Abstract
Persistent methicillin-resistant Staphylococcus aureus (MRSA) endovascular infections represent a significant clinical-therapeutic challenge. Of particular concern is antibiotic treatment failure in infections caused by MRSA that are “susceptible” to antibiotic in vitro. In the current study, we investigate specific purine biosynthetic pathways and stringent response mechanism(s) related to this life-threatening syndrome using genetic matched persistent and resolving MRSA clinical bacteremia isolates (PB and RB, respectively), and isogenic MRSA strain sets. We demonstrate that PB isolates (vs RB isolates) have significantly higher (p)ppGpp production, phenol-soluble-modulin expression, polymorphonuclear leukocyte lysis and survival, fibronectin/endothelial cell (EC) adherence, and EC damage. Importantly, an isogenic strain set, including JE2 parental, relP-mutant and relP-complemented strains, translated the above findings into significant outcome differences in an experimental endocarditis model. These observations indicate a significant regulation of purine biosynthesis on stringent response, and suggest the existence of a previously unknown adaptive genetic mechanism in persistent MRSA infection.
Keywords: MRSA, stringent response, (p)ppGpp, purine biosynthesis, persistence, endovascular infection
Persistent endovascular infections due to methicillin-resistant Staphylococcus aureus (MRSA) is a significant and growing public health threat. We demonstrate that the stringent response plays a key role in the persistent MRSA endovascular infection outcome to vancomycin through purine biosynthesis.
Infective endocarditis (IE) is a life-threatening syndrome often caused by methicillin-resistant Staphylococcus aureus (MRSA) [1]. Despite the use of gold-standard anti-MRSA antibiotics, (vancomycin [VAN] or daptomycin [DAP]), treatment failures associated with these syndromes remain unacceptably high (~ 30%) [2, 3]. Persistent MRSA bacteremia (PB; defined as ≥ 7 days of positive blood cultures despite appropriate antibiotic therapy) occurs in 15%–30% of such infections. This phenomenon is very worrisome because many PB strains are typically “susceptible” in vitro to standard-of-care anti-MRSA agents such as VAN and DAP as determined by Clinical and Laboratory Standards Institute breakpoints [3, 4]. Therefore, PB outcomes pose an urgent and unmet therapeutic challenge to public health. Understanding the specific molecular mechanisms of PB is essential to develop novel anti-MRSA strategies to prevent or minimize the persistent outcomes.
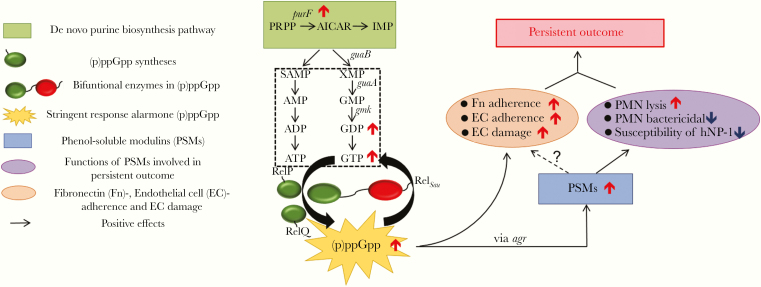
The stringent response is a highly conserved adaptation mechanism to stressful conditions (eg, antibiotic pressure), initiated by the rapid synthesis of a bacterial alarmone, guanosine 3′-diphosphate-5′-di(tri)phosphate [(p)ppGpp] [5–8]. Staphylococcus aureus possesses 3 (p)ppGpp synthetases: the bifunctional RelSau protein, with hydrolase and synthetase functions, and 2 small alarmone synthetases, RelP (RelPSau) and RelQ (RelQSau) [9, 10] (Figure 1). While the stringent response has been well studied in other organisms, particularly in gram-negative bacteria [5, 11], neither its impact on the PB outcomes nor its intersection with purine biosynthesis has been evaluated in MRSA endovascular infections.
Figure 1.
Hypothesized model of the role of purine biosynthesis and the stringent response alarmone, (p)ppGpp, in persistent methicillin-resistant Staphylococcus aureus (MRSA) endovascular infection. De novo purine biosynthesis pathway mediates the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) to inosine monophosphate (IMP), which subsequently become purines ATP and GTP. The stringent response alarmone, guanosine 3′-diphosphate-5′-di(tri)phosphate [(p)ppGpp], is mainly produced from GDP/GTP, and its production is controlled by a bifunctional synthetase RelSau protein, and 2 other monofunctional synthetases RelP and RelQ in S. aureus [5–10]. The elevated expression of purF gene, encoding a rate-limited enzyme ATPase (PRPP→PRA), in the purine biosynthesis pathway leads to increased GDP/GTP and subsequent (p)ppGpp levels. Increased (p)ppGpp leads to up-regulation (agr-dependent) of persistent-related factors, such as (1) phenol-soluble modulins (PSMs), which increase S. aureus–induced Neutrophils (PMN) lysis and support S. aureus to overcome PMN- and human neutrophil peptide 1 (hNP-1)–mediated bactericidal killing; and (2) MRSA–endothelial cell (EC) interactions, including enhanced fibronectin (Fn)/EC adherence and EC damage, and eventually facilitating persistent MRSA bacteremia outcomes.
De novo purine biosynthesis represents a basis for nucleotide metabolism and is critical for bacterial cell growth through nucleobase syntheses (eg, GDP and GTP) [12]. (p)ppGpp is synthesized from GDP/GTP [10, 13] and could also provide feedback control of GTP levels by inhibiting GTP biosynthesis enzymes [14, 15]. We recently demonstrated a critical role of purine biosynthesis in the PB outcomes [16]. Those studies led us to hypothesize that the purine biosynthesis impact on GDP/GTP affects (p)ppGpp synthesis, which in turn provides a link to the stringent response in adaptive survival mechanisms of MRSA (Figure 1). The current investigation was designed to test these hypotheses by studying the interrelationships among purine biosynthesis, the stringent response, and PB vs resolving MRSA clinical bacteremia (RB, defined as initial bacteremia resolved within 2–4 days to therapy) outcomes both in vitro and in an experimental IE model.
METHODS
Bacterial Strain and Plasmids
Bacterial strains and plasmids used in the current study are listed in Table 1. Eight representative clinical MRSA isolates (1 PB and 1 RB strain for each of the 4 most common clonal complex [CC] and agr types) were obtained from a multinational clinical trial collection and other sources [4, 17]. All of the MRSA isolates were VAN-susceptible (MICs ≤ 2 μg/mL) and had neither VAN in vitro tolerance nor VAN heteroresistant subpopulations [4, 17, 18]. JE2 (USA300 LAC derivative cured of 3 plasmids [19]) and its isogenic purF and relP mutant strains from Nebraska transposon mutant library were also used in this study. The relP mutant was complemented by transforming plasmid pSK236::relP as described previously [20].
Table 1.
Bacterial Strains and Plasmids Used in This Study
| Strains and Plasmids | Description | Vancomycin MIC, μg/mL | References |
|---|---|---|---|
| Clinical MRSA isolates | |||
| PB: 300-169 | agr-I, SCCmec IV, CC45, fast-growth ratea, early global regulators onsetb, nonresponderc | 0.5 | [4, 16] |
| RB: 301-188 | agr-I, SCCmec IV, CC45, slow-growth rate, late global regulators onset, responderd | 0.5 | [4, 16] |
| PB: 300-246 | agr-II, SCCmec I, CC5, fast-growth rate, early global regulators onset, nonresponder | 0.5 | [4, 16] |
| RB: 010-016 | agr-II, SCCmec II, CC5, slow-growth rate, late global regulators onset, responder | 0.5 | [4, 16] |
| PB: 31082 | agr-I, SCCmec IV, CC8, fast-growth rate, early global regulators onset, nonresponder | 2.0 | [16, 18] |
| RB: 30568 | agr-I, SCCmec IV, CC8, slow-growth rate, late global regulators onset, responder | 2.0 | [16, 18] |
| PB: 33367 | agr-III, SCCmec II, CC30, fast-growth rate, early global regulators onset, nonresponder | 1.0 | [16, 18] |
| RB: 22033 | agr-III, SCCmec II, CC30, slow-growth rate, late global regulators onset, responder | 1.0 | [16, 18] |
| LAC* | Erm sensitive community-acquired MRSA LAC strain (USA300) | … | [23] |
| Laboratory MRSA strains | |||
| JE2 | LAC, USA300 derivative that was cured of its 3 plasmids (agr-I, SCCmec IV, CC8) | 2.0 | [19] |
| LAC*rshsyn | LAC variant that lacked 3 conserved amino acids in the (p)ppGpp synthetase domain of RelSau | … | [23] |
| JE2 ∆relP | Transposon mutant with insertion in USA300_2446 | 2.0 | NTML |
| JE2 ∆purF | Transposon mutant with insertion in USA300_0972 | 2.0 | NTML |
| JE2 ∆relP/prelP | JE2 ∆relP complemented with plasmid pSK236::relP | 2.0 | This study |
| JE2 ∆purF/ppurF | JE2 ∆purF complemented with plasmid pSK236::purF | 2.0 | [16] |
Abbreviations: CC, clonal complex; MIC, minimum inhibitory concentration; MRSA, methicillin-resistant Staphylococcus aureus; NTML, Nebraska Transposon Mutant Library; PB, persistent methicillin-resistant Staphylococcus aureus bacteremia; RB, resolving methicillin-resistant Staphylococcus aureus bacteremia.
aTSB medium, aerobic, 37°C [16].
cNonresponder, the isolates in which < 1.5-log10 colony-forming units (CFU) mean reductions per gram of vegetation, kidney, and spleen were observed due to vancomycin treatment in a rabbit infective endocarditis (IE) model [4].
dResponder, the isolates in which vancomycin treatment caused a ≥ 5-log10 CFU mean reduction per gram of vegetation and ≥ 3-log10 CFU mean reduction/g of kidney or spleen in IE model [4].
Determination of VAN MICs
MICs of VAN were determined by a standard Etest method (bioMérieux, LABalme-les-Grottes, France).
Detection of (p)ppGpp Levels
The intracellular (p)ppGpp levels of the study MRSA strains were detected by using a fluorescent chemosensor, PyDPA, as previously described [21, 22]. In brief, exponential phase (3 hours’ incubation) MRSA cells were harvested and adjusted at optical density (wavelength at 600 nm; OD600nm) of 1.000. This time point was used based on our previous studies showing earlier-onset activation (2–4 hours’ incubation) of key global regulators in the PB vs RB strains [16, 17]. After adding lysis reagent (100% methanol), the supernatant was collected and concentrated by using a lyophilizer. The dried extracts were then suspended in HEPES buffer (1 mM, pH 7.4 containing 16% dimethyl sulfoxide [v/v]), and mixed with PyDPA (40 μM) [21, 22]. Fluorescence was measured using a Perkin-Elmer LS-55 fluorescence spectrometer with Ex 344 nm/Em 470 nm for (p)ppGpp levels [21, 22]. The samples extracted from LAC* strain treated with mupirocin and LAC*rshsyn strain were used as positive and negative controls, respectively [23].
Detection of GDP and GTP Levels
The intracellular GDP levels of the study MRSA cells from exponential phase growth (3 hours’ incubation) were detected by using a Transcreener GDP FI kit (BellBrook Labs) [24]. GDP levels were expressed as fluorescence intensity. GTP levels were quantified by a luciferase-based assay using Promega BacTiter Glo and GTPase-Glo Kinase Kits (Promega) [25–27]. In brief, samples from the (p)ppGpp detection assay were mixed with an equal volume of GTPase-Glo reagent to convert GTP to ATP. After 30 minutes of incubation at room temperature, total ATP including the residual ATP and that generated in this reaction was detected using a luciferin/luciferase-based ATP detection reagent. In parallel, the residual ATP levels in extracted samples were measured, samples from the (p)ppGpp detection assay were mixed with an equal volume of BacTiter-Glo reagent and incubated for 5 minutes at room temperature, and ATP levels were determined by measuring luminescence levels. GTP levels (luminescence intensity) were presented as the total luminescence levels minus residual ATP luminescence levels [25, 27].
RNA Isolation and Target Gene Expression by Reverse-Transcription–Quantitative Polymerase Chain Reaction
Total RNA of the study MRSA strains from the exponential phase (3 hours’ incubation) was isolated by using a RNeasy Kit (Qiagen) [28]. One microgram of DNase-treated RNA was transcribed into complementary DNA. Real-time quantitative polymerase chain reaction (qPCR) was performed using a SYBR green PCR master kit (Applied Biosystems). The amplification of psmα1–4, psmβ1,2 and gyrB was performed using primers as described previously [4, 29]. gyrB was used to normalize the transcript quantification. The relative quantification of gene expression was calculated by the ΔΔCT method [4].
Neutrophils (PMN) Bactericidal Activity and MRSA-Induced PMN Lysis
Human neutrophil–mediated killing was performed as described previously [29, 30]. In brief, exponential phase MRSA cells were washed and adjusted to OD600nm of 1.000 in Hanks’ balanced salt solution (HBSS). Frozen human neutrophils obtained from Astarte Biologics were thawed in 37°C water bath and gently resuspended in HBSS [31]. The bacterial suspension was mixed to 106 colony-forming units (CFU)/mL neutrophils with the initial ratios 10:1 (bacteria:neutrophils). The percentage of bacterial survival was expressed as the percentage of the initial inoculum that survived with neutrophils exposure. Following the PMN bactericidal activity experiment above, lysis of the human neutrophils was determined with a standard release of lactate dehydrogenase assay according to the manufacturer’s recommended protocols (Cytotoxicity Detection kit, Roche Applied Sciences) [29, 32].
In Vitro Susceptibility to Human Neutrophil Peptide 1
The human neutrophil peptide 1 (hNP-1) susceptibility was detected by exposing 105 CFU/mL MRSA cells from exponential phase growth to hNP-1 (Peptides International) at 1.25 μg/mL according to the method described previously [33, 34]. Survival rates were expressed as the survival cells compared to the initial inoculum.
In Vitro Susceptibility to VAN
A starting inoculum of 108 CFU/mL of MRSA cells at exponential phase were exposed to 15 μg/mL of VAN (mimic targeted trough serum level of VAN treatment for severe MRSA infections in human) in cation-adjusted MHB for 24 hours [4, 18]. Survival rates were expressed as the survival cells compared to the initial inoculum.
Adherence to Fibronectin
A starting inoculum of 5 × 103 CFU/well of exponential phase MRSA cells were added to the 6-well plates coated with purified human fibronectin (Fn) (50 μg/mL, Sigma Chemicals) and incubated for 1 hour at 37°C. The adherence was expressed as the percentage of the initial inoculum bound [35].
Endothelial Cell Adherence
Human microvascular endothelial cells (HMEC-1) were obtained from the Centers for Disease Control and Prevention and maintained as previously described [36, 37]. MRSA exponential phase cells were added to plate confluent with HMEC-1 cells at a final inoculum of 5 × 103 CFU/well and incubated for 1 hour at 37°C [38]. The adherence rate was expressed as a percentage of the initial inoculum [38].
Endothelial Cell Damage
A well-established 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to test the endothelial cell (EC) damage [39]. In brief, exponential phase MRSA cells were added to HMEC-1 in 24-well plates (5 × 105 ECs/well) corresponding to a multiplicity of infection of 50 (as established by pilot studies). After 3 hours of incubation, the wells were washed with HBSS and medium containing lysostaphin (10 μg/mL, Sigma Chemicals) was added to lyse extracellular MRSA cells [39]. After 18 hours of incubation, 100 μL of MTT (5 mg/mL, Sigma Chemicals) was added and incubated for 2 hours. The medium was replaced with 150 μL of 0.04 M HCl to stop the reaction. Absorbance was measured at OD560nm. HMEC-1 without MRSA exposure was considered 0% damage control. The damage was presented as a percentage of 1 – (OD560nm values of samples / OD560nm values of control) [39].
Experimental IE Model in Rabbits
A well-characterized rabbit model of catheter-induced aortic valve endocarditis was used to validate the in vivo role of stringent response in persistent MRSA outcomes [33, 40]. The Institutional Animal Care and Use Committee of the Lundquist Institute for Biomedical Innovation at Harbor–University of California, Los Angeles Medical Center approved all animal study protocols. After 72 hours of catheterization, animals were infected intravenously of JE2 parental, its isogenic relP mutant or relP-complemented strain at 105 CFU/animal, an 95% infective dose (ID95) as established previously [28, 40]. At 24 hours postinfection, animals were randomized to receive no therapy (controls) or VAN (3.75 mg/kg, intravenously, twice daily for 3 days). VAN-treated animals were sacrificed at 24 hours after the last treatment dose to avoid VAN carryover effect. At sacrifice, cardiac vegetation, kidney, and spleen were sterilely removed and quantitatively cultured [28, 40]. MRSA counts in the target tissues were given as the mean log10 CFU/g of tissue (± standard deviation).
Statistical Analysis
All in vitro experiments were performed in triplicate and repeated 3 times. Statistical significance values were obtained by performing 2-tailed Student t test, or 1-way analysis of variance with Tukey multiple comparisons test (no adjustment). P values of < .05 were considered statistically significant.
RESULTS
PB Strains Exhibit Significantly Higher (p)ppGpp, GDP, and GTP Levels Than RB Strains
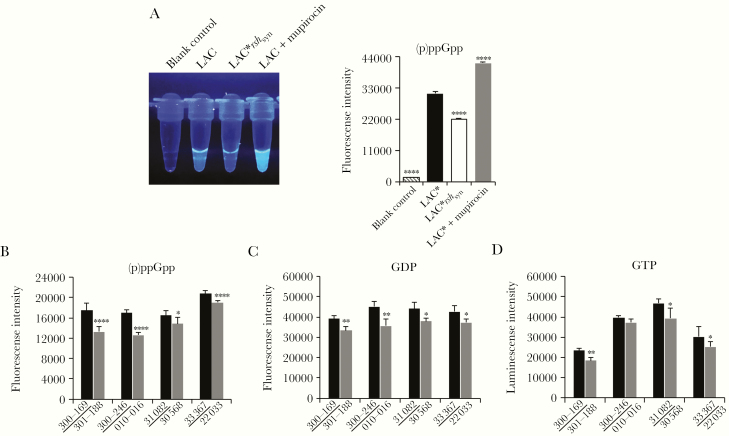
(p)ppGpp levels were tested by using a fluorescent chemosensor PyDPA method [21, 22]. We first validated this assay by demonstrating significant lesser and greater fluorescence intensities representing (p)ppGpp levels in the negative (LAC*rshsyn mutant strain) and positive (LAC* + mupirocin) controls vs LAC* parental strain, respectively (Figure 2A; P < .0001). Significantly higher (p)ppGpp levels were observed in PB strains vs their respective matched RB strains (Figure 2B; P < .05). We hypothesized that higher levels of (p)ppGpp would plausibly be due to elevated GDP/GTP levels in PB vs RB isolates. Indeed, significantly higher intracellular GDP and GTP levels were observed in PB strains vs their respective RB strains (Figure 2C and 2D).
Figure 2.
Increased (p)ppGpp associated with higher GDP and GTP levels in clinical persistent methicillin-resistant Staphylococcus aureus (MRSA) bacteremia (PB; underlined) vs resolving MRSA bacteremia (RB) strains. A, (p)ppGpp levels (fluorescence intensity) in buffer alone (blank control), LAC*, LAC*rshsyn (negative control), and mupirocin-treated LAC* (positive control). Levels of (p)ppGpp (B), GDP (C), and GTP (D) in PB vs RB clinical strains. Data are representative of 3 independent experiments. *P < .05, **P < .01, ***P < .001, ****P < .0001 vs the parental strain LAC* or their respective matched PB strain by Student t test. Error bars represent standard deviations.
PB Strains Express Higher psm Levels Than RB Strains
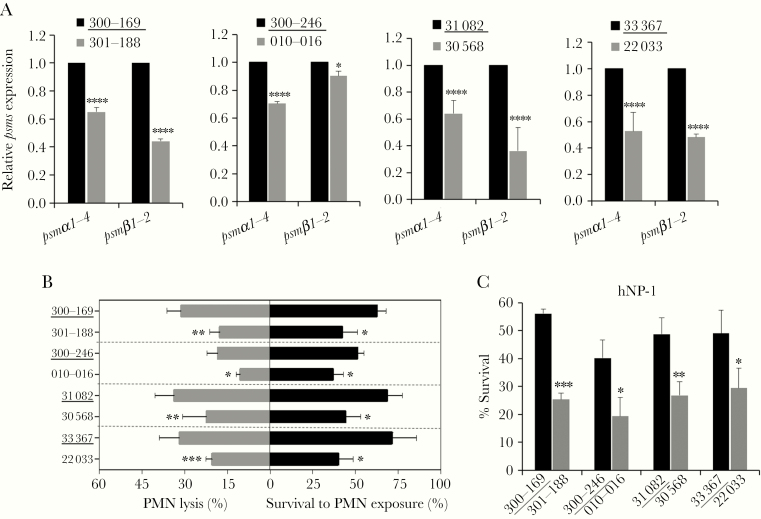
The stringent response from (p)ppGpp can regulate the production of phenol-soluble modulins (PSMs), which have been shown to play key roles in lysis of PMN [29, 41], thus contributing to the disruption of the host innate immune defense. We demonstrated that PB isolates exhibited significantly higher expression of psmα1–4 (1.4- to 1.9-fold; P < .0001), the more active form of PSM, relative to their respective matched RB comparators (Figure 3A). Additionally, 3 of the 4 PB isolates showed substantial higher psmβ1–2 expression (2.1- to 2.8-fold) vs their respective RB counterparts (Figure 3A; P < .0001).
Figure 3.
psms expression and its impacts on persistent methicillin-resistant Staphylococcus aureus (MRSA) bacteremia (PB) outcomes related phenotypes. Relative expression of psmα1–4 and psmβ1,2 (A); Neutrophils (PMN) lysis activity (B, left side) and survival to PMN exposure (B, right side); and susceptibilities to human neutrophil peptide 1 (hNP-1) (C) in clinical PB (underlined) vs resolving MRSA bacteremia strains. Data are representative of 3 independent experiments. *P < .05, **P < .01, ***P < .001, ****P < .0001 vs their respective PB strain by Student t test. Error bars represent standard deviations.
PSMs Correspond to Phenotypes Related to PB Outcomes
PSMs have been shown to mediate PMN lysis and dysfunction to promote S. aureus survival [41]. We noted that PB strains caused significantly greater PMN lysis (range, 18.4%–33.6%) than their respective matched RB strains (range, 10.4%–22.3%; Figure 3B), paralleling the increased S. aureus survival in PMN killing assay (51%–71% and 37%–44% in PB and RB strains, respectively; Figure 3B). As defensin contributes to the bactericidal activity within the phagolysosomes of PMNs, we also evaluated the sensitivity of PB vs RB isolates. As shown in Figure 3C, PB strains had significantly higher survival upon exposure to hNP-1 as compared to their respective RB strains.
Isogenic purF Mutants Validate the Causal Relationship Among Adaptive Purine Biosynthesis Regulation, Stringent Response, and PB Phenotypes
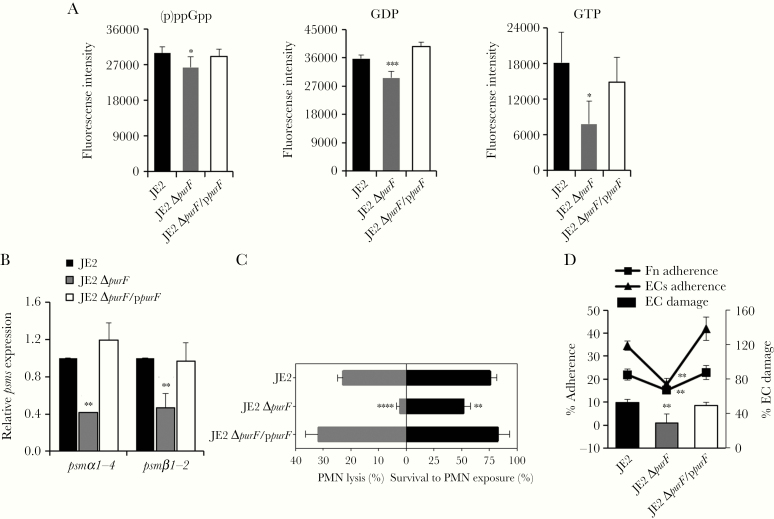
To verify that the above findings are linked to purine synthesis, MRSA parental strain JE2, its isogenic purF-mutant and purF-complemented strains were examined. Consistently, the purF mutant (a surrogate for RB clinical strains [16]) had significantly decreased (p)ppGpp, GDP, and GTP levels (Figure 4A; P < .05) vs its isogenic parental and purF-complemented strains. In addition, the purF mutant exhibited significantly reduced psmα1–4 and psmβ1,2 transcription (Figure 4B; P < .01), in association with lower PMN lysis and survival vs its isogenic parental and purF-complemented strains (Figure 4C; P < .05). Also, greater hNP-1–induced killing was observed in the purF mutant (survival rate, 27.5%) vs its isogenic parental and purF-complemented strains (survival rate, 49.2% and 44.0%, respectively; P < .05). To establish persistence, S. aureus must adhere to matrix ligands (eg, Fn) on host cells such as ECs, and damage host cells to facilitate disease and dissemination [38, 42]. We demonstrated that the purF mutant had significantly less Fn/EC adherence, and reduced EC damage vs its isogenic parental and purF-complemented strains (Figure 4D; P < .01).
Figure 4.
Interaction of the purine biosynthesis and stringent response. Levels of (p)ppGpp, GDP, and GTP (A); relative expression of psmα1–4 and psmα1,2 (B); Neutrophils (PMN) lysis activity (C, left side) and survival to PMN exposure (C, right side); and fibronectin (Fn) and endothelial cell (EC) adherence and EC damage (D) in JE2 parental, its isogenic purF mutant, and purF-complemented strains. Data are representative of 3 independent experiments. *P < .05, **P < .01, ***P < .001, ****P < .0001 vs JE2 parental and purF-complemented strains by Student t test. Error bars represent standard deviations.
(p)ppGpp Positively Impacts Coordinated Virulence Phenotypes Involved in Persistence
To assess the role of (p)ppGpp in the regulation of PSMs and its impacts on the phenotypes related to the PB outcomes, the parental strain JE2, its isogenic relP mutant, and relP-complemented strain set was used (Table 1). A significantly lower (p)ppGpp level was demonstrated in the relP mutant vs its parental strain and restored in the relP-complemented strain (Figure 5A; P < .0001). Importantly, akin to the RB clinical isolates and the purF mutant strain, the relP mutant of JE2 exhibited multiple phenotypes associated with RB: (1) significantly decreased psmα1–4 and psmβ1,2 expression (Figure 5B); (2) reduced PMN lysis and less survival in PMNs exposure (Figure 5C; P < .05); (3) decreased at least 0.5-fold survival rates against hNP-1 and VAN (P < .01); and (4) less Fn/EC adherence accompanied by reduced EC damage (Figure 5D; P < .01) vs its isogenic JE2 parental and relP-complemented strains.
Figure 5.
(p)ppGpp mediates psms expression and its impacts on persistent methicillin-resistant Staphylococcus aureus bacteremia (PB) outcome-related phenotypes. (p)ppGpp levels (A), relative expression of psmα1–4 and psmβ1,2 (B), Neutrophils (PMN) lysis activity (C, left side) and survival to PMN exposure (C, right side); and fibronectin (Fn) and endothelial cell (EC) adherence and EC damage (D) in JE2 parental, its isogenic relP mutant, and relP-complemented strains. Data are representative of 3 independent experiments. *P < .05, **P < .01, ***P < .001, ****P < .0001 vs JE2 parental and relP-complemented strains by Student t test. Error bars represent standard deviations.
Stringent Response Alters the Efficacy of VAN in an Experimental IE Model
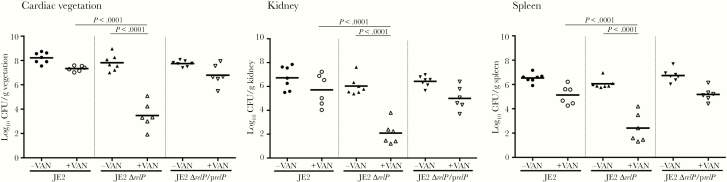
To affirm the impact of the stringent response in a relevant in vivo context, the role of the relP-mediated stringent response relative to PB outcome was investigated in a rabbit IE model. Results demonstrated that rabbits infected with the JE2 parent, relP mutant, or relP-complemented strain had equivalent virulence in the absence of VAN treatment, reflected in similar MRSA densities in key target tissues (Figure 6). Upon VAN treatment, rabbits infected with the relP mutant strain became hypersusceptible, with significantly reduced MRSA densities in all target tissues as compared to the parent and relP-complemented strains (Figure 6).
Figure 6.
The stringent response had a significant impact on the efficacy of vancomycin (VAN) in a rabbit infective endocarditis (IE) model. Densities of methicillin-resistant Staphylococcus aureus (MRSA) in target tissues in the IE model due to 105 colony-forming unit (CFU) challenges of JE2 parental and its isogenic relP-mutant or -complemented strains with/without VAN treatment. Each dot represents 1 animal. Horizontal black bars indicate means of MRSA densities. One-way analysis of variance with Tukey multiple comparisons test (no adjustment) was used to analyze the tissue MRSA counts between different groups.
DISCUSSION
The stringent response is a highly conserved adaptive survival mechanism in bacteria that is activated in response to various environmental stresses [10, 22]. We recently demonstrated that enhanced purine biosynthesis triggers a faster growth phenotype in PB vs RB strains [16]. Kriel et al observed that elevated GTP levels might result in increased ppGpp levels in Bacillus subtilis [14]. We thus postulated that augmented purine biosynthesis with enhanced growth generates key intermediates (eg, GDP and GTP) that are critical to the synthesis of (p)ppGpp [14], thereby directly linking purine biosynthesis and (p)ppGpp in clinical PB isolates. Consistent with our hypothesis, the present investigation demonstrated that PB clinical strains (vs genetically matched RB clinical strains) exhibit significantly higher (p)ppGpp production and corresponding higher GDP/GTP levels. In parallel, mutants defective in purine synthesis (ie, purF mutant) exhibited lower (p)ppGpp level vs its parent and purF-complemented strain.
It has been reported that the stringent response positively regulates the expression of cytotoxic PSMs, especially the PSMα [29]. In turn, S. aureus utilizes these PSMs to modulate PMN killing by lysing these host immune cells [29, 41]. Notably, the transcription of psm is positively regulated by AgrA, the response regulator in the quorum-sensing agr system [29] and inhibited by agr-specific inhibitors in MRSA strain LAC* [43]. Of great interest, our previous studies showed significantly earlier and higher agrA expression as a signature event in PB but not RB clinical isolates [4, 17]. In the current study, the data also revealed significantly increased (p)ppGpp production, accompanied by higher psmα1–4 and psmβ1,2 expression in PB vs RB clinical strains. These themes were confirmed in a genetically defined strain set, JE2 parental and its isogenic purF and relP mutants. These findings are consistent with the results of Geiger et al, who demonstrated marked declines in psmα and psmβ expression in 2 (p)ppGpp-defective S. aureus rel mutants [29]. Thus, these results add credence to the plausibility that the accumulation of (p)ppGpp can lead to elevated psm transcription.
PSMs have an extraordinary ability to lyse and alter the functionality of many eukaryotic cell types including endothelium, erythrocytes, and most importantly for innate host defense, PMNs [41]. We found here that higher psm expression in PB vs RB strains correlated with greater PMN lysis and enhanced survival in PMN and upon exposure to PMN defensin hNP-1. In support of our data, Yang et al recently reported that a chromosomal mutation of purB (an adenylosuccinate lyase of purine biosynthesis) in JE2 background significantly attenuated neutrophil lysis and reduced virulence in a zebrafish embryo infection model [44]. Collectively, these results further validate a unique interconnection between purine biosynthesis and survival in PMN among a cadre of connected events in PB vs RB strains—purine biosynthesis, (p)ppGpp production, psm expression, and PMN killing/lysis—which represent a novel and coordinated mechanism promoting PB outcomes in MRSA endovascular infections.
Given the recent perspective that MRSA persistence promotes the evolution of antibiotic resistance [45], this important finding further underscores an urgent need to investigate the specific mechanism(s) of persistence. In the current study, VAN showed significantly greater staphylocidal activity against a relP mutant variant as compared to its parental JE2 strain in vitro. These data are in accordance with Geiger et al showing that a relP mutant in HG001 was more susceptible to VAN-mediated killing than its parental strain [10]. Similarly, the reduced VAN resistance was also reported in relA and/or relQ mutants in Enterococcus faecium and Enterococcus faecalis strain backgrounds [46, 47]. The linkage of the stringent response to sensitivity of cell-wall active agent presumably may be due to changes in cell wall biosynthesis and/or cell wall turnover process [10]. Therefore, as purine biosynthesis positively regulates the stringent response [(p)ppGpp], the resultant increase in (p)ppGpp accumulation in PB vs RB strains appears to confer survival advantages to antibiotic challenges and immune subversion.
Most importantly, the in vitro genotypic and phenotypic findings as detailed above were translatable into significant differences in VAN treatment outcomes in the experimental IE model. For instance, animals infected with the relP mutant were significantly more susceptible to VAN treatment as compared to animals infected with the JE2 parental strain. Based on corroborating results generated in the current study, the PSM-related phenotypes may contribute to this in vivo outcome through at least 2 mechanisms (Figure 1). (1) As part of the first-line innate host defense, PMNs phagocytose bacteria followed by killing via oxidative burst [48]; in response, PB strains secrete higher PSMs, inducing greater PMN lysis [29, 41]. (2) Host defense peptides (eg, hNP-1) produced from PMN granules generally provide rapid and efficient killing of invading intracellular pathogens [48]; as part of the stringent response pathway, PB strains becomes more resistant to hNP-1, thus favoring S. aureus survival during the initial bacteremic stage of endovascular infection [3, 38]. We previously demonstrated that higher in vitro EC damage positively correlated with poor VAN responsiveness in the same IE model due to MRSA [38, 42]. In the current study, the purF and relP mutants also showed significantly less Fn and EC adherence, and EC damage vs the isogenic parental strain, which might contribute to the hypersusceptibility to VAN treatment outcome in the IE model. In agreement with our findings, Goncheva et al recently reported that mutation of purR, a transcriptional repressor of purine biosynthesis, enhanced Fn adherence, and virulence in a murine bacteremia model of S. aureus [49]. Last, the role of the purine biosynthesis pathway and the stringent response in altering the susceptibility of VAN likely contribute to the treatment outcomes of PB MRSA infections [10, 16].
We recognize some limitations of our current study. Most importantly, in clinical settings, the phenotype of PB in patients is likely to represent a complex interaction between host and pathogen, and thus it is multifactorial. In some cases, patients with PB may have longer duration of fever [50], while in other PB patients, all signs of sepsis and leukocytosis have resolved, especially in patients who have bacteremia for more than a week or so. Thus, it is likely that our current model system reflects some, but not all of the pathways to MRSA persistence. Taken together, persistent MRSA endovascular infection is complicated, and no single system is likely to fully recapitulate the complexity of clinical PB phenotype.
In summary, the current findings support the existence of a previously unrecognized adaptive mechanism intersecting purine biosynthesis and the stringent response in persistent endovascular infections caused by MRSA strains, and provide valuable insights into how MRSA adaptively utilizes purine biosynthesis and the stringent response to evade antibiotic action and subvert immune responses to cause PB outcomes.
Notes
Financial support. This work was supported by the National Institutes of Health (grant numbers R01 AI139244 to Y. Q. X; R01 AI130056 to A. S. B; and U01 AI124319 to M. R. Y).
Potential conflicts of interest. M. R. Y. is a founder of NovaDigm Therapeutics, Inc. In addition, M. R. Y. has the following patents issued: “Antimicrobial Kinocidins and Methods”; “Peptides and Methods Inducing PCD”; “Antimicrobial Metapeptides”; “Context-Activated Protides”; and “Anti-Infective Phenyl-OH Benzoates.” All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fowler VG Jr, Miro JM, Hoen B, et al. ICE Investigators Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 2005; 293:3012–21. [DOI] [PubMed] [Google Scholar]
- 2. Fowler VG Jr, Sakoulas G, McIntyre LM, et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 2004; 190:1140–9. [DOI] [PubMed] [Google Scholar]
- 3. Xiong YQ, Fowler VG, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis 2009; 199:201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2011; 55:5631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 2015; 197:1146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 2015; 13:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 2015; 24:72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol 2016; 101:531–44. [DOI] [PubMed] [Google Scholar]
- 9. Geiger T, Goerke C, Fritz M, et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 2010; 78:1873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol 2014; 196:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol 2005; 13:236–42. [DOI] [PubMed] [Google Scholar]
- 12. Christopherson RI, Lyons SD, Wilson PK. Inhibitors of de novo nucleotide biosynthesis as drugs. Acc Chem Res 2002; 35:961–71. [DOI] [PubMed] [Google Scholar]
- 13. Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature 1972; 238:381–4. [DOI] [PubMed] [Google Scholar]
- 14. Kriel A, Bittner AN, Kim SH, et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 2012; 48:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang B, Dai P, Ding D, et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol 2019; 15:141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L, Abdelhady W, Donegan NP, et al. Role of purine biosynthesis in persistent methicillin-resistant Staphylococcus aureus infection. J Infect Dis 2018; 218:1367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdelhady W, Chen L, Bayer AS, et al. Early agr activation correlates with vancomycin treatment failure in multi-clonotype MRSA endovascular infections. J Antimicrob Chemother 2015; 70:1443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abdelhady W, Bayer AS, Seidl K, et al. Reduced vancomycin susceptibility in an in vitro catheter-related biofilm model correlates with poor therapeutic outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57:1447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spentzas T, Kudumula R, Acuna C, et al. Role of bacterial components in macrophage activation by the LAC and MW2 strains of community-associated, methicillin-resistant Staphylococcus aureus. Cell Immunol 2011; 269:46–53. [DOI] [PubMed] [Google Scholar]
- 20. Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 2004; 186:5267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhee HW, Lee CR, Cho SH, et al. Selective fluorescent chemosensor for the bacterial alarmone (p)ppGpp. J Am Chem Soc 2008; 130:784–5. [DOI] [PubMed] [Google Scholar]
- 22. Gao W, Chua K, Davies JK, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 2010; 6:e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corrigan RM, Bellows LE, Wood A, Gründling A. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in gram-positive bacteria. Proc Natl Acad Sci U S A 2016; 113:E1710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verhelst J, Spitaels J, Nürnberger C, et al. Functional comparison of Mx1 from two different mouse species reveals the involvement of loop L4 in the antiviral activity against influenza A viruses. J Virol 2015; 89:10879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mempin R, Tran H, Chen C, Gong H, Kim Ho K, Lu S. Release of extracellular ATP by bacteria during growth. BMC Microbiol 2013; 13:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McElroy WD, DeLuca MA. Firefly and bacterial luminescence: basic science and applications. J Appl Biochem 1983; 5:197–209. [PubMed] [Google Scholar]
- 27. Mondal S, Hsiao K, Goueli SA. A Homogenous bioluminescent system for measuring GTPase, GTPase activating protein, and guanine nucleotide exchange factor activities. Assay Drug Dev Technol 2015; 13:444–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L, Cheung A, Bayer AS, et al. The global regulon sarA regulates β-Lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J Infect Dis 2016; 214:1421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geiger T, Francois P, Liebeke M, et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 2012; 8:e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kraus D, Herbert S, Kristian SA, et al. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 2008; 8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katzenmeyer KN, Szott LM, Bryers JD. Artificial opsonin enhances bacterial phagocytosis, oxidative burst and chemokine production by human neutrophils. Pathog Dis 2017; 75. doi:10.1093/femspd/ftx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alazami AM, Patel N, Shamseldin HE, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 2015; 10:148–61. [DOI] [PubMed] [Google Scholar]
- 33. Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob Agents Chemother 2005; 49:3114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaili S, Cheung AL, Bayer AS, et al. The GraS sensor in Staphylococcus aureus mediates resistance to host defense peptides differing in mechanisms of action. Infect Immun 2016; 84:459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiong YQ, Bayer AS, Yeaman MR, Van Wamel W, Manna AC, Cheung AL. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect Immun 2004; 72:1832–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ades EW, Candal FJ, Swerlick RA, et al. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol 1992; 99:683–90. [DOI] [PubMed] [Google Scholar]
- 37. Zell C, Resch M, Rosenstein R, Albrecht T, Hertel C, Götz F. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int J Food Microbiol 2008; 127:246–51. [DOI] [PubMed] [Google Scholar]
- 38. Seidl K, Bayer AS, Fowler VG Jr, et al. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob Agents Chemother 2011; 55:575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seidl K, Zinkernagel AS. The MTT assay is a rapid and reliable quantitative method to assess Staphylococcus aureus induced endothelial cell damage. J Microbiol Methods 2013; 92:307–9. [DOI] [PubMed] [Google Scholar]
- 40. Abdelhady W, Bayer AS, Seidl K, et al. Impact of vancomycin on sarA-mediated biofilm formation: role in persistent endovascular infections due to methicillin-resistant Staphylococcus aureus. J Infect Dis 2014; 209:1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 2007; 13:1510–4. [DOI] [PubMed] [Google Scholar]
- 42. Seidl K, Bayer AS, McKinnell JA, Ellison S, Filler SG, Xiong YQ. In vitro endothelial cell damage is positively correlated with enhanced virulence and poor vancomycin responsiveness in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Cell Microbiol 2011; 13:1530–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou Y, Niu C, Ma B, et al. Inhibiting PSMalpha-induced neutrophil necroptosis protects mice with MRSA pneumonia by blocking the agr system. Cell Death Dis 2018; 9:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang D, Ho YX, Cowell LM, Jilani I, Foster SJ, Prince LR. A genome-wide screen identifies factors involved in S. aureus-induced human neutrophil cell death and pathogenesis. Front Immunol 2019; 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020; 367:200–4. [DOI] [PubMed] [Google Scholar]
- 46. Honsa ES, Cooper VS, Mhaissen MN, et al. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. MBio 2017; 8. doi:10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abranches J, Martinez AR, Kajfasz JK, Chávez V, Garsin DA, Lemos JA. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol 2009; 191:2248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol 2006; 6:173–82. [DOI] [PubMed] [Google Scholar]
- 49. Goncheva MI, Flannagan RS, Sterling BE, et al. Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat Commun 2019; 10:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hawkins C, Huang J, Jin N, Noskin GA, Zembower TR, Bolon M. Persistent Staphylococcus aureus bacteremia: an analysis of risk factors and outcomes. Arch Intern Med 2007; 167:1861–7. [DOI] [PubMed] [Google Scholar]