Abstract
Faba bean (Vicia faba L.) is an important protein-rich fodder crop, which is widely cultivated in temperate areas. However, antinutritional compounds such as condensed tannins, limit the use of this protein source in monogastric feed formulations. Previous studies demonstrated that two recessive and complementary genes, zt1 and zt2, control absence of tannin and white flower colour in faba bean. An ortholog of the Medicago WD40 transcription factor TTG1 was reported to encode the zt1 phenotype, but the responsible gene for zt2 is still unknown. Here we used a candidate gene approach combined with linkage mapping, comparative genomics and gene expression to fine map the zt2 genomic region and to identify the regulatory gene controlling both traits. Seventy-two genes, including 23 MYB and bHLH regulatory genes predicted to be associated with anthocyanin expression together with WRKY proteins, were screened and genotyped in three mapping populations. The linkage groups constructed identified the regulatory gene, TRANSPARENT TESTA8 (TT8), encoding a basic helix-loop-helix (bHLH) transcription factor, as the candidate for zt2. This finding was supported by qPCR analysis and further validated in different genetic backgrounds. Accordingly, VfTT8 was downregulated in white flowered types while showing high levels of expression in wild genotypes. Our results provide new insights on the regulatory mechanisms of tannin biosynthesis in faba bean and will facilitate the development of an ultimate zt2 diagnostic marker for the fast generation of new value-added cultivars free of tannins and with improved nutritional value.
Subject terms: Genetics, Agricultural genetics, Gene expression, Genetic linkage study, Genetic markers, Genome, Genomics, Genotype, Haplotypes, Plant breeding, Plant genetics, Sequencing
Introduction
Since the beginnings of agriculture, legume species with high protein levels in seeds and foliage have been essential sources of dietary protein for humans and animals. Faba bean (Vicia faba L.) is a worldwide cultivated grain legume, known for its great yield potential and high protein content of 25–40%1. Nevertheless, the successful deployment of this protein rich fodder crop in feedstuffs for monogastric animals is severely limited by the presence of tannins, a class of polyphenols primarily located in the seed coat2,3, which are present at different concentrations depending on the variety, maturity, location and growth conditions4. Although tannins play an important role in plant growth and reproduction, providing protection against biotic and abiotic stresses5, they have a negative impact on nutritive value and digestibility, being responsible for decreases in feed intake, growth rate, feed efficiency, net metabolizable energy and protein digestibility in experimental animals6. Therefore, low-tannin content generally results in higher protein and energy digestibility for monogastric animals7. Consequently, the development of faba bean tannin-free cultivars is a key breeding objective to enhance the nutritional quality and broaden the use of this legume in the livestock feed industry.
Flavonoids are the most common group of polyphenolic compounds in the plant kingdom8. Thanks to the easily detectable mutant phenotypes in flower and seed pigmentation, flavonoid biosynthesis is one of the best-studied secondary metabolic pathways. These compounds play important roles in the interactions of plants with the environment, serving as pigments, signalling molecules, protectants against biotic and abiotic stresses as well as UV protection9,10. Proanthocyanidins (condensed tannins) and anthocyanins are major flavonoid end-products of a well conserved family of aromatic molecules, and both compounds are produced by related branches of the flavonoid pathway which utilize the same metabolic intermediates. Anthocyanin and tannin biosynthesis requires both the structural genes encoding the enzymes that participate in the formation of different flavonoids, as well as the regulatory genes that control transcription of the structural genes11. Thus, mutations in either structural or regulatory genes lead to loss of pigmentation in several plant species12–15.
Both structural and regulatory anthocyanin and tannin biosynthesis genes have been identified and cloned in model plants such as Arabidopsis, maize (Zea mays), snapdragon (Antirrhinum majus) and Petunia9,16–18. Highly conserved structural genes in this pathway include chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H) and dihydroflavonol 4-reductase (DFR). Among the regulatory genes, DNA-binding R2R3-MYB transcription factors, basic-Helix-Loop-Helix (bHLH) transcription factors, and WD40 repeat proteins are known to form MYB-bHLH-WD repeat (MBW) complexes, which activate transcription of the structural genes in the anthocyanin pathway19.
In previous studies, we tested whether the faba bean genes responsible for the white flower and low tannin phenotypes function in the anthocyanin biosynthetic pathway. BLAST and multiple sequence alignment tools were used to design primers for amplification of the conserved key structural genes CHS, CHI, F3H and DFR, with the aim of identifying polymorphisms that might account for functional gene changes in contrasting faba bean populations. However, neither linkage analysis nor expression studies by RT-PCR pointed towards any of these genes as possible regulators of the zt-1 and zt-2 phenotypes20.
Studies in pea revealed, that the absence of flower pigmentation results from mutations in loci A and A2, encoding a bHLH transcription factor and a WD40 protein, respectively21. In lentils, the zero-tannin trait is controlled by a single recessive gene (tan) that encodes a bHLH transcription factor homologous to the A gene from pea22. In faba bean, zero-tannin (zt) is a simple character, which is governed independently by two complementary recessive genes, zt1 and zt22,23. These genes lead to a defect in synthesis of anthocyanins or their precursors at different steps in the pathway, resulting in plants with white flowers and zero tannin content3,24. Given the partial outcrossing behaviour of faba bean, crosses between zt1 and zt2 individuals will produce coloured plants with tannins that might contaminate the crop. For this reason, it is important to know which of the two genes is present in each tannin-free accession, in order to select the appropriate genitors in breeding programs.
Recently, a gene ortholog of the Medicago WD40 transcription factor Transparent Testa Glabra 1 (TTG1), located on faba bean chromosome (chr.) II, was shown to encode the zt1 phenotype25–28. However, although several studies assigned the locus zt2 to the distal part of chr. III26,28,29, the responsible gene is still unknown. More recently, the latter authors29 reported that a KASP (Kompetitive Allele‐Specific PCR) marker (SNP marker Vf_Mt7g100500_001), located at 10.5 cM from the zt2 locus, could be used as a reliable marker to discriminate low‐tannin faba bean plants carrying zt2. However, the relatively high genetic distance from the zt2 locus bears the risk of false selection, due to possible recombination events. Therefore, marker enrichment of the genomic region is needed to identify the underlying gene and allow a fully reliable marker-assisted selection in breeding programs.
The goal of the present study was to saturate the zt2 genomic region and to identify the candidate gene for the white flower trait, using a combination of genetic linkage, association studies and comparative genomics with the model legume species Medicago truncatula (https://www.medicagohapmap.org/fgb2/gbrowse/mt35/). Upon confirmation of the chromosomal region, we exploited the extensive collinearity between faba bean and Medicago to saturate the target region and mine candidate genes potentially associated with the trait. This approach pinpointed a number of MYB and bHLH regulatory genes potentially associated with the anthocyanin expression. Once the gene sequences were identified, we searched for mutations associated with the target phenotypes, using different mapping populations and genetic backgrounds.
In summary, here we describe a detailed approach for fine mapping, identification and validation of the gene underlying zt2 in faba bean. This result will allow the development of a highly efficient diagnostic marker for the selection of zt2 cultivars free of anti-nutritional compounds.
Material and methods
Plant material and sample collection
Three mapping populations (two F2 and one F7) segregating for tannin content (TC) and flower colour (FC), wild spotted type vs. white flower, were used in this study: (1) M × D: 50 F2 individuals originated from the cross of MAYA (M), with spotted flowers and high TC and DISCO (D), a white flower/zero tannin inbred line, carrying the zt2 gene, (2) W × D: 56 F2 individuals derived from the cross of WIZARD (W), a wild-type flower line with high TC and DISCO (D) and, (3) Vf6 × zt2: 62 F7 individuals, where the maternal parent (Vf6) has spotted flowers and high TC while zt2 is a tannin-free white flowered line. The populations were selfed at the IFAPA of Córdoba, Spain. Plants were grown in insect proof cages to avoid outcrossing. Young leaves were collected from all samples and DNA was isolated according32. The concentration and purity of DNA was measured by a NanoDrop ND-1000 spectrophotometer. The final concentrations of all DNA samples were adjusted to 10 ng/µl for high-throughput genotyping.
Phenotypic evaluation
F2 plants from crosses M × D and W × D were selfed to the F3 generation and the FC was recorded at each F3 family (15 plants per line) to infer their corresponding genotype (homozygous or heterozygous). In the RIL population (Vf6 × zt2), FC was scored in all the lines self-pollinated from F2 to F7. Segregation ratios were tested against the expected ratio using a chi-square analysis for goodness of fit. Phenotypic data on FC in the three populations are available in the Table 1.
Table 1.
Segregation of flower colour (FC) in the three populations used in this study.
| Population | Generation | Individuals evaluated | Observed ratio | Expected ratio | χ2 | p value* |
|---|---|---|---|---|---|---|
| M × D | F2:3 | 50 | 12:30:8 | 1:2:1 | 2.64 | 0.27 |
| W × D | F2:3 | 56 | 18:24:14 | 1:2:1 | 1.71 | 0.42 |
| Vf6 × zt2 | F7 | 62 | 36:26 | 1:1 | 1.61 | 0.20 |
Observed and expected ratios correspond to the homozygous wild type: heterozygous: homozygous white flower colour in F2:3 generations and homozygous wild type: homozygous white flower colour in F7 RIL population.
*α = 0.5.
Genomic location and saturation methodology
Previous linkage map approaches ascribed the target zt2 region in the distal part of faba bean chr. III between the ESTs markers Pis-Gen-8-1-1 and LG3826,28,30,31. Using the sequence information deposited in the M. truncatula Genome Database (MTGD) (https://www.medicagogenome.org/), these genes corresponded to Medtr1g040675 and Medtr1g087900, respectively, spanning 24.3 Mb.
For marker saturation and candidate gene identification we performed a detailed genomic analysis of this syntenic region. Seventy-two genes, including regulatory genes (MYB and bHLH) predicted to be associated with anthocyanin expression together with WRKY proteins were selected to be screened for polymorphism in the three mapping populations (Supplementary Table S1).
Sequencing and genotyping technique
The 72 genes selected include six ESTs markers previously assigned to the target interval26,31 and 23 newly designed M. truncatula markers associated with the anthocyanin expression. The remaining 42 genes were genotyped using the Kompetitive Allele-Specific PCR (KASPar) assays developed by Webb27 and provided by LGC genomics (https://www.biosearchtech.com/products/pcr-kits-and-reagents/genotyping-assays/kasp-genotyping-chemistry) (Supplementary Table S1).
Different tools, including BLASTn and multiple sequence alignment analysis were used to design primers suitable for DNA amplification. The sequences were BLASTed against an in-house faba bean transcriptome33 to obtain the orthologous sequence in faba bean. For intron–exon boundary prediction and primer design, alignment between sequences were implemented in Geneious v.7.1.5 (https://www.geneious.com) The information about primer sequence, annealing temperature, marker type, orthologous sequence in faba bean, locus name and annotation in M. truncatula of the 23 new markers are available in Supplementary Table S2.
To obtain the sequences from DNA samples (parental lines and two contrasting individuals from each population), PCR reactions were conducted in a total volume of 25 µl, using 5 µl DNA of each sample, 200 nM of each primer, 2 mM MgCl2, 200 µM dNTP and 0.6 U Taq polymerase Biotools (B&M Labs, S.A., Madrid, Spain). Amplification products were purified using a standard protocol for DNA precipitation with sodium acetate and ethanol (1/10 3 M sodium acetate, 2 v/v ethanol) (https://www.thermofisher.com/es/es/home/references/protocols/nucleic-acid-purification-and-analysis/dna-protocol/sodium-acetate-precipitation-of-small-nucleic-acids.html#comergent_product_list_15878). PCR products were sequenced by Sanger at STABVIDA (Caparica, Portugal). For each sample, four PCRs were done, and the repeated products were then mixed for sense and antisense strand sequencing.
Sequence analysis and identification of polymorphisms were conducted using Geneious v.7.1.5. For each gene, the sense and antisense sequences were aligned and the consensus sequence further analyzed with BLASTn against the M. truncatula genome (Mt4.0) to assign the percent-consensus-identity. Sequence polymorphisms between the samples were transformed to CAPS (Cleaved Amplified Polymorphism) markers by restriction enzyme digestion of the PCR products. The digestions were carried out in a 15 µl volume of PCR using 5 µl of the amplified products and 3U of the respective restriction endonuclease (ThermoFisher Scientific). The digestions were incubated overnight at their respective temperature. The restricted fragments were separated in 2% agarose gel. Detailed information of the specific gene-based markers designed in this study together with the KASPar markers assayed, is shown in the Supplementary Table S1.
Linkage maps calculation
The segregation of each marker was tested for goodness of fit to the expected segregation ratio (1:2:1 in the two F2s families and 1:1 ratio in the RIL population), using the chi-square test. Linkage maps were constructed using JoinMap 4.134. Markers were grouped using regression mapping algorithm option at a minimum LOD score threshold of 4 and maximum recombination frequency of 0.4 as general linkage criteria to establish linkage groups (LG). Recombination fractions were converted to centimorgans (cM) using the mapping function of Kosambi35.
qPCR samples, RNA isolation, cDNA synthesis and primer design
The most important guidelines of the MIQE checklist36 were considered according to the practical approach for quantitative real-time PCR (qPCR) experiments proposed by Taylor37. The expression profile of candidate genes was analysed in pigmented and white flowers from the Vf6 × zt2 and M × D populations at two different developmental stages. Stage 1 (S1) included immature flowers buds of approximately 1.5 cm length while the stage 2 (S2) included young flowers of 2 cm length and black colour apparent at the top of the petal in the wild types. CYP2 and ELF1A, previously reported as the most stable genes for normalization of the gene expression in the TC experiment were used as reference38.
Only petal tissue was used for total RNA extraction. In order to minimise variation in gene expression among individual plants and stages, petal tissue from three individuals were collected and pooled together for RNA isolation. Finally, three pools of biological replicates were used per individual genotypes. Each sample was frozen in liquid nitrogen and stored at – 80 °C until RNA extraction. In short, the experimental design consisted in a total of 24 samples (2 genotype × 2 stages of development × 2 technical repetitions × 3 biological repetitions).
The Direct-zol RNA Kit (Zymo Research) was used to isolate a high-quality RNA directly from samples in TRIzol Reagent. RNA concentration was determined by measuring the optical density using a NanoDrop spectrophotometer. Only the RNA samples with A260/A280 ratio between 1.9 and 2.1 and A260/A230 greater than 2.0 were used in the analysis. cDNA of each sample was obtained by iScript cDNA Synthesis kit (Bio-Rad, Hercules, California) and diluted to a concentration of 20 ng/µl. A pooled sample comprising all samples considered in the experiment was included for each gene as inter-run calibrator to detect and correct inter-run variation. No-template controls were also included.
Specific primer pairs were designed using the following criteria: Tm of 60 ± 1 °C and PCR amplicon lengths of 80–120 bp, yielding primer sequences with lengths of 19–26 nucleotides and GC contents of 40–80%. Designed primers were synthesized by Integrated DNA Technologies (Leuven, Belgium). Detailed information on the primers designed for RT-qPCR is shown in Table 2.
Table 2.
Information on RT-qPCR primers used in this study.
| GenBank accession | Medicago locus | Annotation | Primer sequence (5′–3′) | Gene position | PCR product size (bp) | PCR efficiency | PCR product Tm (°C) |
|---|---|---|---|---|---|---|---|
| KP006493.1 | MTR_1g022445 | Vicia faba dihydroflavonol reductase 1 (DFR1) | TCTTGTTTGAGCATATGGAAGTTGAG | Exon 5 | 111 | 1.94 ± 0.06 | 74.12 ± 0.06 |
| GGACATTGTACTCTGGAAATTTTGCA | |||||||
| XM_003590588.3 | MTR_1g072140 | Transducin/WD40 repeatprotein | TCCGACTCCCACTACGTCTGC | Exon 1 | 108 | 1.93 ± 0.03 | 84.69 ± 1.01 |
| GATGTGTGTATGGCCACGGAG | |||||||
| XM_024786476.1 | MTR_1g072320 | Transcription factor bHLH (TT8) | CGGAGAGACAAATCCGCCAAC | Exon 2 | 117 | 1.94 ± 0.02 | 78.99 ± 0.35 |
| CCGACACCAGGAGGAAATGAG | |||||||
| XM_003590628.3 | MTR_1g072530 | Transcription factor bHLH66 | CAAGCTTACATACTGGACGACATC | Exon 5–6 | 110 | 1.91 ± 0.03 | 75.90 ± 0.27 |
| ACTAGTGGTATAGCATTTGAGTCA | |||||||
| XM_013613397.2 | MTR_1g077640 | bHLH transcription factor | TAGGAAGTGTGATCAAATATGCAGAC | Exon 7–8 | 120 | 1.94 ± 0.03 | 77.56 ± 0.31 |
| CTTCAAATGCCCATGTAGCACC |
GenBank accession, medicago locus, annotation NCBI, primer sequences, gene position and amplicon size. Primer PCR efficiency and PCR product Tm data represent mean values ± sd.
Real time qPCR assays
The qPCR was carried out using the iTaq Universal SYBR Green Supermix on an ABI PRISM 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Experiments were performed in 96-well optical reaction plates (P/N 4306737) with MicroAmp optical adhesive film (P/N 4311971) from Applied Biosystems. A master mix with total volume of 11 µl for each PCR run was prepared, containing 5 µl of diluted cDNA (10 ng/µl), 5 µl of iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, California) and a primer pair with a concentration of 0.22 µM each. The PCR conditions were 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The specificity of the of the PCR products was confirmed by single and sharp peaks at the melting curves analysis after 40 amplification cycles.
Fluorescence was analyzed using 7500 Software v2.0.1. All amplification plots were analyzed using a threshold values of 0.2 to obtain Cq (quantification cycle) values for each gene-cDNA combination. PCR efficiency of each primer pair was determined for all samples by LinRegPCR program v.1139, using raw normalized (Rn) fluorescence as input data.
The relative gene expression (RGE) was calculated using the advanced quantification model with efficiency correction, multiple reference genes normalization and use of error propagation rules described by Hellemans40 (Eq. 1), where RQ = E∆Ct, being E = PCR efficiency for each primer used to amplified each target gene (TG) and Ct = the number of cycles needed to reach 0.2 arbitrary units of fluorescence. The two reference genes (RG) used for data normalization were CYP2 and ELF1A38.
| 1 |
The RGE values were log-transformed and the significance values to determine differences of the expression of genes were obtained by the ANOVA test using the R programming language.
Results
Segregation analysis of flower colour (FC)
Of the 50 F2:3 plants screened from the M × D population, 12 had spotted flower, 30 were heterozygous/spotted and 8 showed white flower, fitting a 1:2:1 ratio. Further, the 56 F2:3 families from cross W × D, also exhibited a good fit for the expected 1:2:1 (spotted: segregating: white) ratio. Similarly, the 62 RILs evaluated fit the expected 1:1 (spotted: white) ratio (Table 1). The results from FC segregation in these populations further confirmed the monogenic control of the gene with a dominant ZT2 (spotted flower) and a recessive zt2 (white flower) allele.
Genotyping and linkage analysis
The collinearity between faba bean and the model Medicago was exploited to develop new markers and to fine map zt2, in the distal region of faba bean chr. III. Seventy-two gene markers, including candidate genes encoding MYB, bHLH and WRKY proteins, were surveyed in the interval flanked by markers Medtr1g040675 and Medtr1g087900 (Supplementary Table S1). Six of these were ESTs previously used by Satovic et al.31. Moreover, we designed 23 new primer pairs in genes putatively associated with anthocyanin expression, while the remaining 43 genes were genotyped using the KASPar technique.
Thirty out of the 72 markers tested (23 KASP-SNPs, 6 CAPs, 1 Amplified Length Polymorphism/ALP) revealed polymorphisms in at least one of the three faba bean populations, and 12 of these were anchor markers common among populations (Supplementary Table S1). Five of the 23 candidate genes potentially associated with anthocyanin expression exhibited polymorphisms and were thus further genotyped and mapped. The most polymorphic population was M × D with 20 markers, followed by Vf6 × zt2 with 15 markers and by W × D with 13 markers.
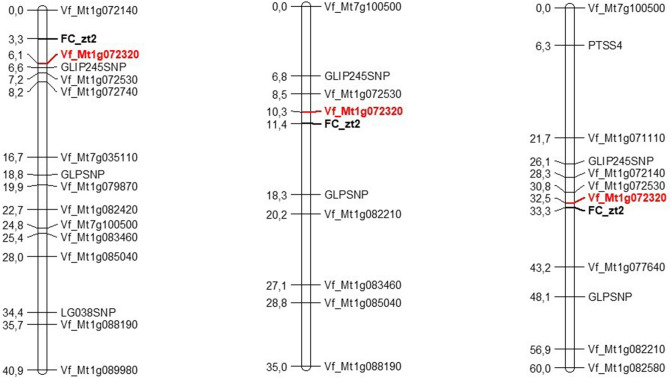
Three linkage groups were constructed (Fig. 1). Fine mapping analysis showed that in cross M × D, zt2 is localized in a window of 6.1 cM between markers Vf_Mt1g072140 and Vf_Mt1g072320, whereas in W × D, zt2 is in a window of 8 cM between Vf_Mt1g072320 and GLPSNP (MTR_1g079490). Finally, in the RIL population (Vf6 × zt2), zt2 was flanked by Vf_Mt1g072320 and Vf_Mt1g077640 (Fig. 1). We found high collinearity between faba bean chr. III and M. truncatula chr. I, except for two markers Vf_Mt7g035110 and Vf_Mt7g100500 which revealed some chromosomal rearrangements due to insertions from Mt Chr. 7. All markers maintained a similar mapping order in the three populations. The closest marker to FC in the three populations (2.8 cM, 1.1 cM and 0.8 cM, respectively) was Vf_Mt1g072320, corresponding to the bHLH transcription factor TRANSPARENT TESTA842 henceforth referred to as VfTT8. We thus selected VfTT8 as the best candidate for zt2 in V. faba.
Figure 1.
Linkage maps from three faba bean populations segregating for flower colour and carrying the gene zt2. M × D F2 population (left), W × D F2 population (middle) and Vf6 × zt2 RIL population (right). Marker names are on the right and the estimated map distances are on the left. Recombinant fractions were converted to centiMorgans using the mapping function of Kosambi.
Haplotype analysis
During fine mapping, 168 individuals (50 F2:3 from M × D, 56 F2:3 from W × D and 62 RILs from cross Vf6 × zt2) were screened to identify recombinants among flanking markers (Supplementary Table S3). Two individuals in W × D and Vf6 × zt2, or three in M × D, showed at least one recombination event between zt2 and the closest marker Vf_Mt1g072320. A number of 5–9 crossovers were also observed in the rest of flanking markers and crosses, except for Vf_Mt7g100500, which was previously reported by Zanotto29 as a reliable marker for the selection of zt2. Strikingly, marker Vf_Mt7g100500 revealed the highest number of misclassified individuals in the three populations (17, 10 and 20, respectively). This finding contradicts the results of the previous report29 and precludes the reliable use of this marker in faba bean breeding programs. We concluded that, according to these recombination events, Vf_Mt1g072320 was the best candidate to discriminate low tannin individuals carrying the zt2 gene.
In the case of Vf_Mt1g072320, we designed two primer pairs in different genomic regions in order to ensure amplification in different genetic backgrounds and to optimize resolution of the banding patterns (Supplementary Table S2). Using primer pair 1, the recessive allele linked with the zt2 locus in crosses W × D and Vf6 × zt2 is G. Therefore, considering identity by descent in the germplasm analysed, and without recombination between the marker and the zt2 locus, white flower genotypes are expected to be GG whereas wild flower genotypes are expected to be AG or AA. Primer pair 2 was only used in cross M × D, and the corresponding genotypes were CC in white flower zt2 genotypes and TC or TT in wild individuals (Supplementary Table S3).
Expression analysis of candidate genes
To corroborate that VfTT8 corresponds to the zt2 gene, RT-qPCR analysis was performed on flower tissue of two RIL individuals derived form from cross Vf6 × zt2 with contrasting FC. The candidate genes assayed were those most closely linked to FC_zt2 in the three linkage maps (see Fig. 1): the DFR1 gene MTR_1g022445 (reported by Ray41), the WD40 protein MTR_1g072140, and the bHLH transcription factors MTR_1g07230 (VfTT8), MTR_1g072530 and MTR_1g077640. The cDNAs from RIL6 (pigmented flower) and RIL2 (white flower) obtained from two different developmental stages (S1 and S2) were analysed.
To confirm the sensitivity and specificity of RT-qPCR, we tested the presence of genomic DNA (gDNA) contamination in the cDNA samples using primer pairs for MTR_1g072530 and MTR_1g077640, which span an exon-exon junction (Table 2). The first primer pair amplified a 195 bp product using gDNA as template or a 110 bp band using cDNA, while the second primer amplified a 224 bp or a 120 bp band using gDNA or cDNA, respectively. None of the cDNAs tested produced bands corresponding to residual gDNA, confirming the purity of samples. The dissociation curve analysis confirmed that all the primer pairs used produced a single and specific PCR product (Table 2). Mean PCR efficiency for all genes was up to 95.5%, while that for the reference genes CYP2 and ELF1A was 1.88 ± 0.009 and 1.88 ± 0.01 (mean ± sd), respectively.
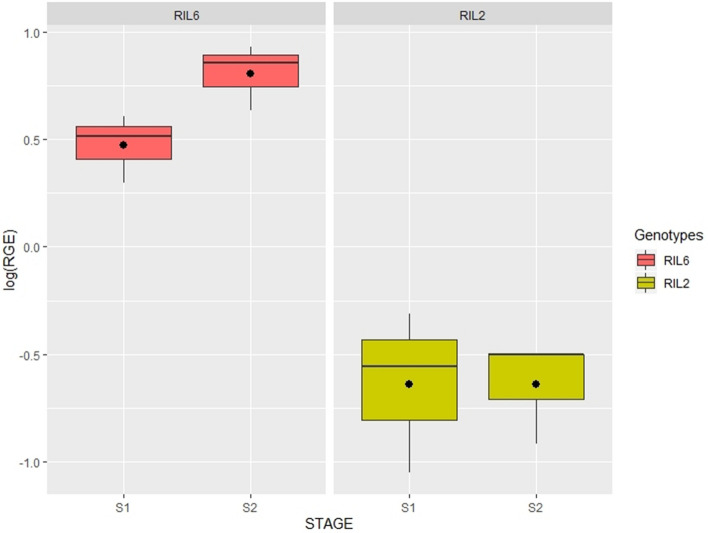
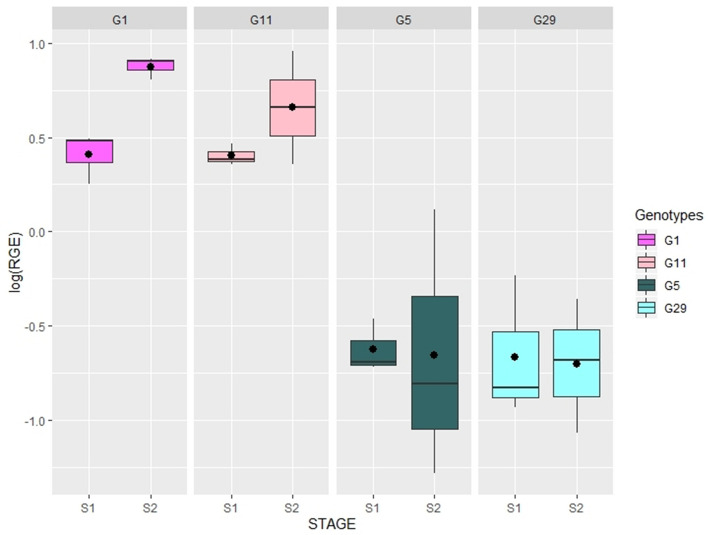
The overall expression pattern of MTR_1g072320 in both flowering stages was significantly higher (p < 0.001) in the V6 × zt2 wild-type line (RIL6) compared to RIL2, being strongly down-regulated in this white flowered genotype (Table 3 and Fig. 2). To validate these results, the expression of MTR_1g072320 was measured in a second faba bean population (M × D), using cDNA from two F2 individuals (G1 and G11) with pigmented and two (G5 and G29) with white flowers. The expression pattern of MTR_1g072320 matched that obtained in the Vf6 × zt2 RILs, showing highly significant differences in the expression level (p < 0.001) between the contrasting FC genotypes. Collectively, these results revealed a strong down-regulation of MTR_1g072320 in white flowered genotypes, further confirming that VfTT8 is the gene responsible for loss of flower pigmentation and absence of tannins in the zt2 genotypes (Table 4; Fig. 3).
Table 3.
Gene expression ratios in RIL population Vf6 × zt2.
| Medicago locus | RIL population Vf6 × zt2 | |||
|---|---|---|---|---|
| RIL6 (wild type) | RIL2 (zt2) | |||
| Stage 1 | Stage 2 | Stage 1 | Stage 2 | |
| MTR_1g022445 | 0.08 | 0.28 | − 0.18 | − 0.19 |
| MTR_1g072140 | − 0.49 | − 0.09 | − 0.42 | − 0.16 |
| MTR_1g072320 | 0.47 | 0.81 | − 0.64 | − 0.64 |
| MTR_1g072530 | − 0.03 | 0.08 | − 0.02 | − 0.04 |
| MTR_1g077640 | 0.35 | − 0.50 | 0.06 | 0.09 |
Values indicate log average expression ratios of three biological replicates from each of the genotypes used in the study. RIL6 (wild type) and RIL2 (mutant type) are F7 individuals from the Vf6 × zt2 population. Statistically significant differences in expression (p < 0.001) are indicated in bold.
Figure 2.
Boxplots showing transcript levels of MTR_1g072320 in RIL6 (pigmented flower) and RIL2 (white flower) from cross V6 × zt2, in two developmental stages: immature flowers (S1) and young flowers (S2). Relative gene expression was log-transformed as detailed in “Material and methods” section. Expression values are shown as median (horizontal line), mean (asterisk), upper and lower quartiles (box) and range (whiskers).
Table 4.
Expression of the candidate gene MTR_1g072320 in four F2 genotypes from the M × D population.
| Medicago locus | F2 population Maya × Disco (MxD) | |||||||
|---|---|---|---|---|---|---|---|---|
| G1 (wild type) | G11 (wild type) | G5 (zt2) | G29 (zt2) | |||||
| Stage 1 | Stage 2 | Stage 1 | Stage 2 | Stage 1 | Stage 2 | Stage 1 | Stage 2 | |
| MTR_1g072320 | 0.41 | 0.88 | 0.40 | 0.66 | − 0.63 | − 0.66 | − 0.67 | − 0.71 |
Values indicate log average expression ratios of three biological replicates from each of the genotypes G1 and G11 (wild type); and G5 and G29 (mutant). All the samples showed statistically significant differences in regulation (p < 0.001).
Figure 3.
Validation of MTR_1g072320 expression values in the F2 population M × D. Boxplots show transcript levels in two individuals with pigmented flower (G1, G11) and two white flowers (G5, G29), in two developmental stages: immature flowers (S1) and young flowers (S2). Relative gene expression was log-transformed as detailed in “Material and methods” section. Expression values are shown as median (horizontal line), mean (asterisk), upper and lower quartiles (box) and range (whiskers).
Discussion
Faba bean is a valuable protein crop with a high potential for the feed industry to shift towards more sustainable raw materials. However, despite its prominent nutritional value, faba beans contain antinutrients such as condensed tannins, which are present in the seed coats of coloured-flowering faba bean varieties, reducing the bioavailability of proteins and minerals. The zero-tannin (zt) trait is governed independently by the two complementary recessive genes zt1 and zt2. Gene TTG1 encodes the zt1 phenotypes25–28, but the gene underlying zt2 was still unknown. Thus, identifying markers closely linked to zt2 has been a key objective to increase the accuracy and efficiency of selection and facilitate marker-assisted breeding of faba bean tannin-free cultivars. Previous studies had adscribed this locus to the distal part of chr. III26,28,29, and the latter authors also reported KASP SNP marker Vf_Mt7g100500_001, located at 10.5 cM from flower colour, as being successfully used to discriminate low tannin plants, even though the closest flanking marker Vf_Mt1g072740_001, which is only at 3 cM distance, failed to distinguish these genotypes in the validation panel29. In contrast to the previous report, we found here that Vf_Mt7g100500_001 produced the highest number of misclassified individuals for any of the faba bean populations assayed, thus preventing its use in marker assisted breeding.
We set out to identify candidate transcription factors responsible for the unpigmented zt2 genotypes using marker enrichment of the target region. For the first time, a set of 23 MYB, bHLH and WD40 genes present in the collinear M. truncatula region were assayed. Because not all markers are applicable across populations due to lack of polymorphism, the use of multiple mapping populations of diverse genetic backgrounds is important for accurate fine-mapping and, ultimately, target gene identification. To maximize genetic diversity, we analysed three faba bean segregating populations, thereby increasing the level of polymorphisms and narrowing down the search of potential candidates to a much shorter segment of the M. truncatula genome. This analysis identified Vf_Mt1g072320, encoding a TT8 transcription factor, as the candidate gene controlling zero tannin zt2 in V. faba.
In Arabidopsis, TT8 encoding a bHLH domain transcription factor, TTG1 encoding a WDR protein, and TT2 encoding a R2R3 MYB domain protein, have been identified as key determinants for proanthocyanidin accumulation in developing seeds42,43. The MYB–bHLH–WD40 (MBW) complex TT2–TT8–TTG1 activates the transcription of structural genes in the anthocyanin pathway and thus controls the accumulation of condensed tannins in the Arabidopsis seed coat19. In Medicago, both WD40 repeat proteins and bHLH transcription factors (MtTT8) are involved in the regulation of anthocyanin and proanthocyanidin biosynthesis42–45. In pea, the absence of pigmentation in the flower is the result of mutations in loci A and A2, encoding a bHLH transcription factor and a WD40 protein, respectively21. A single recessive gene, tan, encoding a bHLH transcription factor homologous to that encoded by the A gene in pea22, controls white flower in lentils, while in faba bean the WD40 transcription factor VfTTG1 is responsible for the zt1 phenotypes26,28,29.
The broad conservation of the MBW complex controlling anthocyanin pigmentation in Eudicots44,46 suggests that other MYB or bHLH transcription factor should control the zt2 gene in faba bean. In the present study, MtTT8 (Mt1g072320), a Medicago ortholog of the A and tan genes of pea and lentil, respectively, was assayed as a promising candidate for determining low TC in faba bean. VfTT8 (Vf_Mt1g072320) was polymorphic in the three faba bean populations tested, and strongly cosegregated with white flower lines, supporting the identity of the candidate. These results were further confirmed by RT-qPCR analyses in different populations and genetic backgrounds, revealing that VfTT8 expression levels were very low in white flowered types, in contrast to high transcript levels detected in the wild genotypes.
From the data presented here, we conclude that VfTT8 is the gene responsible for the zt2 phenotypes in faba bean. Although further analysis is necessary to elucidate the mechanistic basis for the observed down-regulation of the gene in white flowered types, our results open the door to the development of an ultimate diagnostic marker based on the allelic variant causing this phenotypic effect. This study therefore increases our understanding of the regulatory mechanisms underlying tannin biosynthesis in this crop and will allow the development of a fast and reliable tool for the generation of value-added faba bean cultivars with optimized flavonoid content.
Supplementary information
Acknowledgements
This research was supported by funding from the EU Project EUCLEG (No. 727312-2), as well as from the Projects RTA2017-00041 and PP.AVA.AVA2019.030, all of them co-financed by ERDF and the latter by the regional government of Andalucía. The authors would like to thank Dr. A Di Pietro for his careful reading of the manuscript and helpful comments and suggestions, Dr. I Casimiro-Soriguer for her valuable assistance in the statistical analysis and Dr. C De Miguel Rojas for her valuable laboratory assistance.
Author contributions
N.G., C.M.A. and A.M.T. conceived and designed the study and lab experiments. N.G. performed the experiments, the analysis and the interpretation of the data. N.G., C.M.A. and A.M.T. wrote the manuscript.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on request.
Competing of interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71070-2.
References
- 1.Matthews, P. & Marcellos, H. Faba bean. NSW Agriculture. Agfact P4.2.7, Agdex 164 (2003).
- 2.Picard J. Aperçu sur l’héredité du caractère absence de tannins dans les graines de féverole. Annales de l’Amélioration des Plantes. 1976;26:101–106. [Google Scholar]
- 3.Bond DA. In vitro digestibility of the testa in tannin-free field beans (Vicia faba L.) J. Agric. Sci. 1976;86:561–566. doi: 10.1017/S0021859600061104. [DOI] [Google Scholar]
- 4.Acamovic T, Brooker JD. Biochemistry of plant secondary metabolites and their effects in animals. Proc. Nutr. Soc. 2005;64:403–412. doi: 10.1079/PNS2005449. [DOI] [PubMed] [Google Scholar]
- 5.Edreva, et al. Stress-protective role of secondary metabolites: diversity of functions and mechanisms. Gen App Plant Physiology. 2008;34:67–78. [Google Scholar]
- 6.Griffiths DW. The inhibition of digestive enzymes by extracts of field bean (vicia faba) J. Sci. Food Agric. 1979;30:458–462. doi: 10.1002/jsfa.2740300503. [DOI] [PubMed] [Google Scholar]
- 7.Crépon K. Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crops Res. 2010;115:329–339. doi: 10.1016/j.fcr.2009.09.016. [DOI] [Google Scholar]
- 8.Tohge T, de Souza LP, Fernie AR. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017;68:4013–4028. doi: 10.1093/jxb/erx177. [DOI] [PubMed] [Google Scholar]
- 9.Winkel-Shirley B. Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002;5:218–223. doi: 10.1016/S1369-5266(02)00256-X. [DOI] [PubMed] [Google Scholar]
- 11.Jaakola L, et al. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002;130:729–739. doi: 10.1104/pp.006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodrich J, Carpenter R, Coen ES. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–964. doi: 10.1016/0092-8674(92)90038-E. [DOI] [PubMed] [Google Scholar]
- 13.Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell. 2002;14:2121–2135. doi: 10.1105/tpc.003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwinn K, et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 16.Hichri I, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 17.Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.2307/3870058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3:212–217. doi: 10.1016/S1360-1385(98)01242-4. [DOI] [Google Scholar]
- 19.Baudry A, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 20.Gutiérrez, N. Desarrollo de marcadores moleculares asociados a factores antinutritivos Vicia faba L. Tesis doctoral. Universidad de Córdoba, 2008. Dialnet scientific databases. https://dialnet.unirioja.es/servlet/tesis?codigo=54702.
- 21.Hellens RP, et al. Identification of Mendel’s white flower character. PLoS ONE. 2010;5:e13230. doi: 10.1371/journal.pone.0013230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirali M, et al. Genetics and biochemistry of zero-tannin lentils. PLoS ONE. 2016;11:e0164624. doi: 10.1371/journal.pone.0164624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrera A, Martín A. Genetics of tannin content and its relationship with flower and testa colours in Vicia faba. J Agric Sci. 1989;113:93–98. doi: 10.1017/S0021859600084665. [DOI] [Google Scholar]
- 24.Cabrera A, Martin A. Variation in tannin content in Vicia faba L. J. Agric. Sci. 1986;106:377–382. doi: 10.1017/S0021859600063978. [DOI] [Google Scholar]
- 25.Gutierrez, N., Ávila, C. M. & Torres, A. M. A TTG1 transcription factor regulates the white flower and the absence of tannins in faba beans (zt1 gene). in V Meeting AEL (ed. De Ron, A. M.) 175–176 (Spanish Association for Legumes (AEL), 2015).
- 26.Gutierrez, N., Avila, C. M. & Torres, A. M. Genomic localization and candidate genes of zt-2, the second gene controlling zero tannin content in faba bean (Vicia faba L.). in iclgg2019.com (2019).
- 27.Webb A, et al. A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.) Plant Biotechnol. J. 2016;14:177–185. doi: 10.1111/pbi.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez N, Torres AM. Characterization and diagnostic marker for TTG1 regulating tannin and anthocyanin biosynthesis in faba bean. Sci. Rep. 2019;9:16174. doi: 10.1038/s41598-019-52575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanotto S, Vandenberg A, Khazaei H. Development and validation of a robust KASP marker for zt2 locus in faba bean (Vicia faba) Plant Breed. 2019 doi: 10.1111/pbr.12772. [DOI] [Google Scholar]
- 30.Gutierrez N, Avila C, Moreno MT, Torres AM. Development of SCAR markers linked to zt-2, one of the genes controlling absence of tannins in faba bean. Aust. J. Agric. Res. 2008;59:62–68. doi: 10.1071/AR07019. [DOI] [Google Scholar]
- 31.Satovic Z, et al. A reference consensus genetic map for molecular markers and economically important traits in faba bean (Vicia faba L.) BMC Genom. 2013;14:932. doi: 10.1186/1471-2164-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres AM, Weeden NF, Martín A. Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theor. Appl. Genet. 1993;85:937–945. doi: 10.1007/BF00215032. [DOI] [PubMed] [Google Scholar]
- 33.Ocaña S, et al. Large-scale transcriptome analysis in faba bean (Vicia faba L.) under Ascochyta fabae infection. PLoS ONE. 2015;10:e0135143. doi: 10.1371/journal.pone.0135143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ooijen, J. W. Joinmap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. (Kyazma BV, 2006).
- 35.Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 36.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 37.Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods. 2010;50:S1–5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez N, Giménez MJ, Palomino C, Avila CM. Assessment of candidate reference genes for expression studies in Vicia faba L. by real-time quantitative PCR. Mol. Breed. 2011;28:13–24. doi: 10.1007/s11032-010-9456-7. [DOI] [Google Scholar]
- 39.Ruijter JM, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray H, Bock C, Georges F. Faba bean: transcriptome analysis from etiolated seedling and developing seed coat of key cultivars for synthesis of proanthocyanidins, phytate, raffinose family oligosaccharides, vicine, and convicine. Plant Genome. 2015;8:1. doi: 10.3835/plantgenome2014.07.0028. [DOI] [PubMed] [Google Scholar]
- 42.Nesi N, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang Y, et al. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151:1114–1129. doi: 10.1104/pp.109.144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, et al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016;210:905–921. doi: 10.1111/nph.13816. [DOI] [PubMed] [Google Scholar]
- 46.Albert NW, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;26:962–980. doi: 10.1105/tpc.113.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on request.