Abstract
Currently, no specific drug and vaccine are available for the new coronavirus SARS-CoV-2, and nutritional supplementation should be helpful. This study tried to provide reference for protein supplementation. Specifically, in silico method was employed to simulate protein degradation by gastrointestinal enzymes and to produce a large number of active peptides, then, the binding ability of these peptides to SARS-CoV-2 spike protein receptor-binding domain (RBD) was evaluated. The results showed that wheat-derived alpha/beta-gliadin, oat-derived avenin, and ribulose bisphosphate carboxylase small chain of different origin could be good protein source in generating potent binders to SARS-CoV-2 spike RBD. In addition, some high-affinity oligopeptides (such as PISCR, VQVVN, PQQQF, etc.) were identified as potential binders of SARS-CoV-2 spike RBD. In summary, a number of plant proteins could be helpful for COVID-19 patients when supplemented with these proteins, the identified oligopeptides could be used as lead compound to design potential entry inhibitors against SARS-CoV-2.
Keywords: COVID-19, Spike, Protein supplementation, Oligopeptides, In silico evaluation
Introduction
Since the end of December 2019, a new coronavirus broke out in Wuhan of China, and then spread all over China and many countries in the world. This new virus, termed as SARS-CoV-2 by the World Health Organization (WHO), was found to cause disease named as COVID-19, including severe pneumonia and other symptoms like fever, dyspnea and asthenia [1]. According to the record of August 15, 2020, 89,695 and 21,179,695 persons were infected, and 4708 and 761,264 persons died, in China and other 200+ countries in the world, respectively. The genome analysis indicated that SARS-CoV-2 has as high as 79.5% of sequence identity with SARS-CoV, which breakout in Beijing 17 years ago [2]. It is well known that SARS-CoV infects human host cells by the interaction between its spike glycoprotein and the receptor ACE2 (angiotensin converting enzyme 2) on human cells [3]. Similar to SARS-CoV, SARS-CoV-2 was considered to use the same receptor ACE2 as virus entry into human [4].
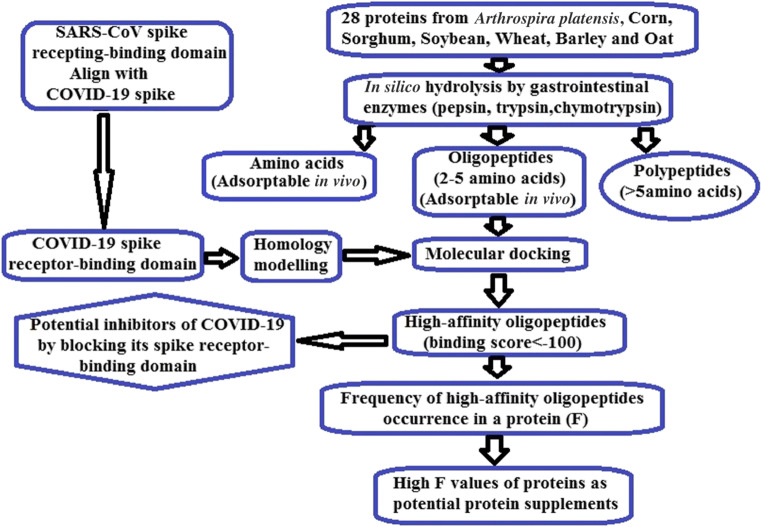
To date, no specific drug and vaccine are available for this virus. In this situation, on the basis of drug treatment, it should be helpful to improve immunity through nutrition strengthening at the same time. Protein is the main “building material” of all parts of the body. Immunoglobulin, cytokines and other components needed for disease resistance need to be synthesized with protein. If there is a lack of protein, the disease resistance will be reduced. So, protein supplementation is usually a good option. It is noted that the protein, after ingestion, first denatured under the action of hydrochloric acid secreted by the gastric wall, the three-dimensional structure was destroyed, and the peptide bond was exposed. Then, under the action of pepsin, intestinal trypsin and chymotrypsin, the protein macromolecules were degraded into amino acids, oligopeptides or polypeptides [5, 6]. The purpose of this paper is to provide reference for protein supplementation. Briefly, in silico method was employed to simulate protein degradation by gastrointestinal enzymes and to produce a large number of active peptides, then, the binding ability of these peptides to SARS-CoV-2 spike protein receptor-binding domain (RBD) was evaluated. It is expected that proteins which release peptides with high binding ability can bring potential inhibition on SARS-CoV-2 while being used as supplements. Figure 1 displayed the strategy for this study.
Fig. 1.
Strategy for in silico evaluation
Materials and Methods
Construction of 3D Model for SARS-CoV-2 Spike RBD
SARS-CoV-2 was considered to use the same receptor ACE2 as SARS coronavirus, i.e., the coronavirus enters host by the binding of spike protein to ACE2 and infection happens. The amino acids sequence of SARS spike RBD was retrieved from PDB database (https://www.rcsb.org/structure/2AJF), and the sequence of SARS-CoV-2 spike was retrieved from NCBI database (https://www.ncbi.nlm.nih.gov/protein/QHO60594.1). The retrieved two sequences with 73.9% of identity were aligned (Fig. 2) to obtain the sequence of SARS-CoV-2 spike RBD. Then, the RBD was subjected to homology modeling using SWISS-MODEL (https://swissmodel.expasy.org), and the 3D model for SARS-CoV-2 spike RBD was constructed.
Fig. 2.
Alignment between the receptor-binding domains of SARS and SARS-CoV-2 spike proteins
In Silico Hydrolysis of Plant Proteins by Gastrointestinal Enzymes
Twenty-eight proteins (Table 1) sequences from Arthrospira platensis, corn, sorghum, soybean, wheat, barley and oat (which are most widely used cereal grains or microalgae in functional foods) were obtained from NCBI database, available at https://www.ncbi.nlm.nih.gov/protein/. Subsequently, these sequences were in silico digested by gastrointestinal enzymes (chymotrypsin, trypsin, pepsin) using the BIOPEP-UWM database [7]. The theoretical degree of hydrolysis (TDH) (proteins with larger TDH produce more oligopeptides) was calculated using the following equation: TDH = d/D*100%, where d is number of hydrolyzed peptide bonds and D is total number of peptide bonds in a protein chain.
Table 1.
Sequences and frequencies of high-affinity oligopeptides from plant proteins digested in silico by gastrointestinal enzymes
| Protein names [amino acids length] | Accession number | Sequences of high-affinity oligopeptides (length < 6 and docking score < −100) binding to receptor-binding domain of SARS-CoV-2 spike RBD [Binding score] | Frequency of high-affinity oligopeptide in a protein (F) |
|---|---|---|---|
| Arthrospira platensis | |||
| Phycocyanin alpha chain [162] | ABD64608.1 | TTQM[−103.3],DISY[−123.9],SPSW[−131.2],IEAL[−100.4] | 0.0247 |
| Phycocyanin beta chain [172] | ABD64607.1 | ITSN[−108.4] | 0.0058 |
| Phycobiliprotein lyase [174] | WP_006618130.1 | DIVEF[−125],APDH[−109.5],PDQQR[−136.8],GSAVL[−107.8],TSIVN[−138.4],CSEIR[−115] | 0.0345 |
| Carotenoid biosynthesis protein [306] | WP_014274510.1 | SSEL[−100.4],GSSW[−119.1],DPAM[−100.7], SQAPF[−131.2],PIVL[−119.2] | 0.0163 |
| Photosynthesis system II assembly factor Ycf48r [334] | WP_006616832.1 | IAVAL[−121.1],PDVSY[−135],DIDF[−110],IVGSN[−127.3],TSVSF[−133.4],PGAPN[−124.6], SAAEM[−100.3],VGAIY[−135.4],GGEIR[−112.2],DPEEW[−130.3],SDPIK[−103.5],STSW[−131.9],TSASS[−103.8] | 0.0389 |
| Corn (Zea mays) | |||
| Ribulose bisphosphate carboxylase large chain[476] | P00874.2 | TPEY[−122.1],VTPQL[−124.5],TTVW[−138],VAY[−102.9], PPAY[−132.8],QGPPH[−125.1],GCTIK[−118.6],TGGF[−108.5],AVIDR[−125.4], SGGDH [−101.4],EGER[−107],EITL[−107.5],TQDW[−122.6],TEIF[−111.3],EIIK[−100] | 0.0315 |
| Ribulose bisphosphate carboxylase small chain[170] | P05348.1 | APTVM[−120.7],STASL[−109.6],PVAR[−135.6],QVW[−133],QVDY[−135.8],IPCL[−108.7],QEAIK[−106.7],PDAF[−131.1],VIGF[−111.7],IAY[−106.1],PPGSD[−127.8] | 0.0647 |
| Sorghum (Sorghum vulgare) | |||
| Ribulose bisphosphate carboxylase large chain[476] | A1E9T2.1 | ASVGF[−121.9],TPEY[−122.1],VTPQL[−124.5],TTVW[−138],VAY[−102.9],PPAY[−132.8],QGPPH[−125.1],GCTIK[−118.6],TGGF [−108.5],AVIDR[−125.4],SGGDH [−101.4],EGER[−107],EITL[−107.5],TQDW[−122.6],TEIF [−111.3],EIIK[−100] | 0.0336 |
| Ribulose biophosphate carboxylase small subunit[169] | BAJ40065.1 | APTVM[−120.7],STATL[−104.9],PVAR[−135.6],STTSF[−126.6],QVW[−133],QVDY[−135.8],VPCL[−100.1],IAY[−106.1],PAGSE[−117] | 0.0533 |
| Wheat (Triticum aestivum) | |||
| Alpha/beta-gliadin[286] | P02863.2 | PVPQL[−133],QPQN[−114.9],P SQL[−102],QPF[−113.9],PQPQL[−133.1],PQQPY[−146.1],QQIL[−111.9],IPCM[−110.4],DVVL[−103.9],QQSTY[−134.8],CCQH[−119.8],VVH[−106.8],AIIL[−117.5],QQQK[−105.4],QQQQ[−120.3],QQPL[−107.5],QQY[−111.9],GQGSF[−129.1],PSQQN [−134.8],EEIR[−111.1],IPPY [−133.1],GIF[−100.3] | 0.0769 |
| Glutenin, high molecular weight subunit[660] | P08488.1 | QCER [−111.1],QESSL[−115.9],EACR [−103.4],CCQQL[−131.8],DVSAK [−101.3],GGSF[−117.1],QQGIF[−145.4],QGSY [−123],PTSL[−126.2],PTSL[−126.2],TGQR[−111.7],QQGY[−124.8],PASL[−119],PTSL[−126.2],PTSL[−126.2],PTSL[−126.2],PASL[−119],PTSL[−126.2],DSPY[−118.2],PTVCR[−143] | 0.0303 |
| Glutenin, low molecular weight subunit[307] | P10386.1 | CIPGL[−112.1],QQQPL[−120.1],PQQPL[−126],SQQQP[−115.7],PSIL[−118.7],EAIR[−110],AIIY[−127.1],SIIL[−108.3],AQGTF[−124.7],QIAQL[−121.3],TSIAL[−112.4],CSVN [−107.8] | 0.0391 |
| Ribulose bisphosphate carboxylase large chain[477] | P11383.2 | AGVGF[−116],TPEY[−122.1],TTVW[−138],VAY[−102.9],PPTY[−137.2],QGPPH [−125.1],GCTIK[−118.6],TGGF[−108.5],AVIDR[−125.4],SGGDH[−101.4],EGER[−107],TQDW[−122.6],TEIF[−111.3],EIIR[−109] | 0.0294 |
| Ribulose bisphosphate carboxylase small chain[175] | P26667.1 | APAVM[−116],STAGL[−100.7],PISCR [−154],SSVSN [−117.7],QVW[−133],QVDY[−135.8],VPCL[−100.1],SSPGY[−126.3],PDAY[−131],VIGF[−111.7] | 0.0571 |
| Soybean (Glycine hispida) | |||
| Basic 7S globulin[427] | P13917 | PVQN [−118.6],QVPVL[−116.1],CEQQY[−132.2],QAPF[−122.3],PGCH[−119.4],APISL [−119.4],TTCL[−104.1],PTSK[−124],GAIIF[−132.4],QDIF[−128.7],TITL[−111.6],QGEY[−115.3],QQSVY[−135.9],TQVF[−131.5],AQQL[−104.4],QAQVK [−129.9],SVAPF[−125.7],PSVDL [−125.2],AEITL[−110.6],STSSL[−110.4] | 0.0468 |
| Glycinin G5[516] | P04347 | SSACF[−132.9],ECQL[−105.9],EPDH[−109.9],IETW[−132.5],GAIGF[−118.1],SQQQL [−123.5],QDSH[−106.9],EGDVL [−100.1],VIPL[−102.7],GVPY[−118.6],PDIEH [−125.8],PETM[−109.6],QGQH[−114.7],AQSF[−116.2],QEQED[−110.7],TPSY[−119.7],PPQR[−124.9],GCQTR[−121.1],ISTL[−102.7],SAQY[−115.6],SPDW[−121.1],SVTM[−108.9],CQGN[−102.7],VVPQN [−129.4], AVSSY [−129.4] | 0.0484 |
| Beta-conglycinin beta subunit[439] | P25974 | GTVF[−119.9],SPQL[−103.7],IVQF[−121.1],ADADF[−111.7],AIPVN[−117.4],ETSF[−121.5],EQIR[−109.3],SSSR[−105.4],AIVIL[−137.7],EQQQK[−120.8],EVQR[−106],QIER[−114.7],QVQEL[−130.3],PSIL[−118.7] | 0.0319 |
| Beta-conglycinin alpha subunit[605] | P0DO16 | GIAY [−109.4],QSCN[−107.2],QACH[−111.8],GEIPR[−111],PIPF[−137],PQPR[−124.2],QEEEH [−106.5],EEQEW[−137.7],SPQL[−103.7],ADADY[−106.1],GTAIL[−108.8],AIPVN [−117.4],EASY[−116.5],EQIR[−109.3],SSSR[−105.4],DPIY[−121.4],SIVDM[−114.3],AIVIL[−137.7],AIGIN[−110.1] | 0.0314 |
| Beta-conglycinin alpha’ subunit[621] | P11827 | GVVF[−116.4],GIAY[−109.4],QACH[−111.8],QEEEH[−106.5],QDER[−116.3],ADADY[−106.1, IVIL[−110.4], AIPVN[−117.4],EASY[−116.5],SSSR[−105.4],DPIY[−121.4],SVVDM[−124.1],AIVVL[−126.7],EQQQR[−142.7],PVVVN [−129.6],ATSDL[−102.1],PGSAK[−116.4] | 0.0274 |
| Barley (Hordeum vulgare) | |||
| B hordein precursor[290] | Q40026 | PQQPF[−147.4],PQQPF[−147.4],PQQP[−112.1],PQQPF[−147.4],QQPIL[−125.5],PQGQL[−126.8],QIPY[−128.9],PSIL[−118.7],EAIR[−110],AIVY[−116.2],QIAQL[−121.3],PPDF[−131.5] | 0.0414 |
| D hordein precursor[707] | Q40054 | TTAER[−114.5],QCER[−111.1],QESSL[−115.9],EACR[−103.4],VGQL[−100.8],PVAL[−119.7],SQVVR[−129.1],GGSF[−117.1],QQGGW[−149.7],GTSVK[−102.2],QQSW[−144.4],PGSTF[−132.6],PSATF[−141],QPGQW[−146.5],PSATF[−141],QQGSY[−136.7],GQQL[−103.3],TSPH[−113.1],TQQK[−103.9],PGQGY[−138.8],QPF[−113.9],GQQSN[−112.7],GSPY[−115.6],AQQL[−104.4] | |
| Gamma-hordein-3[289] | P80198.1 | PSGL[−105.7],PQQL[−129],PQQQF[−152.2],PQQM[−119.5],PQQM[−119.5],PQQK[−115.2],QQPL[−107.5],TQQPY[−141.1],QQCTL[−120],ISQQN [−126.6],EQSR[−114.3],AIVM[−104],VQSQL[−119],PIQL[−115.9],VVGSL[−111.3],VIQTL [−125.9],VPPY[−138.5],CSPF[−120.1] | 0.0623 |
| Ribulose bisphosphate carboxylase large chain[479] | P05698.2 | AGVGF[−116],QAGVK[−110.5],TPEY[−122.1],TTVW [−138],VAY[−102.9],PPTY[−137.2],QGPPH[−125.1],GCTIK[−118.6],TGGF[−108.5],AVIDR[−125.4],SGGDH[−101.4],EGER[−107],TQDW [−122.6],TEIF[−111.3],EIIR[−109] | 0.0313 |
| Ribulose bisphosphate carboxylase small chain[174] | Q40004 | APTVM[−120.7],STAGL [−100.7],PVSR[−128.8],QVW[−133],QVDY[−135.8],VPCL[−100.1],ASPGY[−122.1],IIGF[−122.5] | 0.0460 |
| Oat (Avena sativa) | |||
| Avenin[214] | P27919.1 | AQY[−117.3],CQQH[−121],DSCR[−101],VAER[−106.7],CTTM[−102.8],PITW[−143.5],ECCQL[−110.9],PSECR[−135],CDAIW[−130],SIQR[−114.3],SAISP[−106.1],IAGK[−101.7],PVDVL[−115.2],[QEAR[−105.2],TQTSF[−137.2] | 0.0701 |
| 11S globulin precursor[551] | Q38779 | ATTSF[−122.3],QSSR[−124.6],QQQF[−130],QPF[−113.9],DQSQF [−142.2],QVGK[−108.3],STPY[−116.1],DQSF[−106],ADTY[−115],IVQM[−105.1],IQGH[−123.4],VQVVN[−152.4],GQTVF[−146.1],IVPQH[−139.1],EGCQY[−137],IAGK[−101.7],PIDVL[−116.4],QEAR[−105.2],GEEF[−100.4],TQTGF[−130.5] | 0.0363 |
| Ribulose bisphosphate carboxylase large chain[477] | P48684.1 | ASVGF[−121.9],QAGVK[−110.5],TPEY[−122.1],TTVW[−138],VAY[−102.9],PPAY[−132.8],QGPPH[−125.1],GCTIK[−118.6],ITGGF[−125.6],AVIDR[−125.4],SGGDH[−101.4],EGER[−107],TQDW[−122.6],TEIF[−111.3],EIIR[−109] | 0.0314 |
| Ribulose bisphosphate carboxylase small subunit[165] | Q9ZWG4 | STAGL[−100.7],PVGR[−121],QVW[−133],QIDF[−128.3],VPCL[−100.1],PDAY[−131],IIGF[−122.5],PPGCE[−127.7] | 0.0485 |
Molecular Docking and Evaluation of Nutritional Supplement Potential
The peptides generated by in silico hydrolysis of 28 proteins were docked to SARS-CoV-2 spike RBD using HPEPDOCK [8]. The potential of various plant proteins to serve as nutritional supplement of COVID-19 patients was quantified based on the frequency of occurrence of oligopeptides (2 < =peptide length < =5) with high binding affinity (docking score < −100, the binding threshold (−100) just depends on the number of high-binding scores) relative to the length of the protein chain: F=N/L, where F is the occurrence frequency, N is the number of high-affinity oligopeptides within the protein chain, and L is the length of the protein chain (i.e., number of amino acid residues). The binding residues of the selected high-affinity oligopeptides to SARS-CoV-2 spike RBD were determined by LigPlot [9].
Results and Discussion
In Silico Hydrolysis by Gastrointestinal Enzymes
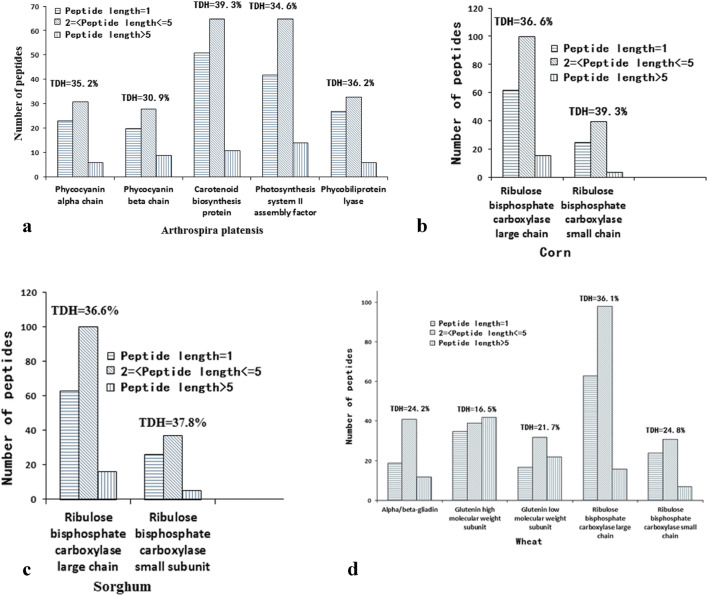
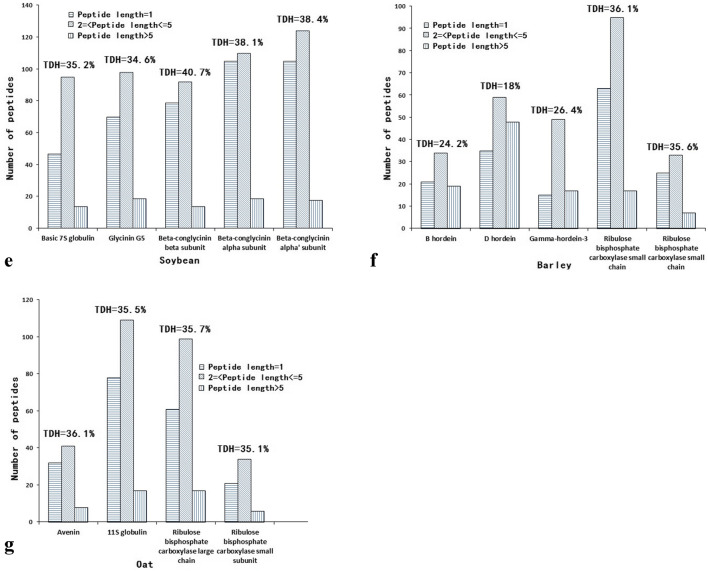
After in silico proteolysis (chymotrypsin, trypsin and pepsin) of 28 proteins from various sources by BIOPEP tools, bioactive peptides were obtained (Fig. 3). For Arthrospira platensis, the theoretical degree of hydrolysis (TDH) was within the range of 30.9 to 39.3%, and 25–65 oligopeptides (with 2–5 amino acids) were generated from five selected proteins. For corn, ribulose bisphosphate carboxylase large and small chains released 100 and 40 oligopeptides with 36.6 and 39.3% of TDH, respectively. Similarly, for sorghum, 100 and 37 oligopeptides were obtained from ribulose bisphosphate carboxylase large and small chains with 36.6 and 37.8% of TDH, respectively. For wheat, the TDH values were varied from 16.5 to 36.1%, and 31–98 oligopeptides were generated from five selected proteins. Likewise, for soybean, the TDH values were within the scope of 34.6 to 40.7%, and 92–124 oligopeptides were generated from five selected proteins. For barley, the TDH values were within the scope of 18.6 to 36.1%, and 33–95 oligopeptides were released from five selected proteins. Finally, for oat, the TDH values were around 35%, and 34–109 oligopeptides were yielded from four selected proteins. In general, the majority of the released peptides were oligopeptides with length of 2–5 amino acids through in silico digest by gastrointestinal enzymes.
Fig. 3.
In silico hydrolysis of Arthrospira platensis (a), corn (b), Sorghum (c), wheat (d), soybean (e), barley (f) and oak (g) proteins
Potential Binding Ability of the Released Peptides to SARS-CoV-2 Spike RBD
In order to evaluate the binding ability of plant proteins in silico digested by gastrointestinal enzymes to SARS-CoV-2 spike RBD, molecular docking was performed and docking scores were obtained. The potential of plant proteins as nutritional supplement was assessed by the frequency (F) of high-affinity (binding score < −100) oligopeptide (length 2–5 amino acids) in a protein. Table 1 showed the F values of high-affinity oligopeptides. The results indicated that the last two proteins with smaller F values were Phycocyanin beta chain (0.0058) and carotenoid biosynthesis protein (0.0163) from Arthrospira platensis; among the better binders were the four proteins ribulose bisphosphate carboxylase small chain (0.0647) from corn, gamma-hordein-3(0.0623) from barley, ribulose bisphosphate carboxylase small chain (0.0571) from wheat and ribulose biophosphate carboxylase small subunit (0.0533) from sorghum; and the top two proteins with larger F values were alpha/beta-gliadin (0.0769) from wheat and Avenin (0.0701) from oat.
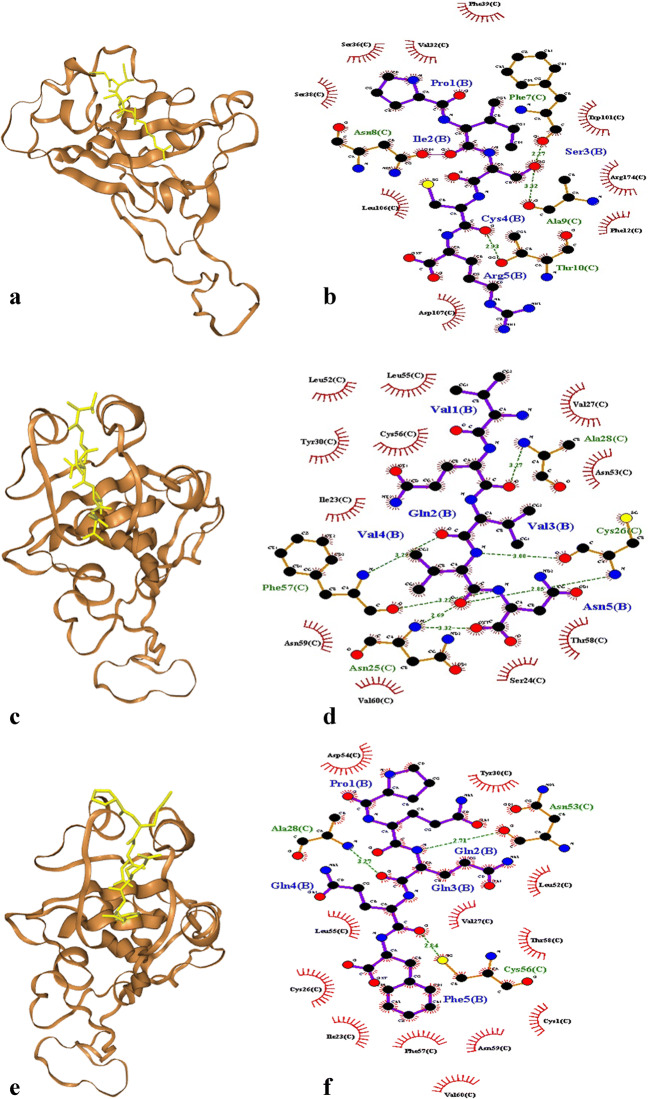
The top high-affinity (binding score < −140) oligopeptides binding to SARS-CoV-2 spike RBD were presented in Table 2. There were seven peptides from barley, four peptides from oat and two peptides from wheat. The most potent binders were PISCR from wheat, VQVVN from oat and PQQQF from barley with binding scores −154, −152.4 and − 152.2, respectively. These peptides can properly bind to hydrophobic groove of SARS-CoV-2 spike RBD (Fig. 4a, c, e). For PISCR, three hydrogen bonds were formed in the following amino acids pairs (peptide vs spike RBD): Ser 3 vs Phe 7, Ser 3 vs Ala 9, and Cys 4 vs Thr 10 (Fig. 4b). For VQVVN, the formed seven hydrogen bonds included: Gln 2 vs Ala 28, Val 3 vs Cys 26, Val 4 vs Cys 26, Val 3 vs Phe 57, Asn 5 vs Phe 57, Val 4 vs Asn 25, and Asn 5 vs Asn 25 (Fig. 4d). For PQQQF, there also existed three hydrogen bonds: Gln 2 vs Asn 53, Gln 3 vs Ala 28, and Gln 4 vs Cys 56 (Fig. 4f).
Table 2.
Top high-affinity oligopeptides binding to SARS-CoV-2 spike RBD
| Peptides | Source | Protein | Docking score |
|---|---|---|---|
| PQQQF | Barley | D hordein | −152.2 |
| QQGGW | Barley | D hordein | −149.7 |
| PQQPF | Barley | B hordein | −147.4 |
| QPGQW | Barley | D hordein | −146.5 |
| QQSW | Barley | D hordein | −144.4 |
| TQQPY | Barley | D hordein | −141.1 |
| PSATF | Barley | D hordein | −141 |
| VQVVN | Oat | 11S globulin | −152.4 |
| GQTVF | Oat | 11S globulin | −146.1 |
| DQSQF | Oat | 11S globulin | −142.2 |
| PITW | Oat | Avenin | −143.5 |
| PISCR | Wheat | Ribulose bisphosphate carboxylase small chain | −154 |
| PQQPY | Wheat | Alpha/beta-gliadin | −146.1 |
| EQQQR | Soybean | Beta-conglycinin alpha’ subunit | −142.7 |
Fig. 4.
Binding of SARS-CoV-2 Spike receptor-binding domain to the identified peptides and the interacting residues. PISCR (a, b), V QVVN (c, d), PQQQF (e, f)
In order to exert in vivo biological activities after oral administration, food derived bioactive peptides have to be absorbed into blood circulation and delivered to targets. It is widely accepted that di- and tri-peptides can be easily absorbed in the intestine by specific peptide transport systems via PepT1 or PepT2 [10], while tetra- and penta-peptides can also be transported through the paracellular tight-junction pathway [11]. Therefore, in this study, after in silico proteolysis by gastrointestinal enzymes, only oligopeptides with peptide length between two and five amino acids were investigated for their binding ability to SARS-CoV-2 spike RBD. The results demonstrated that wheat-derived alpha/beta-gliadin and oat-derived avenin generated highest frequency of high-affinity oligopeptides occurrence, with F values of 0.0769 and 0.0701, respectively; while ribulose bisphosphate carboxylase small chain possessed higher F values, whatever its source, with F values 0.0647 (corn), 0.0533 (sorghum), 0.0571 (wheat), 0.0460 (barley), 0.0485 (oat). It seems that barley and oat become the protein sources generating more potent binders, in which 7 and 4 high-affinity (binding score < −140) oligopeptides were generated respectively. However, it is not sure that barley and oat varieties are superior to other varieties, due to the fact that only limited number of proteins in each variety were evaluated. Anyway, the present data suggests that wheat-derived alpha/beta-gliadin, oat-derived avenin, and ribulose bisphosphate carboxylase small chain of different origin could be good protein source in generating potent binders to SARS-CoV-2 spike RBD. Thus, it could be helpful for COVID-19 patients when supplemented with these proteins.
Previous studies have shown that peptides can act as modulators in viral diseases. For example, the peptides with the positive WWIHS (Wimley-White interfacial hydrophobicity scale) values have been shown to inhibit various viruses, such as SARS-CoV and MERSCoV by interfering with fusion of host cellular and viral glycoprotein membranes [12, 13]. Struck et al. [14] demonstrated that the hexapeptide YKYRYL was specific against SARS-CoV by attaching to its ACE2 receptor. Zhao et al. [15] found that a peptide, NGAICWGPCPTAFRQIGNCGHFKVRCCKIR, exhibited potent antiviral effects against SARS-CoV and MERS-CoV. In this study, a number of high-affinity oligopeptides were identified as potential binders of SARS-CoV-2 spike RBD (Table 2), including PISCR, VQVVN, PQQQF, etc. These peptides should possess good safety due to their source of diet, and can be used as lead compound to design potential entry inhibitors against SARS-CoV-2.
Acknowledgements
No funding is provided.
Compliance with Ethical Standards
Declarations of Interest
The authors report no declarations of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhen Luo and Keying Su contributed equally to this work.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooke FJ, Shapiro DS. Global outbreak of severe acute respiratory syndrome (SARS) Int J Infect Dis. 2003;7:80–85. doi: 10.1016/S1201-9712(03)90001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimitrov DS. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Hannah KW, Krüger N et al (2020) The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 10.1101/2020.01.31.929042
- 5.Cian RE, Garzón AG, Martínez-Augustin O, Botto CC, Drago SR (2018) Antithrombotic activity of brewers' spent grain peptides and their effects on blood coagulation pathways. Plant Foods Hum Nutr 73(3):241–246 [DOI] [PubMed]
- 6.Zhang M, Huang TS, Mu TH (2019) Production and in vitro gastrointestinal digestion of antioxidant peptides from enzymatic Hydrolysates of sweet potato protein affected by pretreatment. Plant Foods Hum Nutr 74(2):225–231 [DOI] [PubMed]
- 7.Minkiewicz P, Iwaniak A, Darewicz M. BIOPEP-UWM database of bioactive peptides: current opportunities. Int J Mol Sci. 2019;20:5978. doi: 10.3390/ijms20235978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Jin B, Li H, Huang S-Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018;46:W443–W450. doi: 10.1093/nar/gky357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 10.Vermeirssen V, Van Camp J, Verstraete W (2004) Bioavailability of angiotensin I converting enzyme inhibitory peptides. Bri J Nutr 92:357–366 [DOI] [PubMed]
- 11.Hong SM, Tanaka M, Koyanagi R, Shen W, Matsui T (2016) Structural design of oligopeptides for intestinal transport model. J Agric Food Chem 64:2072–2079 [DOI] [PubMed]
- 12.Sainz B, Mossel EC, Gallaher WR, et al. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chia TJ, Wu YC, Chen JY, Chi SC (2010) Antimicrobial peptides (AMP) with antiviral activity against fish nodavirus. Fish Shellfish Immun 28:434–439 [DOI] [PubMed]
- 14.Struck AW, Axmann M, Pfefferle S, Drosten C, Meyer B (2012) A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antivir Res 94:288–296 [DOI] [PMC free article] [PubMed]
- 15.Zhao H, Zhou J, Zhang K, et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]