Gene inactivation is considered to be an important driver of orthopoxvirus evolution. Whereas cowpox virus contains intact orthologs of genes present in each orthopoxvirus species, numerous genes are inactivated in all other members of the genus. Inactivation of additional genes can occur upon extensive passaging of orthopoxviruses in cell culture leading to attenuation in vivo, a strategy for making vaccines. Whether inactivation of multiple viral genes enhances replication in the host cells or has a neutral effect is unknown in most cases. Using an experimental evolution protocol involving serial passages of an attenuated vaccinia virus, rapid acquisition of inactivating frameshift mutations occurred. After only 10 passage rounds, the starting attenuated vaccinia virus was displaced by viruses with one fixed mutation and one or more additional mutations. The high frequency of multiple inactivating mutations during experimental evolution simulates their acquisition during normal evolution and extensive virus passaging to make vaccine strains.
KEYWORDS: experimental evolution, frameshift mutations, gene inactivation, poxvirus, vaccinia virus evolution, virus evolution
ABSTRACT
Unlike RNA viruses, most DNA viruses replicate their genomes with high-fidelity polymerases that rarely make base substitution errors. Nevertheless, experimental evolution studies have revealed rapid acquisition of adaptive mutations during serial passage of attenuated vaccinia virus (VACV). One way in which adaptation can occur is by an accordion mechanism in which the gene copy number increases followed by base substitutions and, finally, contraction of the gene copy number. Here, we show rapid acquisition of multiple adaptive mutations mediated by a gene-inactivating frameshift mechanism during passage of an attenuated VACV. Attenuation had been achieved by exchanging the VACV A8R intermediate transcription factor gene with the myxoma virus ortholog. A total of seven mutations in six different genes occurred in three parallel passages of the attenuated virus. The most frequent mutations were single-nucleotide insertions or deletions within runs of five to seven As or Ts, although a deletion of 11 nucleotides also occurred, leading to frameshifts and premature stop codons. During 10 passage rounds, the attenuated VACV was replaced by the mutant viruses. At the end of the experiment, virtually all remaining viruses had one fixed mutation and one or more additional mutations. Although nucleotide substitutions in the transcription apparatus accounted for two low-frequency mutations, frameshifts in genes encoding protein components of the mature virion, namely, A26L, G6R, and A14.5L, achieved 74% to 98% fixation. The adaptive role of the mutations was confirmed by making recombinant VACV with A26L or G6R or both deleted, which increased virus replication levels and decreased particle/PFU ratios.
IMPORTANCE Gene inactivation is considered to be an important driver of orthopoxvirus evolution. Whereas cowpox virus contains intact orthologs of genes present in each orthopoxvirus species, numerous genes are inactivated in all other members of the genus. Inactivation of additional genes can occur upon extensive passaging of orthopoxviruses in cell culture leading to attenuation in vivo, a strategy for making vaccines. Whether inactivation of multiple viral genes enhances replication in the host cells or has a neutral effect is unknown in most cases. Using an experimental evolution protocol involving serial passages of an attenuated vaccinia virus, rapid acquisition of inactivating frameshift mutations occurred. After only 10 passage rounds, the starting attenuated vaccinia virus was displaced by viruses with one fixed mutation and one or more additional mutations. The high frequency of multiple inactivating mutations during experimental evolution simulates their acquisition during normal evolution and extensive virus passaging to make vaccine strains.
INTRODUCTION
Vertebrate poxvirus DNA genomes generally contain between 150 and 300 genes, of which 90 are conserved in all members of this subfamily (1) and, with only one known exception (2), are essential for replication. The high numbers of remaining genes have diverse functions related to host interactions and virulence (3). Cowpox virus, a member of the orthopoxvirus (OPXV) genus, contains orthologs of genes present in every OPXV, whereas all other species of the genus have inactivating mutations in individual genes, suggesting their loss during evolution from a cowpox virus-like ancestor. Indeed, gene loss or inactivation has previously been suggested to be the major evolutionary process that drives the divergence of OPXV species (4). Although loss of genes may restrict the ability to infect diverse hosts, gene number does not directly correlate with pathogenicity for a specific species since variola virus, the causative agent of smallpox, has the lowest number of genes of any natural OPXV. Inactivation of additional genes commonly occurs upon passaging OPXVs in cell culture leading to alterations in host range in vitro and attenuation in vivo, a strategy employed in making vaccines. An example of such a vaccine is the severely host-restricted modified vaccinia virus Ankara (MVA); following more than 500 passages in chicken embryo fibroblasts, MVA has the smallest number of intact genes of any OPXV (5, 6). Whether loss or inactivation of viral genes enhances replication in the cell lines used for passaging or has a neutral effect is unknown in most cases. Great progress in DNA sequencing has allowed the study of experimental evolution of vaccinia virus (VACV) in real time. These studies are facilitated by starting with an attenuated virus and screening for adaptive mutations (7–12). Such investigations have led to an amplification-mutagenesis or accordion model in which a gene copy number first increases, providing some adaptation and effectively enhancing the mutation rate in the amplified gene, followed by base substitutions and, finally, contraction of the number of gene copies (7, 13).
Previously, we described a scheme in which adaptation occurred during passaging of an attenuated VACV strain (vA8myx) that had the A8R intermediate transcription factor gene replaced with the myxoma virus ortholog (12). The object of those experiments was to find adaptive mutations in the transcription apparatus that would compensate for the heterologous A8 protein. Replication of the hybrid vA8myx in monkey BS-C-1 cells was reduced by approximately 15-fold relative to virus expressing the VACV A8R gene. However, after serial passaging, large plaques were apparent and increased in number during successive steps though they did not completely displace the parent vA8myx by 10 rounds. At the tenth round, viruses forming large plaques in parallel passage experiments were clonally purified. Remarkably, the virus yields in BS-C-1 cells of the cloned viruses approached that of virus expressing homologous VACV A8R. Whole-genome sequencing of individual clones revealed five different point mutations in the two largest RNA polymerase subunits or point mutations that optimized the Kozak translation initiation sequence regulating the myxoma virus A8R gene. Their phenotypes were corroborated by inserting individual mutations into vA8myx by homologous recombination. The finding of mutations in the transcription apparatus met our expectations, but the absence of mutations in the intermediate transcription factors themselves was surprising to us. The present study was initiated in order to find additional mutations that enhance replication of vA8myx. Besides identifying another point mutation in the large subunit of RNA polymerase and a mutation enhancing the Kozak translation initiation sequence of the myxoma A8R gene, the dominant adaptive mutations arose by single nucleotide insertions or deletions, mainly in runs of identical nucleotides, leading to frameshifts that inactivated genes with no apparent relationship to transcription. A similar example of adaptation by gene inactivation was recently reported in an experimental evolution study in which deletion of the B1 kinase was complemented by inactivation of the B12 pseudokinase (14). Thus, gene inactivation, in addition to gene duplication and base substitution, has the potential of enhancing virus replication.
RESULTS
Frameshift mutations arise during experimental evolution.
The previous experimental evolution strategy in which BS-C-1 cells were infected with vA8myx followed by serial passaging (12) was adopted with two modifications: the vA8myx stock was subjected to plaque purification again prior to amplification in BS-C-1 cells, and the virus multiplicity for each round of infection was reduced to 0.01 PFU/cell in order to accelerate the fixation of adaptive mutations. The rationale was that a high-fitness genotype might rescue a low-fitness one when a cell is infected with two or more infectious particles, thereby precluding fixation of the beneficial mutation. Three parallel passage experiments starting with vA8myx were carried out, and some virus was saved at each round for analysis and DNA sequencing. Numerous large plaques were evident after just a few rounds, and they appeared to have almost entirely replaced the small plaques by the tenth round. Whole-genome sequencing of the virus populations of the three parallel passage experiments was carried out after rounds 2, 3, 4, 5, 8, and 10 followed in each case by an additional expansion in BS-C-1 cells at >1 PFU/cell to obtain sufficient DNA without further selection. In total, seven different mutations were found in six genes. One mutation optimized the Kozak translation initiation sequence of the A8myx gene in experiment A, and in experiment B a point mutation in J6R encoding Rpo147, the largest RNA polymerase subunit (Table 1), recapitulated results obtained in our previous study (12). The mutation in the Kozak sequence was the same as that found previously. However, the point mutation in J6R was G1145E, whereas the previous J6R mutations were M424I and A1212V. Neither of these mutations came close to fixation, and the two are not further analyzed here. Mutations in A26L, G6R, A14.5L, and F10L were also found in the present study (Table 1) but not in the previous one.
TABLE 1.
Evolutionary dynamics of mutations in the course of 10 virus passage roundsa
| Passage | % mutation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population A |
Population B |
Population C |
|||||||||
| A26 | G6 | Kozak | A26 | G6 | G6Δ11 | J6 | A26 | G6 | A14.5 | F10 | |
| P2 | 9.3 | 0 | 0 | 10.9 | 0 | 0 | 0 | 11.8 | 0 | 0.4 | 0 |
| P3 | 22.9 | 0.4 | 0 | 24.5 | 0 | 2.6 | 1.0 | 23.8 | 0 | 1.1 | 1.3 |
| P4 | 51.1 | 14.0 | 0 | 54.6 | 0.3 | 4.8 | 2.5 | 52.8 | 1.2 | 7.9 | 1.7 |
| P5 | 82.3 | 32.0 | 2.4 | 64.1 | 1.8 | 15.7 | 13.0 | 60.0 | 2.8 | 20.0 | 7.5 |
| P8 | 95.1 | 60.3 | 7.1 | 94.0 | 12.3 | 35.7 | 32.5 | 94.8 | 5.9 | 31.1 | 60.0 |
| P10 | 96.0 | 91.5 | 16.0 | 98.2 | 15.8 | 61.0 | 18.1 | 98.0 | 12.6 | 74.0 | 8.0 |
The percentage of each mutation is presented for each passage round number (P2 to P10). The mutations of A26, G6, A14.5, and F10 were single nucleotide deletions or insertions, except for G6Δ11, as shown in Fig. 1. The mutation of J6 (Rpo147) was G for E at position 1145, and the Kozak mutation was C for A at position −3 of the A8myx ORF.
In experiments A, B, and C, a frameshift mutation was detected in 9% to 12% of the copies of the A26L gene in round 2, increasing to more than 50% at round 4 and to 96% to 98% at round 10 (Table 1). In each case, there was a deletion of one A in the same run of seven As (Fig. 1). The identical location of the mutation as well as the kinetics suggested that the mutation might have occurred during the expansion of the starting virus clone and might have been present at low frequency at the start of serial passaging. The mutation caused a frameshift at amino acid 37 of the predicted 500-amino-acid open reading frame (ORF), following which were 22 heterologous amino acids and then a stop codon.
FIG 1.
Nucleotide deletions and insertions. Nucleotide and translated sequences of wild-type (WT) A26L, G6R, and A14.5L ORFs and mutated forms are shown. Homopolymer runs and the 11-nucleotide sequence present before the deletions are shown in red in the WT sequence. Frameshift amino acids in the mutated sequences are shown in blue. The sequence preceding the F10L ORF with a T run in the putative promoter is colored red.
Two mutations were found in the G6R ORF (Fig. 1). One was a deletion of a T in a run of seven Ts that occurred in all three passages. However, the rounds of detection differed and the prevalence at round 10 ranged from 12.6% to 91.5% (Table 1). The other G6R mutation was a deletion of 11 nonrepeating nucleotides between an ATC sequence and a TAG sequence (Fig. 1) that was found only in passage B. The latter mutation occurred earlier than the frameshift and reached a prevalence of 61% at round 10. Despite the proximity of the two types of mutations in G6R, no individual 150-nucleotide read had both mutations, suggesting that they occurred independently in different genomes. The single nucleotide deletion led to a frameshift after amino acid 67 of the predicted 165-amino-acid ORF, which was followed by 11 heterologous amino acids and then a stop codon, whereas the 11-nucleotide deletion in G6R resulted in a frameshift at amino acid 46 followed by one heterologous amino acid and a stop codon.
A deletion of one T in a run of five Ts occurred in the A14.5L gene in experiment C (Fig. 1), which reached 74% penetrance by round 10 (Table 1). The A14.5 protein was predicted previously to be only 53 amino acids in length (15), and the frameshift at amino acid 40 resulted in an alternative reading frame that was slightly longer than the original (Fig. 1). The mutations in A26L and G6R occurred in the longest homopolymeric runs in their ORFs. However, in the A14.5L ORF, there were two other homopolymeric runs of equal length closer to the N terminus, suggesting that the mutation in the distal one was selected. An insertion of one T into a stretch of five Ts occurred in the promoter region of F10R in population C (Fig. 1).
The A26L, G6R, A14.5L, and J6R mutations occurred within the ORFs and likely altered protein function, whereas the mutation within the Kozak sequence of A8myx and the promoter of F10L likely affected gene expression.
Construction and analysis of A26L and G6R deletion mutants.
In experiments A and B, the majority of genomes had mutations in both A26L and G6R, whereas in experiment C, most had mutations in A26L and A14.5L. Because interpretation of the data obtained by passaging the virus was complicated by multiple mutations, we engineered single vA8myx mutations by homologous recombination. We focused on A26L and G6R as mutated forms occurred in each of the separate passages and the locations and short lengths of the frameshifted ORFs led to the prediction that the proteins would be inactive. If adaptation of vA8myx was due to the absence of functional A26 and G6 proteins, then a deletion mutant should have a phenotype similar to those of the frameshift mutants. Single and double deletion mutants of vA8myx were constructed by replacing the entire A26L ORF with the DsRed gene and by replacing G6R, except for the 3′-terminal 33-bp sequence, with the enhanced green fluorescent protein (EGFP) gene, with regulation in each case under the control of a VACV promoter. The fluorescent proteins facilitated the isolation of the mutants as well as competition experiments, which are described below. After clonal purification, whole-genome sequencing of each of the recombinant viruses did not reveal mutations in any other genes. To prepare equivalent titer stocks of vA8myx and the newly created recombinant viruses, we used RK13 cells that expressed the two VACV intermediate transcription factors A8 and A23. This cell line, RK13-A8/A23, was previously shown to complement A8R and A23R deletion mutants of VACV (16), and we found that vA8myx, vA8myxΔA26, vA8myxΔG6, and vA8myxΔA26ΔG6 and the parental virus VTF7-3, which we refer to as the wild type (WT) because it has the VACV A8R gene, all replicated to similar titers (Fig. 2A).
FIG 2.
Plaque formation and replication of vA8myx and A26L and G6R deletion mutants. (A) Replication of viruses in RK13-A8/A23 cells. Cells were infected in duplicate with 0.01 PFU/cell of vA8myx (gray), vA8myxΔA26 (green), vA8myxΔG6 (light blue), vA8myxΔA26ΔG6 (red), or the WT virus (dark blue). At the indicated hours postinfection (pi), virus titers were determined by plaque assay on BS-C-1 cells. (B) Plaque sizes. Crystal violet-stained plates (upper panel) and plaque areas (lower panel) are shown. (C) Virus spread. BS-C-1 cells were infected in triplicate with 0.01 PFU/cell of viruses (data are color coded as described for panel A). At the indicated times, virus titers were determined by plaque assay. (D) Virus yields. BS-C-1 cells were infected in triplicate with 5 PFU/cell of viruses (data are color coded as described for panel A). At the indicated times, virus titers were determined by plaque assay.
Plated on BS-C-1 cells, the deletion mutants formed plaques 1.6-fold to 2.2-fold larger than those of vA8myx (P < 0.0001), though they were smaller than those of the WT virus (Fig. 2B). The sizes of vA8myxΔA26ΔG6 and vA8myxΔG6 plaques were not significantly different from each other but were larger than those of vA8myxΔA26 (P < 0.006). The increased plaque sizes of the deletion mutants might be explained by higher virus yield per cell or by more-efficient spread to neighboring cells or by a combination of the two. We infected BS-C-1 cells at a multiplicity of 0.01 PFU/cell to measure spread (Fig. 2C) and a multiplicity of 5 PFU/cell to determine virus yield under one-step conditions (Fig. 2D). At the low multiplicity of infection, the virus yields for vΔA26, vΔG6, and vΔA26ΔG6 were 5-fold, 8-fold, and 60-fold higher than that seen with vA8myx at 48 h. Although the differences narrowed at 72 h, the differences between the deletion mutants and vA8myx remained statistically significant (P < 0.0085). At the latter time point, the titer of the double mutant was significantly higher than that of either of the single mutants (P = 0.001) and not significantly lower than that of the WT (P = 0.126). In the high-multiplicity experiment, the increase seen with each mutant was significantly greater than that seen with vA8myx at all times, with P values of ≤0.003 at 24 h. At the latter time, the virus yields determined for vΔA26, vΔG6, and vΔA26ΔG6 were 13-fold, 16-fold, and 40-fold higher than that determined for vA8myx. The titer of vA8myxΔA26ΔG6 was significantly higher than that of vA8myxΔA26 or vA8myxΔG6 (P ≤ 0.006) and was not significantly lower than that of the WT (P = 0.59). Taken together, the data indicated that deletions of A26L and G6R individually enhanced replication compared to the parental attenuated VACV and that replication of the double deletion mutant was higher and approached that of the WT.
Mutations in the predicted catalytic site of G6 confer the same phenotype as a deletion.
The G6 protein belongs to the NLPC/P60 superfamily, which consists of proteins with a papain-like fold with known or predicted protease, amidase, lipase, or acyltransferase activity (17). Although G6 orthologs are present in all poxviruses, a previous study had shown that deletion of the VACV gene did not impair replication in a variety of cultured cells and that the attenuated phenotypes of the deletion and active site mutations were similar in mice (2). To determine whether mutation of the catalytic cysteine would have the same beneficial effect on the phenotype of vA8myx as the deletion, we replaced EGFP in vA8myxΔG6 with G6(C109S), in which serine occupied the position of the invariant cysteine 109 in the predicted catalytic site. We also constructed two control viruses from vA8myxΔG6. In one, EGFP was replaced with intact G6R, making it a vA8myxΔG6 revertant (vA8myxΔG6rev). In the other (vA8myxG6stop), the EGFP was replaced with the G6R ORF with stop codons at positions 24 and 25. The plaque sizes of the ΔG6, G6stop, and G6(C109S) variants of vA8myx were significantly larger than those seen with vA8myx and vΔG6rev (P < 0.0001), but the plaque sizes of those three variants were not significantly different from each other (Fig. 3A). The yields of v8myxG6(C109S) and vA8myxΔG6 were similar, indicating that inactivation of the putative enzymatic activity of G6 produced the same phenotype as a deletion (Fig. 3B).
FIG 3.
Plaque formation and replication of vA8myxG6(C109S) and other viruses. (A) Plaque sizes. vA8myxΔG6rev (red), vA8myx (gray), vA8myxΔA26 (green), vA8myxG6C109S (yellow), vA8myxG6stop (light blue), and the WT virus (dark blue) were plated on BS-C-1 cells. After 48 h, the plates were stained with crystal violet. Upper panel, images of plaques; lower panel, areas of plaques. (B) Virus spread assay. BS-C-1 cells were infected in duplicate with 0.01 PFU/cell of viruses color coded as described for panel A. At the indicated times, virus titers were determined by plaque assay.
A26L and G6R deletion mutants outcompete vA8myx.
We carried out direct competition between vA8myx and the ΔA26L and ΔG6R mutants to mimic the conditions present during successive rounds of the original passage experiment. For this analysis, BS-C-1 cells were coinfected with a total of 0.01 PFU per cell of vA8myxΔA26 or vA8myxΔG6 or vA8myxΔA26ΔG6 together with 10-fold more vA8myx in each case. The vA8myxΔA26 strain expressed DsRed, vA8myxΔG6 expressed EGFP, vA8myxΔA26ΔG6 expressed both fluorescent proteins, and vA8myx expressed neither. After each round, the cells were lysed, dilutions were plated on BS-C-1 cells, and plaques were visualized by fluorescent and visible light microscopy. Red and green plaques and total plaques stained with crystal violet were enumerated. By the third round, the single mutants as well as the double mutant almost completely replaced vA8myx (Fig. 4A). A fluorescent image of the competition between vA8myxΔG6 and vA8myx is shown in Fig. 4B. In our original passages, the G6R mutation was always present in the background of the A26L mutation since the latter occurred earlier. Thus, these experiments performed with vA8myxΔG6 were important to demonstrate that inactivation of G6R, even in the presence of intact A26L, was sufficient for competition with vA8myx.
FIG 4.
Competition between vA8myx and A26L and G6R single and double deletion mutants. (A) BS-C-1 cells were coinfected in triplicate with a total of 0.01 PFU per cell of vA8myxΔA26, vA8myxΔG6, or vA8myxΔA26ΔG6 together with 10-fold more vA8myx in each case. vA8myxΔA26 expressed DsRed, vA8myxΔG6 expressed EGFP, vA8myxΔA26ΔG6 expressed both fluorescent proteins, and vA8myx expressed neither. After each round, the red and green plaques and total plaques stained with crystal violet were enumerated. (B) Image of fluorescent and total virus plaques. Results of competition between vA8myx and vA8myxΔG6 are shown.
Comparisons of viral mRNA and protein expression.
It could be reasonably expected that replacement of the VACV small-subunit A8R intermediate transcription factor gene by an ortholog from another poxvirus genus would affect intermediate and late transcription. To analyze transcription, BS-C-1 cells were infected with 5 PFU per cell of each virus. RNA was extracted at 4 or 8 h and reverse transcribed, and the numbers of transcripts of an early (I3) and the predominantly intermediate (D13) and predominantly late (A3) gene were determined by digital PCR using gene-specific primers (Fig. 5). The I3 transcripts were more abundant at 4 h than at 8 h as expected but differed less than 2-fold for the different viruses. Expression of intermediate and late transcripts follows DNA replication and can usually be detected at a low level at 4 h and at a greatly increased level at 8 h. D13 and A3 transcripts were a few fold higher in cells infected with the WT virus at 4 h, but this was no longer true at 8 h. Moreover, there were relatively modest differences between the levels of intermediate and late mRNAs in cells infected with vA8myx, with the deletion mutants, and with the control virus with the homologous VACV A8 transcription factor. These data suggested that the attenuation of vA8myx and the adaptation by the deletion mutants were not due to a general effect on viral transcription. However, mRNA levels are determined by rates of synthesis and degradation, which are dynamic processes that vary during the course of infection; thus, the time points represent only snapshots.
FIG 5.
Quantitation of early, intermediate, and late mRNAs. BS-C-1 cells were infected in triplicate with 5 PFU/cell of vA8myx, vA8myxΔA26, vA8myxΔG6, vA8myxΔA26ΔG6 or WT virus. At 4 and 8 h postinfection (hpi), the cells were harvested and RNA extracted. Following reverse transcription, the cDNAs were quantitated by ddPCR using primers specific for I3, D13, and A3 mRNAs.
Western blotting was used to analyze expression of representative VACV proteins. E3 is an early protein, and A17 and A3 are both late proteins. The early protein provides information regarding entry and immediate gene expression. The A17 and A3 proteins undergo proteolytic processing during membrane and core formation, respectively, and might be able to provide information regarding morphogenesis as well as expression. As shown in Fig. 6, there were similar levels of E3 at 10 h in vA8myx, the deletion mutants, and the WT virus. Overall, the levels of A17 and A3 were similar, although the intensities of the bands from the vA8myxΔG6 were slightly lower than in the other samples in two separate 10-h experiments, as shown in Fig. 6 for one of the two. At 10 h, there was greater processing of A17 and A3 in cells infected with the WT than in those infected with vA8myx or the mutant viruses as determined by the relative intensities of the upper and lower bands (Fig. 6). However, by 24 h, A17 processing had greatly increased in all samples whereas A3 processing remained low in vA8myx and vΔA26 but had increased to WT levels in vΔG6 and vΔA26ΔG6 (Fig. 6). These data indicated a delay and decrease in processing, particularly of the A3 core protein in cells infected with vA8myx, which increased at most slightly for vΔA26 but strongly for vΔG6 and vΔA26ΔG6. Decreased processing of A3 is consistent with effects on morphogenesis.
FIG 6.
Synthesis and processing viral proteins. BS-C-1 cells were infected with 5 PFU/cell of vA8myx, vA8myxΔA26, vA8myxΔG6, or vA8myxΔA26ΔG6 or the WT virus. After 10 h and 24 h, the cells were harvested and the lysates analyzed by Western blotting using antibodies specific for E3, A17, and A3 proteins. The relative amounts of the upper (unprocessed) and lower (processed) A17 and A3 bands were determined. At 10 h, the proportions of processed A17 were 43%, 45%, 48%, 49%, and 61% and those of A3 were 22%, 20%, 24%, 29%, and 45% for vA8myx, vΔA26, vΔG6, vΔA26ΔG8, and the WT virus, respectively. At 24 h, the proportions of processed A3 were 26%, 29%, 47%, 51%, and 46% for vA8myx, vΔA26, vΔG6, vΔA26ΔG8, and the WT virus, respectively.
VACV morphogenesis.
Transmission electron microscopy was used to examine the morphogenesis of vA8myx, the A26L and G6R single and double deletion mutants, and WT virus. All stages of development, including from crescent membranes to immature virions (IVs), mature virions (MVs), wrapped virions (WVs), and enveloped virions (EVs), were discerned in cells infected with each virus. The similar appearances of IVs and MVs in infected cells are shown in Fig. 7. The numbers of the various viral forms per 50 cell sections at 12 h and 24 h are presented in Table 2. Although the numbers are not highly quantitative because of cell-to-cell variation as shown in the duplicates for the 12-h vΔG6 samples (Fig. 7), the trend was a higher number of particles with mature-looking cores (MV plus WV plus EV) in cells infected with vA8myxΔG6, vA8myxΔA26ΔG6, and the WT compared to vA8myx and vA8myxΔA26, particularly at the 24-h time point, consistent with the efficiency of processing the A3 core protein (Fig. 6). There was also a higher number of WVs than of MVs in vΔA26 compared to vA8myx and the other mutants at both 12 and 24 h.
FIG 7.
Transmission electron microscopy. BS-C-1 cells were infected with 10 PFU/cell of vA8myx, vA8myxΔA26, vA8myxΔG6, or vA8myxΔA26ΔG6 or the WT virus. After 24 h, the cells were fixed and thin sections imaged by transmission electron microscopy. The low and high magnifications are indicated by bars at the lower right corners. Abbreviations: C, crescent, IV, immature virion; MV, mature virion; nu, IV with nucleoid.
TABLE 2.
Enumeration of viral formsa
| Time point and viral form | No. of viral forms per 50 cells | |||||
|---|---|---|---|---|---|---|
| 12 h | vA8myx | ΔA26 | ΔG6 | ΔG6 | ΔA26ΔG6 | WT |
| Crescent | 676 | 836 | 411 | 350 | 538 | 290 |
| IV | 824 | 917 | 574 | 514 | 561 | 676 |
| MV | 117 | 96 | 521 | 331 | 182 | 174 |
| WV | 15 | 123 | 30 | 48 | 53 | 279 |
| EV | 44 | 45 | 77 | 57 | 47 | 146 |
| MV+WV+EV | 176 | 264 | 628 | 436 | 282 | 599 |
| 24 h | ||||||
| Crescent | 1,231 | 734 | 748 | 691 | 411 | |
| IV | 1,858 | 1,963 | 1,293 | 1,830 | 1,420 | |
| MV | 321 | 318 | 1,299 | 689 | 934 | |
| WV | 49 | 167 | 122 | 227 | 731 | |
| EV | 42 | 126 | 106 | 133 | 172 | |
| MV+WV+EV | 412 | 611 | 1,527 | 1,049 | 1,837 | |
The numbers of viral forms in 50 cells are shown. For the data presented in the second 12-h column (corresponding to ΔG6 virus), 100 cells from another cell section were analyzed and the numbers divided by 2.
Comparison of infectivity of virus particles.
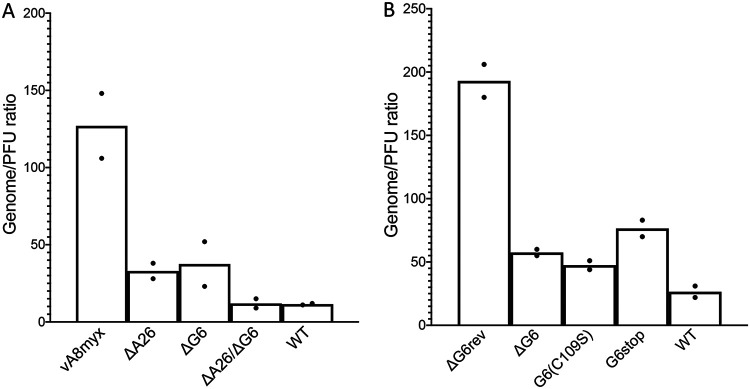
A wide range in the percentages of VACV particles that form plaques has been found in studies reported from several laboratories, probably as a consequence of the differing methods used as well as of the use of different virus strains. A genome-to-PFU ratio of 10 to 20 for sucrose gradient-purified VACV was determined by a droplet digital PCR (ddPCR) method developed in our laboratory (18). The ratios that were obtained for crude cell lysates by degrading accessible DNA with Benzonase nuclease and then quantifying the amount of protected DNA, which is presumed to be present in particles, by ddPCR were similar to those obtained for purified virus. Using the same technique for cell lysates, we found that the particle/PFU ratio determined for vA8myx was up to 10-fold higher than the ratios obtained for WT virus or the A26L and G6R deletion mutants, indicating lower infectivity (Fig. 8A). The virus with a G6 active site mutation, vA8myxG6(C109S), had a particle-to-PFU ratio that was similar to the ratios measured for the G6R deletion mutant vA8myxΔG6 and for the G6 stop mutant vA8myxG6stop but that was lower than that of virus vA8myxG6rev, in which G6 had been repaired (Fig. 8B). Data from three separate experiments were combined, with results showing that the difference in particle/PFU ratios between vA8myx and the mutants was significant (P = 0.034, 0.037, and 0.024 for ΔA26, ΔG6, and ΔA26ΔG6, respectively). Taken together, the electron microscopy and ddPCR data suggested that the adaptive mutations increased the number and infectivity of virus particles.
FIG 8.
Particle (genome)/PFU ratios of vA8myx and mutants. (A) BS-C-1 cells were infected in duplicate with 5 PFU/cell of vA8myx, vA8myxΔA26, vA8myxΔG6, or vA8myxΔA26ΔG6 or the WT virus. After 24 h, the cells were harvested and lysates prepared. Samples of the lysates were used to determine PFU levels in BS-C-1 cells, and additional samples were treated with Benzonase. Resistant DNA was quantified by ddPCR using VACV-specific primers. (B) BS-C-1 cells were infected with vA8myxΔG6rev, vA8myxΔG6, vA8myxG6R(C108S), vA8myxG6stop, or the WT virus as described for panel A. PFU and DNA levels were determined as described for panel A.
DISCUSSION
Several previous VACV experimental evolution studies highlighted adaptation resulting from gene copy number expansion followed by base substitutions (7–10). Of the seven adaptive mutants identified in the present study, five were indels (insertions or deletions) and only two were base substitutions, and the latter did not reach fixation. Four of the indels occurred within ORFs causing reading frameshifts, and one was in a putative promoter region. Three of the frameshifts resulted in premature stop codons, whereas one occurred near the end of an ORF, producing an altered and extended sequence. The indels were single nucleotide insertions or deletions within runs of five to seven As or Ts except for one caused by deletion of 11 nucleotides located between ATC and TAG sequences. Indels in VACV have also been found in other studies. In a previous experimental evolution experiment in which the VACV B1 kinase gene was deleted, a single nucleotide addition or deletion in a run of eight As leading to frameshift mutations in the B12 pseudokinase ORF was found to enhance replication (14). An analysis of thymidine kinase-negative mutants revealed single nucleotide insertions; a C was added to a run of four Cs in one case, whereas a nucleotide was added that was identical to the preceding one in another two mutants (19). In addition, single nucleotide deletions in runs of four to six Gs or Cs have been shown to promote instability of recombinant genes in the attenuated modified VACV Ankara vector and the elimination of these runs is recommended (20). Single nucleotide insertions or deletions are common in many systems and are thought to result from strand misalignments occurring during replication (21). Every DNA polymerase examined produces deletion or insertion errors, which can equal or exceed nucleotide substitution rates. Insertions or deletions commonly occur at homopolymer runs due to slippage of the primer and template strands. Continued polymerization then leads to a deletion if the unpaired nucleotide is in the template strand or to an insertion if it is in the primer strand. There are several ways in which the misalignment can be initiated. One way is by the DNA polymerase and newly synthesized strand transiently dissociating from the template, an event the rate of whose occurrence is inversely proportional to the processivity of the enzyme.
VACV is A/T-rich, and runs of identical nucleotides occur in many VACV genes such that frameshifting potentially could occur in any of them. However, mutations that prevent expression of an essential gene are immediately lost and those that prevent expression of a nonadaptive gene are not selected. Therefore, the deletions in A26L, G6R, and A14.5L were likely random but the adaptation resulting from frameshifts was specific. Of the seven mutations detected, the A26L mutation and one of the two G6R mutations occurred in all three parallel passages. The other mutations were unique to individual passage series but were detected by round three or four, indicating their rapid occurrence. The A26L frameshift was present in 96% to 98% of the sequence reads at round 10 of each passage. The presence of mutations in other genes means that the dominant A26L mutation viruses had two to four mutated genes. Whether the multiple gene mutations arose in separate genomes and came together in a single genome by recombination or occurred successively cannot be differentiated by the Illumina 150-bp read sequencing employed. In either model, the increase in the levels of most mutations between rounds 8 and 10 suggests that each contributed to selection. The two exceptions were the mutations in J6R and F10L, whose levels decreased, consistent with clonal interference (22) that may have been enhanced by bottlenecks due to virus dilutions after each round.
Because the viruses that dominated the final passages of the experimental evolution experiment had multiple mutations, it was necessary to produce viruses with single mutations for further study. We elected to make deletions instead of frameshifts to study the A26 and G6 mutations because of their simple construction and isolation as well as to confirm that the phenotype was due to absence of the protein. The deletion mutants exhibited higher replication rates than vA8myx and outcompeted the latter within three passage cycles. Moreover, the double vA8myxΔA26ΔG6 virus replicated at a higher rate than the mutants with individual mutations. Although we can readily envision how these frameshift mutations occurred, the basis for their selection is more difficult to comprehend. We verified that the defect of vA8myx was due to the replacement of the VACV intermediate transcription factor gene with an ortholog as genome sequencing did not reveal any other changes that might impair replication. Furthermore, replication of vA8myx was rescued in a complementing cell line that expressed the VACV intermediate transcription factors. The finding of adaptive RNA polymerase mutations in the previous experimental evolution study and again in the present one also suggests adaptation to a perturbation in transcription. Nevertheless, a defect in transcription was not suggested by PCR snapshots of representative intermediate or late RNAs at 8 h after infection with vA8myx compared to a WT control. Nor was transcription significantly affected by mutations in A26 or G6. If only specific RNAs and the timing of their synthesis are affected, then intensive transcriptome sequencing (RNA-seq) and ribosome profiling studies would be necessary to reveal this. The replication defect of vA8myx manifested most clearly as a reduction in the infectivity of virus particles, which was considerably increased by the A26 and G6 mutations. Western blotting showed that processing of the A3 core protein was delayed and decreased in vA8myx and increased particularly in the G6 and double A26-plus-G6 mutants. Processing of A3 occurs during morphogenesis (23, 24), and electron microscopy demonstrated increased numbers of particles with defined cores in the cells infected with the vA8myxΔA26, vA8myxΔG6, and vA8myxΔA26ΔG6 mutants compared to vA8myx.
Previous studies of A26 and G6 provided little insight into how deletions of these proteins could enhance morphogenesis of vA8myx. The VACV A26 protein appears to have multiple roles. Although the protein lacks a hydrophobic domain, it localizes to the surface of MVs (25, 26) in association with the A27 protein and the A17 transmembrane protein (27–29). The cowpox virus ortholog of A26 is required for embedding virus particles in A-type inclusions (30, 31), a process that can also be accomplished by VACV A26 (32). A26 also has a role in attachment of MVs to the plasma membrane by binding to laminin (33) and acts as a fusion suppressor by binding to entry-fusion proteins, though deletion of A26L from the VACV Western Reserve (WR) strain had no effect on virus yields in HeLa cells (34, 35). In BS-C-1 cells, deletion of A26L had little effect on entry or replication (36). A hint that deletion of A26L might have a positive effect on virus spread comes from a study reporting that A26 is absent in EVs, leading to a suggestion that it negatively interferes with wrapping of MVs by Golgi membranes (37). This could explain why deletion of A26L from vA8myx increased the numbers of WVs and EVs. However, deletion of the A26L gene from VACV WR had no effect on the ratio of MVs to EVs in either HeLa or RK13 cells (27).
Little is known about the role of the G6 protein, although orthologs are present in all poxviruses. G6 belongs to the NLPC/P60 superfamily that consists of proteins with a papain-like fold and with known or predicted protease, amidase, lipase, or acyltransferase activity (17). Thus far, however, none of these activities have been described for any of the viral G6 orthologs. Expression of VACV G6 and association with virus particles, possibly between the core and the viral membrane, were reported previously (2). Despite its conservation, deletion of the VACV gene or mutation of the predicted catalytic cysteine did not impair replication in cultured cells, although both mutants were shown to be attenuated in a mouse respiratory infection model (2). A14.5, the third frameshifted protein, is associated with the membranes of IVs and MVs (15). Like G6, an A14.5 deletion mutant replicated well in cell culture but was attenuated in the mouse model. The three proteins A26, G6, and A14.5 seem to have little in common except for their expression late in infection and association at or below the membrane of MVs. One hypothesis is that subtle perturbations in gene expression dysregulate aspects of virion assembly, which is a complex process involving at least 80 proteins.
In contrast to several previous studies in which adaptation was achieved by increases in gene copy numbers and base substitutions, we demonstrated the rapid appearance and fixation of frameshift mutations resulting in gene inactivation, which is an important feature of OPXV evolution.
MATERIALS AND METHODS
Cells and virus.
BS-C-1 cells (ATCC CCL-26) were maintained in Eagle’s minimum essential medium (EMEM) supplemented with 10% fetal bovine serum, 100 units/liter penicillin, 100 μg/liter streptomycin, and 2 mM l-glutamine. RK13-A8/A23 cells (16) were maintained in supplemented EMEM containing 100 μg/ml Zeocin. The recombinant VACV strains were all derived from the Western Reserve strain (WR; ATCC VR-1354).
Serial passaging of vA8myx.
The parental vA8myx virus used for passaging was described previously (12). Briefly, this attenuated virus was constructed from vTF7-3, a Western Reserve (WR) strain of VACV encoding the T7 RNA polymerase (38), by replacing the gene for the small A8 subunit of the intermediate transcription factor by the ortholog from myxoma virus, encoding a V5 tag at the N terminus. Prior to passaging, vA8myx was plaque purified and expanded once in BS-C-1 cells and aliquots were used to infect parallel cultures of BS-C-1 cells at a multiplicity of infection of 0.01 PFU/cell. After 48 h, the infected cells were harvested and the lysates diluted for a repeat infection at the same multiplicity. The experiment was stopped after 10 consecutive rounds.
Whole-genome sequencing.
DNA was extracted from 5 × 106 infected BS-C-1 cells. Briefly, DNA was extracted from the cytoplasm with proteinase K and phenol and then subjected to ethanol precipitation. Sequencing was performed with an Illumina MiSeq-2 system with a 150-bp configuration by Genewiz or the NIAID core facility.
Construction of recombinant viruses.
The vA8myxΔA26 and vA8myxΔG6 viruses were constructed by replacing the corresponding gene with DsRed or EGFP under the control of VACV p11 or an early-late synthetic VACV promoter, respectively. The double deletion mutant vA8myxΔA26ΔG6 was constructed similarly. The reporter genes were flanked by ∼500-bp sequences adjacent to the replaced genes, and the PCR products were transfected into BS-C-1 cells that were infected with vA8myx at a multiplicity of 2 PFU/cell. The entire A26 ORF was deleted, whereas 33 bp at the 3′ terminus of G6 was retained so as to not interfere with expression of the adjacent gene. After 20 h, the cells were harvested and the recombinant viruses were plaque purified. Genome sequencing confirmed the insertions/deletions but did not reveal any other mutations. Revertant virus vA8myxΔG6rev was constructed by replacing the EGFP gene in vA8myxΔG6 by the VACV G6R gene from a PCR product and isolating nonfluorescent plaques. Similarly, vA8myxG6(C109S) and vA8myxG6stop were constructed using PCR products. For all subsequent experiments, vA8myx and all its derivatives were grown in RK13-A8/A23 cells (16) and PFU levels determined on BS-C-1 cells.
Competition experiments.
BS-C-1 cells were infected in triplicate with 0.01 PFU/cell of a 10-to-1 mixture of vA8myx and vA8myxΔA26, vA8myxΔG6, or vA8myxΔA26ΔG6. The mixture of viruses was subjected to three to four consecutive passages at a multiplicity of infection of 0.01 for 48 h for vA8myxΔA26 and vA8myxΔA26ΔG6 and 24 h for vA8myxΔG6. The viruses from each passage were analyzed by plaque assay, and the numbers of fluorescent plaques were compared to the numbers of all plaques stained by crystal violet with an Evos M7000 system (Thermo Fisher).
Growth curves and plaque assays.
For multistep spread and one-step growth assays, BS-C-1 cells were infected in triplicate with the corresponding viruses at 0.01 PFU/cell or 5 PFU/cell, respectively. After 1 h at room temperature, the inocula were aspirated and the plates washed three times with medium. Incubations were continued at 37°C for the indicated times, and the cells were dislodged by scraping. Approximately 2.5 × 105 cells were resuspended in 500 μl of EMEM, freeze-thawed three times, and sonicated. The titer was determined by plaque assay on BS-C-1 cells after 48 to 72 h of incubation at 37°C and staining with crystal violet. Plaque areas were calculated by ImageJ after visualizing the images with the Evos system.
Quantitation of transcription of VACV genes.
BS-C-1 cells were infected in triplicate with 5 PFU/cell of different viruses at room temperature. After 1 h, the inocula were aspirated and the plates washed three times with fresh medium. The incubation was continued at 37°C for 4 and 8 h, and RNA was extracted with an RNeasy kit (Qiagen), subjected to reverse transcription with a SuperScript IV first-strand synthesis system containing ezDNase with a mixture of oligo(dT) and random primers (Invitrogen). The transcripts of VACV genes I3, D13, and A3 were quantified by ddPCR performed with specific primers using an automated droplet generator and a QX200 droplet reader (Bio-Rad) as described previously (18, 39).
Transmission electron microscopy.
BS-C-1 cells were infected with virus at 10 PFU/cell. After 12 or 24 h, cells were fixed, dehydrated, and embedded with Embed 812 resin (Electron Microscopy Sciences, Hatfield, PA) as described previously (40). Samples were imaged with a FEI Tecnai Spirit transmission electron microscope (FEI, Hillsboro, OR). Viral structures were manually counted on sections of 50 or 100 cells.
Particle (genome)/PFU ratios.
Particle/PFU ratios were determined as described previously (18). BS-C-1 cells were infected for 24 to 48 h in duplicate with 5 PFU/cell of virus, and 100 μl of lysate was mixed with 100 μl of a mixture containing 50 mM Tris-HCl (pH 8.0), 2 mM MgCl2, and 0.1 mg/ml bovine serum albumin. Samples were incubated at 37°C for 1 h with 20 units of Benzonase nuclease. The Benzonase was deactivated with 5 mM EDTA for 20 min at room temperature, and DNA was extracted by the use of a DNA blood kit (Qiagen) and quantified by ddPCR. The genome/PFU ratio was calculated by dividing the number of genomes by the number of infectious particles counted in the plaque assay.
Statistical analysis.
Student's t test (unpaired, parametric) was performed using Prism 8 (GraphPad) if three or more biological samples were obtained.
Data availability.
Data are available at the Sequence Read Archive under BioProject accession number PRJNA636166.
ACKNOWLEDGMENTS
We thank Catherine Cotter for maintaining cell lines and all members of our laboratory for helpful discussions. Craig Martens, Kishore Kanakabandi, Dan Bruno, and Dan Sturtevant performed DNA sequencing.
This research was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol 77:7590–7600. doi: 10.1128/jvi.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. 2008. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology 374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haller SL, Peng C, McFadden G, Rothenburg S. 2014. Poxviruses and the evolution of host range and virulence. Infect Genet Evol 21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrickson RC, Wang CL, Hatcher EL, Lefkowitz EJ. 2010. Orthopoxvirus genome evolution: the role of gene loss. Viruses 2:1933–1967. doi: 10.3390/v2091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoine G, Scheiflinger F, Dorner F, Falkner FG. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 6.Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, Hausmann J. 2007. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J Gen Virol 88:3249–3259. doi: 10.1099/vir.0.83156-0. [DOI] [PubMed] [Google Scholar]
- 7.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. 2014. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog 10:e1004002. doi: 10.1371/journal.ppat.1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan G, Kitzman JO, Shendure J, Geballe AP. 2015. Experimental evolution identifies vaccinia virus mutations in A24R and A35R that antagonize the protein Kknase R pathway and accompany collapse of an extragenic gene amplification. J Virol 89:9986–9997. doi: 10.1128/JVI.01233-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cone KR, Kronenberg ZN, Yandell M, Elde NC. 2017. Emergence of a viral RNA polymerase variant during gene copy number amplification promotes rapid evolution of vaccinia virus. J Virol 91:e01428-16. doi: 10.1128/JVI.01428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasani TA, Cone KR, Quinlan AR, Elde NC. 2018. Long read sequencing reveals poxvirus evolution through rapid homogenization of gene arrays. Elife 7:e35453. doi: 10.7554/eLife.35453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart CA, Zhivkoplias EK, Senkevich TG, Wyatt LS, Moss B. 2018. RNA Polymerase Mutations Selected during Experimental Evolution Enhance Replication of a Hybrid Vaccinia Virus with an Intermediate Transcription Factor Subunit Replaced by the Myxoma Virus Ortholog. J Virol 92:e01089-18. doi: 10.1128/JVI.01089-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci U S A 99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson AT, Wang ZG, Rico AB, Wiebe MS. 2019. A poxvirus pseudokinase represses viral DNA replication via a pathway antagonized by its paralog kinase. PLoS Pathog 15:e1007608. doi: 10.1371/journal.ppat.1007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betakova T, Wolffe EJ, Moss B. 2000. Vaccinia virus A14.5L gene encodes a hydrophobic 53-amino acid virion membrane protein that enhances virulence in mice and is conserved amongst vertebrate poxviruses. J Virol 74:4085–4092. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren RD, Cotter C, Moss B. 2012. Reverse genetic analysis of poxvirus intermediate transcription factors. J Virol 86:9514–9519. doi: 10.1128/JVI.06902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol 4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Americo JL, Earl PL, Moss B. 2017. Droplet digital PCR for rapid enumeration of viral genomes and particles from cells and animals infected with orthopoxviruses. Virology 511:19–22. doi: 10.1016/j.virol.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weir JP, Moss B. 1983. Nucleotide sequence of the vaccinia virus thymidine kinase gene and the nature of spontaneous frameshift mutations. J Virol 46:530–537. doi: 10.1128/JVI.46.2.530-537.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt LS, Earl PL, Xiao W, Americo JL, Cotter CA, Vogt J, Moss B. 2009. Elucidating and minimizing the loss by recombinant vaccinia virus of human immunodeficiency virus gene expression resulting from spontaneous mutations and positive selection. J Virol 83:7176–7184. doi: 10.1128/JVI.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel TA, Bebenek K. 2000. DNA replication fidelity. Annu Rev Biochem 69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 22.Gerrish PJ, Lenski RE. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103:127–144. doi: 10.1023/A:1017067816551. [DOI] [PubMed] [Google Scholar]
- 23.Katz E, Moss B. 1970. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci U S A 66:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss B, Rosenblum EN. 1973. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol 81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 25.Sarov I, Joklik WK. 1972. Studies on the nature and location of the capsid polypeptides of vaccinia virions. Virology 50:579–592. doi: 10.1016/0042-6822(72)90409-6. [DOI] [PubMed] [Google Scholar]
- 26.Katz E, Margalith E. 1973. Location of vaccinia virus structural polypeptides on the surface of the virus particle. J Gen Virol 18:381–384. doi: 10.1099/0022-1317-18-3-381. [DOI] [PubMed] [Google Scholar]
- 27.Howard AR, Senkevich TG, Moss B. 2008. Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein. J Virol 82:12384–12391. doi: 10.1128/JVI.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ching YC, Chung CS, Huang CY, Hsia Y, Tang YL, Chang W. 2009. Disulfide bond formation at the C termini of vaccinia virus A26 and A27 proteins does not require viral redox enzymes and suppresses glycosaminoglycan-mediated cell fusion. J Virol 83:6464–6476. doi: 10.1128/JVI.02295-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang DR, Hsiao JC, Wong CH, Li GC, Lin SC, Yu SSF, Chen WL, Chang W, Tzou D. 2014. Vaccinia viral protein A27 is anchored to the viral membrane via a cooperative interaction with viral membrane protein A17. J Biol Chem 289:6639–6655. doi: 10.1074/jbc.M114.547372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shida H, Tanabe K, Matsumoto S. 1977. Mechanism of virus occlusion into A-type inclusion during poxvirus infection. Virology 76:217–233. doi: 10.1016/0042-6822(77)90298-7. [DOI] [PubMed] [Google Scholar]
- 31.McKelvey TA, Andrews SC, Miller SE, Ray CA, Pickup DJ. 2002. Identification of the orthopoxvirus p4c gene, which encodes a structural protein that directs intracellular mature virus particles into A-type inclusions. J Virol 76:11216–11225. doi: 10.1128/jvi.76.22.11216-11225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard AR, Weisberg AS, Moss B. 2010. Congregation of orthopoxvirus virions in cytoplasmic A-type inclusions is mediated by interactions of a bridging protein (A26p) with a matrix protein (ATIp) and a virion membrane-associated protein (A27p). J Virol 84:7592–7602. doi: 10.1128/JVI.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu WL, Lin CL, Yang MH, Tzou DLM, Chang W. 2007. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J Virol 81:2149–2157. doi: 10.1128/JVI.02302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang SJ, Chang YX, Izmailyan R, Tang YL, Chang W. 2010. Vaccinia virus A25 and A26 proteins are fusion suppressors for mature virions and determine strain-specific virus entry pathways into HeLa, CHO-K1, and L cells. J Virol 84:8422–8432. doi: 10.1128/JVI.00599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang HW, Yang CH, Luo YC, Su BG, Cheng HY, Tung SY, Carillo KJD, Liao YT, Tzou DM, Wang HC, Chang W. 2019. Vaccinia viral A26 protein is a fusion suppressor of mature virus and triggers membrane fusion through conformational change at low pH. PLoS Pathog 15:e1007826. doi: 10.1371/journal.ppat.1007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bengali Z, Satheshkumar PS, Moss B. 2012. Orthopoxvirus species and strain differences in cell entry. Virology 433:506–512. doi: 10.1016/j.virol.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulaeto D, Grosenbach D, Hruby DE. 1996. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J Virol 70:3372–3375. doi: 10.1128/JVI.70.6.3372-3377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu R, Moss B. 2018. Vaccinia virus C9 ankyrin repeat/F-box protein is a newly identified antagonist of the type I interferon-induced antiviral state. J Virol 92:e00052-18. doi: 10.1128/JVI.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruri-Avidal L, Domi A, Weisberg AS, Moss B. 2011. Participation of vaccinia virus L2 protein in the formation of crescent membranes and immature virions. J Virol 85:2504–2511. doi: 10.1128/JVI.02505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available at the Sequence Read Archive under BioProject accession number PRJNA636166.