Abstract
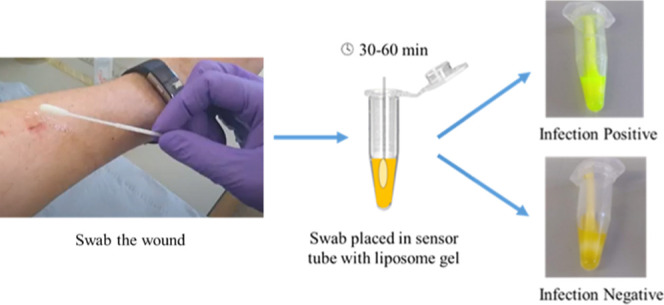
Wound infection is commonly observed after surgery and trauma but is difficult to diagnose and poorly defined in terms of objective clinical parameters. The assumption that bacteria in a wound correlate with infection is false; all wounds contain microorganisms, but not all wounds are clinically infected. This makes it difficult for clinicians to determine true wound infection, especially in wounds with pathogenic biofilms. If an infection is not properly treated, pathogenic virulence factors, such as rhamnolipids from Pseudomonas aeruginosa, can modulate the host immune response and cause tissue breakdown. Life-threatening sepsis can result if the organisms penetrate deep into host tissue. This communication describes the sensor development for five important clinical microbial pathogens commonly found in wounds: Staphylococcus aureus, P. aeruginosa, Candida albicans/auris, and Enterococcus faecalis (the SPaCE pathogens). The sensor contains liposomes encapsulating a self-quenched fluorescent dye. Toxins, expressed by SPaCE infecting pathogens in early-stage infected wounds, break down the liposomes, triggering dye release, thus changing the sensor color from yellow to green, an indication of infection. Five clinical species of bacteria and fungi, up to 20 strains each (totaling 83), were grown as early-stage biofilms in ex vivo porcine burn wounds. The biofilms were then swabbed, and the swab placed in the liposome suspension. The population density of selected pathogens in a porcine wound biofilm was quantified and correlated with colorimetric response. Over 88% of swabs switched the sensor on (107–108 CFU/swab). A pilot clinical study demonstrated a good correlation between sensor switch-on and early-stage wound infection.
Keywords: bacterial infection, point-of-care, wounds, biofilm, infection detection, liposomes, fluorescent dye
Wound infection at early stages is difficult to diagnose. The primary clinical indicators include peri-wound redness, wound heat, and pain—all of which can be equally caused by an inflammatory response to the injury itself.1,2 This matters, since the earlier infection is diagnosed, the greater the chance of improving healing and avoid scarring or a potentially serious systemic infection which, in turn, could lead to sepsis. Importantly, it will also allow a greater likelihood of avoiding overtreatment of wound infection with costly in-hospital investigations and unnecessary antibiotic management, potentially worsening the risks for antimicrobial resistance.
The classical model of infection goes back to the 19th century and Robert Koch’s observations that an infection illness can be reproduced in an uninfected animal by the transfer of bacteria from the diseased animal. It is undoubtedly true that microorganisms are required to cause infection. It is not true, however, that the mere presence of a microorganism in a wound means that there is an infection requiring active treatment. All wounds, from the time the skin incision or burn is made, become colonized with bacteria from the air, surrounding skin, and any material involved in the initial wounding—such as shrapnel, gravel, etc. Most wounds heal without infection: host immune response, wound cleaning, and topical antisepsis are generally sufficient to prevent initial bacterial colonization developing into a full infection. The difficulty is in differentiating between those wounds that are in the early phases of healing (with some associated inflammation) and those that are in the early phases of infection.
A further problem with detecting wound infection is the absence of specific clinical diagnostic criteria for wound infection. For example, the American Burns Association claim that a wound biopsy with >105 colony forming units/g of tissue defines wound infection.3 However, wound biopsies are rarely used in the U.K. and Europe, and this arbitrary definition of bacterial concentration is of little practical value.4
Diagnosis of infection requires clinicians to make a judgment based on clinical indications that are subjective, nonspecific, and based on clinician experience. The clinical indicators currently in use include signs such as pyrexia, symptoms such as increased local pain, and laboratory tests of inflammation such as a raised white blood cell count or C-reactive protein (CRP). By the time wound infection is unambiguous, it is likely that the infection is firmly established and harder to treat. Wounds suspected to be infected are swabbed, and the swab sent to centralized hospital microbiology laboratories for microbial culture along with, in some cases, PCR or mass-spectrometric identification of organisms. However, the time taken to obtain a test result (typically 24–48 h) means that suspect wounds are often treated as infected and antibiotics prescribed before diagnosis. Patients are, therefore, at high risk of infection with resistant bacteria, e.g., antibiotics were given to 50% of 238 adults with burns in a Scandinavian study, including 26% without infection.5 This also has resource implications for healthcare providers, including admissions to hospitals that are longer than necessary.
In the last 20 years, our understanding of the nature of wound infection has improved, with an emerging understanding of the crucial role of the bacterial biofilm in the pathogenesis of wound infection. Culture-independent techniques, such as next-generation DNA sequencing of wounds, have revealed the complex microbiome of multispecies wound biofilms, constituting both bacterial and fungal species. It is now clear that the biofilm state is fundamental to our understanding of wound infection.6,7 Our view is that bacterial biofilms (defined as a confluent layer of bacteria) are the “natural state” of bacteria in the wound matrix, since a biofilm gives bacteria some protection from immune clearance and antisepsis/antibiotic action.8
In 2016, the International Wound Infection Institute (IWII) revised and updated the Wound Infection Continuum (WIC) through consensus voting (Delphi process).9 WIC defines the progression of infection in wounds and acknowledges the impact and the critical role of biofilm in wound infection. The continuum (Figure 1) was divided into five key phases, from wound contamination to systemic infection, with the addition of the fate of wounds—which follow two trajectories, either healing or infection.10 Treatment is not normally required for the first two phases, where the wound has an insignificant wound bioburden and is considered to not be infected. The clinically infected condition requiring treatment begins during the phase of local infection, when the microbial population exponentially increases through to the last two phases: of spreading and systemic infections.
Figure 1.
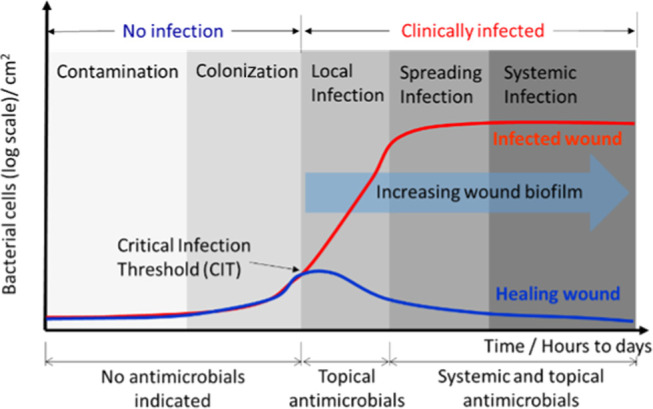
Graphical depiction of wound infection continuum that relates the state of wound infection to the bacterial cell-dependent virulence factors and increasing wound biofilm. Critical infection threshold (CIT) at the end of the colonization phase is the onset of the exponential proliferation of bacteria. The CIT is an important transition, at which point clinical intervention is necessary before the wound condition proceeds to spreading and systemic infection.
In a wound that has passed all of the clinically infected phases, the growing biofilm requires aggressive treatment using superficial debridement and systemic and topical antimicrobials.8,9 A timeline that describes the transition from colonization through the local infection phases is termed the critical infection threshold (CIT). The fate of the wound is solely decided by the wound bioburden and the associated virulence factors produced at this CIT.11,12 In wounds entering the infection phase, the exponential rise in wound bioburden, with increasing virulence, overwhelms the immune defense, triggers the immune response, disrupts healing, and irreversibly damages the local tissues. The wound biofilm gradually matures and undermines the effectiveness of systemic and topical treatments in the later phases of infection. If systemic infection is missed or left untreated, sepsis, septic shock, organ failure, and even death could potentially occur. There is evidence that some wound biofilms downwardly modulate the immune response, for example, some Pseudomonas aeruginosa biofilms secrete alkaline proteases and elastase, which can destroy proteins involved in the complement immune response.13 In the healing wound scenario, the wound bioburden is usually maintained and then reduces to a healthy level of colonization, where the normal cycle of wound healing is reinstated. This reduction of bioburden in healing wounds is triggered either by resuming the immune system, or by the timely treatment with antimicrobials in the early phase of local infection.
Clinicians have no point-of-care (PoC) diagnostic for clinically-relevant acute wound infections. Given an alert of local infection, triggered by the wound bioburden at CIT, clinicians could quickly act and treat the wound, or feel more confident in waiting prior to prescribing (potentially unnecessary) antibiotics. This could be achieved by a real-time sensor that prompts a visible signal in response to the presence of infecting microorganisms (bacteria and fungi) of pathogenic nature.14,15 An intelligent wound dressing was recently developed using the same liposome technology and demonstrated to be able to detect the pathogenic bacteria infection on a porcine wound model.16
Most competing technologies measure the presence of bacteria via various methods, including real-time polymerase chain reaction (PCR) and mass-spectrometry (MS). Two technologies are currently commercially available that claim to aid wound infection diagnosis: Woundchek measures the presence of elevated protease activity following swabbing but has only recently been evaluated by the U.K.’s National Institute for Clinical Excellence (NICE) for its utility in chronic wounds.17 MolecuLight uses UV light and a camera to illuminate a wound and record fluorescence emission from bacterially secreted pyocyanin. While the mechanism allowing detection of P. aeruginosa biofilms is clear (expressed pyocyanin is fluorescent), its role in early-stage wound infection diagnosis is uncertain, as few early colonizing bacteria secrete fluorescent molecules. A small one-arm study has recently been completed, with just 30 patients, who all had chronic wounds. Early results suggest the technology may have utility in chronic wound management.18 The test of this technology is on a per wound basis, where bacteria will fluoresce red and samples are obtained from the discrete red locations. Convatec Ltd. submitted a patent application (2010) proposing infection detection via enzymes including lysozyme, elastase, cathepsin G, and myeloperoxidase, which may have utility in chronic wound management, if developed.19
In this work, we developed a technology that can quickly detect the virulence factors expressed by bacteria in early-stage wound infection, via an easy to read, visible, color change. It utilizes a swab, from a wound that is suspected to be infected, which is simply dipped in the sensor tube, and incubated at body temperature for an hour. We demonstrated that this simple and cost-effective technology detected most wound infections associated with bacteria, including Staphylococcus aureus, P. aeruginosa, Candida albicans, Candida auris, and Enterococcus faecalis species. This work led to a small pilot study, carried out in three U.K. hospitals, using in vivo wound swabs, which resulted in a good correlation between the sensor activation and the retrospective, independently carried out, clinical diagnosis of wound infection.
Materials and Methods
Preparation of Signaling Liposomes
1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) were synthetic lipids (purity >99%) purchased from Avanti Polar Lipids. Cholesterol (>99% purity), 5(6)-carboxyfluorescein, agarose, and 10,12-tricosadiyonic acid (TCDA) were supplied by Sigma-Aldrich, U.K. All sodium chloride, sodium hydroxide, and N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) used in the preparation of buffer solution were high-purity reagent grade. Two types of buffers were prepared. Buffer 1 was prepared using 6.24 g of NaCl, 224 mg of NaOH, and 2.382 g of HEPES in 1 L of MilliQ water, while buffer 2 was made of 1.877 g of 5(6)-carboxyfluorescein, 58.4 mg of NaCl, 540 mg of NaOH, and 238.4 mg of HEPES in 100 mL of MilliQ water. Both buffers were heat sterilized and stored at room temperature before use. For the preparation of vesicles, lipids, cholesterol, and TCDA were individually prepared in HPLC grade chloroform (Chemosolve from Sigma-Aldrich, U.K.) for the concentration of 100 mmol dm–3. Then, 2.12 mL of DPPC, 1 mL of TCDA, 0.8 mL of cholesterol, and 0.08 mL of DPPE were mixed in a glass vial. Using nitrogen gas, the lipid mixture was dried to form a lipid film that was further dried in a vacuum desiccator (at 2 mbar) for an hour before it was rehydrated in buffer 2. Lipid film in buffer 2 was then placed in a hot water bath heated at 70 °C for 10 min before it was frozen in liquid nitrogen and thawed three times in a hot water bath. The final lipid film was then extruded three times through 200 nm diameter nanoporous polycarbonate membrane (Whatman track-etch nucleopore membranes from Sigma-Aldrich, U.K.) using liposome extruder (Liposofast LM-50 extruder, Avanti Polar Lipids) heated at 55 °C. NAP-25 gel chromatography columns (DNA grade, Sephadex columns, GE Healthcare, U.K.) were prewashed with buffer 1. The nonencapsulated dyes from the extruded liposomes were removed by passing through prewashed gel columns, and the purified liposomes were stored at 4 °C until further use. The operation of liposome extrusion was carried out inside a clean class II safety cabinet, to avoid microbial contamination.
SPaCE Swab Sensor Preparation
Typically, 0.2% (w/v) agarose was prepared in buffer 1 and heat sterilized. One milliliter of liposomes was brought to 37 °C before gently mixing with 1 mL of (0.2%) agarose gel, which was maintained at 40 °C. Three hundred microliters of liposome–agarose gel mixture was pipetted into each sterilized Eppendorf tube. Sensor tubes were then sealed in a plastic bag using a vacuum sealer and stored at 4 °C before use. This operation was also aseptically carried out in a class II safety cabinet, using sterilized appliances to minimize the microbial contamination.
Microbiology
The clinical strains (n = 83) studied in this work were as follows: 20 S. aureus, 20 P. aeruginosa, 20 C. albicans, 3 C. auris, and 20 E. faecalis. S. aureus was cultured in tryptic soy broth (TSB), while P. aeruginosa and E. faecalis strains were cultured in Luria Broth (LB) media. Both C. albicans and C. auris were grown in yeast extract–peptone–dextrose (YPD) broth. All bacteria were picked from single colonies and cultured in a shaker incubator (200 rpm, 37 °C) for 18 h, before the cell density was readjusted in sterilized deionized water for the initial inoculation on the porcine burn wound. The list of bacterial strains is given in Table 1.
Table 1. List of Clinical Microbial Pathogens Used in This Studya.
| S. aureus | P. aeruginosa | C. albicans | C. auris | E. faecalis |
|---|---|---|---|---|
| 2, 3, 10, 16, 21*, 25, 38, 49, 52, 56, 67, 69*, 82, 101, 112, 114, 126, 160, 253, 295 | 259, 260, 734, 854, 855, 856, 887, 889, 927, 935, 936, 937, 45124#, 45291#, 45311#, 45400#, 45468#, 45498#, 45506#, 45701# | 2620, 2621, 2622, 2628, 3560, 3563, 3564, 3567, 3568, 3663, 3665, 3666, 3667, 3668, 4153, 4158†, 4160‡, 4350±, 4539, 4540 | 8971, 8977, 8984 | 41, 42, 43, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 64, 66, 67, 68, 69 |
Note: S. aureus: All were MSSA strains recovered from infected acute wounds treated at John Radcliffe Hospital, Oxford, U.K. (*methicillin-resistant S. aureus (MRSA) strains). P. aeruginosa: All strains were from infected chronic wounds, AmpliPhi Biosciences Corporation, U.K. Strains marked (#) were extracted from patients with infected acute wounds or blood agar, Southmead Hospital, North Bristol NHS Trust, Bristol, U.K. C. albicans: All were from the human host with high vaginal swab (HVS) isolation, broncheo-alveolar washings, blood, kidney, pus from the chest, endocarditis (blood), and category 2 pathogens, Microbiology Department, University of Bath, Bath, U.K. (†NCPF 3090, ‡NCPF 3122, ±NCPF 3206). C. auris: All were collected from the human host with assigned NCPF numbers, acquired from Public Health England (PHE). E. faecalis: All strains were obtained from Queen Victoria Hospital, East Grinsted, U.K.
Porcine Wound Biofilm Model
The ex vivo porcine wound model was previously developed at the University of Bath.8 Fresh porcine skin was locally sourced from an organic pig farm. Hair and excess fat were removed before the skin was used as a substrate for creating burn wounds. First, the skins were thoroughly washed in deionized water before being cut into uniform square pieces (approximate size of 4 × 4 cm2). UV disinfection of the skin was carried out using a commercial UV source (Hamamatsu, Japan) equipped with a 254 nm UV lamp for flood UV exposure. Each skin quadrant was subjected to three cycles of a 10 min UV exposure (with a 10 min break after each exposure). The effectiveness of this disinfection method was validated using nutrient agar on which microorganisms were attempted to be grown via comprehensive swabbing of each disinfected skin quadrant. The skin, disinfected using the above method, grew no microorganisms on nutrient agar plates after incubation for 12–18 h. Each skin piece was then aseptically transferred onto a three-ply cotton mesh presoaked with 0.15% Kathon (bactericide) in each Petri dish. Sterilized brass blocks, with a contact area of 1.9 × 1.9 cm2, were dry-heated on a hotplate to 170 °C, before being brought into contact with each skin piece for 20 s, to create a burn wound. The wounds had an appearance of second-degree partial-thickness burns, bearing blisters, and tissue damage. Finally, each burn wound was inoculated with 50 μL of bacteria/yeast (approx. 105 CFU/mL) in sterilized water. Each Petri dish containing skin pieces was covered, wrapped in a wet towel, sealed in a zippered plastic bag, and then incubated at 37 °C for 24 h. The whole procedure was aseptically carried out under the flow cabinet using sterilized appliances.
Evaluation of Sensor Performance
Using a disinfected cotton swab presoaked in sterilized HEPES buffer, biofilms from each porcine wound were swabbed (using the “Essen spiral” swab method)20 and placed inside a SPaCE sensor tube. With lids closed, the sensor tubes were incubated at 37 °C for 1 h before being viewed under the UV light (Woods lamp) to visualize the fluorescent response. The sensor tube incubation period for all study species was optimized, and the 1 h postincubation period was observed to be a sufficient length of time for the activation of swabs from the infected wound biofilms. The porcine wounds without the bacterial infection were also swabbed and tested as negative controls, while the swabs soaked in Triton (detergent) were used as the positive controls. Swabs from porcine wounds infected with the selected strains of bacteria (n = 23) were dispersed in sterile deionized water, serially diluted and plated on selected agars for incubation, and colony counting for quantitative analysis.
SPaCE-Pilot Clinical Study (IRAS project ID: 260069; REC Reference: 19/WM/0200)
The SPaCE-Pilot clinical study design and results will be published separately. The SPaCE-Pilot study was run as a multicenter pilot across three U.K. hospital burn services. The aim of the pilot was to assess the ability of the SPaCE technology in aiding diagnosis, via the indication of clinically important burn wound infection, through a color change (giving an early indication of, for instance, the requirement for antibiotics, a possible delay in healing, and/or unexpected need for surgical intervention). Although the study was not powered to give definitive quantitative data, the sensitivity, specificity, positive predictive value, negative predictive value, and test accuracy were calculated.
A total of 33 patients (with a burn wound) were recruited with their consent (for protocol details, please see the Supporting Information). For every consenting patient, a photograph was taken of each wound, and the designated research nurse aseptically swabbed the wound in precleaned and again in postcleaned states. Each swab was placed inside a 2 mL tube containing 300 μL of the sensor liposomes, which was then incubated at 37 °C for an hour. Following incubation, each sensor tube was observed for the degree of activation. The wound infection condition of each wound was clinically judged by senior wound care clinicians. Routine clinical microbiology qualitatively determined the bacterial species involved in each wound. The clinical decision of wound infection was independently made and was not based on the colorimetric response of the tested SPaCE swab sensor.
Results
In Vitro (Porcine Wound Biofilm) Testing
The performance of the lab-prototype SPaCE swab sensor was evaluated using the listed clinical strains collected from human hosts. Each strain was grown into a 24 h biofilm on an ex vivo porcine burn wound. The biofilm swab results, after 2 h incubation, are shown in Figure 2. The response of each sensor tube was determined by comparing it with the positive and negative controls. Following the swab insertion and incubation, the majority of the SPaCE sensor tubes changed color to a fluorescent green under UV light. All swabs taken from wounds infected with S. aureus indicated a positive response (Figure 2a), while the remaining tested species showed varying degrees of positive response (89%). These included P. aeruginosa (100%), C. albicans (75%), C. auris (67%), and E. faecalis (80%), as shown in Figure 2b–e.
Figure 2.
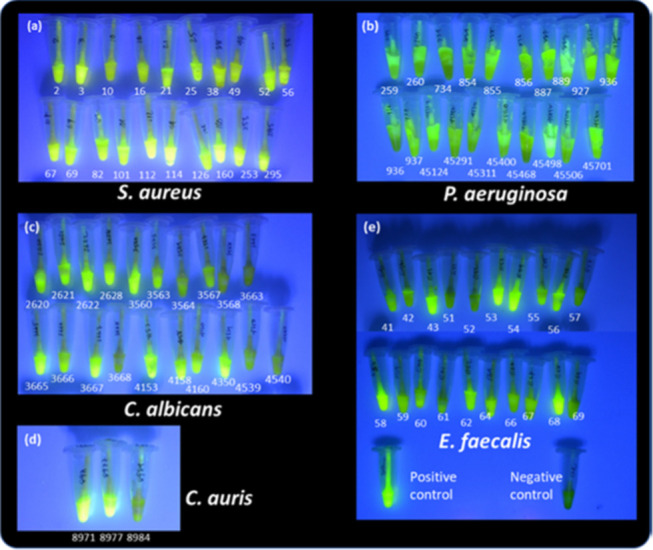
SPaCE swab sensor tubes, seen under UV light, after incubation at 37 °C for 2 h with wound swab from (a) S. aureus, (b) P. aeruginosa, (c) C. albicans, (d) C. auris, and (e) E. faecalis biofilms (with positive and negative controls) grown on a porcine wound for 24 h.
Compared to the positive control, some P. aeruginosa (734, 936, 45468), C. albicans (3568, 3663, 3668, 4539, 4540), C. auris (8984), and E. faecalis strains (51, 52, 57, 69) were not as strongly responsive as a positive infection over the same incubation period. Considering that all of these strains were recovered from infected human hosts, the inconsistency in a degree of positive response could be due to the inconsistent colonization and growth condition on a porcine wound. This prompted us to make a determination of the population density of the infecting species on each wound. The swabs taken from a 24 h biofilm on each wound, infected with 23 selected strains, provided the wound-associated cell density, which was directly correlated with the sensor response. Figure 3 depicts the population cell density picked from each wound to be between 106 and 108 CFU/swab. It was generally observed that the higher the cell density carried by the swab, the stronger the response of the sensor. The weak fluorescent response was observed in some bacterial strains, such as P. aeruginosa (734) and E. faecalis (51 and 52) with a relatively lower cell density of 107 CFU/swab, although this was not the case in viral pathogens of C. albicans and C. auris. In fact, C. albicans strains only required a cell density of around 107 CFU/swab to give a positive response (Figure 3).
Figure 3.
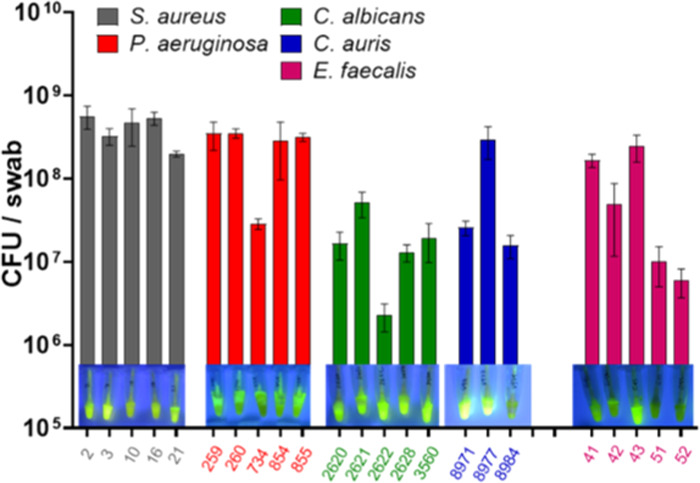
CFU per swab of the 23 selected SPaCE pathogens grown on the porcine wound at 37 °C for 24 h (inset photos: respective SPaCE tubes showing fluorescent activation after inoculation with the swabs at 37 °C for 2 h). Swabs carrying the relatively lower biofilm cell count (i.e., ≤107 CFU/swab) are linked to a relatively weaker fluorescent signal (such as P. aeruginosa (734) and E. faecalis (51 and 52) strains, above), demonstrating the cell density-dependent activation of SPaCE swab sensor.
Clinical Study: SPaCE-Pilot
A full write up and discussion of the SPaCE-Pilot clinical study will be published in due course. The central challenge of calculating swab accuracy data from a clinical study is the absence of a definitive “gold standard” reference for the presence—or absence—of wound infection, since aided by clinical microbiology and patient response to antibiotics.21
Test sensitivity was 57% (8/14) and specificity 71% (12/17). However, this hides the fact that all of the responses classified by the test as being infected were also described by the clinical team as being infected, giving a positive predictive value of 100%, a negative predictive value of 86%, and a test accuracy of 92%. The statistical analysis also assumed that the gold standard of the clinical judgment is infallible, whereas it does contain an element of human error and judgment, and thus complete agreement would not be expected.
One of the important conclusions from the clinical study was the degree of subjectivity in translating the observed color of the SPaCE indicator solution to a number (Figure 4). This created a further degree of uncertainty in test result interpretation. The next phase of the development of the test will be to replace the color observation with a fluorometric measure of the test solution (using a proprietary low-cost fluorimeter).
Figure 4.
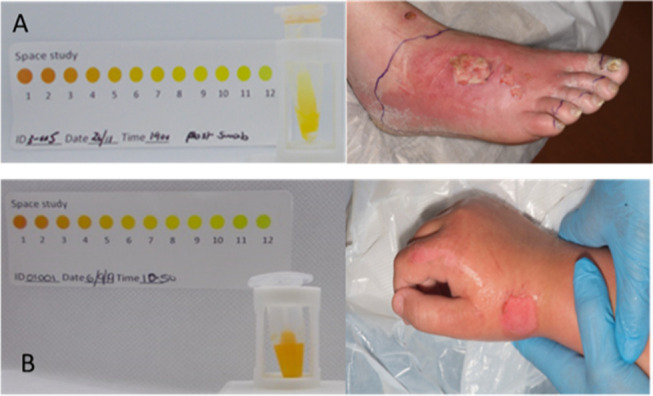
(A) SPaCE swab positive response (left) and clear evidence of burn wound infection (right): infection diagnosed by burn peri-wound redness, patient reported heat and pain, and clinical microbiology (S. aureus positive). (B) SPaCE pathogen negative result: no clinical concern of infection, no patient reported heat or pain, no peri-wound redness.
Discussion
Previously published research into the liposomes used in this sensor show that they are activated by expressed virulence factors from S. aureus and P. aeruginosa.22−24 The cell density-dependent activation of cytolytic Phenol Soluble Modulins (PSMs) from S. aureus and rhamnolipids from P. aeruginosa is understood at a gene level, and this knowledge has been instrumental in developing our qualitative picture of the progression of wound infection, shown in Figure 1.22,23 The mechanism of action from C. albicans and C. auris has not yet been studied in terms of liposome activation, but C. albicans/auris expression toxins have been reported in the wider literature. A 2016 study identified the peptide Ece1-III62–92K, named Candidalysin expressed by C. albicans as its cytolytic agent.25 It is likely that this is the peptide that lyses the liposomes used in the SPaCE sensor. What is unclear is, whether it is genetically regulated by the quorum-sensing gene control system in the manner of the S. aureus toxins. C. auris is an emerging, multidrug resistant, Candida species associated with high mortality rates.26 The principal expressed virulence factors appear to be proteases and phospholipases, the latter being the likely liposome activating moiety. E. faecalis is a gut-living pathogen that expresses a two-component cytolysin and has an established lytic activity. It is likely that the latter is the liposome-active moiety.27 The SPaCE pathogens cover most of the principal microbes found in burn wounds.28
Conclusions
True wound infection is a complex, multifactorial process preceded by a continuum of microbial contamination, colonization, and early to late-stage infection. The key point here is that the presence of a microbial pathogen in a wound does not necessarily mean that the wound is actually infected by that organism: infection requires a critical density of bacteria in a wound matrix, failure of the immune system to adequately respond to the emerging microbial load, and failure of systemic or topical antibiotics or antisepsis, if used. This would make the development of a wound infection sensor, based purely on the presence of bacteria, to be likely of low accuracy.
The swab sensor developed, tested, and described in this communication has a unique approach to wound infection detection. By measuring the presence of the expressed cytolytic virulence factors expressed by the colonizing organisms at a point during the infection continuum, we believe it is possible to indicate early wound infection with high sensitivity (few false-negative results and high specificity; few false-positive results). Ultimately, the only way to determine the efficacy of the system would be in a large, statistically powered, clinical study. Such a study is planned to commence in 2022. Moreover, in this future study, the utility of using a fluorimeter to better quantify the sensor output will be evaluated in parallel to clinician-reported color change, thus determining whether better test accuracy would result from this approach.
Acknowledgments
The authors thank the Scar Free Foundation for support via the Children’s Burns Research Center, The Annette Charitable Trust, Engineering and Physical Sciences Research Council (EPSRC), and Medical Research Council (MRC).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.0c01265.
SPaCE-pilot study protocol (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
EPSRC Research Grant: EP/R51164X/1 Medical Research Council Research Grant: R/N006496/1
The authors declare no competing financial interest.
Supplementary Material
References
- Weinstein R. A.; Mayhall C. G. The epidemiology of burn wound infections: Then and now. Clin. Infect. Dis. 2003, 37, 543–550. 10.1086/376993. [DOI] [PubMed] [Google Scholar]
- Collier M. Recognition and management of wound infections.. World Wide Wounds 2004, 7, 8–14. [Google Scholar]
- Greenhalgh D. G.; Saffle J. R.; Holmes J. H.; Gamelli R. L.; Palmieri T. L.; Horton J. W.; Tompkins R. G.; Traber D. L.; Morzingo D. W.; Deitch E. A.; et al. American Burn Association Consensus Conference to define sepsis and infection in burns. J. Burn Care Res. 2007, 28, 776–790. 10.1097/BCR.0b013e3181599bc9. [DOI] [PubMed] [Google Scholar]
- Kallstrom G. Are Quantitative Bacterial Wound Cultures Useful?. J. Clin. Microbiol. 2014, 52, 2753–2756. 10.1128/JCM.00522-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelgren P.; Björnhogen V.; Bragderyd K.; Jonsson C. E.; Ransjö U. A prospective study of infections in burn patients. Burns 2002, 28, 39–46. 10.1016/S0305-4179(01)00070-5. [DOI] [PubMed] [Google Scholar]
- Percival S. L.; Hill K. E.; Williams D. W.; Hooper S. J.; Thomas D. W.; Costerton J. W. A review of the scientific evidence for biofilms in wounds. Wound Repair Regener. 2012, 20, 647–657. 10.1111/j.1524-475X.2012.00836.x. [DOI] [PubMed] [Google Scholar]
- Davis S. C.; Ricotti C.; Cazzaniga A.; Welsh E.; Eaglstein W. H.; Mertz P. M. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regener. 2008, 16, 23–29. 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Thet N. T.; Wallace L.; Wibaux A.; Boote N.; Jenkins A. T. A. Development of a mixed-species biofilm model and its virulence implications in device related infections. J. Biomed. Mater. Res., Part B 2019, 107, 129–137. 10.1002/jbm.b.34103. [DOI] [PubMed] [Google Scholar]
- International Wound Infection Institute (IWII) Wound Infection in Clinical Practice Wounds Int. 2016.
- Haesler E.; Ousey K. Evolution of the wound infection continuum. Wounds Int. 2018, 9, 6–10. [Google Scholar]
- Cooper R. A. The contribution of virulence to wound infection. Br. J. Community Nurs. 2013, 7, 10–14. 10.12968/bjcn.2002.7.sup4.12615. [DOI] [Google Scholar]
- Hamood A. N.; Griswold J. A.; Duhan C. M. Production of extracellular virulence factors by Pseudomonas aeruginosa isolates obtained from tracheal, urinary tract, and wound infections. J. Surg. Res. 1996, 61, 425–432. 10.1006/jsre.1996.0140. [DOI] [PubMed] [Google Scholar]
- Kharazmi A. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol. Lett. 1991, 30, 201–205. 10.1016/0165-2478(91)90026-7. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Loftus A. L.; Mulley G.; Jenkins A. T. A. A thin detection/response system for pathogenic bacteria. J. of Amer. Chem. Soc. 2010, 132, 6566–6570. 10.1021/ja101554a. [DOI] [PubMed] [Google Scholar]
- Thet N. T.; Hong S. H.; Marshall S.; Laabei M.; Jenkins A.; et al. Visible, colorimetric dissemination between pathogenic strains of Staphylococcus aureus and Pseudomonas aeruginosa using fluorescent dye containing lipid vesicles. Biosens. Bioelectron. 2013, 41, 538–543. 10.1016/j.bios.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Thet N. T.; Alves D. R.; Bean J. E.; Booth S.; Nzakizwanayo J.; Young A. E. R.; Jones B. V.; Jenkins A. T. A. Prototype development of the intelligent hydrogel wound dressing and its efficacy in the detection of model pathogenic wound biofilms. ACS Appl. Mater. Interfaces 2016, 8, 14909–14919. 10.1021/acsami.5b07372. [DOI] [PubMed] [Google Scholar]
- Medtech innovation briefing nice.org.uk/guidance/mib83 (published 5 Oct, 2016).
- https://clinicaltrials.gov/ct2/show/study/NCT02682069 (accessed April 1, 2020).
- Linnane P. G.; Shaw H. L.. Wound dressing. U.S. Patent. US7,723,559B22010.
- Al Ghazal P.; Korber A.; Klode J.; Schmid E. N.; Buer J.; Dissemond J. Evaluation of the Essen Rotary as a new technique for bacterial swabs: results of a prospective controlled clinical investigation in 50 patients with chronic leg ulcers. Int. Wound J. 2014, 11, 44–49. 10.1111/j.1742-481X.2012.01036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A.; Spickett-Jones F.; Jenkins A. T. A.; Young A. E. A systematic review of intervention studies demonstrates the need to develop a minimum set of indicators to report the presence of burn wound infection. Burns 2020, 10.1016/j.burns.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Laabei M.; Jamieson W. D.; Yang Y.; Elsen J.; Jenkins A. T. A. Investigating the lytic activity and structural properties of Staphylococcus aureus phenol soluble modulin (PSM) peptide toxins,. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 3153–3161. 10.1016/j.bbamem.2014.08.026. [DOI] [PubMed] [Google Scholar]
- Laabei M.; Jamieson W. D.; Lewis S. E.; Diggle S. P.; Jenkins A. T. A. A new assay for rhamnolipid detection – importance virulence factors of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2014, 98, 7199–7209. 10.1007/s00253-014-5904-3. [DOI] [PubMed] [Google Scholar]
- Laabei M.; Jamieson W. D.; Massey R. C.; Jenkins A. T. A. Staphylococcus aureus interaction with phospholipid vesicles – a new method to accurately determine accessory gene regulator (agr) activities. PLoS One 2014, 9, e87270 10.1371/journal.pone.0087270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes D. L.; Wilson D.; Richardson J. P.; Mogavero S.; Tang S. X.; Wernecke J.; Höfs S.; Gratacap R. L.; Robbins J.; Runglall M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin E.; Hager C.; Chandra J.; Mukherjee P. K.; Retuerto M.; Salem I.; Long L.; Isham N.; Kovanda L.; Borroto-Esoda K.; et al. The emerging pathogen Candida auris: Growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chemother. 2017, 61, e02396-16 10.1128/AAC.02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D.; Martin M. J.; Gilmore M. S. Structure, function and biology of the Enterococcus faecalis cytolysin. Toxins 2013, 5, 895–911. 10.3390/toxins5050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D.; Elsayed S.; Reid O.; Winston B.; Lindsay R. Burn wound infections. Clin. Microbiol. Rev. 2006, 19, 403–434. 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


