Abstract
Background
M184V/I cause high-level lamivudine (3TC) and emtricitabine (FTC) resistance and increased tenofovir disoproxil fumarate (TDF) susceptibility. Nonetheless, 3TC and FTC (collectively referred to as XTC) appear to retain modest activity against human immunodeficiency virus-1 with these mutations possibly as a result of reduced replication capacity. In this study, we determined how M184V/I impacts virus load (VL) in patients failing therapy on a TDF/XTC plus nonnucleoside reverse-transcriptase inhibitor (NNRTI)-containing regimen.
Methods
We compared VL in the absence and presence of M184V/I across studies using random effects meta-analysis. The effect of mutations on virus reverse-transcriptase activity and infectiousness was analyzed in vitro.
Results
M184I/V was present in 817 (56.5%) of 1445 individuals with virologic failure (VF). Virus load was similar in individuals with or without M184I/V (difference in log10 VL, 0.18; 95% confidence interval, .05–.31). CD4 count was lower both at initiation of antiretroviral therapy and at VF in participants who went on to develop M184V/I. L74I was present in 10.2% of persons with M184V/I but absent in persons without M184V/I (P < .0001). In vitro, L74I compensated for defective replication of M184V-mutated virus.
Conclusions
Virus loads were similar in persons with and without M184V/I during VF on a TDF/XTC/NNRTI-containing regimen. Therefore, we did not find evidence for a benefit of XTC in the context of first-line failure on this combination.
Keywords: antiretroviral, compensatory mutation, drug resistance, HIV, lamivudine
Lamivudine is a cornerstone antiretroviral whose efficacy has been ascribed to high fitness cost of the lamivudine resistance mutation M184V. However, here we demonstrate elevated viral loads in the context of M184V, likely attributable to compensatory mutations such as L74I.
(See the Editorial Commentary by Kuritzkes, on pages 1067–9.)
The global scale up of antiretroviral therapy (ART) using a public health approach with limited viral load (VL) monitoring has been accompanied by high prevalence of drug resistance to nonnucleoside reverse-transcriptase inhibitor (NNRTI)-containing regimens among individuals with virological failure (VF) in low- and middle-income countries (LMICs) [1–6].
The cytosine analogs lamivudine (3TC) and emtricitabine (FTC), collectively referred to as XTC, are components of first- and second-line regimens recommended by the World Health Organization (WHO). However, high-level XTC resistance can be conferred and selected by single amino acid changes at position 184 of reverse transcriptase (RT) in the highly conserved (Y183, M184, D185, D186) amino acid domain that includes the active (catalytic) site of the p66 polymerase subunit of RT [7]. M184V/I are the most commonly occurring drug-resistant mutations in persons with acquired resistance to first-generation NNRTI-containing regimens [1–6].
Several lines of evidence suggest that in addition to causing high-level reductions in XTC susceptibility in vitro and modestly increased tenofovir disoproxil fumarate (TDF) susceptibility, viruses with these mutations retain some in vivo susceptibility to XTC possibly because of their reduced replication capacity [8–10]. For example, early studies showed that in patients receiving 3TC monotherapy, or dual therapy with AZT/3TC, VL did not return to baseline despite the development of M184V [9, 11–14]. In addition, discontinuation of 3TC during combination ART (cART) was associated with a modest increase in VL [15–17]. By contrast, the COLATE study, a randomized controlled trial conducted in Europe in the early 2000s, showed that there was no effect of removal of 3TC from a failing regimen in which the endpoint was viral suppression to <200 copies/mL or VL change of 1.4 log10 [18].
Therefore, to understand the relationship between M184I/V and VL in the era of tenofovir-based cART in which thymidine analog mutations (TAMs) were not present, and also in the context of limited or no access to VL monitoring, we studied individuals failing the WHO-recommended first-line regimen of TDF/XTC/NNRTI across a range of settings [19].
METHODS
The study population has previously been described and is presented in Supplementary Table 1 [20–41]. Patients treated with TDF plus 3TC/FTC and nevirapine/efavirenz (EFV) were included when documented VF and RT sequence data from codons 40 to 240 were available. Virologic failure was locally determined, and the threshold for LMICs was 1000 copies/mL. Human immunodeficiency virus (HIV)-1 RT sequences were determined by standard Sanger sequencing at individual study sites.
Mutations were defined as amino acid differences at positions 1 to 240 between each sequence and the consensus subtype B amino acid reference sequence. Because some individuals may have been exposed to thymidine analogs before TDF-containing regimens [5], we excluded individuals with sequences containing TAMs—M41L, D67N, K70R, L210W, T215Y/F, and K219Q/E.
Each sequence was subtyped as previously described, and sequence quality control measures were taken to identify sequences with APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) G-to-A hypermutation [20]. Duplicate sequences were removed. All patients reported that they were antiretroviral (ARV) naive at baseline. The primary outcome was VL at VF; hence, patients without this outcome were excluded.
Statistical Analysis
We graphically compared the distribution of log10 VLs according to presence of M184I/V mutation both within and across studies. To quantify the impact of M184I/V on VL, we calculated mean log10 VL in each study according to M184I/V. Differences were pooled across studies using random-effects meta-analysis. Estimates of the standard error in each study were calculated by dividing the pooled estimate of the standard deviation by the square root of the number of patients with/without M184I/V in any given study. We repeated this process in subgroups of patients defined by several baseline characteristics: presence of K65R mutation, presence of major NNRTI mutations, choice of NRTI, choice of NNRTI, categories of baseline CD4 count (< and >200 cells/mm3), and categories of baseline VL (< and >100 000 copies per mL). We used the same methods for analyses of CD4 count and treatment failure. To assess whether M184I/V was associated with VL failure independently of other mutations, we performed a separate analysis in which we used a mixed linear regression model adjusting for study as a random effect and other mutations associated with increased VL (which were identified by forward stepwise variable selection). Next, we used Fisher’s exact test to identify mutations associated with M184I/V. We used 2-sided P values and Stata version 15.1 for all statistical analyses.
In Vitro Analyses
A patient-derived pol sequence was identified with mutations of interest, and the gag-PR-RT-IN region was amplified by polymerase chain reaction with flanking restriction sites inserted into primers. After cloning into an expression plasmid, site-directed mutagenesis was performed to revert isoleucine back to leucine at RT amino acid 74, valine back to methionine at RT amino acid 184, or both. Plasmids expressing gag-pol were cotransfected into 293T cells along with a vesicular stomatitis virus-G envelope expressing plasmid and a vector encoding luciferase expressed from a long terminal repeat promoter as previously described [42]. Supernatant containing virus was harvested 2 days later and used to infect fresh 293T cells. Luminescence as a read out of infection was read by luminometry 2 days later. Viral p24 abundance in supernatants was estimated using Western blot, using a p24 antibody as previously described [43].
RESULTS
Among 2873 participants included in the initial group, 1445 from 32 study groups across 15 countries had an available failure VL measurement, and M184I/V was present in 817 (56.5%) of these (Table 1 and Supplementary Table 1). Participants were from sub-Saharan Africa (55.4%), Asia (19.2%), Europe (16.2%), and North America (9.3%). All participants were on TDF, most of them were also treated with EFV (75.2%) and 3TC (64.5%), and participants harboring M184I/V-mutated virus were significantly more likely to have high-level tenofovir and NNRTI resistance (Table 2). Participants harboring M184I/V were also more likely to have multiple NNRTI mutations.
Table 1.
Baseline Characteristics of Participants by Geographic Region
| Region | M184 I/V | Patients | EFV | 3TC | Baseline CD4 Count | Baseline Log10 Viral Load | ||
|---|---|---|---|---|---|---|---|---|
| N With Data | N With Data | |||||||
| Overall | No | 628 | 523 (83.3%) | 350 (55.7%) | 351 | 180.0 (82.0 to 288.0) | 253 | 5.0 (4.5 to 5.5) |
| Yes | 817 | 564 (69.0%) | 582 (71.2 %) | 385 | 88.0 (36.0 to 165.0) | 187 | 5.2 (4.7 to 5.7) | |
| Sub-saharan Africa | No | 257 | 198 (77.0%) | 204 (79.4%) | 142 | 148.0 (69.0 to 264.0) | 43 | 5.3 (4.5 to 5.7) |
| Yes | 543 | 356 (65.6%) | 430 (79.2%) | 270 | 77.0 (35.0 to 138.0) | 71 | 5.3 (4.7 to 5.7) | |
| Asia | No | 136 | 112 (82.4%) | 110 (80.9%) | 0 | - | 0 | - |
| Yes | 141 | 121 (85.8%) | 122 (86.5%) | 4 | 69.5 (33.5 to 159.0) | 5 | 4.7 (4.6 to 5.9) | |
| Europe | No | 146 | 127 (87.0%) | 25 (17.1%) | 138 | 199.5 (84.0 to 304.0) | 136 | 5.0 (4.6 to 5.5) |
| Yes | 88 | 53 (60.2%) | 23 (26.1%) | 77 | 157.0 (62.0 to 232.0) | 76 | 5.1 (4.8 to 5.7) | |
| North America | No | 89 | 86 (96.6%) | 11 (12.4%) | 71 | 204.0 (98.0 to 351.0) | 77 | 4.7 (4.3 to 5.3) |
| Yes | 45 | 34 (75.6%) | 7 (15.6%) | 34 | 67.5 (27.0 to 156.0) | 35 | 5.2 (4.8 to 5.6) |
Abbreviations: 3TC, lamivudine; EFV, efavirenz.
Table 2.
Summary of Drug Resistance Characteristics of Participants at Virological Failure With Tenofovir + Cytosine Analog + NNRTI by Geographical Region
| Region | M184 I/V | TDF Resistance, n (%) | At Least One Major NNRTI Mutation, n (%) | Number of NNRTI Mutations, Mean (SD) | Failure Log10 Viral Load | Failure CD4 Count | |
|---|---|---|---|---|---|---|---|
| N With Data | Median (IQR) | ||||||
| Overall | No | 137 (21.8%) | 380 (60.5%) | 1.2 (1.3) | 4.3 (3.4 to 5.0) | 237 | 263.0 (121.0 to 382.0) |
| Yes | 539 (66.0%) | 792 (96.9%) | 2.9 (1.3) | 4.7 (4.1 to 5.3) | 211 | 104.0 (29.0 to 236.0) | |
| Sub-saharan Africa | No | 80 (31.1%) | 175 (68.1%) | 1.5 (1.4) | 4.7 (3.9 to 5.2) | 29 | 262.0 (180.0 to 360.0) |
| Yes | 400 (73.7%) | 531 (97.8%) | 2.9 (1.3) | 4.8 (4.1 to 5.3) | 52 | 137.0 (20.0 to 219.0) | |
| Asia | No | 30 (22.1%) | 91 (66.9%) | 1.3 (1.4) | 4.8 (4.1 to 5.3) | 119 | 188.0 (71.0 to 355.0) |
| Yes | 82 (58.2%) | 130 (92.2%) | 2.9 (1.5) | 4.9 (4.2 to 5.3) | 118 | 87.5 (29.0 to 229.0) | |
| Europe | No | 20 (13.7%) | 65 (44.5%) | 0.7 (1.0) | 3.4 (2.7 to 4.6) | 32 | 323.0 (238.0 to 387.0) |
| Yes | 38 (43.2%) | 86 (97.7%) | 2.6 (1.4) | 4.2 (3.8 to 4.8) | 12 | 242.5 (122.0 to 345.0) | |
| North America | No | 7 (7.9%) | 49 (55.1%) | 0.8 (0.9) | 3.4 (2.4 to 4.3) | 57 | 312.0 (198.0 to 476.0) |
| Yes | 19 (42.2%) | 45 (100.0%) | 2.8 (1.4) | 4.2 (3.7 to 4.7) | 29 | 173.0 (42.0 to 329.0) |
Abbreviations: IQR, interquartile range; NNRTI, nonnucleoside reverse-transcriptase inhibitor; SD, standard deviation; TDF, tenofovir disoproxil fumarate.
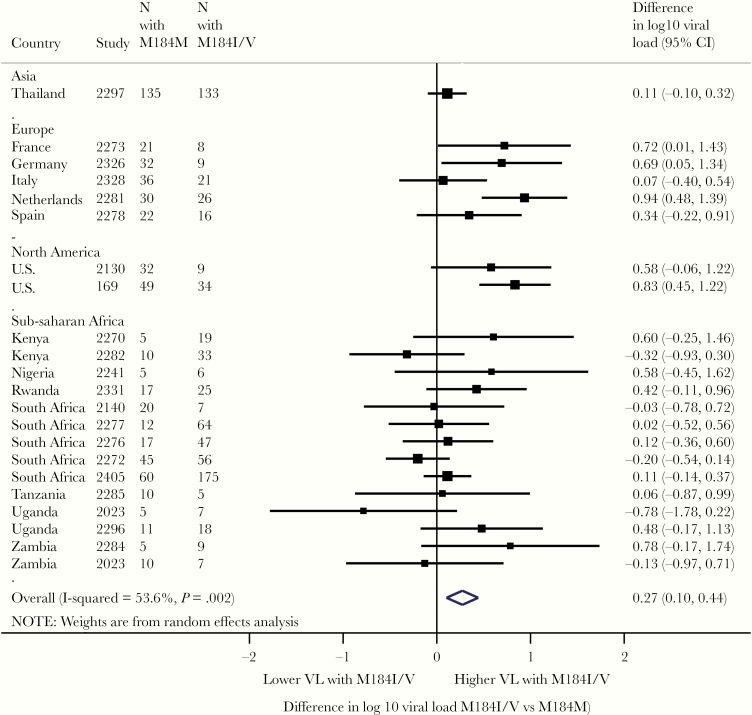
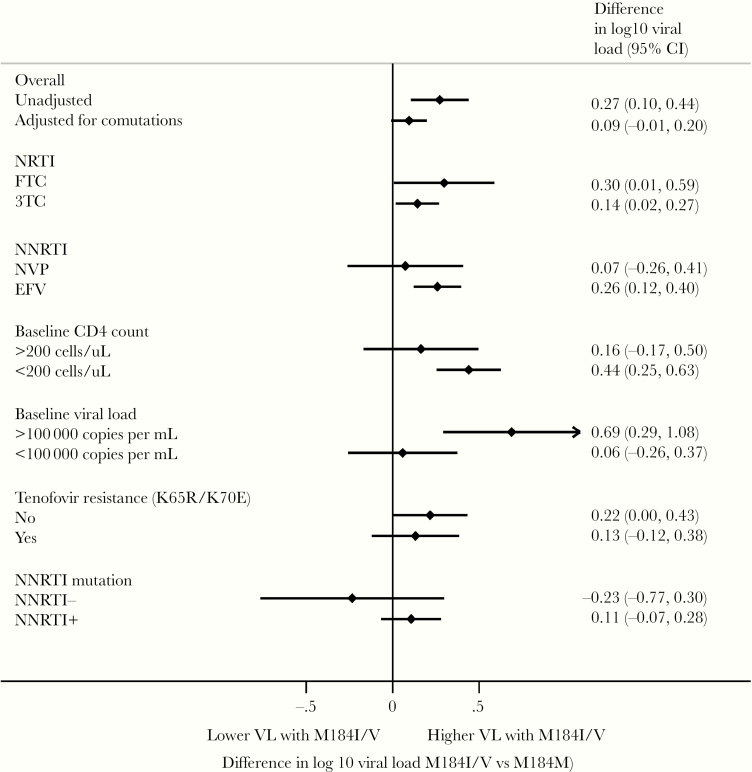
In a crude comparison of VL failure, patients with M184I/V present had a higher median log10 VL (4.7; interquartile range [IQR], 3.4–5) than patients without M184I/V (median 4.3; IQR, 4.1–5.3). When restricting analyses to comparisons of patients within the same study, the estimated difference in VL was nonsignificant in the vast majority of studies (Figure 1). When within-study differences were pooled across studies, there was a marginally higher VL in patients with M184I/V present compared with absent (pooled difference in log10 VL, 0.18; 95% confidence interval [CI], .05–.31) (Figure 2). After statistical adjustment for other mutations independently associated with increased VL, M184I/V was no longer significantly associated with VL failure. However, the estimated difference and 95% CI (0.09; 95% CI, −.01 to .20) excluded any meaningful decrease in VL failure associated with M184I/V. There was no evidence that relationship between M184I/V and VL failure was modified by choice of NRTI, choice of NNRTI, or drug resistance to NNRTI or tenofovir (Figure 2).
Figure 1.
Difference in viral load by mutations at reverse-transcriptase position 184 in study groups with 95% confidence interval (CI) using random-effects meta-analysis. Boxes represent means, lines represent 95%. Estimates to the right indicate higher viral load in the presence of M184V/I, and estimates to the left indicate lower viral load in presence of M184V/I.
Figure 2.
Association of M184V/I mutation with log10 viral load across subgroups. Diamonds represent means, lines represent 95%. Estimates to the right indicate higher viral load in the presence of M184V/I. 3TC, lamivudine; EFV, efavirenz; FTC, emtricitabine; NNRTI, nonnucleoside reverse-transcriptase inhibitor; NRTI, nucleoside reverse-transcriptase inhibitor; NVP, nevirapine.
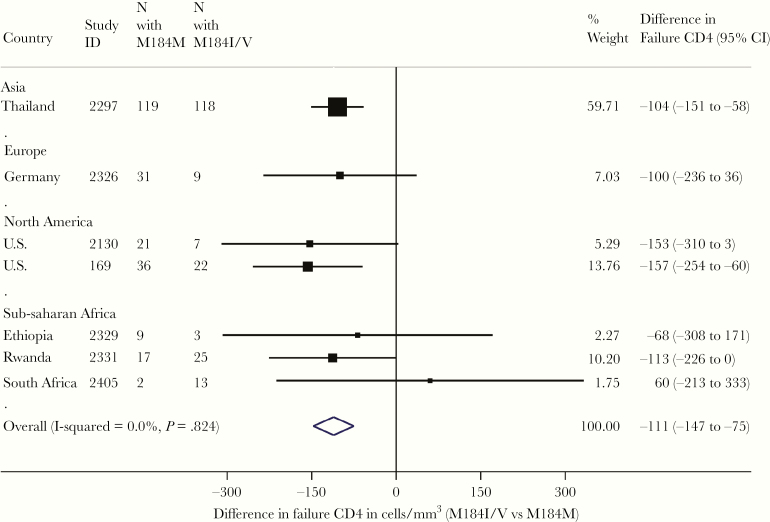
We next explored the relationship between detection of M184I/V failure and CD4 count, noting that the duration of VF was likely longer in LMIC regions. Mean baseline CD4 was significantly lower among patients who went on to develop M184I/V by treatment failure compared with those who did not (88 vs 180, P < .0001). CD4 count at VF was also lower in patients with M184V/I than those without (Figure 3). Between baseline and treatment failure, CD4 count increased to a similar extent in patients with and without M184I/V (median increase, 79 vs 48 cells/mm3; P = .55).
Figure 3.
Differences in CD4 count during virological failure within studies by presence and absence of M184V/I. Boxes represent means, lines represent 95%. Estimates to the left of center line indicate lower CD4 count in participants with M184V/I.
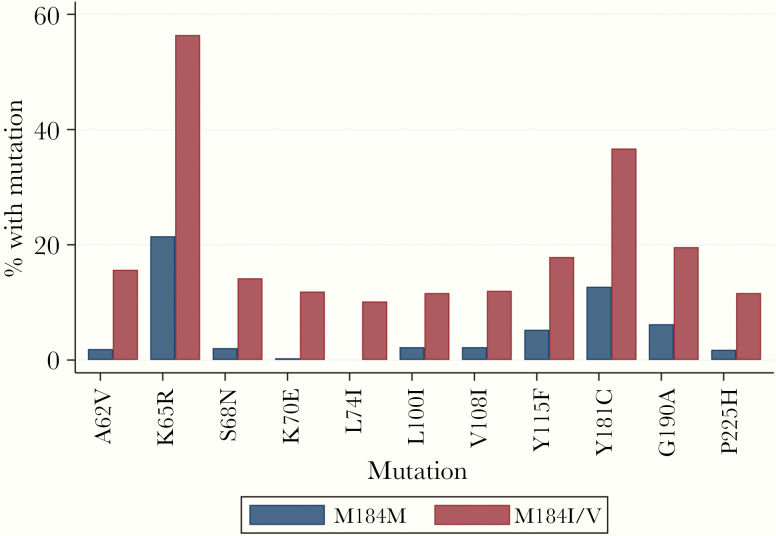
We then examined NRTI mutations associated with M184V/I that might play a compensatory role for M184I/V. We looked for associations in the dataset between M184V/I and RT amino acid positions known to be associated with drug exposure. Figure 4 shows mutations with strong evidence of an association with M184I/V. Many of these mutations have previously been associated with drug resistance to tenofovir, either directly (K65R, K70E) or as compensatory mutations for K65R (A62V, S68N, F155Y). The following NNRTI mutations were also associated: A98G, L100I, K103R, V108I, Y181C, Y188L, G190A, P225H, L228R, and M230L.
Figure 4.
Human immunodeficiency virus reverse-transcriptase inhibitor resistance-associated mutations enriched in virologically failing participants (n = 1445) with M184V/I. Mutations are shown that occurred in at least 10% of individuals with M184V/ at a significance level of <.001.
Of note, L74I was the only mutation to be exclusively associated with M184V/I, occurring in 83 (10.2%) of patients with M184I/V, but not in 628 patients in which M184I/V was absent (P for association <.0001). L74I was observed in 11.7% of subtype C-infected participants with M184I/V at VF and in 14.4% of CRF01_AE participants with M184I/V at VF (Supplementary Table 2).
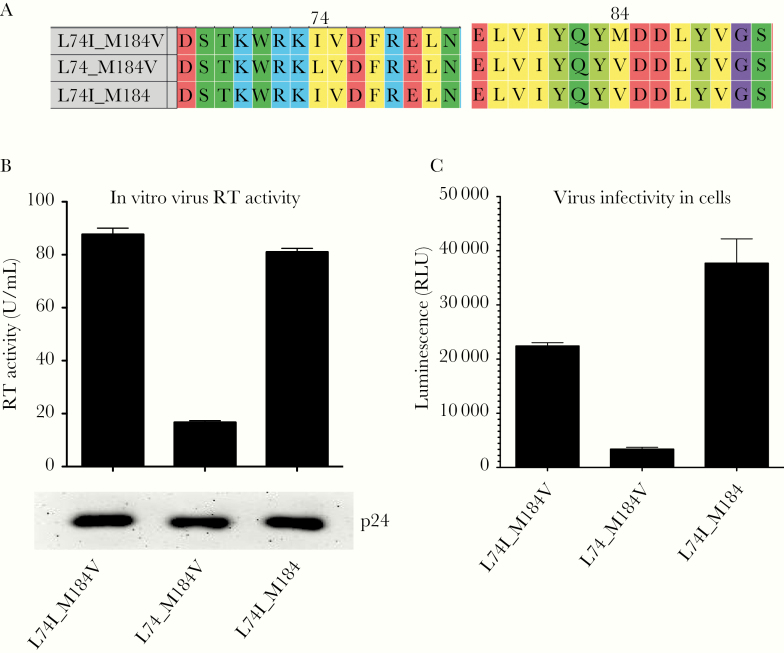
A previous study reported that L74I can restore replication to a virus with the K65R mutation without conferring drug resistance [44]; therefore, we sought to test the hypothesis that L74I could restore replication “fitness” to a M184V mutant virus, thereby explaining the higher than expected VLs. Molecular characterization of virus with the mutations M184V and L74I was undertaken. The viral isolate tested also had NNRTI resistance mutations A98G, K103N, and P225H. Site-directed mutagenesis was performed to revert isoleucine back to leucine at 74 and revert valine to methionine at 184 (Figure 5A). However, we did not assess the impact of M184I. We measured (1) infectivity of these viruses and (2) RT efficiency in a single-round replication assay (Figure 5). We found that removing the L74I mutation significantly decreased the efficiency of reverse transcription (Figure 5B, compare left bar with middle bar), whereas virus abundance was not affected, as determined by Western blot of viral p24 abundance in supernatants (Figure 5B, bottom panel). Infectivity was also significantly decreased by reversion of the compensatory mutation (Figure 5C, compare left bar with middle bar). Mutation of M184V back to M, leaving a virus with only L74I, had no impact on RT efficiency and a minor effect on infectivity (Figure 5B and C, compare left and right bars).
Figure 5.
In vitro replication measurement of lamivudine-resistant subtype C clinical isolate containing M184V and L74I and revertant mutations. (A) Amino acid multiple sequence alignment of clinical isolate and revertant mutants generated by site-directed mutagenesis. Numbering is relative to strain HXB2. (B) In vitro reverse-transcription (RT) efficiency contained in pelleted single-round virus from cells producing clinical human immunodeficiency virus (HIV) isolate RT sequence and mutants. Bottom panel shows Western blot of corresponding virus-associated p24 in supernatants from cells. (C) Single-round infection of target HEK 293T cells by equal quantities of luciferase expressing vesicular stomatitis virus-G pseudotyped HIV viruses from B. Data in B and C were performed in replicate, and means are presented with error bars corresponding to standard deviation. RLU, relative light units.
DISCUSSION
Despite having a low genetic barrier to drug resistance, 3TC has retained importance and a central role in both first- and second-line ART [45]. Therefore, a complete understanding of 3TC efficacy is important, particularly given reports suggesting that 3TC use confers VL benefit despite high-level resistance to the drug in the form of the M184V/I.
Our primary finding that VL was similar in participants with and without M184V/I at the time of VF was robust across baseline CD4 count, baseline VL, gender, and different NNRTI and NRTI drugs in the first-line treatment regimen. We observed lower baseline and VF CD4 counts in individuals with M184V/I, although rate of change of CD4 did not differ based on M184V/I status. Lower baseline CD4 count is known to be associated with higher VF rates and a higher probability of drug resistance at VF [6, 46]. A possible explanation for this finding is that the antiviral effect of a competent immune system is important in limiting replication and emergence of resistance in tissue compartments where ARV drug penetration is suboptimal. A lower CD4 count at VF in the group with M184V/I further argues against this mutation being “protective” or “benign.” These data are also consistent with reports of the pathogenic potential of M184V-containing viruses in both humans [47] and animal models [48].
We identified L74I as being specifically enriched in individuals with M184V and not present at all in those without M184V/I. We observed significant prevalence of L74I in subtypes C and CRF01_AE, although limited numbers of participants across subtypes hindered a full understanding of subtype distribution. In vitro experiments demonstrated that L74I restores replication efficiency to a virus with the M184V mutation over a single round of infection, and that enhancement was due to efficiency of HIV reverse transcription in viral particles.
The emergence of L74I exclusively in patients with M184V/I suggests an in vivo selection advantage of L74I + M184V replication over M184V alone, at least in some individuals. L74I was first reported as a mutation associated with exposure to abacavir or less commonly tenofovir [49, 50], and it appeared more common in patients with TAMs [50]. Correlation with M184V/I has not been made to date, and in vitro experiments have not been performed with L74I + M184V/I-containing viruses.
Because L74I was observed only in approximately 10% of those with M184V/I, we postulate that alternative mutations, less strongly linked to M184V/I or perhaps outside the region of the pol gene sequenced in this study, could have similar effects as L74I in participants with M184V/I. Data from our study support the transmission potential of M184V/I-containing viruses in the context of prolonged VF and accumulated coevolved mutations in RT that occurs under “real-world” conditions.
Limitations of this study include its retrospective cross-sectional design, absence of drug levels or adherence data, and unknown duration of VF for participants. Our study was not designed to provide a mechanistic understanding of the relationship between M184 and fitness: it was designed to understand the pathogenic potential of M184V-containing viruses in patients treated in the real world. Finally, there was heterogeneity between population groups, and, to account for this, analyses were conducted within study. It should also be noted that stratification by tenofovir or NNRTI resistance resulted in small numbers for subanalyses.
CONCLUSIONS
In summary, we show that 3TC-resistant and 3TC-susceptible viruses show similar VLs in patients failing NNRTI-based ART containing 3TC, tenofovir, and NNRTI, likely in part due to viral evolution of compensatory changes that maintain replication efficiency of M184V/I-containing viruses. These data reinforce the importance of effective VL monitoring to limit HIV drug resistance and disease progression in the face of suboptimal drug pressure, particularly in low-resource settings. Finally, given that we did not find benefit of 3TC in patients failing first-line treatment, a prospective clinical trial could determine whether there is benefit for including XTC in second-line regimens for the treatment of persons whose viruses develop M184I/V after VF on a first-line treatment regimen.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the participants. We also thank Petra Mlcochova for assistance with Western blotting.
Financial support. This study was funded by the Wellcome Trust.
Potential conflicts of interest. R. K. G. has acted as ad hoc consultant for Gilead Sciences and ViiV. C. F. P. has acted as ad hoc consulant for Gilead Sciences, Merck, Janssen, Theratechnologies, and ViiV. R. S. S. has received research funding from Janssen. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gupta RK, Hill A, Sawyer AW, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis 2009; 9:409–17. [DOI] [PubMed] [Google Scholar]
- 2. Goodall RL, Dunn DT, Nkurunziza P, et al. ; DART Virology Group Rapid accumulation of HIV-1 thymidine analogue mutations and phenotypic impact following prolonged viral failure on zidovudine-based first-line ART in sub-Saharan Africa. J Antimicrob Chemother 2017; 72:1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boender TS, Kityo CM, Boerma RS, et al. Accumulation of HIV-1 drug resistance after continued virological failure on first-line ART in adults and children in sub-Saharan Africa. J Antimicrob Chemother 2016; 71:2918–27. [DOI] [PubMed] [Google Scholar]
- 4. Ciaffi L, Koulla-Shiro S, Sawadogo AB, et al. ; MOBIDIP study group Boosted protease inhibitor monotherapy versus boosted protease inhibitor plus lamivudine dual therapy as second-line maintenance treatment for HIV-1-infected patients in sub-Saharan Africa (ANRS12 286/MOBIDIP): a multicentre, randomised, parallel, open-label, superiority trial. Lancet HIV 2017; 4:e384–92. [DOI] [PubMed] [Google Scholar]
- 5. Gregson J, Kaleebu P, Marconi VC, et al. Occult HIV-1 drug resistance to thymidine analogues following failure of first-line tenofovir combined with a cytosine analogue and nevirapine or efavirenz in sub Saharan Africa: a retrospective multi-centre cohort study. Lancet Infect Dis 2017; 17:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. TenoRes Study Group. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 2016; 16:565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tisdale M, Kemp SD, Parry NR, Larder BA. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3’-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci U S A 1993; 90:5653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J, Kuritzkes DR. A novel recombinant marker virus assay for comparing the relative fitness of HIV-1 reverse transcriptase variants. J Acquir Immune Defic Syndr 2001; 27:7–13. [DOI] [PubMed] [Google Scholar]
- 9. Larder BA, Kemp SD, Harrigan PR. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 1995; 269:696–9. [DOI] [PubMed] [Google Scholar]
- 10. Paredes R, Sagar M, Marconi VC, et al. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J Virol 2009; 83:2038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Randomised trial of addition of lamivudine or lamivudine plus loviride to zidovudine-containing regimens for patients with HIV-1 infection: the CAESAR trial. Lancet 1997; 349:1413–21. [PubMed] [Google Scholar]
- 12. Eron JJ, Benoit SL, Jemsek J, et al. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N Engl J Med 1995; 333:1662–9. [DOI] [PubMed] [Google Scholar]
- 13. Pluda JM, Cooley TP, Montaner JS, et al. A phase I/II study of 2’-deoxy-3’-thiacytidine (lamivudine) in patients with advanced human immunodeficiency virus infection. J Infect Dis 1995; 171:1438–47. [DOI] [PubMed] [Google Scholar]
- 14. Kuritzkes DR, Quinn JB, Benoit SL, et al. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. AIDS 1996; 10:975–81. [DOI] [PubMed] [Google Scholar]
- 15. Deeks SG, Hoh R, Neilands TB, et al. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J Infect Dis 2005; 192:1537–44. [DOI] [PubMed] [Google Scholar]
- 16. Campbell TB, Shulman NS, Johnson SC, et al. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clin Infect Dis 2005; 41:236–42. [DOI] [PubMed] [Google Scholar]
- 17. Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods 2007; 146:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fox Z, Dragsted UB, Gerstoft J, et al. ; COLATE study group A randomized trial to evaluate continuation versus discontinuation of lamivudine in individuals failing a lamivudine-containing regimen: the COLATE trial. Antivir Ther 2006; 11:761–70. [PubMed] [Google Scholar]
- 19. Steegen K, Bronze M, Papathanasopoulos MA, et al. HIV-1 antiretroviral drug resistance patterns in patients failing NNRTI-based treatment: results from a national survey in South Africa. J Antimicrob Chemother 2017; 72:210–9. [DOI] [PubMed] [Google Scholar]
- 20. Rhee SY, Varghese V, Holmes SP, et al. Mutational correlates of virological failure in individuals receiving a WHO-recommended tenofovir-containing first-line regimen: an international collaboration. EBioMedicine 2017; 18:225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Theys K, Vercauteren J, Snoeck J, et al. HIV-1 subtype is an independent predictor of reverse transcriptase mutation K65R in HIV-1 patients treated with combination antiretroviral therapy including tenofovir. Antimicrob Agents Chemother 2013; 57:1053–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunt GM, Dokubo EK, Takuva S, et al. Rates of virological suppression and drug resistance in adult HIV-1-positive patients attending primary healthcare facilities in KwaZulu-Natal, South Africa. J Antimicrob Chemother 2017; 72:3141–8. [DOI] [PubMed] [Google Scholar]
- 23. Rokx C, Fibriani A, van de Vijver DA, et al. ; AIDS Therapy Evaluation in the Netherlands National Observational Cohort Increased virological failure in naive HIV-1-infected patients taking lamivudine compared with emtricitabine in combination with tenofovir and efavirenz or nevirapine in the Dutch nationwide ATHENA cohort. Clin Infect Dis 2015; 60:143–53. [DOI] [PubMed] [Google Scholar]
- 24. Hoffmann CJ, Ledwaba J, Li JF, et al. Resistance to tenofovir-based regimens during treatment failure of subtype C HIV-1 in South Africa. Antivir Ther 2013; 18:915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sobrino-Vegas P, Gutiérrez F, Berenguer J, et al. ; CoRIS [The Cohort of the Spanish HIV Research Network (CoRIS) and its associated biobank; organizational issues, main findings and losses to follow-up]. Enferm Infecc Microbiol Clin 2011; 29:645–53. [DOI] [PubMed] [Google Scholar]
- 26. Kaleebu P, Kirungi W, Watera C, et al. ; HIV Drug Resistance Working group Virological response and antiretroviral drug resistance emerging during antiretroviral therapy at three treatment centers in Uganda. PLoS One 2015; 10:e0145536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Neogi U, Häggblom A, Santacatterina M, et al. Temporal trends in the Swedish HIV-1 epidemic: increase in non-B subtypes and recombinant forms over three decades. PLoS One 2014; 9:e99390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brooks K, Diero L, DeLong A, et al. Treatment failure and drug resistance in HIV-positive patients on tenofovir-based first-line antiretroviral therapy in western Kenya. J Int AIDS Soc 2016; 19:20798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sunpath H, Wu B, Gordon M, et al. High rate of K65R for antiretroviral therapy-naive patients with subtype C HIV infection failing a tenofovir-containing first-line regimen. AIDS 2012; 26:1679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Etiebet MA, Shepherd J, Nowak RG, et al. Tenofovir-based regimens associated with less drug resistance in HIV-1-infected Nigerians failing first-line antiretroviral therapy. AIDS 2013; 27:553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang WL, Kouyos RD, Scherrer AU, et al. ; Swiss HIV Cohort Study (SHCS) Assessing efficacy of different nucleos(t)ide backbones in NNRTI-containing regimens in the Swiss HIV Cohort Study. J Antimicrob Chemother 2015; 70:3323–31. [DOI] [PubMed] [Google Scholar]
- 32. Ugbena R, Aberle-Grasse J, Diallo K, et al. Virological response and HIV drug resistance 12 months after antiretroviral therapy initiation at 2 clinics in Nigeria. Clin Infect Dis 2012; 54(Suppl 4):S375–80. [DOI] [PubMed] [Google Scholar]
- 33. Neogi U, Engelbrecht S, Claassen M, et al. Mutational heterogeneity in p6 Gag late assembly (L) domains in HIV-1 subtype C viruses from South Africa. AIDS Res Hum Retroviruses 2016; 32:80–4. [DOI] [PubMed] [Google Scholar]
- 34. Van Zyl GU, Liu TF, Claassen M, et al. Trends in genotypic HIV-1 antiretroviral resistance between 2006 and 2012 in South African patients receiving first- and second-line antiretroviral treatment regimens. PLoS One 2013; 8:e67188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dinesha TR, Gomathi S, Boobalan J, et al. Genotypic HIV-1 drug resistance among patients failing tenofovir-based first-line HAART in South India. AIDS Res Hum Retroviruses 2016; 32:1234–6. [DOI] [PubMed] [Google Scholar]
- 36. Lam EP, Moore CL, Gotuzzo E, et al. Antiretroviral resistance after first-line antiretroviral therapy failure in diverse HIV-1 subtypes in the SECOND-LINE Study. AIDS Res Hum Retroviruses 2016; 32:841–50. [DOI] [PubMed] [Google Scholar]
- 37. Skhosana L, Steegen K, Bronze M, et al. High prevalence of the K65R mutation in HIV-1 subtype C infected patients failing tenofovir-based first-line regimens in South Africa. PLoS One 2015; 10:e0118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ndahimana Jd, Riedel DJ, Mwumvaneza M, et al. Drug resistance mutations after the first 12 months on antiretroviral therapy and determinants of virological failure in Rwanda. Trop Med Int Health 2016; 21:928–35. [DOI] [PubMed] [Google Scholar]
- 39. Sigaloff KC, Hamers RL, Wallis CL, et al. ; PharmAccess African Studies to Evaluate Resistance (PASER) Unnecessary antiretroviral treatment switches and accumulation of HIV resistance mutations; two arguments for viral load monitoring in Africa. J Acquir Immune Defic Syndr 2011; 58:23–31. [DOI] [PubMed] [Google Scholar]
- 40. Jiamsakul A, Sungkanuparph S, Law M, et al. ; TREAT Asia Studies to Evaluate Resistance – Monitoring Study (TASER-M) HIV multi-drug resistance at first-line antiretroviral failure and subsequent virological response in Asia. J Int AIDS Soc 2014; 17:19053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Riddler SA, Haubrich R, DiRienzo AG, et al. ; AIDS Clinical Trials Group Study A5142 Team Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med 2008; 358:2095–106.18480202 [Google Scholar]
- 42. Gupta RK, Kohli A, McCormick AL, Towers GJ, Pillay D, Parry CM. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in vitro assays. AIDS 2010; 24:1651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gupta RK, Mlcochova P, Pelchen-Matthews A, et al. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci U S A 2009; 106:20889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chunduri H, Rimland D, Nurpeisov V, Crumpacker CS, Sharma PL. A Leu to Ile but not Leu to Val change at HIV-1 reverse transcriptase codon 74 in the background of K65R mutation leads to an increased processivity of K65R+L74I enzyme and a replication competent virus. Virol J 2011; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection Recommendations for a public health approach - Second edition Available at: http://www.who.int/hiv/pub/arv/arv-2016/en. Accessed 14 April 2017.
- 46. Mollan K, Daar ES, Sax PE, et al. ; AIDS Clinical Trials Group Study A5202 Team HIV-1 amino acid changes among participants with virologic failure: associations with first-line efavirenz or atazanavir plus ritonavir and disease status. J Infect Dis 2012; 206:1920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Linder V, Goldswain C, Adler H, et al. Lamivudine monotherapy: experience of medium-term outcomes in HIV-infected children unable to adhere to triple therapy. Pediatr Infect Dis J 2016; 35:e199–205. [DOI] [PubMed] [Google Scholar]
- 48. Van Rompay KK, Matthews TB, Higgins J, et al. Virulence and reduced fitness of simian immunodeficiency virus with the M184V mutation in reverse transcriptase. J Virol 2002; 76:6083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wirden M, Roquebert B, Derache A, et al. Risk factors for selection of the L74I reverse transcriptase mutation in human immunodeficiency virus type 1-infected patients. Antimicrob Agents Chemother 2006; 50:2553–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wirden M, Lambert-Niclot S, Marcelin AG, et al. Antiretroviral combinations implicated in emergence of the L74I and L74V resistance mutations in HIV-1-infected patients. AIDS 2009; 23:95–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.