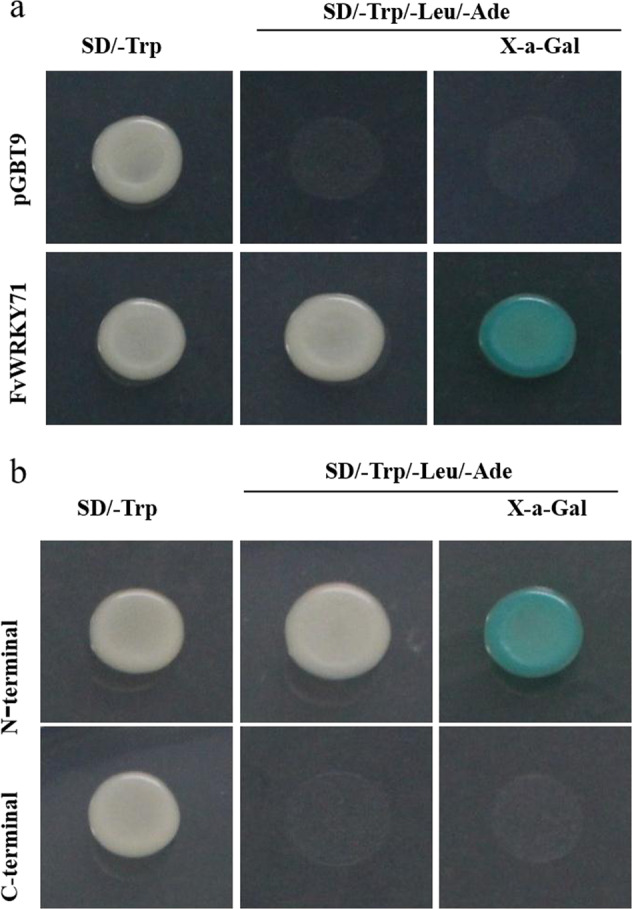
The construct of pBD-FvWRKY71 was transformed into yeast Y2H Gold cells, and the cells were then examined on SD/-Trp and SD/-Trp/-Leu/-Ade/X-α-gal plates. a Transactivation activation analysis of FvWRKY71. b Transactivation activation analysis of the N-terminus and C-terminus of FvWRKY71
Overexpression of FvWRKY71 in Arabidopsis promotes flowering
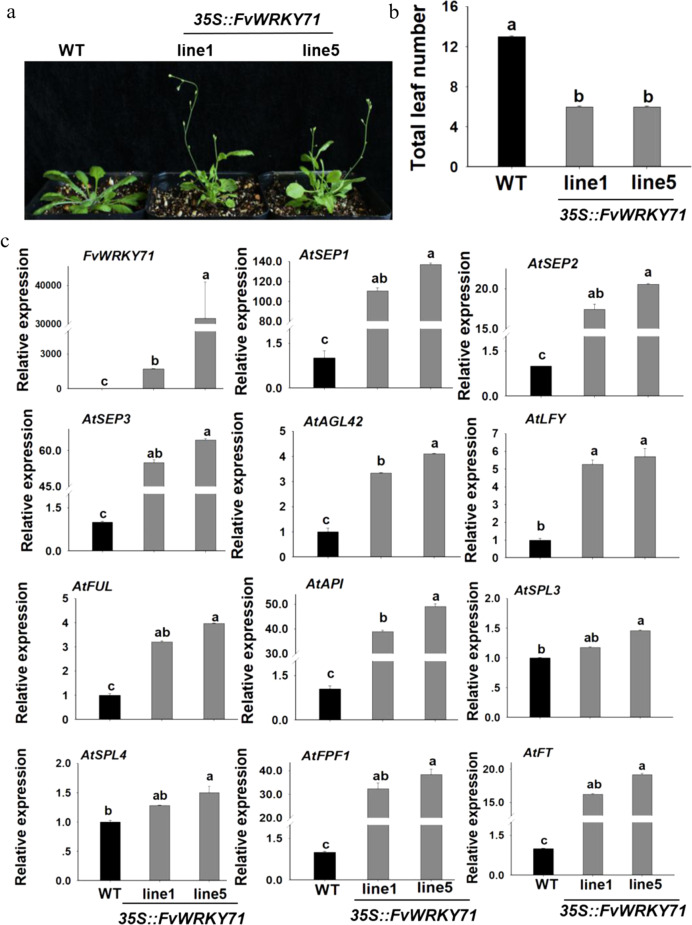
To investigate the biological function of the FvWRKY71 gene, we constructed a p35S::FvWRKY71 overexpression vector (Fig. S1a) and transformed it into Arabidopsis to obtain transgenic plants. The transgenic Arabidopsis lines were examined by PCR to determine on DNA levels (Fig. S1b) and by RT-qPCR to determine on RNA levels (Fig. 4c). Two T3 homozygous positive transgenic lines were selected for further phenotypic analysis. By observing transgenic lines 1 and 5, we found that the ectopic expression of the FvWRKY71 gene in Arabidopsis induced an early flowering phenotype compared with wild-type plants (Fig. 4a). Under long (16 h) daylight conditions, FvWRKY71-overexpressing transgenic plants started bolting at 19 days after sowing, whereas wild-type plants started bolting at 33 days after sowing. In addition, the number of rosette leaves at flowering in the transgenic lines was approximately seven, while the number of rosette leaves at flowering in the wild-type plants was approximately thirteen (Fig. 4b). These data indicate that the overexpression of FvWRKY71 in Arabidopsis can promote flowering.
Fig. 4. The phenotypes of Arabidopsis FvWRKY71-overexpressing plants and gene expression analysis.
a The phenotypes of Arabidopsis FvWRKY71-overexpressing transgenic lines and wild-type control plants. b The number of rosette leaves at flowering in transgenic plants and wild-type plants. c RT-qPCR analysis of FvWRKY71 and genes involved in flowering, including AtSEP1, AtSEP2, AtSEP3, AtAGL42, AtAP1, AtLFY, AtFUL, AtSPL3, AtSPL4, AtFPF1, and AtFT, between wild-type and transgenic Arabidopsis plants. Values are the mean ± SD from three independent experiments with three biological replicates. Different letters indicate significant differences (P < 0.05, based on Duncan’s multiple range test)
To investigate whether the overexpression of FvWRKY71 in Arabidopsis affected the transcript levels of flowering-related genes for the early flowering phenotype. We searched the Arabidopsis flowering database (http://www.phytosystems.ulg.ac.be/florid/) for candidate genes responsible for accelerating flowering in FvWRKY71-overexpressing transgenic lines. Twelve genes involved in the determination of flowering time and flower development were identified by RT-qPCR analysis. The expression levels of SEPALLATA1 (SEP1), SEPALLATA2 (SEP2), SEPALLATA3 (SEP3), AGMOUS-LIKE 42 (AGL42), AP1, LFY, FUL, FLOWERING PROMOTING FACTOR 1 (FPF1), and FT were shown to be significantly increased compared with those in wild-type plants (Fig. 4c). The expression level of FvWRKY71 in transgenic line 5 was higher than that in transgenic line 1, and the expression of downstream genes was also higher in transgenic line 5 than in transgenic line 1 (Fig. 4c).
Overexpression of FvWRKY71 in woodland strawberry accelerates flowering
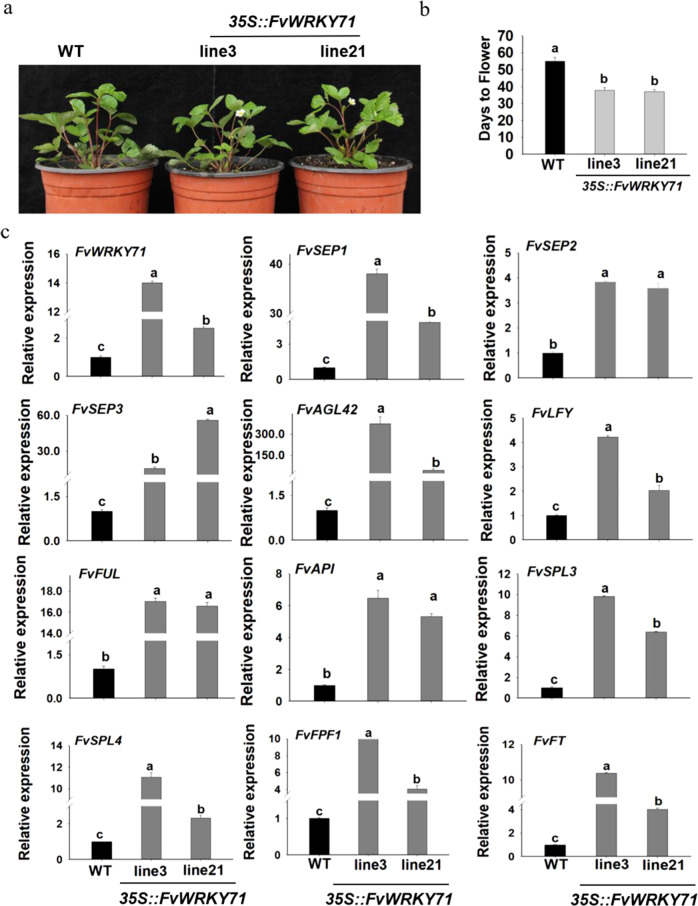
To further investigate the function of FvWRKY71, we also overexpressed FvWRKY71 in ‘Ruegen’ strawberry. The transgenic lines were first examined by PCR to detect DNA levels (Fig. S1c). Next, we investigated transgenic lines 3 and 21 by RT-qPCR (Fig. 5c). The transcript level of FvWRKY71 was significantly increased in transgenic lines 3 and 21 compared with the non-transgenic control plants (Fig. 5c). Interestingly, we observed that the FvWRKY71-overexpressing transgenic lines promoted flowering in woodland strawberry relative to wild-type plants (Fig. 5a), and the early flowering phenotype was consistent with the results of FvWRKY71 overexpression in Arabidopsis. We found that transgenic line 3, transgenic line 21, and wild-type plants took 38, 37, and 55 days, respectively, to start flowering under the same conditions (Fig. 5b). Next, we tested the expression levels of SEP1, SEP2, SEP3, AGL42, AP1, LFY, FUL, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SPL3), SPL4, FPF1, and FT in FvWRKY71-overexpressing strawberry transgenic lines by RT-qPCR. We found that the transcript abundance of these flowering-related genes was significantly increased in transgenic strawberry lines compared with wild-type control plants (Fig. 5c), which was in agreement with the results of the overexpression of FvWRKY71 in Arabidopsis (Fig. 5c). We also analyzed the expression of FvSOC1 and FvTFL1 in transgenic strawberry plants by RT-qPCR. We found that the expression of FvSOC1 and FvTFL1 was significantly reduced in transgenic line 3 compared with that in wild-type plants, whereas there was no considerable difference between transgenic line 21 and wild-type plants (Fig. S3). Taken together, these data reveal that FvWRKY71 promotes flowering and regulates the expression of flowering-related genes.
Fig. 5. The phenotypes of FvWRKY71-overexpressing transgenic woodland strawberry lines and gene expression analysis.
a The phenotypes of FvWRKY71-overexpressing woodland strawberry lines and wild-type plants. b Days to flowering between woodland strawberry transgenic lines and wild-type plants after transfer from culture medium. c RT-qPCR analysis of FvWRKY71 and genes involved in flowering between wild-type and transgenic woodland strawberry plants, including FvSEP1, FvSEP2, FvSEP3, FvAGL42, FvAP1, FvLFY, FvFUL, FvSPL3, FvSPL4, FvFPF1, and FvFT. Values are the mean ± SD from three independent experiments with three biological replicates. Different letters indicate significant differences (P < 0.05, based on Duncan’s multiple range test)
FvWRKY71 accelerates the flowering of woodland strawberry mainly via the direct activation of FvFUL, FvSEP1, FvAGL42, FvLFY, and FvFPF1
To further identify the genes directly regulated by FvWRKY71 for early flowering in woodland strawberry, we investigated the promoter sequences of FvSEP1, FvSEP2, FvSEP3, FvAGL42, FvAP1, FvLFY, FvFUL, FvSPL3, FvSPL4, FvFPF1, and FvFT to determine whether they contained W-boxes (TTGACT/C). We found that FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4 exhibit W-boxes in these promoter regions (Supplemental File S1). A yeast one-hybrid experiment was first conducted to detect whether FvWRKY71 could bind to the promoters of FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4. pAD-FvWRKY71 was cotransformed into yeast Y1H strains in combination with proFvFUL-pAbAi, proFvSEP1-pAbAi, proFvAGL42-pAbAi, proFvLFY-pAbAi, proFvFPF1-pAbAi, or proFvSPL4-pAbAi. The first five transformed strains were able to grow on SD/-Leu selective medium containing aureobasidin A (ABA) (100, 100, 150, 300, and 300 μg/L, respectively), whereas that transformed with FvSPL4 (100 μg/L ABA) could not (Fig. 6), which indicates that FvWRKY71 can bind to the promoters of FvFUL, FvSEP1, FvAGL42, FvLFY, and FvFPF1.
Fig. 6. Yeast one-hybrid assay to test whether FvWRKY71 could directly bind to the promoters of FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4. pAD-FvWRKY71 was cotransformed into yeast Y1H strains in combination with proFvFUL-pAbAi, proFvSEP1-pAbAi, proFvAGL42-pAbAi, proFvLFY-pAbAi, proFvFPF1-pAbAi, or proFvSPL4-pAbAi.
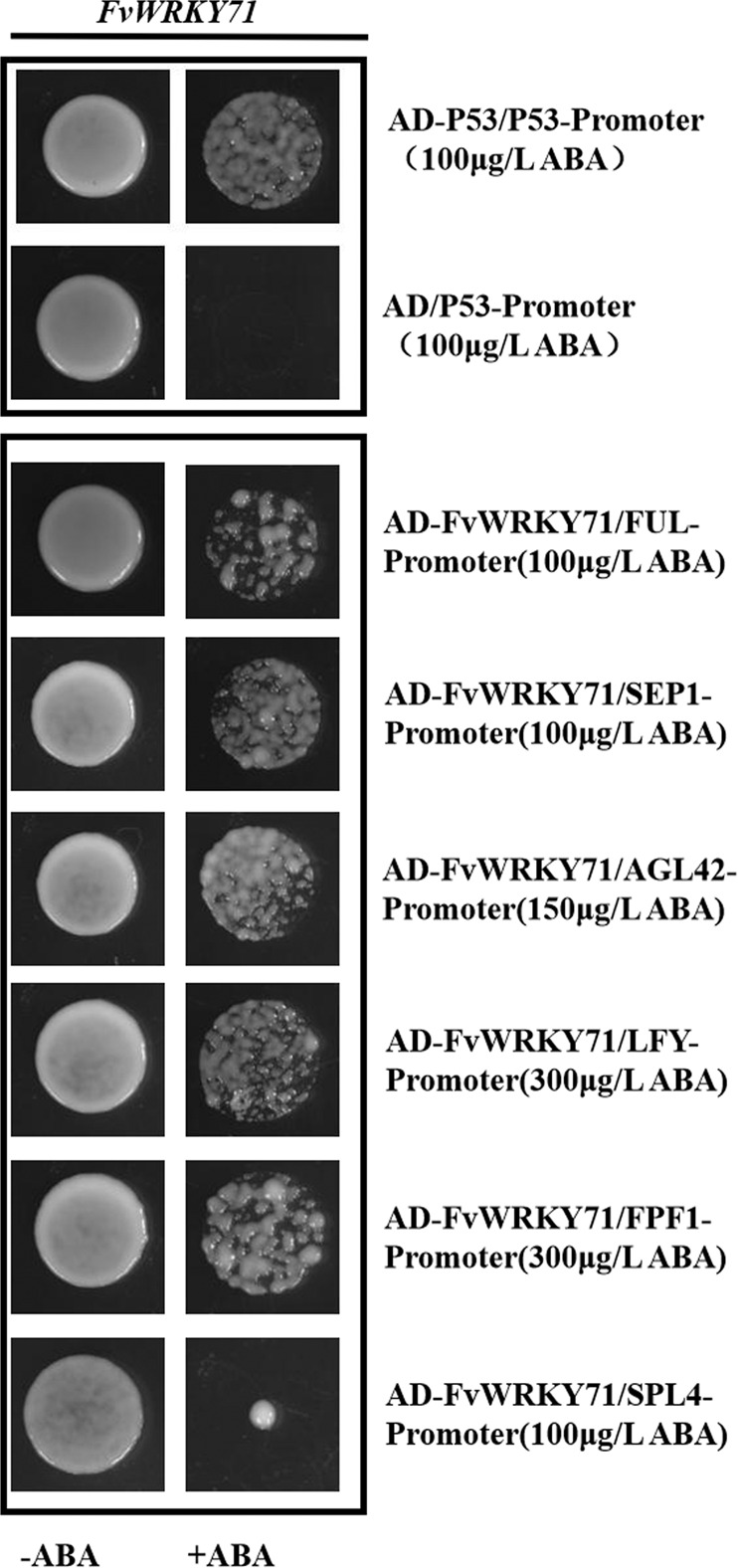
The transformed strains all grew on SD/-Leu selective medium containing aureobasidin A (ABA) (100, 100, 150, 300, 300, and 100 μg/L, respectively)
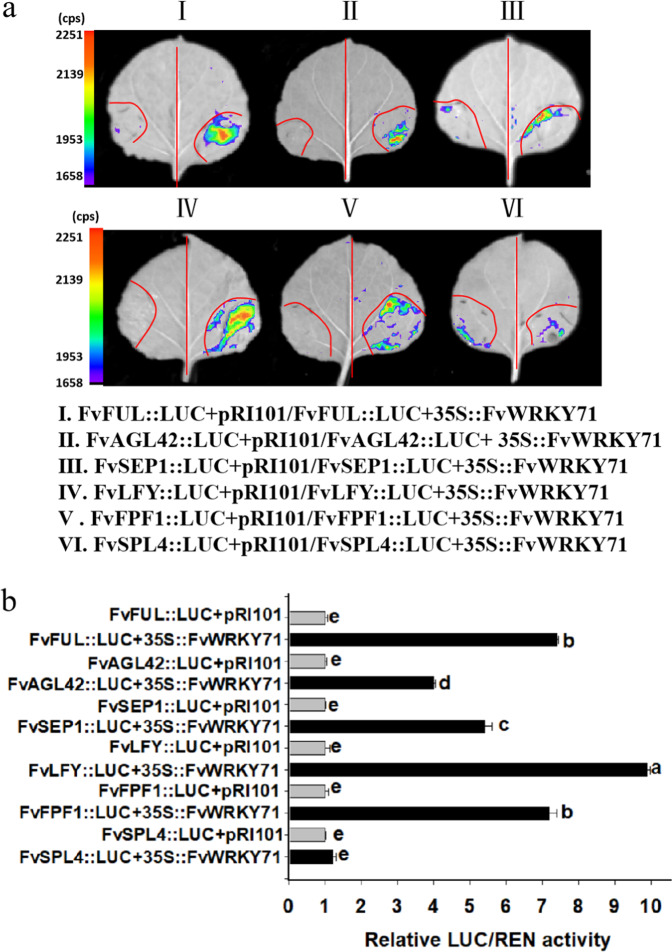
A luciferase assay is an important method for detecting the binding of transcription factors and specific domain sites in the promoters of target genes35. To clarify the effect of FvWRKY71 on the promoters of FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4, we performed luciferase reporter assays in N. benthamiana leaf cells. We constructed proFvFUL-LUC, proFvSEP1-LUC, proFvAGL42-LUC, proFvLFY-LUC, proFvFPF1-LUC and proFvSPL4-LUC reporters and the effector plasmid 35S:FvWRKY71 (Fig. S1). As expected, FvWRKY71 exerted activation effects on FvFUL, FvSEP1, FvAGL42, FvLFY, and FvFPF1 in these LUC reporter assays, but not on FvSPL4 (Fig. 7a), and FvWRKY71 activated the expression of the FvFUL, FvAGL42, FvSEP1, FvLFY, and FvFPF1 genes to various levels (7.4-, 4.0-, 5.4-, 9.9-, and 7.2-fold, respectively) compared with the corresponding control (Fig. 7b). Together, these data suggest that FvWRKY71 accelerates the flowering of woodland strawberry mainly through the direct activation of FvFUL, FvAGL42, FvSEP1, FvLFY, and FvFPF1.
Fig. 7. Luciferase activity assay for determining whether FvWRKY71 could directly bind to the promoters of FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4.
a Tobacco transient expression assay. p35S::FvWRKY71 was cotransformed into N. benthamiana in combination with proFvFUL::LUC, proFvAGL42::LUC, proFvSEP1::LUC, proFvLFY::LUC, proFvFPF1::LUC, or proFvSPL4::LUC. b The comparison of luciferase activity. The transcriptional activity of these infiltrated tobacco leaves based on the ratio of LUC to REN was investigated with a dual luciferase reporter gene assay kit. Different letters indicate significant differences (P < 0.05, based on Duncan’s multiple range test)
Discussion
Flowering is a key step in plant growth and development. Early flowering is an important trait for Rosaceae crops such as strawberry, apple, and peach. Here, we identified the transcription factor WRKY71 in woodland strawberry and found that FvWRKY71 was expressed in all examined organs, especially in the shoot apex and red fruit. In Arabidopsis, the transcript level of AtWRKY71 gradually increased with seedling development from 4 to 16 days and reached the highest level at 16 days, which is consistent with the floral transition22. The higher expression of FvWRKY71 in the shoot apex is consistent with the floral transition. Functional analysis by overexpressing FvWRKY71 in Arabidopsis and strawberry revealed that FvWRKY71 plays an important role in accelerating flowering.
Flower induction is crucial for plant growth and development, and can promote the shift from vegetative growth to reproductive growth36,37. Flower induction is regulated by many exogenous and endogenous factors. In the model plant Arabidopsis, at least five flowering pathways have been found, including the photoperiod, gibberellin, vernalization, autonomous, and age pathways38–40. These pathways are related to various signals, such as photoperiod, age, temperature, abiotic stress and hormones, and regulate the expression of factors involved in flowering. These flowering pathways can be regulated independently from each other or in an interrelated manner, thus forming a complex flowering regulation network. The functional analysis of genes representing various flowering pathways did not reveal large differences between diploid woodland strawberry and cultivated strawberry25,26,41.
In perennial plants, molecular studies on flowering have focused on the FT, LFY, TFL1, and SOC1 genes. As a key component of flowering pathways, the expression of FT is influenced by many factors, some of which act as positive regulators and others as repressors42–47. The signal is passed on to FvAP1 and FvFUL to effect the transition from the vegetative to the reproductive stage29,48. The FT gene is highly expressed in the leaves in cultivated strawberry and is induced by long days and high temperatures26. Decreasing or increasing LFY transcript abundance can delay or accelerate flowering, respectively49. It has been reported that Arabidopsis WRKY75 can promote flowering via FT and SOC1 but not LFY. The promoter of FT contains some W-box cis-elements, and FT is the direct target of AtWRKY75. In addition, AtWRKY75 partially controls GA signaling in the regulation of flowering time19. Arabidopsis WRKY12 and WRKY13 modulate flowering in opposite directions under short-day conditions. AtWRKY12 and AtWRKY13 can interact with two DELLA proteins, AtGAI and AtRGL1, to affect the transcriptional activity of AtWRKY12 and AtWRKY13, which regulate the expression of AtFUL, a direct downstream target gene of AtWRKY12 and AtWRKY13, to control flowering20. The transcription levels of AP1 and LFY are higher in WRKY71-1D than in wild-type plants, and AtWRKY71 promotes flowering through the direct activation of FT and LFY22. In our study, the relative expression levels of FT, LFY, and FUL in the Arabidopsis and strawberry transgenic lines were also higher than those in the wild-type (Figs. 4c and 5c). We identified W-boxes in the LFY promoter region, while we could not find a W-box in the FT promoter in woodland strawberry. There are three W-boxes in the Arabidopsis FT promoter region22. Therefore, FvWRKY71 could not directly regulate FT gene expression by binding to W-box elements in woodland strawberry and might affect the transcript level of FT in other unidentified ways. The overexpression of FUL in Arabidopsis can promote flowering50,51. Interestingly, we found that the strawberry FUL gene presented two W-boxes in its promoter region (Supplemental File S1) and showed that FvWRKY71 could interact with the FvFUL promoter to modulate FvFUL expression. These findings indicate that different WRKY proteins regulate the transcript levels of different floral meristem identity genes or floral integrators to control their distinct flowering pathways and that some WRKY proteins may be involved in controlling the timing of GA-mediated flowering.
TFL1 and SOC1 function as major repressors of flowering in perennial species, including strawberry and rose52–54. We found that the expression levels of FvTFL1 and FvSOC1 were significantly reduced in woodland strawberry transgenic line 3 compared with wild-type plants, which is consistent with the early flowering phenotype of FvWRKY71-overexpressing plants. However, there were no large differences between transgenic line 21 and wild-type plants (Fig. S3). The higher expression of FvWRKY71 in transgenic line 3 (14.0-fold higher than wild-type control) than transgenic line 21 (2.5-fold higher than wild-type control) may result in lower expression of FvTFL1 and FvSOC1 in transgenic line 3. In Arabidopsis, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) mediates both endogenous and exogenous signals to accelerate flowering30. These findings reveal that the functions of important flowering genes may not be completely the same between annual and perennial plants.
During the transition to flowering, FPF1 regulates flowering through an independent pathway that is parallel to that of LFY and AP155. We found W-boxes in the promoter sequences of the FPF1 gene in woodland strawberry, and we showed that FvWRKY71 can directly regulate FvFPF1 expression by binding to the W-boxes of the FvFPF1 promoter. The SPL3 and SPL4 transcription factors can also directly bind to the promoter of the LFY gene55. SPL3 directly activates LFY, FUL, and AP1 expression in Arabidopsis56. The expression levels of SPL3 and SPL4 are significantly increased in FvWRKY71-overexpressing lines, and a W-box exists in the FvSPL4 promoter. However, FvWRKY71 cannot bind to the W-box of the FvSPL4 gene (Figs. 6 and 7). In Arabidopsis, the promoter region of the AP1 gene contains five W-boxes, whereas AtWRKY71 cannot bind to any of the AP1 W-boxes22. These findings indicate that the FvSPL4 and AtAP1 genes contain W-box elements but that they are not targets of WRKY71 in woodland strawberry and Arabidopsis. WRKY71 might regulate the expression of these genes to accelerate flowering in indirect ways.
MADS domain transcription factors play important roles in flower development, the control of flowering time and fruit development57. In our study, we observed that many MADS-domain proteins, such as AP1, AGL42, SEP1, SEP2, and SEP3, exhibited different expression levels between FvWRKY71-overexpressing transgenic plants and wild-type plants (Figs. 4c and 5c). It has been reported that the MADS-domain proteins AP1, AP3, PI, AG, and SEP3 play important roles in floral tissues to modulate flower development58. SEP1, SEP2, and SEP3 encode MADS box transcription factors, which are a group of organ identity genes that are required for flower development. LFY directly activates SEP1, SEP2, and SEP3 in Arabidopsis59. AGL42 encodes MADS-box transcription factors and is closely related to SOC1 in Arabidopsis60. Here, we found that the upstream regulatory region of woodland strawberry AP1 does not contain a W-box, while there are five W-boxes in the Arabidopsis AP1 promoter. Additionally, we found that the promoter regions of FvAGL42 and FvSEP1 contain W-boxes (Supplemental File S1) and that FvWRKY71 is able to directly control the expression of FvAGL42 and FvSEP1 by binding to W-boxes in the promoters of FvAGL42 and FvSEP1 (Figs. 6 and 7). Therefore, these results indicate that FvWRKY71 promotes flowering through the direct activation of different flowering-related genes between strawberry and Arabidopsis.
Together, our results show that FvWRKY71 acts as a positive regulator of flowering in woodland strawberry. The FUL, SEP1, AGL42, LFY, and FPF1 gene homologs of the flowering pathway genes of Arabidopsis were analyzed in strawberry. We found that these genes were highly expressed in FvWRKY71-overexpressing transgenic plants. The transcriptional regulatory cascade of FvWRKY71 involved in accelerating flowering in woodland strawberry (Fig. 8) differs from the regulatory mechanism of WRKY71 in promoting flowering in Arabidopsis. FvWRKY71 promotes flowering via the direct modulation of FvFUL, FvSEP1, FvAGL42, FvLFY, and FvFPF1 expression. These data will enrich the known regulatory network of WRKY71 in different plants and provide information on the promotion of flowering on Rosaceae plants.
Fig. 8. Proposed transcriptional regulatory cascade whereby FvWRKY71 regulates flowering in woodland strawberry.
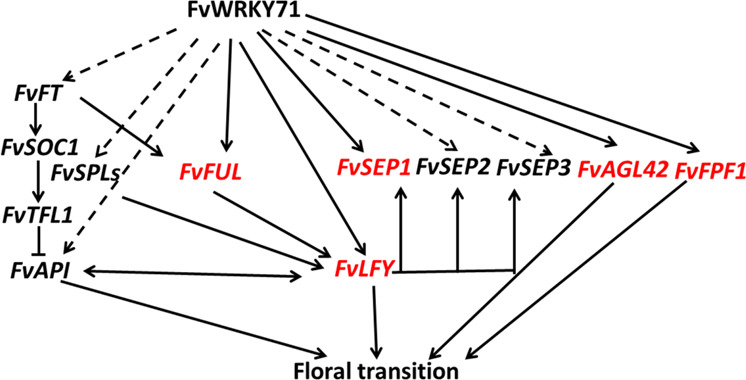
FvWRKY71 accelerates flowering via the direct activation of FvFUL, FvLFY, FvFPF1, FvAGL42, and FvSEP1 (red font) and the indirect regulation of FvFT, FvSPL3, FvSPL4, FvAP1, FvSEP2, and FvSEP3 (dotted line). The other solid lines indicate regulatory relationships reported in the literature29,48,51,52,55,59,60. Arrows represent activation, and bars represent repression
Materials and methods
Plant materials and growth conditions
The material used in this study was the diploid woodland strawberry (Fragaria vesca) ‘Ruegen’. The strawberries were grown in the greenhouse at Shenyang Agricultural University, China. FvWRKY71-overexpressing transgenic strawberry plants and wild-type plants were transferred from culture medium to the soil substrate and cultivated in an incubator at 23 °C (16 h/8 h, light/dark). Thirty days later, the plants were transplanted to the greenhouse of Shenyang Agricultural University in June. The days to flowering were recorded in the transgenic plants (six for line 3 and six for line 21) and six wild-type plants in the greenhouse after transfer from culture medium.
Wild-type (WT) Arabidopsis (Columbia) seeds were synchronized at 4 °C for 2 days and then placed at 22 °C ± 2 °C under long-day (16 h/8 h, light/dark) conditions.
The tobacco species used for transient expression analysis was Nicotiana benthamiana, which was grown at 23 °C under a long-day photoperiod (16 h/8 h, light/dark) for 1 month.
Phylogenetic analysis and multiple-sequence alignment
The full-length sequences of WRKY71 proteins from different plants were aligned by using ClustalW (http://www.clustal.org/), and the phylogenetic tree was constructed using MEGA6.0 software (http://www.megasoftware.net/)61. The neighbor-joining (NJ) method was used to produce a phylogenetic tree with the following parameters: bootstrap analysis (1,000 replicates; random seeds), Poisson correction and pairwise deletion61. The FvWRKY71 orthologs used for phylogenetic tree construction came from Arabidopsis thaliana, Prunus persica, Malus x domestica, Pyrus bretschneideri, Prunus mume, Prunus avium, Vitis vinifera, Theobroma cacao, Gossypium arboreum, and Carica papaya. DNAMAN software was used to carry out amino acid sequence alignments between FvWRKY71 and different species (version 6.0; Lynnon Biosoft, Quebec, Canada).
Subcellular localization
The coding region without the stop codon of FvWRKY71 was amplified by RT-PCR from the red fruit of ‘Ruegen’ based on woodland strawberry genome information62,63. The FvWRKY71-F and FvWRKY71-R primers are shown in Supplemental Table S1. The products were inserted into the pRI101-GFP vector with EcoRI and KpnI to generate the plasmid p35S::FvWRKY71-GFP. The pRI101-GFP and p35S::FvWRKY71-GFP plasmids were transformed into Agrobacterium tumefaciens strain EHA105. The leaves of 1-month-old tobacco (N. benthamiana) plants were infiltrated with A. tumefaciens cells harboring pRI101-GFP and pRI101-FvWRKY71-GFP. The GFP fluorescence signal was observed by confocal fluorescence microscopy (Leica DMi8 A, Wetzlar, Germany) 2 days after transient transformation.
Transactivation assay
The coding region of FvWRKY71 was amplified by RT-PCR from the red fruit of ‘Ruegen’ using specific primers (Supplemental Table S1) and then inserted into the pGBT9 vector. The pGBT9-FvWRKY71 plasmid and the pGBT9 vector were introduced into Y2H Gold cells using the PEG/LiAc method64, and the cells were grown on SD/-Trp agar plates. The pGBT9 vector was used as a negative control. After 3 days, single transformed yeast colonies were diluted with 10 μL sterile water, and a 3-μL aliquots were dropped onto SD/-Trp and SD/-Trp/-Leu/-Ade/X-a-Gal plates to observe yeast growth at 30 °C for 3–4 days. Next, we amplified the coding sequences of the N-terminus and C-terminus of FvWRKY71 by RT-PCR from the red fruit of ‘Ruegen’ (Supplemental Table S1), and the PCR products were cloned into the pGBT9 vector to identify the activation area at the N-terminus or C-terminus.
Overexpression vector construction and plant transformation
The full-length coding region of FvWRKY71 was amplified by RT-PCR from the red fruit of ‘Ruegen’ and cloned into pRI101-AN to generate the overexpression plasmid p35S::FvWRKY71. The PCR primer sets are shown in Supplemental Table S1. The placsmid was confirmed by sequencing. The plasmid was introduced into A. tumefaciens strain GV3101 via the freeze-thaw method and transformed into Arabidopsis via the floral dip method65. Positive transgenic plants were screened on plates containing 1/2 MS medium with 30 mg/L kanamycin. Furthermore, two T3 generation transgenic Arabidopsis lines (Line 1 and Line 5) were detected by RT-qPCR for gene expression analysis. The numbers of rosette leaves were analyzed in each plant after the first inflorescence emergence. Ten plants of each line were used for analyzing the numbers of rosette leaves.
To further elucidate the function of FvWRKY71, we introduced the overexpression plasmid p35S::FvWRKY71 into ‘Ruegen’ strawberry as described previously66. Explants were placed on medium supplemented with 250 mg/L timetin and 250 mg/L cefotaxime and 10 mg/L kanamycin for selection culture in the dark. When adventitious buds appeared, they were placed under light. The concentration of kanamycin was increased from 10 to 30 mg/L in subsequent subcultures. After ~8 months of selective culture, we obtained transgenic plants and transferred them to the greenhouse, where they were grown under short-day conditions. The positive transgenic strawberry plants were evaluated by PCR to detect DNA levels and RT-qPCR for gene expression analysis.
RNA isolation and expression analysis
To test the expression of representative genes involved in flowering, young leaves of the transgenic plants and wild-type plants were chosen. Total RNA was isolated from three mixed samples using the modified CTAB method as described previously67. Each mixed sample was obtained from six transgenic plants or wild-type plants. Total RNA (1 μg) was reverse-transcribed in a 20-μL reaction using a PrimeScriptTM RT reagent Kit (Takara, Japan). The 10-μL reaction for removing genomic DNA contained 2.0 μL of 5× gDNA Eraser Buffer, 1.0 μL gDNA Eraser, 1 μg RNA, and RNase-free ddH2O to 10 μL. The 20-μL reverse transcription reaction contained 10 μL of the previous reaction product, 1.0 μL PrimeScript RT Enzyme Mix1, 1.0 μL RT Primer Mix, 4.0 μL of 5× PrimeScript Buffer 2, and 4.0 μL of RNase-free ddH2O. The following thermal profile was used for reverse transcription: 42 °C for 5 min, followed by 37 °C for 30 min and 85 °C for 5 s. RT-qPCR was performed in an ABI 7500 system (Applied Biosystems, Foster City, CA, USA). All cDNA samples were diluted with ddH2O (V:V; 1:4). RT-qPCR was performed in a volume of 10 μL containing 5 μL UltraSYBR Green Mixture reagent (ComWin Biotech, Beijing), 0.5 μL cDNA, 1 μL of the primer set, and 3.5 μL ddH2O, and relative expression levels were calculated using the 2–ΔΔCt method, taking the WT plants as a control. Each sample was quantified in triplicate with three biological replicates. The reaction conditions for RT-qPCR consisted of an initial step (50 °C for 2 min, 95 °C for 10 min), followed by 40 cycles (95 °C for 15 s, 60 °C for 1 min) and a denaturing step (95 °C for 15 s, 60 °C for 15 s, 95 °C for 15 s). We performed normalization first with Arabidopsis 18S rRNA as a control and then with strawberry 26S rRNA as a control68. The gene primers used for the RT-qPCR experiments are listed in Supplemental Table S1.
Yeast one-hybrid assay
The coding sequence of FvWRKY71 was obtained through RT-PCR with specific primers from the red fruit of ‘Ruegen’. Then, the fragment was digested with EcoRI and BamHI and inserted into the pGAD424 vector to obtain pAD-FvWRKY71. The promoter regions of FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4 were amplified by PCR from the genomic DNA of the leaves of ‘Ruegen’, verified by sequencing and cloned into the pAbAi vector with the EcoRI and BamHI restriction enzymes. The primers used for the yeast one-hybrid analysis are listed in Supplemental Table S1.
The plasmids proFUL-pAbAi, proSEP1-pAbAi, proAGL42-pAbAi, proLFY-pAbAi, proFPF1-pAbAi, and proSPL4-pAbAi were introduced into the Y1H Gold yeast strain using the PEG/LiAC method61 and grown on SD/-Ura agar plates. After 3 days, we transferred positive yeast to agar plates containing SD/-Ura/+AbA (50–500 μg/L) and conducted screening to determine the minimum inhibitory concentration of AbA in relation to yeast growth. Next, we transformed pAD-FvWRKY71 into yeast strains containing proFUL-pAbAi, proSEP1-pAbAi, proAGL42-pAbAi, proLFY-pAbAi, proFPF1-pAbAi, and proSPL4-pAbAi and plated them in SD/-Leu/and SD/-Leu/+AbA plates. Finally, we observed the growth of the yeast on agar plates. The Yeast Protocols Handbook (Clontech) was used to perform the above yeast experiments.
Dual-luciferase reporter system
Two kilobase sequences of the FvFUL, FvSEP1, FvAGL42, FvLFY, FvFPF1, and FvSPL4 promoters containing W-boxes (TTGACT/C) were amplified from the genomic DNA of the leaves of ‘Ruegen’. The fragments were digested with HindΙΙΙ and BamHI and inserted into the pGreenII0800-LUC vector as reporter plasmids35. The overexpression vector p35S::FvWRKY71 was used as an effector plasmid.
These plasmids were introduced into A. tumefaciens strain GV3101 via the freeze-thaw method. Agrobacterium strain GV3101 carrying the effector plasmid (empty vector or p35S::FvWRKY71) and the specific reporter plasmid (ProFvFUL::LUC, ProFvSEP1::LUC, ProFvAGL42::LUC, ProFvLFY::LUC, ProFvFPF1::LUC, and ProFvSPL4::LUC) was cultured to OD600 = 1.0 at 28 °C. The cells were harvested and resuspended in medium (1 M MgCl2, 100 mM acetosyringone and 1 M MES, pH 5.6). Then, the solution was placed at room temperature without shaking for 2 h and infiltrated into 1-month-old N. benthamiana leaves. The infiltrated tobacco plant was placed in the dark for 24 h and then placed under light for 48 h. We tested luciferase signaling with a living fluorescence imager (Lb985, Berthold, Germany). The transcriptional activity in the infiltrated tobacco leaves indicated by the ratio of LUC to REN was investigated with a dual luciferase reporter gene assay kit (Beyotime, China). Six biological repeats were measured for each sample. The primers used for the LUC/REN activity analysis are listed in Supplemental Table S1.
Accession numbers
AT5G62165 (AGL42), AT1G69120 (AP1), AT5G24860 (FPF1), AT1G65480 (FT), AT5G60910 (FUL), AT5G61850 (LFY), AT5G15800.2 (SEP1), AT3G02310 (SEP2), AT1G24260 (SEP3), AT2G33810 (SPL3), AT1G53160 (SPL4), FvH4_7g28740.1 (AGL42), FvH4_4g29600.1 (AP1), FvH4_2g30770.1 (FPF1), FvH4_6g00090.1 (FT), FvH4_5g13500.1 (FUL), FvH4_5g09660.1 (LFY), FvH4_7g28690.1 (SEP1), FvH4_6g46420.1 (SEP2), FvH4_4g23530.1 (SEP3), FvH4_3g08880 (SPL3), FvH4_3g19650.1 (SPL4), and FvH4_7g12700.1 (SOC1). GenBank: JN172097.1 (FvTFL1),
Supplementary information
Acknowledgements
We thank Yuepeng Han from the Wuhan Botanical Garden of the Chinese Academy of Sciences for kindly providing the pGreenII0800-LUC vector. This work was supported by the National Key R&D Program of China (2019YFD1000200), National Natural Science Foundation of China (31601730), China Postdoctoral Science Foundation (2017M611264), Key R&D and Technology Transfer Program (Z17–0–035), Shenyang Young and Middle-aged Science and Technology Innovation Talents Support Plan (RC190446) and LiaoNing Revitalization Talents Program (XLYC1902069).
Author contributions
Y.L. and J.Z. designed the experiments. Y.S., Y.L., B.W., H.D., and S.Y conducted the experiments. J.Z., H.L., and Y.L. analyzed the data. Y.S., Y.L., Z.Z., and J.Z. wrote and modified the manuscript. All authors involved in this study read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Yingying Lei, Yiping Sun
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00355-4).
References
- 1.Ishiguro R, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 2.Rushton PJ, Somssich IE, Patricia R, Shen QJ. WRKY transcription factors. Plant Signal. Behav. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Bakshi M, Oelmüller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014;9:e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, A. & Roychoudhury, A. WRKY proteins: signaling and regulation of expression during abiotic stress responses Scientific World J.2015, 807560 (2015). [DOI] [PMC free article] [PubMed]
- 5.Zheng Z, et al. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2010;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 6.Birkenbihl RP, Diezel C, Somssich IE. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012;159:266–285. doi: 10.1104/pp.111.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Dong Q, Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185:288–297. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Deyholos M. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 9.Ren X, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, et al. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, et al. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013;74:730–745. doi: 10.1111/tpj.12159. [DOI] [PubMed] [Google Scholar]
- 12.Silke R, Imre ES. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl Acad. Sci. USA. 2010;107:22338–22343. doi: 10.1073/pnas.1016436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo D, et al. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell. 2015;27:3112–3127. doi: 10.1105/tpc.15.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016;171:1427–14429. doi: 10.1104/pp.15.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu D, et al. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- 17.Qiu Y, Yu D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009;65:35–47. [Google Scholar]
- 18.Wu X, et al. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, et al. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018;176:790–803. doi: 10.1104/pp.17.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, et al. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant. 2016;9:1492–1503. doi: 10.1016/j.molp.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y, et al. MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 2013;212:1–9. doi: 10.1016/j.plantsci.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016;85:96–106. doi: 10.1111/tpj.13092. [DOI] [PubMed] [Google Scholar]
- 23.Huang RW, et al. CpWRKY71, a WRKY transcription factor gene of Wintersweet (Chimonanthus praecox), promotes flowering and leaf senescence in Arabidopsis. Int. J. Mol. Sci. 2019;20:5325. doi: 10.3390/ijms20215325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darrow G. The Strawberry: History, Breeding and Physiology. NY: Holt, Rinehart and Winston; 1966. [Google Scholar]
- 25.Rousseau-Gueutin M, et al. Comparative genetic mapping between octoploid and diploid Fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics. 2008;179:2045–2060. doi: 10.1534/genetics.107.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima R, et al. Molecular cloning and expression analyses of FaFT, FaTFL, and FaAP1 genes in cultivated strawberry: their correlation to flower bud formation. Biol. Plant. 2014;58:641–648. [Google Scholar]
- 27.Whitaker VM, et al. A roadmap for research in octoploid strawberry. Hortic. Res. 2020;7:33. doi: 10.1038/s41438-020-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown T, et al. Genetical control of everbearing habit and 3 other characters in varieties of Fragaria vesca. Euphytica. 1965;14:97–112. [Google Scholar]
- 29.Koskela EA, et al. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 2012;159:1043–1054. doi: 10.1104/pp.112.196659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouhu K, et al. The Fragaria vesca homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 represses flowering and promotes vegetative growth. Plant Cell. 2013;25:3296–3310. doi: 10.1105/tpc.113.115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaston A, et al. PFRU, a single dominant locus regulates the balance between sexual and asexual plant reproduction in cultivated strawberry. J. Exp. Bot. 2013;64:1837–1848. doi: 10.1093/jxb/ert047. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, et al. Genome-wide analysis of the expression of wrky family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLoS ONE. 2016;11:e0154312. doi: 10.1371/journal.pone.0154312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei W, et al. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 2016;105:129–144. doi: 10.1016/j.plaphy.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 35.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard A. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 37.Gu X, et al. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013;4:1947. doi: 10.1038/ncomms2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amasino RM, Michaels SD. The timing of flowering. Plant Physiol. 2010;154:516–520. doi: 10.1104/pp.110.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 40.Wang JW. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014;65:4723–4730. doi: 10.1093/jxb/eru246. [DOI] [PubMed] [Google Scholar]
- 41.Mouhu K, et al. Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant. Biol. 2009;9:122. doi: 10.1186/1471-2229-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu JY, Meaux JD. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell. 2014;26:2024–2037. doi: 10.1105/tpc.114.124685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartmann U, et al. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2010;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- 44.Markus S, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 45.Cao S, et al. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CQ, et al. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell. 2014;26:3589–3602. doi: 10.1105/tpc.114.130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawa M, Kay SA. GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickland DP, Hanzawa Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant. 2015;8:983–997. doi: 10.1016/j.molp.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Blázquez MA, et al. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- 50.Ferrándiz C, et al. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 51.Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- 52.Iwata H, et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012;69:116–125. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- 53.Randoux M, et al. Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot. 2012;63:6543–6554. doi: 10.1093/jxb/ers310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rantanen M, et al. Strawberry homolog of TERMINAL FLOWER1 integrates photoperiod and temperature signals to inhibit flowering. Plant J. 2015;82:163–173. doi: 10.1111/tpj.12809. [DOI] [PubMed] [Google Scholar]
- 55.Melzer S, et al. FPF1 modulates the competence to flowering in Arabidopsis. Plant J. 1999;18:395–405. doi: 10.1046/j.1365-313x.1999.00461.x. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi A, Wu ML. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL and APETALA1. Dev. Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker A, Theiben G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 58.Dornelas MC, et al. MADS: the missing link between identity and growth? Trends Plant Sci. 2011;16:89–97. doi: 10.1016/j.tplants.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Pelaz S, et al. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 60.Dorca-Fornell C, Gregis V, Grand. V. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 2011;67:1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edger PP, et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience. 2017;7:gix124. doi: 10.1093/gigascience/gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gietz RD, Schiest RH. High-efficiency yeast transformation using the LiAC/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 65.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2010;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 66.Li WJ, et al. FveRGA1, encoding a DELLA protein, negatively regulates runner production in Fragaria vesca. Planta. 2018;247:941–951. doi: 10.1007/s00425-017-2839-9. [DOI] [PubMed] [Google Scholar]
- 67.Chang L, et al. Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J. Phytopathol. 2010;155:431–436. [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.