Abstract
Hepatitis C virus (HCV) remains a global public health problem with high prevalence rates and chronicity of infection. Present work aimed to describe the main mutations in the NS3 region of the HCV genome related to the resistance of patients to the currently available direct-acting antivirals (DAAs). To guide the study description, the preferred items in the PRISMA protocol for systematic review were used. The data collected were HCV genotypes and subtypes and mutations in HCV NS3, general and stratified by continent. The 10 papers selected for this systematic review reported studies in seven countries, on three continents, and generated data of 2937 patients. The most frequent HCV subtype was 1a. Prevalence of genotypes suggested that there were few demographic regions reached by the studies, since there were regional variations in the type of genotypes reported in the available bibliographies. Of the total study population, 35.3% (n = 1037) had mutations in the NS3 gene region of HCV, suggesting a high rate of resistance to DAAs and a low sustained virologic response among those who used some therapeutic option. Ten major mutations were identified: Q80K, V170I, S122G, V36L, T54S, D168Q, A156S, Q80G, S122R, and V55A. The Q80K mutation was the highlight of the study, appearing not only with greater representativity (61.6%) but also as the only one described in the three continents analyzed. This systematic review reinforces the need to carry out more studies of detection of these mutations to fill in all information gaps that might help in optimization of treatment.
Keywords: Hepatitis C virus, NS3 gene, HCV subtype, Q80K mutation, Direct-acting antivirals
Introduction
The hepatitis C virus (HCV) has been known since 1975, when researchers concluded that most cases of post-transfusion hepatitis were not caused by infections of hepatitis A or hepatitis B viruses but by those of another unknown infectious agent [6]. In 1989, Choo et al. [3] decoded the complementary DNA of the HCV RNA using samples from experimentally infected chimpanzees. In the years that followed, there was an evolution of HCV studies that allowed for the association of the virus with liver diseases, including hepatocellular carcinoma, and the development of diagnostic tests and effective treatments for infected patients [3].
Currently, the high prevalence of HCV infections makes the disease a global public health problem. In 2015, it was estimated that 1% of the worldwide population lived with the disease, representing approximately 71 million infected individuals. Differences in the disease distribution among the various countries in the world occur mainly because the increase in prevalence is driven by low income, health professional negligence, and injection drug use [24].
In Brazil, the current prevalence of HCV is 1.6%, a rate similar to that of other Latin American countries, such as Argentina, Venezuela, Mexico, and Peru [21]. There are many regional variations in the types of HCV genotypes identified. In Brazil, genotype 1 is predominant (74.3%), followed by genotype 3 (14.2%) and genotype 2 (10.4%). The remaining 1.1% of genotypes are represented by a mixture of genotypes 4, 5, and 6 [24]. It is very important to know these genotype variations since they are responsible for the variability in the disease chronicity and impact the treatment response [21].
HCV belongs to the genus Hepacivirus of the family Flaviviridae. Its genome consists of a single-stranded, positive-sense RNA with a single open reading frame (ORF) that is 9400 nucleotides in length and flanked at the ends by 30 or 50 untranslated regions known as 5′ non-translated region (NTR) and 3′ NTR [28]. Translation of the virus ORF region produces a polypeptide that, after cleavage, produces structural proteins (CORE, E1, and E2), a transmembrane protein (p7), and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) [18].
Elucidation of the HCV genomic structure is key to understanding the virus life cycle for allowing the further identification of effective drugs to fight hepatitis and the existence of mutations that give rise to resistance to these drugs. The treatment of HCV infection aims to achieve a sustained virologic response (SVR), which corresponds to the persistence of undetectable viremia (viral RNA) at 6 months after the end of treatment [18]. The main targets of the drugs are the nonstructural proteins, because they are involved in polyprotein processing and viral RNA replication. Treatment is expected to decrease the incidence of chronic hepatitis C and improve the quality of life of infected patients [34].
Following the evolution and development of such new direct-acting antivirals (DAAs), the main concerns have been about the therapeutic responses presented by the different genotypes and subtypes of the virus, the gene mutations that cause drug resistance, and the high costs of the drugs. The resistance toward DAAs appears primarily because of amino acid substitutions that produce conformational changes in the NS3, NS5A and NS5B proteins, thereby preventing the drug from interacting with its protein target [16], which causes the virus to be resistant to treatment and the SVR rate to decrease [14, 15, 32]. These resistance-associated variants (RAVs) of HCV have been identified during the virus life cycle or even during DAA therapy, presenting different occurrence rates according to the patient’s genotype [33] and association with high resistance to different protease inhibitors [23].
Because of the virus characteristics and the current state of the disease, as well as the fact that the amino acid composition at different drug resistance sites reduces treatment effectiveness, information on mutation patterns and resistance to antiviral agents is also increasingly needed. In addition, such information is important for guiding decisions about the therapeutic combination strategies to be used for each genotype, and is relevant to actions that can assist patients who develop resistance to DAAs.
Although studies have evaluated the mutations related only to drugs, it is necessary to analyze the effects of DAAs above virus genotypes. The effectiveness of the DAAs therapies revealed greater differences between the genotypes and as known, HCV genotype frequencies vary geographically. The emergence of antiviral resistance and suboptimal activity of the DAAs therapies against diverse HCV genotypes associated to the high cross resistance among DAAs hinders the effective control of HCV spread worldwide. Therefore, the goal of this systematic review was to identify the major mutations in the NS3 region of the HCV genome that may be related to the resistance of patients to the DAAs currently used as therapeutic options. Detection of these mutations might help to select the most optimized treatment option.
Materials and methods
Systematic review attempts to answer a specific research question objectively and impartially. For that, it uses systematic and defined methods a priori in the identification and selection of studies, data extraction and analysis of results. Its implementation consists of a set of steps to be rigorously followed, with strict inclusion criteria, as follows: research question definition and formulation; search of studies in different sources that address the question to be investigated; critical evaluation of studies based on well-defined inclusion and exclusion criteria; collection of data from each study and construction of a database; analysis and data presentation; data interpretation and calculation of results; and review improvement and updating [4]. To guide the study description, the preferred items in the PRISMA (preferred reporting items for systematic reviews and meta-analyses) protocol for systematic review were used [20]. The protocol is registered in the PROSPERO (prospectively registered systematic reviews in health and social care) registry.
Search and retrieval of studies
The first step was to define and formulate the study question, following the anagram PICOS, where “P” represents the population studied (i.e., the patients infected with HCV), “I” represents the intervention (i.e., the treatment with DAAs described in the Clinical Protocol and Therapeutic Guidelines of the Ministry of Health, Brazil), “C” represents the comparison (i.e., the presence of different gene mutations in the NS3 region of the virus genome), “O” represents the outcome (i.e., the identification of these mutations described in this NS3 region), and “S” represents the type of study (i.e., the experimental ones).
Later, in April 2018, the articles were searched from three scientific databases (PubMed Central, Web of Science, and Scopus), without determination of publication date interval, to select papers that addressed the presence of mutations in the NS3 region of HCV associated with resistance to DAAs. The following descriptors were used: “hepatitis C,” “mutation,” “NS3,” and “resistance.” This first search identified 2627 articles. Given this high number, we chose to include two other descriptors (viz., “PCR” and “RAV”) to delimit the number of articles and decrease the bias. The new search identified 53 relevant articles.
Study selection according to inclusion and exclusion criteria
Some inclusion and exclusion criteria were used in the critical evaluation of each article found. To make the process more efficient and to decrease the heterogeneity among the studies, the evaluation was divided into three phases: Filter 1, Filter 2, and Filter 3.
In the Filter 1 phase, the title, abstract, and keywords of each article were read. The inclusion criteria were patients infected with HCV and experimental studies. The exclusion criteria were evaluations of other components (e.g., CD4 + cells) and literature reviews. As a result, 24 of the 53 articles remained to be evaluated in the second filter.
The Filter 2 phase involved reading the introduction and conclusion. The inclusion criteria were patients infected only with HCV and treatment-naive patients. The exclusion criteria were co-infected patients or evaluations performed in patients with other diseases, such as hemophilia. Following this phase, 15 articles remained to be evaluated in the final filter.
In the Filter 3 phase, complete reading of the article was done. The inclusion criteria of this last phase were the specific parameters of the analytical methodology used in each study; namely, PCR and sequencing. The exclusion criteria were poorly described prevalence data and/or unclear information. As a result, 10 articles remained [5, 10, 11, 14, 15, 22, 27, 29, 31, 32] to be analyzed after data tabulation.
References from revision papers and consensus were manually collected in order to ensure the inclusion of all relevant papers. Study quality was assessed using the Grading of Recommendations, Assessment, Development, and Evaluations (GRADE). The quality of evidence of the studies was classified into four categories: high, moderate, low, or very low quality. We also analyzed the influence of possible conflicts of interest and any information on ethical approval of the studies. No contact was made with clinical investigators in order to verify possible research in progress.
Data extraction
Between April 7 and 15, 2018, data collection from the 10 articles was performed independently by three researchers in order to construct a database to identify possible differences among the data and minimize errors in their analysis. The information tabulated in the database consisted of the scientific database used, first author, year of publication, country of study, total number of patients in the study, name of mutation found, percentage of the mutation found, and HCV genotype carried by the patients.
Statistical analysis
Descriptive statistics were calculated for the HCV genotypes and subtypes and stratified by continent. In addition, the types of mutations in the NS3 region were determined for clustering analysis and stratification by continent. Calculations of absolute and relative percentage frequencies were done.
Results
Literature search results
For this systematic review, 2627 articles were initially found in scientific databases. After using the descriptors “hepatitis C,” “mutation,” “NS3,” “resistance,” “PCR,” and “RAV,” this number dropped to 53 articles. Of these, 43 were discarded because they did not meet the study inclusion criteria. Therefore, a total of 10 articles published between 2013 and 2018 that identified gene mutations in the NS3 region of HCV were finally chosen for the systematic review.
General description of the data
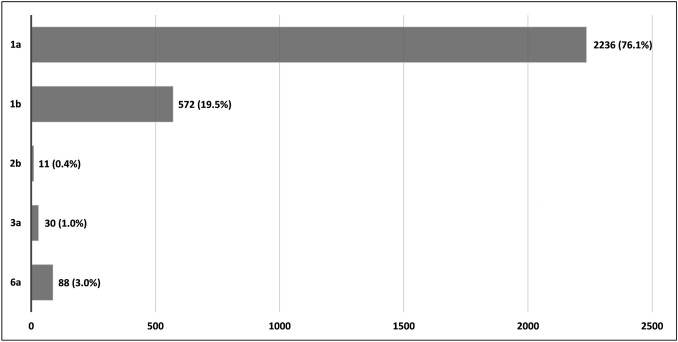
The 10 selected articles reported studies in seven countries from three continents, generated data of 2937 patients. Of the 2937 patients analyzed, 2236 (76.1%) carried HCV subtype 1a, 572 (19.5%) carried subtype 1b, 11 (0.4%) carried subtype 2b, 30 (1.0%) carried subtype 3a, and 88 (3.0%) carried subtype 6a (Fig. 1). Of the total patients (n = 2937), 1037 (35.3%) presented mutations in the NS3 region of the HCV genome. Of the 1037 patients that presented mutations, 514 (49.6%) were from the European, 345 (33.3%) from the Asian, and 178 (17.2%) from the American continents.
Fig. 1.
Distribution of different hepatitis C virus subtypes among 2937 infected patients. Systematic review, 2013–2018
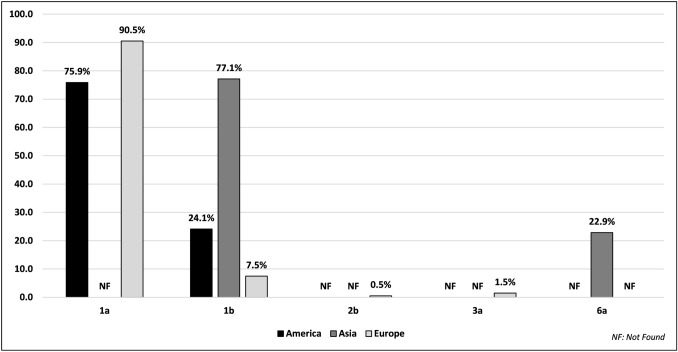
For the American continent, the prevalence of subtypes 1a and 1b were 75.9% and 24.1%, respectively. For the European continent, subtypes 1a, 1b, 2b, and 3a occurred at a frequency of 90.5%, 7.5%, 0.5%, and 1.5%, respectively. On the Asian continent, the prevalence of subtypes 1b and 6a was 77.1% and 22.9%, respectively (Fig. 2).
Fig. 2.
Distribution of different hepatitis C virus subtypes among 2937 infected patients, stratified by continent. Systematic review, 2013–2018
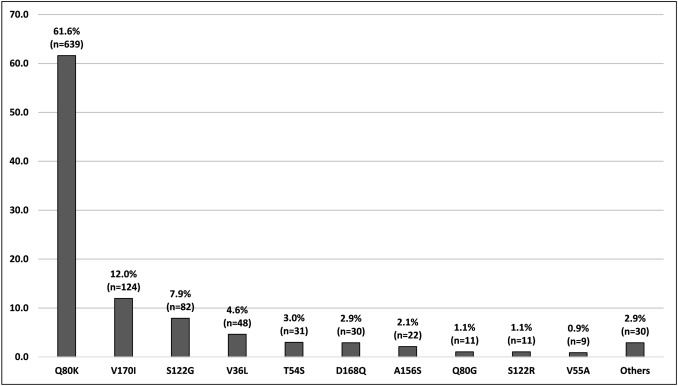
Among the 1037 patients who presented genetic alterations in the NS3 region of HCV, 10 main mutations were found: Q80K, V170I, S122G, V36L, T54S, D168Q, A156S, Q80G, S122R, and V55A. The other mutations that presented with low frequencies (i.e., Q80R, T54A, D168E, and V36M) or that were associated (e.g., Q80K/R) for a better understanding were classified in this systematic review as “others.” The most frequent mutations were Q80K (61.6%), followed by V170I (12.0%) and S122G (7.9%) (Fig. 3).
Fig. 3.
Distribution of the mutation types found in the 1037 HCV-infected patients who presented alterations in the NS3 region of the virus genome. Systematic review, 2013–2018
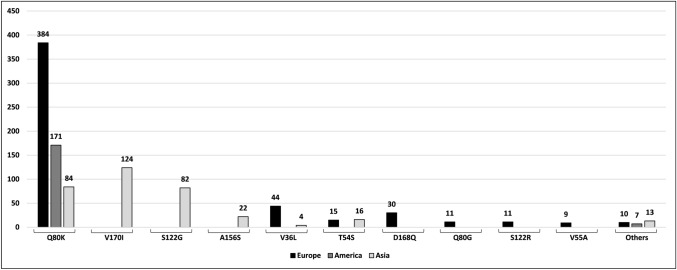
Analysis of the geographic distribution of these mutations present in the 1037 infected patients (Fig. 4) revealed that Q80K was the most representative, since it was the only one described on all three continents analyzed. The V170I, S122G, and A156S mutations were identified exclusively on the Asian continent, whereas the D168Q, Q80G, S122R, and V55A mutations were identified exclusively on the European continent. The V36L and T54S mutations were identified in Europe and Asia, while V36L was found mostly in Europe and T54S was found in the same proportion on both continents. The other poorly described mutations grouped into “others” were found on all three continents.
Fig. 4.
Frequency of each mutation type on each continent. Systematic review, 2013–2018
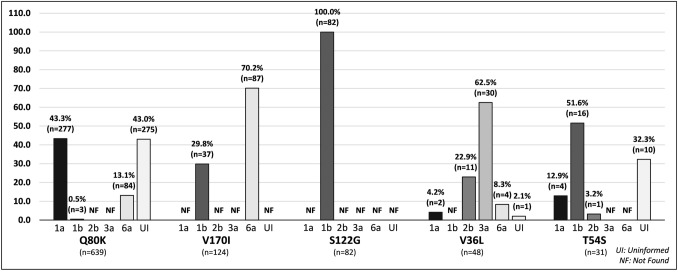
Among the 10 major mutations identified in this study, Q80K mutation was found in subtypes 1a, 1b and 6a at a frequency of 43.3%, 0.5% and 13.1%, respectively (Fig. 5). V170I mutation appeared in subtypes 1b (29.8%) and 6a (70.2%). The presence of S122G mutation was identified in the subtypes 1b (100%) and V36L was presented in subtypes 1a (4.2%), 2b (22.9%), 3a (62.5%) e 6a (8.3%). Finally, T54S mutation was identified in subtypes 1a (12.9%), 1b (51.6%) and 2b (3.2%) (Fig. 5). The Q80K, V36L and T54S mutations had a frequency of 43%, 2.1% and 32.3%, respectively, in uniformed subtypes (Fig. 5).
Fig. 5.
Frequency of each mutation type by viral genotype. Systematic review, 2013–2018
Discussion
For many years, the only treatment option available for hepatitis C was interferon-alpha (IFNα), a therapy that had little effectiveness because of its low SVR rates (ranging from 10 to 15%) in treated patients [34]. Subsequently, a double therapy that combined pegylated IFNα with ribavirin was made available. Despite being a more efficient therapeutic option by providing a cure rate of 45–60% in patients with genotypes 1 and 4 and 70–80% in patients with genotypes 2 and 3, it had many adverse reactions [35].
For these and other reasons, the development of more effective antiretroviral drugs is important. In 2011, the first generation of DAAs, which act primarily on some of the nonstructural proteins of the virus, including the NS3 region, were released and had higher cure rates; for example, ranging from 65 to 75% for genotype 1. After 2 years, in 2013, the second generation of DAAs brought new benefits to patients because they were less toxic and more effective, increasing the SVR and raising cure rates to 90–100% [35].
After the evolution and development of DAAs, the main concerns have been about the therapeutic responses presented by different subtypes of the virus and the gene mutations that cause resistance to the drugs. The pathogenesis of the virus, and the regional variations of its genotypes and genetic variability, characterized by amino acid changes within target proteins, are very important determinants of the effectiveness of DAAs [13, 16, 24, 35].
Our systematic literature review included all articles found in the PubMed Central, Web of Science, and Scopus databases, from the descriptors selected according to the topic of interest. A total of 2627 articles were found on the presence of mutations in the HCV NS3 region that were associated with low rates of SVR in infected patients. After applying the inclusion and exclusion criteria, 10 articles were finally used in this systematic review.
This study provides information on NS3 region mutations on three continents and seven countries. Given that HCV infection affects about 71 million people around the world, the need for research on this topic in more demographic regions is obvious [25].
The systematic review revealed that most of the studies were conducted in Asia and Europe, and less frequently on the American continent. This result was compatible with the prevalence of HCV infection in the general population described by region by the World Health Organization in 2017, which presented a higher prevalence of the disease in the Asian and European continents than in the Americas [25]. On the other hand, a 2015 research on the global prevalence of DAA resistance associated with variants, using data published in GenBank, showed that the presence of at least one resistance variant occurred mainly in Asia, followed by Africa, America, and Europe, confirming the importance of further studies on this topic on the American continent [2].
The most frequent HCV subtype among the patients analyzed in this study was 1a (at 76.1%), followed by 1b, 6a, 3a, and 2b (at 19.5%, 3.0%, 1.0%, and 0.4%, respectively). These data are consistent with the available epidemiologic references, which show that the most frequent genotype worldwide is 1 (49.1%), followed by 3 (17.9%), 4 (16.8%), 2 (11.0%), and 5 and 6. Studies also described a greater presence of genotypes in the following order: 1, 3, 2, 6, 4, and 5 in Asia; 1, 3, 2, 4, 5, and 6 in Europe; and 1, 3, 2, 4, 6 and 5 in the Americas. That is, genotype 1 always presented with the highest prevalence [24], as shown in the data of this systematic review.
Although HCV genotype 1 and 3 infections are the most found globally, the other genotypes may be prevalent in some countries because of the genetic variability of the virus [19, 24]. The fact that genotype 6 was the third most present in this systematic review can be explained by the inclusion of a study that described the presence of mutations in a representative number of patients of this genotype in China [13, 17, 24].
Ten major mutations were found distributed among the three continents. The presence of mutations V170I, S122G, V36L, T54S, A156S, S122R, and V55A was well cited in studies and related to resistance to DAAs. Nevertheless, the studies analyzed in this systematic review did not offer sufficient information to compare the presence of the mutations with the patient’s genotype/subtype and to associate their presence with resistance to some specific DAA [1, 2, 8, 14–16, 26, 30].
On the other hand, the D168Q and Q80G mutations were less commonly reported, and when cited, other mutations at the same position were identified, such as D168A, E, H, T, or V and Q80K, L, or R [8]. It was not possible to carry out substantial associations with these two mutations from the data of this study.
The most important finding in this systematic review was the presence of the Q80K mutation only on the American continent, since it was the region with the lowest number of studies. In addition, it was the most frequent among all the mutations described. Many studies have described the presence of Q80K in patients carrying subtype 1a and have related its presence with a significantly lower SVR rate than that in patients without this HCV mutation [1, 2, 7–9].
An European study conducted in 2016 represented the profile of studies collected and analyzed in this systematic review, since it characterized the presence of mutations in treatment-naive patients. It also confirmed the result of this systematic review because it had also identified the Q80K variant in a greater proportion of patients, mostly in those carrying subtype 1a [12].
The studies included in this systematic review analyzed data from treatment-naive patients, who were likely to be patients with a primary infection. These pretreatment studies are important because, unlike patients already treated, the treatment-naive patients were probably infected with the already-mutated virus. The development of studies in patients of this profile can therefore optimize the first choice of therapeutic option to be used, since these patients already have resistance due to the mutation.
One limitation of this systematic review is that, owing to the scope of the topic, two new descriptors had to be inserted in the search. In addition, studies reporting changes in the NS5A and NS5B regions, which did not use the same molecular analysis methodology, could not be included. Another important point is that many studies were excluded because they did not clearly present essential information, such as the relationship of each mutation to the absence of SVR in the patient, whether this association was present only in patients with primary infections and/or treament-naïve patients or in patients already under treatment, and in which each genotype mutation was identified.
From this systematic review, it is possible to claim that the most frequent genotype worldwide is genotype 1, being present in 95.6% of the total patients in the study. The frequent presence of the Q80K mutation in this study, comprising 61.6% of the total number of mutations identified in the three continents, is supported by studies that have already described the presence of this mutation in patients carrying genotype 1. The profile of the studies gathered in this systematic review confirms the presence of mutations in treatment-naive patients in all continents, with the Q80K variant being in a greater proportion, mostly in those carrying HCV subtype 1a.
Finally, this systematic review reinforces the need to carry out more studies on this topic and to fill in all information gaps, including identifying the presence of mutations in each nonstructural region of the virus and associating them with the genotypes and subtypes, the patient’s demographic region, the patient’s treatment status (i.e., whether treatment-naïve or undergoing therapy), and whether there was a SVR, and if not, what DAA was used. These investigations are fundamental and will allow a clearer understanding of the aspects that involve advances in the eradication of the virus and its diseases.
Acknowledgements
The authors gratefully acknowledge the financial support of Foundation for the Support of Research in the State of Goiás (FAPEG) (Grant No. 04/2017 PROGRAMA PESQUISA PARA O SUS GESTÃO COMPARTILHADA EM SAÚDE FAPEG/SES-GO/CNPq/MS-DECIT/2017 – PPSUS/GO to Irmtraut Araci Hoffmann Pfrimer). The authors also gratefully acknowledge the financial support of Coordination for the Advancement of Higher Education Staff (CAPES) through fellowship to Ana Elisa de Figueiredo Miranda Mundim.
Compliance with ethical standards
Conflict of interest
No potential conflicts of interest.
Ethical standards
All studies involving human participants, included on present systematic review, were in accordance with the ethical standards of the institutional and/or national research committee, following national ethical guidelines of their respectively country, and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed A, Felmlee DJ. Mechanisms of hepatitis C viral resistance to direct acting antivirals. Viruses. 2015;7(12):6716–6729. doi: 10.3390/v7122968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 2016;6:20310. doi: 10.1038/srep20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.de Sousa MR, Ribeiro ALP. Systematic review and meta-analysis of diagnostic and prognostic studies: a tutorial. Arq Bras Cardiol. 2009;92:241–251. doi: 10.1590/S0066-782X2009000300013. [DOI] [PubMed] [Google Scholar]
- 5.Dietz J, Susser S, Berkowski C, Perner D, Zeuzem S, Sarrazin C. Consideration of viral resistance for optimization of direct antiviral therapy of hepatitis C virus genotype 1-infected patients. PLoS ONE. 2015 doi: 10.1371/journal.pone.0134395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med. 1975;292(15):767–770. doi: 10.1056/NEJM197504102921502. [DOI] [PubMed] [Google Scholar]
- 7.Fourati S, Pawlotsky JM. Virologic tools for HCV drug resistance testing. Viruses. 2015;7(12):6346–6359. doi: 10.3390/v7122941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iio E, Shimada N, Abe H, Atsukawa M, Yoshizawa K, Takaguchi K, et al. Efficacy of daclatasvir/asunaprevir according to resistance-associated variants in chronic hepatitis C with genotype 1. J Gastroenterol. 2016;52(1):94–103. doi: 10.1007/s00535-016-1225-x. [DOI] [PubMed] [Google Scholar]
- 9.Ikram A, Obaid A, Awan FM, Hanif R, Naz A, Paracha RZ, et al. Identification of drug resistance and immune-driven variations in hepatitis C virus (HCV) NS3/4A, NS5A and NS5B regions reveals a new approach toward personalized medicine. Antivir Res. 2017;137:112–124. doi: 10.1016/j.antiviral.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez-Sousa MÁ, Gutiérrez-Rivas M, Álvaro-Meca A, García-Álvarez M, Harrigan PR, Fedele CG, et al. NS3 Resistance-associated variants (RAVs) in patients infected with HCV genotype 1a in Spain. PLoS ONE. 2016;1(9):e0163197. doi: 10.1371/journal.pone.0163197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirst ME, Li EC, Wang CX, Dong HJ, Liu C, Fried MW, et al. Deep sequencing analysis of HCV NS3 resistance-associated variants and mutation linkage in liver transplant recipients. PLoS ONE. 2013;8(7):16–18. doi: 10.1371/journal.pone.0069698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kliemann DA, Tovo CV, Da Veiga ABG, De Mattos AA, Wood C. Polymorphisms and resistance mutations of hepatitis C virus on sequences in the European hepatitis C virus database. World J Gastroenterol. 2016;22(40):8910–8917. doi: 10.3748/wjg.v22.i40.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavanchy D. Chronic viral hepatitis as a public health issue in the world. Best Pract Res Clin Gastroenterol. 2008;22(6):991–1008. doi: 10.1016/j.bpg.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Zhang Y, Liu Y, Shao X, Luo Q, Cai Q, et al. Naturally occurring drug resistance associated variants to hepatitis C virus direct-acting antiviral agents in treatment-naive HCV genotype 1b-infected patients in China. Med (Baltim) 2017;96(19):e6830. doi: 10.1097/MD.0000000000006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Liu Y, Zhang Y, Shao X, Luo Q, Guo X, et al. Naturally occurring resistance-associated variants to hepatitis C virus direct-acting antiviral agents in treatment-naive HCV genotype 6a-infected patients. Biomed Res Int. 2017;2017:9849823. doi: 10.1155/2017/9849823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Cai Q, Li Z, Shao X, Luo Q, Zhang X, et al. Effect of drug resistance mutations on antiviral agents in HCV patients. Antivir Ther. 2016;21(5):369–375. doi: 10.3851/IMP2852. [DOI] [PubMed] [Google Scholar]
- 17.Martins T, Narciso-Schiavon JL, Schiavon LL. Epidemiology of hepatitis C virus infection. Rev Assoc Med Bras. 2010;57:107–112. doi: 10.1590/s0104-42302011000100024. [DOI] [PubMed] [Google Scholar]
- 18.Mellor J, Holmes EC, Jarvis LM, Yap PL, Simmonds P. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J Gen Virol. 1995;76(Pt 10):2493–2507. doi: 10.1099/0022-1317-76-10-2493. [DOI] [PubMed] [Google Scholar]
- 19.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowruz F, Shaheen S, Mujtaba G, Noreen S. An overview on hepatitis C virus genotypes and its control. Egypt J Med Hum Genet. 2015;16:291–298. doi: 10.1016/j.ejmhg.2015.05.003. [DOI] [Google Scholar]
- 22.Palanisamy N, Danielsson A, Kokkula C, Yin H, Bondeson K, Wesslén L, et al. Implications of baseline polymorphisms for potential resistance to NS3 protease inhibitors in Hepatitis C virus genotypes 1a, 2b and 3a. Antivir Res. 2013;99:12–17. doi: 10.1016/j.antiviral.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Pessôa MG, Mazo DFC, de Carvalho IMVG, Carrilho FJ. Mutações de resistência aos inibidores de protease em pacientes com Hepatite C crônica no Brasil. Rev Panam Infectol. 2014;16:57–61. [Google Scholar]
- 24.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22(34):7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puchades RL, Berenguer M. Introduction to hepatitis C virus infection: overview and history of hepatitis C virus therapies. Hemodial Int. 2018;22(Suppl 1):S8–S21. doi: 10.1111/hdi.12647. [DOI] [PubMed] [Google Scholar]
- 26.Qiu P, Sanfiorenzo V, Curry S, Guo Z, Liu S, Skelton A, et al. Identification of HCV protease inhibitor resistance mutations by selection pressure-based method. Nucl Acids Res. 2009;37(10):e74. doi: 10.1093/nar/gkp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd SJ, Abdelrahman T, MacLean AR, Thomson EC, Aitken C, Gunson RN. Prevalence of HCV NS3 pre-treatment resistance associated amino acid variants within a Scottish cohort. J Clin Virol. 2015;10(8):e0134395. doi: 10.1016/j.jcv.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. Molecular biology of hepatitis C virus. J Gastroenterol. 2007;42:411–423. doi: 10.1007/s00535-007-2030-3. [DOI] [PubMed] [Google Scholar]
- 29.Takeda H, Ueda Y, Inuzuka T, Yamashita Y, Osaki Y, Nasu A, et al. Evolution of multi-drug resistant HCV clones from pre-existing resistant-associated variants during direct-acting antiviral therapy determined by third-generation sequencing. Sci Rep. 2017;7:45605. doi: 10.1038/srep45605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trucchi C, Orsi A, Alicino C, Sticchi L, Icardi G, Ansaldi F. State of the art, unresolved issues, and future research directions in the fight against hepatitis C virus: perspectives for screening, diagnostics of resistances, and immunization. J Immunol Res. 2016;2016:1412840. doi: 10.1155/2016/1412840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Rao HY, Xie XW, Wei L. Direct-acting antiviral agents resistance-associated polymorphisms in Chinese treatment-naive patients infected with genotype 1b hepatitis C virus. Chin Med J (Engl) 2015;128:2625–2631. doi: 10.4103/0366-6999.166038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang GP, Terrault N, Reeves JD, Liu L, Li E, Zhao L, et al. Prevalence and impact of baseline resistance-associated substitutions on the efficacy of ledipasvir/sofosbuvir or simeprevir/sofosbuvir against GT1 HCV infection. Sci Rep. 2018;8(1):3199. doi: 10.1038/s41598-018-21303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeminian LB, Padovani JL, Corvino SM, Silva GF, de Pardini MIMC, Grotto RMT. Variability and resistance mutations in the hepatitis C virus NS3 protease in patients not treated with protease inhibitors. Mem Inst Oswaldo Cruz. 2013;108:13–17. doi: 10.1590/s0074-02762013000100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, et al. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993–999. doi: 10.1016/j.jhep.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X. Direct anti-HCV agents. Acta Pharm Sin B. 2016;6(1):26–31. doi: 10.1016/j.apsb.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]