Abstract
The avian influenza A(H7N9) virus has caused high mortality rates in humans, especially in the elderly; however, little is known about the mechanistic basis for this. In the current study, we used nonhuman primates to evaluate the effect of aging on the pathogenicity of A(H7N9) virus. We observed that A(H7N9) virus infection of aged animals (defined as age 20–26 years) caused more severe symptoms than infection of young animals (defined as age 2–3 years). In aged animals, lung inflammation was weak and virus infection was sustained. Although cytokine and chemokine expression in the lungs of most aged animals was lower than that in the lungs of young animals, 1 aged animal showed severe symptoms and dysregulated proinflammatory cytokine and chemokine production. These results suggest that attenuated or dysregulated immune responses in aged animals are responsible for the severe symptoms observed among elderly patients infected with A(H7N9) virus.
Keywords: Aging, influenza, immune senescence, dysregulated immunity, nonhuman primate
Aged cynomolgus macaques experienced greater symptom severity than younger animals after A(H7N9) virus infection because of immune senescence or dysregulated immune responses.
The first reported human case of infection with avian-origin influenza A(H7N9) virus was in February 2013 [1]. In the 2016–2017 influenza season, highly pathogenic H7N9 virus was detected in both humans and chickens [2, 3]. These viruses were shown to possess amino acid mutations important for mammalian adaptation and transmission between ferrets via respiratory droplets [2, 4–9]. As of June 2019, 1568 laboratory-confirmed human cases, including 615 deaths, had been reported [10]. Epidemiological studies revealed that A(H7N9) infection in elderly people (≥60 years old) was more frequent than in young people [11–14]. Older patients also had a high risk of death, with odds ratios of 1.84 (age 60–74 years) and 2.28 (age ≥75 years) when the odd ratio for patients aged 16–59 years was adjusted to 1 [14]. Several possible explanations for this difference have been proposed [15, 16]. Individuals born before 1968 experienced first infections with so-called hemagglutinin (HA) group 1 viruses, seasonal H1N1, or H2N2 viruses, resulting in their being less susceptible to A(H5N1) HA group 1 virus infection but susceptible to A(H7N9) HA group 2 virus infection because of lifelong immunological imprinting [17]. However, it has not yet been elucidated how aging per se contributes to the severe symptoms caused by A(H7N9) virus infection.
In general, as people age, they become more susceptible to infectious diseases due to hypofunction of the immune system [18]. The seasonal influenza–associated mortality rate is higher among elderly persons (≥75 years) than among other age groups [19]. This higher mortality rate may be caused by the exacerbation of underlying medical problems by the infection or by immunosenescence [19]. Numerous studies of the innate immune system in aged animals have revealed that the functions of various innate immune cells (eg, neutrophils, natural killer cells, and macrophages) are impaired in aged humans and mice [20]. However, the mechanism responsible for the high susceptibility of the elderly to A(H7N9) virus remains unclear. Recent studies of A(H7N9) virus, including our findings, have demonstrated that nonhuman primates (eg, cynomolgus macaques) are a valid infection model of A(H7N9) virus infection in humans [4, 21]. In addition, cynomolgus macaques have been shown to be useful as an aging animal model of virus infection [22]. In this study, we therefore used aged cynomolgus macaques to analyze the intrinsic effect of aging on the severity of A(H7N9) virus infection.
METHODS
Viruses and Cells
A/Anhui/1/2013 (H7N9) was kindly provided by Yuelong Shu (World Health Organization Collaborating Center for Reference and Research on Influenza, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China) and was propagated in embryonated chicken eggs. Madin-Darby canine kidney cells were maintained in Eagle’s minimum essential medium containing 5% newborn calf serum. All experiments with A(H7N9) viruses were performed in enhanced biosafety level 3 containment laboratories in Japan at the University of Tokyo, Shin Nippon Biomedical Laboratories (SNBL), Kagoshima, and the Institute for Virus Research in Kyoto University (IVRKU), Kyoto, which are approved for such use by the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Experimental Infection of Cynomolgus Macaques
Aged (age 20–26 years) and young (age 2–3 years) female cynomolgus macaques (Macaca fascicularis) were obtained from the National Institute of Biomedical Innovation, Japan Laboratory Animal, and SNBL, and tested to ensure that they were serologically negative by neutralization against A/Osaka/1365/2009 (H1N1pdm09), A/Kawasaki/UTK-4/2009 (seasonal H1N1), A/Kawasaki/UTK-20/2008 (H3N2), B/Tokyo/UT-E2/2008 (type B), and A/duck/Hong Kong/301/78 (H7N2) viruses. In this study, we compared aged animals with young animals that were part of our group’s study published in 2013 [4]. The health condition and characteristics of each animal before the initiation of experiments are described in Supplementary Table 1.
These macaques were intramuscularly anesthetized with ketamine and xylazine or medetomidine and inoculated with a suspension containing 107 plaque-forming units/mL of Anhui/1 through a combination of intratracheal (4.5 mL), intranasal (0.5 mL per nostril), ocular (0.1 mL per eye), and oral (1 mL) routes, resulting in a total infectious dose of 107.8 plaque-forming units. Macaques were monitored twice daily, once in the morning and once in the evening, for changes in rectal temperature, appetite (food consumption), and general conditions. One macaque died on day 4 (animal 4) and underwent pathological and virological analyses; the other macaques were euthanized on day 3 or 6 after infection for pathological and virological analyses. Virus titers in the lungs and various other organs were determined by means of plaque assays in Madin-Darby canine kidney cells.
Pathological Examination
Excised macaque tissues were preserved in 10% phosphate-buffered formalin. The tissues were then processed for paraffin embedding and cut into 3-μm-thick sections. The sections of each tissue sample were stained using a standard hematoxylin-eosin procedure. Each serial section was processed for immunohistological staining with a mouse monoclonal antibody for type A influenza nucleoprotein antigen (prepared in our laboratory). Specific antigen–antibody reactions were visualized with DAB staining, using the DAKO Envision system (DAKO Cytomation), as described elsewhere [4].
Microarray Analysis
Total RNA was extracted from lung samples and blood by using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol. RNA was labeled with cyanine 3 dye with the Quick Amp labeling kit (Agilent Technologies) and hybridized to the Rhesus Macaque Gene Expression Microarray (Agilent microarray design identification no. 026806; Agilent Technologies), as described elsewhere [23]. Individual microarrays were performed for each lung sample collected from naive and infected animals.
Statistical analysis was performed using the LIMMA package. The log2 of the intensity of each probe was background corrected and normalized between arrays (using the quantile method). Duplicate probes were averaged. Hierarchical clustering was used to review biological replicate quality. Next, 1-way or 2-way analyses of variance were constructed, and the moderated t statistic was calculated to determine the significance between the means. The duplicateCorrelation function was used to control for multiple samples from the same animal. All P values were Benjamini-Hochberg corrected to control the false discovery rate (FDR). Probes were considered differentially expressed if they had a fold change ≥2 and an FDR <0.01. Primary gene expression data are available in the Gene Expression Omnibus (series no. GSE152406) in accordance with proposed Minimum Information About a Microarray Experiment (MIAME) guidelines. Macaque gene annotations for the array were provided by Agilent. To increase coverage, the array was reannotated to the human genome using Agilent’s Earray (performed 28 March 2014). Gene symbols were merged between the 2 annotations, with preference given to gene annotations that successfully aligned to the macaque genome.
Gene Functional Enrichment Analysis
Functional enrichment analysis was performed with ToppCluster software and included the following categories: gene ontology, domain, pathway transcription factor binding site, drug, and disease. An enrichment score >2 (corresponding to an FDR <0.01) was the minimum required for enrichment. Ingenuity Pathway Analysis was used to identify possible gene expression regulators (eg, transcription factors) and to provide additional insight into pathways associated with differentially expressed genes.
Cytokine Assays
For cytokine and chemokine measurements, macaque lungs were treated with the Bio-Plex Cell Lysis Kit (Bio-Rad Laboratories) according to the manufacturer’s instructions, and measured with the MILLIPLEX MAP Non-Human Primate Cytokine/Chemokine Panel–Premixed 23-Plex (Millipore). Array analysis was performed using the Bio-Plex Protein Array system (Bio-Rad Laboratories).
Study Approval
All animal experiments in this study were approved by the Institutional Animal Care and Use Committee of SNBL (approval nos. IACUC814-002, and IACUC814-006) and the Committee on the Ethics of Animal Experiments of IVRKU, and they were performed in accordance with the animal welfare bylaws of the Drug Safety Research Laboratories, SNBL, which are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International, and the guidelines for animal experiments at IVRKU.
RESULTS
Severe Symptoms in A(H7N9) Virus–Infected Aged Animals
Epidemiological studies of human cases of A(H7N9) virus infection indicate that elderly individuals who are infected with A(H7N9) virus experience severe disease compared with young individuals. To determine whether aging influences the severity of disease caused by A(H7N9) virus infection in nonhuman primates, we infected 7 aged (aged 20–26 years) and 6 young (aged 2–3 years) cynomolgus macaques with A/Anhui/1/2013 (Anhui/1) and monitored their clinical symptoms (Table 1). Three of the aged animals (nos. 1, 2, and 3) did not show any symptoms before their scheduled autopsies at 3 days after infection. Three other aged animals (nos. 4, 5, and 7) showed respiratory and/or general symptoms (eg, cough, activity and appetite loss, and cramp) from 2–3 days after infection, of which 1 died (animal 4 at day 4 after infection), and 1 was euthanized (animal 7 at day 3) before the scheduled autopsy on day 6. In contrast, with the exception of 4 of the 6 young animals showing fever on day 1 after infection, the young animals were asymptomatic until at least 6 days after infection. These findings indicate that A(H7N9) virus infection causes more severe symptoms in aged cynomolgus macaques than in younger ones.
Table 1.
Symptoms of Animals After A(H7N9) Infectiona
| Animal No.(Age, y) | Symptoms by Time After Infection | |||||
|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | |
| 1 (24) | − | − | − | … | … | … |
| 2 (23) | − | − | − | … | … | … |
| 3 (21) | − | − | − | … | … | … |
| 4 (26) | − | − | Cough; loss of activity | Death | … | … |
| 5 (22) | − | Cough | Cough | Loss of activity | Cough; decreased activity and appetite | Decreased activity and appetite |
| 6 (20) | − | − | − | − | − | − |
| 7 (21) | − | Decreased activity | Recumbency; cramp | … | … | … |
| 11 (2) | Fever | − | − | … | … | … |
| 12 (2) | Fever | − | − | … | … | … |
| 13 (2) | − | − | − | … | … | … |
| 14 (2) | Fever | − | − | − | − | − |
| 15 (2) | Fever | − | − | − | − | − |
| 16 (3) | − | − | − | − | − | − |
aAutopsy was performed on day 3 in animals 1–3, 7, and 11–13, and on day 6 in animals 5. 6, and 14–16. Minus signs indicate asymptomatic status.
Virus Titers in A(H7N9) Virus–Infected Animals
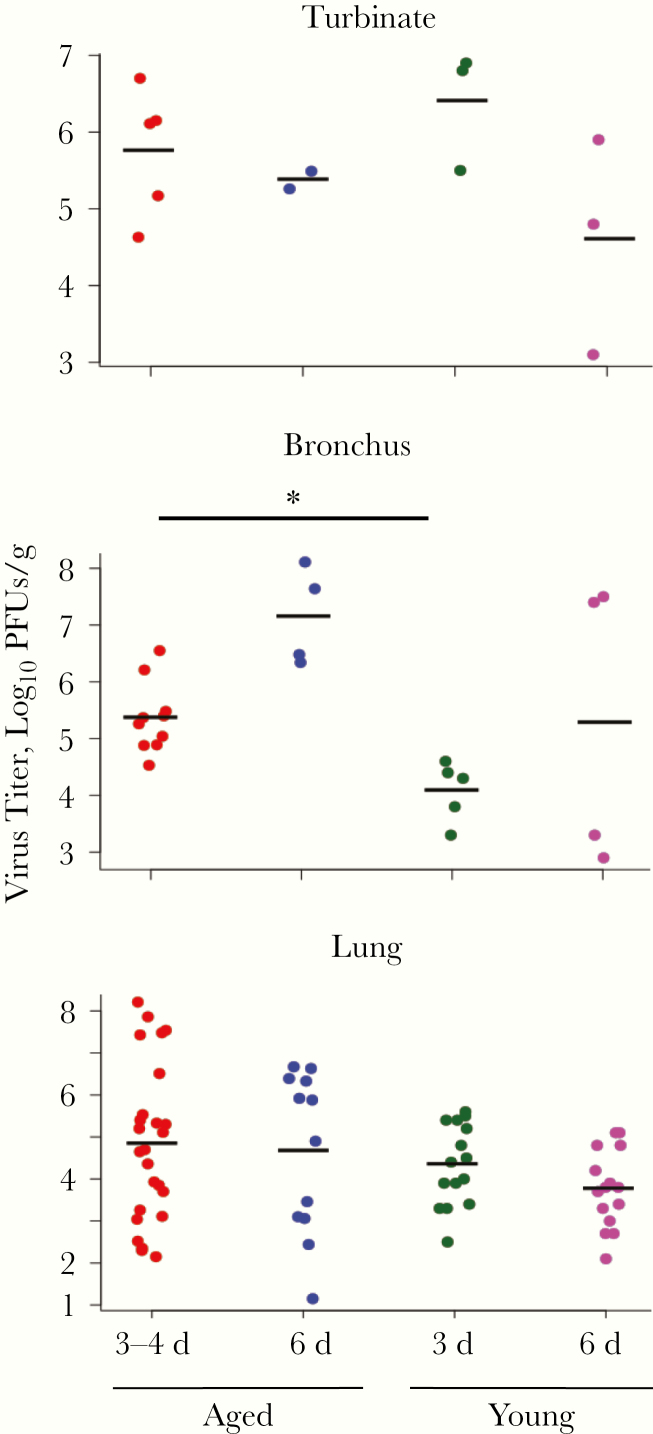
We next measured virus titers in the respiratory and systemic organs of the infected animals. In both young and aged animals, virus was detected in the upper and lower respiratory organs (Table 2). Virus titers in the bronchus from aged animals at 3–4 days after infection were significantly higher than those from young animals (Figure 1), indicating that the virus was not efficiently eliminated from the respiratory organs of the aged animals. Of note, the aged animals that showed severe symptoms at days 3–4 (animals 4 and 7) had significantly higher virus titers in their lungs than did young animals (Table 2; P < .01) or aged animals without severe symptoms (Table 2; P < .01).
Table 2.
Virus Titers in the Respiratory Organs Collected From A(H7N9) Virus–Infected Animalsa
| Organ | Viral Titer by Animal No. and Time After Infection, Log10 PFUs/g | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aged Group | Young Group | ||||||||||||
| Day 3 | Day 4 | Day 6 | Day 3 | Day 6 | |||||||||
| 1 | 2 | 3 | 7 | 4 | 5 | 6 | 11 | 12 | 13 | 14 | 15 | 16 | |
| Turbinate | 6.2 | 5.2 | 6.1 | 4.6 | 6.7 | 5.5 | 5.3 | 5.5 | 6.8 | 6.9 | 3.1 | 4.8 | 5.9 |
| Trachea | 4.5 | 5.2 | 5.4 | 6.1 | 5.1 | 6.9 | 5.8 | 4.9 | 4.2 | 3.7 | 4.7 | ND | 5.6 |
| Bronchus | |||||||||||||
| Right | 4.9 | 4.5 | 5.4 | 6.2 | 5.4 | 6.5 | 7.6 | 4.6 | 3.3 | 3.8 | ND | 2.9 | 7.5 |
| Left | 5.0 | 4.9 | 5.3 | 6.6 | 5.5 | 6.3 | 8.1 | ND | 4.4 | 4.3 | 3.3 | ND | 7.4 |
| Right lung | |||||||||||||
| Upper | 3.7 | 6.5 | 4.4 | 3.9 | 4.7 | 6.6 | 3.5 | ND | 4.4 | 3.9 | 2.7 | 3.7 | 4.2 |
| Middle | 3.9 | 5.3 | ND | 7.5 | 4.7 | 6.4 | 3.1 | 3.3 | 4.5 | 5.2 | 3.0 | 3.4 | 5.1 |
| Lower | 5.4 | 5.5 | 2.4 | 7.9 | 3.1 | 6.7 | 5.9 | ND | 5.5 | 5.4 | 2.1 | 2.7 | 5.1 |
| Left. lung | |||||||||||||
| Upper | 2.3 | 2.5 | 3.0 | 7.4 | 5.3 | 6.3 | 1.2 | 3.3 | 2.5 | 4.8 | ND | 3.8 | 4.8 |
| Middle | ND | 3.3 | ND | 8.2 | 5.1 | 4.9 | 3.1 | ND | 4.0 | 3.4 | ND | 3.8 | 3.9 |
| Lower | 2.2 | ND | ND | 7.5 | 5.2 | 5.9 | 2.4 | 3.9 | 5.4 | 5.6 | ND | 3.3 | 4.8 |
Abbreviations, ND, not detected/ PFUs, plaque-forming units.
aAged and young cynomolgus macaques were inoculated with 107.8 PFUs (6.1 mL) of A(H7N9) through multiple routes. One, 2, or 3 macaques per group were euthanized at 3, 4, and 6 days after infection for virus titration.
Figure 1.
Virus titers in the respiratory organs of A(H7N9) virus–infected animals. Turbinate, bronchus, and lung tissues were collected from aged and young animals on days 3, 4, and 6 after infection with 107.8 plaque-forming units (PFUs) (6.1 mmol/L) of A(H7N9) (Anhui/1). Virus titers were measured by means of plaque assays in Madin-Darby canine kidney cells. Sample numbers are indicated in Table 2; samples were compared using 2-tailed unpaired t tests, with P values adjusted using the Holm method. *P = .02.
We also measured virus titers in the systemic organs such as brain, lymphoid tissues, heart, kidney, liver, and digestive organs of the A(H7N9) virus–infected animals. In several extrapulmonary organs of some aged animals, we detected low to modest levels of virus (Supplementary Table 2), but virus antigens were detected only in the tonsils and mediastinal lymph nodes (data not shown). Notably, virus was detected in several portions of the brain of 2 young animals (nos. 12 and 16); in animal 16, virus was also detected in several other systemic organs. These findings demonstrate that A(H7N9) virus can spread to systemic organs. such as the central nervous system and lymphoid organs, in nonhuman primates. However, enhanced systemic distribution of A(H7N9) virus in aged animals was not observed.
Pathological Analysis of A(H7N9) Virus–Infected Animals
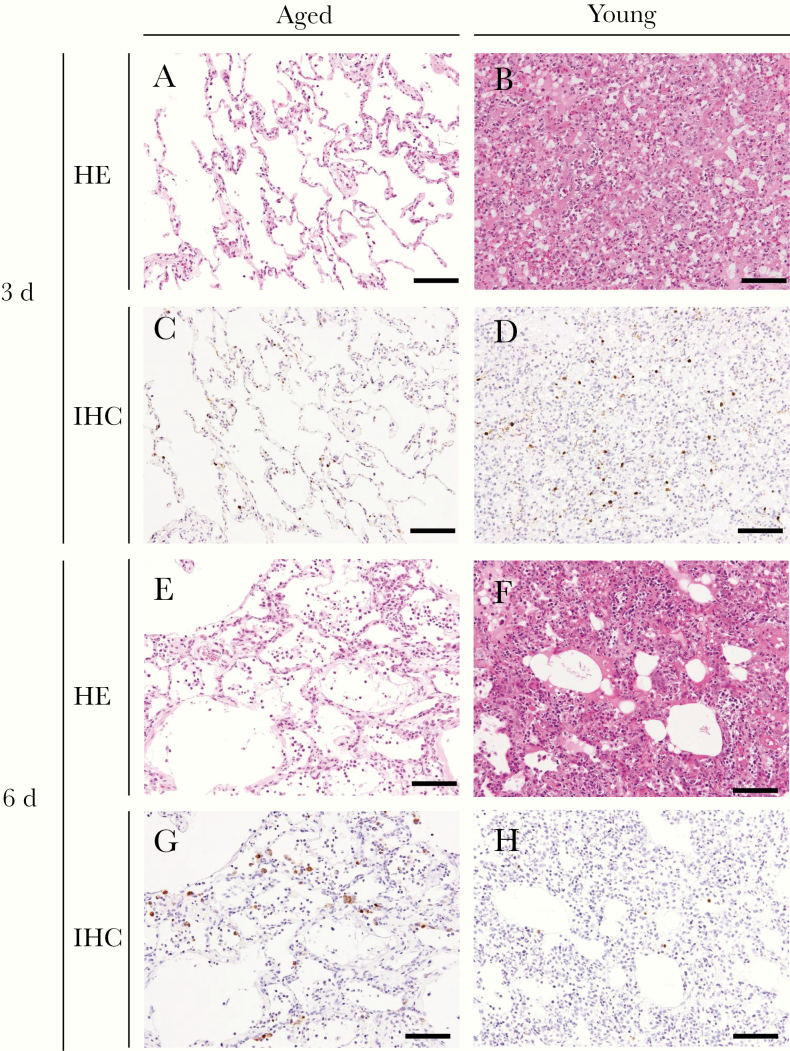
Because the virus titers in the respiratory organs tended to be high in aged animals, we compared the pathological changes between aged and young animals. Gross pathological analysis of the aged animal lungs revealed edema in a single lobe or several lobes (Supplementary Table 3). The histopathological findings from lung tissues of A(H7N9) virus–infected aged animals were different from those in young animals (Figure 2). In aged animals, areas with mild inflammation and many viral antigen-positive cells were observed even at 6 days after infection (Figures 2E and 2G). In contrast, lung inflammation had already progressed by 3 days after infection in young animals (Figure 2B), and the number of viral antigen-positive cells in young animals at day 6 was lower than that at day 3 (Figures 2D and 2H). Pathological scores of inflammation for each lung section were obtained by using hematoxylin-eosin staining (Table 3). The scores were significantly lower for the aged animals than for the young animals at both 3–4 and 6 days after infection (Table 3 and Supplementary Figure 1; P < .05).
Figure 2.
Pathology of the lungs of A(H7N9) virus–infected animals. Shown are representative pathological findings in the lungs of animals infected with A(H7N9) at 3 (A, B, C, D) and 6 (E, F, G, H) days after infection with hematoxylin-eosin (HE) staining (A, B, E, F) and immunohistochemistry (IHC) for influenza nucleoprotein antigen (C, D, G, H). Findings are shown for animals 2 (A, C), 12 (B, D), 5 (E, G), and 15 (F, H). Brown color indicates influenza nucleoprotein antigens (C, D, G, H). Scale bars represent 100 µm.
Table 3.
Pathology Scores for Lung Tissue Collected from A(H7N9) Virus–Infected Animalsa
| Lung Tissue | Pathology Score by Animal No. and Day After Infection | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aged Group | Young Group | ||||||||||||
| Day 3 | Day 4 | Day 6 | Day 3 | Day 6 | |||||||||
| 1 | 2 | 3 | 7 | 4 | 5 | 6 | 11 | 12 | 13 | 14 | 15 | 16 | |
| Right lung | |||||||||||||
| Upper | 3 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 3 | 2 | 3 |
| Middle | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 1 | 2 | 2 | 3 | 2 | 3 |
| Lower | 2 | 1 | 1 | 2 | 2 | 2 | 3 | 2 | 3 | 3 | 3 | 3 | 3 |
| Left lung | |||||||||||||
| Upper | 1 | 0 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Middle | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 |
| Lower | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 3 | 3 |
aThe inflammatory scoring system defined scores as follow: 0, normal; 1, mild (a few inflammatory cells); 2, moderate (inflammatory cells infiltrate, with edema and/or alveolar hemorrhage in <50% of section); and 3, severe (inflammatory cells infiltrate, with edema and/or alveolar hemorrhage in ≥50% of section).
Microarray Analysis of Lungs and Blood from A(H7N9) Virus–Infected Animals
Because lung inflammation was relatively mild considering the high virus titers in the lung tissues and the severity of the symptoms experienced by the aged animals, we hypothesized that aging might attenuate the inflammatory response against A(H7N9) virus infection. To address the effect of aging on the development of inflammation in A(H7N9) virus–infected animals, we performed a transcriptional analysis of lungs from naive and A(H7N9) virus–infected young and aged animals. Transcripts that were differentially expressed between A(H7N9) virus–infected aged and young animals on either day (day 3 or 6) were clustered, and each cluster was analyzed with ToppCluster software to identify enriched biological functions. The expression of each transcript in aged naive, aged infected, or young infected animals relative to that in young naive animals is shown in Supplementary Figures 2 and 3.
In the lungs of aged infected animals at 3 days after infection, the interleukin 6 (IL-6)–mediated signaling pathway was down-regulated by A(H7N9) virus infection compared with naive aged animals, whereas it was up-regulated in the lungs of young infected animals compared with naive young animals (Supplementary Figure 2). In contrast, the type I interferon (IFN) response, cytokine signaling, phosphoinositide 3-kinase (PI3K) signaling, and integrin signaling were up-regulated in aged infected animals at 6 days after infection, whereas these transcripts in young infected animals were not affected or down-regulated (Supplementary Figure 2).
Next, to assess the effect of aging on the systemic antiviral response, we performed a similar differential expression analysis using whole-blood samples collected from naive and A(H7N9) virus–infected aged and young animals at 3 and 6 days after infection. We found that several thousand transcripts were differentially expressed in aged animals (Supplementary Figure 3). Genes enriched for immune processes (eg, Toll-like receptor signaling and innate immunity) were up-regulated in the blood of aged naive animals compared with young naive animals (Supplementary Figure 3), consistent with reports in elderly humans [24]. In young animals, Toll-like receptor signaling and innate immunity were up-regulated with infection at day 3. In contrast, genes associated with interleukin 10 signaling and IFN regulatory factor 7 were up-regulated in both aged and young infected animals on infection at 3 days after infection (Supplementary Figure 3). At 6 days after infection, negative regulators of viral genome replication, including innate immune responses, were strongly up-regulated in the blood of aged infected animals, whereas their expression returned to normal in young infected animals (Supplementary Figure 3). Taken together, these results demonstrate that the immunological responses against A(H7N9) infection of aged animals differ from those of young animals.
Analysis of Cytokine and Chemokine Production in A(H7N9) Virus–Infected Animals
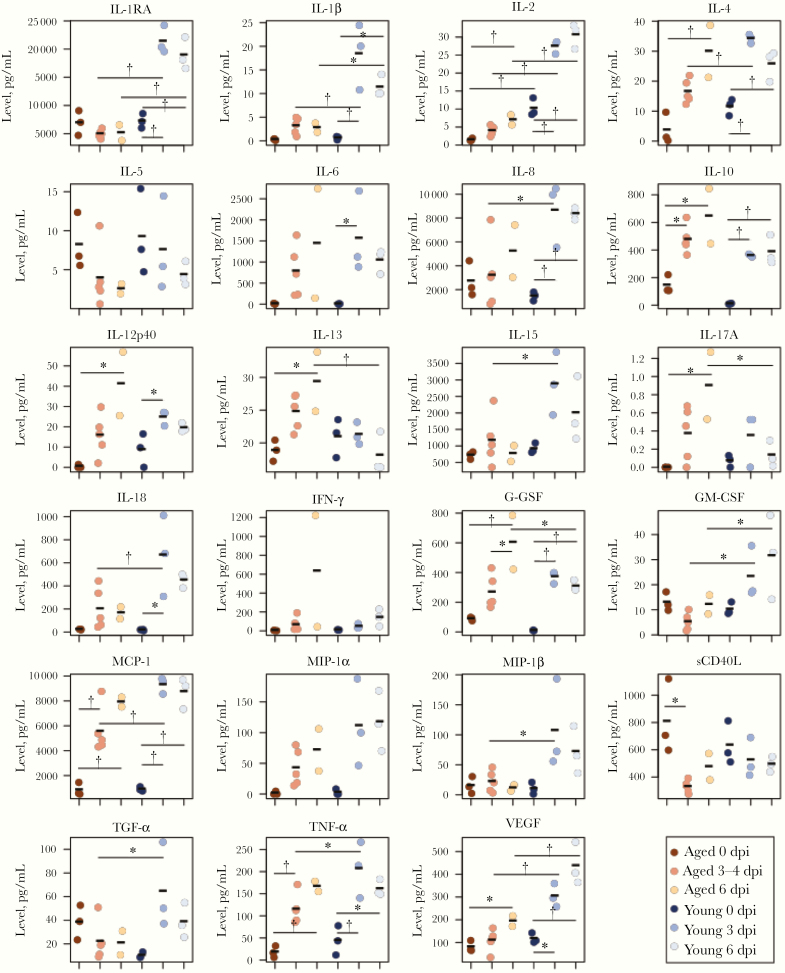
Since the immunological responses to A(H7N9) virus infection in aged animals differed from those in young animals, we evaluated cytokine and chemokine production in aged animals on A(H7N9) infection by means of a Bioplex analysis of lung tissues and blood. As expected, the expression of cytokines and chemokines, including interleukin 1 receptor antagonist, interleukin 8 (IL-8), interleukin 1β, 2, 4, 15, and 18, granulocyte-macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein 1, macrophage inflammatory protein (MIP) 1β, transforming growth factor α, tumor necrosis factor α, and vascular endothelial growth factor, was significantly higher in the lungs of the infected young animals than in those of infected aged animals (Figure 3). In contrast, the expression of interleukin 13 (IL-13), interleukin 17A, and granulocyte colony-stimulating factor (G-CSF) in infected young animals was lower than that in infected aged animals (Figure 3). These data suggest that the lower cytokine and chemokine production in the lungs of aged animals compared with young animals may be involved in the attenuation of inflammation in aged animals on A(H7N9) infection. However, the transcriptome analysis showed that cytokine signaling and the type I IFN response were greater in aged than in young animals at day 6 after infection (Supplementary Figure 2). Protein production and gene expression of some cytokines were not correlated among the animals; this discrepancy requires further study.
Figure 3.
Multiplex analysis of cytokines and chemokines in lung tissues. Lung tissues were isolated from young and aged animals. Cytokines and chemokines in lung tissue samples were measured using a multiplex immunoassay. Samples were obtained from aged naive animals (day 0; n = 3), aged animals at days 3–4 (n = 5) and day 6 (n = 2), young naive animals (day 0; n = 3), and young animals at days 3 (n = 3) and 6 (n = 3); samples were compared using 1-way or 2-way analysis of variance (*P < .05; †P < .01). Data on young animals were adapted from those published by Watanabe et al [4]. Abbreviations: G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p40, IL-13, IL-15, IL-17A, and IL-18, interleukin 1β, 2, 4, 5, 6, 8, 10, 12p40, 13, 15, 17A, and 18; IL-1RA, interleukin 1 receptor antagonist; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; sCD40L, soluble cD40L; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
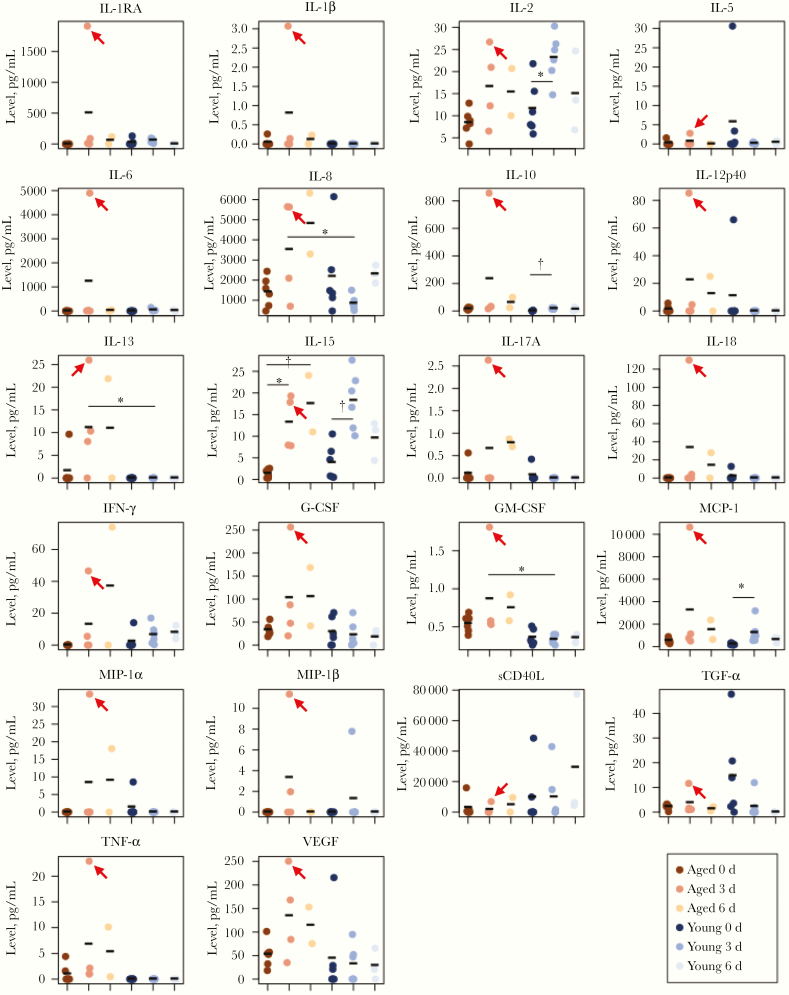
To understand the systemic cytokine and chemokine production, we also measured these regulators in the serum collected from the A(H7N9) virus–infected animals. Although A(H7N9) virus infection induced significantly higher levels of IL-8, IL-13, and GM-CSF in the serum of infected aged animals compared with infected young animals, overall the cytokine and chemokine levels in the serum of aged animals was similar to that of young animals (Figure 4). In contrast, we found that several cytokines and chemokines (eg, interleukin 1 receptor antagonist, IL-6, IL-8, IL-13, interleukin 1β 10, 12, 17A, and 18, IFN-γ, G-CSF, GM-CSF, monocyte chemoattractant protein 1, MIP-1α, MIP-1β, and tumor necrosis factor α) were extremely up-regulated in the serum of 1 aged animal (no. 7) with severe symptoms (red arrows in Figure 4). This result indicates that the expression of cytokines and chemokines in serum on A(H7N9) infection does not differ greatly between aged and young animals, except for animal 7 with severe symptoms. Taken together with the microarray data and bioplex analysis findings, our results show that cytokine and chemokine production is attenuated in the lung of aged infected animals and that their production in serum is dysregulated in an aged animal with severe symptoms.
Figure 4.
Multiplex analysis of cytokines and chemokines in blood. Serum was isolated from the blood of young and aged animals. Cytokines and chemokines in the serum were measured by using a multiplex immunoassay. Samples were obtained from aged naive animals (0 days after infection; n = 6), aged animals at days 3 (n = 4) and 6 (n = 2), young naive animals (0 days; n = 6), and young animals at days 3 (n = 6), and 6 (n = 3); samples were compared using 1-way or 2-way analysis of variance (*P < .05; †P < .01). Data on young animals were adapted from those published by Watanabe et al [4]. Red arrows indicate values from an aged animal with severe clinical symptoms (animal 7). Abbreviations: G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL-1β, IL-2, IL-5, IL-6, IL-8, IL-10, IL-12p40, IL-13, IL-15, IL-17A, and IL-18, interleukin 1β, 2, 5, 6, 8, 10, 12p40, 13, 15, 17A, and 18; IL-1RA, interleukin 1 receptor antagonist; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; sCD40L, soluble cD40L; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
DISCUSSION
A(H7N9) virus is reported to infect a high proportion of elderly people [11–14], although the precise reason for this remains unclear. In this study, we experimentally infected aged nonhuman primates with the A(H7N9) virus and found that aged animals showed more severe symptoms than young animals, which indicates that aged nonhuman primates may be a useful model for the study of severe disease in the elderly infected with A(H7N9) virus. Our analyses showed that the inflammatory response such as cytokine and chemokine production in the lungs of aged animals was attenuated at 3 and 6 days after infection compared with young animals even though similar or higher viral replication was detected in respiratory organs of aged animals compared with young animals. Furthermore, both microarray and bioplex analyses revealed that IL-6 production was impaired in aged animals. IL-6 is a key cytokine for protecting animals against influenza virus infection by enhancing both recruitment and the phagocytic activity of macrophages [25]. Therefore, the attenuated immunological response including low IL-6 production, so-called immunosenescence, may be responsible for the high pathogenicity of A(H7N9) virus in aged animals.
High levels of cytokines and chemokines were detected in the serum of an A(H7N9) virus–infected aged animal (animal 7) that showed severe symptoms, indicating that aberrant production of proinflammatory cytokines and chemokines with virus replication might synergistically enhance the severity of the infection. Because other aged animals showed attenuated immunological responses to A(H7N9) virus infection, the mechanism responsible for the immune response dysregulation in aged animals remains unclear.
However, this dysregulated immune response, the so-called cytokine storm, has been reported in many human cases: A(H7N9) virus causes multiple organ dysfunction with aberrant systemic immune responses [26], and disease severity for patients with A(H7N9) virus infection was positively correlated with their serum levels of cytokines and chemokines [27]. Moreover, the dysregulated immune response is also associated with the high pathogenicity of H5N1 and 1918 pandemic H1N1 viruses [28–30]. Although a defined trigger that causes the dysregulated immune response has not been revealed, pulmonary endothelial cells were implicated as key players in the early stage of the response to stimulate proinflammatory cytokine expression through IFN-α release [31, 32]. Therefore, aged nonhuman primates might be a good model to study the mechanism responsible for the immune dysregulation that is caused by avian-origin human influenza viruses and to evaluate treatments for it.
The microarray data indicate that genes associated with cytokine signaling (Supplementary Figure 2), type 1 IFN response (Supplementary Figure 2), and negative regulators of viral genome replication (Supplementary Figure 3) were up-regulated in the lungs and whole blood of aged animals at 6 days after infection, compared with young animals. However, the protein expression levels of cytokines and chemokines in the lungs of aged animals at 6 days after infection were significantly lower than those of young animals. These data indicate that the expression of messenger RNAs and proteins in response to virus infection was not consistent. One possible explanation for this discrepancy is that the messenger RNA response might not be successfully translated to the protein response in aged animals because aging down-regulates protein synthesis [33].
In summary, our analysis suggests that an inadequate antiviral response in aged animals causes the failure to eliminate viruses efficiently from the lungs during early infection. Our findings also suggest that high levels of proinflammatory cytokines and chemokines might be involved in the fatal outcome of an aged animal. Taken together, we suggest 2 types of outcome (immunosenescence and aberrant immune response) from aged animals with A(H7N9) virus infection. Our data provide considerable information about the effect of aging on the pathogenicity of A(H7N9) virus, which will be of value in the development of new and effective treatments for elderly patients with A(H7N9) virus infection.
Supplementary Material
Notes
Acknowledgments. For provision of the A/Anhui/1/2013 (H7N9) virus, we thank Yuelong Shu, director of the World Health Organization Collaborating Center for Reference and Research on Influenza, director of the Chinese National Influenza Center, deputy director of the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China, and dean of the School of Public Health (Shenzhen) of Sun Yat-sen University. We also thank Susan Watson for editing the manuscript; Yasuhiro Yasutomi and Hiroaki Shibata, Tsukuba Primate Research Center, National Institutes of Biomedical Innovation, Health and Nutrition, for providing aged cynomolgus macaques; and Naomi Fujimoto, Izumi Ishikawa, Yuko Sato, and Sara Takasaki for technical assistance.
Author contributions. S. F., K. I. H., M. K., H. K., Y. T., T. M., and T. W. carried out the experiments. N. N. and H. H. carried out the pathological analysis and immunohistochemistry. Y. K. supervised the experiments. S. F., K. I. H., M. K., H. K., Y. T., T. M., T. W., S. Y., and Y. K. analyzed the data. R. W. G. and J. E. S. analyzed the microarray data. T. J. d. S. L. carried out the statistical analyses. S. F., K. I. H., M. K., N. N., R. W. G., H. K., Y. T., T. M., T. J. d. S. L., T. W. J. E. S., and H. H. drafted figures and parts of the manuscript. S. Y. and Y. K. wrote the manuscript.
Financial support. This research was supported by Strategic Basic Research Programs from the Japan Science and Technology Agency, Leading Advanced Projects for medical innovation (grant JP19am001007), the Japan Initiative for Global Research Network on Infectious Diseases (grant JP19fm0108006), a Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (grants JP19fk0108104 and JP19fk0108058), the Ministry of Education, Culture, Science, Sports, and Technology of Japan (grants-in-aid for scientific research on innovative areas 16H06429, 16K21723, and 16H06434), and the Center for Research on Influenza Pathogenesis, funded by the National Institute of Allergy and Infectious Diseases (contract HHSN272201400008C).
Potential conflicts of interest. Y K. has received speaker’s honoraria from Toyama Chemical and Astellas; has received grant support from Chugai Pharmaceuticals, Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Otsuka Pharmaceutical, Denka Seiken, and Shionogi & Co; and is a cofounder of FluGen. All other authors report no conflicts of interest. The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888–97. [DOI] [PubMed] [Google Scholar]
- 2. Li Y, Qi W, Qiao J, Chen C, Liao M, Xiao C. Evolving HA and PB2 genes of influenza A (H7N9) viruses in the fifth wave—increasing threat to both birds and humans? J Infect 2017; 75:184–6. [DOI] [PubMed] [Google Scholar]
- 3. Zhang F, Bi Y, Wang J, et al. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect 2017; 75:71–5. [DOI] [PubMed] [Google Scholar]
- 4. Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013; 501:551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mok CK, Lee HH, Lestra M, et al. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 2014; 88:3568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamayoshi S, Yamada S, Fukuyama S, et al. Virulence-affecting amino acid changes in the PA protein of H7N9 influenza A viruses. J Virol 2014; 88:3127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamayoshi S, Fukuyama S, Yamada S, et al. Amino acids substitutions in the PB2 protein of H7N9 influenza A viruses are important for virulence in mammalian hosts. Sci Rep 2015; 5:8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai M, Watanabe T, Kiso M, et al. A highly pathogenic avian H7N9 influenza virus isolated from a human is lethal in some ferrets infected via respiratory droplets. Cell Host Microbe 2017; 22:615–26 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamayoshi S, Kiso M, Yasuhara A, Ito M, Shu Y, Kawaoka Y. Enhanced replication of highly pathogenic influenza A(H7N9) virus in humans. Emerg Infect Dis 2018; 24:746–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Influenza at the human-animal interface. https://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_09_04_2019.pdf?ua=1 [Google Scholar]
- 11. Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 2013; 368:2277–85. [DOI] [PubMed] [Google Scholar]
- 12. Li Q, Zhou L, Zhou M, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 2014; 370:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huo X, Chen L, Qi X, et al. Significantly elevated number of human infections with H7N9 virus in Jiangsu in eastern China, October 2016 to January 2017. Euro Surveill 2017; 22:30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X, Jiang H, Wu P, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 2017; 17:822–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bui C, Bethmont A, Chughtai AA, et al. A systematic review of the comparative epidemiology of avian and human influenza A H5N1 and H7N9—lessons and unanswered questions. Transbound Emerg Dis 2016; 63:602–20. [DOI] [PubMed] [Google Scholar]
- 16. Ji H, Gu Q, Chen LL, et al. Epidemiological and clinical characteristics and risk factors for death of patients with avian influenza A H7N9 virus infection from Jiangsu Province, Eastern China. PLoS One 2014; 9:e89581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 2016; 354:722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int 2009; 22:1041–50. [DOI] [PubMed] [Google Scholar]
- 19. Quandelacy TM, Viboud C, Charu V, Lipsitch M, Goldstein E. Age- and sex-related risk factors for influenza-associated mortality in the United States between 1997–2007. Am J Epidemiol 2014; 179:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol 2013; 13:875–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Itoh Y, Shichinohe S, Nakayama M, et al. Emergence of H7N9 influenza A virus resistant to neuraminidase inhibitors in nonhuman primates. Antimicrob Agents Chemother 2015; 59:4962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smits SL, de Lang A, van den Brand JM, et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog 2010; 6:e1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muramoto Y, Shoemaker JE, Le MQ, et al. Disease severity is associated with differential gene expression at the early and late phases of infection in nonhuman primates infected with different H5N1 highly pathogenic avian influenza viruses. J Virol 2014; 88:8981–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Franceschi C, Capri M, Monti D, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev 2007; 128:92–105. [DOI] [PubMed] [Google Scholar]
- 25. Yang ML, Wang CT, Yang SJ, et al. IL-6 ameliorates acute lung injury in influenza virus infection. Sci Rep 2017; 7:43829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diao H, Cui G, Wei Y, et al. Severe H7N9 infection is associated with decreased antigen-presenting capacity of CD14+ cells. PLoS One 2014; 9:e92823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo J, Huang F, Liu J, et al. The serum profile of hypercytokinemia factors identified in H7N9-infected patients can predict fatal outcomes. Sci Rep 2015; 5:10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z, Zhang A, Wan Y, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A 2014; 111:769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Short KR, Kedzierska K, van de Sandt CE. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect Microbiol 2018; 8:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo XJ, Thomas PG. New fronts emerge in the influenza cytokine storm. Semin Immunopathol 2017; 39:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manickam C, Shah SV, Lucar O, Ram DR, Reeves RK. Cytokine-mediated tissue injury in non-human primate models of viral infections. Front Immunol 2018; 9:2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teijaro JR, Walsh KB, Cahalan S, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 2011; 146:980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rattan SI, Derventzi A, Clark BF. Protein synthesis, posttranslational modifications, and aging. Ann N Y Acad Sci 1992; 663:48–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.