Abstract
Red sea bream iridovirus (RSIV) and Viral hemorrhagic septicemia virus (VHSV) are the most important viral marine pathogens in South Korea because RSIV and VHSV infect and cause high mortality rates in major fish species such as Paralichthys olivaceus and Sebastes schlegelii. These viruses can be transmitted both vertically and horizontally, and early diagnosis is imperative. In this research, RSIV and VHSV viral genomes are detected by PCR-lateral flow assay (LFA). PCR-LFA is sensitive, capable of detecting a viral genome at a concentration of 2–200 fg/µL. Development of this detection method is very meaningful because LFA is simple, requiring a minimum of personnel training to perform. Additionally, LFA requires less time than other detection methods and can be an immediate detection tool that is indispensable in preventing rapid viral spread.
Keywords: Red sea bream iridovirus (RSIV), Viral hemorrhagic septicemia virus (VHSV), PCR, Lateral flow assay
Introduction
Viral hemorrhagic septicemia virus (VHSV) is classified in the Novirhabdovirus genus from the Rhabdoviridae family [4]. Various marine and freshwater fish species are known to be infected by VHSV [11]. This virus can cause systemic hemorrhaging in the internal organs, skin, muscle, and other tissues of fish [8]. VHSV can cause major damage in marine and freshwater fish farms [11, 12]. In recent years, fish farms have become commonplace for breeding many fish species, and VHSV is able to be transmitted horizontally through water transfer [1, 8]. Severe VHSV outbreaks have occurred in Europe, the Great Lakes of the United States, and other regions worldwide [1, 5, 16].
Red sea bream iridovirus (RSIV) is one of megalocytiviruses, Viruses in the Iridoviridae family have large dsDNA genomes that can be infectious to diverse vertebrates and invertebrates [15]. Infection with RSIV has been found in freshwater and marine fish species in Korea [13]. Like VHSV, RSIV is transmitted horizontally through water [17], causing a high infection rate [13, 15].
In recent years, molecular biological methods like PCR or other amplification tools and serological methods based on antibodies, for example, enzyme-linked immune-sorbent assays (ELISA), have been used to detect most pathogens or microorganisms quickly [1, 10]. Many molecular biological methods have been developed for detection of marine pathogens [1, 7–10, 14]. Normal PCR is a reliable, classical, molecular biological method for detecting marine pathogens. However, PCR products must be visualized by gel electrophoresis; this can be time-consuming and technologically cumbersome [3, 18].
Lateral flow assay (LFA) is a technology that uses paper-based devices to detect target materials in mixtures, and the results can be displayed within a short time (5–30 min) [22, 23]. PCR-LFA is a method that combines PCR and LFA to detect PCR products rapidly. The PCR products are double-labeled by gene-specific primer sets to generate sandwich assays [19, 20]. Detection is performed by trapping the labeled PCR products on the surface of a lateral flow strip possessing test and control lines [20, 22]. PCR-LFA has several advantages. This methodology is simpler than conventional PCR-gel electrophoresis, requiring minimal personnel training. Also, PCR-LFA allows for point-of-care testing, allowing faster and easier detection of pathogens in the field environment [19–22].
In this study, we developed a molecular biological method that combines PCR and LFA for the rapid and sensitive detection of two marine viruses, RSIV and VHSV.
Materials and methods
Preparation of target genes
The coat protein (CP) is one of the crucial and common proteins in many viruses, hence the CP gene was used for RSIV detection using PCR-LFA [2, 6]. The complete CP genes from a number of RSIV strains have been sequenced, and a conserved region was used for the target gene (Fig. 1, representative reference: NCBI GenBank Accession Number AY310918.1). This gene was synthesized by Macrogen (Seoul, Korea). After synthesis, the RSIV CP gene was cloned into and secured in the pGEM T-easy vector (Promega Inc, Madison, WI, USA). For VHSV, the glycoprotein (G) gene was used for detection using PCR-LFA. A number of G gene sequences from various VHSV strains have been analyzed, and a highly conserved region was used for the target gene (Fig. 1, representative reference: JQ651388.1). This gene was synthesized by Bioneer (Daejeon, Korea), and then cloned into and secured in the pGEM T-easy vector. Partial genes for other 17 viruses [NNV, Nervous necrosis virus (CP gene, KF841612.1); EHNV, Epizootic haematopoietic necrosis virus (CP gene, AY187045.1); IHNV, Infectious haematopoietic necrosis virus (G gene, X89213.1); ISAV, Infectious salmon anemia virus (G gene, NC_006499.1); KHV, Koi herpesvirus (CP gene, DQ177346.1); SAV, Salmon alphavirus (CP gene, JQ799139.1); SVC, Spring viraemia of carp (G gene, NC_002803.1); IHHNV, Infectious hypodermal and haematopoietic necrosis virus (G gene, NC_002190.2); WSSV, White spot syndrome virus (Outer membrane protein gene, AY126666); YHV, Yellow head virus 1 (G gene, EU487200.1); MrNV, Macrobrachium rosenbergii nodavirus (CP gene, AY222840.1); LSNV, Laem-singh virus (RdRP gene, DQ127905.1); CCV, Channel catfish virus (CP gene, NC_001493.2); GIV, Grouper iridovirus (CP gene, KX284838); AbHV, Abalone herpesvirus (Hypothetical protein gene, NC_018874.1); OsHV, Osteroid herpesvirus 1 (G gene, NC_005881.2); and BP, Baculovirus penaei (Unknown protein gene, DQ496179.1)] were also synthesized by Macrogen (Seoul, Korea) and cloned into the the pGEM T-easy vector for using in sensitivity tests as templates.
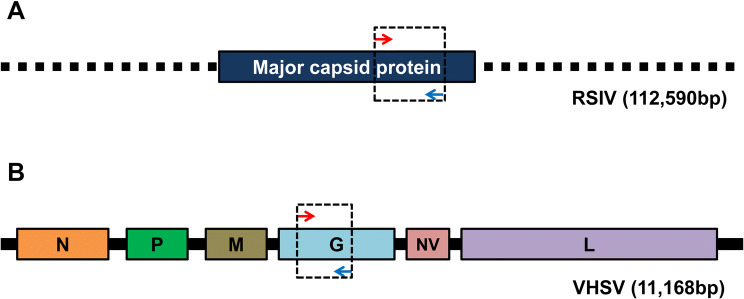
Fig. 1.
Genome structure of two marine viruses and position of primers for diagnosis of each virus. a RSIV partial genome structure and position of primers. b VHSV genome structure and position of primers
Plasmid DNA isolation
E. coli colonies containing RSIV and VHSV target genes were cultured in LB broth with 1% of ampicillin at 37 °C in a vibrating incubator for 15 h. After a cultured cell was obtained, its colonies were extracted by AccuPrep® Nano-Plus Plasmid Mini Extraction Kit (Bioneer). Secured plasmid-containing elution buffer was used for the PCR and PCR-LFA template.
Primer design and optimization of PCR-LFA conditions
RSIV CP and VHSV G genes were amplified by specific primer sets (Table 1). The reaction mixture for the PCR was prepared by mixing the following reagents: 10 µL of AccuPower® PCR PreMix & Master Mix (Bioneer, Korea), 7 µL of distilled water, 1 µL of template DNA, and 10 pmol of each primer, resulting in a total of 20 µL. The cycling conditions for PCR were: initial denaturation at 95 °C for 5 min; 35 cycles at 95 °C for 30 s, held at 55 °C for 30 s, then 72 °C for 30 s; and a final extension at 72 °C for 5 min. The PCR products were electrophoresed in 1% agarose gel containing ethidium bromide (EtBr) for detection of the amplified target size. After agarose gel electrophoresis, the PCR products were confirmed by sequencing. The sequencing was performed by Macrogen, and the sequencing results were compared with known sequences of RSIV and VHSV genes in NCBI GenBank.
Table 1.
Marine virus-specific primers used in this study
| Primer name | Primer sequence | Accession no. |
|---|---|---|
| RSIV F | 5′-FITC-aacgtaaccagtgggttca tc-3′ | AY310918.1 |
| RSIV R | 5′-Biotin-atgggcaaattaaggtaggcg-3′ | |
| VHSV F | 5′-FITC-aagggcttggtctctgtcc-3′ | JQ651388.1 |
| VHSV R | 5′-Biotin-aggcagtgatatcactgtgg-3′ |
LFA was performed with Genline Hybridetect 1 strips (Milenia Biotec, Gießen, Germany). This strip contains gold nanoparticle (AuNP)-conjugated anti-FITC Antibody (Ab) for fluorescence, immovable anti-Biotin ligand to combine with biotin for the test line, and immovable anti-Rb Ab to combine with AuNP::Ab for the control line of reaction. For PCR-LFA, forward primers were labeled with FITC, and reverse primers were labeled with biotin at 5’ ends. After PCR, the PCR products were double-labeled with FITC and biotin; and used for LFA testing to confirm results. The PCR products for the LFA were prepared by adding 1 μL of PCR products to 99 μL of Dipstick Assay Buffer (Milenia Biotec, Gießen, Germany). After mixing, the strip was dipped into the mixture and incubated at room temperature for 10 min. The results of the PCR-LFA were confirmed by examining the test and control lines with the naked eye (Fig. 2).
Fig. 2.
Principle of PCR-LFA
PCR-LFA reactions from fish samples
Olive flounder (Paralichthys olivaceus) samples were provided by Korean Institute of Ocean Science and Technology (KIOST, Busan, Korea) and used for application of introduced diagnosis methods in fish tissue samples. Viral genomes were isolated by viral gene-spin™ viral DNA/RNA extraction kit (iNtRON Biotechnology, Seongnam, Korea). RSIV viral DNA was used as template for direct PCR-LFA diagnosis using aforementioned PCR conditions. For VHSV, viral RNA was used as template for SuPrimeScript RT-PCR Premix (GeNetbio, Daejeon, Korea). The reaction mixture for VHSV RNA was prepared with the following reagents: 7 µL of distilled water, 1 µL of template DNA, and 10 pmol of each primer in a final 20 µL of reaction mixture. VHSV cDNA synthesis occurred during incubation of the mixture at 50 °C for 30 min. The remainder of the process was the same as for the conventional PCR method. The results are shown with both agarose gel electrophoresis and lateral flow assay.
Results
Specificity of primer sets in PCR results
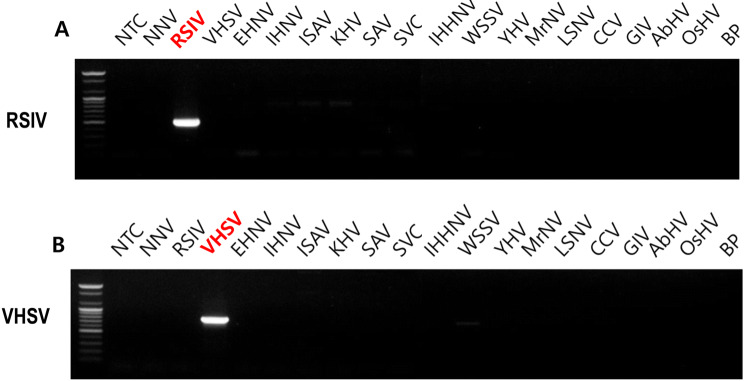
The amplified PCR products were detected on 1% agarose gel. DNA bands were visualized by EtBr and were confirmed by expected band size. The expected target band was 319 bp for RSIV and 302 bp for VHSV detection. Clear, appropriate-sized bands were observed for both RSIV and VHSV (Fig. 3). These PCR results demonstrate that the primer sets have specificity for each target virus gene, have no cross-reactivity to 18 other marine virus genes, and are appropriate for future research.
Fig. 3.
Specificity of diagnose of RSIV and VHSV among 19 marine virus genes with conventional PCR and agarose gel electrophoresis. a RSIV detection with RSIV specific primer sets. b VHSV detection with VHSV specific primer sets. NNV, Nervous necrosis virus; EHNV, Epizootic haematopoietic necrosis virus; IHNV, Infectious haematopoietic necrosis virus; ISAV, Infectious salmon anemia virus; KHV, Koi herpesvirus; SAV, Salmon alphavirus; SVC, Spring viraemia of carp; IHHNV, Infectious hypodermal and haematopoietic necrosis virus; WSSV, White spot syndrome virus; YHV, Yellow head virus 1; MrNV, Macrobrachium rosenbergii nodavirus; LSNV, Laem-singh virus; CCV, Channel catfish virus; GIV, Grouper iridovirus; AbHV, Abalone herpesvirus; OsHV, Osteroid herpesvirus 1; and BP, Baculovirus penaei
Specificity of PCR-lateral flow assay
To test PCR-LFA, 5′-FITC-labeled forward primers and 5′-biotin-labeled reverse primers were used. The same PCR method as described above was used for PCR-LFA, and 1 µL of PCR products was mixed with 99 µL of dipstick assay buffer. The results were visible 10 min after the LFA strip was dipped into the mixture (Fig. 4). The PCR-LFA results were as reliable as those of conventional PCR.
Fig. 4.
Specificity of diagnose of RSIV and VHSV among 19 marine virus genes with PCR-LFA. a RSIV detection with RSIV specific primer sets and LFA strips. b VHSV detection with VHSV specific primer sets and LFA strips. NNV, Nervous necrosis virus; EHNV, Epizootic haematopoietic necrosis virus; IHNV, Infectious haematopoietic necrosis virus; ISAV, Infectious salmon anemia virus; KHV, Koi herpesvirus; SAV, Salmon alphavirus; SVC, Spring viraemia of carp; IHHNV, Infectious hypodermal and haematopoietic necrosis virus; WSSV, White spot syndrome virus; YHV, Yellow head virus 1; MrNV, Macrobrachium rosenbergii nodavirus; LSNV, Laem-singh virus; CCV, Channel catfish virus; GIV, Grouper iridovirus; AbHV, Abalone herpesvirus; OsHV, Osteroid herpesvirus 1; and BP, Baculovirus penaei
Sensitivity of PCR-lateral flow assay
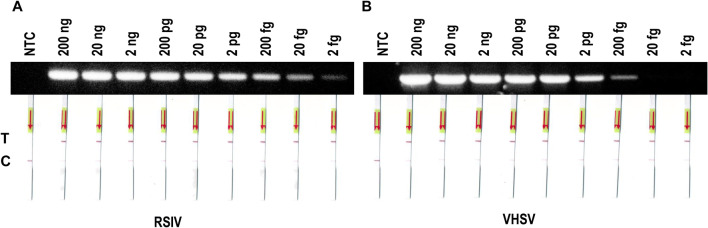
Next, tenfold serial dilutions from 200 ng to 2 fg of each viral gene-containing plasmid template were prepared. These dilutions were used for comparison of PCR-LFA sensitivity to that of conventional PCR. Conventional PCR could detect 2 fg of RSIV template and 200 fg of VHSV template. PCR-LFA could detect 2 fg of both RSIV and VHSV templates (Fig. 5). These results indicate that PCR-LFA with these two viruses is at least as sensitive as conventional PCR, and depending on the situation, may be more sensitive.
Fig. 5.
Sensitivity of diagnose using PCR products with conventional agarose gel electrophoresis and LFA. a Sensitivity of RSIV diagnoses. b Sensitivity of VHSV diagnoses. T, test line and C, control line
Diagnosis in virus-infected fish
To check diagnostic applications in fish tissues infected with viruses, healthy, RSIV-infected, and VHSV-infected olive flounder were used. Muscle tissues from each fish were obtained, and viral DNA and RNA were isolated. PCR (for RSIV) and RT-PCR (for VHSV) were performed and agarose gel electrophoresis and LFA were followed for visualization. Overall, these results demonstrate that diagnosis using PCR-LFA is reliable not only in viral gene-containing plasmids, but also in infected fish samples. According to results, the reactions were confirmed only in infected samples for both viruses (Fig. 6).
Fig. 6.
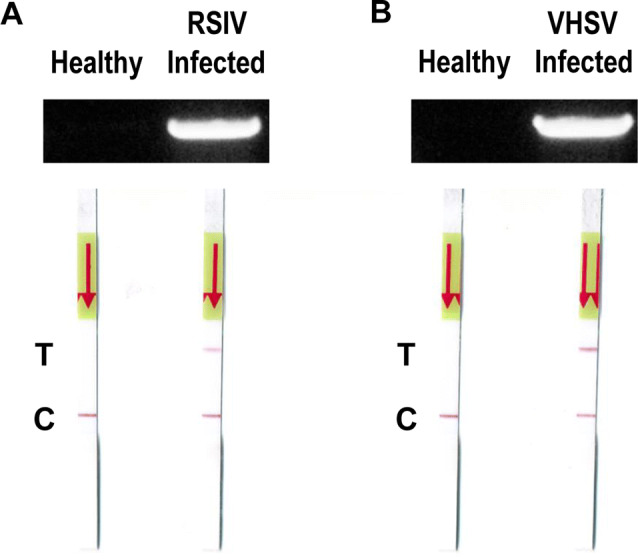
Validation of PCR-LFA diagnosis with genome derived from healthy and virus-infected fish. a Diagnose using PCR and PCR-LFA with DNA derived from healthy and RSIV-infected fish. b Diagnose using PCR and PCR-LFA with RNA derived from healthy and VHSV-infected fish. T, test line and C, control line
Discussion
Viral infections by RSIV and VHSV can cause a major problem to the marine aquaculture industry and significant economic losses. Therefore it is important to diagnose viral diseases rapidly to prevent further disease transmission [6]. Though many methods have been developed to diagnose marine viruses and other pathogens, but there is no on-site diagnostic assay for RSIV and VHSV. In this study, a combined method called PCR-LFA was used for diagnostic assay for RSIV and VHSV in synthesized genes and also fish samples. This PCR-LFA combined high sensitivity of PCR, simplicity and rapidness of LFA. According to our results, low amounts of both RSIV and VHSV viral genes were detectable by the PCR-LFA method; the sensitivity of PCR-LFA is same or higher than conventional PCR and agarose gel electrophoresis (Fig. 5).
In conclusion, PCR–LFA methods for RSIV and VHSV were developed for easy, fast, and point-of-care diagnosis. Based on our results, PCR-LFA can be used as a reliable and sensitive method for detection of two marine virus, RSIV and VHSV, infections in both field and laboratory settings.
Acknowledgement
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (NRF-2017M3A9E407275).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Al-Hussinee L, Lumsden J. Detection of VHSV IVb within the gonads of Great Lakes fish using in situ hybridization. Dis Aquat Organ. 2011;95(1):81–86. doi: 10.3354/dao02338. [DOI] [PubMed] [Google Scholar]
- 2.Beachy RN, Loesch-Fries S, Tumer NE. Coat protein-mediated resistance against virus infection. Annu Rev Phytopathol. 1990;28:451–474. doi: 10.1146/annurev.py.28.090190.002315. [DOI] [Google Scholar]
- 3.Duncan R, Kourout M, Grigorenko E, Fisher C, Dong M. Advances in multiplex nucleic acid diagnostics for blood-borne pathogens: promises and pitfalls. Expert Rev Mol Diagn. 2016;16(1):83–95. doi: 10.1586/14737159.2016.1112272. [DOI] [PubMed] [Google Scholar]
- 4.Einer-Jensen K, Ahrens P, Forsberg R, Lorenzen N. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J Gen Virol. 2004;85(5):1167–1179. doi: 10.1099/vir.0.79820-0. [DOI] [PubMed] [Google Scholar]
- 5.Emmenegger EJ, Moon CH, Hershberger PK, Kurath G. Virulence of viral hemorrhagic septicemia virus (VHSV) genotypes Ia, IVa, IVb, and IVc in five fish species. Dis Aquat Organ. 2013;107(2):99–111. doi: 10.3354/dao02671. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann B, Beer M, Schütze H, Mettenleiter TC. Fish rhabdoviruses: molecular epidemiology and evolution. In: Fu ZF, editor. The World of rhabdoviruses. Berlin: Springer; 2005. pp. 81–117. [DOI] [PubMed] [Google Scholar]
- 7.Hwang J, Suh S-S, Park M, Cho S-H, Lee S, Lee T-K. Rapid and sensitive detection of iridovirus by loop-mediated isothermal amplification (LAMP) Afr J Microbiol Res. 2015;9(6):382–387. doi: 10.5897/AJMR2014.7013. [DOI] [Google Scholar]
- 8.Hwang J, Park SY, Suh S-S, Park M, Lee S, Lee T-K. Efficient detection of pathogen virus in sand dabs, Paralichthys olivaceus using loop-mediated isothermal amplification (LAMP) Acta Oceanol Sin. 2016;35(8):44–50. doi: 10.1007/s13131-016-0889-7. [DOI] [Google Scholar]
- 9.Hwang JY, Lee S, Priyathilaka TT, Yang H, Kwon H, Kwon MG, et al. Phylogenetic analysis and duplex RT-PCR detection of viral hemorrhagic septicemia virus in olive flounder (Paralichthys olivaceus) from Korea. Aquaculture. 2018;484:242–249. doi: 10.1016/j.aquaculture.2017.11.038. [DOI] [Google Scholar]
- 10.Inada M, Handayani CR, Kawato Y, Mekata T, Yuasa K, Miwa S. Modification of PCR program for detection of infectious myonecrosis virus. Fish Pathol. 2016;51(4):210–212. doi: 10.3147/jsfp.51.210. [DOI] [Google Scholar]
- 11.Kim R, Faisal M. Experimental studies confirm the wide host range of the Great Lakes viral haemorrhagic septicaemia virus genotype IVb. J Fish Dis. 2010;33(1):83–88. doi: 10.1111/j.1365-2761.2009.01093.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim W-S, Jung S-J, Kim J-O, Kim D-W, Kim J-H, Oh M-J. Genetic positioning of Korean viral hemorrhagic septicemia virus (VHSV) from cultured and wild marine fishes. J Fish Pathol. 2011;24(1):1–9. doi: 10.7847/jfp.2011.24.1.001. [DOI] [Google Scholar]
- 13.Kim K, Hwang S, Cho M, Jung S, Kim Y, Jeong H. A natural infection by the red sea bream iridovirus-type Megalocytivirus in the golden mandarin fish Siniperca scherzeri. J Fish Dis. 2018;41(8):1229–1233. doi: 10.1111/jfd.12815. [DOI] [PubMed] [Google Scholar]
- 14.Kim HJ, Cuenca A, Olesen NJ. Validation of a novel one-step reverse transcription polymerase chain reaction method for detecting viral haemorrhagic septicaemia virus. Aquaculture. 2018;492:170–183. doi: 10.1016/j.aquaculture.2018.03.047. [DOI] [Google Scholar]
- 15.Kurita J, Nakajima K, Hirono I, Aoki T. Polymerase chain reaction (PCR) amplification of DNA of red sea bream iridovirus (RSIV) Fish Pathol. 1998;33(1):17–23. doi: 10.3147/jsfp.33.17. [DOI] [Google Scholar]
- 16.Lumsden JS, Morrison B, Yason C, Russell S, Young K, Yazdanpanah A, et al. Mortality event in freshwater drum Aplodinotus grunniens from Lake Ontario, Canada, associated with viral haemorrhagic septicemia virus, Type IV. Dis Aquat Organ. 2007;76(2):99–111. doi: 10.3354/dao076099. [DOI] [PubMed] [Google Scholar]
- 17.Oh S-Y, Oh M-J, Nishizawa T. Potential for a live red seabream iridovirus (RSIV) vaccine in rock bream Oplegnathus fasciatus at a low rearing temperature. Vaccine. 2014;32(3):363–368. doi: 10.1016/j.vaccine.2013.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Poddar SK. Influenza virus types and subtypes detection by single step single tube multiplex reverse transcription-polymerase chain reaction (RT-PCR) and agarose gel electrophoresis. J Virol Methods. 2002;99(1):63–70. doi: 10.1016/S0166-0934(01)00380-9. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Chen J, Li J, Xu Y, Jin H, Xu N, et al. Novel approach based on one-tube nested PCR and a lateral flow strip for highly sensitive diagnosis of tuberculous meningitis. PLoS ONE. 2017;12(10):e0186985. doi: 10.1371/journal.pone.0186985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taboada L, Sánchez A, Pérez-Martín RI, Sotelo CG. A new method for the rapid detection of Atlantic cod (Gadus morhua), Pacific cod (Gadus macrocephalus), Alaska pollock (Gadus chalcogrammus) and ling (Molva molva) using a lateral flow dipstick assay. Food Chem. 2017;233:182–189. doi: 10.1016/j.foodchem.2017.04.087. [DOI] [PubMed] [Google Scholar]
- 21.Tu P-A, Shiu J-S, Lee S-H, Pang VF, Wang D-C, Wang P-H. Development of a recombinase polymerase amplification lateral flow dipstick (RPA-LFD) for the field diagnosis of caprine arthritis-encephalitis virus (CAEV) infection. J Virol Methods. 2017;243:98–104. doi: 10.1016/j.jviromet.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Wu W, Zhao S, Mao Y, Fang Z, Lu X, Zeng L. A sensitive lateral flow biosensor for Escherichia coli O157: H7 detection based on aptamer mediated strand displacement amplification. Anal Chim Acta. 2015;861:62–68. doi: 10.1016/j.aca.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 23.Yetisen AK, Akram MS, Lowe CR. Based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13(12):2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]