Abstract
Many dinoflagellates species, especially of the Alexandrium genus, produce a series of toxins with tremendous impacts on human and environmental health, and tourism economies. Alexandrium tamutum was discovered for the first time in the Gulf of Naples, and it is not known to produce saxitoxins. However, a clone of A. tamutum from the same Gulf showed copepod reproduction impairment and antiproliferative activity. In this study, the full transcriptome of the dinoflagellate A. tamutum is presented in both control and phosphate starvation conditions. RNA-seq approach was used for in silico identification of transcripts that can be involved in the synthesis of toxic compounds. Phosphate starvation was selected because it is known to induce toxin production for other Alexandrium spp. Results showed the presence of three transcripts related to saxitoxin synthesis (sxtA, sxtG and sxtU), and others potentially related to the synthesis of additional toxic compounds (e.g., 44 transcripts annotated as “polyketide synthase”). These data suggest that even if this A. tamutum clone does not produce saxitoxins, it has the potential to produce toxic metabolites, in line with the previously observed activity. These data give new insights into toxic microalgae, toxin production and their potential applications for the treatment of human pathologies.
Keywords: dinoflagellates, Alexandrium tamutum, transcriptomics, toxin producing enzymes, harmful algal blooms
1. Introduction
Harmful algal blooms (HABs) are intense algal proliferations that can profoundly disrupt the habitat where they occur via the production of high biomass accumulation, which causes oxygen depletion and the production of high amounts of toxins [1,2]. HABs induce detrimental effects on marine organisms, humans, environments and economies [1,3]. They are often associated to toxin-producing freshwater cyanobacteria and marine dinoflagellates [4]. Dinoflagellates, together with diatoms, are the main primary producers in aquatic ecosystems. Alexandrium spp. is one of the most common marine dinoflagellates known to be responsible for seasonal HABs; there are more than 40 species, half of which are known to produce toxins [5]. Microalgae are known to produce a series of compounds, generally derived from secondary metabolism, known to regulate and control algal biology, species succession, communication and competition, and predator deterrence [6]. The main toxins produced by Alexandrium spp. are saxitoxins and their analogues (57 natural saxitoxin analogues are known to date [4]), and neurotoxic alkaloids responsible for paralytic shellfish poisoning (PSP), such as neosaxitoxin, gonyautoxin, spirolides and goniodomines [5].
HABs are known to be responsible for shellfish-borne toxicity, fish kills, as well as human intoxications (generally due to bioaccumulation by filter-feeding bivalves and fish, and subsequent transfer through the food web), and even death [5,7,8]. Saxitoxin and its analogues have the ability to bind to voltage-gated sodium, calcium and potassium channels, and lead to death via respiratory paralysis [9]. Algal toxins have been reported to cause 50,000–500,000 human intoxications per year, with an overall global mortality rate of 1.5% [9]. The number of human deaths has stimulated studies on dinoflagellate toxins and their synthesis, which is still not clear, their toxicity on marine organisms and humans, as well as the development of new technologies for toxin detection and HAB monitoring [10]. Toxicity is highly variable between different clones of the same species [11]. This is the case, for example, of A. minutum sampled along the Irish coast, where clones collected on the southern coast were toxic, while clones collected on the western coast were not [12].
Alexandrium tamutum was described for the first time by Montresor and colleagues [13] from the Gulf of Naples, Mediterranean Sea. Their clone was also chemically analysed, and known toxins were not detected, classifying the strain as non-toxic. In addition to the Mediterranean Sea, A. tamutum was also identified in Scottish [14], Malaysian [15] and Chinese waters [16]. The Scottish strain was found negative for PSP toxins as well. Ianora et al. [6] isolated from the Gulf of Naples another clone of A. tamutum, named at that time as A. tamarense FE107, whose ingestion by the copepod Temora stylifera induced reproduction impairment via a dramatic reduction in copepod egg production and hatching success, with egg viability dropping to 0% after 24 h of feeding. Chemical analyses were also performed and toxins were not detected (HPLC analysis excluded a cytotoxicity related to saxitoxins). The FE107 clone was successively characterized by 18S sequencing genotyping and identified as A. tamutum [17]. Lauritano and co-workers also tested the anti-proliferative activity of A. tamutum FE107 on human hepatocellular liver carcinoma (HepG2, ATCC HB-8065TM) after 24 h exposure, as well as on normal human lung fibroblast (MRC-5, ATCC CCL-171TM)), and human melanoma cells (A2058, ATCC CRL-11147TM) after 72 h exposure. Results showed that there was a strong anti-proliferative activity on MRC-5 cells incubated with 50 μg/mL A. tamutum FE107 extract, and on A2058 cells when incubated with 100, 50, 25, 12.5 and 10 μg/mL A. tamutum FE107 extract. On the contrary, 24 h FE107 exposure did not show any effect on HepG2 cells [17].
Considering that A. tamutum showed toxic effects on copepod reproduction and anti-proliferative activity on human cells, it was hypothesized that it may produce some toxic compounds other than saxitoxins which were not detected by the chemical analyses. In this study, the full transcriptome of the dinoflagellate A. tamutum FE107 is reported and in silico identification of transcripts that can be involved in the synthesis of toxic compounds is performed. Until now, various studies have focused on characterizing the saxitoxin biosynthetic pathway [18,19], however, currently available data have not fully identified it. Different studies have used “omics” technologies to shed light on natural product biosynthesis. However, due to their huge genome sizes (3–245 Gbp [4]), molecular resources for dinoflagellates are still scarce [20], and mainly related to transcriptomic data.
Based on bioinformatic predictions merged from different studies, the putative pathway for saxitoxin biosynthesis in cyanobacteria begins with sxtA (with four catalytic domains named sxtA1, sxtA2, sxtA3 and sxtA4), followed by sxtG, sxtB, sxtC, sxtD, sxtS, sxtU, sxtH/T, sxtV and sxtW [4]). Saxitoxin analogues are then synthesized starting from saxitoxin to sxtI, sxtJ, sxtK, sxtL, sxtN, sxtO and sxtX. In addition, there are a series of regulatory genes that allow the regulation of saxitoxin production in response to environmental stimuli and/or modification of the production at the post-transcriptional level [4]. In the transcriptome of various saxitoxin-producing dinoflagellates, several of these genes have also been identified [21,22,23]. Previous studies have shown that not all Alexandrium spp. have the same sxt genes, and differences were mainly assigned comparing toxic versus non-toxic strains. For example, Hackett [19] showed that the C-terminal region of sxtA (referring to the cyanobacterial domain sxtA4) was exclusively found in saxitoxin-producing strains. Zhang et al. [24] found the same domain in the genome of non-toxic mutants of Alexandrium catenella, but it was not found expressed (by both RNA-seq and qPCR analyses). These data suggest that sxtA4 gene is present in the genome but it is not transcribed in the studied conditions. More ambiguous evidence appeared regarding the sxtG gene. This gene, previously considered another marker of saxitoxin-producing strains, was also found in three non-saxitoxin-producing Alexandrium strains (even if the absence in such transcripts of a stop codon could cause its non-functionality) [23]. Moreover, as previously mentioned, a few Alexandrium species also showed the production of other toxins, whose biosynthetic pathways are less known, precisely spirolides (SPXs) (e.g., Alexandrium ostenfeldii, [25]), gymnodimines (GYMs) (e.g., Alexandrium ostenfeldii [26,27]) and goniodomins (e.g., Alexandrium hiranoi [28,29]). Finally, some cases were reported of strains capable of producing an unidentified ichthyotoxin, but not PSP [30], showing that the toxicological profile of Alexandrium spp. still needs to be fully explored.
Considering that Alexandrium spp. have been shown to increase the production of toxins under phosphate starvation (P-starvation) culturing condition [31,32,33], the transcriptome sequencing of A. tamutum cultured under phosphate starvation has also been performed in this study in order to stimulate the transcription of sxt or toxin-related genes. Finally, differential expression analysis between the two conditions (control and P-starvation) was also performed in order to identify (1) sxt genes, (2) metabolic changes induced by P-starvation, and (3) possible other sequences involved in toxin synthesis, or compounds of biotechnological interest.
2. Results and Discussion
2.1. Transcriptome Sequencing and De Novo Assembly
RNA-sequencing (RNA-seq) from six samples, three cultured in control culturing condition and three in P-starvation, yielded 164,128,287 and 62,180,610 total raw and normalized fragments, respectively. Normalised RNA-seq reads were assembled with a de novo approach, because no available reference genome of A. tamutum was available, obtaining a raw assembly of 293,633 transcripts grouped into 220,519 genes. The mean GC content was 65.23%. The average and the median contig length were 776.87 bp and 472 bp, respectively. The N50 was 1202 bp. Controls were performed on the raw transcriptome assembly in order to check for its quality. Transcripts were translated into proteins with Transdecoder, obtaining a total of 182,156 protein sequences (minimum length 50aa). Among these, 26,506 (14.55%) were complete (with a methionine and a stop codon), 12,782 (7.01%) started with a methionine, but lacked a stop codon, 72,875 (40%) only had a stop codon, and 69,993 (38.42%) did not start with a methionine and did not have a stop codon. Protein sequences were blasted against two datasets of the core eukaryotic genes [34] to verify completeness of the assembly, including 248 and 458 protein sequences; respectively 248 out of 248 (100%) and 458 out of 458 (100%) could be detected in the assembly. The assembly protein length was also compared to the length of the core eukaryotic genes. About 460 of the proteins covered more than 90% of the length of the corresponding core eukaryotic proteins and 663 covered more than 80% of the length of the corresponding core eukaryotic protein. Considering that RNA-seq can suffer from the contamination of organisms that are not the target of the sequencing, transcript sequences were blasted against the NCBI database of bacteria and Archaea in order to remove possible contaminations. By this procedure, 3156 transcripts were detected and removed. The distribution of the GC content was also analysed in the dataset; it followed a normal distribution with a mean value of 65.1% and a standard deviation of 3.48. By using a z-test, 60,299 sequences were identified to have a GC content significantly different from the observed mean (p < 0.01). These sequences were blasted against the NCBI (NR) database to look for contaminants, and 803 sequences were removed due to matching with bacterial or metazoan sequences. The obtained filtered assembly was composed by 28,9674 transcripts, grouped into 21,7725 genes. The mean GC content was 65.31%. The average and the median contig lengths were 780.16 bp and 475 bp, respectively, and the N50 was 1206 bp.
2.2. Functional Annotation
The sequences of the assembled transcripts were translated into proteins by using Transdecoder (minimum length of 50aa). If multiple translations were possible, the longest complete ORF was kept, and if a complete ORF was not detected, the longest sequence was maintained. The sequences were also analysed for the presence of repetitive elements (i.e., DNA transposons, retroelements, satellites, rRNA, etc.) with Repeat Masker (version open-4.0.5, [35]) that were then removed. In order to associate a function to the assembled transcripts, Blast2GO software was used. There were 180,552 proteins to be annotated, and 31,286 of which had associated blast hits and GO terms. During the blast step, it was realized that 1294 proteins had bacterial hits. These sequences were removed from downstream analyses. At the end, the obtained filtered assembly was composed by 288,380 transcripts, grouped into 216,911 genes. The mean GC content was 65.31%. The average and the median contig lengths were 778.28 bp and 803 bp, respectively. The N50 was 1204 bp. The final dataset was then translated into proteins (minimum length 50aa) to obtain a total of 179,258 protein sequences. Among these, 26,230 (14.63%) were complete (with a methionine and a stop codon), 12,559 (7.00%) started with a methionine but lacked a stop codon, 71,911 (40.11%) only had a stop codon, and 68,558 (38.24%) did not start with a methionine and did not have a stop codon. Alexandrium tamutum transcriptome assembly statistics are reported in Table 1.
Table 1.
Alexandrium tamutum transcriptome assembly statistics.
| Number of Genes | 216,911 |
|---|---|
| Number of transcripts | 288,380 |
| Percent GC content | 65.31 |
| Contig N50 | 1204 |
| Median contig length | 474 |
| Average contig length | 778.28 |
| Number of proteins | 179,258 |
| Number of complete proteins | 26,230 |
| Number of partial proteins | 153,028 |
2.3. Differential Expression Analysis
Differential expression analysis identified 415 transcripts with significant expression variations (|LogRealFC| > 4; p-value adjusted ≤0.01) in P-starvation conditions relative to control (i.e., A. tamutum cultured in complete K medium). Among the 415 differentially expressed genes (DEGs; of which 231 were up-regulated and 184 down-regulated) 266 transcripts had no NCBI NR assignment (of which 151 were up-regulated and 115 were down-regulated), while the remaining 149 transcripts included 80 up-regulated and 69 down-regulated genes. Functional classification analysis showed that the top GO represented classes among DEGs were ribosomes and membranes as cellular components, oxidation-reduction processes as biological process, and structural constituents of ribosomes, hydrolase activity and binding as molecular function (Figure S1). The full list of DEGs and their log2-fold changes, adjusted P values (padj), and GO annotations are reported in Table S1.
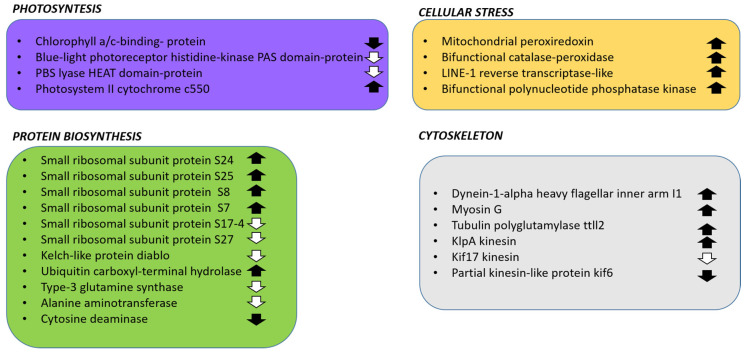
The most up-regulated annotated DEGs were the f-box/wd-40 domain-containing protein cdc4 (padj = 9.55 × 10–5), involved in the recognition and ubiquitination of target proteins [36]); an inosine-5-monophosphate dehydrogenase (padj = 8.79 × 10–5), involved in the guanine biosynthetic pathway [37]; a partial interferon-induced guanylate-binding protein 1-like (padj = 2.08 × 10–4), which is a large GTPase with capability of GMP production [38]. The most down-regulated annotated DEGs were a chloroplast light harvesting complex protein (padj = 3.09 × 10–3); the arginine n-methyltransferase 7 isoform 2 (padj = 4.43 × 10–2), involved in genetic imprinting [39]; a partial heat repeat-containing pbs protein (padj = 1.68 × 10–2), associated with phycobilisome complexes [40]. Major functional groups differentially expressed are related to photosynthesis, protein synthesis, cellular stress responses and cytoskeleton structure and functioning. Figure 1 summarizes these results, while details are reported in the following paragraphs.
Figure 1.
Summary of the main results for differentially expressed genes (DEGs) in the dinoflagellate Alexandrium tamutum cultured in phosphate starvation conditions. Up-regulated transcripts were found related to photosynthesis, cellular stress, protein biosynthesis and cytoskeleton organization biological processes; down-regulated transcripts were found related to photosynthesis, protein biosynthesis and cytoskeleton organization biological processes.
2.3.1. DEGs Involved in Photosynthesis
A number of transcripts involved in photosynthetic activity were differentially expressed in P-starvation: a transcript associated to a chlorophyll a/c-binding protein, a PAS domain protein constituent of the blue-light photoreceptor and a PBS lyase HEAT domain-containing protein were significantly down-regulated (-18.24-, -4.12- and -4.31-fold, respectively), while a transcript of the photosystem II cytochrome c550 was up-regulated (8.12 fold). Chlorophyll a/c-binding protein constitutes a key component of the light-harvesting complex (LHC) [41]. PAS-domain proteins act as cytosolic sensors to overall energy level of the cell [42], and PBS lyases participate to the construction of the light-harvesting phycobilisomes (PBS) [43]. The down-regulation of photosynthesis-related transcripts in P-starvation was also observed in the model diatom Phaeodactylum tricornutum [44], which is in general coherent with the lowered photosynthetic efficiency that was observed in green algae [45], and in higher plants [46]. On the other hand, the redox and photoprotective role of cytochrome c550 [47] can explain its up-regulation.
2.3.2. DEGs Involved in Protein Synthesis
P-starvation induces a drastic proteomic reprogramming of stressed organisms as observed in the marine green alga Micromonas commoda [48]. In the current study, many transcripts associated with amino acid synthesis and ribosomal elements were found differentially expressed. The following transcripts associated to structural proteins belonging to the small ribosomal subunit were found up-regulated: S24 (9.54-fold), S25 (4.24-fold), S8 (7.91-fold), S7 (5.10-fold); proteins S17-4 and S27 were instead down-regulated (−4.61- and −5.43-fold respectively). Similarly, P-starvation culturing condition for the dinoflagellate Karenia brevis induced both over-expression and down-expression of different ribosomal proteins [49]. Specifically, the ribosomal proteins S18, L16 and S4 showed a gradual decrease in expression over the 48 h of the experiment; ribosomal proteins L2, L5 and L15 were strongly down-regulated at 48 h; ribosomal proteins S2, S5 and P0 were down-regulated over all the course of the experiment; finally, ribosomal protein S4 showed a gradual increase in expression. On the contrary, nitrogen starvation did not affect ribosomal proteins in the green alga Botryosphaerella sudeticus [50], while it affected the green alga Nannochloropsis gaditana [51]. However, various studies have also suggested extra-ribosomal functions for ribosomal proteins [52,53], but this is still unknown for microalgae.
In addition, protein degradation was also affected by phosphate deficiency. In fact, a transcript annotated as the Kelch diablo, an adaptor protein involved in the ubiquitination pathway [54], was found down-regulated (-4.48-fold); moreover, a transcript coding a ubiquitin carboxyl-terminal hydrolase, involved in the ubiquitin monomer processing from polyubiquitin chains [55], was found up-regulated (-4.71-fold). Finally, regarding amino acids biosynthesis, two transcripts related to glutamine synthesis called type-3 glutamine synthase (-11.51- and -5.61-fold), a transcript related to alanine aminotransferase (-5.08-fold), and a transcript related to cytosine deaminase (-4.47-fold) were found down-regulated, suggesting an optimization of cellular resources, avoiding the synthesis of certain amino acids in nutrient deficiency.
2.3.3. DEGs Related to Cellular Stress
The overexpression of oxidative stress-response enzymes such as superoxide dismutase (SOD), catalase (CAT) and peroxidases (POD) is often associated with the early response to P-starvation in plants [46]; on the other hand, in microalgae, the expression of these genes in stressful conditions is highly variable and there is no common trend across species [56]. In this study, P-starvation induced the overexpression of transcripts coding for a mitochondrial peroxiredoxin (10.26-fold), a bifunctional catalase-peroxidase (5.34-fold), a LINE-1 reverse transcriptase-like (4.25-fold) and a bifunctional polynucleotide phosphatase kinase (6.42-fold). Peroxiredoxin reduces hydrogen peroxide via the oxidation of a redox-active cysteine residue [57] and its expression levels were up-regulated by light-induced stress in the raphidophyte Chattonella marina [58] and increased in Chlamydomonas reinhardtii in iron starvation conditions [59]. Bifunctional catalase-peroxidase, while mainly found in bacteria [60], was also characterized in some microalgae, such as the dinoflagellate Prorocentrum minimum [61] and the green alga Volvox carteri f. nagariensis (GenBank accession no. XP_002956382).
The LINE-1 reverse transcriptase is associated with LINE-1 retrotransposon propagation [62] and, while not directly involved in stress response, it could have a role in microalgal genetic adaptation to stressful environments, as observed in other eukaryotes [63]. For the same reason, the bifunctional polynucleotide phosphatase kinase could be involved in DNA repair [64], and was found to be over-expressed in A. tamutum.
2.3.4. DEGs Related to Cytoskeleton
While phosphate deficiency is well studied in higher plants, where it causes stunting [50], there is less evidence of the effects of such stressful conditions for microalgae. In this transcriptome, P-starvation induced the up-regulation of transcripts coding for a dynein annotated as (“dynein-1-alpha heavy flagellar inner arm I1”), a myosin G, a tubulin polyglutamylase ttll2 and a klpA kinesin (4.47-, 4.84-, 4.29-, 8.16-fold, respectively). Dyneins are microtubular motor proteins and contain a variable number of heavy chains [65]; furthermore, in the dinoflagellate Prorocentrum donghaiense, 16 dynein heavy chain (DHC) genes were substantially up-regulated [66] in P-starvation, suggesting a promotion of organelle traffic and cellular motility. Myosin G transcripts codes for a motor domain of the actin-based microtubular protein myosin [67]; tubulin polyglutamylase ttll2 is involved in post-transcriptional tubulin polyglutamylation, which regulates the electrostatic interaction between microtubules and MAPs [68]); finally, kplA is a bidirectional microtubule motor protein [69]. P-starvation also induced the down-regulation of transcripts coding a kif17 kinesin (-13.53-fold) and a kif6 kinesin (-4.05-fold); kinesin kif17 is a motor protein involved in intraflagellar transport [70], while the kif6 family is involved in intracellular organelle transport [71]. These transcriptional variations suggest a plasticity of the A. tamutum cytoskeleton.
2.4. Sequences Coding Enzymes Involved in Toxin Synthesis
In order to search in the transcriptome of A. tamutum for sequences related to toxin synthesis, the transcripts related to the saxitoxins biosynthetic pathway found by Hackett and colleagues in A. tamarense CCMP1598 (Group IV) ([19] deposited in GenBank with the accession numbers JV310009-JV310320) were used as queries. Of the 258 contigs found by Hackett et al. (related to the cyanobacteria genes sxtA, sxtB, sxtD, sxtF/M, sxtG, sxtH/T, sxtI, sxtL, sxtN, sxtS, sxtU and sxtX), only 18 sequences related to sxtA, sxtG and sxtU genes were found in the current A. tamutum transcriptome. SxtA is the starting gene of SXT-synthesis in cyanobacteria and has a polyketide synthase (PKS)-like structure. It is characterized by four catalytic domains: S-adenosyl-methionine- (SAM) dependent methyltransferase (sxtA1), which adds a methyl group to acetyl ACP forming propionyl ACP, GCN5-related N-acetyltransferase (sxtA2), acyl carrier protein (sxtA3), and a class II aminotransferase (sxtA4) [18,72]. The sxtA4 domain catalyses a Claisen condensation between arginine and propionyl-ACP, producing the first ‘A’ intermediate [4,72]. Following the reactions catalysed by sxtA, sxtG catalyses the transfer of a guanidine group from a second arginine to the growing SXT backbone forming a ‘B intermediate’. Finally, sxtU encodes for a short-chain alcohol dehydrogenase. This enzyme is thought to reduce the terminal aldehyde group on C1 of the STX precursor (tricyclic alcohol intermediate) in the SXT biosynthetic pathway that was theorized in cyanobacteria and dinoflagellates [73,74]. The 18 matching sequences in the transcriptome are listed in Table 2. Each transcript was further analyzed by using the NCBI Conserved Domain (CDD) database, confirming the identity of the encoded domains found in A. tamarense CCMP1598. In addition, none of the identified sequences showed differential expression rates under P-starvation.
Table 2.
Sxt genes found in A. tamutum transcriptome. For each gene the encoded protein, the relative A. tamarense query from [19], the transcript ID and the best matching NCBI BLAST results are listed.
| Gene | Encoded Protein | Original Sequence Code in Hackett, 2013 | Sequence ID and Internal BLAST Results | NCBI BLAST Best Match and Accession Number |
|---|---|---|---|---|
| sxtA | Phosphopantetheine attachment site (ACP in PKS) | sxtA, N terminus Contig93306 | >TR16074|c0_g1_i1 Length = 3230 Score = 402 bits (203), Expect = e-110 Identities = 443/523 (84%) Strand = Plus/Minus |
Alexandrium catenella strain CS319 sxtA-like (sxtA) gene, partial sequence; KM100452.1 |
| sxtA | Aspartate aminotransferase | sxtA, C terminus Aspartate aminotransferase Contig106704 | >TR2168|c0_g2_i2 Length = 1724 Score = 371 bits (187), Expect = e-101 Identities = 352/407 (86%) Strand = Plus/Plus |
Aminobacter sp. MSH1 chromosome, complete genome; CP026265.1 |
| sxtG | Amidinotransferase | sxtG, Amidinotransferase Contig22175 | >TR24523|c0_g4_i1 Length = 3296 Score = 1292 bits (652), Expect = 0.0 Identities = 1483/1760 (84%) Strand = Plus/Plus |
Ensifer adhaerens strain Casida A plasmid pCasidaAA, complete sequence; CP015881.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig1416 | >TR101104|c3_g5_i1 Length = 1221 Score = 712 bits (359), Expect = 0.0 Identities = 704/819 (85%) Strand = Plus/Minus |
Polyangium brachysporum strain DSM 7029, complete genome; CP011371.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig22159 | >TR119378|c0_g1_i1 Length = 1982 Score = 696 bits (351), Expect = 0.0 Identities = 648/747 (86%) Strand = Plus/Minus |
Sandaracinus amylolyticus strain DSM 53668, complete genome; CP011125.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig22852 | >TR140478|c0_g2_i1 Length = 1469 Score = 381 bits (192), Expect = e-104 Identities = 381/444 (85%) Strand = Plus/Minus |
Emiliania huxleyi CCMP1516 hypothetical protein mRNA; XM_005787014.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig22852 | >TR140478|c0_g1_i1 Length = 1457 Score = 381 bits (192), Expect = e-104 Identities = 381/444 (85%) Strand = Plus/Minus |
Emiliania huxleyi CCMP1516 hypothetical protein mRNA; XM_005787014.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig24779 | >TR3419|c0_g1_i1 Length = 1019 Score = 559 bits (282), Expect = e-158 Identities = 585/686 (85%) Strand = Plus/Minus |
Emiliania huxleyi CCMP1516 hypothetical protein partial mRNA; XM_005768652.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig31067 | >TR32351|c0_g1_i1 Length = 691 Score = 394 bits (199), Expect = e-108 Identities = 415/487 (85%) Strand = Plus/Minus |
Stigmatella aurantiaca DW4/3-1, complete genome; CP002271.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig34277 | >TR63446|c0_g4_i1 Length = 1023 Score = 533 bits (269), Expect = e-150 Identities = 551/645 (85%) Strand = Plus/Plus |
Caulobacter mirabilis strain FWC 38 chromosome, complete genome; CP024201.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig34756 | >TR70482|c0_g1_i1 Length = 1181 Score = 486 bits (245), Expect = e-136 Identities = 587/701 (83%) Strand = Plus/Minus |
Anopheles gambiae str. PEST AGAP008667-RA (AgaP_AGAP008667), partial mRNA; XM_314766.4 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig44170 | >TR142607|c0_g1_i1 Length = 1418 Score = 593 bits (299), Expect = e-168 Identities = 638/751 (84%) Strand = Plus/Minus |
Chromobacterium vaccinii strain XC0014 chromosome, complete genome; CP022344.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig44865 | >TR84807|c0_g1_i1 Length = 1081 Score = 422 bits (213), Expect = e-117 Identities = 450/529 (85%) Strand = Plus/Minus |
Phenylobacterium zucineum HLK1, complete genome; CP000747.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig64321 | >TR80533|c0_g3_i1 Length = 989 Score = 472 bits (238), Expect = e-131 Identities = 490/574 (85%) Strand = Plus/Plus |
Bradyrhizobium diazoefficiens DNA, complete genome, strain: NK6; AP014685.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig64321 | >TR80533|c0_g2_i1 Length = 916 Score = 472 bits (238), Expect = e-131 Identities = 490/574 (85%) Strand = Plus/Plus |
Bradyrhizobium diazoefficiens DNA, complete genome, strain: NK6; AP014685.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig86383 | >TR142098|c0_g1_i2 Length = 1193 Score = 448 bits (226), Expect = e-124 Identities = 427/494 (86%) Strand = Plus/Minus |
PREDICTED: Aegilops tauschii subsp. tauschii momilactone A synthase-like (LOC109773470), mRNA; XM_020332162.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig86383 | >TR142098|c0_g1_i3 Length = 936 Score = 424 bits (214), Expect = e-117 Identities = 424/494 (85%) Strand = Plus/Minus |
PREDICTED: Aegilops tauschii subsp. tauschii momilactone A synthase-like (LOC109773470), mRNA; XM_020332162.1 |
| sxtU | Short-chain alcohol dehydrogenase | sxtU, Short-chain alcohol dehydrogenase Contig97277 | >TR33997|c0_g1_i3 Length = 1012 Score = 555 bits (280), Expect = e-157 Identities = 616/728 (84%) Strand = Plus/Plus |
Stenotrophomonas maltophilia strain AA1, complete genome; CP018756.1 |
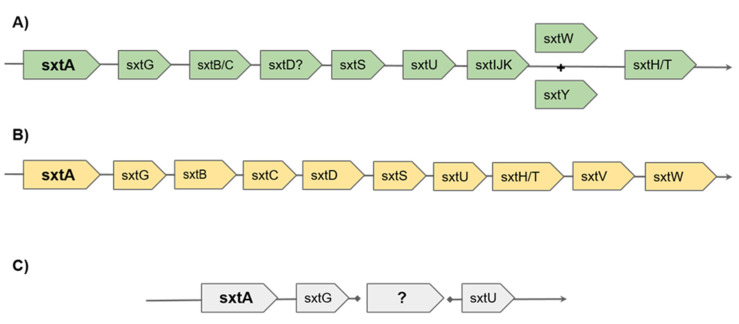
In dinoflagellates, the biosynthesis of SXT involves the genes sxtA, sxtG, sxtB, sxtD, sxtS, sxtU, sxtH/T and sxtI (as recently reviewed by Akbar et al; Figure 2). The absence of transcripts homologous to sxtB, sxtD, sxtS, sxtH/T and sxtI in this transcriptome could explain the absence of SXT detection in the studied clone [6]. However, such data cannot exclude the synthesis of a toxic intermediate. Regarding the expression levels of sxtA, sxtG and sxtU, these genes did not change at different growth stages in A. catenella [75], but were down-regulated in nitrogen and phosphate deficiency in A. minutum [76].
Figure 2.
Sxt biosynthetic pathway: (A) sxt biosynthetic pathway as theorized by Kellmann in the cyanobacteria Cylindrospermopsis raciborskii T3 [72]; (B) biosynthetic pathway as theorized by Zhang et al. [75] and reviewed by Akbar et al. in dinoflagellates [4]; (C) sxt incomplete pathway as observed from the transcripts present in the current A. tamutum transcriptome.
However, Alexandrium spp. are known to harbour intracellular amounts of phosphate [5,77], and many authors observed an increased toxicity and toxin production rate in P-starvation: for example, Han et al. [78] found that P-starvation increased the toxicity of A. pacificum up to 20 times in comparison to nitrogen starvation cultures. In the literature, various studies reported that nutrient starvation induced an increase in toxicity of other microalgae, but such effects vary among species: toxicity increased in nitrogen starvation (N-starvation) in the dinoflagellate Karenia brevis [79], whereas it decreased in the dinoflagellate Protogonyaulax tamarensis and increased dramatically in P-starvation [80]. Similar results have also been observed for other microalgae, such as the diatom Pseudo-nitzschia sp. with increased toxicity when cultured under starvation of different micro- and macro-nutrients [81].
2.5. Structure Prediction of Proteins Encoded by sxt Genes
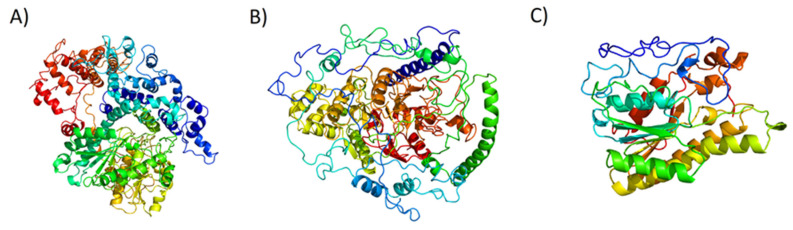
In order to further investigate sxt genes at the protein structure levels, in silico prediction of their three-dimensional structure was performed using the fold recognition approach: the in silico modelling of the three-dimensional structures of the sxt gene products found in the A. tamutum transcriptome were performed by using the PHYRE2 program [82]. The modelled structures had PHYRE2 confidence scores of 100%. The transcript homologous for sxtA C-terminus possessed an incomplete amino acid sequence, and was excluded from further analyses. The results of the in silico modelling are reported in Table 3 and in Figure 3.
Table 3.
Report of PHYRE2 modelling. The A. tamutum sxt genes (and transcript code), template (protein of known structure used for the in silico modelling), its protein data bank (PDB) code, confidence (probability that the sequence and template are homologous), % id (% of identity) and NCBI Conserved Domain (CDD) search output are reported.
| A. tamutum sxt Putative Protein | Template (PDB Code) | Confidence | % id | CDD Search Output |
|---|---|---|---|---|
| sxtA N-Terminus; TR16074|c0_g1_i1 | Methyltransferase (6B3A) | 100% | 24% | Phosphopantetheine attachment site |
| sxtG Amidinotransferase; TR24523|c0_g4_i1 | Arginine deaminase (1RXX) | 100% | 32% | Aminotransferase superfamily; Arginine deiminase |
| sxtU Short-chain alcohol dehydrogenase; TR101104|c3_g5_i1 | Glucose dehydrogenase (1GEE) | 100% | 46% | Rossmann-fold NAD(P)(+)-binding proteins |
Figure 3.
PDB sxt protein models: (A) sxtA N-Terminus; (B) sxtG Amidinotransferase; (C) sxtU Short-chain alcohol dehydrogenase. Individual PDB models are available in Files S1, S2 and S3.
Although the genes involved in sxt biosynthesis have been found in many cyanobacteria and dinoflagellates, there are few three-dimensional protein models. The crystallographic structure of an amidinotransferase annotated as sxtG in the cyanobacterium Microseira wollei has been recently published [83], highly different in amino acid number and sequence from the sxtG found in the present work. To our knowledge, these are the first in silico predictions of three-dimensional structures of sxt gene products in dinoflagellates.
2.6. Other Toxin-Related Transcripts
Other genes putatively related to toxin compound production were annotated in the A. tamutum transcriptome (Table 4). Two transcripts related to aflatoxin metabolism (one annotated as “aflatoxin b1 aldehyde reductase member 4-like”, the other as “aflatoxin b1 aldehyde reductase member 2”), one as “gliotoxin biosynthesis protein, two annotated as “toxicos en levadura”, and three transcripts simply annotated as “toxin biosynthesis protein” were found. These sequences were manually checked on NCBI databases in order to unravel their potential role.
Table 4.
Potential toxin-related genes found in the A. tamutum transcriptome. For each gene, the automatic transcript annotation, the transcript ID, the output of CDD search and their potential function are listed.
| Transcript Automatic Annotation | Transcript ID | CDD Search Output | Potential Function |
|---|---|---|---|
| aflatoxin b1 aldehyde reductase member 2 | TR29768|c0_g1_i1 | Aldo-keto reductase (AKR) | Metabolites detoxification; carbon metabolism |
| aflatoxin b1 aldehyde reductase member 4-like | TR93512|c0_g1_i1 | Aldo-keto reductase (AKR) | Metabolites detoxification; carbon metabolism |
| gliotoxin biosynthesis protein | TR57386|c0_g1_i1 | GGCT-like domain | Metabolites biosynthesis; glutathione homeostasis |
| “toxicos en levadura” | TR119771|c0_g1_i1 | HRD ubiquitin ligase complex | Ubiquitination; triggered immunity |
| “toxicos en levadura” | TR119771|c0_g2_i1 | HRD ubiquitin ligase complex | Ubiquitination; triggered immunity |
| toxin biosynthesis protein | TR47414|c1_g1_i2 | 2OG-Fe(II) oxygenase superfamily | Toxins/Metabolites production |
| toxin biosynthesis protein | TR120505|c0_g1_i1 | 2OG-Fe(II) oxygenase superfamily | Toxins/Metabolites production |
| toxin biosynthesis protein | TR120505|c0_g1_i2 | 2OG-Fe(II) oxygenase superfamily | Toxins/Metabolites production |
Both aflatoxin-related transcripts showed a conserved domain associated to Aldo-keto reductases that reduce aldehydes and ketones to primary and secondary alcohols [84]. Such enzymes are produced by many organisms, with a broad range of substrates and functions (biosynthesis of hormones and small metabolites, detoxification mechanisms), and were suggested as potential chemopreventive agents [85]. Another aldehyde reductase has also been reported in the green algae Nannochloropsis gaditana [86], but to our knowledge this is the first case of an aldehyde reductase of this family in dinoflagellates.
GGCT (gamma-glutamyl cyclotransferase)-like domains in the transcript annotated as “gliotoxin biosynthesis protein” that are involved in the metabolism of different molecules from gamma-glutamyl dipeptides, and in glutathione homeostasis [87] were also found. Such domains have also been reported in the green algae Coccomyxa subellipsoidea [88], but never in a dinoflagellate.
The two “toxicos en levadura” transcripts (that shared a 96% identity) possessed a conserved domain related to the HRD ubiquitin ligase complex (involved in quality control ubiquitination and protein turnover [89], expressed in Arabidopsis thaliana after exposure to chitin or inactivated crude cellulase preparations, probably as part of its triggered immunity (original Gene ID: 820924).
Finally, the three sequences annotated as “toxin biosynthesis protein” possess a domain related to the 2-Oxoglutarate-Fe(II) oxygenase superfamily. In plants, these enzymes are known to be the most versatile in nature, catalysing a large diversity of biologically relevant reactions, including the biosynthesis of toxins and other metabolites [90,91]. Prolyl 4-hydroxylases (P4Hs, catalyses the hydroxylation of peptidyl prolines) belong to this superfamily that was previously characterized in Chlamydomonas reinhardtii [92], but the variety of substrates of these enzymes in microalgae needs further exploration. None of these transcripts showed differential expression under P-starvation. The analyzed sequences, their CDD outputs and potential functions are shown in Table 4.
Moreover, other sequences annotating polyketide synthases (PKS) or nonribosomal peptide synthases (NRPS) were searched in the transcriptome. Both PKS and NRPS produce secondary metabolites (polyketides and peptides, respectively) via their modular enzymatic assembly lines. Their products have already been suggested to be related to antibacterial, antifungal, antipredator, allelopathic and anticancer activities [21,93,94,95]. In the studied transcriptome, a total of 44 transcripts were annotated as “polyketide synthase”, and 5 annotated as “nonribosomal peptide synthetase polyketide synthase hybrid” were found. None of these showed differential expression under P-starvation.
All PKS share different essential domains: acyltransferase domain (AT), β-ketosynthase domain (KS), β-ketoacyl reductase (KR), enoyl reductase (ER), methyl transferases (MT), thioesterases (TE), dehydrogenase (DH) and acyl carrier protein (ACP) domains. NRPS are composed by a variable number of modules, containing an adenylation (A), a peptide acyl-carrier (T), and a condensation (C) domain, plus different tailoring domains [96]. Finally, PKS-NRPS hybrid genes produce peptide–polyketide metabolites, in which polyketides are fused to an amino acid by an amide bond [97].
According to our phylogenetic analysis, most of the transcripts annotated as polyketide synthases were type I PKS (41 out of 44) and clustered together with other dinoflagellate PKS sequences in several highly supported clades (red dots, Figure S2). Despite the fact that type I PKSs are typically multifunctional enzymes organized into modules, each harbouring a set of distinct domains, we only found single domain type I PKSs, as reported for other toxic dinoflagellate studies [98,99,100]. The three remnant PKS transcripts were found in the same clade containing type III PKS sequences (chalcone and stilbene synthases, Figure S2), recently reported in other dinoflagellate transcriptomes [101]. This study constitutes the first report for type III PKSs in A. tamutum. The full list of PKS found in A. tamutum transcriptome is reported in Table S2.
A CDD search of PKS/NRPS hybrid-annotated transcripts evidenced two “Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II”, two “adenylation domain of nonribosomal peptide synthetases (NRPS)” and one “non-ribosomal peptide synthetase component F + Phosphopantetheine attachment site”. PKS genes have been identified in several microalgae (e.g., Azadinium spinosum, Gambierdiscus spp., Amphidinium carterae, Tetraselmis suecica [95,102,103,104]), while metabolites theorized to derive from hybrid NRPS/PKS gene clusters have been reported in Karenia brevis [105]. The wide range of suggested enzymatic reactions and possible natural products that may be produced, make these microalgal species interesting targets for future studies with biotechnological applications [106].
3. Materials and Methods
3.1. Cell Culturing and Harvesting
Alexandrium tamutum (strain FE107 from the Stazione Zoologica culture collection) was cultured in Keller medium (K) [107]. Experimental culturing for both control and P-starvation conditions was performed in 2 litre polycarbonate bottles (each condition was performed in biological triplicates), constantly bubbled with air filtered through 0.2 µm membrane filters. For the control condition, normal K medium was used, while for the P-starvation experiment, the K medium was prepared with low concentrations of phosphate (0.5 μM PO42− rather than 36 μM PO42− of the control condition). Cultures were kept in a climate chamber at 19 °C on a 12:12 h light:dark cycle at 100 µmol photons m−2 s−1. Initial cell concentration for each bottle was about 5000 cells/mL. Culture growth was monitored daily by sampling 2 mL of culture, and fixing with one drop of Lugol (final concentration of about 2%, v/v) and counting cell numbers in a Bürker counting chamber under an Axioskop 2 microscope (20×) (Carl Zeiss GmbH, Oberkochen, Germany) (as in Elagoz et al. [108]). The growth curve of Alexandrium tamutum (in control and P-starvation conditions) is available in Figure S3. Culture aliquots (50 mL) were sampled during the stationary phase on the fifth growth day (on the same day and at the same time of day for each replicate and for each condition, in order to avoid possible interference due to circadian rhythms), and centrifuged for 15 min at 4 °C at 1900× g (Eppendorf, 5810R, Hamburg, Germany). For RNA extractions, pellets (triplicates for each condition) were re-suspended in 500 µL of TRIZOL© (Invitrogen, Carlsbad, CA), incubated for 2–3 min at 60 °C until completely dissolved, and kept at −80°C.
3.2. RNA Extraction
For RNA extraction, A. tamutum cells, previously frozen in TRIZOL®, were lysed using half a spatula of glass beads (about 200 mg; Sigma-Aldrich, Milan, Italy) for each 2 mL tube, incubating and mixing tubes for 10 min at 60 °C, and then at maximum speed in the Thermo Shaker BS100 (Biosan, Rīga, Latvia). The RNA was then extracted following TRIZOL® manufacturer’s instructions. RNA quantity and purity were assessed by Nano-Drop (ND-1000 UV-Vis spectrophotometer; NanoDrop Technologies, Thermo Fisher Scientific, Waltham, MA, USA) measuring the absorbance at 260 nm, and the 260/280 nm and 260/230 nm ratios (both ratios were about 2.0). RNA quality was further evaluated by gel electrophoresis that showed intact RNA, with sharp ribosomal bands. Finally, total RNA quality was evaluated by measuring the RNA integrity number (RIN) with Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). High quality (RIN > 8) RNA was used for RNAseq for both control and P-starvation conditions.
3.3. Library Preparation and Sequencing
RNA sequencing, including sample quality control, was performed by Genomix4life S.R.L. (Baronissi, Salerno, Italy). Indexed libraries were prepared from 2 ug/ea purified RNA with TruSeq Stranded mRNA Sample Prep Kit (Illumina, CA, USA), according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies) and were then pooled so that each index-tagged sample was present in equimolar amounts (final concentration of the pooled samples was 2nM). The pooled samples were subjected to cluster generation and sequencing using an Illumina HiSeq 2500 System (Illumina, CA, USA) in a 2 × 100 paired-end format at a final concentration of 8 pmol. The raw sequence files generated (fastq files) underwent quality control analysis using FastQC [109].
3.4. Transcriptome Assembly and Annotation
Illumina paired-end 100 bp reads were processed to produce the transcriptome assembly. Reads are freely available under the series entry PRJNA632001 in the Sequence Read Archive (SRA) NCBI database [110]. Raw reads were trimmed and clipped with BBDuk [111] setting a minimum Phred-like quality of 25 and a minimum length of 35 nucleotides. The quality of the reads before and after trimming was checked with the software FASTQC [109]. High quality reads were then normalized with Trinity [112] using the options: -SS_lib_type RF -pairs_together -max_cov 50. De novo transcriptome assembly was then performed with Trinity using the options: -SS_lib_type RF -no_normalize_reads -min_kmer_cov 1 -KMER_SIZE 32. Transcriptome redundancy was removed with CD-HIT-EST [113] using the following options: -r 0 -g 1. A filter for contaminants was performed by BLASTing the transcripts against the NCBI nr database, discarding all the sequences having a significant hit (e value ≤ 0.0001) against bacteria or metazoa. The completeness of the assembly was checked against the core eukaryotic genes database [34]. The distribution of the GC content was evaluated with a z-test, performed with the z.test function in R. In silico translation was performed with TransDecoder [114] and the presence of repetitive elements was analysed with Repeat Masker (Statistics in Table S3), while functional annotation was performed with Blast2GO software [115].
3.5. Transcriptome Expression Quantification and Differential Expression Analysis
Transcript expression quantification was performed using Express (v 1.5.1) [116] after mapping the reads against the assembly with STAR [117]. Posterior counts were used as input to perform transcript differential expression analysis with EBSeq [118]. Transcripts with expression variations with |LogRealFC| > 4 and p value adjusted ≤0.01 were considered significant.
3.6. In Silico Protein Modelling
The NCBI CDD database search interface (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was first used to identify the conserved protein domains of the amino acid sequences. PHYRE2 [82] was used for the three-dimensional (3D) in silico protein modelling of the sxt genes: sxtA N-terminus, sxtG and sxtU.
3.7. Phylogenetic Tree
For the transcripts (44) that were annotated as polyketide synthases by B2GO analysis, a phylogenetic tree was inferred to assess their evolutionary relatedness. We included in our analysis type I and type III PKS sequences of other dinoflagellates from the literature (Table S4). Transcripts were aligned using COBALT [119] and poorly aligned regions were removed with trimAl v1.2 [120] using the automated1 option. The final alignment included 64 sequences and 815 aa (File S4). A maximum likelihood phylogenetic tree was then inferred in PhyML [121] using the evolution model (LG+G+F) suggested by Smart Model Selection (SMS) [122]. Support to nodes was calculated using the Shimodaira–Hasegawa-like (aLRT SH-like) procedure [123]. The resulting tree was visualised and graphically edited in FigTree v1.4.3 [124].
4. Conclusions
Dinoflagellates are known to produce various toxins and to give rise to harmful algal blooms, with important consequences for human health and economies. The increase in the frequency and intensity of harmful algal blooms and the toxins these produce has been related to global warming [3]. Hence, there is an increased need to study toxin biosynthetic pathways and the enzymes involved in their metabolism, in order to rapidly detect harmful algal blooms, monitor their distribution and toxicity, contrast these occurrences, and when possible, reduce their detrimental effects.
In this study, we focused on a clone of the dinoflagellate A. tamutum which has previously shown reproductive impairment on crustacean copepods [6] and antiproliferative activities on human cancer cell lines [17], but which did not produce saxitoxins. We nonetheless performed an in silico search of enzymes that can be involved in toxin synthesis (i.e., saxitoxin synthesis-related genes) or other enzymes that can be involved in the synthesis of potentially toxic compounds (e.g., polyketide synthases). Results showed the presence of transcripts related to only three genes of sxt synthesis (i.e., sxtA sxtG and sxtU), but also the presence of other enzymes that can be involved in toxin production (e.g., aldo-keto reductases, 2-Oxoglutarate-Fe(II) oxygenases, PKS and PKS/NRPS-related transcripts), with or without exposure to stressful nutrient starvation.
These data suggest the presence of possible toxic compounds, other than saxitoxins, which may be responsible for the toxic and antiproliferative activity of A. tamutum, as reported for other Alexandrium spp. which produced “uncommon” toxins [28]. Such findings indicate that the chemistry of toxin production in dinoflagellates is very complex and requires in depth new studies in both the ecological and drug discovery fields in order to identify novel chemical mediators in the marine environment and new lead compounds that can be developed as pharmaceuticals. Microalgae have already shown to have several bioactivities for the treatment of human pathologies, such as anticancer, anti-inflammatory, anti-diabetes, antioxidant, anti-tuberculosis, anti-epilepsy, anti-hypertensive, anti-atherosclerosis and anti-osteoporosis activities [95,125,126,127,128]. In particular, Alexandrium minutum has been shown to be active on human lung cancer cells [129], Alexandrium andersoni induces cell death in lung and colorectal tumour cell lines [130], while Alexandrium tamutum was active on human melanoma cell lines [17]. Altogether, these data suggest that Alexandrium spp. may produce metabolites that can have anticancer applications and are worthy of further investigation.
Acknowledgments
The authors thank Servier Medical Art (SMART) website (https://smart.servier.com/) by Servier for the elements of the Graphical Abstract. SMART is licensed under a Creative Commons Attribution 3.0 Unported License. We thank Massimo Perna and Mariano Amoroso for their technical support. We also thank Luca Ambrosino and Marco Miralto from the BIOINforMA group of the Stazione Zoologica Anton Dohrn for technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/8/386/s1, Table S1: Differential Expressed Genes; Table S2: PKS genes; Table S3: RepeatMasker statistics results; Table S4: List of dinoflagellate PKS sequences from the literature utilized for phylogenetic inference. Figure S1: Histograms of GO classifications showing sequence distribution of the annotated differentially expressed genes within the cellular component (A), biological process (B) and molecular function (C). The y-axis indicates the number of sequences for each category; Figure S2: Maximum likelihood phylogenetic tree of transcripts identified as polyketide synthases by Blast 2GO analysis. Support to nodes (coloured circles) was inferred using the Shimodaira–Hasegawa-like test; Figure S3: A. tamutum growth curve; File S1: PDB file of sxtA; File S2: PDB file of sxtG; File S3: PDB file of sxtU; File S4: Alignment of PKS amino acid sequences.
Author Contributions
Conceptualization, C.L.; methodology, C.L.; formal analysis, G.M.V., D.Š., P.D.L., D.D.L., C.L.; data curation, G.M.V., C.L.; writing—original draft preparation, G.M.V., C.L; writing—review and editing, G.M.V., D.Š., P.D.L., A.I., D.D.L., C.L. All authors have read and agreed to the published version of the manuscript.
Funding
G.M.V. was supported by a Stazione Zoologica Ph.D. fellowship via the Open University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Glibert P.M., Berdalet E., Burford M.A., Pitcher G.C., Zhou M. Harmful Algal Blooms and the Importance of Understanding Their Ecology and Oceanography. Springer; Cham, Switzerland: 2018. pp. 9–25. [Google Scholar]
- 2.Díaz P.A., Álvarez G., Varela D., Pérez-Santos I., Díaz M., Molinet C., Seguel M., Aguilera-Belmonte A., Guzmán L., Uribe E., et al. Impacts of harmful algal blooms on the aquaculture industry: Chile as a case study. Perspect. Phycol. 2019;6:39–50. doi: 10.1127/pip/2019/0081. [DOI] [Google Scholar]
- 3.Kudela R., Berdalet E., Enevoldsen H., Pitcher G., Raine R., Urban E. GEOHAB–The Global Ecology and Oceanography of Harmful Algal Blooms Program: Motivation, Goals, and Legacy. Oceanography. 2017;30:12–21. doi: 10.5670/oceanog.2017.106. [DOI] [Google Scholar]
- 4.Akbar M.A., Yusof N.Y.M., Tahir N.I., Ahmad A., Usup G., Sahrani F.K., Bunawan H. Biosynthesis of saxitoxin in marine dinoflagellates: An omics perspective. Mar. Drugs. 2020;18:103. doi: 10.3390/md18020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson D.M., Alpermann T.J., Cembella A.D., Collos Y., Masseret E., Montresor M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae. 2012;14:10–35. doi: 10.1016/j.hal.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ianora A., Guisande C., Fontana A., Romano G., Carotenuto Y., Esposito F., Turner J., d’Ippolito G., Miralto A. Copepod egg production and hatching success is reduced by maternal diets of a non-neurotoxic strain of the dinoflagellate Alexandrium tamarense. Mar. Ecol. Prog. Ser. 2007;280:199–210. doi: 10.3354/meps280199. [DOI] [Google Scholar]
- 7.Etheridge S.M. Paralytic shellfish poisoning: Seafood safety and human health perspectives. Toxicon. 2010;56:108–122. doi: 10.1016/j.toxicon.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Broadwater M.H., Van Dolah F.M., Fire S.E. Harmful Algal Blooms. John Wiley & Sons, Ltd.; Chichester, UK: 2018. Vulnerabilities of Marine Mammals to Harmful Algal Blooms; pp. 191–222. [Google Scholar]
- 9.Cusick K.D., Sayler G.S. An overview on the marine neurotoxin, saxitoxin: Genetics, moleculartargets, methods of detection and ecological functions. Mar. Drugs. 2013;11:991–1018. doi: 10.3390/md11040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Agostino P., Moffitt M., Neilan B. Toxins and Biologically Active Compounds from Microalgae, Volume 1. CRC Press; Boca Raton, FL, USA: 2014. Current Knowledge of Paralytic Shellfish Toxin Biosynthesis, Molecular Detection and Evolution; pp. 251–280. [Google Scholar]
- 11.Martens H., Tillmann U., Harju K., Dell’Aversano C., Tartaglione L., Krock B. Toxin Variability Estimations of 68 Alexandrium ostenfeldii (Dinophyceae) Strains from The Netherlands Reveal a Novel Abundant Gymnodimine. Microorganisms. 2017;5:29. doi: 10.3390/microorganisms5020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Touzet N., Franco J.M., Raine R. Morphogenetic diversity and biotoxin composition of Alexandrium (Dinophyceae) in Irish coastal waters. Harmful Algae. 2008;7:782–797. doi: 10.1016/j.hal.2008.04.001. [DOI] [Google Scholar]
- 13.Montresor M., John U., Beran A., Medlin L.K. Alexandrium tamutum sp. nov. (Dinophyceae): A new nontoxic species in the genus Alexandrium. J. Phycol. 2004;40:398–411. doi: 10.1111/j.1529-8817.2004.03060.x. [DOI] [Google Scholar]
- 14.Collins C., Graham J., Brown L., Bresnan E., Lacaze J.P., Turrell E.A. Identification and toxicity of Alexandrium tamarense (dinophyceae) in Scottish waters. J. Phycol. 2009;45:692–703. doi: 10.1111/j.1529-8817.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- 15.Fong N., Soon K., Hii T., Hie L., Teen P., Pin C. First Record of Marine Dinoflagellate, Alexandrium tamutum (Dinophyceae ) from Malaysia. Malaysian J. Sci. 2013;32:81–88. [Google Scholar]
- 16.Gu H., Zeng N., Liu T., Yang W., Müller A., Krock B. Morphology, toxicity, and phylogeny of Alexandrium (Dinophyceae) species along the coast of China. Harmful Algae. 2013;27:68–81. doi: 10.1016/j.hal.2013.05.008. [DOI] [Google Scholar]
- 17.Lauritano C., Andersen J.H., Hansen E., Albrigtsen M., Escalera L., Esposito F., Helland K., Hanssen K.Ø., Romano G., Ianora A. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016;3:68. doi: 10.3389/fmars.2016.00068. [DOI] [Google Scholar]
- 18.Stüken A., Orr R.J.S., Kellmann R., Murray S.A., Neilan B.A., Jakobsen K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE. 2011;6:e20096. doi: 10.1371/journal.pone.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackett J.D., Wisecaver J.H., Brosnahan M.L., Kulis D.M., Anderson D.M., Bhattacharya D., Gerald Plumley F., Erdner D.L. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 2013;30:70–78. doi: 10.1093/molbev/mss142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauritano C., Ferrante M.I., Rogato A. Marine Natural Products from Microalgae: An -Omics Overview. Mar. Drugs. 2019;17:269. doi: 10.3390/md17050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellmann R., Stüken A., Orr R.J.S., Svendsen H.M., Jakobsen K.S. Biosynthesis and Molecular Genetics of Polyketides in Marine Dinoflagellates. Mar. Drugs. 2010;8:1011–1048. doi: 10.3390/md8041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeling P.J., Burki F., Wilcox H.M., Allam B., Allen E.E., Amaral-Zettler L.A., Armbrust E.V., Archibald J.M., Bharti A.K., Bell C.J., et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray S.A., Diwan R., Orr R.J.S., Kohli G.S., John U. Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates. Mol. Phylogenet. Evol. 2015;92:165–180. doi: 10.1016/j.ympev.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Zhang S.F., Lin L., Wang D.Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs. 2014;12:5698–5718. doi: 10.3390/md12115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillmann U., Kremp A., Tahvanainen P., Krock B. Characterization of spirolide producing Alexandrium ostenfeldii (Dinophyceae) from the western Arctic. Harmful Algae. 2014;39:259–270. doi: 10.1016/j.hal.2014.08.008. [DOI] [Google Scholar]
- 26.Harju K., Koskela H., Kremp A., Suikkanen S., De La Iglesia P., Miles C.O., Krock B., Vanninen P. Identification of gymnodimine D and presence of gymnodimine variants in the dinoflagellate Alexandrium ostenfeldii from the Baltic Sea. Toxicon. 2016;112:68–76. doi: 10.1016/j.toxicon.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 27.Zurhelle C., Nieva J., Tillmann U., Harder T., Krock B., Tebben J. Identification of novel gymnodimines and spirolides from the marine dinoflagellate Alexandrium ostenfeldii. Mar. Drugs. 2018;16 doi: 10.3390/md16110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruana A.M.N., Amzil Z. Microalgae in Health and Disease Prevention. Elsevier; Amsterdam, The Netherlands: 2018. Microalgae and toxins; pp. 263–305. [Google Scholar]
- 29.Harris C.M., Reece K.S., Stec D.F., Scott G.P., Jones W.M., Hobbs P.L.M., Harris T.M. The toxin goniodomin, produced by Alexandrium spp., is identical to goniodomin A. Harmful Algae. 2020;92:101707. doi: 10.1016/j.hal.2019.101707. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y.Z., Kong L., Holmes M.J. Dinoflagellate Alexandrium leei (Dinophyceae) from Singapore coastal waters produces a water-soluble ichthyotoxin. Mar. Biol. 2007;150:541–549. doi: 10.1007/s00227-006-0396-z. [DOI] [Google Scholar]
- 31.Frangópulos M., Guisande C., DeBlas E., Maneiro I. Toxin production and competitive abilities under phosphorus limitation of Alexandrium species. Harmful Algae. 2004;3:131–139. doi: 10.1016/S1568-9883(03)00061-1. [DOI] [Google Scholar]
- 32.Yang I., Beszteri S., Tillmann U., Cembella A., John U. Growth- and nutrient-dependent gene expression in the toxigenic marine dinoflagellate Alexandrium minutum. Harmful Algae. 2011;12:55–69. doi: 10.1016/j.hal.2011.08.012. [DOI] [Google Scholar]
- 33.Sampedro N., Franco J.M., Zapata M., Riobó P., Garcés E., Penna A., Caillaud A., Diogène J., Cacho E., Camp J. The toxicity and intraspecific variability of Alexandrium andersonii balech. Harmful Algae. 2013;25:26–38. doi: 10.1016/j.hal.2013.02.003. [DOI] [Google Scholar]
- 34.Parra G., Bradnam K., Korf I., Bateman A. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 35.Smit A.F., Hubley R., Green P. RepeatMasker Home Page. [(accessed on 25 June 2020)]; Available online: http://www.repeatmasker.org/
- 36.CDC4-Cell division control protein 4-Saccharomyces cerevisiae (strain ATCC 204508/S288c) (Baker’s yeast)-CDC4 gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/P07834.
- 37.IMPDH-Inosine-5’-monophosphate dehydrogenase-Homo sapiens (Human)-IMPDH gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/B4DNJ7.
- 38.GBP1-Guanylate-binding protein 1 precursor-Homo sapiens (Human)-GBP1 gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/P32455.
- 39.PRMT7-Protein arginine N-methyltransferase 7-Homo sapiens (Human)-PRMT7 gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/Q9NVM4.
- 40.PBS lyase HEAT-like repeat (IPR004155)-InterPro entry-InterPro. [(accessed on 18 June 2020)]; Available online: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR004155/
- 41.Hiller R.G., Wrench P.M., Sharples F.P. The light-harvesting chlorophyll a-c-binding protein of dinoflagellates: A putative polyprotein. FEBS Lett. 1995;363:175–178. doi: 10.1016/0014-5793(95)00297-M. [DOI] [PubMed] [Google Scholar]
- 42.Taylor B.L., Zhulin I.B. PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao K.H., Deng M.G., Zheng M., Zhou M., Parbel A., Storf M., Meyer M., Strohmann B., Scheer H. Novel activity of a phycobiliprotein lyase: Both the attachment of phycocyanobilin and the isomerization to phycoviolobilin are catalyzed by the proteins PecE and PecF encoded by the phycoerythrocyanin operon. FEBS Lett. 2000;469:9–13. doi: 10.1016/S0014-5793(00)01245-X. [DOI] [PubMed] [Google Scholar]
- 44.Alipanah L., Winge P., Rohloff J., Najafi J., Brembu T., Bones A.M. Molecular adaptations to phosphorus deprivation and comparison with nitrogen deprivation responses in the diatom Phaeodactylum tricornutum. PLoS ONE. 2018;13:e0193335. doi: 10.1371/journal.pone.0193335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theodorou M.E., Elrifi I.R., Turpin D.H., Plaxton W.C. Effects of phosphorus limitation on respiratory metabolism in the green alga Selenastrum minutum. Plant Physiol. 1991;95:1089–1095. doi: 10.1104/pp.95.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo J., MacPherson C.R., Liu J., Wang H., Kiba T., Hannah M.A., Wang X.J., Bajic V.B., Chua N.H. The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 2012;12:62. doi: 10.1186/1471-2229-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roncel M., Kirilovsky D., Guerrero F., Serrano A., Ortega J.M. Proceedings of the Biochimica et Biophysica Acta-Bioenergetics. Volume 1817. Elsevier; Amsterdam, The Netherlands: 2012. Photosynthetic cytochrome c550; pp. 1152–1163. [DOI] [PubMed] [Google Scholar]
- 48.Guo J., Wilken S., Jimenez V., Choi C.J., Ansong C., Dannebaum R., Sudek L., Milner D.S., Bachy C., Reistetter E.N., et al. Specialized proteomic responses and an ancient photoprotection mechanism sustain marine green algal growth during phosphate limitation. Nat. Microbiol. 2018;3:781–790. doi: 10.1038/s41564-018-0178-7. [DOI] [PubMed] [Google Scholar]
- 49.Morey J.S., Monroe E.A., Kinney A.L., Beal M., Johnson J.G., Hitchcock G.L., Van Dolah F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genom. 2011;12:1–18. doi: 10.1186/1471-2164-12-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D., Zhu J., Fang L., Zhang X., Chow Y., Liu J. De novo transcriptome profiling uncovers a drastic downregulation of photosynthesis upon nitrogen deprivation in the nonmodel green alga Botryosphaerella sudeticus. BMC Genom. 2013;14:1–18. doi: 10.1186/1471-2164-14-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corteggiani Carpinelli E., Telatin A., Vitulo N., Forcato C., D’Angelo M., Schiavon R., Vezzi A., Giacometti G.M., Morosinotto T., Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 2014;7:323–335. doi: 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- 52.De Las Heras-Rubio A., Perucho L., Paciucci R., Vilardell J., Lleonart M.E. Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev. 2014;33:115–141. doi: 10.1007/s10555-013-9460-6. [DOI] [PubMed] [Google Scholar]
- 53.Zhou X., Liao W.J., Liao J.M., Liao P., Lu H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.dbo-Kelch-like protein diablo-Drosophila virilis (Fruit fly)-dbo gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/B4LIG6.
- 55.Larsen C.N., Price J.S., Wilkinson K.D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 56.Cirulis J.T., Scott J.A., Ross G.M. Management of oxidative stress by microalgae. Can. J. Physiol. Pharmacol. 2013;91:15–21. doi: 10.1139/cjpp-2012-0249. [DOI] [PubMed] [Google Scholar]
- 57.Cox A.G., Winterbourn C.C., Hampton M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 58.Mukai K., Shimasaki Y., Qiu X., Kato-Unoki Y., Chen K., Khanam M.R.M., Oshima Y. Effects of light and hydrogen peroxide on gene expression of newly identified antioxidant enzymes in the harmful algal bloom species Chattonella marina. Eur. J. Phycol. 2019;54:1–11. doi: 10.1080/09670262.2019.1576062. [DOI] [Google Scholar]
- 59.Glaesener A.G., Merchant S.S., Blaby-Haas C.E. Iron economy in Chlamydomonas reinhardtii. Front. Plant Sci. 2013;4:337. doi: 10.3389/fpls.2013.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varnado C.L., Hertwig K.M., Thomas R., Roberts J.K., Goodwin D.C. Properties of a novel periplasmic catalase-peroxidase from Escherichia coli O157:H7. Arch. Biochem. Biophys. 2004;421:166–174. doi: 10.1016/j.abb.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 61.Guo R., Ki J.S. Characterization of a novel catalase-peroxidase (KATG) gene from the dinoflagellate Prorocentrum minimum. J. Phycol. 2013;49:1011–1016. doi: 10.1111/jpy.12094. [DOI] [PubMed] [Google Scholar]
- 62.Sciamanna I., De Luca C., Spadafora C. The reverse transcriptase encoded by LINE-1 retrotransposons in the genesis, progression, and therapy of cancer. Front. Chem. 2016;4:6. doi: 10.3389/fchem.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chénais B., Caruso A., Hiard S., Casse N. The impact of transposable elements on eukaryotic genomes: From genome size increase to genetic adaptation to stressful environments. Gene. 2012;509:7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- 64.Pnkp-Bifunctional polynucleotide phosphatase/kinase-Mus musculus (Mouse)-Pnkp gene & protein. [(accessed on 18 June 2020)]; Available online: https://www.uniprot.org/uniprot/Q9JLV6.
- 65.Dynein heavy chain (IPR026983)-InterPro entry-InterPro. [(accessed on 18 June 2020)]; Available online: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR026983/
- 66.Shi X., Lin X., Li L., Li M., Palenik B., Lin S. Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 2017;11:2209–2218. doi: 10.1038/ismej.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartman M.A., Spudich J.A. The myosin superfamily at a glance. J. Cell Sci. 2012;125:1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tubulin-tyrosine ligase/Tubulin polyglutamylase (IPR004344)-InterPro entry-InterPro. [(accessed on 18 June 2020)]; Available online: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR004344/
- 69.Popchock A.R., Tseng K.F., Wang P., Karplus P.A., Xiang X., Qiu W. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat. Commun. 2017;8:13999. doi: 10.1038/ncomms13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Z., Liang Y., Meng D., Wang L., Pan J. Microtubule-Depolymerizing Kinesins in the Regulation of Assembly, Disassembly, and Length of Cilia and Flagella. Int. Rev. Cell Mol. Biol. 2015;317:241–265. doi: 10.1016/bs.ircmb.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 71.KIF6 Gene-GeneCards|KIF6 Protein|KIF6 Antibody. [(accessed on 18 June 2020)]; Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KIF6.
- 72.Kellmann R., Mihali T.K., Young J.J., Pickford R., Pomati F., Neilan B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mihali T.K., Kellmann R., Neilan B.A. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 2009;10:8. doi: 10.1186/1471-2091-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verma A., Barua A., Ruvindy R., Savela H., Ajani P.A., Murray S.A. The Genetic Basis of Toxin Biosynthesis in Dinoflagellates. Microorganisms. 2019;7:222. doi: 10.3390/microorganisms7080222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y., Zhang S.F., Lin L., Wang D.Z. Whole transcriptomic analysis provides insights into molecular mechanisms for toxin biosynthesis in a toxic dinoflagellate Alexandrium catenella (ACHK-T) Toxins (Basel) 2017;9:213. doi: 10.3390/toxins9070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meng F.Q., Song J.T., Zhou J., Cai Z.H. Transcriptomic Profile and Sexual Reproduction-Relevant Genes of Alexandrium minutum in Response to Nutritional Deficiency. Front. Microbiol. 2019;10:2629. doi: 10.3389/fmicb.2019.02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tatters A.O., Van Wagoner R.M., Wright J.L.C., Tomas C.R. Regulation of spiroimine neurotoxins and hemolytic activity in laboratory cultures of the dinoflagellate Alexandrium peruvianum (Balech & Mendiola) Balech & Tangen. Harmful Algae. 2012;19:160–168. [Google Scholar]
- 78.Han M., Lee H., Anderson D.M., Kim B. Paralytic shellfish toxin production by the dinoflagellate Alexandrium pacificum (Chinhae Bay, Korea) in axenic, nutrient-limited chemostat cultures and nutrient-enriched batch cultures. Mar. Pollut. Bull. 2016;104:34–43. doi: 10.1016/j.marpolbul.2016.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ransom Hardison D., Sunda W.G., Wayne Litaker R., Shea D., Tester P.A. Nitrogen limitation increases brevetoxins in Karenia brevis (dinophyceae): Implications for bloom toxicity. J. Phycol. 2012;48:844–858. doi: 10.1111/j.1529-8817.2012.01186.x. [DOI] [PubMed] [Google Scholar]
- 80.Boyer G.L., Sullivan J.J., Andersen R.J., Harrison P.J., Taylor F.J.R. Effects of nutrient limitation on toxin production and composition in the marine dinoflagellate Protogonyaulax tamarensis. Mar. Biol. 1987;96:123–128. doi: 10.1007/BF00394845. [DOI] [Google Scholar]
- 81.Macintyre H.L., Stutes A.L., Smith W.L., Dorsey C.P., Annabraham, Dickey R.W. Environmental correlates of community composition and toxicity during a bloom of Pseudo-nitzschia spp. in the northern Gulf of Mexico. J. Plankton Res. 2011;33:273–295. doi: 10.1093/plankt/fbq146. [DOI] [Google Scholar]
- 82.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lukowski A.L., Mallik L., Hinze M.E., Carlson B.M., Ellinwood D.C., Pyser J.B., Koutmos M., Narayan A.R.H. Substrate Promiscuity of a Paralytic Shellfish Toxin Amidinotransferase. ACS Chem. Biol. 2020;15:626–631. doi: 10.1021/acschembio.9b00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Penning T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015;234:236–246. doi: 10.1016/j.cbi.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ellis E.M., Slattery C.M., Hayes J.D. Characterization of the rat aflatoxin B1 aldehyde reductase gene, AKR7A1. Structure and chromosomal localization of AKR7A1 as well as identification of antioxidant response elements in the gene promoter. Carcinogenesis. 2003;24:727–737. doi: 10.1093/carcin/bgg016. [DOI] [PubMed] [Google Scholar]
- 86.NGA_0683200-Aldehyde reductase i-Nannochloropsis gaditana (strain CCMP526) (Green microalga)-NGA_0683200 gene & protein. [(accessed on 26 June 2020)]; Available online: https://www.uniprot.org/uniprot/K8Z9U8.
- 87.Gamma-glutamyl cyclotransferase-like (IPR013024)-InterPro entry-InterPro. [(accessed on 26 June 2020)]; Available online: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR013024/
- 88.COCSUDRAFT_18395-Gamma-glutamylcyclotransferase-Coccomyxa subellipsoidea (strain C-169) (Green microalga)-COCSUDRAFT_18395 gene & protein. [(accessed on 26 June 2020)]; Available online: https://www.uniprot.org/uniprot/I0YPY5.
- 89.Gardner R.G., Shearer A.G., Hampton R.Y. In vivo action of the HRD ubiquitin ligase complex: Mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farrow S.C., Facchini P.J. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 2014;5:524. doi: 10.3389/fpls.2014.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herr C.Q., Hausinger R.P. Amazing Diversity in Biochemical Roles of Fe(II)/2-Oxoglutarate Oxygenases. Trends Biochem. Sci. 2018;43:517–532. doi: 10.1016/j.tibs.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koski M.K., Hieta R., Böllner C., Kivirikko K.I., Myllyharju J., Wierenga R.K. The active site of an algal prolyl 4-hydroxylase has a large structural plasticity. J. Biol. Chem. 2007;282:37112–37123. doi: 10.1074/jbc.M706554200. [DOI] [PubMed] [Google Scholar]
- 93.Kobayashi J. Amphidinolides and Its Related Macrolides from Marine Dinoflagellates. J. Antibiot. (Tokyo) 2008;61:271–284. doi: 10.1038/ja.2008.39. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J., Du L., Liu F., Xu F., Hu B., Venturi V., Qian G. Involvement of both PKS and NRPS in antibacterial activity in Lysobacter enzymogenes OH11. FEMS Microbiol. Lett. 2014;355:170–176. doi: 10.1111/1574-6968.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lauritano C., De Luca D., Ferrarini A., Avanzato C., Minio A., Esposito F., Ianora A. De novo transcriptome of the cosmopolitan dinoflagellate Amphidinium carterae to identify enzymes with biotechnological potential. Sci. Rep. 2017;7:11701. doi: 10.1038/s41598-017-12092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nielsen M.R., Sondergaard T.E., Giese H., Sørensen J.L. Advances in linking polyketides and non-ribosomal peptides to their biosynthetic gene clusters in Fusarium. Curr. Genet. 2019;65:1263–1280. doi: 10.1007/s00294-019-00998-4. [DOI] [PubMed] [Google Scholar]
- 97.Gallo A., Ferrara M., Perrone G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins (Basel) 2013;5:717–742. doi: 10.3390/toxins5040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Monroe E.A., Van Dolah F.M. The Toxic Dinoflagellate Karenia brevis Encodes Novel Type I-like Polyketide Synthases Containing Discrete Catalytic Domains. Protist. 2008;159:471–482. doi: 10.1016/j.protis.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 99.Pawlowiez R., Morey J.S., Darius H.T., Chinain M., Van Dolah F.M. Transcriptome sequencing reveals single domain Type I-like polyketide synthases in the toxic dinoflagellate Gambierdiscus polynesiensis. Harmful Algae. 2014;36:29–37. doi: 10.1016/j.hal.2014.04.013. [DOI] [Google Scholar]
- 100.Van Dolah F.M., Kohli G.S., Morey J.S., Murray S.A. Both modular and single-domain Type I polyketide synthases are expressed in the brevetoxin-producing dinoflagellate, Karenia brevis (Dinophyceae) J. Phycol. 2017;53:1325–1339. doi: 10.1111/jpy.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Luca D., Lauritano C. In silico identification of type III PKS chalcone and stilbene synthase homologs in marine photosynthetic organisms. Biology (Basel) 2020;9:110. doi: 10.3390/biology9050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lauritano C., De Luca D., Amoroso M., Benfatto S., Maestri S., Racioppi C., Esposito F., Ianora A. New molecular insights on the response of the green alga Tetraselmis suecica to nitrogen starvation. Sci. Rep. 2019;9:3336. doi: 10.1038/s41598-019-39860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meyer J.M., Rödelsperger C., Eichholz K., Tillmann U., Cembella A., McGaughran A., John U. Transcriptomic characterisation and genomic glimps into the toxigenic dinoflagellate Azadinium spinosum, with emphasis on polykeitde synthase genes. BMC Genom. 2015;16:27. doi: 10.1186/s12864-014-1205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kohli G.S., Campbell K., John U., Smith K.F., Fraga S., Rhodes L.L., Murray S.A. Role of Modular Polyketide Synthases in the Production of Polyether Ladder Compounds in Ciguatoxin-Producing Gambierdiscus polynesiensis and G. excentricus (Dinophyceae) J. Eukaryot. Microbiol. 2017;64:691–706. doi: 10.1111/jeu.12405. [DOI] [PubMed] [Google Scholar]
- 105.López-Legentil S., Song B., DeTure M., Baden D.G. Characterization and localization of a hybrid non-ribosomal peptide synthetase and polyketide synthase gene from the toxic dinoflagellate Karenia brevis. Mar. Biotechnol. 2010;12:32–41. doi: 10.1007/s10126-009-9197-y. [DOI] [PubMed] [Google Scholar]
- 106.Vingiani G.M., De Luca P., Ianora A., Dobson A.D.W., Lauritano C. Microalgal Enzymes with Biotechnological Applications. Mar. Drugs. 2019;17:459. doi: 10.3390/md17080459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keller M.D., Selvin R.C., Claus W., Guillard R.R.L. MEDIA FOR THE CULTURE OF OCEANIC ULTRAPHYTOPLANKTON. J. Phycol. 1987;23:633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x. [DOI] [Google Scholar]
- 108.Elagoz A.M., Ambrosino L., Lauritano C. De novo transcriptome of the diatom Cylindrotheca closterium identifies genes involved in the metabolism of anti-inflammatory compounds. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-61007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Babraham Bioinformatics-FastQC A Quality Control tool for High Throughput Sequence Data. [(accessed on 18 June 2020)]; Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 110.Leinonen R., Sugawara H., Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.BBMap download|SourceForge.net. [(accessed on 26 June 2020)]; Available online: https://sourceforge.net/projects/bbmap/
- 112.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li W., Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 114.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conesa A., Götz S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008;2008:1–12. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., Kendziorski C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Papadopoulos J.S., Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 120.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guindon S., Gascuel O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 122.Lefort V., Longueville J.-E., Gascuel O. SMS: Smart Model Selection in PhyML | Molecular Biology and Evolution | Oxford Academic. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 124.FigTree. [(accessed on 26 June 2020)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 125.Martínez Andrade K., Lauritano C., Romano G., Ianora A. Marine Microalgae with Anti-Cancer Properties. Mar. Drugs. 2018;16:165. doi: 10.3390/md16050165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giordano D., Costantini M., Coppola D., Lauritano C., Núñez Pons L., Ruocco N., di Prisco G., Ianora A., Verde C. Advances in Microbial Physiology. Volume 73. Academic Press; Cambridge, MA, USA: 2018. Biotechnological Applications of Bioactive Peptides From Marine Sources; pp. 171–220. [DOI] [PubMed] [Google Scholar]
- 127.Brillatz T., Lauritano C., Jacmin M., Khamma S., Marcourt L., Righi D., Romano G., Esposito F., Ianora A., Queiroz E.F., et al. Zebrafish-based identification of the antiseizure nucleoside inosine from the marine diatom Skeletonema marinoi. PLoS ONE. 2018;13:e0196195. doi: 10.1371/journal.pone.0196195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Riccio G., Lauritano C. Microalgae with immunomodulatory activities. Mar. Drugs. 2020;18:2. doi: 10.3390/md18010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Galasso C., Nuzzo G., Brunet C., Ianora A., Sardo A., Fontana A., Sansone C. The marine dinoflagellate Alexandrium minutum activates a mitophagic pathway in human lung cancer cells. Mar. Drugs. 2018;16:502. doi: 10.3390/md16120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sansone C., Nuzzo G., Galasso C., Casotti R., Fontana A., Romano G., Ianora A. The Marine Dinoflagellate Alexandrium andersonii Induces Cell Death in Lung and Colorectal Tumor Cell Lines. Mar. Biotechnol. 2018;20:343–352. doi: 10.1007/s10126-018-9817-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.