Abstract
Integration of transposable elements into the genome is mutagenic. Mechanisms targeting integrations into relatively safe locations, hence minimizing deleterious consequences for cell fitness, have emerged during evolution. In budding yeast, integration of the Ty1 LTR retrotransposon upstream of RNA polymerase III (Pol III)‐transcribed genes requires interaction between Ty1 integrase (IN1) and AC40, a subunit common to Pol I and Pol III. Here, we identify the Ty1 targeting domain of IN1 that ensures (i) IN1 binding to Pol I and Pol III through AC40, (ii) IN1 genome‐wide recruitment to Pol I‐ and Pol III‐transcribed genes, and (iii) Ty1 integration only at Pol III‐transcribed genes, while IN1 recruitment by AC40 is insufficient to target Ty1 integration into Pol I‐transcribed genes. Swapping the targeting domains between Ty5 and Ty1 integrases causes Ty5 integration at Pol III‐transcribed genes, indicating that the targeting domain of IN1 alone confers Ty1 integration site specificity.
Keywords: genome‐wide de novo insertion sites, integrase genome‐wide occupancy, RNA Polymerases I and III, Ty1 retrotransposon
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination
Despite interacting with a subunit shared by RNA polymerases I and III, a short integrase amino acid motif of yeast retrotransposon Ty1 targets it specifically to Pol III‐transcribed genes.

Introduction
Transposable elements (TEs) are mobile repetitive DNA sequences found in the genomes of most organisms (Huang et al, 2012). TEs are mutagenic and represent a threat to genome integrity, inactivating, or altering host gene expression or inducing large chromosomal rearrangements (Bourque, 2009; Levin & Moran, 2011). In humans, more than hundred heritable diseases have been assigned to de novo TE insertions (Hancks & Kazazian, 2016). TEs also play a role in genome evolution by modifying host functions, phenotypes, and gene regulation and can contribute to the long‐term adaptation of organisms to different environments (Chuong et al, 2016).
Where TEs integrate in the genome will determine their impact on their host. TE distribution, which is rarely random (Sultana et al, 2017), arises from the balance between two processes. First, selection leads to the elimination of strongly deleterious insertions and the maintenance of beneficial ones (Chuong et al, 2016; Cosby et al, 2019). Second, TEs have repeatedly evolved mechanisms that direct their integration into “safe” locations, where insertions will have minimal adverse effect on the organism's fitness (Boeke & Devine, 1998; Spaller et al, 2016; Cheung et al, 2018). These regions often consist of non‐essential repeated sequences, such as telomeric regions, ribosomal DNA arrays, upstream and downstream regions of transfer RNA genes (tDNAs), or non‐essential regions upstream of open reading frames (Zou et al, 1996; Penton & Crease, 2004; Fujiwara et al, 2005; Ye et al, 2005; Naito et al, 2009; Guo & Levin, 2010; Pardue & DeBaryshe, 2011; Baller et al, 2012; Mularoni et al, 2012; Spaller et al, 2016; Kling et al, 2018). Preferential targets have been described for the integration of different classes of TEs, including retroelements (Sultana et al, 2017).
Long terminal repeat (LTR) retrotransposons are retroelements related to retroviruses. They replicate by reverse transcription of their mRNA into a double‐stranded DNA copy (cDNA), which is imported into the nucleus and integrated into the genome by the LTR retrotransposon integrase (IN). The interaction between IN and cellular tethering factors plays a major role in integration site selection by targeting the integrase/cDNA complexes, aka the intasome, to specific chromosome locations. Tethering factors were first identified for Ty3 and Ty5 in Saccharomyces cerevisiae. These LTR retrotransposons integrate at the transcription start site of Pol III‐transcribed genes and in subtelomeric regions, respectively (Kirchner et al, 1995; Xie et al, 2001). The Tf1 LTR retrotransposon of Schyzosaccharomyces pombe and the MLV retrovirus both target the promoter region of Pol II‐transcribed genes (De Rijck et al, 2013; Gupta et al, 2013; Sharma et al, 2013; Hickey et al, 2015; Jacobs et al, 2015), and the HIV‐1 retrovirus targets the gene body of Pol II‐transcribed genes (Cherepanov et al, 2003; Llano et al, 2006). In all cases, tethering factors bind chromatin or have functions related to DNA transcription or replication (Sultana et al, 2017).
Ty1, the most active and abundant LTR retrotransposon in S. cerevisiae laboratory strain S288C, integrates preferentially within a 1 kb window upstream of Pol III‐transcribed genes. It targets nucleosomal DNA near the H2A/H2B interface (Baller et al, 2012; Mularoni et al, 2012). This integration pattern allows Ty1 to replicate while minimizing disruption to the host genome, as most Pol III‐transcribed genes are multicopy tDNAs and thus individually non‐essential. Furthermore, Ty1 insertion has a limiting impact on tDNA expression (Bolton & Boeke, 2003). Targeted integration proximal to tDNAs is a strategy that has been adopted several times by TEs to minimize damage to compact genomes (Cheung et al, 2018; Kling et al, 2018). The integration of Ty1 in these regions requires a functional Pol III promoter in the target gene (Devine & Boeke, 1996) and is influenced by the chromatin‐remodeling factor Isw2 and the Bdp1 subunit of TFIIIB (Bachman et al, 2005).
Recently, we have shown that an interaction between Ty1 IN (IN1) and the AC40 subunit of Pol I and Pol III is a major driver for Ty1 integration upstream of Pol III‐transcribed genes (Bridier‐Nahmias et al, 2015). The study used the S. pombe AC40 ortholog (AC40sp) as a loss‐of‐interaction mutant. The replacement of AC40 by AC40sp severely compromised Ty1 integration upstream of Pol III‐transcribed genes, leading to a redistribution of Ty1 insertions in the genome. IN1 binding to other Pol III subunits was also described in vitro (Cheung et al, 2016). However, it is not clear whether these interactions participate in Ty1 integration site selection.
IN1 has a three‐domain organization common to all retroelement integrases: the Zn2+ coordinating N‐terminal domain (NTD), the catalytic core domain (CCD), and the less conserved C‐terminal domain (CTD) (Fig 1A, top) (Wilhelm et al, 2005). IN1 C‐terminal residues 578–635 are necessary and sufficient to mediate the interaction with AC40 in vivo (Bridier‐Nahmias et al, 2015). This region also contains a bipartite nuclear localization signal (bNLS, residues 596–630; Fig 1A, top) consisting of two Lys‐Lys‐Arg motifs separated by a 29 amino acid linker (Kenna et al, 1998; Moore et al, 1998; Lange et al, 2011). This raises the question of whether IN1 nuclear import and interaction with AC40 could act in concert during Ty1 replication.
Figure 1. Mutations in the IN1 bNLS linker sequence abolish the interaction with AC40.
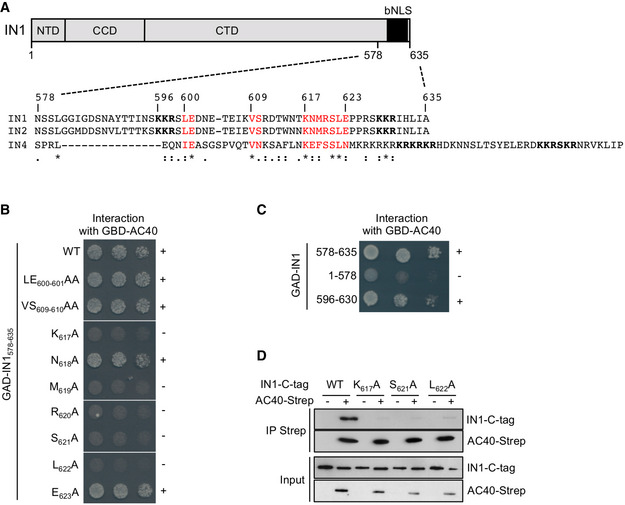
- Top. The Ty1 integrase (IN1) showing N‐terminal and catalytic core domains (NTD and CCD) and the bipartite NLS at the C‐terminus (CTD). Bottom. Alignment of amino acid sequences of Ty1, Ty2, and Ty4 integrase C‐termini (IN1, IN2, and IN4, respectively). In bold: Basic amino acids required for NLS function. In red: Amino acids in the bNLS linker relatively conserved between the three integrases. *, identity; :, high similarity; ., low similarity; ‐, gap in sequence.
- Two‐hybrid interaction between GBD‐AC40 and WT or mutant GAD‐IN1578–635. Alanine substitutions in IN1578‐635 are indicated. Cells were plated in twofold serial dilutions on DO‐Leu‐Trp‐His plates. No growth or protein expression defects were detected (Fig EV1A and B). +, interaction; −, no interaction.
- Two‐hybrid interaction between GBD‐AC40 and different IN1 regions fused to GAD, as indicated. Cells were plated in ten‐fold serial dilutions on DO‐Leu‐Trp‐His plates. No growth or protein expression defects were detected (Fig EV1C and D). +, interaction; −, no interaction.
- In vitro interaction between AC40 and IN1 proteins co‐expressed in E. coli. Co‐precipitation of WT or mutated (K617A, S621A, or L622A) IN1‐C‐tag using AC40‐Twin‐Strep‐tag as bait from bacterial protein extracts co‐expressing (+) or not (−) these proteins. Expected sizes are 41 kDa for AC40‐Twin‐Strep‐tag and 100 kDa for IN1‐C‐tag (WT and mutants).
Source data are available online for this figure.
In this study, we identify a short sequence, namely the Ty1 targeting domain, in the bNLS linker of IN1 that directs the interaction with AC40. Single amino acid substitutions in the Ty1 targeting domain do not affect the frequency of Ty1 retrotransposition but impair the recruitment of IN1 to Pol III‐transcribed genes. Consequently, these IN1 mutations induce the same changes in the Ty1 integration profile as observed in the AC40sp loss‐of‐interaction mutant. While IN1 is also recruited to Pol I‐transcribed genes through its interaction with AC40, Pol I‐transcribed genes are poor targets of Ty1 integration. When the Ty1 targeting domain is used to replace the Ty5 IN sequence responsible for Ty5 integration into heterochromatin, Ty5 integration is redirected to Pol III‐transcribed genes. This work therefore confirms the fundamental role of the IN1‐AC40 interaction in Ty1 integration site selection and reveals that the targeting domain of Ty1 is necessary and sufficient to confer Ty1 integration preference to another retrotransposon.
Results
Mutations in the IN1 bNLS linker sequence abolish the interaction with AC40
Saccharomyces cerevisiae LTR retrotransposons Ty1, Ty2, and Ty4 have the same integration preferences for regions upstream of Pol III‐transcribed genes (Kim et al, 1998; Carr et al, 2012), and the C‐termini of their integrases (IN1, IN2, and IN4, respectively) interact with the Pol III subunit AC40 (Bridier‐Nahmias et al, 2015). To identify conserved amino acids potentially involved in the AC40 interaction, we aligned the C‐terminal sequences of IN1, IN2, and IN4 (Fig 1A, bottom) and observed that IN1 and IN2 are highly similar in this region, whereas IN4 is more divergent. Amino acids at positions 600–601, 609–610, and 617–623 in IN1 were either identical or highly similar in all three INs. We replaced each of these amino acids by alanine, individually or in pairs, in a Gal4‐activating domain GAD‐IN1578–635 fusion protein, checked that protein level was not affected by these mutations (Fig EV1A), and studied the interaction of the mutant fusion proteins with Gal4 binding domain GBD‐AC40 using a two‐hybrid assay. The interaction between IN1578–635 and AC40 was maintained in the presence of mutations LE600–601AA, VS609–610AA, N618A, or E623A and suppressed by single alanine substitution of K617, M619, R620, S621, or L622 (Figs 1B and EV1B). GBD‐AC40 interacted with GAD‐IN1578–635 but not with GAD‐IN11–578 (Fig 1C) as shown previously (Bridier‐Nahmias et al, 2015). Since amino acids K617‐L622 are located in the IN1 bNLS linker sequence, we also tested the interaction between GBD‐AC40 and GAD fused to the entire bNLS sequence (GAD‐IN596–630). Interaction between the two fusion proteins was detected, suggesting that this region of 34 amino acids in IN1 is sufficient for interaction with AC40 (Figs 1C and EV1C and D).
Figure EV1. Mutations in the IN1 bNLS linker sequence abolish the interaction with AC40.
- Validation of the expression of GAD‐IN1 fusion proteins for the two‐hybrid assay shown in Fig 1B. Whole‐cell extract samples were prepared for immunoblotting from 1 DO600 of the cell culture by TCA precipitation. GAD‐IN1 fusion proteins were detected by Western blot with anti‐HA antibody (12CA5, Roche). All constructs harbor an HA‐tag that is present between the GAD and IN1 sequences in the original pACTII vector. Expected sizes of the proteins are 26 kDa for GAD‐IN1578–635 and GAD‐IN1578–635 mutants and 82 kDa for GAD‐IN11–578. “x”, non‐relevant sample.
- Growth control of cell cultures corresponding to the two‐hybrid assay shown in Fig 1B. Twofold serial dilutions of cell cultures, starting from 10−1, were plated on DO‐Leu‐Trp plates to check for growth.
- Growth control of cell cultures corresponding to the two‐hybrid assay shown in Fig 1C. Twofold serial dilutions of cell cultures, starting from 10−1, were plated on DO‐Leu‐Trp plates to check for growth.
- Validation of the expression of GAD‐IN1 fusion proteins for the two‐hybrid assay shown in Fig 1C. Whole‐cell extract samples were prepared for immunoblotting from 1 DO600 of the cell culture by TCA precipitation. GAD‐IN1 proteins were detected by Western blot with anti‐GAD antibody (Santa Cruz Biotechnology). Expected sizes of the proteins are 82 kDa for GAD‐IN11–578, 26 kDa for GAD‐IN1578–635, and 24 kDa for GAD‐IN1596–630. “x”, non‐relevant sample.
Source data are available online for this figure.
To determine whether amino acids required for the two‐hybrid interaction between AC40 and the IN1 C‐terminus were also critical for the interaction between the two full‐length proteins, we co‐expressed AC40‐Strep and WT or mutant IN1‐C‐tag tagged proteins in E. coli and performed co‐precipitation of the two proteins with the cell extracts. We focused on K617, S621, and L622, given their strict conservation in IN1, IN2, and IN4 (Fig 1A, bottom). In the presence of Strep‐Tactin beads, AC40‐Strep co‐immunoprecipitated with WT IN1 but not with the IN1 K617A, S621A, and L622A mutants (Fig 1D). These data strongly suggest that full‐length IN1 and AC40 proteins bind directly to each other, and their interaction depends on residues K617, S621, and L622 located in the IN1 bNLS.
Non‐AC40 binding IN1 mutants do not affect Ty1 integration frequency
Mutations in the IN1 bNLS that induce a substantial or complete loss of IN1 nuclear accumulation reduce the frequency of Ty1 retromobility, as seen when the two Lys‐Lys‐Arg (KKR) motifs are mutated, either individually or simultaneously, but also for mutations of specific acidic residues in the linker region (Kenna et al, 1998; Moore et al, 1998; Lange et al, 2011). To investigate whether the amino acids we identified in the bNLS linker sequence as being necessary for the AC40 interaction were also required for Ty1 nuclear import and retromobility, we introduced K617A, S621A, or L622A mutations into a GFP2‐bNLS fusion protein previously used to assess IN1 bNLS function (McLane et al, 2008). In contrast to the IN1‐bNLSmut construct (596KKR598‐AAA and 628KKR630‐AAA), the three IN1‐bNLS single mutants were still able to target GFP2 into the nucleus (Fig 2A). Thus, amino acids required for the IN1‐AC40 interaction are dispensable for NLS function.
Figure 2. Non‐AC40 binding IN1 mutants do not affect Ty1 integration frequency.
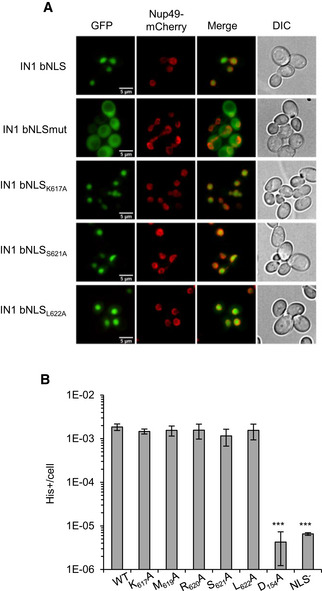
- Localization of GFP2‐IN1 bNLS variants in yeast cells by direct fluorescence microscopy. Nup49‐mCherry signal was used to visualize the location of the nuclear envelope. Corresponding DIC images are shown. Scale bar is 5 μm.
- Retrotransposition frequency (log scale) of pGAL1‐Ty1his3AI bearing the indicated substitutions of conserved residues in a spt3‐101 rad52∆ strain. IN1 catalytic core domain mutant D154A is defective for integration and NLS mutant (KKR628–630GGT) for nuclear import. Values are mean ± SD, n = 4 experiments, each performed with four independent colonies. ***P ≤ 0.001, otherwise not significant (Student t‐test).
Source data are available online for this figure.
The same mutations were introduced individually into a Ty1 element expressed from the GAL1 promoter in a 2‐micron plasmid and containing the retromobility indicator gene his3AI, allowing detection of Ty1‐HIS3 insertion events as His+ prototroph cells (Curcio & Garfinkel, 1991). To determine the frequency of Ty1his3AI integration in the genome, we expressed this plasmid in a spt3‐101 null rad52∆ mutant strain, deficient in endogenous Ty1 expression and homologous recombination. SPT3 is required for Ty1 transcription, and its absence prevents the trans‐complementation of the mutant IN1 by WT IN1 from endogenous Ty1 elements (Curcio & Garfinkel, 1992). RAD52 deletion precludes insertion of the Ty1‐HIS3 cDNA by homologous recombination with preexisting genomic Ty1 copies, a preferred pathway when IN1‐dependent integration is defective (Sharon et al, 1994). The frequency of His+ cells was similar between strains expressing WT or K617A, M619A, R620A, S621A, or L622A mutant Ty1his3AI (Fig 2B). In contrast, mutations that inactivate IN1 nuclear import (Moore et al, 1998) or catalytic activity (Wilhelm & Wilhelm, 2005) caused a substantial decrease in the frequency of His+ cells compared to WT (Fig 2B, mutants NLS‐ and D154A, respectively).
Thus, single amino acid mutations in IN1 that strongly affect the interaction with AC40 do not impair Ty1 retrotransposition. These data confirm that the IN1‐AC40 interaction is not required for Ty1 overall integration frequency (Bridier‐Nahmias et al, 2015).
AC40 recruits IN1 at Pol I and Pol III‐transcribed genes
In addition to AC40, other Pol III subunits have been suggested to mediate the interaction between IN1 and Pol III (Cheung et al, 2016). Pol III is a stable complex, and immunoprecipitation of a specific subunit results in the co‐precipitation of the whole complex (Oficjalska‐Pham et al, 2006; Bhalla et al, 2019). To determine whether IN1 remains associated with Pol III in the absence of the IN1‐AC40 interaction, we immunoprecipitated Pol III in yeast cells expressing hemagglutinin (HA)‐tagged C160, the largest Pol III subunit, and WT or mutant IN1 fused to streptavidin (IN1‐Strep). WT IN1 was associated with Pol III but not the K617A, S621A, or L622A IN1 mutants (Fig 3A). Therefore, the interaction with AC40 is necessary for IN1 binding to Pol III in vivo. Since an interaction was detected in vitro between IN1 and the C34 and C53 proteins (Cheung et al, 2016), we asked whether these associations could be detected in vivo apart from the Pol III complex. This was not the case as both C53 and C34 co‐immunoprecipitated with WT IN1 but not with the IN1 mutant K617A (Fig EV2A and B). These data indicate that in vivo, the interaction of C34 and C53 with IN1 occurs primarily when Pol III is associated with IN1. Altogether, these data show that AC40 is determinant for the binding of IN1 to the Pol III complex.
Figure 3. AC40 recruits IN1 at Pol III‐transcribed genes.
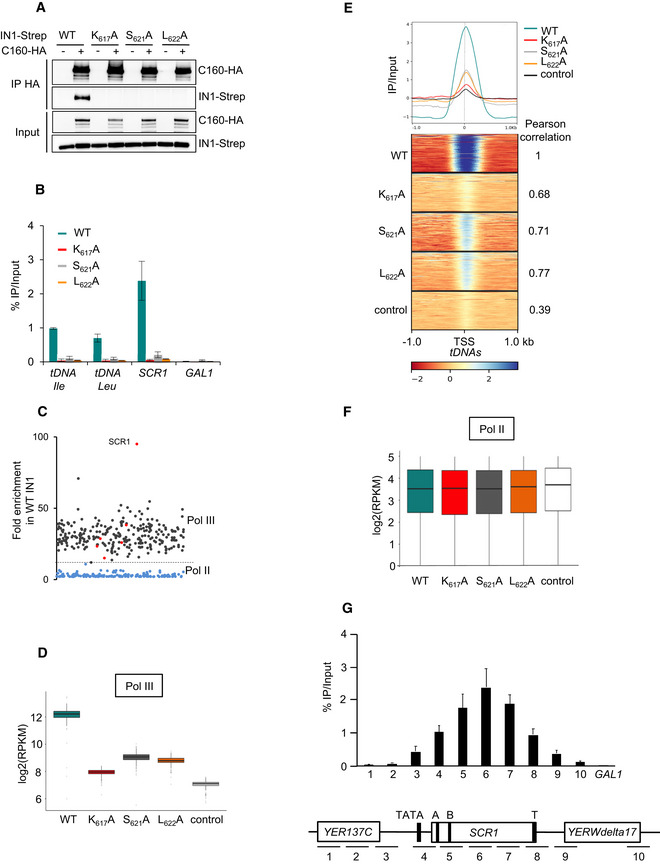
- Co‐immunoprecipitation of ectopic IN1 using C160‐HA as bait, from yeast protein extracts expressing WT or mutated IN1‐Strep (K617A, S621A, or L622A) from the GAL1 promoter in the presence of galactose. Expected sizes are 160 kDa for C160‐HA and 100 kDa for IN1‐Strep (WT and mutants).
- Quantitative ChIP analysis of HA‐IN1 enrichment at Pol III‐transcribed genes. Immunoprecipitated DNA from yeast cells producing ectopic IN1 is expressed as a value relative to that of the input. Pol III‐transcribed genes: tDNA‐Ile and tDNA‐Leu families (16 and 22 genes, respectively) and the unique SCR1 gene. GAL1 ORF serves as a control. Data represent means ± SD (n ≥ 3).
- Fold enrichment of WT HA‐IN1 over input at all the genes where HA‐IN1 has been detected by ChIP‐seq analysis. Each dot represents a gene. Gray, tDNAs; red, other Pol III‐transcribed genes; blue, Pol II‐transcribed genes. The dashed line indicates the separation between Pol II and Pol III‐transcribed genes.
- WT and mutant IN1 association with all Pol III‐transcribed genes. Values obtained from ChIP‐seq analysis have been normalized in log2 RPKM (reads per kilobase per million mapped reads). Control is anti‐HA immunoprecipitation of chromatin extracts in cells expressing IN1‐Strep. Immunopurified DNA from 3–4 independent experiments was combined in one replicate. Data are presented as boxplots showing the median, the first quartile, and the third quartile. Error bars show minimum and maximum values. Regions used for calculation are gene bodies.
- Genome‐wide occupancy profiles (top) and heatmaps (bottom) of WT and HA‐IN1 mutants over a 1 kb window upstream and downstream of all tDNAs. Pearson correlation to WT values corresponding to Fig EV2D is indicated.
- WT and mutant IN1 association to all Pol II‐transcribed genes, respectively. Control, replicates and data presentation, as described for panel D. Regions used for calculation are gene bodies.
- Recruitment of IN1 along the SCR1 gene locus. Top. ChIP‐qPCR analysis as described for panel Fig 3B. Data represent means ± SD (n = 3). Bottom. Schematic representation of the SCR1 locus with DNA amplicon positions. TATA, A and B boxes and transcription terminator (T) are indicated.
Source data are available online for this figure.
Figure EV2. AC40 recruits IN1 at Pol I‐ and Pol III‐transcribed genes.
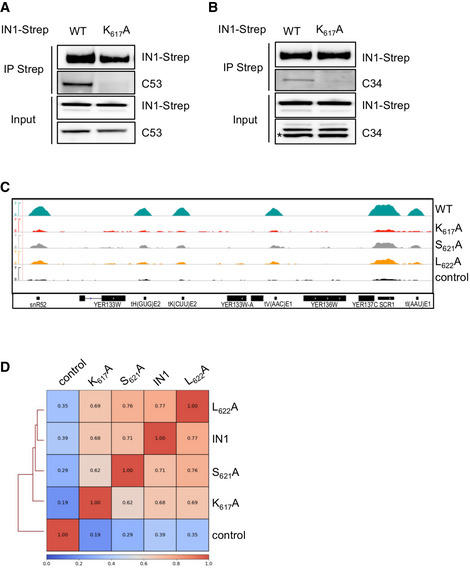
-
A, BCo‐immunoprecipitation of endogenous Pol III subunits (C53 and C34) using ectopically expressed Ty1 integrase (IN1‐Strep) as bait. Panels A and B were two separate lysates from two separate co‐IPs. Expected sizes are 53 kDa for C53, 34 kDa for C34 and 82 kDa for IN1‐Strep (WT and K617A mutant). *; C34.
-
CGenome browser visualization of different IN1‐HA occupancy for chromosome V (coordinates chrV:431129..443275). Control is an anti‐HA immunoprecipitation of chromatin extracts in cells expressing IN1‐Strep. The region contains tH(GUG)E2, tK(CUU)E2, tV(ACC)E1, tI(AAU)E1, SNR52, and SCR1, all transcribed by Pol III.
-
DPearson correlation heatmap at tDNAs between the following ChIP‐seq: IN1 WT, K617A, S621A, L622A and control.
Source data are available online for this figure.
To establish whether AC40 plays a major role in IN1 recruitment to Pol III‐transcribed genes, we developed IN1 chromatin immunoprecipitation (ChIP) experiments to assay the effect of the K617A, S621A, and L622A mutations on recruitment. WT IN1 and the various mutants were tagged at their N‐terminus using a 3xHA epitope tag and expressed from a tetracycline‐off promoter. Quantitative PCR revealed significant enrichment of ectopic WT HA‐IN1 at all tested Pol III‐transcript loci, compared to background level measured on the GAL1 gene promoter (Fig 3B). In contrast, HA‐IN1 mutants that did not interact with AC40 were barely detected at these loci. Thus, recruitment of IN1 to Pol III‐transcribed loci depends on its interaction with AC40.
To assess whether the genome‐wide occupancy of WT IN1 correlates with Ty1 integration site preferences, we performed ChIP sequencing (ChIP‐seq) using the same HA‐IN1 constructs, with a strain expressing IN1‐Strep as control. Analysis of reads mapping to unique sites revealed a strong association of WT HA‐IN1 with most nuclear tDNAs and the Pol III‐transcribed genes SNR6, SNR52, SCR1, RPR1, and RDN5. Very weak or no HA‐IN1 binding was observed for RNA170 and ZOD1, previously shown to have low level of Pol III occupancy (Moqtaderi & Struhl, 2004) (Fig 3C and Table EV1). Three tDNAs, tK(CUU)C, tM(CAU)C, and tD(GUC)N, were not recovered: These genes are either absent or transcriptionally inactive in the laboratory strain we used (Kumar & Bhargava, 2013; Patterson et al, 2019). HA‐IN1 was absent from most Pol II‐transcribed genes (Table EV1) or present at a much lower level than at Pol III‐transcribed genes (Fig 3C). Low HA‐IN1 occupancy at Pol II‐transcribed genes may be an artifact of ChIP‐seq due to the level of expression of these genes (Teytelman et al, 2013). The genome‐wide distribution of ectopic WT IN1 revealed a strong bias for Pol III‐transcribed genes, confirming that the interaction of IN1 with the Pol III is the main driver for targeted integration of Ty1.
ChIP‐seq analysis of the HA‐IN1 mutants that compromise the IN1‐AC40 interaction (K617A, S621A, and L622A) revealed their occupancy was substantially reduced at all Pol III‐transcribed genes, as indicated by the lower number of reads corresponding to Pol III‐transcribed genes with the three mutants, compare to WT HA‐IN1 (Fig 3D), and by a metagene analysis comparing WT and mutant HA‐IN1 binding on all the 275 nuclear tDNAs (Fig 3E). No effect of these mutants was observed at Pol II‐transcribed genes (Fig 3F). Quantification by pair‐wise Pearson correlation between WT and either K617A, S621A, or L622A IN1 confirmed the apparent stronger decrease in Pol III‐transcribed gene occupancy of K617A HA‐IN1 compared to the other mutants (Figs 3E and EV2C and D). The metagene analysis indicated a sharp peak around the transcription start site (TSS), which does not coincide with Ty1 integration sites, normally located upstream of Pol III‐transcribed genes (Baller et al, 2012; Mularoni et al, 2012; Bridier‐Nahmias et al, 2015). Since tDNAs are too small to allow spatial resolution of Pol III localization, we performed ChIP analysis on SCR1, the longest Pol III‐transcribed gene, to determine whether IN1 is associated with the polymerase over the entire gene. WT HA‐IN1 was detected all along the SCR1 gene with maximum binding in the coding region (Fig 3G). This pattern is similar to that observed with Pol III (Ghavi‐Helm et al, 2008), confirming that the interaction with Pol III is determinant for IN1 recruitment to the chromatin. HA‐IN1 occupancy was not completely suppressed in the three mutants, suggesting that the K617, S621, or L622 mutants may have a residual level of interaction with AC40 that would not be detected in experiments where IN1 is overexpressed (i.e., in two‐hybrid or in vitro Co‐IP assays). Alternatively, additional protein–protein interactions, like those previously identified with other Pol III subunits, could contribute to the recruitment of Ty1 integration complex at Pol III‐transcribed genes in the absence of the IN1‐AC40 interaction (Cheung et al, 2016).
AC40 is common to both Pol I and Pol III, suggesting that IN1 may also interact with Pol I. Consistently, we found that Pol I is associated with IN1 in vivo. Pull‐down of A190‐TAP, the largest subunit of Pol I, retained WT IN1‐HBH, whereas no association was detected with the IN1 mutants (Fig 4A). The RDN1 locus is composed of 100‐200 tandem repeats of the 35S‐precursor rDNA (RDN37), transcribed by Pol I, and the 5S rDNA (RDN5), transcribed by Pol III (Dammann et al, 1993). Analysis of ChIP‐seq reads mapping at multiple positions revealed WT HA‐IN1 at both genes (Fig 4B). IN1 occupancy may be overestimated, as reads corresponding to all repeats are aggregated on the two copies that are represented in the S. cerevisiae reference genome (https://www.yeastgenome.org). However, IN1 mutants compromising the IN1‐AC40 interaction—S621A, L622A, and particularly K617A—reduced HA‐IN1 occupancy at both loci. Since ChIP‐seq data result in a poor resolution of the RDN1 locus, we also performed conventional ChIP experiments that showed a clear enrichment of IN1 at both Pol I‐ and Pol III‐transcribed genes but not at the intergenic NTS1 and NTS2 regions (Fig 4C), confirming that the presence of IN1 correlates with that of the two polymerases. WT HA‐IN1 occupancy appeared lower at RDN5 than at other Pol III‐transcribed genes (compare Figs 3B and 4C), probably because Pol III is less associated with RDN5 (Moqtaderi & Struhl, 2004). On the other hand, IN1 binding to the 18S locus transcribed by Pol I decreased in the presence of the K617A, S621A, or L622A mutations (Fig 4D). Altogether, these results support a role of AC40 in the binding of IN1 to the RDN1 repeats.
Figure 4. AC40 recruits IN1 at the RDN1 repeats.
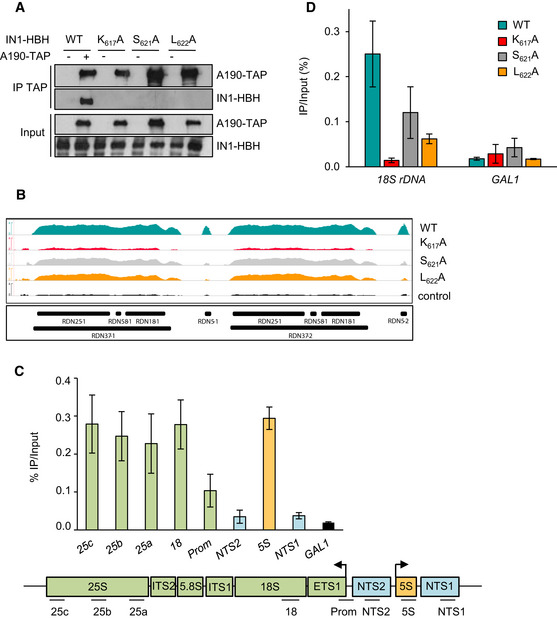
- Co‐immunoprecipitation of ectopic IN1 using TAP‐tagged‐A190 as bait from yeast protein extracts expressing WT or the indicated IN1‐HBH mutants from a pTet‐Off promoter in the absence of doxycycline. Expected sizes are 204 kDa for A190‐HA and 82 kDa for IN1‐Strep (WT and mutants).
- Genome browser visualization of HA‐IN1 occupancy at the RDN1 locus. Occupancy of WT HA‐IN1 and K617A, S621A, and L622A HA‐IN1 mutants is represented in each panel. Control, as described for panel 3D.
- Top. IN1 is recruited at Pol I‐transcribed genes. ChIP‐qPCR analysis as described for panel 3B. Data represent means ± SD (n = 3). Bottom. Schematic representation of the RDN1 locus with DNA amplicon positions.
- Recruitment of WT and mutant HA‐IN1 (K617A, S621A, or L622A) at the 18S rDNA locus transcribed by Pol I. ChIP‐qPCR analysis as described for panel 3B. Data represent means ± SD (n = 3). GAL1 ORF serves as a control.
Source data are available online for this figure.
Finally, the reduced association of HA‐IN1 mutants with Pol I‐ and Pol III‐transcribed genes did not result in a significant increase of IN1 occupancy at other specific loci and particularly at subtelomeres (Table EV1). Collectively, these results indicate that the IN1‐AC40 interaction is necessary for the interaction between IN1 and Pol III and thus its recruitment at Pol III‐transcribed genes. IN1 also interacts with Pol I via AC40 and is present at Pol I‐transcribed genes.
Non‐AC40 binding Ty1 mutants have altered integration profiles
To investigate the integration profile of Ty1 mutants that have an impaired IN1‐AC40 interaction, libraries of His+ selected de novo Ty1 insertion events were generated in cells expressing WT or mutant Ty1his3AI elements from the GAL1 promoter (Barkova et al, 2018). We used an spt3‐101 null rad52∆ mutant strain to avoid both trans‐complementation of the mutant IN1 by endogenous WT IN1 and Rad52‐dependent recombination events. Initially, we performed qualitative PCR to monitor Ty1 insertion events at the SUF16 tRNA gene and the SEO1 subtelomeric gene. These genes were identified as hot spots of Ty1 integration in WT and AC40sp loss‐of‐interaction mutant, respectively (Bridier‐Nahmias et al, 2015). In independent cultures expressing WT Ty1his3AI, we observed multiple bands upstream of the SUF16 tRNA gene, characteristic of Ty1‐HIS3 insertion in the three nucleosomes upstream of tDNA genes (Bachman et al, 2005; Dakshinamurthy et al, 2010) (Fig 5A). This profile was significantly different for Ty1his3AI harboring K617A, S621A, or L622A mutations in IN1, with many fewer integration events upstream of SUF16, and increased insertion at SEO1, compared to WT Ty1his3AI (Fig 5A). This observation suggests that IN1 mutations at K617, S621, and L622 have the same effect on Ty1 integration site targeting as the AC40sp loss‐of‐interaction mutant.
Figure 5. Non‐AC40 binding Ty1 mutants have altered integration profiles.
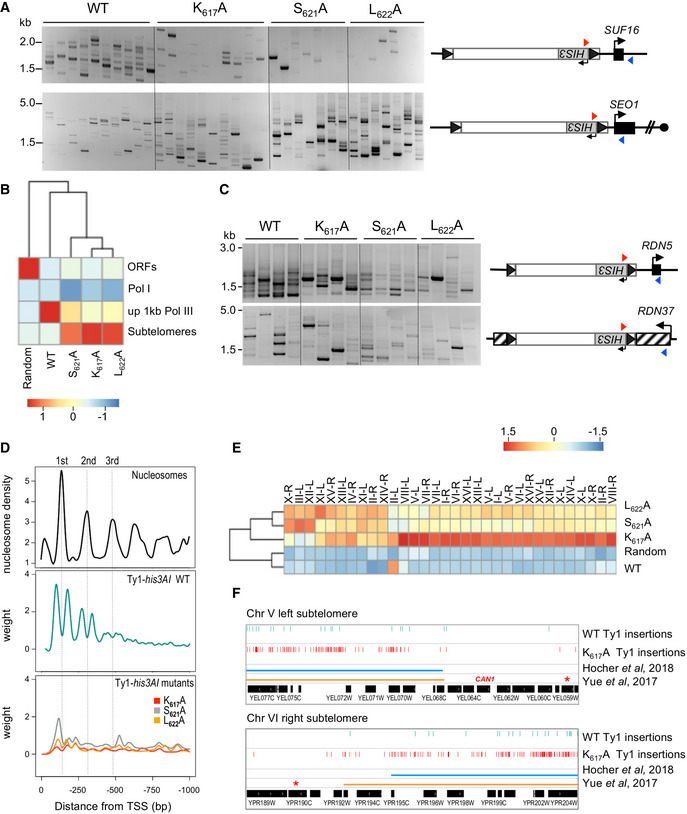
- Detection of de novo Ty1 insertions upstream of the SUF16 and SEO1 genes by PCR using a primer in HIS3 (red triangle) and a primer in the locus of interest (blue triangle). Ty1 retrotransposition was induced in cells transformed with plasmids expressing WT or mutant (IN1 K617A, S621A, and L622A) Ty1his3AI from the GAL1 promoter. Total genomic DNA was extracted from His+ cells obtained from independent cultures.
- Genome‐wide Ty1 insertion frequencies at each genomic feature are clustered in a heatmap. Score is computed in column Z‐score. ORFs, all Pol II‐transcribed genes except genes at subtelomeres; Pol I, one RDN37 copy; up 1 kb Pol III, 1 kb upstream of all Pol III‐transcribed genes; subtelomeres, genomic coordinates corresponding to chromatin covered by Sir2 and Sir3, when they are co‐overexpressed (Hocher et al, 2018); Random, 100,000 random Ty1 computed insertions in the genome.
- Detection of de novo Ty1 insertions upstream of RDN5 and in RDN37 genes by PCR as described for panel 5A. Four total genomic DNA preparations from panel A were randomly tested by PCR.
- Ty1 insertion profile upstream of tDNAs. Total genomic DNA extracted in (B) was prepared for Ty1 de novo integration event sequencing. Ty1 insertions are computed in a 1 kb window upstream of all the 275 nuclear tDNAs (position 0 in the graph). Each position is divided by the number of insertions at this position (weight). The Smoothing curves indicate the general trend. Nucleosome center positions for the three‐first nucleosomes upstream of all tDNAs are from (Brogaard et al, 2012).
- Ty1 insertion frequencies for each left and right subtelomere of chromosomes are clustered in a heatmap. Score is computed in row Z‐score. Random, as described for panel B. High level of WT Ty1‐HIS3 integration in II‐L is due to the presence of the tRNA gene tF(GAA)B.
- Genome browser visualization of WT and IN1 K617A mutant Ty1‐HIS3 insertions into chromosome V left and chromosome VI right subtelomeres compared to the subtelomere boundaries defined by Hocher et al (2018) and Yue et al (2017), respectively. Red stars indicate the first essential gene of each subtelomere. The CAN1 gene is indicated on chromosome V.
Source data are available online for this figure.
To extend our analysis to the entire genome, we characterized Ty1‐HIS3 de novo insertion event libraries using high‐throughput sequencing. We could discriminate Ty1‐HIS3 de novo insertions from endogenous elements using six nucleotides in the 3′ LTR that were specific to the Ty1his3AI element (Baller et al, 2012). By comparing the Z‐score of Ty1 insertions on four non‐overlapping features (Fig 5B), we confirmed that WT Ty1 insertions occurred mainly in a 1 kb window upstream of most Pol III‐transcribed genes (Table EV2), the only exceptions being tDNAs that were absent or not transcribed in our strain. There were significantly fewer Ty1‐HIS3 insertions at Pol III‐transcribed genes with the three IN1 mutants (Fig 5B). However, the preference for Pol III‐transcribed genes was not completely lost, as insertions in these regions were higher than expected if Ty1 targeting was random, supporting the idea that the interaction with Pol III is not fully abolished with these mutants. Using the sequencing reads mapping at multiple positions in the genome, we also detected Ty1‐HIS3 de novo insertion events at RDN5 and RDN37 genes in the RDN1 repeats and in the adjacent RDN5 variants (Fig EV3A). To determine whether these insertions were or not an artifact of aggregating integrations occurring in all RDN1 repeats in the only two repeats of the reference genome, we compared the percentage of Ty1‐HIS3 insertions to random insertions generated in silico in two artificial yeast genomes composed of 20 and 60 RDN1 repeats. We relied on two studies to estimate the actual number of actively transcribed RDN1 repeats. In Ide et al, a yeast strain with only 20 RDN1 copies is viable and displays no growth defect, suggesting that the level of Pol I transcription in WT cells may correspond to only 20 actively transcribed copies (Ide et al, 2010), while Merz et al estimate that, among the 150–200 rDNA repeats present in the genome, about 30–50%, i.e. approximately 60 copies, are actively transcribed (Merz et al, 2008). With this approach, we observed a 7‐ to 20‐fold enrichment of WT Ty1‐HIS3 insertions in a 1‐kb window upstream of RDN5 locus compared to the two situations where these insertions would be randomly distributed (Fig EV3B). In the presence of the K617A, S621A, or L622A mutations, the percentage of insertions decreased but was still higher than the two random situations (Fig EV3B). A similar decrease was observed by qualitative PCR (Fig 5C). This could be explained by the residual recruitment of IN1 mutants still observed by ChIP‐seq (Fig 4B). This confirms that the Pol III‐transcribed gene RDN5 is a hotspot of Ty1 insertions, which depends on the interaction between IN1 and AC40. In contrast, the percentage of WT Ty1‐HIS3 insertion events at RDN37 was close to the random situations, lower than at RDN5 and not decreased by the K617A, S621A, or L622A mutations (Fig 5C and EV3B). These results indicate that Ty1 integration is not preferentially targeted to RDN37 and that the recruitment of IN1 at Pol I‐transcribed genes is not sufficient for subsequent Ty1 integration.
Figure EV3. Ty1 integration profile at the RDN1 repeats.
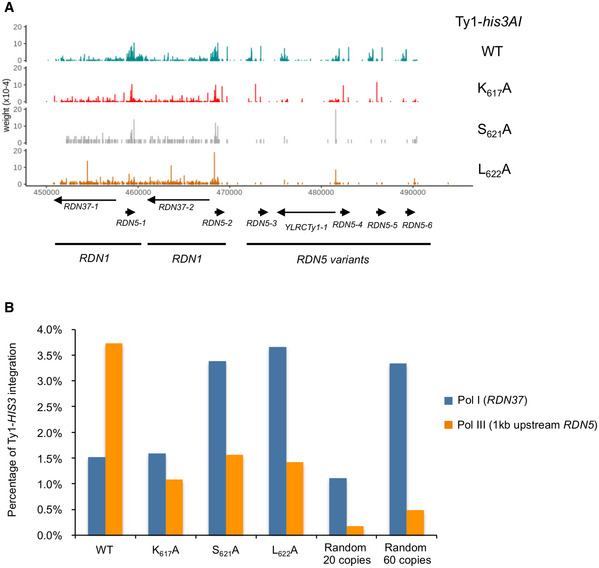
- Visualization of WT and IN1 mutant Ty1‐HIS3 insertions into the RDN1 repeats and adjacent RDN5 variants.
- Quantification of Ty1‐HIS3 insertions events (WT and mutants) at RDN37 (Pol I‐transcribed gene) and in a 1 kb window upstream of RDN5 (Pol III‐transcribed gene). Control cases are 1 × 105 Ty1 random insertions in artificial yeast genomes composed of 20 or 60 RDN1 repeats. Of note, less de novo Ty1 insertion events were recovered for the S621A and L622A mutants than for WT and K617A mutants.
WT Ty1‐HIS3 insertions displayed a periodic profile in the region of the three nucleosomes located upstream of Pol III‐transcribed genes, with two insertion sites per nucleosome, as seen previously (Fig 5D) (Baller et al, 2012; Mularoni et al, 2012; Bridier‐Nahmias et al, 2015). This profile was modified with the three mutants, with the first site of the first nucleosome being less affected with the S621A mutant, suggesting that close proximity to the tDNA is a determinant of integration. The K617A mutant, which had the lowest HA‐IN1 occupancy at Pol III‐transcribed genes (Fig 3D and E), displayed the largest decrease in integration events at Pol III‐transcribed genes (Fig 5B and D). This correlation suggests that the strength of the IN1‐AC40 interaction influences integration efficiency at Pol III‐transcribed genes. Concomitant with the decrease in integration at Pol III‐transcribed genes, the three IN1 mutants showed an increase in integration events at the end of each chromosome arm, a phenotype that was most pronounced for the K617A mutant (Fig 5B and E). These results are consistent with the redistribution of Ty1 insertions in these regions observed in the AC40sp loss‐of‐interaction mutant (Bridier‐Nahmias et al, 2015). Ty1 de novo insertions at the ends of chromosomes were mostly located in regions defined as subtelomeres, based on heterochromatin specificities or loss of synteny between different Saccharomyces strains (Yue et al, 2017; Hocher et al, 2018), suggesting that these subtelomeric regions harbor determinants allowing Ty1 targeting (Fig 5F). In contrast, no increase in Ty1‐HIS3 de novo insertion events was detected in Pol II‐transcribed genes for the three mutants (Fig 5B), as exemplified for the CAN1 gene (Fig 5F).
Altogether, these results further support a major role for the IN1‐AC40 interaction in Ty1 integration targeting at Pol III‐transcribed genes. They also confirm that, when this interaction is compromised, Ty1 insertions are not random but principally occur in subtelomeres. Based on these data, we have named the IN1 sequence that contains the amino acids required for the interaction with AC40, the Ty1 targeting domain of IN1 (TD1).
IN1 targeting domain directs Ty5 integration at Pol III‐transcribed genes
To determine whether TD1 is sufficient to confer Ty1 integration site preferences, we transferred the sequence into the Ty5 retrotransposon, which preferentially integrates into heterochromatin at yeast silent mating loci (HMR and HML) and near telomeres (Zou et al, 1996). Ty5 selectivity relies on an interaction between a hexapeptide (TD5, targeting domain of Ty5) in the C‐terminus of IN5 and the heterochromatin protein Sir4 (Gai & Voytas, 1998; Xie et al, 2001). Exchange of TD5 for the IN1 bNLS containing TD1 in IN5, expressed in a two‐hybrid vector, resulted in IN5 interacting with AC40, but not with Sir4 (Figs 6A and EV4A–D). WT IN5, IN5 lacking TD5 (IN5∆TD5), and IN5∆TD5+bNLS harboring the L622A mutation in TD1, all failed to interact with AC40, demonstrating that the interaction was strictly dependent on TD1.
Figure 6. IN1 targeting domain directs Ty5 integration at Pol III‐transcribed genes.
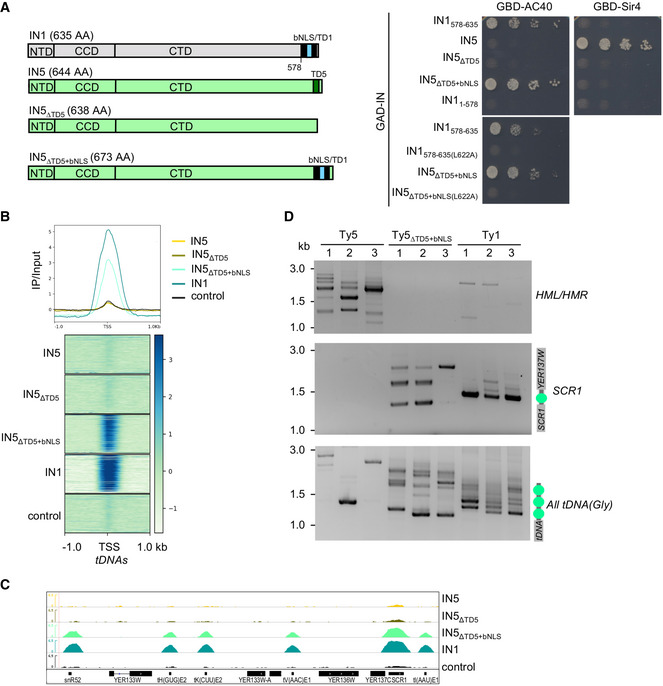
- Two‐hybrid interaction between GBD‐AC40 (left) or GBD‐Sir4 (right) and different GAD‐IN5 or GAD‐IN1 constructions. Cells were plated in fivefold serial dilutions on DO‐Leu‐Trp‐His plates. No growth or protein expression defects were detected (Fig EV4A–D).
- Genome‐wide occupancy profiles (top) and heatmaps (bottom) of IN5, WT, and indicated mutants, ± 1 kb upstream and downstream of all tDNAs.
- Genome browser visualization of different HA‐IN occupancy for chromosome V (chrV:431129..443275). Occupancy of the indicated integrase is represented in each panel. Control, as panel 3D. The region contains tH(GUG)E2, tK(CUU)E2, tV(ACC)E1, tI(AAU)E1, SNR52, and SCR1, all transcribed by Pol III.
- Detection of Ty5, Ty5ΔTD5+bNLS or Ty1 de novo integration events at HMR and HML loci, SCR1, and upstream of all glycine tDNAs, by PCR using a primer in HIS3 and a primer in the locus of interest. Retrotransposition was induced in cells transformed with WT or mutated pGAL1‐Ty5his3AI (Ty5 or Ty5ΔTD5+bNLS) and pGAL1‐Ty1his3AI (Ty1). Total genomic DNA was extracted from His+ cells obtained from independent cultures. Nucleosome position (green circles) is indicated on the right of the panels (Brogaard et al, 2012).
Source data are available online for this figure.
Figure EV4. IN1 targeting domain directs Ty5 integration at Pol III‐transcribed genes.
-
A–D(A and C) Growth control of cell cultures corresponding to the two‐hybrid assay shown in Fig 5A. Ten‐fold serial dilutions of cell cultures, starting from 10−1, were plated on DO‐Leu‐Trp plates to check for growth (B and D). Validation of the expression of GAD‐IN1 and GAD‐IN5 fusion proteins for the two‐hybrid assay shown in Fig 5A. Whole‐cell extract samples were prepared for immunoblotting as for Fig EV1A. Expected sizes of the proteins are 25 kDa for GAD‐IN1578–635 and GAD‐IN1578–635 mutants and 82 kDa for GAD‐IN11–578, around 100 kDa for GAD‐IN5 constructions.
-
EGenome browser visualization of HA‐IN1 or HA‐IN5 occupancy at the RDN1 locus encoding ribosomal RNA genes. Control is an anti‐HA immunoprecipitation of chromatin extracts in cells expressing IN1‐Strep. Values obtained from ChIP‐seq analysis have been normalized for each condition (WT and mutant IN5) to input and adjusted in log2 RPKM.
-
FPearson correlation heatmap at tDNAs between the following ChIP‐seq: IN1, IN5, IN5∆TD5, IN5∆TD5+bNLS, and control.
-
GRetrotransposition frequency, shown on a log scale of pGAL1‐Ty5his3AI and pGAL1‐Ty5his3AI (IN5∆TD5+bNLS) in an spt3‐101 (LV47) or rad52∆ strain (LV172). Values are mean ± SD, n = 2 experiments, each performed with four independent colonies.
Source data are available online for this figure.
To establish whether the interaction with AC40 is sufficient to target IN5∆TD5+bNLS to Pol III‐transcribed genes, we performed ChIP‐seq in strains ectopically expressing HA‐tagged IN5, IN5∆TD5, and IN5∆TD5+bNLS. Metagene analysis of the tagged proteins binding to tDNAs revealed a clear enrichment of IN5∆TD5+bNLS at these loci, not detected for IN5 and IN5∆TD5 (Fig 6B). IN5∆TD5+bNLS was also present at the other Pol III‐transcribed and Pol I‐transcribed genes (Figs 6C and EV4E). Overall, IN5∆TD5+bNLS genome occupancy profile was very similar to that of IN1 (Pearson correlation of R = 0.9 Fig EV4F and Table EV3), confirming that the TD1‐AC40 interaction is sufficient for recruitment at Pol III‐transcribed genes. We did not detect IN5 enrichment at subtelomeric regions bound by Sir4 (Zill et al, 2010), nor at HML and HMR, which are Ty5 integration sites, which may be due to the weak association between Ty5 integration sites and Sir4 occupancy (Baller et al, 2011) and may reflect a loose and dynamic interaction between IN5 and Sir4 proteins difficult to detect by ChIP.
To explore the impact of the TD1‐TD5 exchange on Ty5 integration site selectivity, we introduced IN5∆TD5+bNLS into a functional Ty5his3AI reporter expressed from the GAL1 promoter. This replacement caused a 10‐fold decrease in the frequency of Ty5 retrotransposition but did not inactivate the element. His+ colonies represented bona fide integration events, as a similar colony frequency was observed in the absence of homologous recombination (Fig EV4G). His+ selected de novo insertion events were generated for WT and mutant Ty5his3AI, and the insertion profiles were monitored at specific loci by qualitative PCR (Fig 6D). We used an spt3‐101 null strain that does not express endogenous Ty1 elements, to avoid interference between insertions of endogenous Ty1 and the mutant Ty5 at Pol III‐transcribed genes. The Ty1his3AI profile displayed multiple insertion events at the Pol III‐transcribed SCR1 gene and glycine tDNAs, whereas no insertion was seen at HML and HMR. The WT Ty5 profile displayed multiple insertion events at HML and HMR, whereas no insertions were recovered at SCR1 and very few were seen at the glycine tDNAs. In contrast, Ty5∆TD5+bNLS did not integrate at HML and HMR, whereas multiple insertion events were detected at the Pol III‐reporter genes. The position of nucleosomes over the SCR1 and glycine tDNAs suggests that the difference in Ty1 and Ty5∆TD5+bNLS banding patterns at Pol III‐transcribed genes might be due to Ty1, but not Ty5, preferentially integrating into nucleosomes. Thus, the Ty1 preference for nucleosomes might not depend on interaction with AC40.
Together, these data indicate that TD1 is sufficient to direct the integration of the Ty5 retrotransposon upstream of Pol III‐transcribed genes.
Discussion
Here, we identify the Ty1 targeting domain of IN1 (TD1) and show that it plays a critical role in the interaction with the Ty1 tethering factor AC40. We demonstrate that the interaction with AC40 orchestrates the recruitment of IN1 over the genome and that TD1 can function as an independent module that targets the integration of another related retrotransposon upstream of the typical Ty1 Pol III‐transcribed target genes.
We provide several lines of evidence supporting a major role for AC40 in recruiting Ty1 to both Pol III and Pol I‐transcribed genes. First, IN1 interacts directly with AC40 in the absence of other yeast proteins. Second, mutations in TD1 that reduce the interaction with AC40 abolish IN1 association with both Pol I and Pol III transcription complexes. Third, IN1 is present at both Pol I‐ and Pol III‐transcribed genes and this presence depends on the IN1‐AC40 interaction. Fourth, Ty5 integrase harboring TD1 interacts with AC40 and is recruited to Pol I‐ and Pol III‐transcribed genes. Direct interactions were previously observed in vitro between IN1 and the C31, C34, and C53 Pol III‐specific subunits but their precise role in the recruitment of the IN1 complex in vivo was not determined (Cheung et al, 2016). Here, we show that in vivo, the interaction of IN1 with C53 and C34 within Pol III depends on the IN1‐AC40 interaction. The redistribution of Ty1 integration into subtelomeres, seen in the absence of the IN1‐AC40 interaction ((Bridier‐Nahmias et al, 2015) and this study), was not observed in a rpc53∆2‐280 mutant, which decreases Ty1 integration at the SUF16 tRNA gene (Cheung et al, 2016). This suggests that either C53 secures IN1 binding to Pol III once IN1 has been recruited by AC40 or helps Ty1 integration at a step downstream of IN1 recruitment. Further studies will be necessary to address the role of the Pol III complex, and especially of C31, C34, and C53, in Ty1 integration.
Upon maturation of the Gag‐Pol protein in the VLPs, IN1 is associated with Ty1 cDNA. Under our ChIP experimental conditions, HA‐IN1 was ectopically expressed and was therefore probably absent of the VLPs. Moreover, HA‐IN1 was probably not associated with Ty1 cDNA, since cells were grown at 30°C, a temperature that restricts Ty1 replication resulting in a reduced level of Ty1 cDNA (Lawler et al, 2002; Garfinkel et al, 2005). Therefore, our results suggest that the cDNA might not be necessary for IN1 recruitment at Pol III‐transcribed genes. A recent study reached a similar conclusion for the recruitment of Ty3 integrase at tRNA genes (Patterson et al, 2019).
Previous genome‐wide mapping of Ty1 insertion sites did not reveal a clear pattern of Ty1 insertion into Pol I‐transcribed genes at the RDN1 locus (Baller et al, 2012; Mularoni et al, 2012; Bridier‐Nahmias et al, 2015), likely due to the highly repetitive nature of the RDN1 repeats (Bridier‐Nahmias et al, 2015). Here, we show that Ty1 integration in the RDN1 repeats occurs mostly at Pol III‐transcribed genes and at nearly random levels in Pol I‐transcribed genes. A similar pattern has already been described but was based on a small number of Ty1 insertions and was shown not to be the consequence of mitotic instability of Ty1 insertions in RDN1 (Bryk et al, 1997). Importantly, the recruitment of IN1 by AC40 appears to be insufficient for Ty1 integration into Pol I‐transcribed genes. Consistently, the loss of IN1‐AC40 interaction did not further decrease the number of insertion events. These observations suggest that Ty1 integration into Pol I‐transcribed genes may require additional co‐factors or a particular chromatin structure, like the well‐positioned nucleosomes present upstream of Pol III‐transcribed genes.
The IN1 mutants that disrupt the interaction with AC40 induce the same redistribution of Ty1 insertions at chromosome ends as seen in a AC40sp loss‐of‐interaction mutant (Bridier‐Nahmias et al, 2015). The insertion sites of these Ty1 mutants are scattered throughout subtelomeric regions. Ty5 insertions also occur throughout subtelomeric regions (Baller et al, 2011). This scattered dispersion may explain why we have failed to detect IN1 and IN5 by ChIP‐seq in these regions. High‐resolution mapping of DNA binding sites will be required to address this point (Hafner et al, 2018; Meers et al, 2019).
To date, the specific retargeting of TE integration sites when the main targeting is compromised has only been observed for Ty1. When the interaction between HIV, MLV, and Ty5 INs and their primary tethering factors (LEDGF/p75, BET proteins, and Sir4, respectively) is altered, the integration of these retroelements at their usual targets decreases substantially and becomes random (Gai & Voytas, 1998; Schrijvers et al, 2012; Wang et al, 2012; De Rijck et al, 2013; Sharma et al, 2013). Chromosome ends are preferential targets of several TE families in different organisms (Casacuberta, 2017). In S. cerevisiae, subtelomeres are devoid of essential genes, are rich in stress‐responsive genes, and evolve rapidly in response to stress (Snoek et al, 2014) or domestication (Yue et al, 2017). Targeting of Ty1 integration may have evolved to provide a balance between integration into “safe” genomic regions, i.e., tDNAs, and integration into fast‐evolving regions when adaptation is necessary, i.e., subtelomeres. Accordingly, we propose that the IN1‐AC40 interaction may be regulated by environmental stress. The observations that Ty5 targeted integration requires phosphorylation of the IN5 targeting domain, which is reduced by stress (Dai et al, 2007), and nutrient starvation regulates the Ty1 replication cycle (Morillon et al, 2000; Todeschini et al, 2005)) both lend support this hypothesis.
The pattern of integration of the Ty1 IN1 mutants largely overlaps in subtelomeric domains (Yue et al, 2017; Hocher et al, 2018). This correlation suggests that specific feature(s) in subtelomeres attract or facilitate Ty1 integration in the absence of the IN1‐AC40 interaction. As we failed to detect an interaction between IN1 and Sir4, Ty1 integration into subtelomeres probably involves a mechanism different from that of Ty5. In a cis‐targeting model, IN1 is tethered to subtelomeres through a weak interaction with a subtelomeric specific co‐factor, such that the subtelomeric interaction will only be favored when binding to AC40 is compromised. The dispersed nature of Ty1 insertion sites suggests that the co‐factor could be distributed across subtelomeres, like a histone mark specifically enriched in these regions (Hocher et al, 2018). Alternatively, IN1 co‐factor could be present at a limited number of subtelomeric sites, and after recruitment, the Ty1 intasome would scan for a chromatin environment favorable for integration. Given Ty1 targets stable nucleosomes upstream of Pol III‐transcribed genes (Baller et al, 2012; Mularoni et al, 2012), nucleosome stability could also be a determinant for integration at subtelomeres. A similar two‐step targeting model has been proposed to explain the absence of correlation between Sir4 binding sites and Ty5 integration sites in subtelomeres (Baller et al, 2011). In a trans‐targeting model, the proximity of the subtelomeres with the nuclear pores (Zimmer & Fabre, 2011), through which the Ty1 intasome transits, would facilitate Ty1 integration in these regions, especially when the interaction with AC40 is compromised. Consistent with this hypothesis, mutations in several components of the nuclear pore alter Ty1 integration preferences (Manhas et al, 2018). HIV integration also occurs preferentially in chromatin proximal to the nuclear periphery (Di Primio et al, 2013; Lelek et al, 2015; Marini et al, 2015).
This work demonstrates that the IN1 targeting domain functions as an independent module to target integration at Pol III‐transcribed genes: The addition of this sequence to Ty5 IN is sufficient to direct Ty5 integration to these loci. Entry of the Ty1 intasome into the nucleus involves the classical import machinery and requires an interaction between importin‐α and the IN1 bNLS (McLane et al, 2008). The IN1 bNLS consists of two regions of basic amino acids, which are essential for IN1 nuclear import, separated by a linker sequence that also contributes, although to a lesser extent, to import (Kenna et al, 1998; Moore et al, 1998; Lange et al, 2011). The residues that are required for IN1 interaction with AC40 and Ty1 integration upstream of Pol III‐transcribed genes cluster in a short peptide, namely TD1, within the linker. The IN1 linker has been proposed to induce a conformation that facilitates interaction of the two basic amino acid‐rich regions with two NLS‐binding pockets present in importin‐α (Kosugi et al, 2009; Lange et al, 2011). The conformation of the linker when the basic amino acid‐rich regions are bound to importin‐α could expose TD1 recognized by AC40. Mutations in TD1 substantially reduce the interaction with AC40 but not Ty1 retrotransposition frequency or IN1 nuclear import (this study). Thus, IN1 bNLS and TD1 have independent functions in nuclear import and integration targeting, respectively.
Retroviral vectors have been used in gene therapy to correct various monogenic disorders. However, these vectors rely on the properties of HIV and MLV integrases, whose preferences for either transcribed genes (i.e., HIV) or promoters (i.e., MLV) make them potentially harmful for the genome (Anguela & High, 2019; Goswami et al, 2019). Chimeras between IN from retroviruses and retroelements and chromatin‐binding proteins have already been constructed to channel integration to new positions in the genome (Gao et al, 2008; Ferris et al, 2010). As Ty1 integration upstream of Pol III‐transcribed genes preserves gene integrity (Bolton & Boeke, 2003) and given that Pol III transcription and structure, including the presence of AC40 and nucleosome positioning at Pol III‐transcribed genes (Helbo et al, 2017), are conserved between yeast and humans, adapting Ty1 integration targeting to retroviral integrases could overcome the risk of insertional mutagenesis associated with current MLV and HIV retroviral‐based vectors, allowing the development of safer retrovirus‐based vectors for use in human gene transfer technologies.
Materials and Methods
Growth media, yeast strains, and plasmids construction
Saccharomyces cerevisiae strains used in this study were grown using standard methods and are listed in Table EV4. All plasmids and primers used in this study are reported in Tables EV5 and EV6. Plasmids were constructed using standard molecular biology procedures (details can be obtained on request). Mutations were introduced in plasmids with Q5® Site‐directed mutagenesis (NEB). All the constructs were validated by DNA sequencing (Eurofins Genomics).
Two‐hybrid assays
Two‐hybrid assays were performed in strain PJ69‐4A which contains the HIS3 reporter to detect positive interactions (James et al, 1996). Cultures of transformants were grown overnight at 30°C in synthetic complete medium lacking leucine and tryptophan (SC‐LEU‐TRP) to maintain plasmid selection. Serial dilutions of aliquots of 1 optical density at 600 nm (OD600), washed in 1 ml of H20, were plated on SC‐LEU‐TRP (growth control) or SC‐LEU‐TRP‐HIS (interaction), starting from 10−1. Plates were incubated 4 days at 20°C. Each two‐hybrid assay is a representative example of at least two biological replicates.
Transposition assays
To estimate the frequency of retrotransposition in strains transformed by pGAL1‐Ty1his3AI, pGAL1‐Ty5his3AI, or their derivatives, four independent transformants of each strain were grown to saturation 2 days at 30°C in liquid SC‐URA containing 2% raffinose. Each culture was diluted thousand‐fold in liquid SC‐URA containing 2% galactose and grown 5 days to saturation at 20°C, which is the optimal temperature for Ty1 retrotransposition. Aliquots of cultures were plated on YEPD (100 μl at 10−5) and SC‐HIS (Ty1: 100 μl at 10−2, Ty5: 1 ml). Plates were incubated for 3 days at 30°C and colonies counted to determine the fraction of [HIS+] prototroph. Retrotransposition frequencies were defined as the mean of at least four experiments, and each one performed with four independent transformants.
Direct fluorescence microscopy
BY4742 WT cells were transformed with pMET‐GFP2‐IN1 NLS constructs and pUN100‐Nup49‐mCherry. Cells were grown overnight in SC‐URA‐LEU medium at 30°C. Cells were then washed twice in H2O and resuspended in SC‐URA‐LEU‐MET to induce GFP2‐IN1 NLS expression for 6 h at 30°C. Live cell imaging was done using a widefield microscopy system featuring a Nikon Ti‐E body equipped with the Perfect Focus System and a 100 × oil immersion objective. We used an Andor Neo sCMOS camera, which features a large field of view of 276 × 233 μm at a pixel size of 108 nm. We acquired 3D z‐stacks consisting of 15 frames with z‐steps of 300 nm. We used a dual‐band filter set (eGFP, mRFP) and for each z position acquired two color channels consecutively with an exposure time of 300 ms. The complete imaging system including camera, piezo, LEDs (SpectraX) was controlled by the Andor IQ2 software.
PCR assays for detection of Ty1 and Ty5 integration events
After retrotransposition induction as described above, total genomic DNA was extracted from yeast cultures grown at 20°C for 5 days according to classical procedures (Barkova et al, 2018). Double‐strand DNA (dsDNA) concentration was determined using Qubit™ fluorometric quantification (Thermo Fisher Scientific). De novo Ty1 insertions upstream of tG(GCC)C (SUF16) were amplified with PCR primers O‐AL27 and O‐AB91 and at the SEO1 subtelomeric gene with PCR primers O‐AL27 and O‐AL10. De novo Ty5 insertions at the HML and HMR loci, the SCR1 gene, and all glycine tDNAs were amplified with O‐AL27 combined with O‐CG27, O‐AL64, or O‐AB115, respectively. PCR reaction consisted of 30 ng of dsDNA (or 75 ng for detection of insertions at SEO1), 5 μl Buffer 5×, 0.5 μl dNTP 10 mM, 0.625 μl of each primer at 20 μM, 0,25 μl of Phusion DNA Polymerase (Thermo Scientific) in a 25 μl final volume. Amplification was performed with the following cycling conditions in ProFlex™ PCR System (Life Technologies) cycler: 98°C 2 min, 30× [98°C 10 s, 60°C 30 s, 72°C 1 min], 72°C 5 min, and hold 4°C. PCR products were separated on a 1.5% agarose gel.
Co‐precipitation experiments
For experiments performed with proteins expressed in E. coli, BL21(DE3) Rosetta (Novagen) bacterial cells were transformed with pet28‐6xHis‐IN1‐C‐tag (C‐tag is a four amino acid peptide tag (Djender et al, 2014)) and pACYC‐AC40‐Twin‐Strep‐tag, and streaked on LB agar plates supplemented with ampicillin, chloramphenicol, and 1% glucose. Transformants were inoculated in the same medium overnight at 30°C, diluted to 0,05 OD600 in fresh 100 ml of LB medium with ampicillin and chloramphenicol without glucose and grown at 30°C to an OD600 = 0.5. Cells were transferred at 24°C for 30 min, and protein expression was induced by adding 0.5 mM isopropyl 1‐thio‐β‐d‐galactopyranoside (IPTG) and growing cells for 3 h at 24°C. Cells were harvested at 1,100 g for 15 min at room temperature, washed with water, and resuspended in 1 ml of ice‐cold extraction buffer (20 mM Tris–HCl pH 7.5, 300 mM NaCl, 10% glycerol, 0.1% NP40) before adding 1 μg/ml of lysozyme. The extract was kept 10 min on ice and then lysed by sonication (Q700 sonicator, Qsonica) on ice for 5 cycles (output amplitude 10%, 5 s ON, 40 s OFF). The lysate was centrifuged at 21,000 g at 4°C for 30 min, and the supernatant was collected on a separate tube. A 800 μl of protein extract was incubated with 40 μl of Magstrep XT beads (Iba‐lifescience) for 3 h at 4°C on a wheel and washed twice with 1 ml of extraction buffer. Bound proteins were eluted with 50 μl of 1× Laemmli sample buffer, separated by SDS–PAGE, and analyzed by Western blot using CaptureSelect™ Biotin Anti‐C‐tag conjugate (1/5,000 dilution, Thermo Fisher) and anti‐Strep‐Tactin‐HRP conjugate (1/5,000 dilution, Iba‐lifescience). Experiments were reproduced at least 3 times with independent cultures.
Co‐IP experiments in yeast cells between Pol III (C160‐HA) and IN1‐Strep were performed as followed. MW4415 or LV33 yeast strains expressing HA‐tagged or untagged C160 respectively were transformed with a 2 micron plasmid expressing WT or K617A, S621A, or L622A IN1 mutants from an inducible GAL1 promoter. Overnight cell cultures were grown to saturation at 30°C in SC‐URA to maintain plasmid selection, were diluted at OD600 = 0.001 in 100 ml of SC‐URA and containing 2% galactose, and grown at 30°C. At OD600 = 1, cultures were harvested by centrifugation at 4°C. Cell pellets were resuspended in 500 μl of IP extraction buffer (50 mM HEPES‐KOH pH 7.5, 300 mM NaCl, 1 mM EDTA, 0.05% NP40, 0.5 mM DTT, 5% glycerol) supplemented with protease inhibitors (Thermo Fisher), and lysed with 0.25 ml of glass beads using a vortex (Disruptor Genie®, VWR) for 30 min at 4°C. The whole protein extract (except an aliquot kept for the input control) was incubated with 50 μl of Dynabeads Pan Mouse IgG (Thermo Fisher) coated with anti‐HA antibody (12CA5, Roche) 1 h at 4°C on a wheel. Samples were washed three times with IP extraction buffer. Immunoprecipitated proteins were eluted from beads by boiling samples for 5 min at 95°C with 25 μl of 2× SDS sample buffer, and the entire eluted fraction was analyzed by Western blot. Total lanes correspond to 1/20 dilution of the total input engaged in the IP. Proteins were detected with thousand‐fold diluted primary antibodies (anti‐HA antibody (12CA5, Roche) and anti‐Strep (Qiagen)) and revealed with ECL (Thermo Fisher) and Fusion FX camera (Vilbert‐Lourmat). Experiments were reproduced twice with independent cultures.
Co‐IP experiments in yeast cells between Pol I (A190‐TAP) and ectopically expressed IN1‐HBH (HBH is an histidine biotin tag (Tagwerker et al, 2006)) were performed as described for Pol III but with some minor modifications. All the cultures (MGD353‐13D or MGD353‐13D A190‐TAP transformed by pCM185‐IN1‐HBH) were grown in SC‐TRP containing 2% glucose. Immunoprecipitation was performed with Dynabeads Pan Mouse IgG. Proteins were detected with primary antibodies anti‐TAP (1/5000 dilution, Invitrogen), and anti‐Streptavidin‐HRP (1/15,000 dilution, Pierce). Experiments were reproduced three times with independent cultures.
Co‐IP experiments in yeast cells between ectopically expressed IN1‐Strep and C34 or C53 were done as described for Pol III, except that IP was performed using Dynabeads Pan Mouse IgG (Thermo Fisher) coated with anti‐Strep (Qiagen). C34 and C53 proteins were detected with primary antibodies (Huet et al, 1985), at respectively 1/3,000 and 1/10,000 dilutions.
Chromatin immunoprecipitation and chromatin immunoprecipitation sequencing
Yeast strain LV1689 was transformed with pCM185 expressing 3xHA‐IN1‐C‐tag WT or harboring IN1 K617A, S621A, or L622A mutations. ChIP was performed as previously described (Harismendy et al, 2003), with minor modifications. Briefly, log‐phase cultures (50 ml) in SC‐TRP to maintain plasmid selection were cross‐linked with 1% formaldehyde for 5 min at room temperature. Cells were pelleted by centrifugation and lysed with glass beads using an orbital shaker (IKA; VRX basic Vibrax; 500 g, 40 min, 4°C). The cell lysate was drawn off the beads and then spun for 20 min at 24,000 g in a microcentrifuge at 4°C. The cell pellet was resuspended in sonication buffer (50 mM HEPES‐KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X‐100, 0.1% Na‐deoxycholate, 1 mM PMSF, O‐Complete protease inhibitor (Roche)), placed on a rotative wheel for 1 h at 4°C, and sonicated for 5 cycles (40 s ON at high level and 20 s OFF) in a Bioruptor (Diagenode; Denville, NJ, USA). After an additional 1‐h incubation at 4°C on a rotative wheel, the solubilized chromatin was recovered by centrifugation for 15 min at 7,700 g at 4°C. The preparation of magnetic beads, immunoprecipitation with anti‐HA 12CA5 antibodies, elution from beads, and reversal of cross‐linking were performed as described (Harismendy et al, 2003). Immunoprecipitated DNA was purified using a QIAquick PCR Purification Kit (Qiagen). The purified DNA samples were analyzed by quantitative real‐time PCR using the SYBR® Green PCR master Mix kit and an ABI PRISM 7500 (Applied Biosystems). The results were normalized with the input DNA PCR signals and indicated by relative IP in the graphs. For ChIP‐seq analysis, the immunopurified DNA from 3 to 4 independent experiments was combined after validation by quantitative real‐time PCR of SCR1 and GAL1 genes. DNA sequencing of 40‐nucleotide tags was performed on a GA‐IIx, Hi‐Seq, or Next‐Seq sequencer using the procedures recommended by the manufacturer (Illumina). Input DNA and DNA from ChIP of a strain expressing IN‐Strep were used as negative controls.
ChIP‐seq data analysis
Raw sequence reads quality was controlled with FasqtQC software tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were trimmed from the adapter sequence and matched to reference genome (SacCer3) using Bowtie2 (Langmead & Salzberg, 2012), with default parameters. Peak calling was performed with MACS2 (Zhang et al, 2008) by comparing ChIP to the corresponding input. Enrichment profile and heatmaps of normalized Reads per Kilo Millions of Mapped reads (RPKM) ratio of IP/input in Log2 were plotted with Deeptools (Ramírez et al, 2016). Bigwig files were generated for genome browser visualizations. All figures were prepared using R packages (http://www.R-project.org). Graphics were generated using ggplot2 (Wickham, 2016).
Construction of de novo Ty1 insertion libraries for high‐throughput sequencing
LV174 strain (spt3‐101 rad52∆) was transformed with pGTy1his3AI‐SCUF WT (Baller et al, 2012) or pGTy1his3AI‐SCUF harboring mutations in IN1 sequences (IN1 K617A, S621A, L622A). For each transformant, total genomic DNA was extracted from 70,000 to 100,000 His+ colonies recovered from seven to ten independent cultures grown at 20°C in the presence of galactose as described in (Barkova et al, 2018). Fasteris prepared and sequenced Illumina libraries.
Ty1 integration data analysis
Pipeline described in the ChIP‐seq Data Analysis section was used for quality check and trimming. To detect de novo Ty1 insertions, only reads containing the SCUF sequence and ending with Ty1 LTR sequence were considered. Clean reads were then matched to reference genome (SacCer3) using BWA short aligner (Li & Durbin, 2009) for paired‐end reads. PCR duplicates were removed with MarkDuplicates (Picard tools, http://broadinstitute.github.io/picard/), and ambiguous reads ending by Ty1 LTR were discarded. Only properly aligned paired reads (one start and end coordinates) were retained for further analysis (WT 64 002 paired reads; K617A, 48 845; S621A, 5 052; L622A, 11 654). Ty1 de novo integrations were assigned to the corresponding genomic features, and analyses were performed using in‐house R (http://www.R-project.org) pipelines. To compare the associations of integration profiles (both in vivo and in silico) with selected genomic features, we used a statistical approach in which the enrichment of each association with a specific feature was expressed as a z‐score, which reflects the number of standard deviations from the mean value of the compared features. With this approach, the differences in the total number of insertions recovered in each library do not impact the analysis. Z‐score is calculated as follow: z = (x‐μ)/((x; number of insertions (specific feature)/number of total insertions, μ; mean value (all compared features), (; standard deviation (all compared features). Graphics were generated using ggplot2 (Wickham, 2016).
Author contributions
AA‐L, AB, CC, CG, HF, IA, NP, and RM performed the experiments. AA‐L performed computational analysis. AA‐L, AB, CC, JA, and PL analyzed data and prepared figures AA‐L, AB, JA, and PL wrote the manuscript. JA and PL conceived and supervised the study and secured funding.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank A. Corbett and D. Voytas for plasmids; A. Bridier‐Nahmias and members of the laboratory for stimulating discussions; A. Leseur for contribution during her training; E. Fabre, C. Fernandez‐Tornero, G. Herrada, B. Palancade, J. Reguera for critical reading of the manuscript. This work was supported by intramural funding from Centre National de la Recherche Scientifique (CNRS), the Université Paris Diderot and the Institut National de la Santé et de la Recherche Médicale (INSERM), and from grants from the Fondation ARC pour la Recherche sur le Cancer (PJA 20151203412), the Agence Nationale de la Recherche through the generic call projects ANR‐13‐BSV3‐0012 and ANR‐17‐CE11‐0025. AAL was supported by a post‐doctoral fellowship from Fondation pour la Recherche Médicale (FRM‐SPF20170938755), AB by a post‐doctoral fellowship from the ANR through the initiatives d'excellence (Idex ANR‐11‐IDEX‐0005‐02) and the Labex “Who am I?” (ANR11‐LABX‐0071) and IA by the PhD program from the CEA. This work has benefited from the facilities and expertise of the high‐throughput sequencing platform of I2BC.
The EMBO Journal (2020) 39: e104337
Contributor Information
Joël Acker, Email: joel.acker@cea.fr.
Pascale Lesage, Email: pascale.lesage@inserm.fr.
Data availability
The datasets produced in this study are available in the following databases:
-
•
Chip-Seq data: Array Express E-MTAB-9038 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9038/);
-
•
Ty1 de novo insertion data: Sequence Read Archive PRJNA597319 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA597319/).
References
- Anguela XM, High KA (2019) Entering the modern era of gene therapy. Annu Rev Med 70: 273–288 [DOI] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD (2005) TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev 19: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baller JA, Gao J, Voytas DF (2011) Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5. Proc Natl Acad Sci USA 108: 20351–20356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF (2012) A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res 22: 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkova A, Asif‐Laidin A, Lesage P (2018) Genome‐wide mapping of yeast retrotransposon integration target sites In Methods in enzymology, Carpousis AJ. (ed), pp 197–223. Cambridge, MA; San Diego, CA; Oxford; London: Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Bhalla P, Vernekar DV, Gilquin B, Couté Y, Bhargava P (2019) Interactome of the yeast RNA polymerase III transcription machinery constitutes several chromatin modifiers and regulators of the genes transcribed by RNA polymerase II. Gene 702: 205–214 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Devine SE (1998) Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93: 1087–1089 [DOI] [PubMed] [Google Scholar]
- Bolton EC, Boeke JD (2003) Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res 13: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G (2009) Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev 19: 607–612 [DOI] [PubMed] [Google Scholar]
- Bridier‐Nahmias A, Tchalikian‐Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Saïb A, Werner M, Voytas D, Lesage P (2015) An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348: 585–588 [DOI] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J (2012) A map of nucleosome positions in yeast at base‐pair resolution. Nature 486: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255–269 [DOI] [PubMed] [Google Scholar]
- Carr M, Bensasson D, Bergman CM (2012) Evolutionary genomics of transposable elements in Saccharomyces cerevisiae . PLoS ONE 7: e50978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E (2017) Drosophila: retrotransposons making up telomeres. Viruses 9: 192–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z (2003) HIV‐1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278: 372–381 [DOI] [PubMed] [Google Scholar]
- Cheung S, Ma L, Chan PHW, Hu H‐L, Mayor T, Chen H‐T, Measday V (2016) Ty1‐Integrase interacts with RNA Polymerase III specific subcomplexes to promote insertion of Ty1 elements upstream of Pol III‐transcribed genes. J Biol Chem 291: 6396–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S, Manhas S, Measday V (2018) Retrotransposon targeting to RNA polymerase III‐transcribed genes. Mob DNA 9: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C (2016) Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby RL, Chang N, Feschotte C (2019) Host – transposon interactions: conflict, cooperation, and cooption. Genes Dev 33: 1098–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ (1991) Single‐step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA 88: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ (1992) Posttranslational control of Ty1 retrotransposition occurs at the level of protein posttranslational control of Tyl retrotransposition occurs at the level of protein processing. Mol Cell Biol 12: 2813–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Xie W, Brady TL, Gao J, Voytas DF (2007) Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell 27: 289–299 [DOI] [PubMed] [Google Scholar]
- Dakshinamurthy A, Nyswaner KM, Farabaugh PJ, Garfinkel DJ (2010) BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae . Genetics 185: 1193–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM (1993) Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae . Nucleic Acids Res 21: 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K et al (2013) The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep 5: 886–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine SE, Boeke JD (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 10: 620–633 [DOI] [PubMed] [Google Scholar]
- Di Primio C, Quercioli V, Allouch A, Gijsbers R, Christ F, Debyser Z, Arosio D, Cereseto A (2013) Single‐cell imaging of HIV‐1 provirus (SCIP). Proc Natl Acad Sci USA 110: 5636–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djender S, Beugnet A, Schneider A, de Marco A (2014) The biotechnological applications of recombinant single‐domain antibodies are optimized by the C‐terminal fusion to the EPEA sequence (C Tag). Antibodies 3: 182–191 [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A et al (2010) Lens epithelium‐derived growth factor fusion proteins redirect HIV‐1 DNA integration. Proc Natl Acad Sci U S A 107: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Osanai M, Matsumoto T, Kojima KK (2005) Telomere‐specific non‐LTR retrotransposons and telomere maintenance in the silkworm. Bombyx Mori Chromosom Res 13: 455–467 [DOI] [PubMed] [Google Scholar]
- Gai X, Voytas DF (1998) A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell 1: 1051–1055 [DOI] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF (2008) Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res 18: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Nyswaner KM, Stefanisko KM, Chang C, Moore SP (2005) Ty1 copy number dynamics in Saccharomyces. Genetics 169: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi‐Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J (2008) Genome‐wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22: 1934–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R, Subramanian G, Silayeva L, Newkirk I, Doctor D, Chawla K, Chattopadhyay S, Chandra D, Chilukuri N, Betapudi V (2019) Gene therapy leaves a vicious cycle. Front Oncol 9: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Levin HL (2010) High‐throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe . Genome Res 20: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner‐Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF (2013) Bromo‐ and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol 87: 12721–12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner L, Lezaja A, Zhang X, Lemmens L, Shyian M, Albert B, Follonier C, Nunes JM, Lopes M, Shore D et al (2018) Rif1 binding and control of chromosome‐internal DNA replication origins is limited by telomere sequestration. Cell Rep 23: 983–992 [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH (2016) Roles for retrotransposon insertions in human disease. Mob DNA 7: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Gendrel C‐G, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O (2003) Genome‐wide location of yeast RNA polymerase III transcription machinery. EMBO J 22: 4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbo AS, Lay FD, Jones PA, Liang G, Grønbæk K (2017) Nucleosome positioning and NDR structure at RNA polymerase III promoters. Sci Rep 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey A, Esnault C, Majumdar A, Chatterjee AG, Iben JR, McQueen PG, Yang AX, Mizuguchi T, Grewal SIS, Levin HL (2015) Single nucleotide specific targeting of the Tf1 retrotransposon promoted by the DNA‐binding protein Sap1 of Schizosaccharomyces pombe . Genetics 201: 905–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocher A, Ruault M, Kaferle P, Descrimes M, Garnier M, Morillon A, Taddei A (2018) Expanding heterochromatin reveals discrete subtelomeric domains delimited by chromatin landscape transitions. Genome Res 28: 1867–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CRL, Burns KH, Boeke JD (2012) Active transposition in genomes. Annu Rev Genet 46: 651–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J, Riva M, Sentenac A, Fromageot P (1985) Yeast RNA polymerase C and its subunits: specific antibodies as structural and functional probes. J Biol Chem 260: 15304–15310 [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T (2010) Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696 [DOI] [PubMed] [Google Scholar]
- Jacobs JZ, Rosado‐Lugo JD, Cranz‐Mileva S, Ciccaglione KM, Tournier V, Zaratiegui M (2015) Arrested replication forks guide retrotransposon integration. Science 349: 1549–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two‐hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna MA, Brachmann CB, Devine SE, Boeke JD (1998) Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo . Mol Cell Biol 18: 1115–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8: 464–478 [DOI] [PubMed] [Google Scholar]
- Kirchner J, Connolly CM, Sandmeyer SB (1995) Requirement of RNA polymerase III transcription factors for in vitro position‐specific integration of a retroviruslike element. Science 267: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Kling E, Spaller T, Schiefner J, Bönisch D, Winckler T (2018) Convergent evolution of integration site selection upstream of tRNA genes by yeast and amoeba retrotransposons. Nucleic Acids Res 46: 7250–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto‐Sato E, Tomita M, Yanagawa H (2009) Six classes of nuclear localization signals specific to different binding grooves of importinα. J Biol Chem 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Kumar Y, Bhargava P (2013) A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes. BMC Genom 14: 402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mclane LM, Mills RE, Devine SE, Corbett AH (2011) Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped‐read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler JF Jr, Haeusser DP, Dull A, Boeke JD, Keeney JB, Lawler JF, Irol JV (2002) Ty1 defect in proteolysis at high temperature. J Virol 76: 4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz‐Griffero F, Zimmer C et al (2015) Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6: 6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV (2011) Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM (2006) An essential role for LEDGF/p75 in HIV integration. Science 314: 461–464 [DOI] [PubMed] [Google Scholar]
- Manhas S, Ma L, Measday V (2018) The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration. Nucleic Acids Res 46: 3552–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini B, Kertesz‐Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F et al (2015) Nuclear architecture dictates HIV‐1 integration site selection. Nature 521: 227–231 [DOI] [PubMed] [Google Scholar]
- McLane LM, Pulliam KF, Devine SE, Corbett AH (2008) The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res 36: 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Bryson TD, Henikoff JG, Henikoff S (2019) Improved CUT&RUN chromatin profiling tools. Elife 8: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J (2008) Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high‐mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev 22: 1190–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Rinckel LA, Garfinkel DJ (1998) A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol 18: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K (2004) Genome‐wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol 24: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Springer M, Lesage P (2000) Activation of the Kss1 invasive‐filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae . Mol Cell Biol 20: 5766–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD (2012) Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res 22: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Oficjalska‐Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O (2006) General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A‐mediated dephosphorylation of Maf1. Mol Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- Pardue M‐LL, DeBaryshe PG (2011) Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci USA 108: 20317–20324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Shavarebi F, Magnan C, Chang I, Qi X, Baldi P, Bilanchone V, Sandmeyer SB (2019) Local features determine Ty3 targeting frequency at RNA polymerase III transcription start sites. Genome Res 29: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton EH, Crease TJ (2004) Evolution of the transposable element Pokey in the ribosomal DNA of species in the subgenus Daphnia (Crustacea: Cladocera). Mol Biol Evol 21: 1727–1739 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: a next generation web server for deep‐sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijvers R, Vets S, De Rijck J, Malani N, Bushman FD, Debyser Z, Gijsbers R (2012) HRP‐2 determines HIV‐1 integration site selection in LEDGF/p75 depleted cells. Retrovirology 9: 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS et al (2013) BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci USA 110: 12036–12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Burkett TJ, Garfinkel DJ (1994) Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol 14: 6540–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek T, Voordeckers K, Verstrepen KJ (2014) Chapter 3: subtelomeric regions promote evolutionary innovation of gene families in yeast In Subtelomeres, Louis EJ, Becker MM. (eds), pp 39–70. Berlin; Heidelberg: Springer‐Verlag; [Google Scholar]
- Spaller T, Kling E, Glöckner G, Hillmann F, Winckler T (2016) Convergent evolution of tRNA gene targeting preferences in compact genomes. Mob DNA 7: 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, Zamborlini A, Cristofari G, Lesage P (2017) Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet 18: 292–308 [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, Auer B, Huang L, Kaiser P (2006) HB tag modules for PCR‐based gene tagging and tandem affinity purification in Saccharomyces cerevisiae . Yeast 23: 623–632 [DOI] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, Van Oudenaarden A (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA 110: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini AL, Morillon A, Springer M, Lesage P (2005) Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae . Mol Cell Biol 25: 7459–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jurado KA, Wu X, Shun M‐C, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P et al (2012) HRP2 determines the efficiency and specificity of HIV‐1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75‐binding site integrase inhibitor. Nucleic Acids Res 40: 11518–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2 elegant graphics for data analysis. New York: Springer‐Verlag; [Google Scholar]
- Wilhelm M, Wilhelm F‐X (2005) Role of integrase in reverse transcription of the Saccharomyces cerevisiae retrotransposon Ty1. Eukaryot Cell 4: 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm FX, Wilhelm M, Gabriel A (2005) Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element. Cytogenet Genome Res 110: 269–287 [DOI] [PubMed] [Google Scholar]
- Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol 21: 6606–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Pérez‐González CE, Eickbush DG, Eickbush TH (2005) Competition between R1 and R2 transposable elements in the 28S rRNA genes of insects. Cytogenet Genome Res 110: 299–306 [DOI] [PubMed] [Google Scholar]
- Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Lagomarsino MC et al (2017) Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet 49: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W et al (2008) Model‐based analysis of ChIP‐Seq (MACS). Genome Biol 9: R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill OA, Scannell D, Teytelman L, Rine J (2010) Co‐evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol 8: e1000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Fabre E (2011) Principles of chromosomal organization: lessons from yeast. J Cell Biol 192: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Ke N, Kim JM, Voytas DF (1996) The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev 10: 634–645 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
The datasets produced in this study are available in the following databases:
-
•
Chip-Seq data: Array Express E-MTAB-9038 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9038/);
-
•
Ty1 de novo insertion data: Sequence Read Archive PRJNA597319 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA597319/).


