Understanding the introduction, spread, and establishment of SARS-CoV-2 within distinct human populations as well as the evolution of the pandemics is crucial to implement effective control strategies. In this work, we report that the initial virus strains introduced in Mexico came from Europe and the United States and that the virus was circulating locally in the country as early as mid-March. We also found evidence for early local transmission of strains with a H49Y mutation in the Spike protein, which could be further used as a molecular marker to follow viral spread within the country and the region.
KEYWORDS: SARS-CoV-2, pandemic, phylogenomics
ABSTRACT
The coronavirus disease 2019 (COVID-19) pandemic has affected most countries in the world. Studying the evolution and transmission patterns in different countries is crucial to enabling implementation of effective strategies for disease control and prevention. In this work, we present the full genome sequence for 17 SARS-CoV-2 isolates corresponding to the earliest sampled cases in Mexico. Global and local phylogenomics, coupled with mutational analysis, consistently revealed that these viral sequences are distributed within 2 known lineages, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage A/G, containing mostly sequences from North America, and lineage B/S, containing mainly sequences from Europe. Based on the exposure history of the cases and on the phylogenomic analysis, we characterized 14 independent introduction events. Additionally, three cases with no travel history were identified. We found evidence that two of these cases represented local transmission cases occurring in Mexico during mid-March 2020, denoting the earliest events described for the country. Within this local transmission cluster, we also identified an H49Y amino acid change in the Spike protein. This mutation represents a homoplasy occurring independently through time and space and may function as a molecular marker to follow any further spread of these viral variants throughout the country. Our results provide a general picture of the SARS-CoV-2 variants introduced at the beginning of the outbreak in Mexico, setting the foundation for future surveillance efforts.
IMPORTANCE Understanding the introduction, spread, and establishment of SARS-CoV-2 within distinct human populations as well as the evolution of the pandemics is crucial to implement effective control strategies. In this work, we report that the initial virus strains introduced in Mexico came from Europe and the United States and that the virus was circulating locally in the country as early as mid-March. We also found evidence for early local transmission of strains with a H49Y mutation in the Spike protein, which could be further used as a molecular marker to follow viral spread within the country and the region.
INTRODUCTION
Coronavirus disease 2019 (COVID-19), declared a pandemic by the WHO on 11 March 2020 (1), is caused by a novel betacoronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), detected in December of 2019 in the province of Wuhan in China (2). This is the third outbreak related to zoonotic betacoronaviruses known to have occurred in humans in the last 2 decades, after SARS (severe acute respiratory syndrome) in 2002 and MERS (Middle East respiratory syndrome) in 2012. After its emergence in China, SARS-CoV-2 was spread initially to other parts of the world by people with a travel history to China but gradually shifted to local transmissions (LTs) (3). Viral spread was first detected in Thailand, South Korea, and Japan, and by the second half of January, the first positive cases appeared in the United States and Europe (France, Italy, and Spain). The current SARS-CoV-2 genome analysis provided on the Nextstrain site (4) points out that viral transmission is now mainly community driven (5, 6).
In many countries, despite diagnostic efforts and initial control strategies, SARS-CoV-2 spread went undetected until a critical number of cases requiring hospitalization and intensive care was reached, alerting the authorities in charge. As of 4 May 2020, SARS-CoV-2 had infected more than 3,578,000 people and caused around 251,000 deaths worldwide (3). In Mexico, the first case of SARS-CoV-2 was detected on 27 February 2020, corresponding to a person who had travelled back to Mexico from Italy and who was in direct contact with a confirmed SARS-CoV-2 case. Soon after, additional cases were detected among travelers who had returned from the United States and Europe, with the number of cases increasing every day. By 4 May, there were over 23,400 confirmed cases and 2,150 deaths within the country, indicating local transmission (7). Understanding the introduction, spread, and establishment of SARS-CoV-2 within distinct human populations is crucial to enabling implementation of effective control strategies. In this work, we studied the early introduction dynamics of the first SARS-CoV-2 cases in Mexico. For this, we used a whole-genome (WG) sequencing and phylogenomic approach. We obtained 17 full viral genome sequences, including sequences from the first case detected and sampled within the country. Phylogenomic placement showed that these viruses belong to the A2/G and B/S lineages, two of the three circulating viral lineages reported so far. Our analysis also confirmed that there have been multiple independent introduction events (IEs) in Mexico from travelers abroad. We also found evidence for early local transmission of viral variants possessing the mutation H49Y in the Spike protein, which could be further used as a molecular marker to follow viral spread within the country.
RESULTS AND DISCUSSION
Multiple introduction events of SARS-CoV-2 variants from two different lineages.
A total of 17 full viral genome sequences were obtained from selected Mexican samples representing the earliest sampled cases detected in the country (Fig. 1). From the epidemiological data associated with the Mexican samples, 15 of the cases corresponded to introduction events from travelers returning from abroad that entered the country through Mexico City International Airport, with 5 of them then relocating to other places within the country using local transportation (either aerial or terrestrial). Two additional cases reported no travel history (Table 1). Global phylogenetic analysis (8) confirmed that 8 Mexican variants (samples 8, 17, 19, 24, 27, 28, 30, and 31) grouped within SARS-CoV-2 lineage B (also called lineage S, composed of sequences predominantly from the Americas). The remaining 9 Mexican variants (samples 2, 5, 6, 7, 13, 16, 22, 32, and 33) grouped within lineage A (also called lineage G, which includes sublineages A2 and A2a and is composed of sequences predominantly from Europe) (Fig. 2) (9). Lineage A has been defined as composed of those viruses that share two nucleotide (nt) substitutions (position 8782 in ORF1ab and position 28144 in ORF8), that are closest to the root of the tree, and that are most similar to the virus corresponding to reference sequence Wuhan/WH04/2020 (EPI_ISL_406801). Lineage B viruses share the nucleotide substitution C18060T and are most similar to viruses corresponding to reference sequence Wuhan-Hu-1 as an early representative (9). The letters G (for A) and S (for B) are equivalents assigned in the GSAID/Nexstrain genomic epidemiological report (8). Both the A2/G and B/S lineages are contemporary, as the estimated date of emergence for lineage B/S is 29 December 2019 (95% confidence interval [95% CI], 20 December 2019 to 3 January 2020) whereas that for lineage A2/G is 18 January 2020 (95% CI, 2 January 2020 to 19 January 2020) (8). The collection dates for the Mexican samples that fall within lineage B1 range from 4 March 2020 to 15 March 2020, while those for the Mexican samples that fall within lineage A2 range from 27 February 2020 to 15 March 2020, including the variant corresponding to the first reported case in Mexico (10) (sample 33). This suggests an initial cocirculation of both the A2/G and B/S lineages in Mexico (Fig. 2). Viruses belonging to the third lineage reported, lineage V, were not identified in this study.
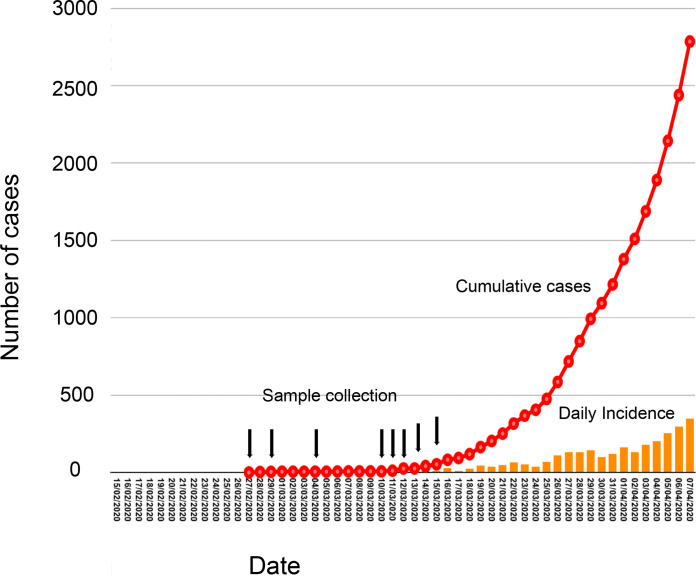
FIG 1.
Epidemiological positioning of the SARS-CoV-2 samples from Mexico. An epidemiological curve representing the early SARS-CoV-2 epidemic in Mexico, dating from late February until early April, is shown. The rise in cumulative cases is indicated in red, while the daily incidence is indicated with orange bars. Dates of collection for the samples used in this study are indicated with black arrows. Any one arrow might indicate the collection of more than two samples (dates shown in Table 1).
TABLE 1.
List of viral genomes derived from Mexican samplesa
| Sample ID | Virus name | GISAID accessionID | Location (country_city) | Age (yrs) | Sex | EH place | Collection date (day/mo/yr) | Date of arrival in Mexico | Port of entry |
|---|---|---|---|---|---|---|---|---|---|
| 2 | Mexico/CDMX-INER_01 | EPI_ISL_424345 | Mexico_CDMX | 42 | M | EH_Spain | 12/03/2020 | 12/03/2020 | Mexico City |
| 5 | Mexico/CDMX-INER_02 | EPI_ISL_424348 | Mexico_CDMX | 55 | F | EH_Spain_France | 13/03/2020 | 12/03/2020 | Mexico City |
| 6 | Mexico/CDMX-INER_03 | EPI_ISL_424625 | Mexico_CDMX | 29 | M | EH_Spain | 13/03/2020 | 11/03/2020 | Mexico City |
| 7 | Mexico/CDMX-INER_04 | EPI_ISL_424626 | Mexico_CDMX | 25 | F | EH_Egypt_Barcelona_Spain | 15/03/2020 | 15/03/2020 | Mexico City |
| 8 | Mexico/CDMX-INER_05 | EPI_ISL_424627 | Mexico_CDMX | 38 | M | EH_None | 15/03/2020 | NA | Mexico City |
| 13 | Mexico/Chihuahua-IMSS_01 | EPI_ISL_424731 | Mexico_Chihuahua | 21 | M | EH_UK_France_USA | 13/03/2020 | 12/03/2020 | Mexico City |
| 16 | Mexico/Chiapas-InDRE_02 | EPI_ISL_424666 | Mexico_Chiapas | 18 | F | EH_Italy | 29/02/2020 | 25/02/2020 | Mexico City |
| 17 | Mexico/EdoMex-InDRE_03 | EPI_ISL_424667 | Mexico_Edomex | 71 | M | EH_Italy | 04/03/2020 | 21/02/2020 | Mexico City |
| 19 | Mexico/Queretaro-InDRE_04 | EPI_ISL_424670 | Mexico_Queretaro | 43 | M | EH_Spain_Holand_USA | 10/03/2020 | 06/03/2020 | Mexico City |
| 22 | Mexico/Puebla-InDRE_05 | EPI_ISL_424672 | Mexico_Puebla | 31 | M | EH_Spain_France | 11/03/2020 | 09/03/2020 | Mexico City |
| 24 | Mexico/CDMX-InDRE_06 | EPI_ISL_424673 | Mexico_CDMX | 63 | F | EH_Denver_USA | 12/03/2020 | 06/03/2020 | Mexico City |
| 27 | Mexico/CDMX-INCMNSZ_01 | EPI_ISL_426361 | Mexico_CDMX | 70 | F | EH_Vail_USA | 12/03/2020 | 08/03/2020 | Mexico City |
| 28 | Mexico/CDMX-INCMNSZ_02 | EPI_ISL_426362 | Mexico_CDMX | 70 | M | EH_Vail_USA | 12/03/2020 | 08/03/2020 | Mexico City |
| 30 | Mexico/CDMX-INCMNSZ_03 | EPI_ISL_426363 | Mexico_CDMX | 59 | M | EH_Madrid_Spain | 08/03/2020 | 12/03/2020 | Mexico City |
| 31 | Mexico/CDMX-INCMNSZ_04 | EPI_ISL_426364 | Mexico_CDMX | 71 | M | EH_Vail_USA_Germany | 10/03/20 | 08/03/20 | Mexico City |
| 32 | Mexico/CDMX-INCMNSZ_05 | EPI_ISL_426365 | Mexico_CDMX | 32 | M | EH_None | 12/03/20 | NA | Mexico City |
| 33 | Mexico/CDMX-InDRE_01 | EPI_ISL_412972 | Mexico_CDMX | 35 | M | EH_Italy | 27/02/20 | NA | Mexico City |
ID, identifier; F, female; M, male; EH, exposure history; CDMX, Mexico City; Edomex, State of Mexico; NA, not available.
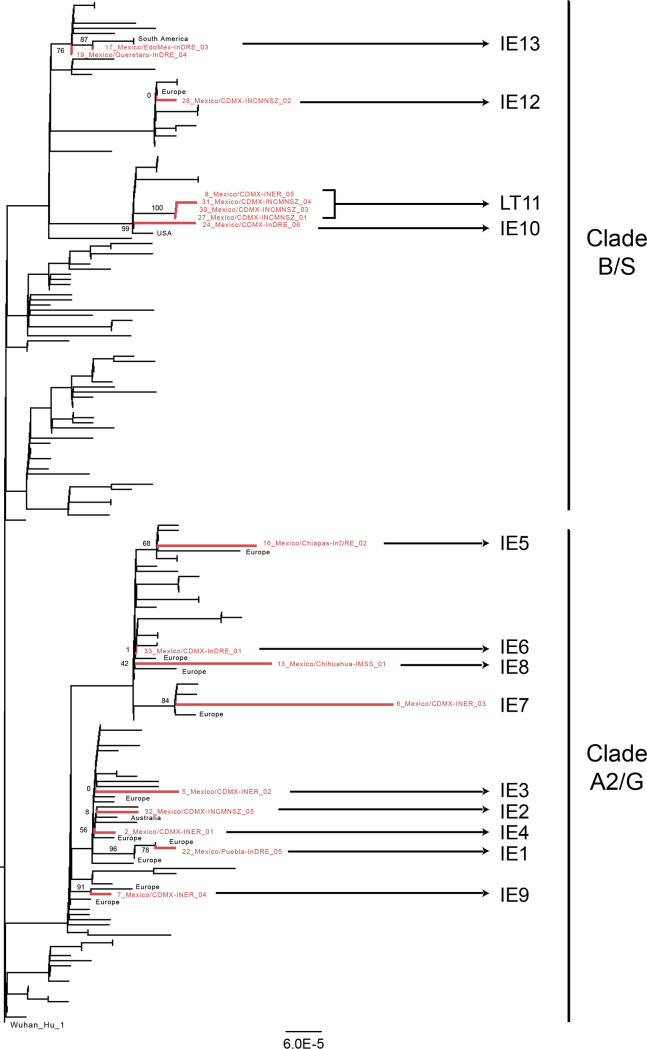
FIG 2.
Phylogenomic positioning of the SARS-CoV-2 variants in Mexico. A RAxML tree estimated from the reduced whole-genome alignment was built using the concatenated main viral ORFs and was rooted using the Wuhan-Hu-01 isolate. Sequences corresponding to the viral isolates collected in Mexico that were sequenced in this work are shown in red, while the region of origin for the identified closest related immediate ancestors and/or sister isolates is indicated in black. Support values for the branches of interest are indicated with numbers next to the branches. The clusters identified in this work representing introduction events (IE1 to IE10, IE12, and IE13) and the local transmission (LT11) are indicated next to the variant names.
Consensus clustering patterns observed within the large-scale and subsampled trees (see Table S2 in the supplemental material), showed eight well-supported independent introduction events (IE1, IE3, IE5, IE7, IE9, IE10, IE12, and IE13; Fig. 2). We were unable to determine the origins and the immediate phylogenetic relatedness of the Mexican sequences for IE2, IE4, IE6, and IE8, due to low support values and inconsistent clustering patterns across trees (Table S2). The current resolution of phylogenomic analyses is limited by the low diversity of the SARS-CoV-2 virus. Thus, for the characterized Mexican viral variants, we were able to determine the geographical origins with confidence only at the regional level and not at country-level resolution. Altogether, these observations suggest that the virus variants identified in this study were more closely related to viral sequences circulating at the time in the United States and Europe than to those circulating in China or South East Asia.
Evidence for early local transmission.
Sequences 27 and 31 correspond to two individuals that shared travel history to Vail, CO, USA, and that were in direct contact with case 28 in the return flight to Mexico. Nonetheless, the distribution of sequence 28 in an independent group (IE12) in relation to sequences 27 and 31 (Fig. 2; see also Table S2) suggests that there were at least two different viral variants cocirculating within that specific location in the United States. On the other hand, sequences 8, 27, 30, and 31 grouped together, representing a local transmission cluster (LT11) (Fig. 2). This observation is supported by the phylogenetic consistency within our local trees and the global tree (8) as well as by the high support value (bootstrap value, 100) observed for LT11 in all cases (Fig. 2; see also Table S2). Case 8 corresponds to an individual with no epidemiological relationship or contact with cases 27, 30, and 31 and with no travel history. Case 30 had a history of travel to Europe but not of direct exposure to cases 27 and 31. For case 30, the possibility of this person acquiring the virus while abroad cannot be ruled out. However, our analysis shows evidence that this person while have contracted the virus while in Mexico. Thus, The data from LT11 strongly support the idea that at least one independent local transmission event had occurred in Mexico City (case 8), as early as the second week of March 2020.
Genetic variation within the Mexican viral genomes.
Compared to the Wuhan-Hu-1 reference genome, the Mexican sequences displayed between 4 and 10 nucleotide substitutions and between 1 and 5 amino acid changes. This is consistent with the reported rate of evolution of ∼8 × 10−4 nucleotide substitutions per site per year, equivalent to ∼2 substitutions per month (11, 12). Collectively, 46 nucleotide substitutions and 20 amino acid residues were identified within the Mexican viral genomes (Table S4 and S5). As expected, the majority of these variants were not conserved through the genomes, and only 15 nucleotide changes and 6 amino acid changes were shared by two or more sequences. These results exclude sequences 6, 13, and 16, which showed considerably lower coverage and depth than the sequences of the remaining 14 high-quality viral genomes obtained. Thus, most of the variability observed could be explained by errors introduced during reverse transcription, PCR amplification, sequencing, or assembly (Table S1 and S4).
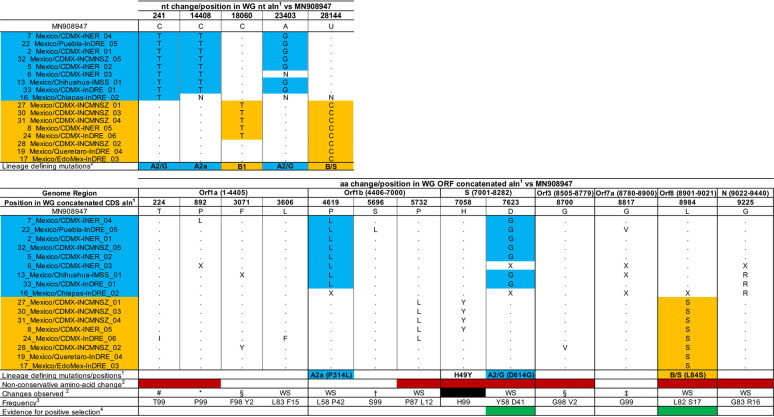
Consistent with our phylogenomic analysis, all Mexican sequences belonging to lineage A2 showed two lineage-specific nucleotide substitutions, C241T and A23403G. A23403G results in a D614G amino acid change in the Spike protein (Table 2). All lineage A2a sequences (including the Mexican isolates) had the additional nucleotide substitution C14408T, resulting in amino acid change P314L in Orf1b (8). Sequences within lineage B had nucleotide substitution T28144C, rendering the L84S amino acid change in the Orf8 protein. Similarly, lineage B1 sequences also showed the C18060T nucleotide substitution (Table 2). No evidence for recombination was found within any of the alignments, in agreement with previous observations (13). Taken together, our results suggest that the Mexican viral sequences display genetic changes corresponding to their phylogenetic placement.
TABLE 2.
Nucleotide and amino acid changes in the Mexican samples compared to the reference strain Wuhan-Hu-1
General lineage-defining mutations are shown in blue and orange, as defined in https://nextstrain.org/ncov/global and in GISAID preliminary analysis summary update 2020-04-07 1500UTC. No changes were observed in the M coding DNA sequence (CDS) (position 8283 to position 8504).
Nonconservative amino acid changes are shown in red. Clusters defining homoplasmic mutation of the Mexican sequences are shown in black. Widespread mutations are indicated under “WS.” Unique changes to the Mexican sequences representing singletons are excluded (extended information is available in Table S5).
Shared with USA/WA_UW153/2020|EPI_ISL_416691|2020_03_13.
Shared with England/SHEF_C0707/2020|EPI_ISL_420270|2020_03_22.
Shared with 45 sequences, mostly from Australia and Spain.
Shared with 5 sequences from Australia, Portugal, Spain, and the United States.
Shared with Portugal/PT0024/2020|EPI_ISL_418009|2020_03_15.
Shared with USA/WA_UW304/2020|EPI_ISL_418872|2020_03_23.
Percent frequency of a given amino acid (aa) observed in the alignment. Only changes present in >1% of the sequences in the alignment are shown.
Sites with a dN/dS value of >1 are shown in green and are compared to the list of sites determined under diverse dN/dS models described elsewhere (http://covid19.datamonkey.org/2020/04/01/covid19-analysis/).
According to the natural selection analyses performed for SARS-CoV-2 and enabled by data from GISAID (Global Initiative on Sharing All Influenza Data) (14–16), most of the variable sites detected in our analyses are likely to be evolving under conditions of negative or neutral evolution, as expected for RNA viruses (17). However, two of these sites (site 614 in the Spike protein and site 84 in Orf8) have been predicted to be evolving under conditions of positive selection (Table 2; see also Table S5). Site 614 in the Spike has been scored as evolving under conditions of pervasive positive selection and belongs to a predicted cytotoxic T lymphocyte (CTL) linear epitope that may be recognized by one or more HLA alleles (14–16). Mutation D/G at this site has been speculated to be involved in increased spread of the virus (16). Similarly, site 84 in Ofrf8 is also evolving under conditions of pervasive positive selection, may show intrahost variation, and, again, belongs to a predicted CTL linear epitope (14–16). Nonetheless, observations on the functional properties of these mutations are still debatable, and the data available to date are as yet inconclusive concerning any changes in biological and/or evolutionary properties of the virus.
H49Y amino acid change in Mexican sequences within LT11.
We further identified within all Mexican sequences grouping with the local transmission cluster (LT11) the C21707T nucleotide substitution (following the whole-genome and nucleotide alignment numbering), which corresponds to an H-to-Y amino acid change in position 49 of the Spike protein. Mapping this nucleotide substitution onto the branches of the global virus phylogeny (8) (with dates ranging from December 2019 to April 2020) revealed that C21707T had occurred with a frequency of 0.4% (20/4533) within the viral genomes available as of 6 May 2020. It first occurred within a single cluster of 14 sequences from China (representative sequence, Jiangsu/JS02/2020 EPI ISL 411952). The estimated date of emergence for this cluster that eventually stopped circulating and had no more descendants was 12 January 2020 (95% CI, 8 January 2020 to 16 January 2020). This mutation emerged again in all the sequences within LT11 from Mexico (Table 1; see also Fig. 2). Since then, C21707T has also appeared independently as a singleton in different virus variants circulating worldwide.
All of the sequences showing this nucleotide substitution within the global phylogeny belong to different viral lineages (A2 or B1 or others), confirming that there is no phylogenetic correlation between them (e.g., no founder effect) and instead supporting the idea of the independent occurrence of this change as a homoplasy. Despite C21707T appearing several times as a singleton (represented by tips on a tree), it has been fixed in only two lineages comprising independent viral subpopulations, occurring on internal branches or nodes of the tree in the China and Mexico sequences. Both global and local phylogenetic analyses show that the viral sequences from abroad that are most closely related to LT11 do not have this nucleotide substitution. Thus, at least for LT11, this change likely originated when this viral variant was initially introduced in the country. We do not know if any variants derived from LT11 continued circulating in the country; therefore, it would be interesting to use genomic surveillance to follow up on the frequency of occurrence of C21707T in viruses currently circulating in Mexico, either to determine if these represent sequencing errors (18) or to learn whether it represents a real although low-frequency homoplasy.
The H49Y mutation resulting from the C21707T nucleotide substitution represents a nonconservative amino acid change located within the N-terminal domain (NTD) of the S glycoprotein trimer, a protein region that has not been fully studied so far (19). No evidence of episodic or directional positive selection was found for that site, as tested under local and global analyses to estimate dN/dS ratios (14, 15). It would be relevant to explore if this change is associated with any biological properties of the virus, as has been shown previously for other virus populations (20). However, further structural biology analysis and experimental data would be needed to determine if this site has any functional impact or to determine its implications, if any, in local transmission dynamics.
MATERIALS AND METHODS
Ethics statement.
All clinical samples were processed at the Instituto de Diagnóstico y Referencia Epidemiológicos (InDRE), following official procedures (21). All samples used for this work are considered part of the national response to COVID-19, and the data collected are directly related to disease control.
Sample collection and diagnostics.
All samples used in this study were collected under Mexican Official Norm NOM-024-SSA2-1994 for prevention and control of acute respiratory infections in the primary health realm, as part of the early diagnostics scheme for SARS-CoV-2 in public health laboratories and hospitals in Mexico City (Red Nacional de Laboratorios Estatales de Salud Pública, RNLSP; Instituto Nacional de Enfermedades Respiratorias, INER; Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, INCMNSZ; and Instituto Mexicano del Seguro Social, IMSS). Oro- and nasopharyngeal swabs were collected and placed in virus transport medium upon collection, following InDRE official procedures (22). A tracheal aspirate was also obtained from one patient and was frozen at –70°C until use. Diagnosis was done using validated protocols for SARS-CoV 2, as approved by InDRE and by the World Health Organization (WHO) (23).
Sample processing and whole-genome sequencing.
All samples were prepared for RNA extraction and amplified as described in previous publications (24, 25). Briefly, centrifuged and filtered supernatants were treated with Turbo DNase and RNase. Nucleic acids were then extracted using a PureLink viral RNA/DNA kit (Thermo Fisher), following the manufacturer’s instructions and using linear acrylamide (Ambion) as the RNA carrier. Total cDNA was synthesized using a SuperScript III reverse transcriptase system (Thermo Fisher) and primer A (5′-GTTTCCCAGTAGGTCTCN9-3′), which has a degenerated 9-mer sequence at the 3′ end. The second strand was generated by two rounds of synthesis with Sequenase 2.0, followed by 15 cycles of amplification using Phusion DNA polymerase and primer B (5′-GTTTCCCAGTAGGTCTC-3), which hybridizes with the 5′ end of primer B. Next, double-stranded DNA was purified using a DNA clean & concentrator kit (Zymo Research) and used as input material to generate whole-metagenome shotgun sequencing libraries, following the instructions provided for Nextera XT DNA library preparation kits (26) (Illumina). Finally, all samples were sequenced on an Illumina NextSeq 500 platform using a 150-cycle high-output kit (v2.5) to obtain paired-end reads of 75 bp. Sequencing yields are reported in Table S1 in the supplemental material.
Bioinformatic analysis. (i) Data quality control and processing.
Read quality control was carried out using FAST-QC (27) and the default parameters. Adapter sequences and low-quality bases were removed using Fastp v0.19 (28). Low-complexity reads, those with a length shorter than 40 bases, and duplicates were excluded using CD-HIT-DUP v.4.6.8 (29). Off-target reads were then filtered out using Bowtie2 v2.3.4.3 (30) with the default parameters against human genome version GRCh38.p13, and the SILVA database (31) as a reference to filter out human DNA and ribosomal sequences.
(ii) Viral genome assembly.
The reads obtained were used as the input to assemble viral genomes using the Wuhan-Hu-1 reference genome sequence (MN908947). For this, the reads obtained for each sample were mapped against the reference using Bowtie2 v2.3.4.3 (30). Aligned reads were then used for de novo assembly with SPAdes v3.14.0 (32). Consensus genome sequences were generated using the majority threshold criterion. Only sequences with a coverage level above 80% and a mean depth of ≥8× were considered for the analyses (Table S1).
Phylogenetic analyses.
(i) Data collation. From 4,698 complete SARS-CoV-2 genomes available in the GISAID (Global Initiative on Sharing All Influenza Data) platform on the morning of 7 April 2020, a total of 3,014 sequences genomes (>29,000 nucleotides [nt]; high coverage only) were downloaded to generate a local database. As the collection dates of the Mexican samples ranged from late February to March, we filtered sequences collected between 1 February 2020 and 31 March 2020 for our database (Table 1). Unique sequences were extracted from those sequences, and those that were identical were collapsed, leaving at total of 2,633 sequences. We then included the 17 consensus viral genomes determined in this study and the Wuhan-Hu-1 reference genome sequence, yielding a total of 2,651 sequences. We aligned the whole-genome nucleotide data set using MUSCLE v3.8 (33) with the default parameters and then used the getorfs script from the EMBOSS suite (34) to extract complete ORFs (open reading frames) above 300 nt in size (Orf1a, Orf1b, Spike, M, Orf3a, Orf7a, Orf8, and N), and those were then individually realigned as described previously (33). Finally, to exclude untranscribed regions (UTRs) and noncoding intergenic regions from the phylogenomic analyses, individual ORFs were concatenated to generate an additional 28,320-nt-long whole-genome (WG) alignment.
(ii) Data subsampling and tree inference. The individual and concatenated alignments were then reversed to nucleotides and used for estimating maximum likelihood (ML) trees using RAxML v8 (35) with the following parameters: -T 2 -f a -x 390 -m GTRGAMMA -p 580 -N 100. All trees were rooted on the Wuhan-Hu-1 reference genome sequence (36). Given that SARS-CoV-2 shows a low degree of genetic variation (37), lineage definition must be based on consensus branching patterns within different trees and on shared nucleotide substitution patterns (8), in addition to bootstrap support values. Based on these criteria, the position of the Mexican sequences was determined within the whole-genome (WG) and individual ORF1a, ORF1b, and S trees (Table S2) and was then confirmed on the global phylogeny available in Nextstrain (38).
To visualize details of the phylogenetic relatedness of the Mexican sequences, we then subsampled the previous large-scale WG alignment in a phylogenetically informed manner on the basis of the position of the Mexican variants within the large-scale trees and by selecting sequences using pairwise genetic distances (20, 39). Briefly, all Mexican sequences were retained together with their immediate ancestors and descendants, and 184 sequences were further selected based on the minor pairwise genetic distance in relation to the Mexican genomes by the use of a threshold value of 99.5% (Table S3). The subsampled WG alignment was scanned for recombinant sequences using the GARD algorithm (40) in the Datamonkey server (13, 41). A total of 201 sequences, including the Wuhan-Hu-1 reference genome, were used to reestimate subsampled trees, as described above.
Global phylogenetic analysis and shared nucleotide substitution patterns (8) were used to confirm the position of the Mexican sequences based on the consensus clustering patterns observed within the large-scale WG tree, the subsampled WG tree, and the individual large-scale ORF trees, using a bootstrap value of >50 for branch support, when possible (Table S2). In general, we observed consistency within the global tree and our large-scale and subsampled trees, as represented by a conserved general structure at an internal branch level. Finally, analysis of the phylogenetic relationship between the Mexican and other viral sequences to identify groups of introduction events (IEs) and local transmissions (LTs) was done based on the following local definition: each IE or LT must include (i) one or more Mexican sequences, (ii) a minimum of one of the most closely related sister sequences, and/or (iii) the immediate common ancestor (42, 43).
(iii) Mutation identification. Snippy software (44) was used to identify all mutations that were unique to the Mexican genomes compared to the reference genome sequence of isolate Wuhan-Hu-1. The large-scale WG alignment in nucleotides (including the UTR and intergenic regions) was used as the input for Table S4, while the large-scale WG concatenated ORF alignment was used as the input for Table S5. The frequency and distribution for nucleotide and amino acid changes were determined using a normalized sequence logo, implemented by JalView (45). Nonconservative amino acid changes were determined based on amino acid properties. Finally, the list of amino acid changes obtained was compared to the list of known sites scored as evolving under conditions of pervasive, episodic, or directional positive selection, as tested using several dN/dS models that had been fine tuned for SARS-CoV-2 data sets, in which the effects of an inflated dN/dS ratio value that is due to the occurrence of intraspecies/intrahost polymorphism that may not be attributable by positive selections can be mitigated by restricting the site-specific analyses to internal branches (14, 15).
Data availability.
The SARS-CoV-2 sequences generated from isolates collected in Mexico can be found in GISAID and in Nextstrain (38). The corresponding GISAID accession numbers are listed in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by grants Epidemiología Genómica de los Virus SARS-CoV-2 Circulantes en México from the National Council for Science and Technology (CONACyT) of Mexico and 057 from the Ministry of Education, Science, Technology and Innovation (SECTEI) of Mexico City to C.F.A. and by grants from the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX-LXI y Comisión de Igualdad de Género de la Legislatura LXII de la H. Cámara de Diputados de la República Mexicana) to S.A.-R. and C.B. M.E.-Z. is supported by a Leverhulme Trust ECR Fellowship (ECF-2019-542).
We thank the Unidad de Secuenciación Masiva y Bioinformática of the Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas (CONACyT no. 260481) for their support in sequencing services. We thank the Instituto de Biotecnologia-UNAM for granting access to its computer cluster and Jerome Verleyen for his computational support. We thank all the staff of the Technological Development and Molecular Research Unit, Virology Department, and Sample Control and Services Department at InDRE for their technical assistance. The findings and conclusions in this report are solely our responsibility and do not necessarily represent those of the institutions involved.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2020. Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinotti F, Di Domenico L, Ortega E, Mancastroppa M, Pullano G, Valdano E, Boelle P-Y, Poletto C, Colizza V. 2020. Lessons learnt from 288 COVID-19 international cases: importations over time, effect of interventions, underdetection of imported cases. medRxiv 10.1101/2020.02.24.20027326. [DOI]
- 4.Nextstrain. 2020. Genomic analysis of nCoV spread. Situation report 2020–01-23. https://nextstrain.org/narratives/ncov/sit-rep/2020-01-23.
- 5.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Liao X, Qian S, Yuan J, Wang F, Liu Y, Wang Z, Wang F-S, Liu L, Zhang Z. 2020. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis 26:1320–1323. doi: 10.3201/eid2606.200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Government of Mexico. 2020. Comunicado Tecnico Diario COVID-19 04/18/2020. https://www.gob.mx/cms/uploads/attachment/file/547271/Comunicado_Tecnico_Diario_COVID-19_2020.04.18.pdf.
- 8.Nextstrain. 2020. Genomic epidemiology of novel coronavirus - global subsampling. (Filtered to 8 April 2020) https://nextstrain.org/ncov/global?dmax=2020-04-08.
- 9.Rambaut A, Holmes EC, Hill V, O’Toole A, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv doi: 10.1101/2020.04.17.046086. [DOI] [PMC free article] [PubMed]
- 10.Garcés-Ayala F, Araiza-Rodríguez A, Mendieta-Condado E, Rodríguez-Maldonado AP, Wong-Arámbula C, Landa-Flores M, del Mazo-López JC, González-Villa M, Escobar-Escamilla N, Fragoso-Fonseca DE, Esteban-Valencia MC, Lloret-Sánchez L, Arellano-Suarez DS, Nuñez-García TE, Contreras-González NB, Cruz-Ortiz N, Ruiz-López A, Fierro-Valdez MA, Regalado-Santiago D, Martínez-Velázquez N, Mederos-Michel M, Vázquez-Pérez J, Martínez-Orozco JA, Becerril-Vargas E, Salas J, Hernández-Rivas L, López-Martínez I, Alomía-Zegarra JL, López-Gatell H, Barrera-Badillo G, Ramírez-González JE. 2020. Full genome sequence of the first SARS-CoV-2 detected in Mexico. Arch Virol 18. doi: 10.1007/s00705-020-04695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedford T, Greninger AL, Roychoudhury P, Starita LM, Famulare M, Huang ML, Nalla A, Pepper G, Reinhardt A, Xie H, Shrestha L, Nguyen TN, Adler A, Brandstetter E, Cho S, Giroux D, Han PD, Fay K, Frazar CD, Ilcisin M, Lacombe K, Lee J, Kiavand A, Richardson M, Sibley TR, Truong M, Wolf CR, Nickerson DA, Rieder MJ, Englund JA, Hadfield J, Hodcroft EB, Huddleston J, Moncla LH, Müller NF, Neher RA, Deng X, Gu W, Federman S, Chiu C, Duchin J, Gautom R, Melly G, Hiatt B, Dykema P, Lindquist S, Queen K, Tao Y, Uehara A, Tong S, et al. . 2020. Cryptic transmission of SARS-CoV-2 in Washington State. medRxiv 10.1101/2020.04.02.20051417. [DOI] [PMC free article] [PubMed]
- 12.Rambaut A, Duchene S, Duplessis L, Volz E. 2020. Phylodynamic analysis. 176 genomes. 6 Mar 2020. Virological http://virological.org/t/phylodynamic-analysis-176-genomes-6-mar-2020/356.
- 13.Robertson DL. 2020. nCoV’s relationship to bat coronaviruses & recombination signals (no snakes) - no evidence the 2019-nCoV lineage is recombinant. Virological https://virological.org/t/ncovs-relationship-to-bat-coronaviruses-recombination-signals-no-snakes-no-evidence-the-2019-ncov-lineage-is-recombinant/331.
- 14.Pond S. 2020. Natural selection analysis of SARS-CoV-2/COVID-19. Observable https://observablehq.com/@spond/natural-selection-analysis-of-sars-cov-2-covid-19.
- 15.Datamonkey.org. 2020. Analyses of SARS-CoV-2 genomic data 04/01/2020. http://covid19.datamonkey.org/2020/04/01/covid19-analysis.
- 16.Campbell KM, Steiner G, Wells DK, Ribas A, Kalbasi A. 2020. Prediction of SARS-CoV-2 epitopes across 9360 HLA class I alleles. bioRxiv 10.1101/2020.03.30.016931. [DOI]
- 17.Pond SLK, Murrell B, Poon A. 2012. Evolution of viral genomes: interplay between selection, recombination, and other forces. Methods Mol Biol 856:239–272. doi: 10.1007/978-1-61779-585-5_10. [DOI] [PubMed] [Google Scholar]
- 18.DeMaio N, Walker C, Borges R, Weilguny L, Slodkowicz G, Goldman N. 2020. Issues with SARS-CoV-2 sequencing data. Virological https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473.
- 19.Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. 2017. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escalera-Zamudio M, Golden M, Gutiérrez B, Thézé J, Keown JR, Carrique L, Bowden TA, Pybus OG. 2019. Parallel Evolution in the Emergence of Highly Pathogenic Avian Influenza A Viruses. bioRxiv 10.1101/370015. [DOI] [PMC free article] [PubMed]
- 21.Secretaría de Salud. 2020. Dirección de Vigilancia Epidemiológica. http://www.gob.mx/salud/acciones-y-programas/sistema-nacional-de-vigilancia-epidemiologica.
- 22.General Directorate of Epidemiology. 2020. Lineamiento estandarizado para la vigilancia epidemiológica y por laboratorio de la enfermedad respiratoria viral. Secretariat of Health, Government of Mexico, Mexico City, Mexico: https://www.gob.mx/cms/uploads/attachment/file/552972/Lineamiento_VE_y_Lab_Enf_Viral_20.05.20.pdf. [Google Scholar]
- 23.WHO. 2020. Diagnostic detection of 2019-nCoV by real-time RT-PCR protocol. https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2.
- 24.Taboada B, Espinoza MA, Aponte FE, Isa P, Arias-Ortiz MA, Monge-Martínez J, Rodríguez-Vázquez R, Díaz-Hernández F, Zárate-Vidal F, Wong-Chew RM, Firo-Reyes V, del Río-Almendárez CN, Gaitán-Meza J, Villaseñor-Sierra A, Martínez-Aguilar G, Salas-Mier MC, Noyola DE, Pérez-Gónzalez LF, López S, Santos-Preciado JI, Arias CF. 2014. Is there still room for novel viral pathogens in pediatric respiratory tract infections? PLoS One 9:e113570. doi: 10.1371/journal.pone.0113570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taboada B, Isa P, Gutiérrez AL, del Ángel RM, Ludert JE, Vázquez-Salvador N, Tapia-Palacios MA, Chávez P, Garrido E, Espinosa AC, Eguiarte LE, López S, Souza V, Arias CF. 2018. The geographic structure of viruses in the Cuatro Ciénegas Basin, a unique oasis in northern Mexico, reveals a highly diverse population on a small geographic scale. Appl Environ Microbiol 84:e00465-18. doi: 10.1128/AEM.00465-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illumina. 2019. Nextera XT DNA sample prep kit documentation. https://support.illumina.com/sequencing/sequencing_kits/nextera_xt_dna_kit/documentation.html.
- 27.Babraham Bioinformatics. 2019. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 28.Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev A, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 35.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan ML, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pond S. 2020. Genomic diversity and divergence of SARS-CoV-2/COVID-19. Observable https://observablehq.com/@spond/current-state-of-sars-cov-2-evolution.
- 38.Nextstrain. 2020. Genomic epidemiology of novel coronavirus - global subsampling. https://nextstrain.org/ncov/global.
- 39.Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. 2017. Defining HIV-1 transmission clusters based on sequence data. AIDS 31:1211–1222. doi: 10.1097/QAD.0000000000001470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost S. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 41.Datamonkey.org. 2020. Datamonkey adaptive evolution server. http://datamonkey.org/.
- 42.Shiino T, Okabe N, Yasui Y, Sunagawa T, Ujike M, Obuchi M, Kishida N, Xu H, Takashita E, Anraku A, Ito R, Doi T, Ejima M, Sugawara H, Horikawa H, Yamazaki S, Kato Y, Oguchi A, Fujita N, Odagiri T, Tashiro M, Watanabe H. 2010. Molecular evolutionary analysis of the influenza A(H1N1)pdm, May–September, 2009: temporal and spatial spreading profile of the viruses in Japan. PLoS One 5:e11057. doi: 10.1371/journal.pone.0011057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baillie GJ, Galiano M, Agapow PM, Myers R, Chiam R, Gall A, Palser AL, Watson SJ, Hedge J, Underwood A, Platt S, McLean E, Pebody RG, Rambaut A, Green J, Daniels R, Pybus OG, Kellam P, Zambon M. 2012. Evolutionary dynamics of local pandemic H1N1/2009 influenza virus lineages revealed by whole-genome analysis. J Virol 86:11–18. doi: 10.1128/JVI.05347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seemann T. 2020. Snippy software. GitHub https://github.com/tseemann/snippy.
- 45.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SARS-CoV-2 sequences generated from isolates collected in Mexico can be found in GISAID and in Nextstrain (38). The corresponding GISAID accession numbers are listed in Table 1.