EBV, a human herpesvirus, is latently present in most nasopharyngeal carcinomas, Burkitt lymphomas, and some gastric cancers. To develop a lytic-induction therapy for treating patients with EBV-associated cancers, we need a way to efficiently reactivate EBV into lytic replication. EBV’s BZLF1 gene product, Zta, usually controls this reactivation switch. We previously showed that HIF-1α binds the BZLF1 gene promoter, inducing Zta synthesis, and HIF-1α-stabilizing drugs can induce EBV reactivation. In this study, we determined which EBV-positive cell lines are reactivated by classes of HIF-1α-stabilizing drugs. We found, unexpectedly, that HIF-1α-stabilizing drugs only induce reactivation when they also induce accumulation of phosphorylated, wild-type p53. Fortunately, p53 phosphorylation can also be provided by drugs such as nutlin-3, leading to synergistic reactivation of EBV. These findings indicate that some HIF-1α-stabilizing drugs may be helpful as part of a lytic-induction therapy for treating patients with EBV-positive malignancies that contain wild-type p53.
KEYWORDS: BZLF1 gene promoter, deferoxamine, nutlin-3, pevonedistat, hypoxia response element, latent-lytic switch
ABSTRACT
We previously reported that the cellular transcription factor hypoxia-inducible factor 1α (HIF-1α) binds a hypoxia response element (HRE) located within the promoter of Epstein-Barr virus’s (EBV’s) latent-lytic switch BZLF1 gene, Zp, inducing viral reactivation. In this study, EBV-infected cell lines derived from gastric cancers and Burkitt lymphomas were incubated with HIF-1α-stabilizing drugs: the iron chelator deferoxamine (Desferal [DFO]), a neddylation inhibitor (pevonedistat [MLN-4924]), and a prolyl hydroxylase inhibitor (roxadustat [FG-4592]). DFO and MLN-4924, but not FG-4592, induced accumulation of both lytic EBV proteins and phosphorylated p53 in cell lines that contain a wild-type p53 gene. FG-4592 also failed to activate transcription from Zp in a reporter assay despite inducing accumulation of HIF-1α and transcription from another HRE-containing promoter. Unexpectedly, DFO failed to induce EBV reactivation in cell lines that express mutant or no p53 or when p53 expression was knocked down with short hairpin RNAs (shRNAs). Likewise, HIF-1α failed to activate transcription from Zp when p53 was knocked out by CRISPR-Cas9. Importantly, DFO induced binding of p53 as well as HIF-1α to Zp in chromatin immunoprecipitation (ChIP) assays, but only when the HRE was present. Nutlin-3, a drug known to induce accumulation of phosphorylated p53, synergized with DFO and MLN-4924 in inducing EBV reactivation. Conversely, KU-55933, a drug that inhibits ataxia telangiectasia mutated, thereby preventing p53 phosphorylation, inhibited DFO-induced EBV reactivation. Lastly, activation of Zp transcription by DFO and MLN-4924 mapped to its HRE. Thus, we conclude that induction of BZLF1 gene expression by HIF-1α requires phosphorylated, wild-type p53 as a coactivator, with HIF-1α binding recruiting p53 to Zp.
IMPORTANCE EBV, a human herpesvirus, is latently present in most nasopharyngeal carcinomas, Burkitt lymphomas, and some gastric cancers. To develop a lytic-induction therapy for treating patients with EBV-associated cancers, we need a way to efficiently reactivate EBV into lytic replication. EBV’s BZLF1 gene product, Zta, usually controls this reactivation switch. We previously showed that HIF-1α binds the BZLF1 gene promoter, inducing Zta synthesis, and HIF-1α-stabilizing drugs can induce EBV reactivation. In this study, we determined which EBV-positive cell lines are reactivated by classes of HIF-1α-stabilizing drugs. We found, unexpectedly, that HIF-1α-stabilizing drugs only induce reactivation when they also induce accumulation of phosphorylated, wild-type p53. Fortunately, p53 phosphorylation can also be provided by drugs such as nutlin-3, leading to synergistic reactivation of EBV. These findings indicate that some HIF-1α-stabilizing drugs may be helpful as part of a lytic-induction therapy for treating patients with EBV-positive malignancies that contain wild-type p53.
INTRODUCTION
Epstein Barr virus (EBV) is a human gammaherpesvirus that infects more than 90% of humans. Infection of preadolescent children is usually asymptomatic, while primary infection later in life frequently results in infectious mononucleosis (IM) (reviewed in reference 1). Following primary infection, EBV establishes a lifelong, asymptomatic, latent infection in a subset of host memory B cells, where the EBV genome is maintained as an episome with very few genes expressed (reviewed in references 2 to 4). Latent EBV infection also contributes to 2% of human cancers worldwide, including many nasopharyngeal carcinomas (NPCs), Burkitt lymphomas (BLs), and ∼10% of gastric cancers (reviewed in reference 5).
To complete its natural life cycle for transmission to other hosts, EBV exits latency and enters its lytic phase of infection. Lytic induction requires expression of the viral immediate early (IE) genes BZLF1 and BRLF1, which encode the proteins Zta and Rta, respectively. These IE proteins trigger the cascade of events that results in viral genome amplification and production of infectious virion particles. We previously reported that hypoxia-inducible factor 1α (HIF-1α) can bind to a hypoxia response element (HRE) located within the BZLF1 gene promoter (Zp), inducing BZLF1 gene expression and, thereby, promoting lytic reactivation of EBV in some EBV-positive (EBV+) epithelial and B-cell lines (6).
The heterodimer of HIF-1α and HIF-1β (also called ARNT) binds HREs to activate transcription of a variety of genes that contribute to cell survival during hypoxia (reviewed in references 7 and 8). While HIF-1β protein is constitutively present in cells, accumulation of HIF-1α protein is mostly regulated at the posttranslational level. During normoxia, HIF-1α is synthesized, but hydroxylation of two specific proline residues in this protein, catalyzed by oxygen- and iron-dependent enzymes called prolyl hydroxylases (PHDs), marks it for ubiquitination followed by proteasomal degradation (reviewed in reference 8).
Pharmacological inhibition of factors involved in this degradation pathway leads to abundant accumulation of HIF-1α protein. Thus, rather than depriving cells of oxygen, we previously stabilized HIF-1α protein in EBV+ cells in vitro with drugs that block steps in this pathway. Deferoxamine (DFO; Desferal) is an FDA-approved iron chelator that inhibits the activity of the PHDs (reviewed in reference 9). MLN-4924 (pevonedistat) is a specific inhibitor of NEDD8-activating enzyme 1 (NAE1), the first step in the pathway leading to ubiquitination of HIF-1α and some other cellular proteins, including p53; it is currently in phase III clinical trials (reviewed in reference 10).
The tumor suppressor protein p53 is a global regulator of cellular metabolism that is frequently mutated or deleted in human cancers, including in a subset of EBV-associated tumors (reviewed in references 11 and 12). p53 activity is affected by a variety of factors, including the specific DNA sequence to which it is bound, the coregulatory factors with which it is complexed, and the posttranslational modifications present on the protein (reviewed in reference 13). Several inducers of EBV reactivation have been shown to require the presence of wild-type (WT) p53 (for examples, see references 14 to 17). However, the mechanism by which p53 activates Zp transcription remained unknown.
In this study, we set out to determine the range of EBV+ cell types induced into lytic EBV replication by incubation with a variety of different types of HIF-α-stabilizing drugs. Unexpectedly, we found that the presence of HIF-1α/HIF-1β (HIF-1α/β) complexes was not sufficient to activate BZLF1 gene expression; rather, phosphorylated, WT p53 was also required, with the latter serving as a coactivator of HRE-bound HIF-1α/β complexes. These findings indicate that lytic-induction therapies involving drugs that stabilize HIF-1α may work efficiently only in EBV+ cancers that contain WT p53; cotreatment with other drugs or procedures (e.g., irradiation) that enhance the level of phosphorylated, WT p53 or convert endogenous mutant p53 to an active form may improve the effectiveness of HIF-1α-stabilizing drugs.
RESULTS
HIF-1α accumulation is not sufficient to induce reactivation of EBV.
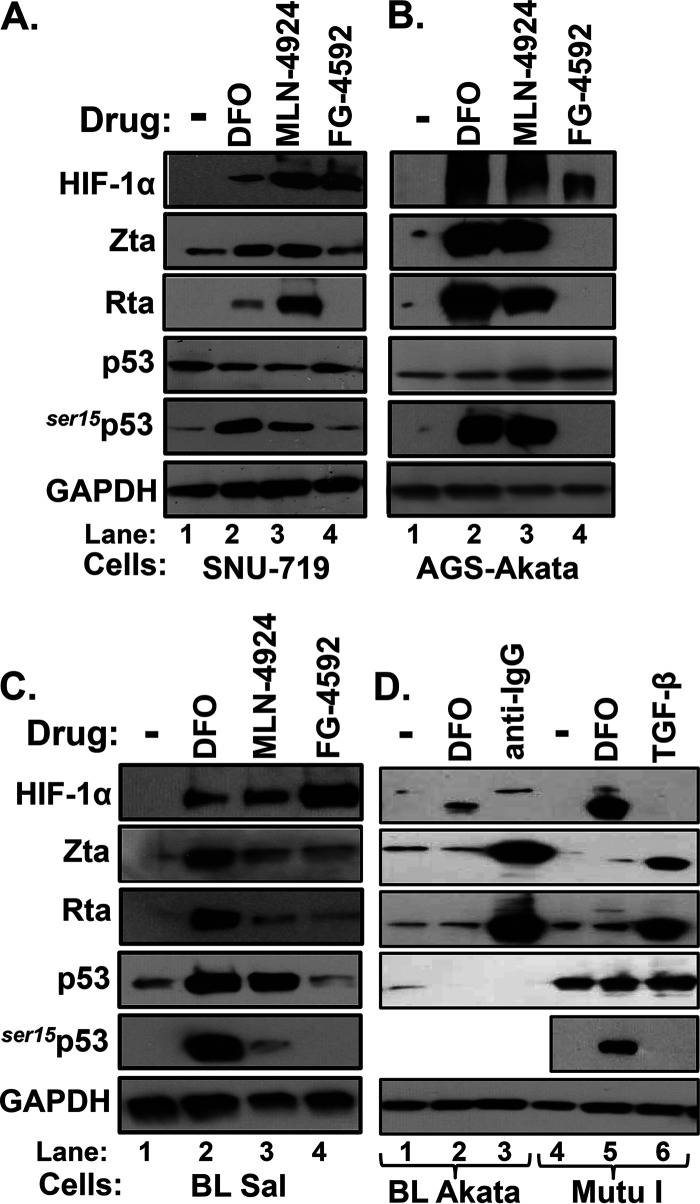
We previously reported that 24-h incubation of the EBV-infected gastric cancer cell lines SNU-719 and AGS-Akata with either DFO or MLN-4924 triggered reactivation of EBV (6), a finding confirmed in this study (Fig. 1A and B). DFO also induced reactivation in the EBV-infected B-cell lines Sal (Fig. 1C) and KemIII (6). Thus, we were surprised to find that DFO failed to induce lytic EBV gene expression in Burkitt lymphoma (BL)-derived Akata and MutuI, other EBV-infected B-cell lines (Fig. 1D). MLN-4924 also failed to induce reactivation within 24 h in Akata and donor 4 lymphoblastoid cell line (LCL) (data not shown); this drug even failed to significantly induce reactivation in Sal cells (Fig. 1C, lane 3 versus lane 1). Nevertheless, as expected, these two drugs stabilized HIF-1α in all of these cell lines (Fig. 1), and EBV reactivation was efficiently induced in Akata and MutuI cells by activation of the B-cell receptor (BCR) or the transforming growth factor β (TGF-β) cellular signaling pathway by incubation with anti-IgG or TGF-β, respectively (Fig. 1D, lanes 3 and 6).
FIG 1.
HIF-1α-stabilizing drugs induce expression of lytic EBV antigens in a cell- and drug-dependent manner. (A to C) Immunoblots showing relative levels of HIF-1α, EBV IE lytic antigens (Zta and Rta), total p53, and p53 phosphorylated on Ser15 (ser15p53) present in SNU-719 (A), AGS-Akata (B), and Sal (C) cells after incubation for 24 h with the drug diluent DMSO (-), 200 μM DFO, 5 μM MLN-4924, or 5 μM FG-4592 as indicated. (D) Immunoblots showing relative levels of the indicated proteins present in BL Akata (lanes 1 to 3) and MutuI (lanes 4 to 6) cells after incubation with the drug diluent PBS (-, 24 h), DFO (200 μM, 24 h), anti-IgG (10 μg/ml, 48 h), or TGF-β (5 μg/ml, 48 h) as indicated. The last two conditions served as positive controls. GAPDH was measured as a loading control. Data are representative of those from two or more independent experiments.
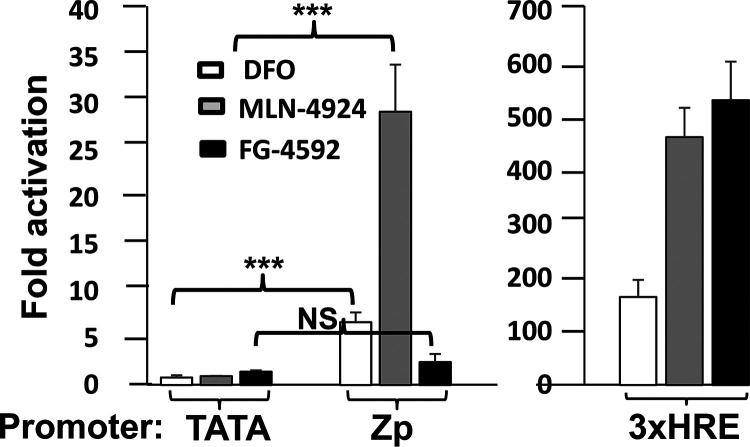
In our search for a highly specific HIF-1α-stabilizing drug that might efficiently induce EBV reactivation, we also tested FG-4592 (roxadustat), a selective inhibitor of PHDs (reviewed in reference 18). Despite stabilizing HIF-1α, FG-4592 failed to induce reactivation in any of the cell lines tested (Fig. 1A to C, lanes 4); it also did not activate transcription from Zp in a reporter assay despite robustly activating transcription from another HRE-containing promoter (Fig. 2). Thus, we conclude that accumulation of HIF-1α is not sufficient to induce lytic EBV reactivation; rather, an additional drug-induced factor(s) or activity is also required.
FIG 2.
Reporter assays showing that FG-4592 can induce transcription from an HRE-containing promoter but not from Zp. AGS cells maintained in 24-well dishes were transfected with 200 ng of DNA/well of (i) pTATA-Luc (a minimal promoter), (ii) p3xHRE-Luc (a reporter plasmid containing three copies of the HRE from the promoter region of the PGK1 gene), or (iii) pZp-Luc. Twenty-four hours later, the drug vehicle (DMSO) or 5 μM FG-4592 was added, and incubation was continued for an additional 24 h prior to harvest and assaying for luciferase activity. Cells were incubated likewise in parallel with DFO (200 μM) and MLN-4924 (5 μM) as positive controls. Data were normalized to the activities observed with the diluent control; they represent means ± SEMs of data obtained from assays performed in triplicate on two independent occasions relative to the values obtained with pTATA-Luc.
Induction of EBV reactivation by HIF-1α-stabilizing drugs correlates with the presence of wild-type, phosphorylated p53.
One hypothesis to explain the above-described findings is that HIF-1α requires a cofactor to induce transcription from Zp: some, but not all, HIF-1α-stabilizing drugs induce accumulation of this cofactor, which may not be present in all cell lines. For example, p53 is known to mediate HRE-directed activation of the VEGFA promoter by HIF-1α/HIF-1β heterodimers (reviewed in reference 19). It is converted into a transcriptionally active cofactor by phosphorylation induced by cellular kinases such as ATM and ATR, including at amino acid residue serine 15 (ser15p53) (reviewed in references 20 and 21). Short-term incubation with DFO induces accumulation of phosphorylated p53 protein (Fig. 1A and B). p53 is known to contribute to EBV reactivation via a variety of lytic inducing agents (16). However, a site to which p53 directly binds in Zp has never been identified, indicating that it may bind indirectly as part of a protein complex, functioning as a coactivator.
Several of our findings are consistent with the hypothesis that phosphorylated, wild-type (WT) p53 protein is the required cofactor for activation of Zp by HIF-1α. First, all four of the cell lines that we previously showed were reactivated by incubation with DFO (SNU-719, AGS-Akata, Sal, and KemIII) (Fig. 1A to C) (6) encode a WT p53 (6, 22). In contrast, DFO failed to reactivate BL Akata cells that make no p53 (23) and MutuI cells that accumulate a mutant variant of p53 (24) (Fig. 1D). Second, we observed an excellent correlation between accumulation of phosphorylated WT p53 and EBV reactivation by the HIF-1α-stabilizing drugs (Fig. 1A to C). For example, whereas DFO and MLN-4924 induced high-level accumulation of both the EBV IE proteins and ser15p53 in AGS-Akata cells, FG-4592 failed to induce accumulation of any of these three proteins (Fig. 1B). Likewise, a 24-h treatment of Sal cells with MLN-4924 induced high-level accumulation of both HIF-1α and WT p53 protein, but neither ser15p53 nor the two EBV IE proteins, while DFO induced accumulation of all five of these proteins (Fig. 1C, lane 3 versus lane 2, respectively).
p53 is required for HIF-1α-dependent induction of Zp.
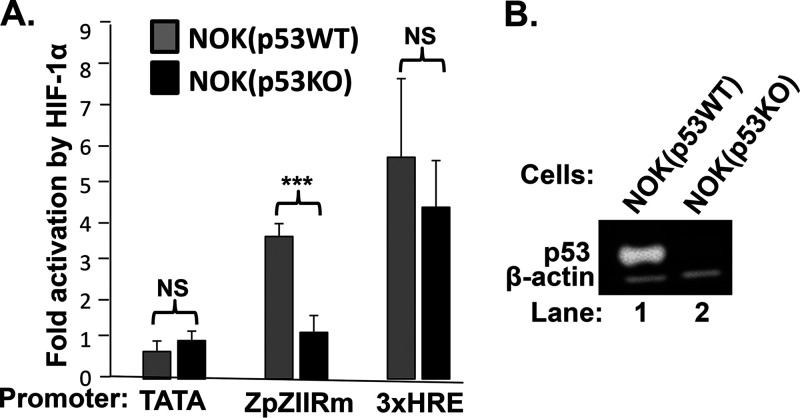
To show directly the requirement of p53 for HIF-1α/β to activate transcription from Zp, we used CRISPR-Cas9 technology to knock out the TP53 genes present in the human normal oral keratinocyte (NOK) cell line, generating NOK(p53KO) cells (Fig. 3B). In reporter assays, activation by HIF-1α/β of Zp, but not 3xHREp, was dependent on the presence of p53 (Fig. 3A). MLN-4924 also failed to activate transcription from Zp in H1299 and HeLa, cell lines in which p53 is missing or largely knocked out by presence of HPV E6 protein, respectively (data not shown). Thus, we conclude that HIF-1α/HIF-1β complexes require the presence of p53 as a cofactor to induce transcription from Zp.
FIG 3.
HIF-1α activation of Zp requires p53. (A) Knockout of the TP53 gene eliminates activation of Zp by HIF-1α/β. NOK(p53WT) and NOK(p53KO) cells were cotransfected in parallel with (i) 50 ng of pcDNA3HA-HIF-1α P402A/P564A (expressing a stable variant of HIF-1α) plus 50 ng of pHIF-1β or 100 ng of pcDNA3 as a normalization control and (ii) 200 ng of the indicated luciferase reporter plasmid. Cells were harvested 48 h later, and luciferase activities were determined. Data were normalized to the activities observed with the pcDNA3 control; they represent means ± SEMs of data obtained from assays performed in triplicate on three independent occasions. A pZp-Luc reporter containing the ZIIR element mutation shown in Fig. 6A was used because of poor transfection efficiency of NOK cells; this mutation has no effect on Zp activation by HIF-1α/β (6). (B) Immunoblots showing relative levels of p53 present in the cells used in the experiment shown in panel A.
p53 is also required for DFO-induced reactivation of EBV.
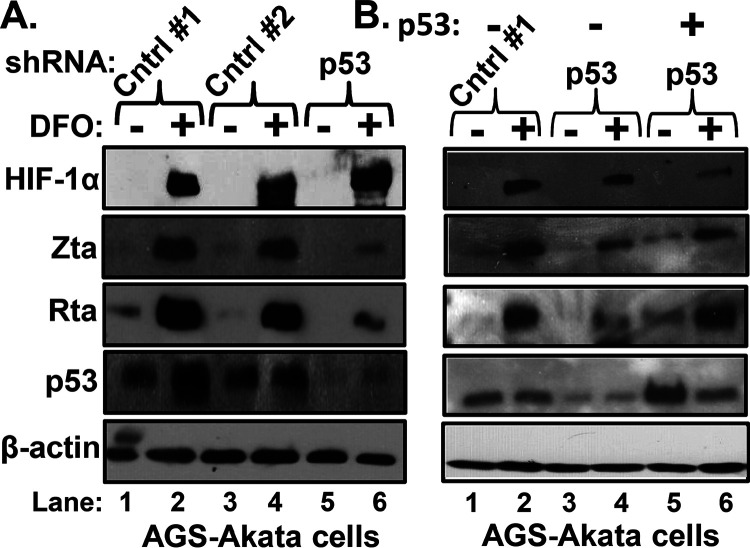
To show that DFO-induced reactivation of EBV requires p53, AGS-Akata cells were transfected with plasmids that express short hairpin RNAs (shRNAs) targeting p53 or scrambled sequences as controls (Cntrl 1 and 2). Two days later, the cells were incubated for 24 h with DFO or its diluent phosphate-buffered saline (PBS). Compared to the control shRNAs, the p53-targeting shRNAs attenuated both p53 and DFO-induced accumulation of lytic EBV antigens by approximately 80% to 90% (Fig. 4A, lane 6 versus lane 2 and lane 4).
FIG 4.
P53 is required for DFO-induced reactivation of EBV. (A) Knockdown of p53 attenuates DFO-directed induction of EBV IE lytic antigens. AGS-Akata cells grown in 10-cm dishes were transduced with lentivirus plasmids that express either of two different nontargeting shRNAs (4 μg/dish; controls 1 and 2) or a mixture of five shRNAs that target p53 (0.8 μg of each one/dish). Two days later, cells were incubated for 24 h in the absence (−) or presence (+) of 200 μM DFO and harvested, and whole-cell extracts were prepared and analyzed for the indicated proteins by immunoblotting. (B) Overexpression of p53 rescues DFO induction of EBV reactivation inhibited by knockdown of p53. AGS-Akata cells were cotransfected with (i) control 1 shRNA or the shRNAs that target p53 as described for panel A and (ii) 100 ng/dish of pcDNA3-WTp53 (lanes 5 and 6) or pcDNA3 (lanes 1 to 4). Two days later, the cells were incubated in the absence or presence of 200 μM DFO for 24 h and processed as described for panel A. Data shown are representative of those from three independent experiments.
To rule out potential off-target effects of the p53 shRNAs, we repeated this experiment while also including a plasmid expressing WT p53 in the transfection mixtures. The presence of this plasmid rescued EBV reactivation by DFO along with p53 expression (Fig. 4B, lane 6). Thus, we conclude that DFO-induced BZLF1 gene expression requires p53 as well as HIF-1α (6).
HIF-1α/β heterodimer recruits p53 to Zp via the HRE.
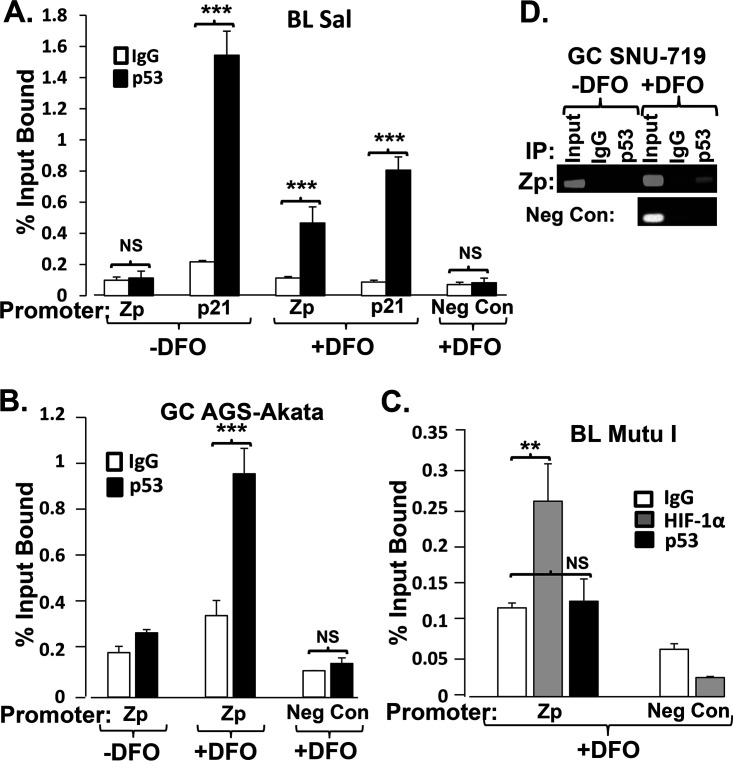
How is p53 required? Does it induce a cellular signaling pathway, leading to synthesis of another factor(s), or act directly in conjunction with HIF-1α/β complexes on Zp? We previously showed that incubation of Sal and SNU-719 cells with DFO promotes binding of HIF-1α to the Zp HRE within the context of whole EBV genomes (6). To determine whether this binding of HIF-1α/β complexes promotes p53 binding to Zp, we performed chromatin immunoprecipitation (ChIP) assays with antibody specific to p53. The promoter region of the cellular CDKN1A gene (p21) served as a positive control for p53 binding, while a region of the EBV genome located 4.8 kbp upstream of Zp served as a negative control (Neg Con). In the absence of DFO, p53 bound the p21 promoter DNA but failed to bind Zp (Fig. 5A). In the presence of DFO, WT p53 bound Zp significantly above the background observed with the IgG control antibody in Sal, AGS-Akata, and SNU-719 cells (Fig. 5A, B, and D, respectively). On the other hand, DFO failed to induce Zp binding of the mutant p53 present in MutuI cells even though it was abundantly present in a phosphorylated form in these cells (Fig. 1D, lane 5) and HIF-1α bound to Zp (Fig. 5C). These findings indicate that DFO induces binding of WT p53 to Zp. Furthermore, HIF-1α binding to Zp can occur independently of p53 binding but is not sufficient to activate transcription from this promoter (Fig. 1D, lane 5), indicating that WT p53 probably serves as its coactivator.
FIG 5.
DFO induces binding of WT, but not mutant, p53 to Zp. (A and B) Quantitative PCR analysis of chromatin obtained from Sal cells (A) and AGS-Akata cells (B) incubated for 24 h with (+) or without (−) 200 μM DFO prior to immunoprecipitation with a p53-specific or anti-IgG control antibody. (C) Quantitative PCR analysis of chromatin obtained from MutuI cells treated for 24 h with 200 μM DFO prior to immunoprecipitation with the above-mentioned antibodies or a HIF-1α-specific antibody. The cellular p21 promoter and a sequence located 4.8 kbp upstream of Zp served as the positive and negative controls (Neg Con) for p53 binding, respectively. Data shown are means ± SEMs of threshold cycle (CT) values from two independent experiments performed in triplicate. (D) Semiquantitative analysis of chromatin obtained from SNU-719 cells incubated as described for Fig. 1A for 24 h with or without 200 μM DFO prior to immunoprecipitation with antibody specific to p53 or anti-IgG antibody. The analysis was performed by 30 cycles of PCR with primers spanning Zp or a region located 4.8 kbp upstream of Zp as a negative control, followed by electrophoresis in a 1.5% agarose gel.
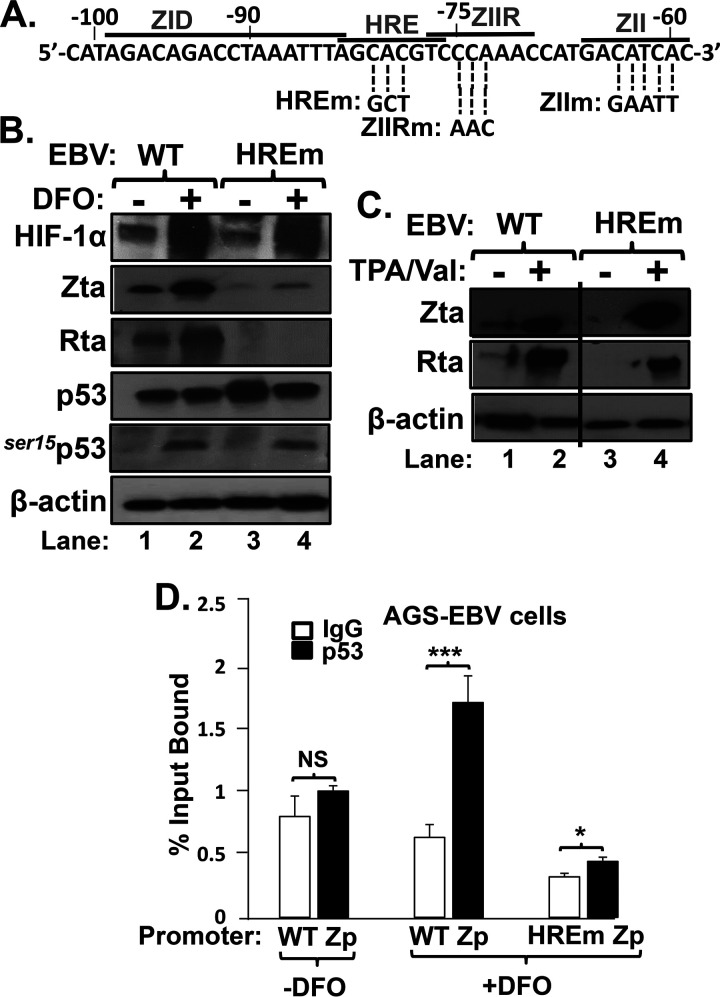
p53 binding to Zp requires the HRE.
To determine whether the HRE mediates p53 binding to Zp, we generated a matched set of AGS-EBV cell lines. EBV-negative AGS cells were infected in parallel with (i) the WT B95.8 strain of EBV present within the p2089 bacterial artificial chromosome (BAC) (25) or (ii) a previously described variant of this EBV BAC containing a 3-bp substitution mutation in the Zp HRE (HREm in Fig. 6A) (6). As expected, DFO induced comparable accumulations of HIF-1α and ser15p53 in the two cell lines but the EBV IE lytic antigens only in the cells containing the WT Zp HRE (Fig. 6B, lane 2 versus lane 4). On the other hand, incubation with the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) plus valproate (TPA/Val) induced these lytic antigens to similar levels in these two cell lines (Fig. 6C). Thus, the failure to reactivate with DFO was due to the HRE mutation. ChIP analysis revealed that p53 bound much better to the Zp containing the WT HRE (Fig. 6D). We conclude that the binding of HIF-1α/β on the Zp HRE enhances binding of p53.
FIG 6.
HRE is required for efficient p53 binding to Zp. (A) Schematic showing the sequence of the nt −102 to nt −59 region of Zp indicating some known cis-acting elements and the base pair substitution mutants in these elements used here. (B) Mutation of the Zp HRE is sufficient to prevent reactivation of EBV by DFO. Immunoblots show the relative levels of the indicated proteins following incubation of AGS-B95.8(WT) versus AGS-B95.8(HREm) cells with 200 μM DFO (+) or its diluent PBS (−) for 24 h. (C) Immunoblots showing that EBV reactivates to similar levels in these two cell lines when incubated for 48 h with a different inducer, TPA (20 ng/ml) plus valproate (1 mM). (D) Quantitative PCR analysis of chromatin obtained from these two cell lines treated with DFO as described for panel B and processed as described in the legend to Fig. 5. Data shown are means ± SEMs of CT values from two independent experiments performed in triplicate.
Phosphorylation of p53 is also required for DFO-induced reactivation of EBV.
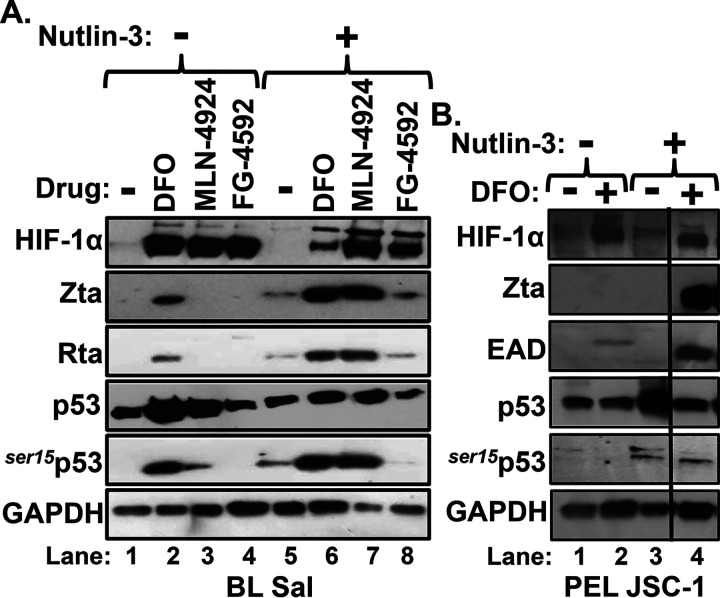
We were initially surprised that a 24-h incubation of Sal cells with MLN-4924 failed to induce significant EBV reactivation given that it induced high-level accumulation of WT p53 as well as HIF-1α (Fig. 1C). The accumulation of p53 was expected because MLN-4924 inhibits proteasomal degradation of both of these proteins as an indirect consequence of inhibiting NEDDylation of Cullin-RING ligases. This finding suggested that appropriate posttranslational modification of p53 to a transcriptionally active form might also be necessary for induction of BZLF1 gene expression by HIF-1α/β. To begin to test this hypothesis, we used nutlin-3, an antagonist of p53’s interaction with the E3 ubiquitin ligase Mdm2, to promote accumulation of phosphorylated p53 (26). Sal cells were pretreated or not for 24 h with nutlin-3, followed by addition of the desired HIF-1α-stabilizing drug. As expected, only DFO induced both high-level phosphorylation of p53 and expression of lytic EBV antigens in the absence of nutlin-3 (Fig. 7A, lane 2 versus lanes 1, 3, and 4). However, in addition to inducing accumulation of phosphorylated p53, nutlin-3 also synergized with MLN-4924 to induce EBV reactivation to significantly higher levels than observed with either drug by itself (Fig. 7A, lane 7 versus lanes 3 and 5, respectively).
FIG 7.
Nutlin-3, an Mdm2 inhibitor, synergizes with DFO and MLN-4924 in inducing reactivation of EBV. (A) Sal cells were incubated for 24 h with or without 2.5 μM nutlin-3, followed by incubation for 24 h with 200 μM DFO, 5 μM MLN-4924, 5 μM FG-4592, or DMSO alone or in combination with nutlin-3 as indicated. Afterward, cells were harvested, and the relative levels of the indicated proteins were determined by immunoblot analyses. (B) JSC-1 cells were incubated and processed likewise with the indicated drugs. The vertical black line indicates a location where an irrelevant lane was deleted.
We also examined a variety of other EBV-positive cell lines for synergistic enhancement of viral reactivation by addition of nutlin-3 (Fig. 7B and data not shown). Interestingly, while DFO usually induced accumulation of phosphorylated p53, it failed to do so in the pleural effusion lymphoma-derived cell line JSC-1 (Fig. 7B, lane 2). In this case, addition of nutlin-3 provided the phosphorylation of p53 needed for HIF-1α to be able to activate BZLF1 gene expression, leading to high-level synthesis of EBV lytic antigens (Fig. 7B, lane 4). Nutlin-3 also enhanced phosphorylation of p53 in AGS-Akata cells, leading to enhanced induction of synthesis of Zta protein by DFO in these cells as well (Fig. 8B, lane 4 versus lane 2). On the other hand, nutlin-3 failed both to alleviate the inability of FG-4592 to induce EBV reactivation and to induce p53 phosphorylation in the presence of FG-4592, indicating that FG-4924 may inhibit the latter activity (Fig. 7A, lane 8). We conclude that nutlin-3-induced phosphorylation of p53 enhances EBV reactivation induced by HIF-1α-stabilizing drugs.
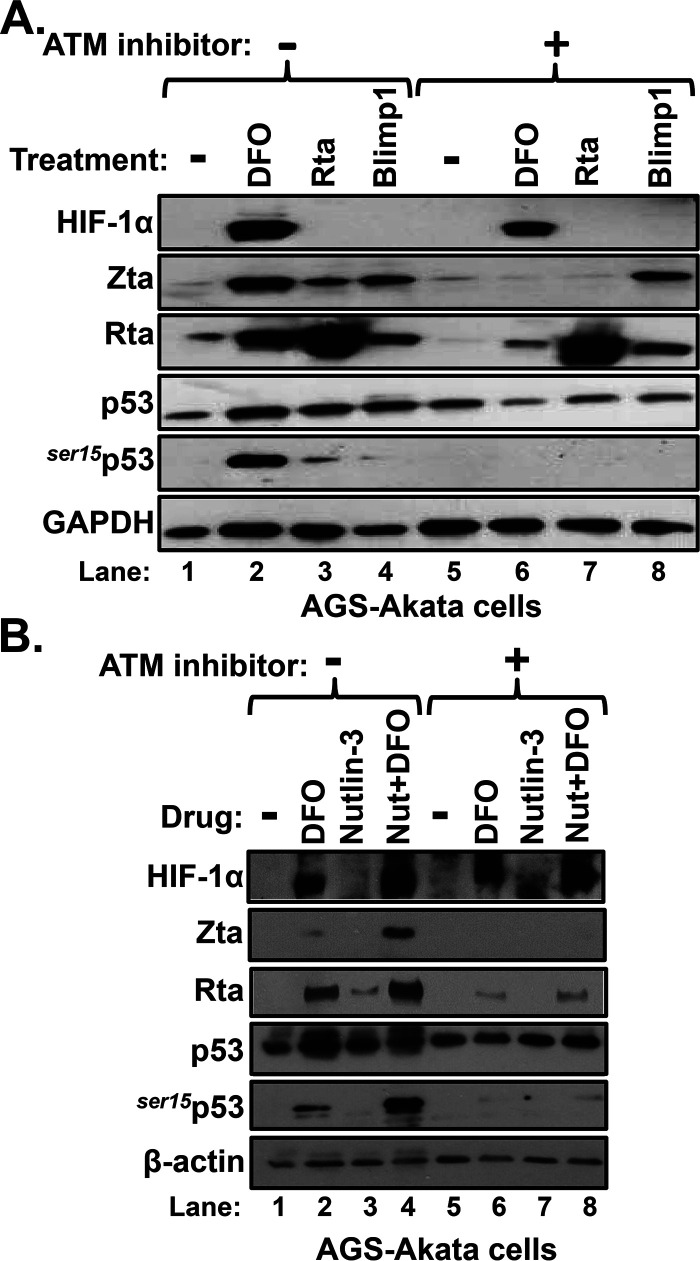
FIG 8.
ATM kinase is also required for DFO-induced reactivation of EBV. (A) AGS-Akata cells grown in 10-cm dishes were incubated for 24 h with or without 10 μM KU-55933, an ATM inhibitor, prior to incubation as well with 200 μM DFO (lanes 2 and 6) or PBS (-; lanes 1 and 5) or transfection with 1.0 μg/dish of pcDNA3-BRLF1 (lanes 3 and 7) or pcDNA3FLAG-BLIMP1 (lanes 4 and 8). Twenty-four hours later, whole-cell extracts were prepared and processed for immunoblot analysis for the indicated proteins. The data shown are representative of those from three independent experiments. (B) ATM kinase is also required for synergistic reactivation of EBV by nutlin-3 plus DFO. AGS-Akata cells were incubated for 24 h with or without 10 μM KU-55933. Concurrently, the cells were incubated with 1.0 μM nutlin-3 where indicated. Afterward, 200 μM DFO or DMSO was added, and incubation was continued for an additional 24 h prior to harvesting and processing for immunoblot analysis.
HIF-1α-dependent reactivation of EBV requires ATM kinase.
To confirm the requirement of phosphorylated p53 for HIF-1α-dependent reactivation of EBV, we used KU-55933, a specific inhibitor of ataxia telangiectasia mutated (ATM). ATM kinase phosphorylates p53 in response to DNA damage and other cellular stressors (reviewed in reference 27). We preincubated AGS-Akata cells without or with KU-55933 for 24 h prior to incubation with DFO. As positive and negative controls for effects of KU-55933, we also transfected the cells in parallel with plasmids expressing Rta and Blimp-1α, respectively, other known activators of BZLF1 gene expression (28, 29). While all three agents induced expression of lytic EBV antigens, only DFO promoted accumulation of HIF-1α, as expected (Fig. 8A, lanes 2 to 4). Treatment with KU-55933 abolished both the synthesis of Zta above the background level and phosphorylation of p53 induced by DFO and Rta (Fig. 8A, lane 6 versus lane 2 and lane 7 versus lane 3, respectively). It also abolished activation of BZLF1 gene expression by DFO alone and DFO plus nutlin-3 (Fig. 8B). Interestingly, Blimp-1α induced synthesis of Zta and Rta, but not ser15p53, regardless of the presence of KU-55933 (Fig. 8A, lane 8 versus lane 4), indicating that it can reactivate EBV via a pathway(s) not dependent upon phosphorylation of p53 by ATM kinase. Thus, we conclude that HIF-1α-dependent reactivation of EBV requires phosphorylation of p53.
EBV reactivation by p53 involves cooperation with HIF-1α/β on the Zp HRE.
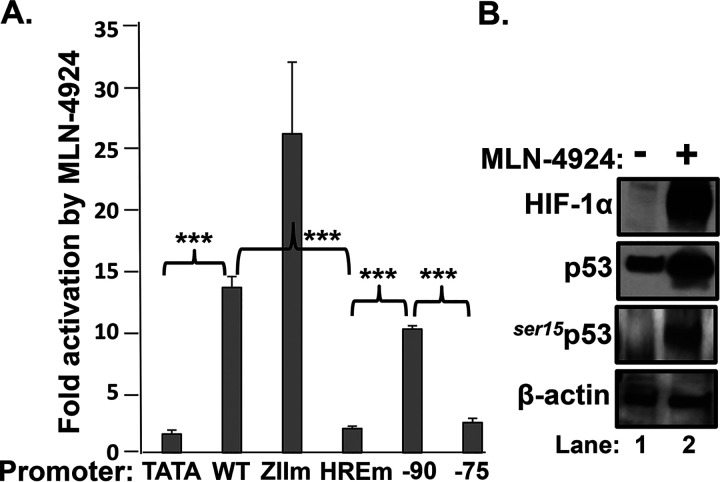
Using luciferase reporter assays, Farhang Ghahremani et al. (30) demonstrated that p53-induced vascular endothelial growth factor (VEGF) expression upon hypoxia exposure requires the binding of both HIF-1α and p53 to their separate HIF and p53 response elements located within the VEGF promoter. To test whether Zp might also contain a p53 response element to which p53 needed to bind for HIF-1α-directed transcription from Zp, we likewise used reporter assays to map the region(s) of Zp required for transcriptional activation of Zp by p53/HIF-1α/β complexes. EBV-negative AGS cells were transfected with variants of the Zp-Luc reporter plasmid containing 5′ deletions or the substitution mutations indicated in Fig. 6A. Twenty-four hours later, MLN-4924 was added, and incubation was continued for another 24 h before assaying for luciferase activity. As expected, the HRE element was required (Fig. 9A), and this drug induced accumulation of both HIF-1α and phosphorylated p53 (Fig. 9B). However, neither the ZID element (largely missing in the variant with the nucleotide [nt] −90 deletion) nor the ZII element was needed for activation by p53/HIF-1α/β. We previously showed that the ZIIR element is also not needed for activation of Zp transcription by HIF-1α/β (6). Thus, we conclude that the p53 requirement for activation of Zp transcription by HIF-1α/β does not map to any nearby known cis-acting element other than the HRE itself; i.e., p53 likely functions as a coactivator component of a p53/HIF-1α/β complex bound to the Zp HRE.
FIG 9.
p53 requirement for activation of Zp transcription by HIF-1α/β maps to the HRE. (A) Reporter assays showing that the HRE is the primary cis-acting element required for Zp activation by MLN-4924. AGS cells maintained in 24-well plates were transfected with 0.20 μg of DNA/well of the indicated luciferase reporter plasmid variants of pZp-Luc. Twenty-four hours later, 5 μM MLN-4924 or DMSO (the drug diluent) was added, and incubation was continued for an additional 24 h prior to harvesting and assaying for luciferase activity. Data were normalized to the values obtained with the vehicle control; they are presented as means ± SEMs from assays performed in triplicate on two independent occasions relative to the values obtained with pTATA-Luc. (B) Immunoblots showing that incubation of AGS cells with 5 μM MLN-4924 for 24 h leads to high-level accumulation of phosphorylated p53 as well as HIF-1α in these cells.
DISCUSSION
We previously reported that HIF-1α induces EBV reactivation via binding to an HRE located within Zp; short-term incubation with iron chelators or a NEDDylation inhibitor can induce lytic EBV gene expression in some EBV-positive epithelial and B-cell lines (6). Others have also recently identified compounds that induce EBV reactivation via iron chelation (31) or effects on Cullin activity (17). In this study, we examined the range of EBV+ cell lines reactivated by DFO and MLN-4924. Importantly, we discovered that phosphorylated, WT p53 serves as an essential cofactor for activation of BZLF1 gene expression by HIF-1α/β (Fig. 1, 4, and 8). Nutlin-3, an Mdm2 inhibitor that induces accumulation of phosphorylated p53 (16, 32), synergized with DFO and MLN-4924 to induce high-level EBV reactivation (Fig. 7). DFO induced binding of WT, but not mutant, p53 protein to Zp (Fig. 5) and only in the presence of an intact HRE (Fig. 6). Furthermore, the requirement for p53 mapped solely to the Zp HRE (Fig. 9). Thus, we conclude that HIF-1α/β induction of BZLF1 gene expression requires phosphorylated, WT p53, with p53 likely functioning as a coactivator for HIF-1α/β complexes on Zp.
Phosphorylated p53 is required for induction of BZLF1 gene expression by HIF-1α/β.
The requirement for phosphorylated, WT p53 for HIF-1α/β-induced reactivation of EBV indicated a need for ATM, one of the cellular kinases that phosphorylates p53. ATM kinase is known to be activated under hypoxic conditions even in the absence of DNA damage (33). However, ATM kinase also phosphorylates HIF-1α at ser696, contributing to its transcriptional activity (34, 35) and, quite possibly, its stability and cellular localization (reviewed in reference 36). There also exists cross talk between p53 and HIF-1α at the level of their abundances via their interactions with Mdm2 (37) and their activities via their interactions with p300 (38).
We initially speculated that appropriate phosphorylation of HIF-1α might be the requirement for activation of Zp transcription by HIF-1α. However, two findings were inconsistent with this hypothesis: DFO induced accumulation of both HIF-1α and phosphorylated mutant p53 in MutuI cells yet failed to reactivate EBV in them (Fig. 1D, lane 5), and FG-4592 induced accumulation of HIF-1α that was transcriptionally active on another promoter, 3xHREp, but induced neither ser15p53 nor transcription from Zp (Fig. 1A to C and Fig. 2). Thus, while ATM kinase-directed phosphorylation of HIF-1α may contribute to its activation of Zp, these posttranslational modifications are not sufficient. Instead, we concluded that phosphorylation of WT p53 was probably the required factor by showing that nutlin-3 enabled MLN-4924- and DFO-induced reactivation of EBV without enhancing the HIF-1α protein level in these cells (Fig. 7).
In some cell lines, DFO and MLN-4924 induced phosphorylation of p53 without the need for nutlin-3 (Fig. 1A to C and Fig. 1A and B, respectively). p53 is known to be phosphorylated by the mitogen-activated protein kinases (MAPKs) p38, Jun N-terminal protein kinases (JNKs), and extracellular signal-regulated kinase 1/2 (ERK1/2), which are themselves activated by phosphorylation (reviewed in reference 39). Phosphorylation of p53 by these drugs likely occurs via several different indirect mechanisms that involve these kinases. For example, iron depletion by incubation with DFO has been shown to lead to increased phosphorylation of p38, JNKs, and ERK1/2, possibly as a consequence of inhibition of the activities of phosphatases that dephosphorylate these kinases (31, 40). Inhibition of ERK1/2 activity was further shown to decrease the ability of iron chelators to induce EBV reactivation (31), providing additional evidence that phosphorylation of p53 is required for activation of Zp by HIF-1α. Likewise, MKK7 phosphorylates JNKs, an activity shown to be reduced by NEDDylation (41). Thus, inhibition of NEDDylation with MLN-4924 may indirectly enhance p53 phosphorylation in part via its effects on MKK7 activity. DFO and MLN-4924 likely also effect phosphorylation of p53 via enhancing accumulation of HIF-1α, with HIF-1α then directly altering the activities of Mdm2, to which p53 also binds (42).
HIF-1α is not the only inducer of BZLF1 gene expression known to require WT p53. Other inducers include hydrogen peroxide (16), histone deacetylase (HDAC) inhibitors (14), and Na, a protein encoded by the EBV BRRF1 gene (15). However, Zp appears to lack any specific p53 response elements. Na activates transcription from Zp via CREB response elements (43). Induction of EBV reactivation by one HDAC inhibitor was shown to involve cooperative interaction of p53 with the cellular transcription factor Sp1, with binding of p53 to Zp requiring the presence of the ZID element (44), a known binding site for Sp1 (45). However, we found that deletion of the ZID element had no effect on MLN-4924-induced activation of Zp transcription by p53/HIF-1α/β complexes (Fig. 9), and binding of p53 induced by DFO mapped to the HRE (Fig. 6). The simplest resolution of these seemingly contradictory findings is that p53 can bind to Zp as a coactivator component of a variety of multiprotein complexes activated via inducers of EBV reactivation; these different complexes include DNA-binding proteins, such as HIF-1α/β, Sp1/Sp3, MEF2s, and AP-1, that sequence-specifically bind to different elements within Zp (reviewed in reference 46).
There also exist inducers of EBV reactivation that require neither WT p53 nor ATM kinase. For example, TGF-β induces EBV reactivation in p53 mutant MutuI cells without activating ATM kinase (as indicated by absence of ser15p53 in Fig. 1D, lane 6); instead, it induces activation of Smad complexes (47, 48). Likewise, Blimp-1α induced EBV IE gene expression even when an ATM inhibitor prevented phosphorylation of p53 (Fig. 8A, lane 8).
HIF-1α/β recruits WT p53 to the Zp HRE.
Involvement of p53 in transcriptional activation by HIF-1α/β is not unique to Zp. For example, induction of VEGFA gene expression induced by acute hypoxia requires both factors. In that case, HIF-1α/β and p53 each bind to their own sequence element located within the VEGFA promoter, with XBP-1s serving as an additional, required factor in the multiprotein complex bound to the HRE (30). In contrast, Zp lacks an identifiable p53 response element, and XBP-1s did not appear to play a role in DFO-induced reactivation of EBV (data not shown). Consistent with these differences, activation of the VEGFA promoter by hypoxia maps to both HREs and p53 response elements (30), while binding of p53 to Zp and transcriptional activation of Zp by p53/HIF-1α/β complexes both mapped to the HRE (Fig. 6 and 9, respectively).
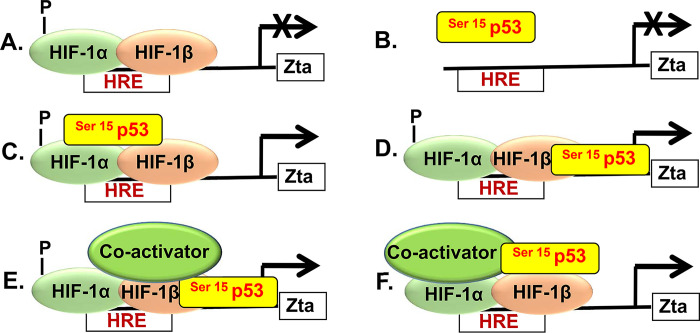
Thus, we propose that the Zp HRE defines a novel class of HREs (Fig. 10). Neither phosphorylated HIF-1α/β by itself (model A) nor HIF-1α/β in complex with XBP-1s (data not shown) can activate transcription from Zp. Phosphorylated p53 by itself is unable to stably bind to and activate Zp (model B). Rather, phosphorylated p53 is recruited to Zp by HIF-1α/β, binding indirectly to Zp in complex with HIF-1α/β (model C) or directly to Zp with its binding stabilized via interaction with HIF-1α/β (model D). Other coactivators, such as p300/CBP, may also be present in the p53/HIF-1α/β complex, further stabilizing the binding of p53 to Zp (models E and F). Given that p53 also induces synthesis of numerous cellular coactivators (reviewed in reference 49), we cannot distinguish among models C to F based upon the data presented here. Among findings of others in support of this model are ones showing coimmunoprecipitation of p53 with HIF-1α after incubation of cells under hypoxic conditions or with DFO or H2O2 (50, 51) and direct binding in vitro of the oxygen-dependent degradation domain (residues 403 to 603) of HIF-1α with p53 tetramers (52), the transcriptionally active form of p53.
FIG 10.
Model for activation of transcription from Zp via p53/HIF complexes. Neither phosphorylated HIF-1α/β nor p53 by itself can activate transcription from Zp (A and B, respectively). Rather, phosphorylated p53 binds indirectly to Zp via HIF-1α/β HIF-1α/β (model C) or directly to Zp, with its binding stabilized via interaction with HIF-1α/β (model D). Other coactivators, such as p300/CBP, may also be present in the p53/HIF-1α/β complex, further stabilizing the direct or indirect binding of p53 to Zp (model E or F, respectively).
Use of HIF-1α-stabilizing drugs in lytic-induction therapy.
A long-term goal of our research is to identify novel classes of drugs for use in lytic-induction therapy for treating patients with a variety of EBV-associated cancer types. Based upon the findings presented here, short-term exposure to drugs that singly or in combination induce accumulation of both HIF-1α and phosphorylated p53 may qualify as good candidates for developing such a therapy. Two of the three HIF-1α-stabilizing drugs examined in this study efficiently induced phosphorylation of p53 within 24 h in most of the cell lines tested. Where needed, other means such as cotreatment with irradiation or nutlin-3 might provide the requirement for p53 phosphorylation.
The cellular gene encoding p53 protein, TP53, is frequently present in a mutant form in cancer cells (12), including in a subset of EBV-positive cancers. Thus, the need for wild-type p53 may limit the EBV-associated cancers in which HIF-α-stabilizing drugs have the potential to work. Fortunately, considerable progress has been made toward the development of drugs that can reverse misfolded oncogenic mutant p53 proteins (reviewed in references 53 to 55), potentially solving this problem. We conclude that HIF-1α-stabilizing drugs, possibly administered in combination with nucleoside analogue prodrugs, such as ganciclovir, that become specifically activated only by herpesvirus-encoded kinases such as the EBV-encoded protein kinase (56), may well turn out to be helpful as part of a lytic-induction therapy for treating many patients with EBV-positive malignancies.
MATERIALS AND METHODS
Cells.
Akata cells (57; gift from Kenzo Takada via Bill Sugden) were derived from an EBV+ Burkitt lymphoma (BL); they retain a type I latency and are p53 negative due to a frameshift mutation and loss of the second allele of the TP53 gene (23). Sal cells (58; gift from Alan Rickinson via Bill Sugden), also derived from an EBV+ BL, are coinfected with WT and EBNA2-deleted EBV genomes; they maintain a Wp-restricted latency (59) and retain WT p53 (6). MutuI cells (60; gift from Alan Rickinson via Shannon Kenney) were also derived from an EBV+ BL; they retain a type I latency and contain a DNA-binding mutant variant of p53 (24). JSC-1 cells (61; obtained from Richard Ambinder via Bill Sugden) were derived from a primary effusion lymphoma (PEL); they are coinfected with both Kaposi’s sarcoma-associated herpesvirus (KSHV) and EBV and retain WT p53 (62). All the above-described cell lines were maintained in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS; HyClone) and 100 U of penicillin and 100 μg of streptomycin (Pen-Strep) (Life Technologies) per ml.
SNU-719 cells (63; gift from Jin-Pok Kim via Bill Sugden) were derived from an EBV+ gastric carcinoma (GC); they retain their original WT EBV genome and WT p53 (6) and were grown in RPMI medium supplemented with 10% FBS and Pen-Strep. Human gastric carcinoma-derived AGS cells (64; obtained from the ATCC) contain WT p53 (22); they were maintained in F12 medium (Life Technologies) supplemented with 10% FBS and Pen-Strep. AGS-Akata/BX1 cells (65; gift from Lindsey Hutt-Fletcher via Shannon Kenney) are AGS cells infected with an Akata strain of EBV that contains genes encoding neomycin resistance and green fluorescent protein under the control of the BXLF1 gene promoter; they were maintained in the above-described medium additionally supplemented with 400 μg/ml of G418. Other latently infected EBV+ derivatives of AGS cells were generated by infection with either the B95.8 strain EBV BAC, p2089 (25), or a variant of it containing a 3-bp substitution mutation in the Zp HRE as previously described (6); they were maintained likewise but additionally supplemented with 100 μg/ml of hygromycin B.
NOK(p53WT) (66; gift from Karl Munger) is a telomerase-immortalized normal oral epithelial keratinocyte line that retains wild-type p53; these cells were maintained in keratinocyte-SFM (K-SFM; Life Technologies, Inc.) supplemented with epidermal growth factor, bovine pituitary extract, and 1% Pen-Strep. NOK(p53KO) cells were derived from NOK(p53WT) cells by CRISPR-Cas9 mutagenesis. In brief, a guide RNA (gRNA) (5′-TCGACGCTAGGATCTGACTG-3′), directed against the second exon of the TP53 gene, was placed into lentiCRISPR v2 (67; gift from Feng Zhang; Addgene plasmid 52961) via double-stranded oligonucleotide insertional mutagenesis. HEK 293FT cells were cotransfected with this plasmid together with psPAX2 and pMD2.G (gifts from Didier Trono [Addgene plasmids 12260 and 12259, respectively]) to generate lentivirus. NOK cells infected with this virus were grown in the above-described K-SFM additionally supplemented with 1.5 μg/ml of puromycin to select for the CRISPR-Cas9-containing cells that were pooled for use.
Plasmids.
Plasmid pcDNA3HA-HIF-1α P402A/P564A-pcDNA3 expresses an amino-terminal hemagglutinin (HA)-tagged, oxygen-insensitive variant of HIF-1α (68); it was obtained from William Kaelin via Addgene (18955). Plasmid pSV-Sport-ARNT (69; gift from Gary Perdew via Christopher Bradfield; called here pHIF-1β) expresses HIF-1β. Plasmid pcDNA3-BLIMP1 (70; gift from Kenneth Wright) expresses an amino-terminal FLAG-tagged version of Blimp-1α. Plasmid pcDNA3-BRLF1 (71; gift from Eric Johannsen) expresses EBV Rta from the cytomegalovirus (CMV) IE promoter. Plasmid pcDNA3-WTp53 (72; Addgene 69003; gift from David Meek) expresses WT p53. The reporter plasmid p3xHRE-Luc (73; Addgene 26731; gift from Navdeep Chandel) contains three copies of the HRE located within the promoter region of the phosphoglycerate kinase 1 (PGK1) gene cloned into pGL2-Basic (Promega). Plasmid pTATA-Luc (74) contains the nt −31 to +31 region relative to the transcriptional start site of the herpes simplex virus thymidine kinase (HSV-TK) gene’s promoter fused to pGL3-Basic (Promega). Plasmid pZpWT-Luc (75; also called pZp-Luc and p-221ZpWT-Luc) contains the nt −221 to +15 region of Zp relative to the transcription initiation site cloned into pGL3-Basic. Plasmids pZpHREm-Luc (6) and pZpZIIRm-Luc (6) are variants of pZpWT-Luc containing the 3-bp substitution mutations in the HRE (6) and ZIIR elements (76, 77) of Zp indicated in Fig. 6A. The plasmid pZpZIIm-Luc, a variant of pZpWT-Luc containing the 5-bp substitution mutations in the ZII element shown in Fig. 6A, was generated by the QuikChange methodology (Stratagene). The 5′-deletion variants of pZpWT-Luc analyzed in Fig. 9 were also generated by the QuikChange methodology.
Chemicals and other agents.
We used three agents known to stabilize HIF-1α: deferoxamine (DFO; Sigma; also called desferrioxamine and Desferal; stock solution prepared in PBS), MLN-4924 (pevonedistat; AdooQ Bioscience; A11260; stock solution prepared in dimethyl sulfoxide [DMSO]), and FG-4592 (roxadustat; AdooQ Bioscience; A11237; stock solution prepared in DMSO). Other agents used were the following: nutlin-3 (Sigma; catalog no. N6287; stock solution prepared in DMSO), KU-55933 (Sigma; SML1109; stock solution prepared in DMSO), recombinant human TGF-β (R&D Systems; 240-B-002), anti-human IgG (Sigma; I5260); 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma; B8319), and valproate (Ben Venue Laboratories, Inc.).
Immunoblot analysis.
Whole-cell extracts were prepared as previously described (6). Proteins were separated by electrophoresis in SDS gels with a 4% to 15% gradient, 7.5% or 10% polyacrylamide (Bio-Rad), and transferred to nitrocellulose membranes (ISC Biosystem). HIF-1α, Zta, BMRF1/EAD, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin were detected as previously reported (6). Rta was detected utilizing a rabbit polyclonal antibody (1:2,000 dilution) generated against the peptide EDPDEETSQAVKALREMAD. To detect p53 or p53 phosphorylated at serine residue 15 (ser15p53), membranes were usually blocked by incubation for 1 h at room temperature in 5% casein in TBST (10 mM Tris-HCl [pH 7.4], 0.15 M NaCl, 0.1% Tween 20), followed by overnight incubation at 4°C in 5% casein in TBST containing a mouse monoclonal antibody specific to p53 (1:1,000; Santa Cruz; DO-1) or ser15p53 (1:1,000; Cell Signaling; 9284), respectively. Afterward, the membranes were washed, incubated for 1 h at room temperature in 5% casein in TBST along with the appropriate corresponding secondary antibody (1:5,000 horseradish peroxidase [HRP]-conjugated goat anti-mouse IgG [Thermo Scientific; G-21040] or 1:5,000 HRP-conjugated donkey anti-rabbit IgG [GE Healthcare; NA-934]), washed again, incubated for 2 min in enhanced chemiluminescence (ECL) (Luminata Crescendo WBLUE0100; Millipore), and exposed to X-ray film (Kodak or GeneMate). In the experiment involving NOK cells, milk replaced casein and the primary p53 antibody was Santa Cruz catalog no. sc-126 (1:200). Afterward, the membrane was incubated with peroxidase-conjugated secondary antibody (1:10,000; Jackson ImmunoResearch; 115-035-003), and the protein was visualized by addition of Clarity ECL reagent with a ChemiDoc MP imaging system (Bio-Rad). All immunoblots shown are representative of two or more independent experiments. All lanes shown for a protein within a figure panel were taken from the same X-ray film, with vertical lines between lanes, when present, indicating where an irrelevant lane(s) was omitted.
Transient transfections.
All transient transfections were performed with TransIT-LT1 transfection reagent according to the manufacturer’s recommendations (Mirus Corp.). Cells were plated in 24-well plates and transfected 24 h later with the amounts of the plasmids indicated in each figure legend. Where indicated, drugs were added 24 h after transfection. Cells were harvested at the indicated times thereafter and lysed with passive lysis buffer (Promega), and luciferase activity was determined according to the manufacturer’s instructions. Plasmid pcDNA3 and drug diluent served as normalization controls for HIF-1α/β and drugs, respectively. All assays were performed in triplicate on two or more occasions.
Knockdown of p53.
AGS-Akata cells grown in 10-cm dishes were cotransfected with 0.8 μg each of five pLKO.1-based lentivirus vector DNAs encoding shRNAs that target WT p53 (Open Biosystems; RHS4533-NM_000546). As controls, cells were transfected with 4 μg of pLKO.1 expressing the nontargeting shRNA 1864 (Cntrl 1; Addgene; 1864) or NT (Cntrl 2; Sigma-Aldrich; SHC002). Forty-eight hours later, cells were incubated for 24 h with 200 μM DFO where indicated, and whole-cell extracts were prepared and probed for the indicated proteins by immunoblot analysis. Where indicated, we also cotransfected the cells with 100 ng/dish of pcDNA3-WTp53 or pcDNA3.
Chromatin immunoprecipitation assays.
ChIP assays were performed with Sal, SNU-719, AGS-Akata, and MutuI cells as previously described (6), but with substitution of 2 μg of mouse anti-p53 antibody (Santa Cruz Biotechnology; DO-1) for 2 μg of mouse anti-HIF-1α antibody (Abcam; ab8366) where indicated and with 2 μg of mouse anti-IgG antibody (Santa Cruz; sc-2025) as a background control. PCR and quantitative PCR (qPCR) analyses were performed as indicated with the previously described Zp and negative-control primer sets, respectively (6). As a positive control for binding of p53, the promoter of the CDKN1A gene (p21) was analyzed utilizing the following primer set: p21 forward, 5′-CTTTCTGGCCGTCAGGAACA-3′, and p21 reverse, 5′-C TTCTATGCCAGAGCTCAACAT-3′. We also performed ChIP analysis with 1 × 107 AGS cells latently infected with either the wild-type version of the p2089 BAC or a variant of it containing a 3-bp substitution mutation in the Zp HRE essentially as described by Bristol et al. (78). In this case, the chromatin was immunoprecipitated with 4 μg of mouse anti-p53 or anti-IgG antibody.
Statistics.
All statistical analyses were performed using Student’s t test (www.physics.csbsju.edu/stats/t-test.html). Statistically significant differences are indicated in figures as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and NS, not significant.
ACKNOWLEDGMENTS
We thank Henri-Jacques Delecluse, Shannon Kenney, Jin-Pok Kim, Karl Munger, Alan Rickinson, and Bill Sugden for cell lines, Christopher Bradfield, Wolfgang Hammerschmidt, Diane Hayward, Eric Johannsen, William Kaelin, Michael Rehli, Bill Sugden, and Ken Wright for plasmids, and members of the Johannsen, Kenney, and Mertz laboratories for suggestions and discussions.
This work was supported by grants PO1 CA22443 and P30 CA14520 from the U.S. National Institutes of Health (to P.F.L. and J.E.M.) and a grant from the National Science and Technology Agency of Thailand (to T.I.).
REFERENCES
- 1.Balfour HH, Dunmire SK, Hogquist KA. 2015. Infectious mononucleosis. Clin Transl Immunology 4:e33. doi: 10.1038/cti.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longnecker RM, Kieff E, Cohen JI. 2013. Epstein-Barr virus, p 1898–1959. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Thorley-Lawson DA. 2015. EBV persistence—introducing the virus. Curr Top Microbiol Immunol 390:151–209. doi: 10.1007/978-3-319-22822-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang M-S, Kieff E. 2015. Epstein-Barr virus latent genes. Exp Mol Med 47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 6.Kraus RJ, Yu X, Cordes B-L, Sathiamoorthi S, Iempridee T, Nawandar DM, Ma S, Romero-Masters JC, McChesney KG, Lin Z, Makielski KR, Lee DL, Lambert PF, Johannsen EC, Kenney SC, Mertz JE. 2017. Hypoxia-inducible factor-1α plays roles in Epstein-Barr virus’s natural life cycle and tumorigenesis by inducing lytic infection through direct binding to the immediate-early BZLF1 gene promoter. PLoS Pathog 13:e1006404. doi: 10.1371/journal.ppat.1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majmundar AJ, Wong WJ, Simon MC. 2010. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semenza GL. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsella M, Borgna-Pignatti C. 2014. Transfusional iron overload and iron chelation therapy in thalassemia major and sickle cell disease. Hematol Oncol Clin North Am 28:703–727, vi. doi: 10.1016/j.hoc.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, Zhang W, Sun Y, Jia L. 2018. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal 44:92–102. doi: 10.1016/j.cellsig.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine AJ, Oren M. 2009. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joerger AC, Fersht AR. 2016. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem 85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 13.Meek DW. 2015. Regulation of the p53 response and its relationship to cancer. Biochem J 469:325–346. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 14.Chang S-S, Lo Y-C, Chua H-H, Chiu H-Y, Tsai S-C, Chen J-Y, Lo K-W, Tsai C-H. 2008. Critical role of p53 in histone deacetylase inhibitor-induced Epstein-Barr virus Zta expression. J Virol 82:7745–7751. doi: 10.1128/JVI.02717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagemeier SR, Barlow EA, Kleman AA, Kenney SC. 2011. The Epstein-Barr virus BRRF1 protein, Na, induces lytic infection in a TRAF2- and p53-dependent manner. J Virol 85:4318–4329. doi: 10.1128/JVI.01856-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagemeier SR, Barlow EA, Meng Q, Kenney SC. 2012. The cellular ataxia telangiectasia-mutated kinase promotes Epstein-Barr virus lytic reactivation in response to multiple different types of lytic reactivation-inducing stimuli. J Virol 86:13360–13370. doi: 10.1128/JVI.01850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tikhmyanova N, Tutton S, Martin KA, Lu F, Kossenkov AV, Paparoidamis N, Kenney S, Salvino JM, Lieberman PM. 2017. Small molecule perturbation of the CAND1-Cullin1-ubiquitin cycle stabilizes p53 and triggers Epstein-Barr virus reactivation. PLoS Pathog 13:e1006517. doi: 10.1371/journal.ppat.1006517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta N, Wish JB. 2017. Hypoxia-inducible factor prolyl hydroxylase inhibitors: a potential new treatment for anemia in patients with CKD. Am J Kidney Dis 69:815–826. doi: 10.1053/j.ajkd.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. 2017. A compendium of proteins that interact with HIF-1α. Exp Cell Res 356:128–135. doi: 10.1016/j.yexcr.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins LMM, Durell SR, Mazur SJ, Appella E. 2012. p53 N-terminal phosphorylation: a defining layer of complex regulation. Carcinogenesis 33:1441–1449. doi: 10.1093/carcin/bgs145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj N, Attardi LD. 2017. The transactivation domains of the p53 protein. Cold Spring Harb Perspect Med 7:a026047. doi: 10.1101/cshperspect.a026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matozaki T, Sakamoto C, Matsuda K, Suzuki T, Konda Y, Nakano O, Wada K, Uchida T, Nishisaki H, Nagao M, Kasuga M. 1992. Missense mutations and a deletion of the p53 gene in human gastric cancer. Biochem Biophys Res Commun 182:215–223. doi: 10.1016/S0006-291X(05)80133-0. [DOI] [PubMed] [Google Scholar]
- 23.Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T. 1991. p53 is frequently mutated in Burkitt’s lymphoma cell lines. EMBO J 10:2879–2887. doi: 10.1002/j.1460-2075.1991.tb07837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiman KG, Magnusson KP, Ramqvist T, Klein G. 1991. Mutant p53 detected in a majority of Burkitt lymphoma cell lines by monoclonal antibody PAb240. Oncogene 6:1633–1639. [PubMed] [Google Scholar]
- 25.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stühmer T, Chatterjee M, Hildebrandt M, Herrmann P, Gollasch H, Gerecke C, Theurich S, Cigliano L, Manz RA, Daniel PT, Bommert K, Vassilev LT, Bargou RC. 2005. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood 106:3609–3617. doi: 10.1182/blood-2005-04-1489. [DOI] [PubMed] [Google Scholar]
- 27.Lakin ND, Jackson SP. 1999. Regulation of p53 in response to DNA damage. Oncogene 18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 28.Ragoczy T, Heston L, Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol 72:7978–7984. doi: 10.1128/JVI.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reusch JA, Nawandar DM, Wright KL, Kenney SC, Mertz JE. 2015. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. J Virol 89:1731–1743. doi: 10.1128/JVI.02781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farhang Ghahremani M, Goossens S, Nittner D, Bisteau X, Bartunkova S, Zwolinska A, Hulpiau P, Haigh K, Haenebalcke L, Drogat B, Jochemsen A, Roger PP, Marine J-C, Haigh JJ. 2013. p53 promotes VEGF expression and angiogenesis in the absence of an intact p21-Rb pathway. Cell Death Differ 20:888–897. doi: 10.1038/cdd.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yiu SPT, Hui KF, Choi CK, Kao RYT, Ma CW, Yang D, Chiang A. 2018. Intracellular iron chelation by a novel compound, C7, reactivates Epstein-Barr virus (EBV) lytic cycle via the ERK-autophagy axis in EBV-positive epithelial cancers. Cancers 10:505. doi: 10.3390/cancers10120505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 33.Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. 2009. ATM activation and signaling under hypoxic conditions. Mol Cell Biol 29:526–537. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol 22:2283–2293. doi: 10.1128/mcb.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cam H, Easton JB, High A, Houghton PJ. 2010. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell 40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kietzmann T, Mennerich D, Dimova EY. 2016. Hypoxia-inducible factors (HIFs) and phosphorylation: impact on stability, localization, and transactivity. Front Cell Dev Biol 4:11. doi: 10.3389/fcell.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. 2000. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev 14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid T, Zhou J, Köhl R, Brüne B. 2004. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1). Biochem J 380:289–295. doi: 10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu GS. 2004. The functional interactions between the p53 and MAPK signaling pathways. Cancer Biol Ther 3:156–161. doi: 10.4161/cbt.3.2.614. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Richardson DR. 2011. Cellular iron depletion stimulates the JNK and p38 MAPK signaling transduction pathways, dissociation of ASK1-thioredoxin, and activation of ASK1. J Biol Chem 286:15413–15427. doi: 10.1074/jbc.M111.225946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu T, Wang J, Pei Y, Wang Q, Wu Y, Qiu G, Zhang D, Lv M, Li W, Zhang J. 2016. Neddylation controls basal MKK7 kinase activity in breast cancer cells. Oncogene 35:2624–2633. doi: 10.1038/onc.2015.323. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, Li M, Luo J, Gu W. 2003. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem 278:13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 43.Hong GK, Delecluse H-J, Gruffat H, Morrison TE, Feng W-H, Sergeant A, Kenney SC. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J Virol 78:4983–4992. doi: 10.1128/jvi.78.10.4983-4992.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chua H-H, Chiu H-Y, Lin S-J, Weng P-L, Lin J-H, Wu S-W, Tsai S-C, Tsai C-H. 2012. p53 and Sp1 cooperate to regulate the expression of Epstein-Barr viral Zta protein. J Med Virol 84:1279–1288. doi: 10.1002/jmv.23316. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Borras AM, Liu P, Suske G, Speck SH. 1997. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 gene promoter. Virology 228:11–18. doi: 10.1006/viro.1996.8371. [DOI] [PubMed] [Google Scholar]
- 46.Kenney SC, Mertz JE. 2014. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol 26:60–68. doi: 10.1016/j.semcancer.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang C-L, Chen J-L, Hsu Y-P, Ou JT, Chang Y-S. 2002. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-β through cooperativity of Smads and c-Jun/c-Fos proteins. J Biol Chem 277:23345–23357. doi: 10.1074/jbc.M107420200. [DOI] [PubMed] [Google Scholar]
- 48.Iempridee T, Das S, Xu I, Mertz JE. 2011. Transforming growth factor beta-induced reactivation of Epstein-Barr virus involves multiple Smad-binding elements cooperatively activating expression of the latent-lytic switch BZLF1 gene. J Virol 85:7836–7848. doi: 10.1128/JVI.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 50.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. 1998. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 51.Parandavar E, Yazdanparast R. 2017. Differential impact of various reactive oxygen species (ROS) on HIF-1α/p53 direct interaction in SK-N-MC neuroblastoma cells. Cell Biosci 7:52. doi: 10.1186/s13578-017-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sánchez-Puig N, Veprintsev DB, Fersht AR. 2005. Binding of natively unfolded HIF-1alpha ODD domain to p53. Mol Cell 17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Babikir HA, Afjei R, Paulmurugan R, Massoud TF. 2018. Restoring guardianship of the genome: anticancer drug strategies to reverse oncogenic mutant p53 misfolding. Cancer Treat Rev 71:19–31. doi: 10.1016/j.ctrv.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Binayke A, Mishra S, Suman P, Das S, Chander H. 2019. Awakening the “guardian of genome”: reactivation of mutant p53. Cancer Chemother Pharmacol 83:1–15. doi: 10.1007/s00280-018-3701-x. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Wang Z, Chen Y, Petersen RB, Zheng L, Huang K. 2019. Salvation of the fallen angel: reactivating mutant p53. Br J Pharmacol 176:817–831. doi: 10.1111/bph.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng Q, Hagemeier SR, Fingeroth JD, Gershburg E, Pagano JS, Kenney SC. 2010. The Epstein-Barr virus (EBV)-encoded protein kinase, EBV-PK, but not the thymidine kinase (EBV-TK), is required for ganciclovir and acyclovir inhibition of lytic viral production. J Virol 84:4534–4542. doi: 10.1128/JVI.02487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takada K. 1984. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int J Cancer 33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 58.Kelly G, Bell A, Rickinson A. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med 8:1098–1104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- 59.Kelly GL, Stylianou J, Rasaiyaah J, Wei W, Thomas W, Croom-Carter D, Kohler C, Spang R, Woodman C, Kellam P, Rickinson AB, Bell AI. 2013. Different patterns of Epstein-Barr virus latency in endemic Burkitt lymphoma (BL) lead to distinct variants within the BL-associated gene expression signature. J Virol 87:2882–2894. doi: 10.1128/JVI.03003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory CD, Rowe M, Rickinson AB. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J Gen Virol 71:1481–1495. doi: 10.1099/0022-1317-71-7-1481. [DOI] [PubMed] [Google Scholar]
- 61.Cannon JS, Ciufo D, Hawkins AL, Griffin CA, Borowitz MJ, Hayward GS, Ambinder RF. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi’s sarcoma herpesvirus-containing supernatant. J Virol 74:10187–10193. doi: 10.1128/jvi.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarek G, Kurki S, Enbäck J, Iotzova G, Haas J, Laakkonen P, Laiho M, Ojala PM. 2007. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest 117:1019–1028. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park JG, Yang HK, Kim WH, Chung JK, Kang MS, Lee JH, Oh JH, Park HS, Yeo KS, Kang SH, Song SY, Kang YK, Bang YJ, Kim YH, Kim JP. 1997. Establishment and characterization of human gastric carcinoma cell lines. Int J Cancer 70:443–449. doi:. [DOI] [PubMed] [Google Scholar]
- 64.Barranco SC, Townsend CM, Quraishi MA, Burger NL, Nevill HC, Howell KH, Boerwinkle WR. 1983. Heterogeneous responses of an in vitro model of human stomach cancer to anticancer drugs. Invest New Drugs 1:117–127. doi: 10.1007/BF00172070. [DOI] [PubMed] [Google Scholar]
- 65.Borza CM, Hutt-Fletcher LM. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med 8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 66.Piboonniyom S, Duensing S, Swilling NW, Hasskarl J, Hinds PW, Münger K. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res 63:476–483. [PubMed] [Google Scholar]
- 67.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan Q, Bartz S, Mao M, Li L, Kaelin WG. 2007. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol 27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai JC, Perdew GH. 1997. Ah receptor nuclear translocator protein heterogeneity is altered after heterodimerization with the Ah receptor. Biochemistry 36:9066–9072. doi: 10.1021/bi970891w. [DOI] [PubMed] [Google Scholar]
- 70.Györy I, Fejér G, Ghosh N, Seto E, Wright KL. 2003. Identification of a functionally impaired positive regulatory domain i binding factor 1 transcription repressor in myeloma cell lines. J Immunol 170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- 71.Heilmann AMF, Calderwood MA, Johannsen E. 2010. Epstein-Barr virus LF2 protein regulates viral replication by altering Rta subcellular localization. J Virol 84:9920–9931. doi: 10.1128/JVI.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loughery J, Cox M, Smith LM, Meek DW. 2014. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res 42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emerling BM, Weinberg F, Liu J-L, Mak TW, Chandel NS. 2008. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a). Proc Natl Acad Sci U S A 105:2622–2627. doi: 10.1073/pnas.0706790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. 2007. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res 5:71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- 75.Kraus RJ, Mirocha SJ, Stephany HM, Puchalski JR, Mertz JE. 2001. Identification of a novel element involved in regulation of the lytic switch BZLF1 gene promoter of Epstein-Barr virus. J Virol 75:867–877. doi: 10.1128/JVI.75.2.867-877.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu P, Liu S, Speck SH. 1998. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J Virol 72:8230–8239. doi: 10.1128/JVI.72.10.8230-8239.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu X, McCarthy PJ, Lim H-J, Iempridee T, Kraus RJ, Gorlen DA, Mertz JE. 2011. The ZIIR element of the Epstein-Barr virus BZLF1 promoter plays a central role in establishment and maintenance of viral latency. J Virol 85:5081–5090. doi: 10.1128/JVI.02615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bristol JA, Djavadian R, Albright ER, Coleman CB, Ohashi M, Hayes M, Romero-Masters JC, Barlow EA, Farrell PJ, Rochford R, Kalejta RF, Johannsen EC, Kenney SC. 2018. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog 14:e1007179. doi: 10.1371/journal.ppat.1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]